Open Access Article
Open Access ArticleCell-selective zwitterionic parylene with intrinsic antifouling, softness, and conformability†
Shouyan
Zhang
a,
Huiqing
Zhao
a,
Sihao
Qian
a,
Yuzhe
Zhai
a,
Shuhua
Zhang
a,
Zhi
Geng
a and
Bo
Zhu
 *abc
*abc
aSchool of Materials Science and Engineering, Shanghai University, 99 Shangda Road, Baoshan, Shanghai 200444, China. E-mail: bozhu@shu.edu.cn
bJoint International Research Laboratory of Biomaterials and Biotechnology in Organ Repair, Ministry of Education, 99 Shangda Road, Baoshan, Shanghai 200444, China
cShanghai Engineering Research Center of Organ Repair, 99 Shangda Road, Baoshan, Shanghai 200444, China
First published on 21st December 2024
Abstract
Parylene is one of the most widely used polymers to fabricate flexible bioelectronic devices due to its flexibility, excellent barrier property, and photolithography-compatible fabrication. However, the extensively presented biofouling and the lack of biofunctionalities on the parylene surface prevent the bioelectronic device from constructing intimate coupling with cells/tissues. We herewith fabricated an intrinsically antifouling and soft parylene thin film featuring specific biointeraction, which consists of a bottom layer of pristine parylene and a top layer of 2-bromoisobutyrate functionalized parylene with ligand conjugated zwitterionic polymers. This layer-by-layer structure helps ensure the encapsulation property while allowing for tuning surface function for biomedical applications. This biomimetic parylene thin film presents an excellent barrier property (<10 pA leakage current after 12 weeks of soaking in 37 °C PBS buffer), a three-orders-of-magnitude reduced surface modulus (∼45 kPa), and exceptional mechanical compliance and conformability, all of which are crucial for constructing stable coupling with cells/tissues. Remarkably, the biomimetic parylene demonstrated a highly selective interaction toward PC12/HL-1 cells in the presence of a much higher density of white blood cells, thanks to the construction of specific cell interaction on a biofouling-resistant background. We envision that this biomimetic parylene material would offer bioelectronic devices a controllable interaction with biological systems, allowing seamless integration with cells/tissues and promoting the practical use of bioelectronic devices in real-life situations.
Introduction
Bioelectronic devices' biocompatibility is influenced significantly by the surface properties and mechanical properties of encapsulation materials.1,2 Developing flexible encapsulation materials resistant to biological fouling is vital for advancing the practical application of thin-film bioelectronic devices.3–6 Parylene serves a dual role as a substrate and an encapsulation layer, featuring outstanding water and gas barrier properties and biocompatibility (classified as a United States Pharmacopeia Class VI polymer).7–9 Its compatibility with photolithography techniques renders it an attractive encapsulation material in bioelectronics, extensively employed in microelectronic insulation and implant coatings,10–13 including flexible neural probes for brain signal recording studies,14–17 multi-channel arrays for recording electrocardiograms of mouse hearts,18 and curvilinear retinal implants.9 However, the challenge of biofouling at these parylene encapsulation materials poses a significant hurdle for the long-term implantation of bioelectronic devices. It results in biofouling in complex biological fluids, mediating bacterial adhesion and proliferation.19 Moreover, it has already been identified as a significant contributor to implant-induced inflammation.20,21 While studies have explored the use of parylene materials functionalized with polyethylene glycol (PEG) to reduce biofouling and improve electrode biocompatibility, it has been observed that hydrophobic segments of PEG, including its backbone (–CH2–CH2–) and methoxy end (–OCH3), would trigger immune responses.22 This correlation revealed the immunogenic risk of PEG,22,23 thus limiting its suitability for long-term implantation applications.24Zwitterionic polymers have garnered significant attention in medical implants in recent years.25,26 The exceptional antifouling properties of zwitterionic MPC polymers are primarily attributed to their electrical neutrality and the formation of a dense hydration barrier.27,28 Previous studies have demonstrated that the unique internal salt formation in the phosphorylcholine (PC) group neutralizes its overall electrical charge, effectively preventing electrostatic interactions with biomolecules.25,29 Additionally, molecular simulations have shown a relatively dense water region near hydrophilic surfaces with a disrupted hydrogen-bonded network, rendering protein adsorption on such surfaces energetically unfavorable.30 Notably, phosphocholine zwitterions boast a bionic membrane phospholipid bilayer structure, endowing them with superb anti-biofouling and biocompatibility.31 Therefore, we recently engineered a zwitterionic parylene film with excellent biofouling resistance by integrating phosphocholine functionalities.32,33 However, the antifouling parylene would indiscriminately repel all cells, preventing the attachment of target cells and hindering the formation of cellular networks surrounding implants. This is detrimental to constructing an intimate communication with target cells/tissues, considering that the encapsulation material occupies the major surface of the electronic device. An attractive alternative encapsulation system should present a solid affinity for targeted cells/tissues while resisting biofouling, thus attracting specific cells/tissues for intimate communication with devices while synchronously reducing the risk of immune response. Introducing specific peptides to antifouling surfaces to create bioactive surfaces is a promising approach to building cell-selective interaction,34 as the antifouling background could prevent the surface from interacting with other cells nonspecifically, thus ensuring the specificity.35–37 Over the past decade, bioactive surfaces integrating specific peptides with anti-biofouling surfaces have been widely applied in tissue engineering materials,38–40 biosensors41–44 biochips,41,45,46 and implantable materials.31,47–49
To address this issue, we fabricated an intrinsically antifouling and soft parylene film featuring specific biointeraction by combining chemical vapor deposition of functionalized parylenes and atom transfer radical polymerization of zwitterionic polymers (Scheme 1). This bioactive parylene film consists of a bottom layer of pristine parylene and a top layer of 2-bromoisobutyrate functionalized parylene grafted with the ligand conjugated copolymer of 2-methacryloyloxyethyl phosphorylcholine (MPC) and methacrylic acid n-hydroxysuccinimide ester (MNHS). The grafting of MPC polymers onto the parylene surface creates a compact hydration layer, offering strong resistance to protein and cell fouling. Meanwhile, MNHS was incorporated as a comonomer for PMPC polymers due to its outstanding efficiency and selectivity in conjugating with RGD peptides, thereby imparting bio-specificity against the antifouling background. This layer-by-layer structure helps ensure the encapsulation property while offering an opportunity to tune surface function for biomedical applications. Further grafting zwitterionic polymers significantly reduces the surface modulus and makes the film mechanically compliant and conformable to complex surfaces, facilitating intimate electrocoupling with cells and tissues. Remarkably, the biomimetic parylene demonstrated cell-selective interaction even in a high density of white blood cells, owing to the specific interaction established against a background resistant to nonspecific interactions. We envision that this biomimetic parylene material would serve as an attractive encapsulation material for producing antifouling and flexible bioelectronic devices with controllable biointeraction toward seamless integration with biological systems, thus promoting the practical application of bioelectronic arrays in real-life scenarios.
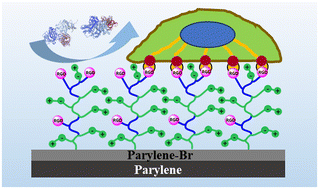 | ||
| Scheme 1 Schematic depicting the antibiofouling and cell-affinity properties of biomimetic zwitterionic parylene films. | ||
Experimental section
Materials
Dichloro-[2,2]-paracyclophane was purchased from Saen Chemical Technology Co., Ltd. Poly((p-xylylene-4-methyl-2-bromoisobutyrate)-co-(p-xylylene)) (parylene-Br) was synthesized by referring to our previous research.32 2-Methacryloyloxyethyl phosphorylcholine (MPC) and methacrylic acid n-hydroxysuccinimide ester (MNHS) were purchased from Joy-Nature Co., Ltd. Tris(2-yridylmethyl)amine (TPMA), cuprous bromide (CuBr), copper(II) bromide (CuBr2) and all solvents were purchased from Adams Reagent, Ltd. S1813 and AZ9260 photoresists were purchased from Dow-Shipley. ZX-238 developer and N-methylpyrrolidone were purchased from Jianghua Microelectronic Materials Co., Ltd.Arginine–glycine–aspartic acid (RGD) peptides were purchased from G.L. Biochem (Shanghai) Ltd. [3-(Methacryloyloxy)propyl] trimethoxysilane (silane A174), bovine serum albumin (BSA) and fetal bovine serum (FBS) were purchased from Sigma-Aldrich. Fibrinogen labeled with isothiocyanate (FTIC-FNG) was purchased from Solarbio. Bovine calf serum (BCA) was purchased from Zhejiang Tianhang Co., Ltd. GlutaMAX™ supplement, sodium pyruvate (100 mM) solution, and MEM non-essential amino acids (100×) were purchased from Gibco. 0.25% trypsin, 0.02% EDTA solution, penicillin–streptomycin (100×), Dulbecco's modified Eagle medium (DMEM), minimum essential media (MEM), Roswell Park Memorial Institute (RPMI) 1640 medium, Claycomb medium, and insulin were purchased from Genom Biomedical Technology Co., Ltd. iFluor 488-labeled phalloidin staining kit was purchased from YEASEN. Red blood cell lysis buffer, 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI Staining Solution), 3,3′-dioctadecyloxacarbocyanine perchlorate cell staining kit (DIO), and 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate cell staining kit (DIL) were purchased from Beyotime Biotechnology. The mouse fibroblast cell line (NIH3T3 cells) and rat pheochromocytoma cell line (PC12 cells) were purchased from Stem Cell Bank, Chinese Academy of Sciences. Highly aggressively proliferating immortalized rat microglial cells (HAPI cells) were purchased from Merck Millipore. The cardiomyocyte cell line (HL-1 cells) was purchased from Procell Life Science & Technology Co., Ltd. Mouse blood was purchased from Beijing Bersee Science and Technology Co., Ltd.
Preparation of zwitterionic and bioactive parylene films
Parylene films were deposited using a chemical vapor deposition (CVD) reactor (Bolimante Test Electric Furnace Co., Ltd., BLMT-1200GAK-3). In the deposition system, dichloro-[2,2]-paracyclophane was vaporized at 90–110 °C and then pyrolyzed at 600–700 °C to produce chloro-p-xylylene. Parylene C films were formed as chloro-p-xylylene condensed onto surfaces at a low substrate temperature (approximately 25 °C). The thickness of the parylene C film was controlled by the initial loading of dichloro-[2,2]-paracyclophane (0.6 μm g−1). Typically, substrates were coated with a 3 μm thick layer of parylene C. Subsequently, a 72 nm thick layer of poly((p-xylylene-4-methyl-2-bromoisobutyrate)-co-(p-xylylene)) (parylene-Br), serving as an initiator for atom transfer radical polymerization (ATRP), was deposited over the parylene films.50 The functionalized films were placed into a glass tube, and the solution containing CuBr (2 mM), CuBr2 (3 mM), TPMA (10 mM), and MPC and MNHS monomers (total concentration of 400 mM) in a degassed mixed solvent (water and dimethyl sulphoxide (DMSO) in a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 volume ratio) was added to the tube. The solution was then heated to 50 °C under nitrogen purging. After 3 hours of reaction time, the film was removed and thoroughly rinsed with water.
5 volume ratio) was added to the tube. The solution was then heated to 50 °C under nitrogen purging. After 3 hours of reaction time, the film was removed and thoroughly rinsed with water.
Subsequently, the films were immersed in a pH 7.4 PBS solution containing 1 mg mL−1 RGD, without the addition of any other chemical agents, and incubated overnight with shaking to facilitate the grafting of RGD peptides onto the parylene film via the grafted copolymer of MPC and MNHS.
Film characterization
X-ray photoelectron spectroscopy (XPS, ESCALAB 250XI, Thermo Scientific, Al K Alpha) measurements were used to determine the composition of the films. The spot size was 500 μm, the energy step size was 0.1 eV, and the standard lens mode was used for the XPS measurement. The XPS data, covering the range of 410–394 eV, was processed using the non-linear least squares fitting (NLLSF) method. The analysis began with importing the raw data into the software, followed by subtracting the background using a linear method. After shift parameters were set, with a maximum shift range of 2 eV and a step size of 0.02 eV, the fitting process was then initiated, with iterative adjustments to the shift parameters until the best fit was achieved. When the optimal fit was reached, the individual spectra components from the overlapping data could be well separated. This enabled the calculation of the atomic ratios for nitrogen species by the areas of the curve-fitted component.The water static contact angle of films was measured using a contact angle measurement system (Attension Theta Flex, Biolin Scientific). Water was added to the film surface at a flow rate of 0.5 μL s−1, resulting in a water droplet volume of 4 μL. Each sample was measured three times to provide an average value.
Scanning electron microscopy (SEM, JSM-7500F, JEOL, operating voltage: 5 kV) was used to observe the surface morphology of films.
The surface roughness of the films was characterized by atomic force microscopy (AFM, Bruker Multimode 8) at tapping mode. Images were sampled at 512 × 512 points, with a scan size of 1 × 1 μm2. The root mean square deviation (Rq) of surface roughness was analyzed using NanoScope Analysis v1.8 software (Bruker).
The infrared spectra of the films were measured using an attenuated total reflection spectrometer (ATR) (Thermo Scientific Nicolet iS20). Spectra were recorded from 500 to 4000 cm−1. A freshly cleaned ZnSe crystal, serving as a single-beam reference spectrum at a 45° incident angle, was used as the background spectrum.
The surface modulus of wetting films was tested on an atomic force microscope (AFM, Bioscope Resolve system, Bruker) equipped with a pre-calibrated PFQNM-LC-A-CAL tip (spring constant: approximately 0.1 N m−1, frequency: 45 kHz) and performed in water at PeakForce QNM mode. Images were sampled at 256 × 256 points, with a scan size of 1 × 1 μm2, and force curves were obtained in water. AFM NanoScope Analysis software v1.8 (Bruker) was used to calculate the root mean square to evaluate the Young's modulus of the films.
QCM measurements
QCM measurements were performed on a quartz crystal microbalance (QCM, Q-Sense AB system, Biolin Scientific) at 25 °C to monitor the interactions of biomolecules with films. The films were deposited on the surface of a QSX 301 sensor crystal (Biolin Scientific) and then placed in the measurement chamber. PBS buffer solution and those containing biomolecules were continuously pumped into the measurement chamber at a flow rate of 30 μL min−1 using an Ismatec ISM597D pump. The resonance frequency change of the sensor, which is related to the mass change arising from biomolecule adsorption, was recorded at a resolution of less than 1 s. The resonance frequencies were measured simultaneously at 5 MHz and its five harmonics (15, 25, 35, 45, 55, and 65 MHz). Changes in the frequency of the third overtone (15 MHz) are presented. The concentrations of the BSA and FBS solutions used in the QCM test were 1 mg mL−1 and 10% (v/v) in PBS buffer, respectively.Fluorescence-labeled protein adsorption assay
FTIC-FNG was used to visualize the films' protein adsorption. The FTIC-FNG solution in PBS buffer (0.1 mg mL−1) was dropped on the thin films and incubated for 1 h, followed by gentle washing with PBS three times. The fluorescence images of samples were recorded using a fluorescence microscope (Olympus, CKX-53). All processes were carried out in darkness.Leakage current test
Leakage current tests were conducted using gold electrodes with a diameter of 200 μm. The electrode preparation process was as follows: spin-coating of photoresist S1813 on electron-grade glass at 3500 rpm for 60 s, followed by baking at 115 °C for 60 s. A mask was placed over the surface and exposed to 365 nm wavelength UV light for 2 seconds under a lithography machine (URE2000/25). Subsequently, the sample was developed in ZX-238 developer for 30 s, rinsed with deionized water, and dried with nitrogen gas. It was then placed in a low-temperature O2 plasma processor (SYDT01E, OPS) for surface treatment at 400 W power for 5 min. Next, 10 nm of chromium and 100 nm of gold were deposited successively using an electron beam evaporation system (MBE-600). The sample was then immersed in N-methylpyrrolidone to remove all photoresists and complete the preparation of the electrode lines. Subsequently, the sample was soaked in a solution containing 100 μL of silane A174, 10 mL of isopropanol, and 10 mL of deionized water for 30 min to enhance the adhesion with the encapsulation layer. A 3 μm layer of parylene C (5 g dichloro-[2,2]-paracyclophane) was deposited on the sample surface, followed by deposition of a layer of parylene-Br (1 g 2-bromoisobutyrate functionalized paracyclophane). The copolymers of MPC and MNHS were then grafted on the surface with parylene-Br as the initiator for the ATRP reaction. The electrode encapsulated by poly(MPC-co-MNHS) grafted parylene was then incubated overnight in 1 mg mL−1 RGD solution of pH 7.4 PBS buffer. The leakage current was measured using a source meter (Keithley 2632B) with a three-electrode setting, where the prepared electrode was the working electrode, and a platinum mesh and Ag/AgCl (3 M KCl solution) electrode were used as the counter and reference electrode, respectively. The electrolyte solution used here was the PBS buffer unless stated otherwise. The leaking current was then measured by applying a voltage of 5 V for 120 s onto the encapsulated electrode. These tests were conducted weekly. After each test, the electrodes were resoaked in 37 °C PBS buffer for continuous testing over 12 weeks.Cell culture
Before culturing the cell, all the films were placed in a 24-well plate (3738-ZX, Corning), where each well was filled with 1 mL of PBS buffer, for 12 hours. Then, 1 mL 75% ethanol solution was added instead to sterilize the films for 30 minutes, followed by three times rinsing with the sterilized PBS buffer. Cells were seeded directly onto the film surfaces at a cell density of 1 × 105 cells per mL, followed by adding 1 mL culture medium to each well.1 mL of mouse blood was lysed at room temperature for 5 min after adding 10 mL red blood cell lysis buffer (1×). Then, 20 mL of PBS was added to quench the lysis, followed by centrifugation at 550 rpm for 5 minutes. The red supernatant was discarded, and the white blood cells (WBCs) were resuspended in DMEM high-glucose medium. WBCs were directly seeded onto the film surfaces at a cell density of approximately 1 × 106 cells per mL.
All the cells were cultured at 37 °C, 5% CO2. The culture medium for all the cells was replaced every two days. The culture medium for NIH 3T3 cells was DMEM high-glucose medium supplemented with 10% fetal bovine serum, 1% GlutaMAX™ supplement, 1% sodium pyruvate solution, 1% MEM non-essential amino acids, and 1% penicillin–streptomycin. The culture medium for HAPI cells was DMEM low-glucose medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. The culture medium for PC12 cells was RPMI 1640 medium supplemented with 1% penicillin–streptomycin (100×), 15% horse serum, and 2.5% FBS. For HL-1 cells; the culture medium for HL-1 cells was Claycomb medium (Gibco) supplemented with 10% FBS, 200 mM L-glutamine, 10 mM norepinephrine, and 1% penicillin–streptomycin (100×).
Cell stain
The cytoskeleton was stained using phalloidin. Initially, cells were fixed by incubating in a 4% paraformaldehyde solution in PBS buffer at 25 °C for 30 minutes. Subsequently, cells were permeabilized by treating them with cytoskeleton buffer containing 0.1% Triton X-100 at 25 °C for 5 minutes. A 1 μL aliquot of 1000× iFluor™ 488 labeled phloropyclic peptide (dissolved in DMSO) was diluted in 1 mL of PBS buffer containing 1% BSA to prepare a 1× working solution. Cells were then incubated in 200 μL of the iFluor 488-labeled phalloidin working solution at 25 °C for 1 hour. Afterward, cell nuclei were counterstained with 200 μL of DAPI staining solution (10 μg mL−1 in ddH2O) for 5 min. Fluorescence observation was performed using a fluorescence microscope, with the cytoskeleton and the cell nuclei exhibiting green fluorescence at 493 nm and the cell nuclei exhibiting blue fluorescence at 364 nm, respectively.For the cell selectivity test, WBCs were stained with 200 μL DIL stain kit (10 μM) for 20 min, while PC12 or HL-1 cells were stained with 200 μL DIO stain kit (10 μM) for 20 min. Fluorescence observation was conducted using a fluorescence microscope. The WBCs exhibited red fluorescence at 549 nm, whereas the PC12 or HL-1 cells exhibited green fluorescence at 484 nm.
Statistical analysis
Statistical differences among data were assessed using one-way analysis of variance (ANOVA). A p-value of less than 0.05 was considered statistically significant (*, p < 0.05).Results and discussion
Synthesis of zwitterionic parylenes
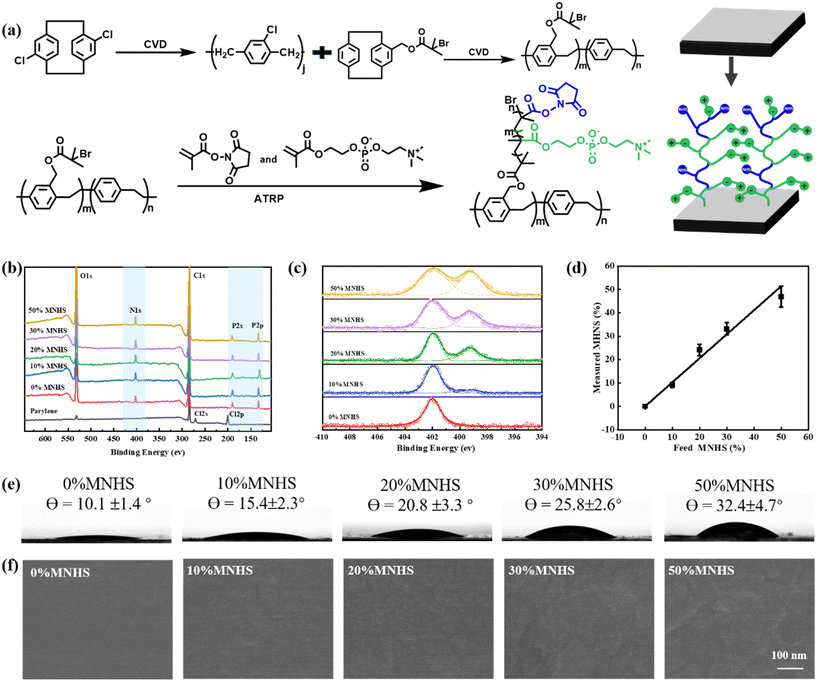
In this paper, we aimed at the peptide-presented zwitterionic polymers grafted parylene film for constructing a cell membrane mimicking parylene film, which presents specific biointeraction on a background to resist biofouling. Typically, we first fabricated a parylene thin film and then placed a thin layer of poly((p-xylylene-4-methyl-2-bromoisobutyrate)-co-(p-xylylene)) (parylene-Br) film over it by chemical vapor deposition (CVD). (Fig. 1a, top). The 2-bromoisobutyrate groups on the top surface then acted as ATRP initiators to graft the copolymer of 2-methacryloyloxyethyl phosphorylcholine and methacrylic acid n-hydroxysuccinimide ester (poly(MPC-co-MNHS)) on the parylene surface (Fig. 1a, below).In this way, we prepared a series of functionalized parylene films grafted with poly(MPC-co-MNHS)s by varying MNHS compositions from 0% to 50% at the feed. Their XPS spectra were registered to determine the composition of poly(MPC-co-MNHS) grafted from parylene films. As illustrated in the XPS spectra in Fig. 1b, compared with the pristine parylene, the poly(MPC-co-MNHS) grafted parylene films exhibited distinct signals of N 1s (402.0 eV), P 2s (190.0 eV), and P 2p (133.0 eV) electrons. The corresponding high-resolution N 1s spectra were further registered to measure the composition of poly(MPC-co-MNHS) on parylene (Fig. 1c). As shown in Fig. 1c, the PMPC grafted parylene showed an N 1s peak of C–N+ from MPC at 402.3 eV, whereas the poly(MPC-co-MNHS) grafted parylene film additionally displayed an N 1s peak of N–O from MNHS at 399.4 eV. In addition, the intensity ratio of the N 1s signal of MNHS to that of MPC increases distinctively with the MNHS composition at feed. Because the two N 1s signals partially overlapped, we used a non-linear least squares fitting approach to calculate the copolymer films' compositions. The monomer composition of the poly(MPC-co-MNHS) grafted parylene film presents a linear correlation with the feed ratios of the monomers (Fig. 1d), indicating this method's excellent polymer composition tunability.
The grafted poly(MPC-co-MNHS) composition might impact the surface characteristics, which are demonstrated to be closely associated with the anti-biofouling properties. Thus, we recorded the water contact angle (Fig. 1e), SEM (Fig. 1f), and AFM (Fig. S2†) morphologies of those parylene films grafted with poly(MPC-co-MNHS) of different compositions. The results indicate that the higher the MNHS composition of the grafted poly(MPC-co-MNHS), the higher the water contact angles of the functionalized parylene films due to the big difference in the affinity to water molecules between the zwitterion group and MNHS group. The SEM and AFM results indicated that surface roughness also correlates with the MNHS composition of the grafted poly(MPC-co-MNHS). As shown in Fig. 1f and S2,† the surface becomes rougher with the increase of the MNHS composition. We considered that this phenomenon is likely due to phase separation in the grafted copolymer at higher MNHS compositions, as the polarities of the PC and NHS groups differ significantly.51 Considering that the hydrophilicity and surface roughness of the surface are crucial for its antifouling performance, we thus investigated how the composition of poly(MPC-co-MNHS) impacts this performance below.
Antifouling of zwitterionic parylenes
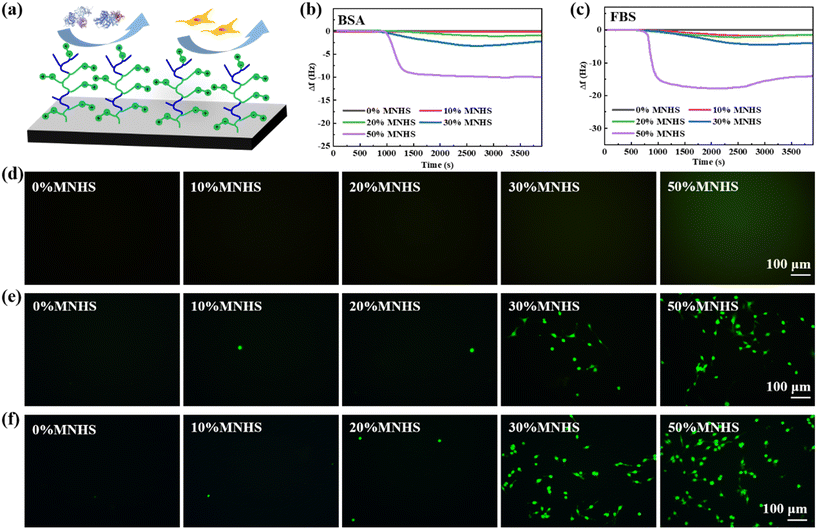
As shown above, investigating the biofouling resistance of the poly(MPC-co-MNHS) grafted parylene films is challenging as the NHS groups can covalently interact with proteins and cells through the NHS ester-mediated covalent conjugation. A previous study revealed that the NHS ester would be hydrolyzed thoroughly after exposure to PBS solution for an extended time, whose length depended on the pH value.52 To explore how long it takes to hydrolyze the NHS esters in the PBS buffer of pH 7.4, we measured the covalent interaction of NHS groups with BSA by a QCM instrument after exposing the poly(MPC-co-MNHS) grafted parylene films (prepared at 20% MNHS feed composition) to the PBS buffer for a series of times (Fig. S1†). The frequency change of the parylene-coated crystals was found to decrease to 0 Hz after exposure to PBS buffer for more than 6 h (Fig. S1†), indicating the NHS esters were entirely hydrolyzed. Therefore, before antifouling tests for the poly(MPC-co-MNHS) grafted parylene films, we immersed the films into the PBS buffer for 12 h to ensure thorough hydrolysis of NHS ester groups, preventing covalent coupling of NHS groups with proteins and cells during the antifouling test. The anti-protein adsorption capacity in poly(MPC-co-MNHS) grafted parylene films was monitored in situ using QCM. The concentrations of the BSA and FBS solutions used in the QCM test were 1 mg mL−1 and 10% (v/v) in PBS buffer, respectively. The results reveal that the poly(MPC-co-MNHS) grafted parylene films feature excellent resistance to the binding of BSA protein (Fig. 2b) and FBS (Fig. 2c) when the NHS group composition is below 20%, offering an MNHS composition window to conjugate the specific ligands. Visualizing the anti-biofouling of the functionalized parylene by FITC-labeled fibrinogen (FITC-FNG) further evidenced this result (Fig. 2d). Due to the strong adhesion and spreading characteristics of the mouse embryonic fibroblast (NIH 3T3) cell line, as well as the role of the rat highly invasive proliferating immortalized (HAPI) microglia cell line as the primary immune cell, both cell lines were selected to test the cell resistance of poly(MPC-co-MNHS)-parylene films. The results indicate that films with the MNHS monomer composition ≤20% also strongly resist the interaction of NIH and HAPI cells. Their remarkable biofouling resistance should be attributed to the phosphorylcholine groups' inherent electro-neutral and hydrophilic nature. In addition, we observed that increasing the ratio of MNHS above 20% compromised the antifouling property of the functionalized parylene films.Bioconjugation of zwitterionic parylene
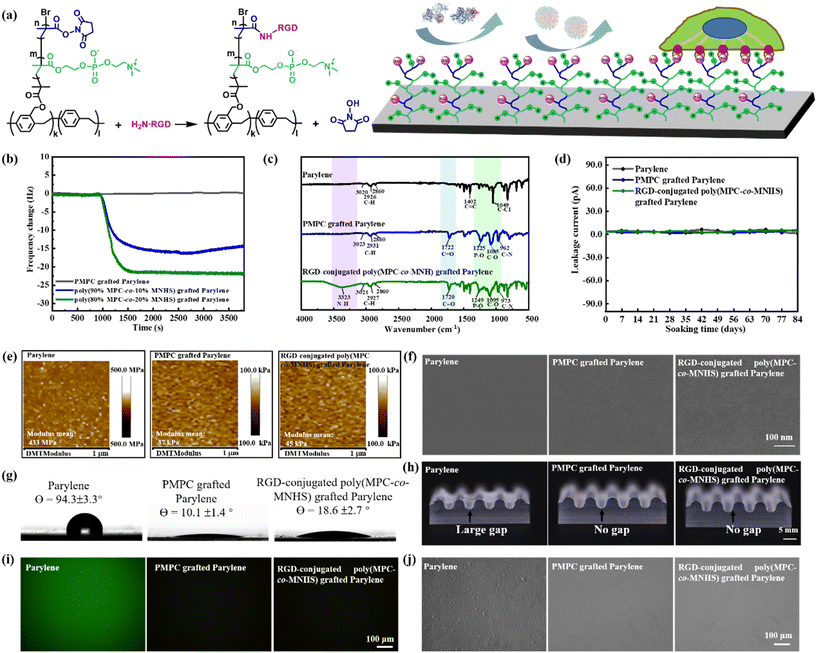
As previously discussed, the functionalized parylene films grafted with poly(MPC-co-MNHS) with MNHS ≤ 20% retain excellent biofouling resistance, offering a valuable range of MNHS composition for ligand conjugation. The arginine–glycine–aspartic acid (RGD) peptide, known for its effective integrin-binding affinity,53 was selected as the model ligand to construct a specific interaction on the functionalized parylene film. We thus conjugated the RGD peptides to the poly(MPC-co-MNHS) polymer chain through an NHS ester-mediated covalent conjugation reaction. The schematic representation for the conjugation of the RGD peptides with poly(MPC-co-MNHS) and their affinity to target cells is illustrated in Fig. 3a. The conjugation of RGD with the poly(MPC-co-MNHS) grafted parylene films was monitored using QCM (Fig. 3b). The QCM crystal coated with the PMPC-grafted parylene has no frequency change when exposed to 1 mg mL−1 RGD solution, whereas that coated with poly(MPC-co-MNHS) grafted parylene presents a significant frequency drop, arising from the conjugation of RGD with the NHS groups. Furthermore, the frequency change due to the RGD conjugation increased with the MNHS composition, supporting this explanation again. We used FT-IR spectrometry to further verify the conjugation of RGD with the poly(MPC-co-MNHS) chains. As shown in Fig. 3c, a new infrared absorption peak at 3323 cm−1, attributed to the N–H symmetric stretching, was observed in the FT-IR spectra of the RGD-conjugated poly(MPC-co-MNHS) grafted parylene films, confirming the successful conjugation of RGD peptides. The bioactive parylene films grafted with RGD-conjugated poly(MPC-co-MNHS) with 20% MNHS would be used later to exhibit the construction of a specific cellular affinity on a biofouling-resistant background.Barrier, softness, compliance, and antifouling of RGD-conjugated zwitterionic parylene
Encapsulation failures reduce the device lifespan and pose the risk of current leakage into the surrounding tissue. Incorporating bromoisobutyrate groups might compromise the dielectric and barrier properties of the parylene film,54,55 thus raising the risk of encapsulation failure. Therefore, we employed a layered parylene film comprising a parylene-Br top layer and a parylene C bottom layer to ensure hermetic performance. The leakage currents of electrodes encapsulated with the pristine parylene, PMPC-grafted parylene, and RGD-conjugated poly(MPC-co-MNHS) grafted parylene in PBS at 37 °C were tested over 12 weeks (84 days). The results indicated no significant difference in the leakage current among the three electrodes encapsulated with these parylene materials (Fig. 3d). Throughout the 84 days, all current values remained below 10 pA. It has demonstrated that the zwitterionic and bioactive parylene encapsulation layer, featuring a layer-by-layer structure, retains parylene's excellent encapsulation performance while achieving functionality.Matching the elastic modulus between the film surface and cells/tissues promotes the biocompatibility of devices, as it efficiently reduces the mechanical tension of cells at the cellular level, which is recognized as one critical factor in activating immune cells.19,56 We herewith used the AFM technique to measure the surface elastic modulus of all the parylene films in water and summarized all the results in Fig. 3e. The parylene in water has a surface elastic modulus of up to 450 MPa, significantly higher than the cell/tissue modulus (less than 100 kPa).57,58 In contrast, the PMPC grafted parylene thin films presented a very small surface modulus of 37 kPa, more than three orders of magnitude lower than that of pristine parylene. We attributed it to the strong hydration capability of the zwitterionic PMPC chains grafted on the surface, which imparts softness to the parylene surface in aqueous environments. Compared to the PMPC grafted parylene films, the surface modulus of the RGD-conjugated poly(MPC-co-MNHS) grafted parylene films is slightly higher but still possesses a low surface modulus of 45 kPa. This means that these zwitterionic and bioactive parylene films could much more closely match the elastic modulus of biological tissues, likely facilitating compatibility with soft biological tissues. Furthermore, the surfaces of parylene, PMPC grafted parylene, and RGD-conjugated poly(MPC-co-MNHS) grafted parylene films were very smooth, as observed by SEM (Fig. 3f). The water contact angles of the films are depicted in Fig. 3g. The contact angle of the parylene film was approximately 94.3 ± 3.3°, which significantly decreased to about 10.1 ± 1.4° after grafting PMPC onto the surface. The RGD-conjugated poly(MPC-co-MNHS) grafted parylene films exhibited a slightly higher water contact angle of approximately 18.6 ± 2.7°.
The conformability of bioelectronic devices is crucial for establishing close integration with biological systems, where the encapsulation materials play a decisive role in the mechanical performance of the entire device. To evaluate the conformability of the parylene film on complex surfaces, we transferred the wet films onto the circular wave grooves of the PDMS substrate with a radius of 1.5 mm. As shown in Fig. 3h, while the 3 μm thick parylene film struggled to conform to the wavy substrate, both PMPC grafted parylene and RGD-conjugated poly(MPC-co-MNHS) grafted parylene films conform well to the complex surface without any interfacial gaps. The wetting-driven conformability of PMPC-parylene and RGD-conjugated poly(MPC-co-MNHS) grafted parylene films should be attributed to the adhesion force between the hydrophilic parylene films and the underlying surface.59,60 Generally, the adhesion force at the contact interface comprises capillary force, van der Waals force, electrostatic force, and chemical bonding force. In this context, the electrostatic and chemical bonding forces should not be the primary factors due to the absence of net charge and saturation of surface chemical bonds.61 We considered the capillary force, a combination of the surface tension and Laplace pressure, to be responsible for the excellent conformability of these hydrophilic parylene films.62
This biomimetic parylene film can effectively eliminate the biofouling on the surface. As shown in Fig. 3i, unlike the pristine parylene film, no fluorescence signal was observed on the PMPC-grafted parylene and RGD-conjugated poly(MPC-co-MNHS)-grafted parylene films after exposure to the FITC-FNG solution. Moreover, it is worth noting that no WBCs could attach to the PMPC grafted parylene and RGD-conjugated poly(MPC-co-MNHS) grafted parylene films, whereas many white blood cells (WBCs) adhered to the surface of the parylene film (Fig. 3j). Its strong resistance to WBC interaction is crucial to ensure its biocompatibility, as WBCs are one of the primary cells that mediate inflammatory responses.63 Clearly, both above results indicated that the RGD-conjugated poly(MPC-co-MNHS) grafted parylene film retains an excellent resistance to nonspecific protein and cell binding, offering an opportunity to construct a ligand-defined specific interaction to targeted cells on a background resisting biofouling.
Cell selectivity of RGD-conjugated zwitterionic parylene
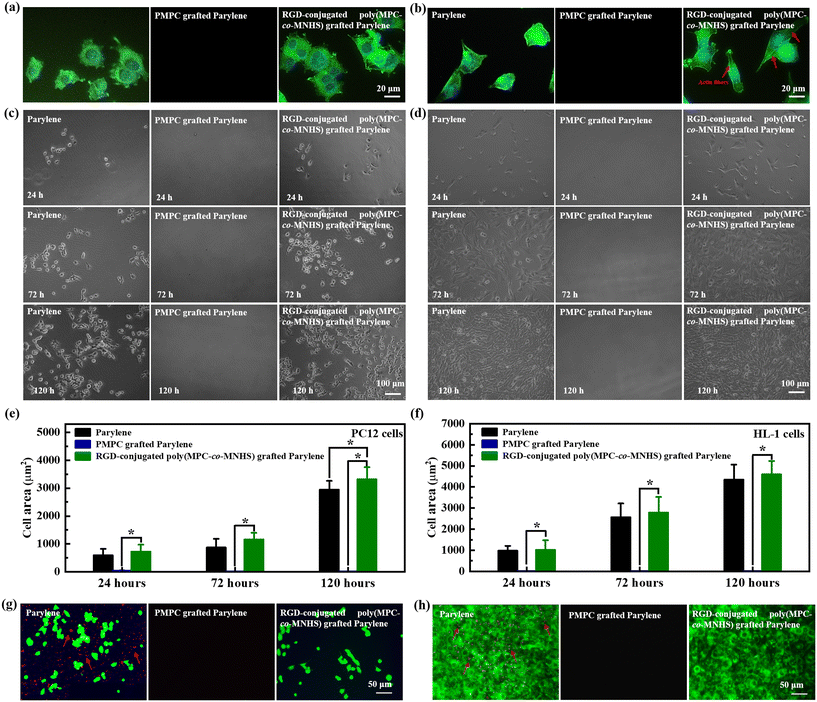
To evaluate its potential as a cell attractive encapsulation material for bioelectronic devices, we cultured neuronal model cells (PC12) and cardiac muscle cells (HL-1) on the parylene film for 24 h. F-actin, a major cytoskeleton component, was stained with phalloidin (green) to assess cell adhesion and spreading. The results demonstrated that both PC12 cells (Fig. 4a) and HL-1 cells (Fig. 4b) could adhere to and extend well on the RGD-conjugated poly(MPC-co-MHNS) grafted parylene film, which is initially bioinert before the RGD conjugation. It also indicated that this strong cell–substrate interaction of the biomimetic parylene originates from the specific ligand–receptor interactions defined by the RGD peptides.We further explored the cell morphologies of PC12 and HL-1 cells to compare the two cell–substrate interactions defined by the nonspecific interaction and the specific ligand–receptor interaction. Compared to pristine parylene, the PC12 cells presented a relatively more extended morphology and a higher spread area (Fig. S3a†) on the RGD-conjugated poly(MPC-co-MHNS) grafted parylene film. The PC12 cells displayed a round morphology with limited filopodial spreading (Fig. 4a). In contrast, the HL-1 cells displayed different morphologies on the two surfaces. The HL-1 cells on the pristine parylene film retained a compact morphology with a slight elongation (Fig. 4b). The HL-1 cells adhered on the RGD-conjugated poly(MPC-co-MHNS) grafted parylene film, developed a more stretched morphology, and showed a more prominent actin stress fiber formation. The area of the cells on the RGD-conjugated poly(MPC-co-MHNS) is slightly larger than that of the cells on the pristine parylene film (Fig. S3b†). In addition, the HL-1 cells plated on the RGD-conjugated poly(MPC-co-MHNS) grafted parylene film were more spread out and elongated than those cultured on the pristine parylene film. This result indicates that the RGD-modified parylene surface can facilitate the formation of a robust focal adhesion for cells comparable to or more robust than that of the pristine parylene by offering ligand-defined specific binding sites for integrin proteins of the cell membrane.
We cultured the PC12 cells (Fig. 4c) and HL-1 cells (Fig. 4d) on the RGD-conjugated poly(MPC-co-MHNS) grafted parylene films for 24, 72, and 120 h, including the pristine parylene film and the PMPC grafted parylene film as control substrates. As shown in Fig. 4e and f, the pristine parylene and the RGD-conjugated poly(MPC-co-MHNS) grafted parylene films could support PC12 and HL-1 cell adhesion and proliferation generously. In contrast, PMPC-grafted parylene films did not support cell adhesion due to the strong hydration and electrical neutrality of the PMPC molecular chains, which effectively prevented cell attachment to the surface. The spreading area of cells for the RGD-conjugated poly(MPC-co-MHNS) grafted parylene films was consistently slightly larger than that for the pristine parylene films and corresponded well with the focal adhesion result. It suggests that either the area covered by a single cell or the proliferation rate of the culture on the RGD-conjugated poly(MPC-co-MHNS) grafted parylene film is larger than that on the pristine parylene. Considering the FDA compliance of pristine parylene, these results likely support the application of the biomimetic parylene material in real-world scenarios, as the above results do not suggest any compromise in the biocompatibility of parylene.
To examine the cell selectivity of these bioactive parylene films, we cultured PC12 and HL-1 cells on the films in the presence of WBCs. To visualize the complex cell–substrate interaction of PC12 and HL-1 cells and WBCs, we stained PC12 and HL-1 cells with green DIO (green fluorescence) and WBCs with red DIL (red fluorescence). In detail, WBCs were cultured on the films at a density of 1 × 106 mL−1 and cultured for 12 h; PC12 cells or HL-1 cells were subsequently seeded at a cell density of 1 × 105 cells per mL for further evaluating the cell–substrate interaction. As shown in Fig. 4g, both PC12 and WBCs cells could adhere to the pristine parylene surface, whereas only PC12 cells could adhere to the RGD-conjugated poly(MPC-co-MHNS) grafted parylene surface. Similar results could be observed in Fig. 4h, where the biomimetic parylene film only interacted with HL-1 cells. The fundamental reason for this phenomenon is that the cell–substrate interaction is defined only by the RGD peptides present on the biomimetic parylene surface since it has intrinsic resistance to biofouling, as demonstrated in the middle picture of Fig. 4g and h. In addition, the RGD-conjugated poly(MPC-co-MHNS) grafted parylene surface still exhibited excellent resistance to WBC adhesion. Similar phenomena have been extensively reported for RGD-conjugated antifouling surfaces, although the underlying reason remains unclear.64–67
The results show that the interaction between cells and the biomimetic parylene film is determined solely by the ligands on the parylene surface. This means that it is possible to use specific biomolecules to create a highly selective interaction with cells on the biomimetic parylene film. Developing a selective cell–substrate interaction in the encapsulation material could improve the ability of bioelectronic devices to attract specific cells and tissues for better electrical communication, while also preventing biofouling with immune proteins and cells. This would ultimately improve the biocompatibility of bioelectronic devices.
Conclusion
In this study, we developed a soft, intrinsically antifouling parylene film with specific biological interactions. The film consists of a pristine parylene bottom layer and a thin top layer of 2-bromoisobutyrate-functionalized parylene, which is used to graft RGD-conjugated zwitterionic polymers further. This layer-by-layer structure ensures effective encapsulation with a low leakage current and enables surface function, enhancing its suitability for biomedical applications. Grafting zwitterionic polymers has provided exceptional biofouling resistance, reduced the surface modulus by three orders of magnitude (∼45 kPa), and significantly enhanced mechanical compliance. Additionally, conjugation with specific peptides imparts excellent cell selectivity to the parylene film, even in a high density of white blood cells. We believe that the design and development of this biomimetic parylene could address the challenges of biological fouling and the absence of cell affinity sites in current parylene materials. It opens up a promising possibility for creating biocompatible flexible bioelectronic devices, enabling seamless integration with cells and tissues.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its ESI.†Conflicts of interest
There are no conflicts of interest associated with this manuscript's publication. As the corresponding author, I confirm that all the authors have read and approved this manuscript for submission.Acknowledgements
B. Z. acknowledges the financial support from the NSFC (22175111, 21474014). Z. G. is grateful for the financial support from the NSFC (21704013).References
- G. Balakrishnan, J. Song, C. Mou and C. J. Bettinger, Adv. Mater., 2022, 34, 2106787 CrossRef CAS PubMed.
- G. Yao, C. Yin, Q. Wang, T. Zhang, S. Chen, C. Lu, K. Zhao, W. Xu, T. Pan, M. Gao and Y. Lin, J. Materiomics, 2020, 6, 397–413 CrossRef.
- M. Kaltenbrunner, T. Sekitani, J. Reeder, T. Yokota, K. Kuribara, T. Tokuhara, M. Drack, R. Schwodiauer, I. Graz, S. Bauer-Gogonea, S. Bauer and T. Someya, Nature, 2013, 499, 458–463 CrossRef CAS PubMed.
- X. Wang, X. Sun, D. Gan, M. Soubrier, H. Y. Chiang, L. Yan, Y. Li, J. Li, S. Yu, Y. Xia, K. Wang, Q. Qin, X. Jiang, L. Han, T. Pan, C. Xie and X. Lu, Matter, 2022, 5, 1204–1223 CrossRef CAS.
- D. H. Kim, J. Viventi, J. J. Amsden, J. Xiao, L. Vigeland, Y. S. Kim, J. A. Blanco, B. Panilaitis, E. S. Frechette, D. Contreras, D. L. Kaplan, F. G. Omenetto, Y. Huang, K. C. Hwang, M. R. Zakin, B. Litt and J. A. Rogers, Nat. Mater., 2010, 9, 511–517 CrossRef CAS PubMed.
- Z. Shi, F. Zheng, Z. Zhou, M. Li, Z. Fan, H. Ye, S. Zhang, T. Xiao, L. Chen, T. H. Tao, Y. L. Sun and Y. Mao, Adv. Sci., 2019, 6, 1801617 CrossRef PubMed.
- M. Golda-Cepaa, K. Engvallb, M. Hakkarainenc and A. Kotarbaa, Prog. Org. Coat., 2020, 140, 1054932 CrossRef.
- J. S. Mandelli, J. Koepp, A. Hama, S. Sanaur, G. A. Rae and C. R. Rambo, Biomed. Microdevices, 2021, 23, 2 CrossRef CAS PubMed.
- D. Rodger, A. Fong, W. Li, H. Ameri, A. Ahuja, C. Gutierrez, I. Lavrov, H. Zhong, P. Menon and E. Meng, Sens. Actuators, B, 2008, 132, 449–460 CrossRef CAS.
- J. M. Hsu, L. Rieth, R. A. Normann, P. Tathireddy and F. Solzbacher, IEEE Trans. Biomed. Eng., 2009, 56, 23–29 Search PubMed.
- J. P. Seymour, Y. M. Elkasabi, H. Y. Chen, J. Lahann and D. R. Kipke, Biomaterials, 2009, 30, 6158–6167 CrossRef CAS PubMed.
- B. J. Kim, J. T. Kuo, S. A. Hara, C. D. Lee, L. Yu, C. A. Gutierrez, T. Q. Hoang, V. Pikov and E. Meng, J. Neural Eng., 2013, 10, 045002 CrossRef CAS PubMed.
- S. A. Hara, B. J. Kim, J. T. Kuo, C. D. Lee, E. Meng and V. Pikov, J. Neural Eng., 2016, 13, 066020 CrossRef PubMed.
- B. J. Kim, B. Chen, M. Gupta and E. Meng, J. Micromech. Microeng., 2014, 24, 065003 CrossRef CAS.
- X. Wang, A. W. Hirschberg, H. Xu, Z. Slingsby-Smith, A. Lecomte, K. Scholten, D. Song and E. Meng, J. Microelectromech. Syst., 2020, 29, 499–513 CAS.
- J. T. Kuo, B. J. Kim, S. A. Hara, C. D. Lee, C. A. Gutierrez, T. Q. Hoang and E. Meng, Lab Chip, 2013, 13, 554–561 RSC.
- D. W. Park, J. P. Ness, S. K. Brodnick, C. Esquibel, J. Novello, F. Atry, D. H. Baek, H. Kim, J. Bong, K. I. Swanson, A. J. Suminski, K. J. Otto, R. Pashaie, J. C. Williams and Z. Ma, ACS Nano, 2018, 12, 148–157 CrossRef CAS PubMed.
- Y. Morikawa, S. Yamagiwa, H. Sawahata, R. Numano, K. Koida, M. Ishida and T. Kawano, Adv. Healthcare Mater., 2018, 7, 1701100 CrossRef PubMed.
- C. F. Guimarães, L. Gasperini, A. P. Marques and R. L. Reis, Nat. Rev. Mater., 2020, 5, 351–370 CrossRef.
- L. Zhang, Z. Cao, T. Bai, L. Carr, J. R. Ella-Menye, C. Irvin, B. D. Ratner and S. Jiang, Nat. Biotechnol., 2013, 31, 553–556 CrossRef CAS PubMed.
- M. D. Swartzlander, C. A. Barnes, A. K. Blakney, J. L. Kaar, T. R. Kyriakides and S. J. Bryant, Biomaterials, 2015, 41, 26–36 CrossRef CAS PubMed.
- M. G. Saifer, L. D. Williams, M. A. Sobczyk, S. J. Michaels and M. R. Sherman, Mol. Immunol., 2014, 57, 236–246 CrossRef CAS PubMed.
- B. Li, P. Jain, J. Ma, J. K. Smith, Z. Yuan, H. C. Hung, Y. He, X. Lin, K. Wu, J. Pfaendtner and S. Jiang, Adv. Sci., 2019, 5, eaaw9562 CrossRef CAS PubMed.
- B. Li, Z. Yuan, H. C. Hung, J. Ma, P. Jain, C. Tsao, J. Xie, P. Zhang, X. Lin, K. Wu and S. Jiang, Angew. Chem., Int. Ed., 2018, 57, 13873–13876 CrossRef CAS PubMed.
- Q. Li, C. Wen, J. Yang, X. Zhou, Y. Zhu, J. Zheng, G. Cheng, J. Bai, T. Xu, J. Ji, S. Jiang, L. Zhang and P. Zhang, Chem. Rev., 2022, 122, 17073–17154 CrossRef CAS PubMed.
- J. G. Wu, J. H. Chen, K. T. Liu, S. C. Luo and S. C. Luo, ACS Appl. Mater. Interfaces, 2019, 11, 21294–21307 CrossRef CAS PubMed.
- A. Erfani, J. Seaberg, C. P. Aichele and J. D. Ramsey, Biomacromolecules, 2020, 21, 2557–2573 CrossRef CAS PubMed.
- J. Yang, Z. Xu, J. Wang, L. Gai, X. Ji, H. Jiang and L. Liu, Adv. Funct. Mater., 2021, 31, 2009438 CrossRef CAS.
- K. Ishihara, Langmuir, 2019, 35, 1778–1787 CrossRef CAS PubMed.
- K. Ishihara, M. Mu, T. Konno, Y. Inoue and K. Fukazawa, J. Biomater. Sci., Polym. Ed., 2017, 28, 884–899 CrossRef CAS PubMed.
- B. Zhu, S. C. Luo, H. Zhao, H. A. Lin, J. Sekine, A. Nakao, C. Chen, Y. Yamashita and H. H. Yu, Nat. Commun., 2014, 5, 4523 CrossRef CAS PubMed.
- S. Qian, S. Zhang, Q. Pan, Q. Sun, R. Song, Y. Zhang, S. Zhang, Z. Geng, J. Huang, L. Wang, Y. He and B. Zhu, Appl. Surf. Sci., 2024, 660, 160032 CrossRef CAS.
- S. Zhang, S. Qian, G. Zhao, Q. Pan, R. Song, T. Zhang, S. Zhang, Z. Geng, J. Huang, L. Wang and B. Zhu, Chem. Eng. J., 2024, 483, 148980 CrossRef CAS.
- Q. Yu, Y. Zhang, H. Wang, J. Brash and H. Chen, Acta Biomater., 2011, 7, 1550–1557 CrossRef CAS PubMed.
- Q. Chen, D. Zhang, J. Gu, H. Zhang, X. Wu, C. Cao, X. Zhang and R. Liu, Acta Biomater., 2021, 126, 45–62 CrossRef CAS PubMed.
- J. Baggerman, M. M. J. Smulders and H. Zuilhof, Langmuir, 2019, 35, 1072–1084 CrossRef CAS PubMed.
- H. Hao, Y. Xue, Y. Wu, C. Wang, Y. Chen, X. Wang, P. Zhang and J. Ji, Bioact. Mater., 2023, 28, 1–11 CAS.
- K. Ye, X. Wang, L. Cao, S. Li, Z. Li, L. Yu and J. Ding, Nano Lett., 2015, 15, 4720–4729 CrossRef CAS PubMed.
- K. A. Kilian and M. Mrksich, Angew. Chem., Int. Ed., 2012, 51, 4891–4895 CrossRef CAS PubMed.
- H. Guo, M. Bai, C. Wen, M. Liu, S. Tian, S. Xu, X. Liu, Y. Ma, P. Chen, Q. Li, X. Zhang, J. Yang and L. Zhang, J. Colloid Interface Sci., 2021, 600, 561–571 CrossRef CAS PubMed.
- T. Kilic, I. Gessner, Y. K. Cho, N. Jeong, J. Quintana, R. Weissleder and H. Lee, Adv. Mater., 2022, 34, e2107892 CrossRef PubMed.
- R. Janissen, P. K. Sahoo, C. A. Santos, A. M. da Silva, A. A. G. von Zuben, D. E. P. Souto, A. D. T. Costa, P. Celedon, N. I. T. Zanchin, D. B. Almeida, D. S. Oliveira, L. T. Kubota, C. L. Cesar, A. P. Souza and M. A. Cotta, Nano Lett., 2017, 17, 5938–5949 CrossRef CAS PubMed.
- F. Sun, H. C. Hung, A. Sinclair, P. Zhang, T. Bai, D. D. Galvan, P. Jain, B. Li, S. Jiang and Q. Yu, Nat. Commun., 2016, 7, 13437 CrossRef CAS PubMed.
- Z. Song, M. Chen, C. Ding and X. Luo, Anal. Chem., 2020, 92, 5795–5802 CrossRef CAS PubMed.
- A. K. Patel, A. D. Celiz, D. Rajamohan, D. G. Anderson, R. Langer, M. C. Davies, M. R. Alexander and C. Denning, Biomaterials, 2015, 61, 257–265 CrossRef CAS PubMed.
- M. Beyer, T. Felgenhauer, F. R. Bischoff, F. Breitling and V. Stadler, Biomaterials, 2006, 27, 3505–3514 CrossRef CAS PubMed.
- J. Chen, J. Cao, J. Wang, M. F. Maitz, L. Guo, Y. Zhao, Q. Li, K. Xiong and N. Huang, J. Colloid Interface Sci., 2012, 368, 636–647 CrossRef CAS PubMed.
- X. Wang, X. Zheng, Y. Duan, L. Ma and C. Gao, ACS Appl. Mater. Interfaces, 2019, 11, 15170–15180 CrossRef CAS PubMed.
- H. A. Lin, B. Zhu, Y. W. Wu, J. Sekine, A. Nakao, S. C. Luo, Y. Yamashita and H. H. Yu, Adv. Funct. Mater., 2018, 28, 1703890 CrossRef.
- T. D. Kozai, N. B. Langhals, P. R. Patel, X. Deng, H. Zhang, K. L. Smith, J. Lahann, N. A. Kotov and D. R. Kipke, Nat. Mater., 2012, 11, 1065–1073 CrossRef CAS PubMed.
- A. Misiura, C. Dutta, W. Leung, O. J. Zepeda, T. Terlier and C. F. Landes, J. Chem. Phys., 2022, 156, 094707 CrossRef CAS PubMed.
- O. Koniev and A. Wagner, Chem. Soc. Rev., 2015, 44, 5495–5551 RSC.
- U. Hersel, C. Dahmen and H. Kessler, Biomaterials, 2003, 24, 4385–4415 CrossRef CAS PubMed.
- J. J. Senkevich, S. B. Desu and V. Simkovic, Polymer, 2000, 41, 2379–2390 CrossRef CAS.
- A. Kahouli, A. Sylvestre, S. Pairis and J. F. Laithier, Polymer, 2012, 53, 3001–3007 CrossRef CAS.
- M. Chakraborty, K. Chu, A. Shrestha, X. S. Revelo, X. Zhang, M. J. Gold, S. Khan, M. Lee, C. Huang, M. Akbari, F. Barrow, Y. T. Chan, H. Lei, N. K. Kotoulas, J. Jovel, C. Pastrello, M. Kotlyar, C. Goh, E. Michelakis, X. Clemente-Casares, P. S. Ohashi, E. G. Engleman, S. Winer, I. Jurisica, S. Tsai and D. A. Winer, Cell Rep., 2021, 34, 108609 CrossRef CAS PubMed.
- A. Lecomte, E. Descamps and C. Bergaud, J. Neural Eng., 2018, 15, 031001 CrossRef PubMed.
- J. Rivnay, H. Wang, L. Fenno, K. Deisseroth and G. G. Malliaras, Sci. Adv., 2017, 3, e1601649 CrossRef PubMed.
- L. Chen and L. Qian, Friction, 2020, 9, 1–28 CrossRef.
- K. Li and S. Cai, Soft Matter, 2014, 10, 8202–8209 RSC.
- J. S. Wexler, T. M. Heard and H. A. Stone, Phys. Rev. Lett., 2014, 112, 066102 CrossRef PubMed.
- J. Qian and H. Gao, Acta Biomater., 2006, 2, 51–58 CrossRef PubMed.
- L. A. Abdulkhaleq, M. A. Assi, R. Abdullah, M. Zamri-Saad, Y. H. Taufiq-Yap and M. N. M. Hezmee, Vet. World, 2018, 11, 627–635 CrossRef CAS PubMed.
- T. Li, N. Li, Y. Ma, Y. J. Bai, C. M. Xing and Y. K. Gong, J. Mater. Chem. B, 2019, 7, 6087–6098 RSC.
- Y. Ma, X. Y. Qiao, Q. Lu, R. Li, Y. J. Bai, X. Li, S. P. Zhang and Y. K. Gong, J. Mater. Chem. B, 2020, 8, 4299–4309 RSC.
- Y. Xiao, M. Wang, L. Lin, L. Du, M. Shen and X. Shi, Nanomedicine, 2019, 14, 183–199 CrossRef CAS PubMed.
- V. A. Tegoulia and S. L. Cooper, J. Biomed. Mater. Res., 2000, 50, 291–301 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lf00289j |
| This journal is © The Royal Society of Chemistry 2025 |