Open Access Article
Open Access ArticleInsights into the anticancer and anti-inflammatory activities of curcumin-loaded quercetin nanoparticles: in vitro bioassays coupled with synchrotron infrared microspectroscopy†
Suhair Sunoqrot *a,
Samah Abusulieha and
Lina A. Dahabiyehb
*a,
Samah Abusulieha and
Lina A. Dahabiyehb
aDepartment of Pharmacy, Faculty of Pharmacy, Al-Zaytoonah University of Jordan, Amman 11733, Jordan. E-mail: suhair.sunoqrot@zuj.edu.jo; Fax: +96264291432; Tel: +96264291511, ext. 471
bDepartment of Pharmaceutical Sciences, School of Pharmacy, The University of Jordan, Amman, Jordan
First published on 14th February 2025
Abstract
Curcumin (CUR) is an important phytochemical with diverse pharmacological activities, particularly in cancer treatment and inflammation management. However, its poor water solubility and rapid metabolism in vivo have limited its therapeutic potential, inspiring the development of delivery systems such as those based on nanotechnology. Previously, we reported that partial oxidation of the plant polyphenol quercetin (QCT) creates an amphiphilic material that self-assembles into discrete nanoparticles (NPs), either alone or with other polymers. Here we developed a nanoparticle (NP) formulation for CUR based on partially oxidized QCT (oxQCT). The NPs were co-formulated with D-α-tocopheryl poly(ethylene glycol) 1000 succinate (TPGS) to enhance their physicochemical properties and potentially synergize with CUR in inducing cancer cell death and alleviating inflammation. CUR was entrapped in oxQCT/TPGS NPs via nanoprecipitation, yielding spherical NPs (87 nm) with 72% loading efficiency, high colloidal stability, and sustained release at physiological pH. The anticancer activity of CUR NPs was evaluated in MCF-7 cells as a model breast cancer cell line, where the oxQCT/TPGS vehicle synergized with free CUR in inducing cell death and promoted its cellular uptake. In RAW 264.7 macrophages, the NPs outperformed free CUR in anti-inflammatory assays by exhibiting stronger reactive oxygen species (ROS) scavenging activities and attenuating the expression of proinflammatory cytokines. We complemented the biological assays with synchrotron-Fourier transform infrared microspectroscopy (SR-μFTIR), which provided insights into biochemical changes in the lipid and protein compositions of MCF-7 and RAW 264.7 macrophages treated with CUR NPs. Our findings present a promising nanoformulation for CUR which can enhance its activity against cancer and inflammation, and highlight the importance of bioanalytical techniques such as SR-μFTIR in discerning the bioactivities of multimodal nanotherapeutics.
Introduction
Turmeric (Curcuma longa L.) has been used throughout history for medicinal purposes in Chinese medicine and Ayurveda.1 Curcumin (CUR) is the principal component of turmeric rhizome and is commonly used for culinary and food coloring purposes. Previous studies have demonstrated the wide range of health benefits for CUR including antioxidant, anti-inflammatory, immunoregulatory, hepatoprotective, neuroprotective, antidiabetic, and anticancer properties.2 In the context of cancer therapy, CUR is considered both as a preventative and as a therapeutic agent with minimal side effects compared to conventional chemotherapy. CUR exerts anticancer effects by interfering with oncogenic transcription factors, growth factors, inflammatory cytokines, and protein kinases involved in cancer progression.3 Collectively, CUR can lead to the induction of cancer cell apoptosis and prevention of metastasis, invasion, as well as angiogenesis.4 CUR's anti-inflammatory activity stems from its ability to modulate inflammatory signaling pathways and downregulate inflammatory mediators, including interleukins, tumor necrosis factor-α (TNF-α), and inducible nitric oxide synthase (iNOS).5 These activities support the use of CUR for treating conditions such as rheumatoid arthritis, inflammatory bowel disease, and neurodegenerative diseases.6Despite its numerous health benefits, CUR has low water solubility, poor bioavailability, and rapid metabolism in vivo, all of which hinder its clinical use.7 Several formulation approaches have been investigated to improve the bioavailability and in vivo performance of CUR, particularly those based on colloidal delivery systems. Examples include micro- and nanoemulsions, liposomes, polymeric nanoparticles (NPs), and polymeric micelles.8–11 Recently, plant polyphenols have gained significant interest as building blocks for functional materials including NP-based drug carriers.12,13 NPs have been synthesized from plant polyphenols by relying on the propensity of these molecules to undergo oxidation-triggered coupling reactions and/or metal coordination.14–16 Their polyphenolic structure enables them to form a variety of intermolecular interactions, including hydrogen bonding, π–π stacking, and van der Waals interactions, offering pharmaceutical scientists with green and easy-to-synthesize nanocarriers for the delivery of a wide range of drug molecules. The polyphenols which have been most extensively investigated in this regard are green tea catechins such as epigallocatechin gallate (EGCG),17–19 tannic acid,20 grape seed extracts,21 and coffee bean extracts.22
Our group recently reported that the ubiquitous plant polyphenol quercetin (QCT) is capable of forming NPs upon oxidation in alkaline media, and the NPs could be used to load small drug molecules. The drug could be incorporated along with surface ligands during23 or after NP formation.24 We also showed that partial oxidation of QCT in an organic solvent such as dimethyl sulfoxide (DMSO) leads to the formation of supramolecular amphiphiles (oxQCT) which can later form NPs by self-assembly and co-assembly with hydrophilic and amphiphilic polymers via traditional nanoprecipitation.25,26 oxQCT NPs represent a novel class of nanomaterials that have the potential to advance the development of nanomedicines for various biomedical applications including cancer and inflammation. However, there is still a gap in our fundamental understanding of the NPs’ interactions with the bio-interface. A thorough understanding of the NPs’ biological interactions with cancer cells and immune cells will be crucial in order to advance them further to pre-clinical and clinical testing. Advanced bioanalytical techniques such as synchrotron-Fourier transform infrared microspectroscopy (SR-μFTIR) can be of tremendous value in this aspect. This technique has emerged as a powerful non-invasive tool that can provide valuable chemical and spatial information on biomolecules within biological samples including blood, tissues, and individual cells, by probing the vibrational motions of their molecular constituents at a microscopic scale.27 SR-μFTIR has been successfully used to identify biomolecular changes associated with cancer progression, cancer cell differentiation pathways, and the mechanisms of cancer cell death.28 In terms of immunomodulation, μFTIR can provide important information about the biochemical changes occurring within immune cells in response to stimuli and treatment.29 Having shown great potential as a drug delivery platform, the main aim of this study was to evaluate oxQCT as a nanocarrier for CUR. Following physicochemical characterization, the anticancer and anti-inflammatory activities of the NPs were evaluated in MCF-7 breast cancer cells and RAW 264.7 macrophages, respectively. In addition to conventional in vitro bioassays such as viability, uptake, apoptosis, and anti-inflammatory assays, we employed SR-μFTIR to provide additional insights into the diverse bioactivities of CUR and CUR-based nanocarriers.
Experimental section
Materials
Curcumin (CUR), quercetin hydrate (QCT), sodium metaperiodate (NaIO4), D-α-tocopheryl poly(ethylene glycol) 1000 succinate (TPGS), 2,2-diphenyl-1-picrylhydrazyl (DPPH), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (USA). Lipopolysaccharide (LPS; E. coli. O55:B5) was obtained from ChemCruz (USA). JC-1 mitochondrial membrane potential dye was procured from Abcam (Cat No. ab141387, UK). Hydrogen peroxide (H2O2) was obtained from Scharlab (Spain). Potassium bromide (KBr) and dimethyl sulfoxide (DMSO) were purchased from Fisher Chemicals (UK). Spectra/Por dialysis membranes with 3.5 and 12–14 kDa molecular weight cut-offs (MWCO) were procured from Repligen (USA). Ultrapure water was prepared using an EMD Millipore Direct-Q 5UV system (USA). Phosphate buffered saline solution (PBS, 10×, pH 7.4) was purchased from EuroClone (Italy). Tween 80 was obtained from RCFL Limited (India).Preparation of CUR-loaded oxQCT NPs
Amphiphilic partially oxidized QCT (oxQCT) was synthesized utilizing a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of QCT
1 molar ratio of QCT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaIO4 as described in our earlier publications (Fig. 1(A)).25,30 Briefly, QCT (1 g; 3.3 mmol) and NaIO4 (708 mg; 3.3 mmol) were reacted in 20 mL DMSO for 24 h at room temperature. The solution was then dialyzed for 5 days against deionized water using a 3.5 kDa MWCO membrane, after which the purified material was lyophilized to obtain oxQCT powder which was used for NP preparation. CUR was loaded into TPGS-modified oxQCT NPs via nanoprecipitation, which involves the gradual addition of polymers and drug molecules dissolved in a water-miscible organic solvent to an aqueous phase under stirring, resulting in the formation of colloidal assemblies.25 Briefly, stock solutions of CUR, TPGS, and oxQCT were prepared at 20 mg mL−1 in DMSO. Then, different amounts of CUR (0.5, 1, and 2 mg) were drawn from the stock solution and mixed with 5 mg of TPGS (250 μL) and 5 mg of oxQCT (250 μL) in a microtube. The organic phase was added dropwise to 2.5 mL of ultrapure water in a 10-mL glass vial protected from light under stirring at 500 rpm for 1 h to obtain a colloidal dispersion. The NPs were then dialyzed (3.5 kDa MWCO membrane) against deionized water for one day to remove the unentrapped drug with regular water changes, after which the NPs were collected and stored at 4 °C for further characterization. For the preparation of blank NPs (without CUR), the CUR solution was replaced with an equivalent volume of DMSO. A schematic representation of the preparation of CUR NPs is presented in Fig. 1(B).
NaIO4 as described in our earlier publications (Fig. 1(A)).25,30 Briefly, QCT (1 g; 3.3 mmol) and NaIO4 (708 mg; 3.3 mmol) were reacted in 20 mL DMSO for 24 h at room temperature. The solution was then dialyzed for 5 days against deionized water using a 3.5 kDa MWCO membrane, after which the purified material was lyophilized to obtain oxQCT powder which was used for NP preparation. CUR was loaded into TPGS-modified oxQCT NPs via nanoprecipitation, which involves the gradual addition of polymers and drug molecules dissolved in a water-miscible organic solvent to an aqueous phase under stirring, resulting in the formation of colloidal assemblies.25 Briefly, stock solutions of CUR, TPGS, and oxQCT were prepared at 20 mg mL−1 in DMSO. Then, different amounts of CUR (0.5, 1, and 2 mg) were drawn from the stock solution and mixed with 5 mg of TPGS (250 μL) and 5 mg of oxQCT (250 μL) in a microtube. The organic phase was added dropwise to 2.5 mL of ultrapure water in a 10-mL glass vial protected from light under stirring at 500 rpm for 1 h to obtain a colloidal dispersion. The NPs were then dialyzed (3.5 kDa MWCO membrane) against deionized water for one day to remove the unentrapped drug with regular water changes, after which the NPs were collected and stored at 4 °C for further characterization. For the preparation of blank NPs (without CUR), the CUR solution was replaced with an equivalent volume of DMSO. A schematic representation of the preparation of CUR NPs is presented in Fig. 1(B).
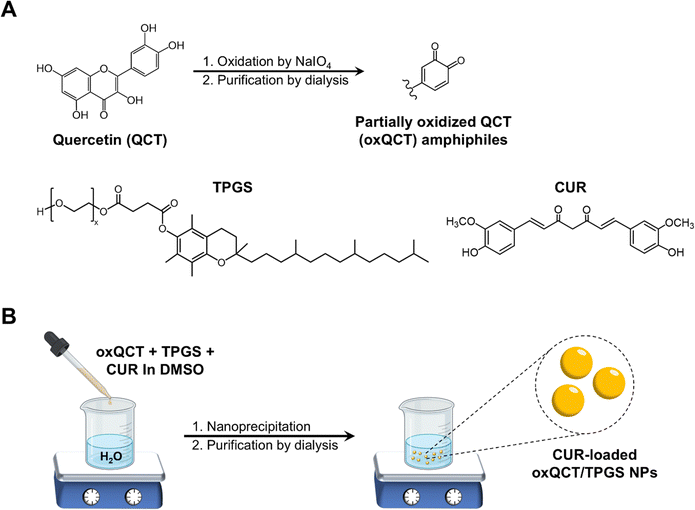 | ||
| Fig. 1 (A) Structure of QCT and its oxidation by NaIO4 to produce partially oxidized oxQCT amphiphiles; (B) preparation of CUR-loaded oxQCT/TPGS NPs by nanoprecipitation. | ||
Characterization of CUR-loaded oxQCT NPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with PBS (pH 7.4), acetate buffer (pH 5.5), or complete Dulbecco's Modified Eagle's Medium (DMEM; EuroClone, Italy) supplemented with 10% fetal bovine serum (FBS; EuroClone). Samples were incubated at 37 °C for 1 h and then the particle size was measured by DLS as described above.
1 with PBS (pH 7.4), acetate buffer (pH 5.5), or complete Dulbecco's Modified Eagle's Medium (DMEM; EuroClone, Italy) supplemented with 10% fetal bovine serum (FBS; EuroClone). Samples were incubated at 37 °C for 1 h and then the particle size was measured by DLS as described above.
 | (1) |
| Qt = ktn | (2) |
In vitro antiproliferative activity of CUR-loaded NPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and allowed to adhere overnight. The next day, cells (n = 5 per treatment group) were treated with different concentrations of CUR (300, 100, 10, 1, and 0.1 μM) in the form of free-CUR (from a 20 mg mL−1 stock solution in DMSO) and CUR-loaded NPs (freshly prepared aqueous dispersions) diluted in the complete medium for 24, 48, and 72 h. Cells were also treated with blank (CUR-free) NPs at concentrations equivalent to CUR-loaded NPs. At the end of each incubation period, the medium in each well was replaced with 100 μL of fresh medium containing 0.5 mg mL−1 MTT and cells were incubated for another 3 h in the CO2 incubator. The supernatant was then carefully removed, and 100 μL of DMSO was added followed by further incubation in an orbital shaker incubator (100 rpm, 37 °C, 10 min) to ensure complete dissolution of formazan crystals. The absorbance intensity of each well which is proportional to the number of viable cells was measured at 540 nm using a Synergy HTX Multi-Mode microplate reader. Cell viability (%) was calculated from the percentage ratio of the absorbance of each treatment group relative to the untreated control group. The half maximal inhibitory concentration (IC50) values were determined by non-linear regression analysis of cell viability versus concentration curves using GraphPad Prism software version 9 (USA). To evaluate potential synergism between CUR and the oxQCT/TPGS NP vehicle, the IC50 of free CUR and that of the empty NPs were used to calculate the combination index (CI) based on the Chou–Talalay method according to eqn (3):33
000 cells per well and allowed to adhere overnight. The next day, cells (n = 5 per treatment group) were treated with different concentrations of CUR (300, 100, 10, 1, and 0.1 μM) in the form of free-CUR (from a 20 mg mL−1 stock solution in DMSO) and CUR-loaded NPs (freshly prepared aqueous dispersions) diluted in the complete medium for 24, 48, and 72 h. Cells were also treated with blank (CUR-free) NPs at concentrations equivalent to CUR-loaded NPs. At the end of each incubation period, the medium in each well was replaced with 100 μL of fresh medium containing 0.5 mg mL−1 MTT and cells were incubated for another 3 h in the CO2 incubator. The supernatant was then carefully removed, and 100 μL of DMSO was added followed by further incubation in an orbital shaker incubator (100 rpm, 37 °C, 10 min) to ensure complete dissolution of formazan crystals. The absorbance intensity of each well which is proportional to the number of viable cells was measured at 540 nm using a Synergy HTX Multi-Mode microplate reader. Cell viability (%) was calculated from the percentage ratio of the absorbance of each treatment group relative to the untreated control group. The half maximal inhibitory concentration (IC50) values were determined by non-linear regression analysis of cell viability versus concentration curves using GraphPad Prism software version 9 (USA). To evaluate potential synergism between CUR and the oxQCT/TPGS NP vehicle, the IC50 of free CUR and that of the empty NPs were used to calculate the combination index (CI) based on the Chou–Talalay method according to eqn (3):33
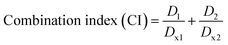 | (3) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well (n = 3), and treated with CUR, CUR NPs, or blank NPs at a concentration equivalent to 20 μM CUR diluted in complete culture medium. After 24 h of treatment, 100 μL of 20 μM JC-1 dye solution was added directly to each well and cells were stained for 10 min at 37 °C. The green fluorescence of JC-1 monomers was detected at 485 nm/528 nm, and the red fluorescence of JC-1 aggregates at 540/620 nm excitation/emission wavelengths using a Synergy HTX Multi-Mode microplate reader. The results were expressed as a ratio of JC-1 monomer/aggregate fluorescence normalized relative to the untreated cells.
000 cells per well (n = 3), and treated with CUR, CUR NPs, or blank NPs at a concentration equivalent to 20 μM CUR diluted in complete culture medium. After 24 h of treatment, 100 μL of 20 μM JC-1 dye solution was added directly to each well and cells were stained for 10 min at 37 °C. The green fluorescence of JC-1 monomers was detected at 485 nm/528 nm, and the red fluorescence of JC-1 aggregates at 540/620 nm excitation/emission wavelengths using a Synergy HTX Multi-Mode microplate reader. The results were expressed as a ratio of JC-1 monomer/aggregate fluorescence normalized relative to the untreated cells.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well (n = 3) overnight, and then treated with 20 μM of CUR, CUR NPs, or blank NPs diluted in complete medium. After 24 h of treatment, 100 μL of the caspase mix was added directly to each well and incubated for 1 h at 37 °C. The red fluorescence of caspase-3 and the green fluorescence of caspase-8 were measured at 540/620 nm and 485/528 nm excitation/emission wavelengths, respectively, using a Synergy HTX Multi-Mode microplate reader. Results were expressed as the mean fold increase in fluorescence intensity of treated cells relative to untreated controls.
000 cells per well (n = 3) overnight, and then treated with 20 μM of CUR, CUR NPs, or blank NPs diluted in complete medium. After 24 h of treatment, 100 μL of the caspase mix was added directly to each well and incubated for 1 h at 37 °C. The red fluorescence of caspase-3 and the green fluorescence of caspase-8 were measured at 540/620 nm and 485/528 nm excitation/emission wavelengths, respectively, using a Synergy HTX Multi-Mode microplate reader. Results were expressed as the mean fold increase in fluorescence intensity of treated cells relative to untreated controls.Anti-inflammatory activity of CUR-loaded NPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) min, then the absorbance was measured at 517
min, then the absorbance was measured at 517![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm using a microplate reader. The experiment was performed in triplicate. The DPPH radical scavenging activity (%) was calculated according to eqn (4):
nm using a microplate reader. The experiment was performed in triplicate. The DPPH radical scavenging activity (%) was calculated according to eqn (4):
 | (4) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in a 96-well plate overnight. The next day, cells (n = 3) were treated with 20 μM CUR, CUR NPs, and blank NPs diluted in complete medium. After 24 h of treatment, the medium in each well was replaced with 100 μL fresh medium containing 20 μM DCFDA and 350 μM H2O2 and the cells were incubated for 45 min at 37 °C. Another group of cells was treated with 100 μL fresh medium containing 20 μM DCFDA only (unstimulated cells). The experiment depends on the oxidation of the non-fluorescent DCFDA by cellular esterases in the presence of intracellular ROS to the highly fluorescent molecule 2′,7′-dichlorofluorescein (DCF). After 45 min of incubation, the medium in each well was replaced with fresh PBS and DCF fluorescence was read with a microplate reader at 485 nm/528 nm excitation/emission wavelengths. Results were expressed as the fold increase in DCF fluorescence relative to untreated and unstimulated cells.
000 cells per well in a 96-well plate overnight. The next day, cells (n = 3) were treated with 20 μM CUR, CUR NPs, and blank NPs diluted in complete medium. After 24 h of treatment, the medium in each well was replaced with 100 μL fresh medium containing 20 μM DCFDA and 350 μM H2O2 and the cells were incubated for 45 min at 37 °C. Another group of cells was treated with 100 μL fresh medium containing 20 μM DCFDA only (unstimulated cells). The experiment depends on the oxidation of the non-fluorescent DCFDA by cellular esterases in the presence of intracellular ROS to the highly fluorescent molecule 2′,7′-dichlorofluorescein (DCF). After 45 min of incubation, the medium in each well was replaced with fresh PBS and DCF fluorescence was read with a microplate reader at 485 nm/528 nm excitation/emission wavelengths. Results were expressed as the fold increase in DCF fluorescence relative to untreated and unstimulated cells.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in a 96-well plate overnight, and then co-treated (n = 3) with 100 μL of 1 μg mL−1 LPS and 20 μM CUR, CUR NPs, or blank NPs for 24 h at 37 °C. The control group received 100 μL of fresh medium (unstimulated and untreated cells). Another group of cells received 100 μL of 1 μg mL−1 LPS only (stimulated and untreated cells). The following day, the supernatant of each well (100 μL) was collected in a separate microtube and stored at −80 °C. The collected supernatants were analyzed to measure the levels of secreted proinflammatory cytokines (TNF-α, Il-1β, and IL-6) using TNF-α (Cat No. BMS622, Invitrogen, USA), Il-1β (BMS630, Invitrogen), and IL-6 (BMS625, Invitrogen) Enzyme-linked Immunosorbent Assay (ELISA) kits following the manufacturer's instructions. Cell supernatants were diluted with the sample diluent provided in the kits (2 times for IL-1β and IL-6 and 20 times for TNF-α) prior to the analyses. The concentration of each proinflammatory cytokine in each sample was then calculated based on its respective standard curve, and the results were expressed as the mean concentration of each cytokine ± SD for each treatment group.
000 cells per well in a 96-well plate overnight, and then co-treated (n = 3) with 100 μL of 1 μg mL−1 LPS and 20 μM CUR, CUR NPs, or blank NPs for 24 h at 37 °C. The control group received 100 μL of fresh medium (unstimulated and untreated cells). Another group of cells received 100 μL of 1 μg mL−1 LPS only (stimulated and untreated cells). The following day, the supernatant of each well (100 μL) was collected in a separate microtube and stored at −80 °C. The collected supernatants were analyzed to measure the levels of secreted proinflammatory cytokines (TNF-α, Il-1β, and IL-6) using TNF-α (Cat No. BMS622, Invitrogen, USA), Il-1β (BMS630, Invitrogen), and IL-6 (BMS625, Invitrogen) Enzyme-linked Immunosorbent Assay (ELISA) kits following the manufacturer's instructions. Cell supernatants were diluted with the sample diluent provided in the kits (2 times for IL-1β and IL-6 and 20 times for TNF-α) prior to the analyses. The concentration of each proinflammatory cytokine in each sample was then calculated based on its respective standard curve, and the results were expressed as the mean concentration of each cytokine ± SD for each treatment group.Cellular uptake of CUR NPs by flow cytometry and fluorescence imaging
The quantitative uptake of CUR NPs by MCF-7 cells and RAW 264.7 macrophages compared to free CUR was carried out using flow cytometry by relying on the intrinsic green fluorescence of CUR. Cells were seeded in 24-well plates at a density of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well (RAW 264.7) or 100
000 cells per well (RAW 264.7) or 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well (MCF-7) in triplicate and incubated with their compatible culture media overnight. The next day, cells were incubated with 20 μM free CUR or CUR NPs in 0.5 mL of serum-free medium for 1 and 4 h. At the end of the incubation period, cells were washed gently with PBS, and detached in 0.1 mL per well PBS using a scraper (RAW 264.7) or incubated with 0.1 mL per well trypsin at 37 °C for 5–10 min (MCF-7). Then, the cells were collected with PBS, centrifuged at 150 × g for 5 min, and resuspended in 0.5 mL PBS. Cell-associated fluorescence was measured immediately using the green fluorescence channel (FL-1) of a BD Accuri C6 Plus Flow Cytometer (BD Biosciences, USA) with the threshold set to 10
000 cells per well (MCF-7) in triplicate and incubated with their compatible culture media overnight. The next day, cells were incubated with 20 μM free CUR or CUR NPs in 0.5 mL of serum-free medium for 1 and 4 h. At the end of the incubation period, cells were washed gently with PBS, and detached in 0.1 mL per well PBS using a scraper (RAW 264.7) or incubated with 0.1 mL per well trypsin at 37 °C for 5–10 min (MCF-7). Then, the cells were collected with PBS, centrifuged at 150 × g for 5 min, and resuspended in 0.5 mL PBS. Cell-associated fluorescence was measured immediately using the green fluorescence channel (FL-1) of a BD Accuri C6 Plus Flow Cytometer (BD Biosciences, USA) with the threshold set to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events. Analysis was performed using BD Accuri C6 Plus software. The results were expressed as the fold increase in fluorescence count relative to untreated cells.
000 events. Analysis was performed using BD Accuri C6 Plus software. The results were expressed as the fold increase in fluorescence count relative to untreated cells.
For visualization of cellular uptake by fluorescence imaging, cells were treated as described above for flow cytometry. At the end of the incubation period, cells were washed gently with PBS and fixed with 4% paraformaldehyde (0.2 mL per well) for 10 min. After washing with PBS, cells were imaged using a ZOE Fluorescent Cell Imager (Bio-Rad Laboratories, USA) using the green channel.
Synchrotron FTIR microspectroscopy (SR-μFTIR)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well on top of 13 mm × 1 mm CaF2 IR transmission windows (Crystran Ltd, Poole, UK) which were placed at the bottom of 24-well plates in duplicate. After 24 h, cells were treated with free CUR, CUR NPs, or blank NPs at a concentration equivalent to 20 μM CUR. In the case of RAW 264.7 macrophages, cells were treated with LPS (1 μg mL−1) alone or co-treated with LPS (1 μg mL−1) + free CUR, CUR NPs, or blank NPs at a concentration equivalent to 20 μM CUR. All treatments lasted for 24 h. At the end of the incubation period, the media was carefully removed and the cells were fixed with 4% paraformaldehyde for 50 min at room temperature. After that, the slides were gently washed with PBS twice and left to dry at room temperature. The slides were then transferred using forceps to dedicated sample holders until measurement.
000 cells per well on top of 13 mm × 1 mm CaF2 IR transmission windows (Crystran Ltd, Poole, UK) which were placed at the bottom of 24-well plates in duplicate. After 24 h, cells were treated with free CUR, CUR NPs, or blank NPs at a concentration equivalent to 20 μM CUR. In the case of RAW 264.7 macrophages, cells were treated with LPS (1 μg mL−1) alone or co-treated with LPS (1 μg mL−1) + free CUR, CUR NPs, or blank NPs at a concentration equivalent to 20 μM CUR. All treatments lasted for 24 h. At the end of the incubation period, the media was carefully removed and the cells were fixed with 4% paraformaldehyde for 50 min at room temperature. After that, the slides were gently washed with PBS twice and left to dry at room temperature. The slides were then transferred using forceps to dedicated sample holders until measurement.Results and discussion
Preparation and characterization of CUR-loaded oxQCT/TPGS NPs
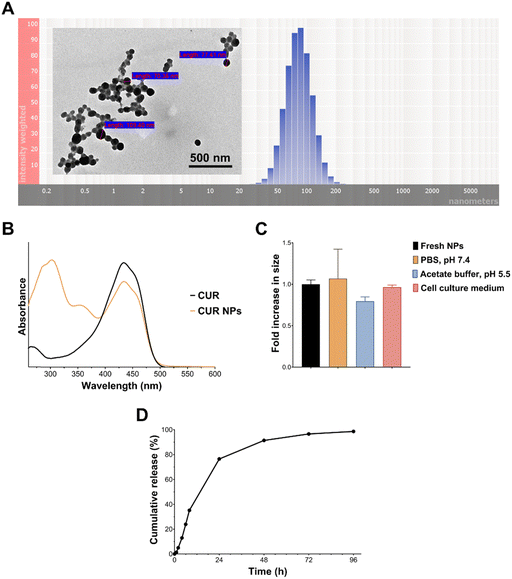
We have previously shown that QCT can undergo oxidative coupling reactions in basic media23,24 or in the presence of oxidizing agents such as NaIO4,30 leading to the formation of an amphiphilic material capable of self-assembly into NPs. The method is similar to the formation of NPs upon oxidation of green tea catechins.44 We have also shown that partially oxidized QCT (oxQCT) can co-assemble with polymers such as poly(ethylene glycol), poloxamers, and TPGS during NP formation by nanoprecipitation. Moreover, co-assembling oxQCT with these polymers could modulate the NPs’ cellular interactions by promoting apoptosis and synergizing with bioactive natural products.25,26 Having shown promise as a drug delivery platform, we decided to explore oxQCT as a nanocarrier for CUR, focusing on CUR's prominent bioactivities in cancer and inflammation. The NPs were co-assembled with TPGS as this polymeric excipient possesses notable antioxidant and anticancer properties,45 which could synergize with CUR and enhance its therapeutic action.As shown in Table 1, CUR-free NPs (blank NPs) exhibited a particle size of 60 nm and a PDI of 0.25. Upon CUR loading, an increase in particle size was noted at all loading levels, most likely due to drug entrapment within the NP matrix. All CUR-loaded NPs exhibited low PDI values ranging between 0.14 and 0.16. CUR-loaded NPs at 0.5 and 1 mg per batch achieved high loading efficiencies of 82 and 72%, respectively. Based on the structure of CUR, QCT, and TPGS (Fig. 1(A)), CUR was most likely entrapped within the NPs by hydrogen bonding, π–π stacking, and/or hydrophobic interactions, contributing to the excellent loading efficiency. However, increasing the CUR amount to 2 mg per batch (20% by weight of oxQCT/TPGS) significantly reduced the loading efficiency to 33%, which could indicate that the amount of CUR exceeded the capacity of the nanocarrier. Based on these results, the formulation CUR NPs_1 was chosen for further experiments as it contained the highest loaded amount of CUR compared to the other formulations.
| Formulation | Particle size (nm) | PDI | Loading efficiency (%) |
|---|---|---|---|
| Blank NPs | 60 ± 1 | 0.25 ± 0.02 | — |
| CUR NPs_0.5 | 81 ± 10 | 0.15 ± 0.04 | 82 ± 1 |
| CUR NPs_1 | 87 ± 4 | 0.14 ± 0.09 | 72 ± 1 |
| CUR NPs_2 | 88 ± 8 | 0.16 ± 0.01 | 33 ± 1 |
As demonstrated in Fig. 2(A), the optimal CUR NPs were associated with a narrow particle size distribution and spherical morphology. The UV-Vis spectrum of CUR (Fig. 2(B)) showed the characteristic peak at λmax = 430 nm, which remained unchanged after loading in CUR NPs, denoting that the nanoprecipitation process did not affect CUR's stability. The NP formulation also displayed excellent colloidal stability under different conditions, such as physiologic (7.4) and acidic pH (5.5), and in the presence of serum-supplemented cell culture media (Fig. 2(C)). As for drug release, a problem which could be encountered in colloidal systems is the burst release effect due to the high surface area-to-volume ratio at the nanoscale. As shown in Fig. 2(D), CUR NPs exhibited slow and sustained release at pH 7.4, with 35% cumulative release after 8 h, and 76–99% cumulative release between 24–96 h. Fitting the release data to the Korsmeyer–Peppas model resulted in a regression coefficient (R2) of 0.995, indicating an excellent fit with the model. Moreover, the release exponent n, which is employed to explain the drug release mechanism, was found to be 1.6. Since this value is greater than 0.85, it is assumed that CUR release followed supercase II transport through a combination of polymer relaxation and swelling.32 The ability of the NP vehicle to sustained CUR release will be valuable in in vivo applications, by protecting the drug from premature elimination while promoting its tissue uptake.
Antiproliferative activity of CUR-loaded NPs
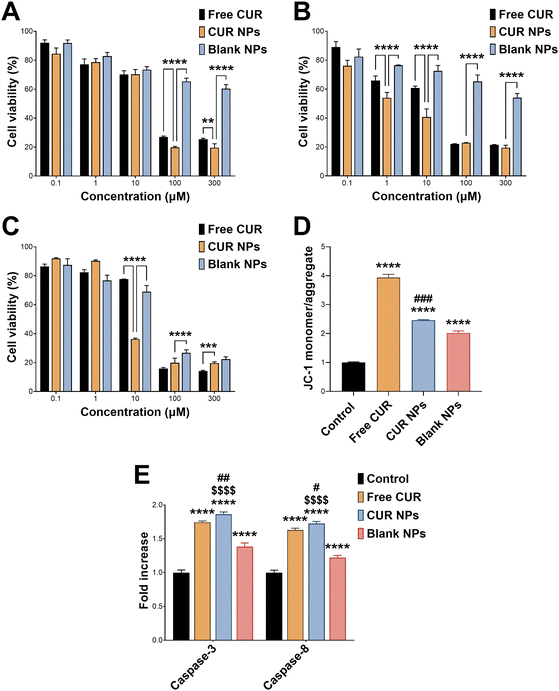
CUR has well-documented anticancer activity against a variety of cancer types, including breast cancer.46,47 Here, we employed MCF-7 cells, a commonly used in vitro model for hormone receptor-positive breast cancer cells, to test the antiproliferative activity of CUR NPs. Cells were treated with various concentrations of CUR, CUR NPs, and blank NPs for 24, 48, and 72 h, followed by performing a cell viability assay. As presented in Fig. 3(A)–(C), all agents including blank NPs exhibited dose- and time-dependent reduction in cell viability. Notably, at 72 h (Fig. 3(C)), CUR NPs were significantly more toxic than free CUR at 10 μM (p < 0.0001; mean difference = 41.4; 95% confidence interval [38.0, 44.8]), and more toxic than blank NPs at 100 μM (p < 0.0001; mean difference = 6.9; 95% confidence interval [3.5, 10.2]). At the highest concentration of 300 μM, CUR NPs and blank NPs demonstrated an equal effect, whereas free CUR had the lowest effect on cell viability (p < 0.001). Upon fitting the 72 h cell viability data by nonlinear regression, the half-maximal inhibitory concentration (IC50) could be determined, which reflects the potency of each treatment. Free CUR was associated with an IC50 of 24.3 μM, consistent with our previous findings.25 Blank NPs demonstrated considerable antiproliferative activity, with an estimated IC50 of 241.5 μM oxQCT/TPGS. On the other hand, CUR NPs achieved an IC50 of 5.7 μM in terms of CUR and 57.8 μM in terms of the oxQCT/TPGS NP vehicle, denoting a marked increase in potency for both the free drug and the NPs. Applying the Chou–Talalay equation (eqn (3)) resulted in a CI of 0.47, which confirms synergism between CUR and the NP vehicle.To further probe the anticancer activity of CUR and CUR NPs, a mitochondrial membrane potential assay was performed by relying on the conformational differences of the JC-1 probe in healthy and depolarized mitochondria. In healthy cells, JC-1 exists as aggregates that emit red fluorescence and are confined to the mitochondria. In cells undergoing apoptosis, depolarization of the mitochondria causes the aggregates to dissociate into monomers that emit green fluorescence as they exit into the cytosol.48 Consequently, the ratio of the JC-1 monomer/aggregate fluorescence intensities serves as a measure of apoptosis induction. MCF-7 cells were treated with free CUR and equivalent concentrations of CUR NPs and blank NPs for 24 h, followed by JC-1 staining. The results shown in Fig. 3(D) confirmed the proapoptotic activity of CUR which resulted in a 3.9-fold rise in JC-1 monomer/aggregate ratio compared to the control (p < 0.0001; mean difference = −2.9; 95% confidence interval [−3.1, −2.8]). Treatment with CUR NPs also resulted in a significant increase in JC-1 monomer/aggregate ratio of 2.5-fold compared to the control (p < 0.0001; mean difference = −1.5; 95% confidence interval [−1.6, −1.3]), although it was lower than the free drug. Nonetheless, CUR NPs maintained a greater increase in JC-1 monomer/aggregate ratio compared to blank NPs (p < 0.001; mean difference = 0.4; 95% confidence interval [0.3, 0.6]), which also demonstrated notable proapoptotic activity with a 2.0-fold increase relative to the control (p < 0.0001; mean difference = −1.0; 95% confidence interval [−1.2, −0.8]). These findings prompted a further investigation of caspases commonly upregulated in cells undergoing apoptosis such as caspase-3 and caspase-8. As shown in Fig. 3(E), all treatments including blank NPs promoted the expression of both caspase-3 and caspase-8. Importantly, CUR NPs were associated with a more significant increase in both caspases compared to the free drug (p < 0.01 and p < 0.05 in the case of caspase-3 and caspase-8, respectively), confirming the proapoptotic activity of the NPs, and that the NP vehicle synergizes with CUR in promoting cancer cell death.
The enhanced anticancer effects of CUR-loaded NPs compared to the free drug could be attributed to the inherent antiproliferative activity of the oxQCT/TPGS NP vehicle. This, in turn, is ascribed to the presence of TPGS, as oxQCT by itself has shown limited cytotoxicity against cancer24,49 and normal cells.30 Previous studies have reported on the anticancer activity of TPGS and TPGS-modified nanocarriers against a variety of cell types. For example, TPGS demonstrated time- and concentration-dependent cytotoxicity against wild-type MCF-7 and doxorubicin-resistant MCF-7/ADR cells. This effect was correlated with mitochondrial damage mediated by increased intracellular Ca2+ and cell cycle arrest in the G2/M phase, leading to apoptosis.50 Furthermore, TPGS-modified bilosomes (bile salt-based vesicles) entrapping CUR displayed an increase in cytotoxicity compared to the plain bilosomes in MCF-7/ADR cells.51 TPGS-polycaprolactone NPs also enhanced the bioactivity of paclitaxel against MCF-7 cells,52 and resveratrol against 4T1 breast cancer cells.53
Uptake of CUR NPs by MCF-7 cells
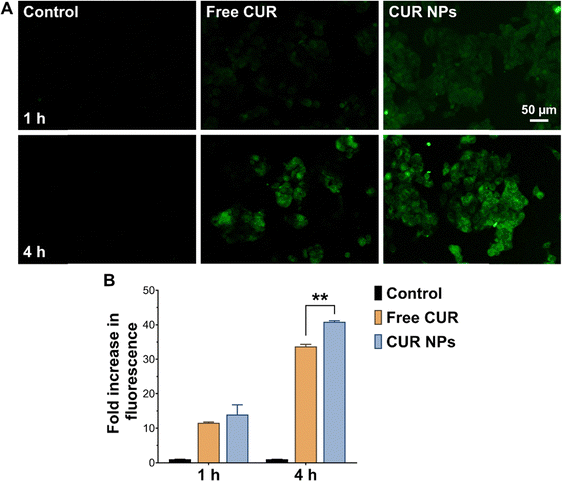
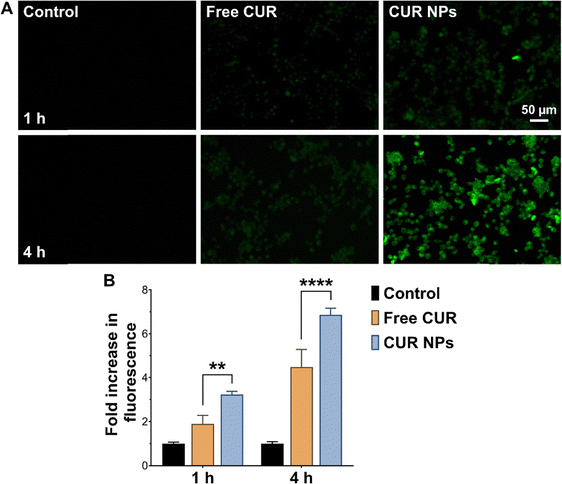
Uptake experiments were performed to determine whether cellular uptake could contribute to the enhanced antiproliferative and proapoptotic activity of CUR NPs. As presented in Fig. 4, MCF-7 cells treated with free CUR up to 4 h exhibited moderate fluorescence, indicating that some CUR was able to internalize into the cell membrane, most likely by passive diffusion. On the other hand, cells treated with CUR NPs demonstrated significantly enhanced fluorescence at both time points as observed by fluorescence microscopy and quantified by flow cytometry, denoting a greater degree of internalization compared to the free drug. The NPs were most likely internalized by endocytosis, similar to other types of polymeric NPs.54 Consequently, the superior anticancer activity of CUR NPs may in part be attributed to the promotion of cellular uptake by the NP formulation.Anti-inflammatory activity of CUR-loaded NPs
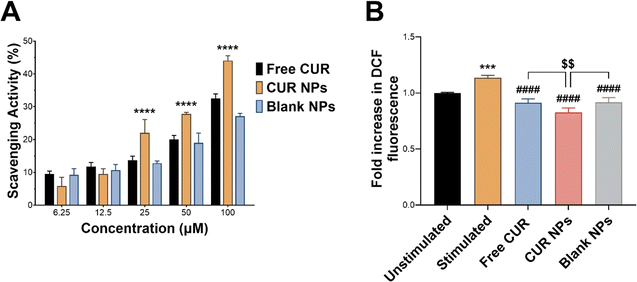
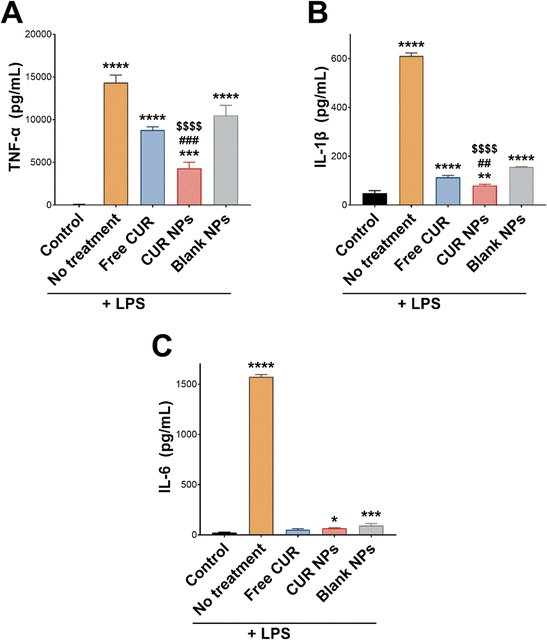
The anti-inflammatory activity of CUR is mainly attributed to its antioxidant activity and its ability to scavenge reactive species and relieve oxidative stress associated with inflammation.5,55 In this work, CUR was incorporated in TPGS-modified oxQCT NPs, where the NP vehicle itself also possesses antioxidant properties. oxQCT is derived from a potent antioxidant and maintains some antioxidant activity after partial oxidation,30 while TPGS displays antioxidant properties due to the vitamin E component.56 The antioxidant activity of CUR, CUR NPs, and blank NPs was experimentally determined by conducting a DPPH radical scavenging assay. As shown in Fig. 5(A), loading CUR in the NPs significantly enhanced its radical scavenging properties starting from 25 μM up to 100 μM (p < 0.0001), which suggests that the NP vehicle based on oxQCT and TPGS could also enhance the anti-inflammatory activity compared to the free drug. To test this hypothesis, an intracellular ROS assay was performed in LPS-stimulated RAW 264.7 macrophages using the DCFDA probe. This macrophage cell line was chosen as an in vitro model for inflammation for its consistent inflammatory response when challenged by stimulants such as LPS.57 LPS stimulation increased intracellular ROS in macrophages by causing a significant rise in DCF fluorescence compared to the unstimulated cells (Fig. 5(B); p < 0.001). Co-treating the cells with LPS and free CUR, CUR NPs, or blank NPs significantly reduced intracellular ROS compared to LPS alone (p < 0.0001), which aligns with the DPPH assay and confirms the antioxidant properties of free CUR and the NPs. Interestingly, treatment with free CUR, CUR NPs, and blank NPs also relieved intrinsic oxidative stress in untreated unstimulated cells as indicated by the reduction in DCF fluorescence (p < 0.05, p < 0.0001, and p < 0.05, respectively). Importantly, CUR NPs demonstrated a superior ability to alleviate oxidative stress in LPS-stimulated macrophages compared to free CUR and blank NPs (p < 0.01), further supporting the ability of the NPs to potentiate the bioactivity of CUR.The anti-inflammatory activity of CUR NPs was further investigated by measuring the proinflammatory cytokines TNF-α, IL-1β, and IL-6 released from LPS-stimulated macrophages. LPS is a potent toxin known to activate immune cells, which in turn secrete proinflammatory cytokines initiating an inflammatory cascade. This action was evident by the significant increase in secreted TNF-α, IL-1β, and IL-6 by LPS-stimulated RAW 264.7 macrophages compared to unstimulated cells (Fig. 6; p < 0.0001). In the case of TNF-α (Fig. 6(A)), co-treating the cells with LPS and CUR, CUR NPs, or blank NPs attenuated the increase in TNF-α secretion to varying degrees, with CUR NPs demonstrating the lowest level of TNF-α compared to free CUR (p < 0.001) and blank NPs (p < 0.0001). The same trend was observed in IL-1β levels (Fig. 6(B)), where free CUR, CUR NPs, and blank NPs inhibited IL-1β secretion upon LPS stimulation, and CUR NPs exhibited significantly lower levels compared to free CUR (p < 0.01) and blank NPs (p < 0.0001). As for IL-6 (Fig. 6(C)), co-treating the stimulated cells with free CUR completely normalized its levels. Although CUR NPs and blank NPs demonstrated statistically higher IL-6 levels compared to the unstimulated cells, no significant difference was detected between them and free CUR. The results of the DPPH, DCFDA, and cytokine assays collectively emphasize the potent anti-inflammatory activity of CUR NPs, and present oxQCT/TPGS NPs as a viable delivery platform that could potentiate CUR's action in inflammatory conditions. Our findings are aligned with previous reports which described the enhanced antioxidant and anti-inflammatory activity of CUR when formulated in inherently-antioxidant nanocarriers such as those derived from green tea polyphenols.58,59
Uptake of CUR NPs by RAW 264.7 macrophages
The superior anti-inflammatory activity of CUR NPs could in part be attributed to the inherent antioxidant properties of the NP vehicle itself. To determine whether cellular uptake may have also played a role as in the case of MCF-7 cells, RAW 264.7 macrophages were imaged by fluorescence microscopy and analyzed by flow cytometry following treatment with free CUR and CUR NPs. Consistent with the uptake results in MCF-7 cells (Fig. 4), the macrophages internalized CUR NPs more readily than free CUR, as evidenced by more prominent fluorescence signals particularly at 4 h (Fig. 7).SR-μFTIR measurements of MCF-7 cells and RAW 264.7 macrophages upon treatment with CUR NPs
In this study, we employed SR-μFTIR to further investigate the anticancer and anti-inflammatory activities of CUR in the developed NP formulation. Two main IR spectral regions were defined and investigated as reported previously.40 The first region (3050–2800 cm−1) corresponds to the distribution of lipids, while the second region (1855–1485 cm−1) corresponds to the distribution of carbonyl (1855–1700 cm−1) and protein bands amide I (1700–1600 cm−1) and amide II (1600–1485 cm−1).60 Note that sample thickness and homogeneity, IR slides, and fixation methods used during sample preparation can all influence the reproducibility and the quality of the obtained IR spectra. In the current work, a standardized published sample preparation protocol was applied to minimize the effect of sample preparation.28 CaF2 IR transmission windows were selected as the growing surface due to their compatibility with IR and their ability to provide excellent cell adherence. The cell volume was carefully optimized to ensure the formation of a homogeneous, monolayer thin film. Fixation was performed using paraformaldehyde, which offers minimal interference in the IR spectrum, ensures uniform fixation, and effectively preserves key biomolecules including lipids and proteins.| Wavenumber (cm−1) | Structure | Alterations compared to the control |
|---|---|---|
| 2955.5 | asCH3: cholesterol esters, triglycerides | Free CUR: increase in signal intensity |
| CUR NPs: decrease in signal intensity | ||
| 2918.0 | asCH2: long chain fatty acids, phospholipids | Free CUR: decrease in signal intensity; blue shift to 2924 cm−1 |
| CUR NPs: increase in signal intensity | ||
| 2872.8 | symCH3: cholesterol esters, triglycerides | CUR NPs: decrease in signal intensity |
| 2850.0 | symCH2: long chain fatty acids, phospholipids | Free CUR: decrease in signal intensity |
| CUR NPs: increase in signal intensity | ||
| 1650.9 | Amide I, α-helix | Free CUR: blue shift to 1652.9 cm−1 |
| CUR NPs: decrease in signal intensity | ||
| Blank NPs: increase in signal intensity | ||
| 1544.5 | Amide II, α-helix | CUR NPs: increase in signal intensity |
| Blank NPs: increase in signal intensity (less than CUR NPs) | ||
| 1740.0 | Lipid carbonyl | Increase in signal intensity in all groups compared to the control |
| Cell line | Treatment | CH2/CH3 |
|---|---|---|
| MCF-7 | Control | 2.06 |
| Free CUR | 1.55 | |
| CUR NPs | 2.80 | |
| Blank NPs | 2.38 | |
| RAW 264.7 | Control | 1.55 |
| LPS | 1.38 | |
| Free CUR | 2.06 | |
| CUR NPs | 1.33 | |
| Blank NPs | 2.53 |
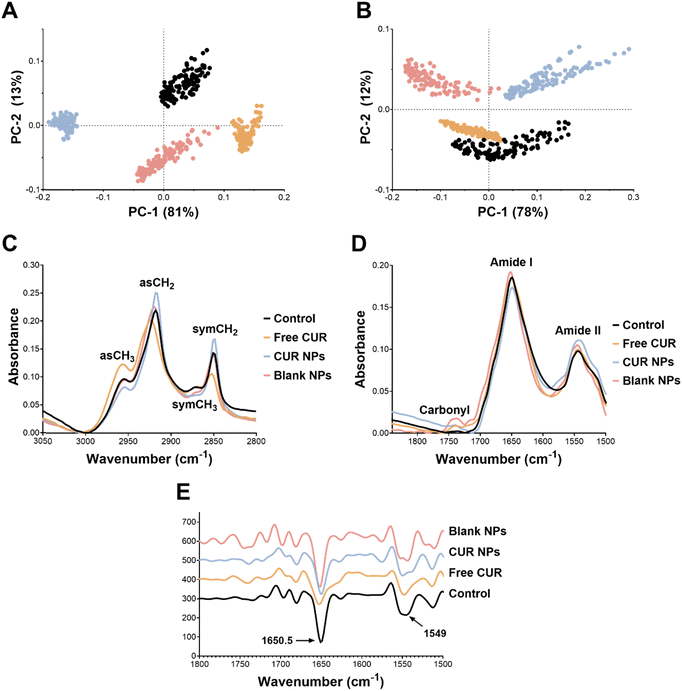
As for the protein-carbonyl region, the PCA score plots also revealed a good separation of the groups (Fig. 8(B)). PC-1 and PC-2 loading of the second derivative protein region (Fig. S1 of the ESI†) was characterized by strong peaks in the area of amide I and amide II. The average raw spectra (Fig. 8(D)) revealed similar signal intensities of amide I and amide II between free CUR and the control. While the blank NPs showed a small increase in amide I and amide II signals, CUR NPs exhibited a decrease in amide I and an increase in amide II signals (more than blank NPs) compared to the control (Table 2). Upon examination of the second derivative spectra of the protein-carbonyl region (Fig. 8(E)), we found that amide I had the strongest signal in the loading plot and, hence, the most important variable for group separation. As seen in Fig. 8(E), the amide I signal at 1650.9 cm−1 in the control, blank and CUR NPs groups was shifted to 1652.9 cm−1 in the free CUR group (Table 2), denoting a possible conformational change in the α-helix protein secondary structure.64,65 Generally, spectral analysis of the protein region revealed that CUR NPs mainly affected the protein profile but had no significant effect on the protein conformation in MCF-7 cells. With regard to the carbonyl region, the three treatments (free CUR, CUR NPs, and blank NPs) caused an increase in the carbonyl signal at 1740 cm−1 (Fig. 8(D) and Table 2). This could be related to an increase in oxidative stress resulting from lipid peroxidation, which contributes to the apoptosis cascade.66,67
| Wavenumber (cm−1) | Structure | Alterations compared to the control |
|---|---|---|
| 2957.0 | asCH3: cholesterol esters, triglycerides | Free CUR, CUR NPs, and blank NPs: decrease in signal intensity |
| 2921.9 | asCH2: long chain fatty acids, phospholipids | Free CUR: increase in signal intensity |
| CUR NPs: decrease in signal intensity | ||
| Blank NPs: increase in signal intensity; red shift to 2918 cm−1 | ||
| 2873.4 | symCH3: cholesterol esters, triglycerides | Free CUR and blank NPs: decrease in signal intensity |
| CUR NPs: increase in signal intensity | ||
| 2852.4 | symCH2: long chain fatty acids, phospholipids | Free CUR: increase in signal intensity |
| Blank NPs: increase in signal intensity; red shift to 2848.6 cm−1 | ||
| 1650.9 | Amide I, α-helix | Free CUR: decrease in signal intensity; blue shift to 1654.8 cm−1 |
| CUR NPs: red shift to 1645.1 cm−1 | ||
| 1541.0 | Amide II, α-helix | Free CUR: increase in signal intensity; blue shift to 1548.9 cm−1 |
| 1740.0 | Lipid carbonyl | No difference between all groups |
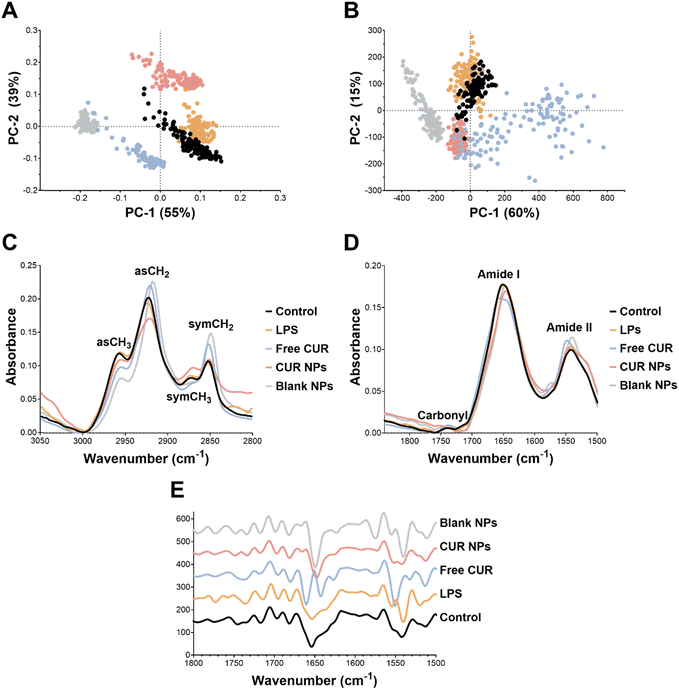
As for the protein-carbonyl region, PC-1 and PC-2 loading plots were characterized by strong amide I and amide II peaks (Fig. S1 of the ESI†). As observed in Fig. 9(D) and Table 4, Among the four groups, free CUR was associated with the most significant changes in the protein spectral region. In terms of signal intensities, the free CUR group exhibited a decrease and an increase in the peak intensities for the amide I and amide II signals compared to the control, respectively. Furthermore, the amide I peak in free CUR was shifted to 1654.8 cm−1 whereas the amide II peak was shifted to 1548.9 cm−1 compared to the control (1650.9 and 1541 cm−1, respectively; Table 4). These findings suggest a possible change in the protein secondary structure (α-helix) to a less ordered structure upon treating the macrophages with free CUR.68 These observations were further corroborated by the second derivative spectra of the protein-carbonyl region (Fig. 9(E)), where the free CUR group exhibited a notable change in the amide I peak compared to the control. Additionally, CUR NPs exhibited a shift in the amide I peak position to 1645.1 cm−1 compared to 1650.9 cm−1 in the control, suggesting a possible change in the protein secondary structure for amide I from α-helix in the control group to the unordered random coil conformation.68 These spectral changes observed in free CUR and CUR NPs may correlate with cytokine secretion.
Taken together, SR-μFTIR analyses corroborated the diverse bioactivities of CUR as a function of the cell line, and how these bioactivities could be modulated by the developed delivery platform oxQCT/TPGS NPs. For example, in MCF-7 cells, most of the biochemical changes detected by SR-μFTIR were related to the lipid profile, suggesting a potential role in affecting plasma membrane permeability and/or lipid peroxidation as they relate to apoptotic events. In the macrophages, the most notable changes were related to the protein composition, which could be ascribed to alterations in cytokine secretion.
Conclusion
In this study, we prepared and characterized a novel nanoformulation for CUR based on an emerging nanocarrier, oxQCT. We showed that co-formulating oxQCT with TPGS significantly improves the anticancer and anti-inflammatory activities of CUR and promotes cellular uptake in MCF-7 and RAW 264.7 macrophages, respectively. MCF-7 cell death was mediated via a mitochondrial and caspase-dependent pathway. In RAW 264.7 macrophages, the NPs significantly attenuated the secretion of proinflammatory cytokines upon LPS stimulation. The promising in vitro results prompt further in vivo validation in appropriate animal models. Furthermore, SR-μFTIR measurements of the treated cell lines provided clues regarding the biochemical changes associated with CUR treatment, both as a free drug and as an NP formulation. This emphasizes the importance of advanced bioanalytical techniques in delineating the biological activities of new NP therapeutics, an important step toward their clinical translation.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Al-Zaytoonah University of Jordan (grant no. 14/08/2021-2022) and the International Centre for Genetic Engineering and Biotechnology (ICGEB; contract no. CRP/22/006). The authors thank Dr Gihan Kamel (IR Beamline Principal Scientist at SESAME) for assistance with SR-μFTIR measurements.References
- J. Sharifi-Rad, Y. E. Rayess, A. A. Rizk, C. Sadaka, R. Zgheib, W. Zam, S. Sestito, S. Rapposelli, K. Neffe-Skocińska, D. Zielińska, B. Salehi, W. N. Setzer, N. S. Dosoky, Y. Taheri, M. El Beyrouthy, M. Martorell, E. A. Ostrander, H. A. R. Suleria, W. C. Cho, A. Maroyi and N. Martins, Front. Pharmacol., 2020, 11, 550909 Search PubMed.
- X.-Y. Xu, X. Meng, S. Li, R.-Y. Gan, Y. Li and H.-B. Li, Nutrients, 2018, 10, 1553 CrossRef.
- A. Allegra, V. Innao, S. Russo, D. Gerace, A. Alonci and C. Musolino, Cancer Invest., 2017, 35, 1–22 CrossRef CAS PubMed.
- M. E. Abd El-Hack, M. T. El-Saadony, A. A. Swelum, M. Arif, M. M. Abo Ghanima, M. Shukry, A. Noreldin, A. E. Taha and K. A. El-Tarabily, J. Sci. Food Agric., 2021, 101, 5747–5762 CrossRef CAS PubMed.
- Y. Peng, M. Ao, B. Dong, Y. Jiang, L. Yu, Z. Chen, C. Hu and R. Xu, Drug Des., Dev. Ther., 2021, 4503–4525 CrossRef CAS PubMed.
- M. C. Fadus, C. Lau, J. Bikhchandani and H. T. Lynch, J. Tradit. Complementary Med., 2017, 7, 339–346 CrossRef PubMed.
- S. Sabet, A. Rashidinejad, L. D. Melton and D. J. McGillivray, Trends Food Sci. Technol., 2021, 110, 253–266 CrossRef CAS.
- M. Kharat and D. J. McClements, J. Colloid Interface Sci., 2019, 557, 506–518 CrossRef CAS.
- Y. Chen, Y. Lu, R. J. Lee and G. Xiang, Int. J. Nanomed., 2020, 3099–3120 CrossRef CAS.
- B. Salehi, M. L. Del Prado-Audelo, H. Cortés, G. Leyva-Gómez, Z. Stojanović-Radić, Y. D. Singh, J. K. Patra, G. Das, N. Martins and M. Martorell, J. Clin. Med., 2020, 9, 746 CrossRef CAS.
- S. Maleki Dizaj, M. Alipour, E. Dalir Abdolahinia, E. Ahmadian, A. Eftekhari, H. Forouhandeh, Y. Rahbar Saadat, S. Sharifi and S. Zununi Vahed, Phytother. Res., 2022, 36, 1156–1181 CrossRef CAS.
- J. Guo, T. Suma, J. J. Richardson and H. Ejima, ACS Biomater. Sci. Eng., 2019, 5, 5578–5596 CrossRef CAS.
- Z. Chen, M. A. Farag, Z. Zhong, C. Zhang, Y. Yang, S. Wang and Y. Wang, Adv. Drug Delivery Rev., 2021, 176, 113870 CrossRef CAS.
- X. Wang, Y. Fan, J. Yan and M. Yang, Chem. Eng. J., 2022, 439, 135661 CrossRef CAS.
- J. Zhou, Z. Lin, Y. Ju, M. A. Rahim, J. J. Richardson and F. Caruso, Acc. Chem. Res., 2020, 53, 1269–1278 CrossRef CAS PubMed.
- D. Wu, J. Zhou, M. N. Creyer, W. Yim, Z. Chen, P. B. Messersmith and J. V. Jokerst, Chem. Soc. Rev., 2021, 50, 4432–4483 RSC.
- S. Xiang, P. Yang, H. Guo, S. Zhang, X. Zhang, F. Zhu and Y. Li, Macromol. Rapid Commun., 2017, 38, 1700446 CrossRef PubMed.
- Z. Yi, G. Chen, X. Chen, X. Ma, X. Cui, Z. Sun, W. Su and X. Li, ACS Appl. Mater. Interfaces, 2020, 12, 33550–33563 CrossRef CAS PubMed.
- Z. Yi, X. Chen, G. Chen, Z. Deng, Q. Tong, Z. Sun, X. Ma, W. Su, L. Ma, Y. Ran and X. Li, ACS Appl. Mater. Interfaces, 2020, 12, 37914–37928 CrossRef CAS.
- P. Chowdhury, P. K. Nagesh, E. Hatami, S. Wagh, N. Dan, M. K. Tripathi, S. Khan, B. B. Hafeez, B. Meibohm and S. C. Chauhan, J. Colloid Interface Sci., 2019, 535, 133–148 CrossRef CAS PubMed.
- T. Wang, Q. Fan, J. Hong, Z. Chen, X. Zhou, J. Zhang, Y. Dai, H. Jiang, Z. Gu, Y. Cheng and Y. Li, Small, 2021, 17, 2102485 CrossRef CAS PubMed.
- S. Sunoqrot, E. Al-Shalabi, A. G. Al-Bakri, H. Zalloum, B. Abu-Irmaileh, L. H. Ibrahim and H. Zeno, ACS Omega, 2021, 6, 2767–2776 CrossRef CAS PubMed.
- S. Sunoqrot, A. G. Al-Bakri, L. H. Ibrahim and N. Aldaken, ACS Appl. Bio Mater., 2022, 5, 5156–5164 CrossRef CAS PubMed.
- S. Sunoqrot, E. Al-Shalabi and P. B. Messersmith, Biomater. Sci., 2018, 6, 2656–2666 RSC.
- S. Sunoqrot, S. Abusulieh and O. H. Abusara, Int. J. Pharm., 2023, 645, 123392 CrossRef CAS PubMed.
- O. Alkhaldi, S. Abusulieh, O. H. Abusara and S. Sunoqrot, Int. J. Pharm., 2024, 665, 124674 CrossRef CAS PubMed.
- A. Refaat and G. Kamel, Appl. Spectrosc. Rev., 2022, 58, 525–544 CrossRef.
- N. Elmadany, E. Khalil, L. Vaccari, G. Birarda, I. Yousef and R. Abu-Dahab, Infrared Phys. Technol., 2018, 95, 141–147 CrossRef CAS.
- B. Dunkhunthod, C. Talabnin, M. Murphy, K. Thumanu, P. Sittisart, T. Hengpratom and G. Eumkeb, J. Evidence-Based Complementary Altern. Med., 2020, 1, 7436920 CrossRef PubMed.
- S. Sunoqrot, E. Al-Shalabi, L. Hasan Ibrahim and H. Zalloum, Molecules, 2019, 24, 3815 CrossRef CAS PubMed.
- R. W. Korsmeyer, R. Gurny, E. Doelker, P. Buri and N. A. Peppas, Int. J. Pharm., 1983, 15, 25–35 CrossRef CAS.
- J. Siepmann and N. A. Peppas, Adv. Drug Delivery Rev., 2012, 64, 163–174 CrossRef.
- T.-C. Chou, Pharmacol. Rev., 2006, 58, 621–681 CrossRef CAS PubMed.
- N. N. Mahmoud, R. Abu-Dahab, L. A. Hamadneh, D. Abuarqoub, H. Jafar and E. A. Khalil, Mol. Pharmaceutics, 2019, 16, 4149–4164 CrossRef CAS PubMed.
- A. R. Althaher, S. A. Oran, M. W. Awadallah, H. H. Ameen, R. F. Shehabi, L. M. S. Bourghli and A. Mastinu, Chem. Biodiversity, 2024, 21, e202400026 CrossRef CAS PubMed.
- E. Al-Shalabi, S. Abusulieh, A. M. Hammad and S. Sunoqrot, Biomater. Sci., 2022, 10, 5504–5519 RSC.
- S. Sunoqrot, M. Alkurdi, A. Q. Al Bawab, A. M. Hammad, R. Tayyem, A. Abu Obeed and M. Abufara, Saudi Pharm. J., 2023, 31, 845–853 CrossRef CAS PubMed.
- G. Kamel, S. Lefrançois, M. Al-Najdawi, T. Abu-Hanieh, I. Saleh, Y. Momani and P. Dumas, Synchrotron Radiat. News, 2017, 30, 8–10 CrossRef.
- G. Kamel, S. Lefrancois, T. Moreno, M. Al-Najdawi, Y. Momani, A. Abbadi, G. Paolucci and P. Dumas, J. Synchrotron Radiat., 2021, 28, 1927–1934 CrossRef CAS PubMed.
- L. A. Dahabiyeh, R. S. H. Mansour, S. S. Saleh and G. Kamel, J. Pharm. Biomed. Anal., 2020, 184, 113186 CrossRef CAS PubMed.
- R. Nimer, G. Kamel, M. A. Obeidat and L. A. Dahabiyeh, Spectrochim. Acta, Part A, 2022, 264, 120259 CrossRef CAS PubMed.
- L. A. Dahabiyeh, R. S. Mansour, W. Darwish, S. S. Saleh and G. Kamel, J. Pharm. Biomed. Anal., 2022, 220, 114981 CrossRef CAS PubMed.
- A. G. Al-Bakri, L. A. Dahabiyeh, E. Khalil, D. Jaber, G. Kamel, N. Schleimer, C. Kohler and K. Becker, Antibiotics, 2022, 11, 1607 CrossRef CAS PubMed.
- D. Liu, X. Chen, Z. Yi, Q. Tong, L. Ma, Y. Tan, X. Cao and X. Li, ACS Appl. Bio Mater., 2024, 7, 1763–1777 CrossRef CAS PubMed.
- S. Rathod, P. Bahadur and S. Tiwari, Int. J. Pharm., 2021, 592, 120045 CrossRef CAS PubMed.
- V. Zoi, V. Galani, G. D. Lianos, S. Voulgaris, A. P. Kyritsis and G. A. Alexiou, Biomedicines, 2021, 9, 1086 CrossRef CAS PubMed.
- K. A. Barcelos, C. R. Mendonça, M. Noll, A. F. Botelho, C. R. D. Francischini and M. A. M. Silva, Cancers, 2022, 14, 2165 CrossRef CAS PubMed.
- F. Sivandzade, A. Bhalerao and L. Cucullo, Bio-Protoc., 2019, 9, e3128 Search PubMed.
- S. Sunoqrot, T. Al-Debsi, E. Al-Shalabi, L. Hasan Ibrahim, F. N. Faruqu, A. Walters, R. Palgrave and K. T. Al-Jamal, ACS Biomater. Sci. Eng., 2019, 5, 6036–6045 CrossRef CAS PubMed.
- L. Tang, K. Huang, W. Jiang, L. Fu, R. Zhang, L. Shen, Z. Ou, Y. Huang and Z. Zhang, Drug Delivery, 2023, 30, 2183830 CrossRef PubMed.
- H. Hegazy, M. M. Amin, W. Fayad and M. Y. Zakaria, Int. J. Pharm., 2022, 619, 121717 CrossRef CAS PubMed.
- T. Chen, F. Xing and Y. Sun, J. Exp. Nanosci., 2024, 19, 2281938 CrossRef.
- P. G. Cavalcante de Freitas, B. Rodrigues Arruda, M. G. Araújo Mendes, J. V. Barroso de Freitas, M. E. da Silva, T. L. Sampaio, R. Petrilli and J. O. Eloy, Cancers, 2023, 15, 2802 CrossRef CAS PubMed.
- L. C. Nelemans and L. Gurevich, Materials, 2020, 13, 366 CrossRef CAS PubMed.
- K. Mokgalaboni, Y. Ntamo, K. Ziqubu, T. M. Nyambuya, B. B. Nkambule, S. E. Mazibuko-Mbeje, K. B. Gabuza, N. Chellan, L. Tiano and P. V. Dludla, Food Funct., 2021, 12, 12235–12249 RSC.
- A. K. Mehata, A. Setia, V. Vikas, A. K. Malik, R. Hassani, H. G. Dailah, H. A. Alhazmi, A. A. Albarraq, S. Mohan and M. S. Muthu, Pharmaceutics, 2023, 15, 722 CrossRef CAS PubMed.
- B. M. Facchin, G. O. dos Reis, G. N. Vieira, E. T. B. Mohr, J. S. da Rosa, I. F. Kretzer, I. G. Demarchi and E. M. Dalmarco, Inflammation Res., 2022, 71, 741–758 CrossRef CAS PubMed.
- X. Chen, Q. Ren, G. Chen, Z. Yi, Q. Tong, Y. Ran, L. Ma, P. Fu, L. Ma and X. Li, ACS Sustainable Chem. Eng., 2023, 11, 7288–7300 CrossRef CAS.
- Q. Pan, L. Xie, H. Zhu, Z. Zong, D. Wu, R. Liu, B. He and Y. Pu, Regener. Biomater., 2024, 11, rbae122 CrossRef CAS PubMed.
- S. Brezillon, V. Untereiner, L. Lovergne, I. Tadeo, R. Noguera, F. X. Maquart, Y. Wegrowski and G. D. Sockalingum, Anal. Bioanal. Chem., 2014, 406, 5795–5803 CrossRef CAS PubMed.
- G. A. Raouf, A. R. L. Al-Malki, N. Mansouri and R. M. Mahmoudi, Life Sci. J., 2011, 8, 453–464 Search PubMed.
- Z. L. Wei, D. J. Jiao and J. X. Xu, J. Spectrosc., 2015, 1, 570190 Search PubMed.
- G. Cakmak-Arslan, H. Haksoy, P. Goc-Rasgele and M. Kekecoglu, Spectrochim. Acta, Part A, 2020, 228, 117719 CrossRef CAS PubMed.
- M. Jackson and H. H. Mantsch, Crit. Rev. Biochem. Mol. Biol., 1995, 30, 95–120 CrossRef CAS PubMed.
- E. Cerf, R. Sarroukh, S. Tamamizu-Kato, L. Breydo, S. Derclaye, Y. F. Dufrêne, V. Narayanaswami, E. Goormaghtigh, J. M. Ruysschaert and V. Raussens, Biochem. J., 2009, 421, 415–423 CrossRef CAS PubMed.
- A. B. Abdelrazzak, A. M. Hezma and G. S. El-Bahy, Biochim. Biophys. Acta, Biomembr., 2021, 1863, 183726 CrossRef CAS PubMed.
- G. Barrera, ISRN Oncol., 2012, 1, 137289 Search PubMed.
- J. Kong and S. Yu, Acta Biochim. Biophys. Sin., 2007, 39, 549–559 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ma01202j |
| This journal is © The Royal Society of Chemistry 2025 |