Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Phosphorous-containing, amphiphilic ABB′ copolymers as siRNA nanocarriers with enhanced stability, reduced in vitro cytotoxicity, and efficient knockdown ability for the treatment of ocular diseases†
Philipp
Weingarten‡
 a,
Molly Tzu-Yu
Lin‡
b,
Moritz
Kränzlein
a,
Molly Tzu-Yu
Lin‡
b,
Moritz
Kränzlein
 a,
Agnes
Fietz
b,
Iris
Kachel
c,
José
Hurst
a,
Agnes
Fietz
b,
Iris
Kachel
c,
José
Hurst
 b,
Sven
Schnichels
b,
Sven
Schnichels
 b and
Friederike
Adams
b and
Friederike
Adams
 *bc
*bc
aWacker-Lehrstuhl für Makromolekulare Chemie, Catalysis Research Center, Technische Universität München, Lichtenbergstraße 4, 85748 Garching bei München, Germany
bCenter for Ophthalmology, University Eye Hospital Tübingen, Elfriede-Aulhorn-Strasse 7, 72076 Tübingen, Germany
cChair of Macromolecular Materials and Fiber Chemistry, Institute of Polymer Chemistry, University of Stuttgart, Pfaffenwaldring 55, 70569 Stuttgart, Germany. E-mail: friederike.adams@ipoc.uni-stuttgart.de
First published on 9th January 2025
Abstract
An RNA interference (RNAi) strategy using siRNA targeting NF-κB could counteract its harmful effects and provide therapeutic benefits for several ocular pathophysiologies. Rare-earth metal-mediated group-transfer polymerization was employed to synthesize precise AB and ABB′ block copolymers from 2-vinylpyridine and dialkyl vinylphosphonates (DAVP, alkyl = ethyl, allyl) as siRNA nanocarriers. Modifying the allyl group of the vinylphosphonate unit transformed these polymers into cationic, amphiphilic copolymers. All polymers showed maximum siRNA encapsulation at a low N/P ratio of 2. The cationic unit distribution along the PDAVP chain influences the encapsulation capacity and the stability of the polyplexes. A less dense distribution of cationic units led to increased amounts of free siRNA, even at higher N/P ratios, but to a better stability of the polyplexes. Diffused calcein signals observed from cells treated with ABB′/siRNA polyplexes revealed an endosomal escape capability while maintaining excellent in vitro cell viability. Ocular cell lines transfected with these polyplexes demonstrated a superior NF-κB/RelA gene silencing efficiency. This study highlights the potential of phosphorous-containing, amphiphilic polymers as nucleic acid carriers with enhanced stability, excellent cytotoxicity profiles, and efficient knockdown ability.
Introduction
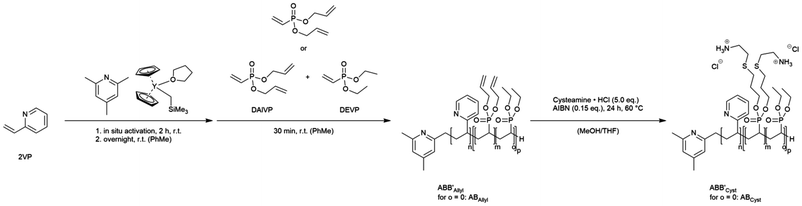
Nuclear factor kappa B (NF-κB) belongs to a family of inducible transcription factors, which, through target genes, regulate key biological processes such as wound healing, angiogenesis, apoptosis, immune, inflammatory, and stress response.1 NF-κB proteins are also known as Rel (REL Proto-Oncogene, NF-κB subunit) family of proteins. Amongst the NF-κB/Rel subunits, the p65/RelA is the most well-studied member, which interacts with NF-κB inhibitor proteins such as nuclear factor of kappa light polypeptide gene enhancer in B-cell inhibitor alpha, or beta (IκBα, IκBβ) in the cytoplasm.2 Various pathological or physiological conditions can trigger the phosphorylation and degradation of IκB, leading to the release of the NF-κB p65 subunit. The free p65 then translocates to the cellular nucleus, activating its regulatory role of various transcription factors and its target genes, including cytokines and chemokines.3 The activation of NF-κB can be induced by oxidative stress which was found to affect ocular tissues such as cornea, iris, lens, and retina.2 On the other hand, the inflammation associated with the activation of NF-κB increased expression of cytokines and chemokines has been reported in several diseases including diabetes and uveitis.4 In fact, increased NF-κB activation has been observed in the retina of diabetic patients and animal models5 and therefore required appropriate therapeutic intervention. However, the downregulation of NF-κB p65 remains challenging due to its cytoplasmic localization. Therefore, RNA interference (RNAi) strategy targeting NF-κB p65 with a respective small interfering ribonucleic acid (siRNA) could represent an attractive approach to preventing its activity, alleviating associated ocular pathophysiological conditions, including neuroinflammation and neovascularization in diabetic eye diseases and ocular surface inflammation.Tailor-made polymers containing different structural elements were designed to obtain delivery systems capable of forming polymer-nucleic acid complexes (polyplexes) and show a perfect balance between sufficient stability and release of the payload.6 Essential structural elements are cationic moieties that electrostatically interact with the anionic phosphate backbone of the nucleic acid. Aliphatic amino pendant groups are commonly used as they provide suitable pKa values to facilitate protonation under physiological conditions and allow the formation of stable polyplexes. After administration of the polyplexes, cellular uptake is a crucial part of the metabolism of siRNA nanocarriers, which can be improved by attaching hydrophobic moieties to the polymer due to better compatibility with the cell membrane.7 Upon cellular uptake, one of the critical factors to obtain a successful siRNA delivery requires endosomal escape to allow the release of polyplexes into the cytosol.8,9 The prevalent hypothesis of the proton sponge effect was commonly observed when polycationic structures are used to deliver nucleic acids. These studies postulated that the buffering capacity from polycationic structures induces an increased osmotic pressure within the maturing endosome and finally leads to its burst.10 As highlighted, the significant role of different cationic moieties or amino groups with pKa values in a broad range is believed to be the primary driving force for the escape from the endosomal compartment. The significance of the interplay between cationic and non-cationic segments in a polymer became more important since such structures became synthetically accessible, either via copolymerization or post-polymerization functionalization.11 Hydrophilic polymer main or side chains could provide general amphiphilicity to a rather hydrophobic polymer, enhance the anti-opsonization properties, and might increase the stealthing properties of polymeric nucleic acid vehicles. In particular, polyethylene glycol (PEG) is a broadly studied segment in polymers and polymeric nanoparticles12 and is currently the standard building block for preventing nanoparticles from aggregation, opsonization, and phagocytosis.13 The concerns of its unintended off-target effects have urged the need for an alternative solution in pharmaceutical applications.14 Phosphoester-containing polymers, in general, already showed promising properties for biomedical purposes.15 Especially, side-chain and in-chain poly(phosphonates) were shown to provide stealthing properties to nanoparticle-based drug delivery systems and would suggest structurally related poly(dialkyl vinylphosphonates) (PDAVP) to have a similar effect on the stability of potential PDAVP-based drug delivery systems.16 PDAVP are hydrolytically stable polymers that could induce an extended half-life within a biological system and can be combined with other polar Michael monomers. The precise polymerization technique of rare-earth metal group-transfer polymerization (REM-GTP) is the only polymerization method that facilitates a controlled (co)polymerization of DAVP-containing monomers.17 First studies on amphiphilic tailor-made block copolymers synthesized via REM-GTP consisting of poly(2-vinylpyridine) (P2VP) and poly(diethyl vinylphosphonate) (PDEVP) showed micellization behavior of these block copolymers that showed low cytotoxicity and a cumulative dual-responsive drug release triggered by pH-value and temperature.18–20 In this study, 2VP was copolymerized via REM-GTP with functional diallyl vinylphosphonate (DAlVP) and hydrophilic diethyl vinylphosphonate (DEVP) using an in situ CH-bond activated yttriocene complex.21 Since REM-GTP enables the controlled polymerization of DAlVP while preserving the integrity of the allyl side chains, the obtained polymers were transformed into polycations under thiol–ene conditions using cysteamine as a functional pendant group. Polymer/siRNA showed promising complexation properties at low N/P ratios while leading to well-defined and stable polyplexes. To explore the potential application in ocular therapeutics, the in vitro cell viability with a broad range concentration of polymers was tested on the human Müller cell line Moorfields/Institute of Ophthalmology-Müller 1 (MIO-M1). Moreover, the capability of the synthesized polymers to exhibit endosomal escape was confirmed, and ABB′ polymers that consisted of additional hydrophilic PDEVP units demonstrated better endosomal escape ability than AB polymers or commonly used poly(ethylenimine) (PEI), which was further approved by a superior NF-κB/RelA gene silencing efficiency by ABB′/siNF-κB polyplexes.
Results and discussion
Polymer synthesis and characterization
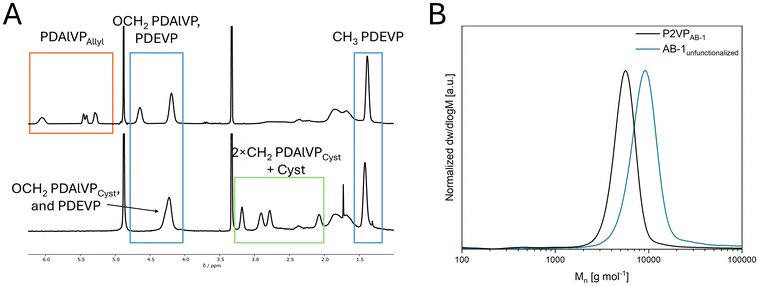
Amphiphilic block copolymer precursors with allyl groups were produced through REM-GTP by adding hydrophobic, pH-sensitive 2VP and hydrophilic dialkyl vinylphosphonates in a sequential manner (Scheme 1). The order of addition for synthesizing these precursors was determined by the coordination strength of the monomers to the metal center, with 2VP exhibiting a weaker coordination than dialkyl vinylphosphonates. The polymerization of 2VP (block A) was initiated after complete C–H bond activation of sym-collidine (sym-col) with Cp2Y(CH2TMS)(thf) (Cp = C5H5, TMS = trimethyl silyl, thf = tetrahydrofuran) to form Cp2Y(sym-col) as the active catalyst.22 The addition of DAlVP (block B) or a pre-mixed mixture of DAlVP and DEVP (B′) is conducted after 2VP was consumed to the full extent.23 The resulting polymers’ composition and block copolymer nature were confirmed using 1H NMR and 1H DOSY NMR spectroscopy (Fig. S1 and S2†) and size-exclusion chromatography (SEC, Fig. 1B and S5†). Various ratios of A/B, A/BB′, and B/B′ were used to synthesize AB and ABB′ type polymers with adjustable molar masses and monomer unit ratios, as detailed in Table 1. All synthesized AB- and ABB′-type block copolymers exhibited narrow molecular weight distributions (Đ < 1.2). A variation of the respective units is expected to influence the polymer properties in terms of their overall hydrophilicity (increase PDEVP) or hydrophobicity (increase P2VP), while the increase of functional poly(diallyl vinylphosphonate) (PDAlVP) leads to a higher cationic charge density upon functionalization. Using the Fineman–Ross technique,24 copolymerization parameters indicate a slight compositional drift from DEVP to DAlVP within the BB′-block (rDEVP = 1.31, rDAlVP = 0.87, see ESI, Fig. S6†).| Entry | P2VP/PDAlVP/PDEVP (A/B/B′)a | M n,NMR,AB(B′)Allyl [kg mol−1] | Đ AB(B′)Allyl | M n,NMR,AB(B′)Cyst [kg mol−1] | CMCe [μmol mL−1] | D h,AB(B′)Cyst [nm] | PDIf | Zeta potentialAB(B′)Cystg [mV] |
|---|---|---|---|---|---|---|---|---|
| a A = P2VP, B = PDAlVP, B′ = PDEVP. BB′ notation indicates statistical copolymerization. Absolute copolymer composition determined via integration of P2VP aromatic signals versus phosphonate signals in 1H NMR spectra of isolated polymers. b Calculated molecular weight of the block copolymer via copolymer composition and Mn,abs,A of block A (absolute molecular weight determination of the P2VP block by SEC in DMF, 30 °C, with 25 mmol L−1 LiBr, triple detection, dn/dc = 0.149 mL g−1, more SEC data: see ESI†). c Determination of polydispersity of block copolymers via SEC in DMF. d Calculated molecular weight of cysteamine-modified polymer via1H NMR spectroscopy and Mn,NMR,AB(B′)Allyl. e Calculated by dividing the obtained CMC [mg mL−1] (Fig. S7–S12†) by the molecular weight Mn,NMR,AB(B′)Cyst. f Determined via DLS. Standard deviation of Z-average given to indicate reproducibility of measurements. Respective size distributions by number and intensity are given in Fig. S19–S24 and Table S1.† g Determined via laser Doppler electrophoresis. Standard deviation given to indicate reproducibility of measurements. h Determination of polydispersity of ABB′-4 via SEC in THF/H2O (1/1) using SEC-MALS. | ||||||||
| AB-1 | 106/44 | 19.4 | 1.14 | 29.4 | 0.014 | 199 ± 2 | 0.31 | 62 ± 1 |
| AB-2 | 52/56 | 16.0 | 1.17 | 28.7 | 0.013 | 88.2 ± 2 | 0.34 | 51 ± 1 |
| ABB′-1 | 85/71/44 | 29.5 | 1.13 | 45.7 | 0.010 | 73.7 ± 1 | 0.40 | 55 ± 1 |
| ABB′-2 | 156/31/59 | 31.9 | 1.12 | 39.0 | 0.011 | 175 ± 1 | 0.12 | 46 ± 1 |
| ABB′-3 | 75/36/77 | 27.3 | 1.17 | 35.5 | 0.011 | 151 ± 1 | 0.28 | 55 ± 0 |
| ABB′-4 | 37/25/123 | 28.8 | 1.07h | 34.5 | 0.025 | 184 ± 2 | 0.20 | 30 ± 1 |
Cationic polymers containing primary amines in the pendant groups were generated under thermal thiol–ene conditions.25 PDAlVP repeating units were functionalized with cysteamine hydrochloride using AIBN as an initiator to obtain successful quantitative conversion and functionalization proven via1H NMR and 1H DOSY NMR spectroscopy (Fig. 1A and Fig. S3 and S4†). Amphiphilic properties were studied by DLS and zeta potential measurements. In the case of AB-type block copolymers, the micelle size of polymers correlates with the molecular weight and composition of the polymer. AB-1 and AB-2 generate well-defined micelles, for which a dependence on the amount of copolymerized hydrophobic P2VP can be observed. The zeta potential of all AB-type block copolymers appears to be in a similar order of magnitude (55 mV), independent from the amount of P2VP or cationic units. In the case of the ABB′-type block copolymers, the copolymerized PDEVP units induce higher degree of complexity in establishing structure–property relationships. ABB′-2 and ABB′-3 mainly differ in the amount of P2VP, which is reflected by the smaller micelle size of ABB′-3, while the obtained micelles of ABB′-2 are significantly less dispersed due to a higher amount of P2VP. The size of ABB′-1 does not align with the previous findings since the larger number of cationic pendant groups compared to uncharged units induces a higher polydispersity of the obtained micelles. All three mentioned ABB′-type block copolymers show a zeta potential in the same order of magnitude as the AB-type block copolymers. However, ABB′-4 showed significantly smaller micelles due to its smaller lipophilic chain, while the lower zeta potential of 30 mV could be attributed to the low cationic density within the BB′ block. The critical micelle concentration (CMC) for AB-1 and AB-2 revealed similar values of approximately 0.013 and 0.014 μmol mL−1 (Table 1, Fig. S7–S12†). ABB′-1–3 showed results in a similar order of magnitude with values at 0.010 μmol mL−1 and 0.011 μmol mL−1 while ABB′-4 exhibited a CMC of 0.025 μmol mL−1 due to the larger amount of hydrophilic PDEVP. The apparent CMC values are significantly higher than those of AB-type P2VP-b-PDEVP block copolymers reported in literature,18,26 mainly due to the increased hydrophilicity induced by cationic units.
The effect of polymers on cellular mitochondrial activity and cell number were investigated using MTS assay and crystal violet staining, respectively (MTS assay, see Fig. 2; crystal violet assay, see Fig. S13†) to display parts of the biocompatibility profiles of AB and ABB′ polymers.27 For the MTS assays, all polymers showed relatively high IC50 in a comparable order of magnitude. The metabolic activity for the cells treated with AB-1 is significantly higher than for AB-2, since the relative number of cationic groups concerning the molecular weight of the polymer is lower. Among the ABB′ block copolymers, the ones that exhibit lower weight fractions of cationic units (ABB′-2 and ABB′-4, Table S1†) induced a less pronounced disruption in the metabolic activity, causing IC50 values of 194 ± 6 μg mL−1 and 288 ± 37 μg mL−1. Nevertheless, the ones with higher relative amounts of cationic repeating units (ABB′-1 and ABB′-3) still showed acceptable metabolic activity at relevant concentrations. Among all the AB and ABB′ polymers, cells subjected to ABB′-1 (65 wt% cationic groups) or AB-2 (81 wt% cationic groups) treatment showed the lowest IC50 values with 57.2 ± 1.7 μg mL−1 and 73.4 ± 3.0 μg mL−1 respectively. The number of cationic repeating units appears to be a key factor in determining its cytotoxic effects (Table 1, Fig. 2). Additionally, the higher molecular weight of the ABB′ copolymers might affect the metabolic activity since AB-1 has a higher IC50 value in comparison to ABB′-1 while having a similar weight fraction of cationic repeating units. The MTS assay gives thus insights into the contribution of each structural motif to the polymer properties. Since hydrophilic poly(vinylphosphonates) are known to show a good biocompatibility, PDEVP portions and also P2VP parts enhanced the cell viability. An increase in the amount of cationic repeating units leads to a decrease in cell viability. Compared to a well-established cationic and amphiphilic copolymer system (PEI-g-PCL-g-PEG), the determined IC50 values are at least an order of magnitude higher, which underlines the application potential of those polymers.28
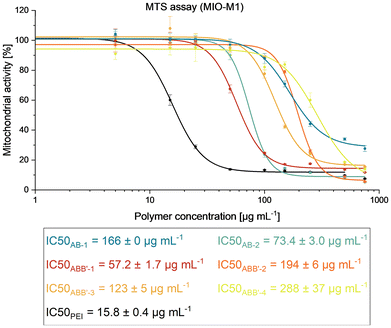 | ||
| Fig. 2 MTS assay of AB-, ABB′- and PEI-treated MIO-M1 cell line at 5, 15, 25, 50, 100, 150, 250, 500 and 750 μg mL−1 and calculation of IC50 values by a sigmoidal plot. | ||
Following MTS assays, cells subjected to polymer treatments were also evaluated with crystal violet assay (Fig. S13†) to examine the healthy adherence cells to circumvent the limitation of the MTS assay, which includes the measurement of metabolic activities of the unhealthy cells.19,29 As crystal violet assay excluded the effect from the detached dead cells, it provides complementary information for the cytotoxicity contributed by the polymer treatment. In general, a similar trend was observed in the MTS results, where the cell number decreased with increasing polymer concentration (Fig. 2, S13†). However, the cell number increased again at higher concentrations from some polymers. Concerning the microscopic images taken (Fig. S14–S18†), it is postulated that these polymers induced cell death and formed cellular debris aggregates at high concentrations, leading to false positive results of high cell numbers detected. Nevertheless, the overall cytotoxicity profiles and interpretations remain similar between the two assays. In conclusion, both assays provide valuable insights for the subsequent determining factors in the applied concentrations for endosomal escape and siRNA transfection investigations with these polymers.
siRNA encapsulation
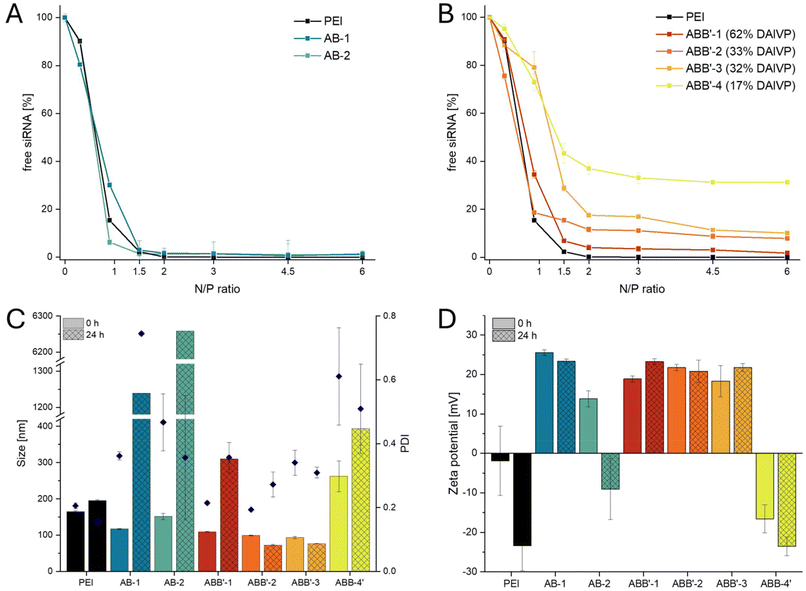
SYBR Gold assay was used to evaluate the ability of the cationic polymers to encapsulate siRNA at various N/P ratios.30 Synthesized block copolymers were compared to hyperbranched PEI (25 kg mol−1) and non-functionalized P2VP-b-PDEVP at different N/P ratios, which is defined herein as the amount of cationic (aliphatic amines) to anionic (phosphates in siRNA backbone) groups. Non-functionalized P2VP-b-PDEVP was mixed with siRNA at similar weight ratios (μgpolymer μgsiRNA−1, Fig. S25†). For P2VP-b-PDEVP, only a weak interaction of siRNA and polymer was observed, proving that pyridyl groups do not contribute to siRNA complexation. At 6-fold gravimetrical excess of polymer, only up to 20% of siRNA were encapsulated, which is due to the general basic character of the P2VP chain, binding of siRNA via minor intermolecular interactions. All other primary amine-containing AB polymers showed complete encapsulation of siRNA at a low N/P ratio of 2 or higher (Fig. 3A). These encapsulation abilities are in the same range as for commonly used hyperbranched PEI – showing the efficient complexation of the newly developed polymers while avoiding a high excess of polymer and, thus, free and potentially toxic amino groups. The siRNA binding pattern of AB-type block copolymers was independent of the size and composition of the polymers, indicating that hydrophobic moieties do not inhibit siRNA complexation when using the same N/P ratios. In contrast, ABB′-type block copolymers showed different encapsulation efficiencies depending on the DAlVP/DEVP ratios (Fig. 3B). While all ABB′ polymers reached a maximum siRNA encapsulation at N/P ratios of 2 or higher, a higher amount of free or surface-bound siRNA was observed with lower amounts of cationic repeating units and thus less dense distribution of primary amines within the poly(vinylphosphonate) block. ABB′-4, bearing 17% of cationic repeating units, appeared to encapsulate with the lowest observed capacity, while ABB′-1 (62% cationic repeating units) encapsulated 95% of the present siRNA, which was almost as efficient as the AB-type block copolymers. For ABB′-2 and ABB′-3, the siRNA encapsulation capacity is almost identical, independent of the molecular weight and the copolymerized P2VP unit, since the number of cationic units in the vinylphosphonate segment is the same. It seems that the cationic charge density and, thus, the amount of statistically distributed cationic repeating units within the vinylphosphonate block limits the capacity of siRNA encapsulation while the non-functional P2VP and PDEVP parts do not significantly contribute to the siRNA binding efficiency.Maximum encapsulation of siRNA was found to occur at a low N/P ratio of 2, which suggested characterization in terms of size and zeta potential at these conditions. For all polymers, sizes were smaller, and zeta potentials were lower than for the free polymers. Bulk mixing of siRNA and polymers AB-1, AB-2, and ABB′-1–3 resulted in 100 to 150 nm polyplex sizes that fit in a range for physiologically relevant applications, while low to moderate polydispersity indices (PDI 0.2–0.5) were observed. ABB′-4 showed no uniform polyplex formation, as indicated by the bigger sizes and polydispersities caused by the inefficient siRNA complexation due to the low charge density shown by the SYBR Gold assay. Stability measurements were performed after 24 hours of storage at ambient temperature in HEPES buffer. Polyplexes of AB-1 and AB-2 agglomerated to particles larger than 1 μm, indicating low stability in solution. In contrast, ABB′-2 and ABB′-3 remained stable after 24 h (Dh = 75 nm) by retaining their polydispersity (PDI = 0.3). It is assumed, that, independent of the size of the hydrophobic chain, a certain amount of hydrophilic DEVP induces polyplex stability. Compared to all AB-type block copolymers, both stable ABB′-2 and ABB′-3 have 67% incorporated DEVP within the vinylphosphonate chain. In contrast, ABB′-4 bearing 83% DEVP suffers from low charge density as the polyplex is large right after preparation and increases its size over time.
The results of the DLS experiments (Fig. 3C) were underlined by considering zeta potential measurements (Fig. 3D). At t = 0 h, the polymers showed comparable zeta potentials between 20 and 30 mV. This was significantly lower than the values measured for the polymers without encapsulated siRNA, indicating the complexation of siRNA. Solely ABB′-4 shows a negative value at t = 0 due to the poor siRNA complexation at the measured N/P ratio (Fig. 3B). Stability measurements after 24 h indicated stable zeta potentials for AB-1 and ABB′-1–3. The agglomeration process of AB-2 leads to a release of siRNA and the poor encapsulation ability of ABB′-4 results in a negative zeta potential. This stability assessment could show that additional hydrophilic vinylphosphonate units, up to a certain extent within the cationic polymer chain, contribute to enhanced polyplex stability. In contrast, the effect of the hydrophobic segments on the stability is not as pronounced (ABB′-2 vs. ABB′-3). In comparison, encapsulation of siRNA using PEI resulted in unstable polymers with a negative zeta potential after 24 h. This indicates the superior solution stability of the new polymers compared to PEI. Since these initial results fit the requirements for nucleic acid delivery, it is a promising application area for the synthesized copolymers obtained from REM-GTP. The synthesized polymers were used for efficient siRNA encapsulation at very low N/P ratios, leading to homogeneous and cationic polyplexes. In comparison with other polymer systems containing hydrophobic, hydrophilic, and cationic parts, the polymers from REM-GTP show lower toxicity while simultaneously also requiring lower N/P ratios for full siRNA condensation.31
Polyplex transfection on ocular cells and siRNA-mediated NF-κB p65 knockdown
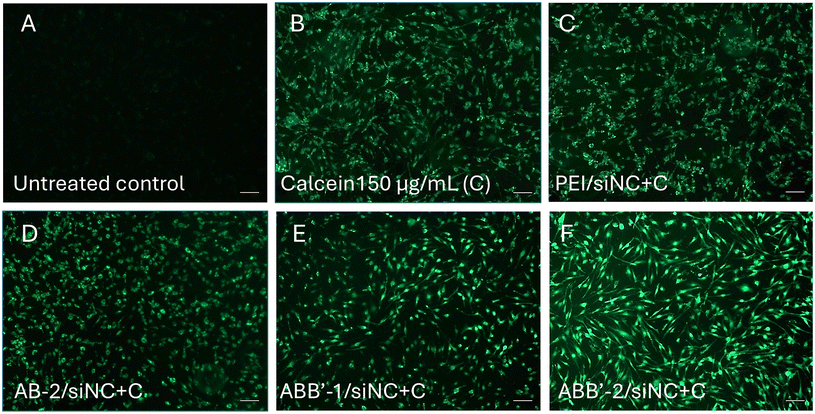
Endosomal escape has remained the most challenging issue for more than four decades in delivering RNA therapeutics for disease treatment.32 Several strategies have been proposed to enhance the endosomal escape for a successful therapeutic delivery. The formed endosome can be destabilized by a polymer-induced proton-sponge effect in the acidic lyso-endosomal environment, which is more effective if a broad range of pKa values between 10 and 4 is covered by the polyamine architecture.33 Cysteamine pendant groups were introduced to the polymers for siRNA complexation due to a high pKa value of the primary amines.18,34 P2VP units should be exclusively converted into hydrophilic protonated monomer units in the pH region below 6. Titration curves with 0.1 M HCl were used to determine the buffering capabilities of AB and ABB′ polymers compared to hyperbranched PEI, which shows a superior siRNA delivery efficiency due to the presence of primary, secondary, and tertiary amines causing efficient complexation of siRNA and destabilization of endosomes. All synthesized polymers showed two different buffering regions in the titration curves (Fig. S26–S28†). At pH 10–8, buffering was due to the protonation of primary amines in the modified vinylphosphonate block, while P2VP exhibited buffering at pH 6.5–5. Thus, the pyridyl groups of the P2VP unit in the newly developed cationic polymers facilitate endosomal escape while destabilizing the micellar properties by destructing the amphiphilic character of the polymers upon protonation for disassembly of the polyplexes and siRNA release. Titration of non-functional P2VP-b-PDEVP confirmed that the modification with cysteamine is essential to facilitate efficient siRNA complexation ability by introducing high pKa values.The preliminary titration experiments were supplemented by calcein assay, which is a widely applied method to examine endosomal escape, assess the release potential of nanocarriers, and study the mechanisms of endosomal escape.35,36 Calcein is a membrane-impermeable fluorescent dye, which, upon endocytosis alone without nanoparticles, is confined within the endosomal/lysosomal compartments with resultant punctate fluorescence signals. On the contrary, when both calcein and nanoparticles enter cells, nanoparticles exhibiting endosomal escape properties would facilitate calcein release into the cytosol, leading to a diffused fluorescence signal. For this purpose, AB-2, ABB′-1, and ABB′-2 were selected based on their different trends in siRNA encapsulation efficiency (Fig. 3A and B), polyplex size, and stability (Fig. 3C and D). Polyplexes of these polymers and negative control siRNA (siNC) were used to investigate their endosomal escape ability in MIO-M1 cells. In addition, to better translate the calcein assay results for subsequent siRNA transfection, Opti-MEM™ and DMEM/F12 medium containing 10% fetal bovine serum (FBS; medium abbreviated with NM) were used as incubation medium for parallel comparison. MIO-M1 cells treated with polyplexes (polymers and siNC) at N/P = 2.0 showed no sign of endosomal escape, regardless of the selection of incubation medium (Fig. S29–S32†). When the N/P ratio was increased to 5, MIO-M1 cells treated with PEI/siNC and AB-2/siNC still exhibited less diffused signals with more punctate patterns (Fig. 4C and D), similar to those observed in calcein-treated cells (Fig. 4B). On the contrary, ABB′-1/siNC- and ABB′-2/siNC-treated cells at this N/P ratio reflected apparent diffused calcein signals (Fig. 4E and F). While ABB′-1 and ABB′-2 contain the same amount of cationic charges at an N/P ratio of 5, ABB′-2 contains nearly doubled the amount of hydrophobic unit 2VP, leading to better cellular uptake and, thus, more noticeable diffused fluorescent signals. In addition, more hydrophilic DEVP present in ABB′-2 could act as an outer shell to potentially better stabilize the polyplex. In short, we have demonstrated that the ABB′ polymers are superior in inducing successful endosomal escape upon cellular uptake via endocytosis. Interestingly, the endosomal escape could be established even though solely primary amines were utilized as cationic siRNA complexing groups, which are generally known to mitigate an endosomal escape.36,37
It is also worth highlighting that the cellular morphology of the polymer/siRNA polyplex (N/P = 5.0) transfected cells subjected to calcein assay remain healthy and are comparable to the untreated control or the calcein-treated only group (Fig. S33–S36†). Moreover, the polymer/siNC complexes at N/P = 5.0 were perfectly sized, ranging from 69 nm to 97 nm with positive surface charge (Fig. S37;† for other size distributions, see Table S2 and Fig. S38–S41†), and were therefore applied in vitro on MIO-M1 cells to further investigate its translational potential for ocular therapeutics.
Based on the promising endosomal escape investigation by calcein assay, human retinal MIO-M1 Müller cells were transfected with PEI, AB-2, ABB′-1, and ABB′-2/siRNA polyplexes complexed with siRNA against NF-κB p65 (siNFκB) or with siNC as negative control at an N/P ratio of 5. For validation purposes, lipoplexes made with commercially available Lipofectamine™ RNAiMAX transfection reagent were used as a positive control. After 72 h of transfection, the silencing efficiency of p65 in three independent samples was more prominent after treatment with ABB′/siNFκB than after treatment with AB/siNFκB or PEI/siNFκB (Fig. 5A, Fig. S42, Table S3†). Specifically, the NF-κB p65 expression on the protein level was significantly downregulated in cells treated with ABB′-1/siNFκB (19.03 ± 3.92%) or ABB′-2/siNFκB (47.55 ± 6.36%) in comparison to the untreated control (Fig. 5). Based on the results from both in vitro NF-κB p65 knockdown experiments and the calcein assay, it is evident that among the tested polymers, including the commonly used branched PEI, the ABB′ polymers exhibited superior siRNA delivery efficiency in vitro while cells treated with polyplexes formed with AB-2 polymers showed neither endosomal escape nor p65 knockdown. Perhaps the presence of hydrophilic moiety (DEVP) with an appropriate proportion of hydrophobic moiety (2VP) might be required to facilitate better micelles formation with more stable polyplexes for better cellular uptake and subsequent transfection efficiency. ABB′/siRNA polyplex showed excellent NF-κB p65 knockdown in an ocular cell line, nearly matching the performance of lipoplexes prepared with the commercially available Lipofectamine™ RNAiMAX transfection reagent (Fig. 5). The obtained results align with other studies on the structure–function relationship, which conveyed the necessity of positive charges on the impact of transfection efficiency.9,33,38 On the other hand, it is noticed that the ABB′-2/siNFκB treated cells showed more apparent diffused calcein signals compared to that of ABB′-1/siNFκB treated group. However, a reverse observation was noted with greater NFκB, p65 knockdown efficiency in the ABB′-1/siNFκB treated group for the siRNA transfection experiments. From the physicochemical composition point of view, ABB′-2 contains almost twice the amount of hydrophobic 2VP with similar hydrophilicity compared to ABB′-1, which likely contributes to a better cellular uptake profile. Furthermore, the difference in siRNA encapsulation efficiency of ABB′-1 and ABB′-2 (Fig. 3) might also contribute to the observed discrepancy in the knockdown effect. Further investigations using qRT-PCR on siRNA transfection at various time points between 4 h and 72 h might help to better elucidate the discrepancy in the results at the molecular level. Nevertheless, our results showed that endosomal escape correlates with knockdown efficiency and is an early indicator of the potential success of siRNA delivery.
Conclusion
Despite the promising properties of phosphorus-containing polymer units, relatively few polymers based on these structures have been investigated for nanomedicinal applications. This is somewhat surprising given the potential benefits that phosphorus-based polymers offer regarding biocompatibility and their ability to interact with biological systems. The limited exploration of these polymers in nanomedicine may be attributed to various factors, including the challenges associated with their synthesis, the need for specialized knowledge in phosphorus chemistry, and the potential regulatory hurdles that novel materials might face in the medical field. Apart from the latter two, the limitations in polymer synthesis by REM-GTP are mainly attributed to the necessary moisture-free conditions for preparing and storing the catalyst and also during polymerization, the limited monomer scope of functional and non-functional monomers, which are restricted to those that are compatible with the highly reactive catalytic system, and the predetermined order of monomer sequences by the coordination strength to the metal center. In addition to that, the synthesized polymers do not provide biodegradable properties of the polymer backbone. Nonetheless, the unique characteristics of phosphorus-containing polymers make them an area of great interest for future research, with the potential to unlock new possibilities for drug delivery, imaging, and therapeutic applications. REM-GTP facilitates the integration of novel phosphonate monomers into nucleic acid delivery systems. Such monomers feature both cationic units and hydrophilic groups, enhancing the ability of polymers to effectively downregulate genes relevant to ocular therapy in combination with other pH-responsive, hydrophobic monomers to form amphiphilic, pH-responsive systems. This development not only underscores the potential of phosphorus-containing polymers in nanomedicine but also paves the way for further exploration of their therapeutic applications, particularly in the area of gene therapy for ocular diseases. Targeting NF-κB, a key transcription factor, using an RNAi strategy could address the associated ocular pathophysiology. For this purpose, the cytotoxicity profiles in vitro and siRNA encapsulation efficiency of the polymers were carefully evaluated before further employing calcein assay to investigate the capability for endosomal escape and siRNA transfection on MIO-M1 cell line. Diffused calcein signals observed from cells treated with ABB′/siRNA polyplexes revealed an endosomal escape capability while maintaining excellent in vitro cell viability. Strong diffused fluorescent signals were observed in cells treated with ABB′-2/siNC, while ABB′-1/siNC polyplex-treated cells led to a partially diffused signal; nonetheless, both indicated successful endosomal escape. As expected, the ocular cell line transfected with ABB′/siNF-κB polyplexes demonstrated a superior NF-κB/RelA gene silencing efficiency after 72 h. While ABB′-2/siRNA showed more apparent endosomal escape than using ABB′-1, ABB′-1/NF-κB demonstrated superior siRNA knockdown efficiency of nearly as effective as the commercially available RNAiMAX transfection reagent. This study highlights the potential of phosphorous-containing, amphiphilic polymers as nucleic acid carriers with enhanced stability, excellent cytotoxicity profiles, and efficient knockdown ability aiming for efficient transfection efficiencies in vivo.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
P. W. gratefully acknowledges funding from the TUM Innovation Network Artificial Intelligence in Material Science (ARTEMIS). M. K. is grateful for the Ph.D. scholarship by the Studienstiftung des deutschen Volkes. F. A. and M. T.-Y. L. are thankful for funding from the Federal Ministry of Education and Research (BMBF) and the Baden-Württemberg Ministry of Science as part of the Excellence Strategy of the German Federal and State Governments. The human Müller cell line Moorfields/Institute of Ophthalmology-Müller 1 (MIO-M1) was obtained from the UCL Institute of Ophthalmology, London, UK.39References
- (a) A. Oeckinghaus and S. Ghosh, Cold Spring Harbor Perspect. Biol., 2009, 1, a000034 CrossRef PubMed; (b) W. Lan, A. Petznick, S. Heryati, M. Rifada and L. Tong, Ocul. Surf., 2012, 10, 137–148 CrossRef PubMed.
- S. K. Srivastava and K. V. Ramana, Exp. Eye Res., 2009, 88, 2–3 CrossRef CAS PubMed.
- S. T. Palayoor, M. Y. Youmell, S. K. Calderwood, C. N. Coleman and B. D. Price, Oncogene, 1999, 18, 7389–7394 CrossRef CAS PubMed.
- (a) J. Hurst, F. Adams and S. Schnichels, Adv. Funct. Mater., 2024, 2407173 Search PubMed; (b) A. Kumar, Y. Takada, A. M. Boriek and B. B. Aggarwal, J. Mol. Med., 2004, 82, 434–448 CrossRef CAS PubMed.
- (a) W.-J. Liang, H.-W. Yang, H.-N. Liu, W. Qian and X.-L. Chen, Life Sci., 2020, 241, 117146 CrossRef CAS; (b) T. Zhang, X. Mei, H. Ouyang, B. Lu, Z. Yu, Z. Wang and L. Ji, J. Nutr. Biochem., 2019, 65, 1–14 CrossRef CAS.
- (a) A. Fire, S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Nature, 1998, 391, 806–811 CrossRef CAS PubMed; (b) R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. van Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Chem. Rev., 2021, 121, 11527–11652 CrossRef CAS PubMed.
- Z. Liu, Z. Zhang, C. Zhou and Y. Jiao, Prog. Polym. Sci., 2010, 35, 1144–1162 CrossRef CAS.
- F. Adams, C. M. Zimmermann, D. Baldassi, T. M. Pehl, P. Weingarten, I. Kachel, M. Kränzlein, D. C. Jürgens, P. Braubach, I. Alexopoulos, M. Wygrecka and O. M. Merkel, Small, 2024, 20, e2308775 CrossRef PubMed.
- T. Bus, A. Traeger and U. S. Schubert, J. Mater. Chem. B, 2018, 6, 6904–6918 RSC.
- (a) N. D. Sonawane, F. C. Szoka and A. S. Verkman, J. Biol. Chem., 2003, 278, 44826–44831 CrossRef CAS PubMed; (b) M. Wojnilowicz, A. Glab, A. Bertucci, F. Caruso and F. Cavalieri, ACS Nano, 2019, 13, 187–202 CrossRef CAS PubMed; (c) S. Yang and S. May, J. Chem. Phys., 2008, 129, 185105 CrossRef PubMed.
- I. Porello, N. Bono, G. Candiani and F. Cellesi, Polym. Chem., 2024, 15, 2800–2826 RSC.
- K. Knop, R. Hoogenboom, D. Fischer and U. S. Schubert, Angew. Chem., Int. Ed., 2010, 49, 6288–6308 CrossRef CAS PubMed.
- J. S. Suk, Q. Xu, N. Kim, J. Hanes and L. M. Ensign, Adv. Drug Delivery Rev., 2016, 99, 28–51 CrossRef CAS PubMed.
- (a) Q. Yang and S. K. Lai, Wiley Interdiscip. Rev.:Nanomed. Nanobiotechnol., 2015, 7, 655–677 CAS; (b) R. Matthes and H. Frey, Biomacromolecules, 2022, 23, 2219–2235 CrossRef CAS.
- (a) J. Li, Y. Hao, H. Wang, M. Zhang, J. He and P. Ni, ACS Appl. Mater. Interfaces, 2024, 16, 51876–51898 CrossRef CAS PubMed; (b) S. Monge, B. Canniccioni, A. Graillot and J.-J. Robin, Biomacromolecules, 2011, 12, 1973–1982 CrossRef CAS PubMed; (c) Y.-C. Wang, Y.-Y. Yuan, J.-Z. Du, X.-Z. Yang and J. Wang, Macromol. Biosci., 2009, 9, 1154–1164 CrossRef CAS PubMed; (d) C. Ornelas-Megiatto, P. R. Wich and J. M. J. Fréchet, J. Am. Chem. Soc., 2012, 134, 1902–1905 CrossRef CAS PubMed.
- (a) J. Simon, T. Wolf, K. Klein, K. Landfester, F. R. Wurm and V. Mailänder, Angew. Chem., Int. Ed., 2018, 57, 5548–5553 CrossRef CAS PubMed; (b) K. N. Bauer, J. Simon, V. Mailänder, K. Landfester and F. R. Wurm, Acta Biomater., 2020, 116, 318–328 CrossRef CAS.
- (a) F. Adams, M. R. Machat, P. T. Altenbuchner, J. Ehrmaier, A. Pöthig, T. N. V. Karsili and B. Rieger, Inorg. Chem., 2017, 56, 9754–9764 CrossRef CAS PubMed; (b) F. Adams, P. Pahl and B. Rieger, Chem. – Eur. J., 2018, 24, 509–518 CrossRef CAS PubMed; (c) A. S. Maier, S. Mansi, K. Halama, P. Weingarten, P. Mela and B. Rieger, ACS Appl. Mater. Interfaces, 2024, 16, 58135–58147 CrossRef CAS PubMed.
- F. Adams, P. T. Altenbuchner, P. D. L. Werz and B. Rieger, RSC Adv., 2016, 6, 78750–78754 RSC.
- P. T. Altenbuchner, P. D. L. Werz, P. Schöppner, F. Adams, A. Kronast, C. Schwarzenböck, A. Pöthig, C. Jandl, M. Haslbeck and B. Rieger, Chem. – Eur. J., 2016, 22, 14576–14584 CrossRef CAS PubMed.
- C. Schwarzenböck, A. Schaffer, P. Pahl, P. J. Nelson, R. Huss and B. Rieger, Polym. Chem., 2018, 9, 284–290 RSC.
- C. Schwarzenböck, P. J. Nelson, R. Huss and B. Rieger, Nanoscale, 2018, 10, 16062–16068 RSC.
- B. S. Soller, S. Salzinger, C. Jandl, A. Pöthig and B. Rieger, Organometallics, 2015, 34, 2703–2706 CrossRef CAS.
- F. Adams, Macromol. Rapid. Commun., 2024, e2400122 Search PubMed.
- M. Fineman and S. D. Ross, J. Polym. Sci., 1950, 5, 259–262 CrossRef CAS.
- K. Halama, A. Schaffer and B. Rieger, RSC Adv., 2021, 11, 38555–38564 RSC.
- M. H. Kravicz, D. T. Balogh, M. Kar, S. Wedepohl, M. V. L. B. Bentley and M. Calderón, Macromol. Biosci., 2019, 19, e1900117 CrossRef PubMed.
- (a) A. Fietz, J. Hurst and S. Schnichels, Int. J. Mol. Sci., 2022, 23(23), 14540 CrossRef CAS PubMed; (b) S. Schnichels, U. Hagemann, K. Januschowski, J. Hofmann, K.-U. Bartz-Schmidt, P. Szurman, M. S. Spitzer and S. Aisenbrey, Br. J. Ophthalmol., 2013, 97, 917–923 CrossRef PubMed.
- M. Zheng, Y. Liu, O. Samsonova, T. Endres, O. Merkel and T. Kissel, Int. J. Pharm., 2012, 427, 80–87 CrossRef CAS PubMed.
- (a) C. Schwarzenböck, S. I. Vagin, W. R. Heinz, P. J. Nelson and B. Rieger, Macromol. Rapid. Commun., 2018, 39, e1800259 CrossRef PubMed; (b) K. Halama, M. T.-Y. Lin, A. Schaffer, M. Foith, F. Adams and B. Rieger, Macromolecules, 2024, 57, 1438–1447 CrossRef CAS; (c) M. Feoktistova, P. Geserick and M. Leverkus, Cold Spring Harb. Protoc., 2016, 2016, pdb.prot087379 CrossRef PubMed.
- N. Hartl, F. Adams, G. Costabile, L. Isert, M. Döblinger, X. Xiao, R. Liu and O. M. Merkel, Nanomaterials, 2019, 9, 986 CrossRef CAS PubMed.
- (a) Y. Liu, J. Nguyen, T. Steele, O. Merkel and T. Kissel, Polymer, 2009, 50, 3895–3904 CrossRef CAS; (b) Y. Liu, O. Samsonova, B. Sproat, O. Merkel and T. Kissel, J. Controlled Release, 2011, 153, 262–268 CrossRef CAS PubMed.
- S. F. Dowdy, R. L. Setten, X.-S. Cui and S. G. Jadhav, Nucleic Acid Ther., 2022, 32, 361–368 CrossRef CAS PubMed.
- A. Akinc, M. Thomas, A. M. Klibanov and R. Langer, J. Gene Med., 2005, 7, 657–663 CrossRef CAS PubMed.
- C. Atallah, C. Charcosset and H. Greige-Gerges, J. Pharm. Anal., 2020, 10, 499–516 CrossRef PubMed.
- F. Hausig-Punke, F. Richter, M. Hoernke, J. C. Brendel and A. Traeger, Macromol. Biosci., 2022, 22, e2200167 CrossRef PubMed.
- A. S. M. Wong, S. K. Mann, E. Czuba, A. Sahut, H. Liu, T. C. Suekama, T. Bickerton, A. P. R. Johnston and G. K. Such, Soft Matter, 2015, 11, 2993–3002 RSC.
- (a) L. M. P. Vermeulen, T. Brans, S. K. Samal, P. Dubruel, J. Demeester, S. C. de Smedt, K. Remaut and K. Braeckmans, ACS Nano, 2018, 12, 2332–2345 CrossRef CAS PubMed; (b) N. Kongkatigumjorn, S. A. Smith, M. Chen, K. Fang, S. Yang, E. R. Gillies, A. P. R. Johnston and G. K. Such, ACS Appl. Nano Mater., 2018, 1, 3164–3173 CrossRef CAS.
- M. A. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259–302 CrossRef CAS PubMed https://pubs.acs.org/doi/pdf/10.1021/cr800409e .
- G. A. Limb, T. E. Salt, P. M. G. Munro, S. E. Moss and P. T. Khaw, Invest. Ophthalmol. Visual Sci., 2002, 43, 864–869 Search PubMed https://arvojournals.org/article.aspx?articleid=2200145 .
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lp00321g |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |