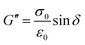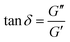Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Unravelling the synergies: effects of hydrogel mechanics and biofunctionalization on mesenchymal stem cell osteogenic differentiation†
Cristina López-Serrano abc,
Murielle Rémya,
Thierry Leste-Lasserred,
Gaétan Laroche‡
abc,
Murielle Rémya,
Thierry Leste-Lasserred,
Gaétan Laroche‡
 *bc and
Marie-Christine Durrieu‡
*bc and
Marie-Christine Durrieu‡ *a
*a
aUniv. Bordeaux, CNRS, Bordeaux INP, CBMN, UMR 5248, Pessac, France. E-mail: marie-christine.durrieu@inserm.fr; Tel: (+33) 05 40 00 30 37
bLaboratoire d’Ingénierie de Surface, Centre de Recherche sur les Matériaux Avancés, Département de Génie des Mines, de la Métallurgie et des Matériaux, Université Laval, Québec, Canada. E-mail: Gaetan.Laroche@gmn.ulaval.ca; Tel: +1 418-656-2131
cAxe Médecine Régénératrice, Centre de Recherche du Centre Hospitalier Universitaire de Québec, Hôpital St-François d’Assise, Québec, Canada
dINSERM, PUMA, U1215, Neurocentre Magendie, Univ. Bordeaux, Bordeaux 33077, France
First published on 23rd May 2025
Abstract
The design of hydrogels for cell culture that mimic the extracellular matrix is complex given the numerous factors involved. This study explores the synergistic effects of mechanical properties and biofunctionalization on mesenchymal stem cell (MSC) differentiation, with a focus on bone regeneration. Although the influence of the elastic and viscoelastic properties of hydrogel matrices on stem cell differentiation is well recognized, the impact of viscoelasticity is still poorly understood. In addition, the synergistic interaction between viscoelastic properties and biofunctionalization remains poorly understood. In this work, poly(ethylene glycol)diacrylate (PEGDA) hydrogels were designed with varying shear moduli and loss tangents. This research uniquely investigates the combined effects of mechanical properties and biofunctionalization, by co-functionalizing the hydrogels with a mixture of RGD and a BMP-2 mimetic peptide. This dual approach allowed for an in-depth analysis of the synergies between mechanics and bioactivity, as well as the impact of different peptide combinations. Our results demonstrated that the mechanical properties of hydrogels are a primary factor in driving differentiation, as biofunctionalization alone is not sufficient to induce this process. Additionally, our study underscores the significant impact of the lesser-known viscoelastic properties on MSC differentiation into an osteoblastic lineage, even after just one week of culture.
Statement of significanceHigh viscoelasticity enhances osteogenic differentiation of hMSCs in hydrogels with optimal stiffness. Functionalization with osteogenic peptides is only able to boost osteodifferentiation in materials with adequate mechanical properties. |
1. Introduction
Mesenchymal stem cells (MSCs) are of great interest for tissue engineering applications since they are multipotent stem cells that can be easily retrieved and they are able to differentiate towards various cell types, including bone, adipose or cartilage cells. Despite their potential and although more than 1000 clinical trials have been registered to investigate their uses,1 their clinical applications remain very limited.2 Some reasons include biological differences among cells from different donors and tissue sources, lack of standardized practices for MSC harvesting and preparation,3 and potential for unwanted differentiation and tumorigenesis.4 Identifying the critical features that guide MSC behaviour and differentiation is key to improving the translation of stem cell-based treatments to the clinic. Advanced culture systems that allow to mimic the complex microenvironments that surround cells in their native tissues are a promising strategy to bridge the gap between in vitro and in vivo findings. Additionally, the rapid obtention of committed cells is of interest for clinical applications and such materials have the potential to elicit fast differentiation thanks to the presence of multiple stimuli.Biological and physical aspects of the cell extracellular matrix (ECM) drive stem cell fate.5 However, the full extent of their individual and synergistic effects are not yet fully understood. Advances in the biomaterials field have been key to enabling the fabrication of materials, such as hydrogels, with tuneable and controlled properties that mimic the native ECM. The differentiation of MSCs towards the osteogenic lineage is known to be influenced by the mechanical properties of the substrate. Substrate stiffness refers to the resistance that a material opposes to deformation and is commonly reported with the measure of elastic modulus. Several studies have aimed to elucidate the optimal substrate stiffness that elicits osteogenesis and the overall trend points towards a Young's modulus between 30 to 50 kPa.6–8 Nevertheless, there is not a clear consensus and some authors have found values outside that range to be more favourable.9,10 Moreover, ECMs and tissues are not purely elastic materials but rather display time-dependent viscoelastic behaviours.11 Few studies have investigated the effects of hydrogel viscoelasticity on osteogenic differentiation. For instance, Cameron et al.12 fabricated polyacrylamide hydrogels with the same elastic modulus but different viscous modulus and concluded that cells spread more and have higher alkaline phosphatase activity on substrates with higher viscoelastic component. Chaudhuri and colleagues also found, using different hydrogels, that overall higher viscoelasticity enhances osteogenecity.13,14 Other authors have recently examined this phenomenon and, although it appears overall that viscoelastic materials favour osteogenecity as opposed to elastic ones, the ideal parameters of viscoelasticity are not clear, suggesting a complex interplay between elasticity and viscoelasticity.10,15
In addition to tuning the mechanical properties of the matrix, functionalizing biomaterials with specific factors that target the desired pathways is crucial for guiding MSC behaviour. The synergy between the mechanical properties and the bioactivity of materials to promote the differentiation of MSCs towards the osteogenic lineage is not addressed in the literature. Peptides like RGD, which promotes cell adhesion, and BMP-2 mimetic peptides, which induce osteogenic differentiation, are commonly used for this purpose.16–19 The synergistic effects of combining these peptides are particularly powerful, as they can enhance integrin signalling and growth factor activity, thereby accelerating the differentiation process.16,20–24 However, the optimal ratios and combinations of these peptides, as well as their interaction with the mechanical properties of the substrate, are not yet fully understood.
Altogether, tuning both the mechanical and biochemical properties of biomaterials may play a key role in hMSC differentiation as they were shown to influence downstream signalling cascades. Indeed, growing evidence shows that biochemical cues and mechanotransduction synergize to drive osteogenic differentiation through the conversion of intracellular signals such as yes-associated protein (YAP)/transcriptional coactivator with PDZ-binding motif (TAZ).25,26
This study aimed to explore these synergies by systematically varying both the mechanical properties and biofunctionalization of PEGDA hydrogels. The mechanical properties, characterized by rheology, were varied by changing the concentration and chain length of the PEG oligomers used to synthesize the hydrogels. Additionally, the hydrogels were covalently grafted with a mixture of RGD and BMP-2 mimetic peptides, controlling the ratios of each peptide. The effects of these combined mechanical and biochemical cues on MSC behaviour were evaluated by assessing cell proliferation and osteogenic differentiation in vitro. Immunocytochemistry and qPCR were employed to analyse early and late osteoblast and osteocyte markers. By examining how different combinations of elasticity, viscoelasticity, and peptide functionalization influence MSC fate, this research aimed to identify not only the optimal conditions for promoting osteogenesis but also which factor most effectively predominantly drives osteogenic differentiation. The potential applications of these innovative hydrogels include their use as a platform for the efficient and large-scale generation of stem cells toward a specific lineage, as well as for implant or scaffold functionalization through an innovative bioactive coating.
2. Materials and methods
2.1. Materials
Poly(ethylene glycol)diacrylate (PEGDA, M.W. 4000 Da), 2-hydroxy-4′-(2-hydroxyethoxy)-2 methylpropiophenone (Irgacure 2959), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), sulfosuccinimidyl 6-(4′-azido-2′-nitrophenylamino)hexanoate (Sulfo-SANPAH), paraformaldehyde, Triton X-100, Tween 20 and bovine serum albumin (BSA) were obtained from Sigma-Aldrich (France). Poly(ethylene glycol)diacrylate (PEGDA, M.w. 400 Da) was obtained from PolyScience (Pennsylvania, USA). Phosphate buffered saline (10×) (PBS), trypsin/EDTA (ethylenediaminetetraacetic acid), penicillin/streptomycin, fetal bovine serum (FBS), Dulbecco's modified Eagle's medium (DMEM), DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride), Alexa Fluor™ 488 phalloidin, and goat anti-mouse lgG (H + L) highly cross-adsorbed secondary antibody Alexa Fluor™ 647 were purchased from Thermo Fisher Scientific (USA). Mouse monoclonal antiosteopontin was obtained from Santa Cruz Biotechnology (USA). Mouse monoclonal antipodoplanin/E11 was obtained from Abnova (UK). Bone marrow-derived hMSCs and MSC osteogenic differentiation medium were obtained from Promocell (Heidelberg, Germany) from a single donor to restrain the number of investigated variables. CKIPKASSVPTELSAISMLYLK(FITC), KRKIPKASSVPTELSAISMLYLC and CG-K(PEG3-TAMRA)-GGRGDS peptides were synthesized by Genecust (Boynes, France). Silicon isolators were obtained from Grace Bio-Labs (Oregon, USA).2.2. Hydrogel fabrication
PEGDA hydrogels were fabricated from long (M.W. = 4000 Da) and short (M.W. = 400 Da) PEGDA chains, at concentrations of 10, 20 or 30% w/v in PBS. The naming of the sample first indicates the proportion of long to short PEGDA used (for example samples labelled 100/0 contain 100% of long PEGDA, while those identified as 0/100 contain only short PEGDA). The third number (10, 20 or 30%) indicates the concentration of total polymer in the solution. The photoinitiator Irgagure2959 was added at a concentration of 0.7 wt% and the solutions were cast in cylindrical moulds of 9 mm diameter and covered with a fluorinated ethylene propylene sheet to achieve a flat surface. The materials were polymerized under UV light (64 W, 365 nm) for 15 minutes. All hydrogels were swollen in PBS for at least 24 hours before any testing. For simplicity, in the results and discussion section, the following naming conventions are followed for the different hydrogel conditions: G1 = 100/0 10%, G2 = 100/0 20%, G3 = 0/100 20% and G4 = 0/100 30%. The compositions were chosen according to their mechanical properties based on a previous screening of multiple PEGDA compositions.272.3. Shear rheology
The mechanical properties of the samples were measured using a TA Discovery Hybrid Rheometer 10 with 8 mm cross-hatched plates. The samples were cut to size using a metallic punch. An initial compression of 0.25 N was initially applied to hold the samples in place, and then frequency sweeps from 0.1 Hz to 10 Hz were carried out at 0.5% strain. Storage modulus (G′), loss modulus (G′′) and loss tangent (tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ) are calculated for a 1 Hz frequency, according to the following formulas, where ε0 is the applied strain (0.5%), σ0 is the induced stressed as measured by the rheometer, and δ is the phase angle between the deformation and the response.
δ) are calculated for a 1 Hz frequency, according to the following formulas, where ε0 is the applied strain (0.5%), σ0 is the induced stressed as measured by the rheometer, and δ is the phase angle between the deformation and the response.At least three samples per condition were measured.
2.4. Peptide functionalization and evaluation
The hydrogels were functionalized according to a previously published protocol.10,27 A comprehensive scheme of the hydrogel functionalization has already been published elsewhere.27 Briefly, the samples were soaked in a 1 mM solution of SulfoSANPAH in HEPES buffer (50 mM, pH 8.5) and immediately exposed to UV light for 15 minutes. The solution was removed and the process was repeated to expose the samples from the other side, followed by rinsing twice with HEPES buffer. The hydrogels were then covered with a solution of either the BMP-2 peptide alone or a mixture of RGD and BMP-2 (0.3 to 0.5 mM in HEPES). To achieve the different grafting ratios for the materials containing the two peptides, the final peptide mixtures were prepared by adding different proportions of each of the peptides while the final concentration was kept constant. Two peptide cocktails, Mix1 and Mix2, were employed, aiming for 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 and 30
50 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 ratios of RGD to BMP-2, respectively. Given the large number of samples required for testing, we had to limit the number of conditions tested and focused on those that seemed to provide promising results based on previous works.24 The gels were rinsed with HEPES for 3 days under agitation, a duration previously shown in our earlier publication to be sufficient for removing non-grafted, adsorbed peptides. In this study, all materials were rinsed until a plateau in fluorescence intensity was reached, confirming the cessation of fluorescently tagged peptide release after 3 days.27 The performance of this technique regarding the success of the covalent bonding and the density and homogeneity of the grafting were equally verified in a previous study through X-ray photoelectron spectroscopy and fluorescence microscopy.27
70 ratios of RGD to BMP-2, respectively. Given the large number of samples required for testing, we had to limit the number of conditions tested and focused on those that seemed to provide promising results based on previous works.24 The gels were rinsed with HEPES for 3 days under agitation, a duration previously shown in our earlier publication to be sufficient for removing non-grafted, adsorbed peptides. In this study, all materials were rinsed until a plateau in fluorescence intensity was reached, confirming the cessation of fluorescently tagged peptide release after 3 days.27 The performance of this technique regarding the success of the covalent bonding and the density and homogeneity of the grafting were equally verified in a previous study through X-ray photoelectron spectroscopy and fluorescence microscopy.27
To evaluate the functionalization density of each peptide, samples were fluorescent peptides according to our previously established protocol:27 RGD-TAMRA and BMP-2-FITC. BMP-2-FITC contains a FITC fluorochrome linked to the C-terminal on the lysine amino acid. The RGD peptide sequence contains a three-unit ethylene glycol spacer linked to the side chain of a lysine amino acid, to which the TAMRA molecule is bound (CG-K(PEG3-TAMRA)-GGRGDS). After grafting and rinsing, fluorescence was evaluated using a Leica DM5500B epifluorescence microscope (Leica Biosystems) equipped with a CoolSnap HQ camera and controlled by Metamorph 7.6 software. Images were acquired at 2.5× magnification, with exposures of 700 ms and 300 ms for TAMRA and FITC respectively. To obtain relative calibration curves, the same microscopy parameters were used to acquire images of a series of drops of 1 μL from peptide solutions ranging from 75 to 1 μM in concentration, containing an equimolar quantity of RGD-TAMRA and BMP-2-FITC fluorescent peptides homogeneously mixed. All images were analysed using ImageJ freeware. Background fluorescence from images of non-functionalized hydrogels was subtracted from the images of the samples. The intensity per area was then correlated with the calibration curves to calculate a relative amount of peptide per area for each peptide and hydrogel condition (n = 3).
2.5. Cell culture
Functionalized hydrogels were sterilized for 5 h with ethanol 70% and subsequently rinsed thoroughly with sterile PBS. The materials were equilibrated in DMEM for 2 h before seeding. hMSCs at passage 5 were seeded at a density of 2500 cells per cm2 onto materials placed in wells of a 48-well plate and kept with only DMEM for the first 4 h of culture. Then, 10% FBS was added and the medium was replaced after 24 hours with osteogenic medium. The medium was replaced every 2 to 3 days and in each case supplemented with 1% penicillin/streptomycin. Cells adhere and proliferate on the surface of the hydrogels. While cells successfully adhered to and proliferated on the hydrogel surfaces, they did not penetrate into the material, thus characterizing this system as a 2D model. A glass surface with osteogenic medium was chosen as relevant control as it has the same culture medium as the experimental conditions, which allows to see the differential effect of the material properties. Of note, cells did not adhere on the pristine hydrogel. For this reason, this material was not considered as control. This culture time was chosen because a fast hMSC differentiation was targeted. Of note, PEGDA does not degrade for such a short exposure to aqueous media, therefore sample mechanical changes during the cell culture experiments are not expected.2.6. Immunocytochemistry analysis
Cells for immunocytochemistry analysis were cultured on hydrogels for 1 week with osteogenic medium. To evaluate cell morphology and protein expression, cells on hydrogels and controls were fixed with 4% PFA for 15 minutes at 4 °C, then permeabilized with 0.5% TritonX-100 for 5 minutes and saturated for 1 hour with 3% BSA. Afterwards, the cytoskeleton was marked by incubating with Alexa Fluor™ 488 phalloidin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 dilution) for 1 hour at 37 °C. Then, the protein of interest was targeted with the corresponding primary antibodies (anti-OPN at a dilution of 1 μg mL−1 and anti-podoplanin at a dilution of 2 μg mL−1) diluted in 1% BSA/PBS for 2 h at 37 °C in a humidified atmosphere. Subsequently, each material was incubated with the secondary antibody (goat anti-mouse lgG (H + L) highly cross-adsorbed secondary antibody Alexa Fluor™ 647), diluted at 5 μg mL−1 in 1% BSA/PBS for 1 h at 37 °C. Finally, cell nuclei were stained using DAPI at 1
400 dilution) for 1 hour at 37 °C. Then, the protein of interest was targeted with the corresponding primary antibodies (anti-OPN at a dilution of 1 μg mL−1 and anti-podoplanin at a dilution of 2 μg mL−1) diluted in 1% BSA/PBS for 2 h at 37 °C in a humidified atmosphere. Subsequently, each material was incubated with the secondary antibody (goat anti-mouse lgG (H + L) highly cross-adsorbed secondary antibody Alexa Fluor™ 647), diluted at 5 μg mL−1 in 1% BSA/PBS for 1 h at 37 °C. Finally, cell nuclei were stained using DAPI at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution. All samples were washed with PBS containing 0.05% Tween 20 between the different incubation steps.
1000 dilution. All samples were washed with PBS containing 0.05% Tween 20 between the different incubation steps.
Samples were imaged Leica DM5500B epifluorescence microscope (Leica Biosystems) equipped with a CoolSnap HQ camera and controlled by Metamorph 7.6 software, at 10× and 40× magnifications, keeping the gain and exposure time constant for all images.
Cell and nuclear morphology were evaluated on 2 samples per condition, with at least 40 cells per condition measured, by analysing DAPI and phalloidin staining on a cell-per-cell basis. The expression of osteogenic markers was evaluated by measuring the integrated density of the fluorescence on the corresponding channel and subtracting the background intensity for an equivalent area. OPN was evaluated in the nucleus only while podoplanin (PDPN) was evaluated in the whole cell. DAPI and phaolloidin staining were used to select the corresponding nuclear or cellular areas for each single cell, and the corresponding regions of interest were measured in the channels for the markers. OPN and PDPN expression was evaluated on 2 samples per condition, with at least 40 cells per condition measured. All image analyses were performed using Fiji freeware.28 Protein expression was evaluated for one experiment, cell morphology was verified on two independent experiments.
2.7. Real-time quantitative polymerase chain reaction (RT-qPCR)
Cells for gene expression analysis were cultured on hydrogels for 1 week with osteogenic medium. Each sample for qPCR consisted of 6 hydrogels and three independent experiments were performed. RNA samples were extracted and processed with the RNeasy micro kit (Qiagen). cDNA was synthesized from 0.1 μg of total RNA using Maxima Reverse Transcriptase (Fisher Scientific) and primed with oligo-dT primers (Fisher Scientific) and random primers (Fisher Scientific). QPCR was performed using a LightCycler 480 Real-Time PCR System (Roche, Meylan, France). QPCR reactions were done once for each sample, using transcript-specific primers, cDNA (4 ng), and LightCycler 480SYBR Green I Master (Roche) in a final volume of 10 μL. Primer sequences are reported in Table 1. For the determination of the reference gene, the RefFinder method was used. Relative expression analysis was normalized against two reference genes, and the peptidylprolyl isomerase A (PPIA) and TATA-box binding protein (TBP) genes were used. The relative level of expression was calculated using the comparative 2ΔΔCT method.| Gene | GeneBank ID | Forward sequence (5′–3′) | Reverse sequence (3′–5′) |
|---|---|---|---|
| PPIA | NM_021130 | CGGGTCCTGGCATCTTGT | CAGTCTTGGCAGTGCAGATGA |
| TBP | NM_003194 | GGGCATTATTTGTGCACTGAGA | GCCCAGATAGCAGCACGGT |
| COL1A1 | NM_000088 | TGGAAGAGTGGAGAGTACTGGATTG | TTGCAGAAGACTTTGATGGCAT |
| RUNX2 | NM_001015051 | TAAGGATTCCCTCAATTCCGA | ATGCTTCGTGTTTCCATGT |
| ALPL | NM_000478 | TCCTGACCCTCCCACTC | GGACAGGGACATGAGCATT |
| PDPN | NM_006474 | GCTCGGCCTCAGATTCC | AACTCATCCAGCTCTTCTCA |
2.8. Statistical analyses
Data are expressed as mean values ± standard deviation. Significance was assessed by one-way ANOVA with Tukey's correction for multiple comparisons, with GraphPad Prism 8.0.1 for Windows. Significant differences were determined for P values < 0.05 (with *, **, *** and **** representing P < 0.05, P < 0.01, P < 0.001 and P < 0.0001 respectively).3. Results
3.1. Hydrogel mechanical properties
The mechanical properties of PEGDA hydrogels with 4 different compositions were evaluated via oscillatory shear rheology. Our objective was to have hydrogels with the same elasticities but different viscoelasticities, and conditions with the same viscoelasticities but different elasticities, and the compositions were chosen based on a previous screening of multiple PEGDA compositions.27 As expected, for a given PEGDA chain length, increasing the concentration of polymer in solution results in an increase in modulus, growing from 12 to 27 kPa in the case of the long chain PEG (100/0 mixtures) and from 24 to 42 kPa in the samples composed of short PEG (0/100 mixtures) (Fig. 1(A)). It is interesting to note that the samples 100/0 20% (G2) and 0/100 20% (G3) result in a storage modulus that is not significantly different. The viscoelastic behaviour of the samples was characterized in the same rheological experiment and reported here as the loss tangent, which is the ratio between the loss and storage moduli and provides a relative indication of the dissipative behaviour with respect to the elastic portion (Fig. 1(B)). The aforementioned samples with equivalent storage modulus (G2 and G3, Fig. 1(A)), due to their different composition in terms of polymer chain length, present different viscoelasticity. The sample G2 has a loss tangent of 0.2 while G3 has an average loss tangent of 0.33. Additionally, the sample G1 has a loss tangent of 0.16, which is not significantly different from the one of G2, while the stiffest sample (G4) has a loss tangent of 0.35 which is equivalent to that of the condition G3. These 4 materials comprise several pairs in which only one property, either the elastic modulus or the viscoelasticity, is changed without affecting the other, which makes these PEGDA hydrogels an interesting platform for mechanobiology studies.3.2. Biofunctionalization of hydrogels
PEGDA hydrogels are not cell adhesive and require functionalization with bioactive molecules to allow cell attachment and proliferation.29 Considering that the aim of this work was to study osteogenic differentiation of MSCs, the PEGDA hydrogels were covalently grafted with an RGD peptide and a BMP-2 mimetic peptide. BMP-2 is a growth factor involved in bone formation and is known to be a strong osteoinductive factor in vitro.30 In this work, an RGD peptide with a three-unit PEG spacer was employed to extend the peptide chain, increasing the distance between the cell-binding site and the material surface.31 This adjustment also made the RGD peptide more comparable in size to the osteogenic peptide. For the BMP-2-mimetic sequence, a peptide derived from the knuckle epitope of the BMP-2 protein was used.32 The materials were functionalized with either BMP-2 alone or a mixture of both peptides. Of note, peptides carrying a fluorophore were employed evaluate the extent of grafting. Evaluating the peptide density in functionalized biomaterials is a challenging task. Here, using fluorescently labeled peptides, we were able to obtain an optical measurement of the number of peptides present in the material. A grafting density can be measured for each peptide and hydrogel condition (Fig. 1(C)) by preparing standard calibration curves that relate to a known amount of peptide molecules with the fluorescence intensity that is emitted (Fig. 1(D) and (E)). It is not straightforward to ascertain that the calculated numbers correspond to the actual number of peptides present on the material surface, but they do provide a relative measurement which allows to compare the ratio of the two peptides among the different conditions. For this reason, the results are discussed in terms of peptide units rather than pmol mm2.In materials functionalized with the combination of RGD and BMP-2, two peptide cocktails were employed, Mix1 and Mix2, which aimed to result in a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 and a 30
50 and a 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 ratio of RGD to BMP-2 respectively. As observed in Fig. 1(C), these ratios were well achieved for G1, G2 and G3, but not for G4. For Mix1, the fractions of RGD and BMP-2 with respect to the total amount of peptide per sample are 47
70 ratio of RGD to BMP-2 respectively. As observed in Fig. 1(C), these ratios were well achieved for G1, G2 and G3, but not for G4. For Mix1, the fractions of RGD and BMP-2 with respect to the total amount of peptide per sample are 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 53, 45
53, 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55, 54
55, 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 and 74
45 and 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 26, for G1, G2, G3 and G4 respectively. In the case of Mix2, the ratios are 30
26, for G1, G2, G3 and G4 respectively. In the case of Mix2, the ratios are 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70, 26
70, 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 74, 38
74, 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 and 68
61 and 68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31. In terms of the total amount of peptide, these vary depending on the hydrogel condition from 2 to 4.5 peptide units per area.
31. In terms of the total amount of peptide, these vary depending on the hydrogel condition from 2 to 4.5 peptide units per area.
3.3. Immunocytochemistry
The immunocytochemistry results presented in Fig. 2, show that the differentiation of MSCs into the osteogenic lineage is strongly influenced by both the mechanical properties of the matrix and its biochemical cues. In this study, cells were cultured on four different hydrogel materials with varying mechanical properties (G1–G4) and functionalized with three distinct peptide combinations (BMP-2, Mix1, and Mix2). The results in Fig. 2 illustrate the morphological analysis and expression levels of two osteogenic markers -OPN, a late osteoblast marker, and PDPN, an early osteocyte marker-, based on immunocytochemistry staining of cells cultured for one week on the materials. Representative images of stained cells with each of the two markers, for G1 to G4 functionalized with Mix1, are presented in the ESI† (Fig. S1 and S2). | ||
| Fig. 2 Analysis of immunocytochemistry staining after 1 week of MSCs cultured on hydrogels (samples per condition = 2; cells per condition > 40). (A) Total cell area. (B) Nuclear area. (C) Class distribution of OPN intensity. X-axis labels represent the bin center in A.U., with alternating labels omitted for improved readability. Statistical analyses are presented in the ESI† (Tables S1–S4). (D) and (E) Representative immunofluorescence images of cells on the different hydrogels after 1 week of culture. Staining of F-actin by phalloidin (green) and nuclei by DAPI (blue). (D) Magnification 10×, scale bar = 100 μm. (E) Magnification 40×, scale bar = 50 μm. | ||
Interestingly, the scatter plot presentation of the cell area (Fig. 2(A)) shows a narrower distribution, with overall smaller cell sizes, in the G1-BMP, G1-Mix 1, G1-Mix 2, G2-BMP, G3-BMP, and G4-BMP conditions (see statistical analyses in Table S1, ESI†). It is important to note that we are observing the cell area data at a 1-week timepoint and it is expected that not all cells are at the same stage of differentiation. This relatively short timempoint was chosen to evidence rapid osteogenic commitment. The distribution of the data points is wider for the G2, G3, and G4 hydrogels, with an even broader distribution observed for the Mix1 and Mix2 gels. The morphological data reveals that the cell spread area increases in materials with medium (G2 and G3 – 26 kPa) and high (G4 – 42 kPa) storage modulus, compared to the softer materials (G1 – 12 kPa). Notably, for conditions from G2 to G4, the increase in cell area is more pronounced in the Mix1 and Mix2 conditions compared to the BMP conditions. However, for the G1 hydrogels, no significant change in cell area is observed regardless of the functionalization (Table S1, ESI†). This suggests that for a soft hydrogel (G1), none of the functionalizations are sufficient to induce osteoblastic differentiation. In hydrogels with a higher storage modulus (G2–G4), a combination of RGD and BMP-2 peptides is necessary for cells to properly form mature focal adhesions and spread. The average cell area on hydrogels G2-Mix1 and G2-Mix2 is higher than in their G3-Mix1 and G3-Mix2 counterparts, despite G3 having a stiffness equivalent to G2 but greater viscoelasticity. The difference between G2-Mix1 and G3-Mix1 is statistically significant, while the one between G2-Mix2 and G3-Mix2 is not (see Table S1, ESI†). G2-Mix1 and G2-Mix2 exhibited the highest nuclear areas, whereas G2-BMP, along with all G3 and G4 conditions, showed comparable intermediate values. All G1 conditions displayed lower nuclear areas. Qualitative observations of cell morphology (Fig. 2(D) and (E)) indicate that cells have acquired large polygonal shapes characteristic of osteoblasts33 on G2, G3 and G4 surfaces functionalized with Mix1 and Mix2. Additionally, in some regions of G3 and G4 hydrogels, cells appear smaller with dendritic extensions, which correlates with the reduced cell spread area measured and may suggest a progression in the differentiation process from osteoblasts to osteocytes.34
To investigate cell differentiation towards the osteogenic lineage, the expression of the osteoblastic marker osteopontin (OPN) was evaluated by immunocytochemistry across all hydrogel conditions (Fig. 2(C) and Fig. S3, ESI†). Although the cell spread area and nuclear area were larger in the G2 condition, the highest expression of OPN was observed in the G3-Mix1 condition (significance P < 0.05, see Table S3), ESI.† The large error bars reflect a broad distribution of values (Fig. S3, ESI†), which is likely due to the different stages of cell development on the materials at this time point, as mentioned earlier. Fig. 2(C) shows the class distribution of OPN protein expression for samples G1 to G4 with the three different functionalizations, which provides a clearer comparison of the cells among surfaces with different mechanical properties but the same functionalization or vice versa.
In all hydrogel conditions functionalized with BMP most cells show an OPN intensity concentrated between 0.2 and 1 A.U., with only the sample G3-BMP having a considerable proportion of cells (20%) between 1.2 and 2.2 A.U. All gels from G1 to G4 functionalized with Mix 1 show a high proportion of cells with OPN intensities between 0.4 and 1 A.U. For the G2-Mix1 and G4-Mix1 conditions, we observe that some cells exhibit OPN overexpression between 1 and 2.5 A.U. Interestingly, for the G3-Mix1 condition, the overexpression is more marked and the average expression of OPN in this condition is significantly higher than that of G2-Mix1, which has the same storage modulus but a lower loss tangent (Fig. 1 and Table S3, ESI†). A similar pattern is observed in all hydrogels functionalized with Mix2, with markedly higher expression of OPN for the sample G3-Mix2, although the condition G3-Mix1 remains the one with the highest expression overall. The expression of the osteocyte related protein PDPN was also evaluated and no significant overexpression was detected in any of the samples (Fig. S3, ESI†).
3.4. Gene expression
From the morphological and protein expression analyses, hydrogel conditions G2 and G3 emerged as particularly favourable for osteogenesis. To further verify these results and uncover more subtle differences between these samples, gene expression analysis was conducted using qPCR on cells cultured on G2 and G3 hydrogels functionalized with Mix1 (Fig. 3). Interestingly, the qPCR results revealed no significant differences in the expression of early osteogenic markers COL1A1, RUNX2 and ALPL across the different hydrogel conditions and the control (cells on glass with osteogenic medium). This indicates that all conditions provide sufficient cues for the initial determination of osteogenic fate after only 1 week of culture. The most striking result was the significant increase in the expression of the podoplanin gene in the G3-Mix1 condition. This is a remarkable finding, indicating that the G3-Mix1 hydrogel condition not only supports osteogenic differentiation but also accelerates the expression of an osteocyte marker after just one week of culture.4. Discussion
Signals from the extracellular environment, including both biochemical and mechanical factors, are widely recognized for their influence on stem cell fate.5,35,36 However, very few studies have explored the interplay between the presence and organization of biomolecules with the mechanical properties in terms of elasticity and viscoelasticity. Very complex relationships govern cell fate and elucidating the roles of each stimulus, and the potential synergistic or antagonistic effects is a challenging task. An interesting approach to exploring these links is to fabricate biomaterials in which one or more properties of interest can be modified independently.While natural hydrogels, such as those made from collagen or alginate, are useful for tissue engineering thanks to their biological properties, batch-to-batch variation and weaker mechanical properties limit their applications.37,38 As an alternative, hydrogels fabricated from synthetic polymers generally offer better control of their chemical and physical properties but lack inherent cell-interactive domains, rendering them bioinert.39 Among synthetic polymers, PEGDA-based hydrogels are great candidates for their use in biomedical applications thanks to their non-cytotoxicity, widely tunable properties and polymerization conditions that are compatible with biofabrication techniques.40,41 In this work, PEGDA hydrogels were fabricated by crosslinking the acrylate groups at the end of the PEGDA chains via photopolymerization. In an ideal network, the acrylate groups at the chain ends become point-like junctions which uniformly link PEG chains. In this model, increasing the polymer concentration in the pre-hydrogel solution results in a denser network which in turn increases the material's elastic modulus, while increasing the molecular weight of the initial PEGDA chains increases the space between crosslinks which causes a decrease in stiffness. However, several studies have shown that photopolymerized PEGDA networks consist of complex structures that deviate from the “ideal” behaviour.40,42 Based on a statistical mechanics model, Levin et al.40 describe PEGDA networks as consisting of stiff polyacrylate (PA) rods that interconnect PEG chains (Fig. 4). In this model, increasing the polymer concentration results in longer PA rods, and it is the relative lengths of the PA rods with respect to the PEG chains 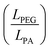 that govern the overall behaviour of the PEGDA hydrogels. LPEG can be estimated by knowing the number of repeat monomers in each PEGDA chain (6 and 88 for 400 and 4000 Da PEGDA, respectively) and the length of one repeat unit (0.3 nm) reported in literature.43 Based on this calculation and the estimations of the length of PA rods calculated for similar hydrogels by Levin et al.,40 we can conclude about the conformation of the hydrogels discussed in this publication. For the conditions G1 and G2, the PEG interconnecting chains are longer than the PA rods
that govern the overall behaviour of the PEGDA hydrogels. LPEG can be estimated by knowing the number of repeat monomers in each PEGDA chain (6 and 88 for 400 and 4000 Da PEGDA, respectively) and the length of one repeat unit (0.3 nm) reported in literature.43 Based on this calculation and the estimations of the length of PA rods calculated for similar hydrogels by Levin et al.,40 we can conclude about the conformation of the hydrogels discussed in this publication. For the conditions G1 and G2, the PEG interconnecting chains are longer than the PA rods  , which means that the response of the hydrogels under deformation is mainly the result of the elastic deformation of the PEG chains (Fig. 4(B), top). Increasing the polymer concentration from 10 to 20% w/v results in the lengthening of the PA rods, which makes the
, which means that the response of the hydrogels under deformation is mainly the result of the elastic deformation of the PEG chains (Fig. 4(B), top). Increasing the polymer concentration from 10 to 20% w/v results in the lengthening of the PA rods, which makes the  ratio closer to 1 in the case of G2, resulting in stiffening of the network. This is in agreement with the calculated storage modulus of these two samples (Fig. 1). On the contrary, the samples with G3 and G4 compositions have short PEG chains compared to the PA rods
ratio closer to 1 in the case of G2, resulting in stiffening of the network. This is in agreement with the calculated storage modulus of these two samples (Fig. 1). On the contrary, the samples with G3 and G4 compositions have short PEG chains compared to the PA rods  (Fig. 4(B), bottom). In this case, the mechanical interactions between PA rods are more frequent and this results in a stiffening of the network, which is again supported by the rheology results measured for these samples (Fig. 1). Similarly to the conclusions about the elastic behaviour of PEGDA hydrogels demonstrated by Levin et al.,40 we propose to use this model to explain their viscoelasticity. Among the samples considered in this study, the loss tangent of the two samples composed of short PEG chains (G3 and G4) is the highest. We hypothesize that this viscoelastic behaviour can be caused by a higher proportion of dangling chain ends from residual acrylate groups44 and the contribution of physical chain entanglements, hydrogen bonding,45 and interactions between PA rods.40 These values of loss tangent, between 0.15 and 0.35, are within the range of those measured for natural tissues.46
(Fig. 4(B), bottom). In this case, the mechanical interactions between PA rods are more frequent and this results in a stiffening of the network, which is again supported by the rheology results measured for these samples (Fig. 1). Similarly to the conclusions about the elastic behaviour of PEGDA hydrogels demonstrated by Levin et al.,40 we propose to use this model to explain their viscoelasticity. Among the samples considered in this study, the loss tangent of the two samples composed of short PEG chains (G3 and G4) is the highest. We hypothesize that this viscoelastic behaviour can be caused by a higher proportion of dangling chain ends from residual acrylate groups44 and the contribution of physical chain entanglements, hydrogen bonding,45 and interactions between PA rods.40 These values of loss tangent, between 0.15 and 0.35, are within the range of those measured for natural tissues.46
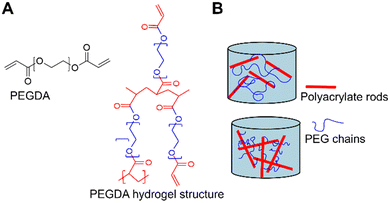 | ||
Fig. 4 Representative schematic of PEGDA hydrogel structure adapted from Levin et al.40 (A) PEGDA oligomer structure and representation of the molecular organization with a polyacrylic backbone and connected PEGDA chains that, in turn, bind to other polyacrylate rods. (B) Schematic representation of two hydrogel compositions where  (top) and (top) and 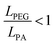 (bottom). (bottom). | ||
This study investigated not only the effects of mechanical properties but also the distinct bioactivities conferred by various peptide surface functionalizations on stem cell differentiation. A previous study demonstrated the feasibility of covalently grafting bioactive peptides to PEGDA hydrogels via the heterobifunctional crosslinker SulfoSANPAH.27 In this work, a step further was made by functionalizing the hydrogels with various peptides: BMP-2 alone or combinations of RGD and BMP-2 in different ratios. Previous works have shown the synergistic effects of RGD and BMP-2 towards promoting osteogenic differentiation.18,47 In a recent publication by Zhang et al.24 it was demonstrated that a 2D material functionalized with 23% RGD and 77% BMP-2 surface coverage promoted the expression of osteogenic markers as compared to surfaces with other ratios. In another study, Ma et al.21 prepared surfaces with orthogonal gradients of the two peptides and found that MSCs experience faster differentiation towards osteoblasts on the surfaces with the highest concentration of both peptides combined, which corresponded to a RGD to BMP-2 ratio of 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 ratio approximately. These discrepancies from one study to the other may arise from the different peptide densities used in each case, as well as the varying time points and culture conditions considered. In the present work, we chose to compare PEGDA functionalized solely with BMP-2 mimetic peptides to PEGDA grafted with 50
25 ratio approximately. These discrepancies from one study to the other may arise from the different peptide densities used in each case, as well as the varying time points and culture conditions considered. In the present work, we chose to compare PEGDA functionalized solely with BMP-2 mimetic peptides to PEGDA grafted with 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 and 30
50 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 RGD:BMP-2 ratios, using peptide densities similar to those demonstrated in previous studies to promote cell differentiation.10,27 These ratios were successfully achieved, as observed with fluorescence microscopy, on samples G1, G2 and G3, while condition G4 always presented a higher content of RGD than BMP-2. Our hypothesis is that the tighter polymer network of the hydrogel G4 prevents the entry of the larger BMP-2 molecules in the bulk of the material, while the smaller RGD peptides can still diffuse inside the hydrogel. The total peptide contents presented slight variations among the different hydrogel conditions, with between 2 and 4.5 peptide units per unit area. These results underscore the inherent challenges in both grafting and accurately quantifying peptides on hydrogel substrates. While many studies often assume that the peptides introduced in the functionalization solution are fully grafted onto the material, the present findings suggest that this assumption may not always be valid, highlighting the need for careful verification in similar experimental setups. An interesting approach to address this, which resolves at least the issue of achieving a constant ratio of two peptides on the biomaterial, is the use of multifunctional peptides containing both sequences of interest within the same peptide backbone.48,49
70 RGD:BMP-2 ratios, using peptide densities similar to those demonstrated in previous studies to promote cell differentiation.10,27 These ratios were successfully achieved, as observed with fluorescence microscopy, on samples G1, G2 and G3, while condition G4 always presented a higher content of RGD than BMP-2. Our hypothesis is that the tighter polymer network of the hydrogel G4 prevents the entry of the larger BMP-2 molecules in the bulk of the material, while the smaller RGD peptides can still diffuse inside the hydrogel. The total peptide contents presented slight variations among the different hydrogel conditions, with between 2 and 4.5 peptide units per unit area. These results underscore the inherent challenges in both grafting and accurately quantifying peptides on hydrogel substrates. While many studies often assume that the peptides introduced in the functionalization solution are fully grafted onto the material, the present findings suggest that this assumption may not always be valid, highlighting the need for careful verification in similar experimental setups. An interesting approach to address this, which resolves at least the issue of achieving a constant ratio of two peptides on the biomaterial, is the use of multifunctional peptides containing both sequences of interest within the same peptide backbone.48,49
The results of this work evidence the synergistic effects of RGD and BMP-2 co-functionalization as well as the effect of mechanical properties on cell differentiation. Fig. 2 shows that cells were unable to fully spread on hydrogels functionalized with BMP-2 alone, regardless of the mechanical properties of the hydrogel. As osteogenic differentiation progresses, cells evolve from an elongated spindle shape to a larger cuboidal morphology, which correlates with an increase in area.50,51 As osteoblasts terminally differentiate into osteocytes, cytoplasmic processes appear conferring the cells a characteristic dendritic shape and the size of the cell body is reduced.52 In this work, cells were the largest on samples G2-Mix1 and G2-Mix2, where they have adopted a polygonal shape. In samples G3-Mix1, G3-Mix2, G4-Mix1 and G4-Mix2, cells are spread but the average area is lower and, in some areas, dendritic processes are clearly visible. There is also an apparent increase in the area of nuclei for cells on samples G2, G3 and G4, as compared to that observed on G1. Overall, the largest nuclei are those on samples G2-Mix1 and G2-Mix2. An increase in nucleus area has been correlated with chromatin organization and protein translocation to the nucleus during osteogenic differentiation.53,54 Regarding the two peptide mixtures, the results do not provide definitive evidence favouring one over the other.
Overall, the highest expression of OPN is found for sample G3, which has a storage modulus of 24 kPa and a loss tangent of 0.33. Keeping in mind that G2 samples have the same storage modulus but lower viscoelasticity (loss tangent of 0.2), these results indicate that higher viscoelasticity on two samples with the same elastic modulus further enhances osteogenic differentiation.
As for COL1A1, RUNX2, and ALPL genes, no overexpression was observed in any of the three tested conditions (positive control, G2-Mix1, and G3-Mix1) (Fig. 3). Nevertheless, a positive point is that these genes were expressed at levels comparable to the positive control commonly reported in the literature (i.e., glass substrate with osteogenic medium). This suggests that our hydrogels are suitable platforms for maintaining osteogenic gene expression.
Although no significant differences in PDPN protein expressions were observed between the hydrogel conditions and the positive control (OM), RT-qPCR analysis revealed a marked overexpression of the PDPN gene in the G3-Mix1 sample compared to G2-Mix1 and the positive control (OM) after only one week of culture. This observation aligns with the general understanding that changes in gene expression typically precede alterations at the protein level, due to the time required for mRNA translation and protein maturation. Therefore, it is consistent to detect both PDPN gene overexpression and OPN protein expression, as these results together suggest the onset of osteogenic differentiation. In particular, the expression of PDPN—a recognized early marker of osteocyte differentiation—on the G3-Mix1 hydrogel after just one week is a highly promising outcome. This reinforces the conclusion that viscoelasticity is a driver of osteogenesis. Few publications have addressed the role of viscoelasticity in osteodifferentiation.10,55,56 A study by Chaudhuri et al.13 found that, on hydrogels with a storage modulus of 17 kPa, faster stress-relaxation (indicative of viscoelasticity) improved various osteogenic markers. However, ideal values of viscoelasticity remain unclear, as some authors have reported conflicting results. For instance, Walker et al.57 measure the highest expression of the early osteogenic marker Runx2 on the matrices with the lowest viscoelasticity (loss tangent of 0.12), as compared to those with higher loss tangent values (0.16 and 0.24). It is important to note that in their study, which focused on achieving chondrogenic differentiation, the hydrogels had a relatively low storage modulus of 4 kPa, which could explain the discrepancy in results.
This study underscores that both elasticity and viscoelasticity are key regulators of cell fate. Notably, high viscoelasticity was favourable for osteogenesis in hydrogels with a 24 kPa storage modulus (G3) but not in stiffer ones (G4, 42 kPa). This conclusion is in agreement with other previously published results.7 A publication by Gong et al.,58 points out along the same lines that viscoelasticity favors cell spread in soft substrates, but does not have an effect on stiff ones. Our findings confirm that the mechanical properties of these PEGDA hydrogels primarily drive MSC differentiation into an osteoblastic lineage. In our system, biofunctionalization alone was insufficient to induce differentiation in softer substrates, highlighting the importance of mechanical context. Furthermore, the data indicate that once the mechanical properties are optimized to promote MSC differentiation toward an osteoblastic lineage, the two considered ratios of adhesion peptides (RGD) to osteogenic differentiation peptides (BMP-2) do not promote significant differences in MSC differentiation. The characterized parameters do not give a strict yes-or-no answer but rather provide values on a scale that comparatively indicate the extent of differentiation. The observed variability, reflected in large error bars, suggests that not all cells are at the same differentiation stage, which is to be expected as they are measured after only one week of culture. Despite this heterogeneity, it is noteworthy that our results show MSC commitment to an osteoblastic lineage after a very short period of one week.
Despite these advances, certain experimental limitations must be acknowledged. Variability in peptide distribution across hydrogel samples may lead to heterogeneous cellular exposure to bioactive cues, potentially influencing differentiation outcomes. To mitigate this, the mechanical properties of PEGDA hydrogels were controlled and their bioactivity quantified—an aspect often overlooked in similar studies. Additionally, donor-to-donor variability in MSC behavior can introduce inconsistencies, as age, sex, and genetic background influence cellular responses. To minimize this source of variability, we used cells derived from a single donor, ensuring a controlled experimental framework. Of course, using cell from a single donor also raise the question of potential hMSC behavior related to donor source, age, and passage number. Finally, the considerable number of samples that had to be synthesized, functionalized, and analyzed to carry out all the assays required for this study prevented us to perform an exhaustive set of biological characterization. For example, Alizarin Red staining for calcium would have help to differentiate between early commitment and full differentiation. However, this issue was circumvented by quantifying genes and proteins expressed at various stage of differentiation.
These findings build upon previous research exploring the role of viscoelasticity in biomaterial design. For instance, Prouvé et al.10 investigated BMP-2-functionalized polyacrylamide hydrogels with tunable viscoelasticity for osteogenic differentiation of MSCs. Viscoelasticity has also been studied in cartilage tissue engineering, as shown by Walker et al.,57 who examined the effect of the viscous modulus on RGD-functionalized hydrogels. One way that viscoelasticity plays a role, particularly in 3D environments, is by enabling cell-mediated remodeling of the matrix, which is essential for processes such as migration and differentiation. In addition to viscoelasticity, researchers also incorporate cell-degradable domains such as the VPM peptide sequence to allow cell remodeling.57 The present study expands on this body of work by specifically focusing on short-term osteoblastic differentiation and incorporating varying bioactivity to evaluate their synergistic effects. There are very few publications exploring the synergy between mechanics and bioactivity on stem cell differentiation. A recent study by Blackford et al.59 demonstrated how substrate stiffness and RGD density cooperate to regulate activity of human pluripotent stem cell-derived hepatocytes. In our work, by integrating mechanical and biochemical factors in a controlled and quantifiable manner, we contributed to the development of biomaterials tailored for bone tissue engineering applications.
This study highlights the unique interplay between mechanical properties and bioactive functionalization in directing MSC differentiation toward the osteogenic lineage and aligns with mechanisms previously described in the literature.25,26 The stiffness, viscoelasticity, and topography of the extracellular environment provide crucial mechanical cues that influence lineage commitment. At the same time, biochemical signals such as growth factors, cytokines, and extracellular matrix composition activate specific intracellular pathways, guiding cell fate. While the synergy between these mechanical and biochemical factors is fundamental for tissue engineering and regenerative medicine, it remains an underexplored aspect in biomaterial design. Our approach of systematically varying both mechanical properties and bioactive peptide presentation addresses this gap, offering a more comprehensive understanding of how these factors interact to regulate osteogenic differentiation.
5. Conclusion
We presented here PEGDA hydrogels suitable for 2D cell culture, in which elasticity and viscoelasticity have been modified independently, within a biologically relevant range. This constitutes a significant achievement, as PEGDA hydrogels were successfully created to maintain consistent elasticity while varying their viscoelasticity, addressing the growing need for materials with these properties. This study also presented surfaces that have been functionalized with different peptide mixtures, which are crucial to fabricating biomimetic microenvironments that optimize osteogenic differentiation in vitro.These findings demonstrated that both elasticity and viscoelasticity play crucial roles in hMSC behaviour and differentiation, proving that cells respond to both types of mechanical properties. This underscores the importance of considering viscoelasticity, alongside elasticity, in the design of hydrogel-based platforms for tissue engineering. The ability to independently control these properties opens new avenues for creating better tissue-mimicking materials, thereby advancing the ability to direct stem cell differentiation and tissue regeneration.
Data availability
The data supporting this article have been included as part of the ESI.† Further experimental data are available upon request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the University of Bordeaux [Cristina Lopez-Serrano's PhD grant] and the Natural Sciences and Engineering Research Council of Canada (NSERC) (GL), as well as the Centre Québécois sur les Matériaux Fonctionnels (CQMF) (GL). The financial support of the ANR (ANR-21-CE06-0031-02 and ANR-20-SFRI-0001) (M. C. D.) is also acknowledged.References
- O. Levy, R. Kuai, E. M. J. Siren, D. Bhere, Y. Milton, N. Nissar, M. De Biasio, M. Heinelt, B. Reeve, R. Abdi, M. Alturki, M. Fallatah, A. Almalik, A. H. Alhasan, K. Shah and J. M. Karp, Shattering Barriers toward Clinically Meaningful MSC Therapies, Sci. Adv., 2020, 6(30), eaba6884, DOI:10.1126/sciadv.aba6884.
- J. Galipeau and L. Sensébé, Mesenchymal Stromal Cells: Clinical Challenges and Therapeutic Opportunities, Cell Stem Cell, 2018, 22(6), 824–833, DOI:10.1016/j.stem.2018.05.004.
- A. C. Bowles-Welch, A. C. Jimenez, H. Y. Stevens, D. A. Frey Rubio, L. E. Kippner, C. Yeago and K. Roy, Mesenchymal Stromal Cells for Bone Trauma, Defects, and Disease: Considerations for Manufacturing, Clinical Translation, and Effective Treatments, Bone Rep., 2023, 18, 101656, DOI:10.1016/j.bonr.2023.101656.
- L. Sensebé, M. Krampera, H. Schrezenmeier, P. Bourin and R. Giordano, Mesenchymal Stem Cells for Clinical Application, Vox Sang., 2010, 98(2), 93–107, DOI:10.1111/j.1423-0410.2009.01227.x.
- R. C. Bretherton and C. A. DeForest, The Art of Engineering Biomimetic Cellular Microenvironments, ACS Biomater. Sci. Eng., 2021, 7(9), 3997–4008, DOI:10.1021/acsbiomaterials.0c01549.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Matrix Elasticity Directs Stem Cell Lineage Specification, Cell, 2006, 126(4), 677–689, DOI:10.1016/j.cell.2006.06.044.
- N. Huebsch, P. R. Arany, A. S. Mao, D. Shvartsman, O. A. Ali, S. A. Bencherif, J. Rivera-Feliciano and D. J. Mooney, Harnessing Traction-Mediated Manipulation of the Cell/Matrix Interface to Control Stem-Cell Fate, Nat. Mater., 2010, 9(6), 518–526, DOI:10.1038/nmat2732.
- O. F. Zouani, J. Kalisky, E. Ibarboure and M.-C. Durrieu, Effect of BMP-2 from Matrices of Different Stiffnesses for the Modulation of Stem Cell Fate, Biomaterials, 2013, 34(9), 2157–2166, DOI:10.1016/j.biomaterials.2012.12.007.
- R. Olivares-Navarrete, E. M. Lee, K. Smith, S. L. Hyzy, M. Doroudi, J. K. Williams, K. Gall, B. D. Boyan and Z. Schwartz, Substrate Stiffness Controls Osteoblastic and Chondrocytic Differentiation of Mesenchymal Stem Cells without Exogenous Stimuli, PLoS One, 2017, 12(1), e0170312, DOI:10.1371/journal.pone.0170312.
- E. Prouvé, M. Rémy, C. Feuillie, M. Molinari, P. Chevallier, B. Drouin, G. Laroche and M.-C. Durrieu, Interplay of Matrix Stiffness and Stress Relaxation in Directing Osteogenic Differentiation of Mesenchymal Stem Cells, Biomater. Sci., 2022, 10(17), 4978–4996, 10.1039/D2BM00485B.
- A. Elosegui-Artola, The Extracellular Matrix Viscoelasticity as a Regulator of Cell and Tissue Dynamics, Curr. Opin. Cell Biol., 2021, 72, 10–18, DOI:10.1016/j.ceb.2021.04.002.
- A. R. Cameron, J. E. Frith and J. J. Cooper-White, The Influence of Substrate Creep on Mesenchymal Stem Cell Behaviour and Phenotype, Biomaterials, 2011, 32(26), 5979–5993, DOI:10.1016/j.biomaterials.2011.04.003.
- O. Chaudhuri, L. Gu, D. Klumpers, M. Darnell, S. A. Bencherif, J. C. Weaver, N. Huebsch, H. Lee, E. Lippens, G. N. Duda and D. J. Mooney, Hydrogels with Tunable Stress Relaxation Regulate Stem Cell Fate and Activity, Nat. Mater., 2016, 15(3), 326–336, DOI:10.1038/nmat4489.
- S. Nam, R. Stowers, J. Lou, Y. Xia and O. Chaudhuri, Varying PEG Density to Control Stress Relaxation in Alginate-PEG Hydrogels for 3D Cell Culture Studies, Biomaterials, 2019, 200, 15–24, DOI:10.1016/j.biomaterials.2019.02.004.
- A. N. Borelli, M. W. Young, B. E. Kirkpatrick, M. W. Jaeschke, S. Mellett, S. Porter, M. R. Blatchley, V. V. Rao, B. V. Sridhar and K. S. Anseth, Stress Relaxation and Composition of Hydrazone-Crosslinked Hybrid Biopolymer-Synthetic Hydrogels Determine Spreading and Secretory Properties of MSCs, Adv. Healthcare Mater., 2022, 11(14), 2200393, DOI:10.1002/adhm.202200393.
- F. R. Maia, S. J. Bidarra, P. L. Granja and C. C. Barrias, Functionalization of Biomaterials with Small Osteoinductive Moieties, Acta Biomater., 2013, 9(11), 8773–8789, DOI:10.1016/j.actbio.2013.08.004.
- C. Chollet, C. Chanseau, M. Remy, A. Guignandon, R. Bareille, C. Labrugère, L. Bordenave and M.-C. Durrieu, The Effect of RGD Density on Osteoblast and Endothelial Cell Behavior on RGD-Grafted Polyethylene Terephthalate Surfaces, Biomaterials, 2009, 30(5), 711–720, DOI:10.1016/j.biomaterials.2008.10.033.
- I. Bilem, P. Chevallier, L. Plawinski, E. D. Sone, M. C. Durrieu and G. Laroche, RGD and BMP-2 Mimetic Peptide Crosstalk Enhances Osteogenic Commitment of Human Bone Marrow Stem Cells, Acta Biomater., 2016, 36, 132–142, DOI:10.1016/j.actbio.2016.03.032.
- O. F. Zouani, C. Chollet, B. Guillotin and M.-C. Durrieu, Differentiation of Pre-Osteoblast Cells on Poly(Ethylene Terephthalate) Grafted with RGD and/or BMPs Mimetic Peptides, Biomaterials, 2010, 31(32), 8245–8253, DOI:10.1016/j.biomaterials.2010.07.042.
- N. M. Moore, N. J. Lin, N. D. Gallant and M. L. Becker, Synergistic Enhancement of Human Bone Marrow Stromal Cell Proliferation and Osteogenic Differentiation on BMP-2-Derived and RGD Peptide Concentration Gradients, Acta Biomater., 2011, 7(5), 2091–2100, DOI:10.1016/j.actbio.2011.01.019.
- Y. Ma, G. M. Policastro, Q. Li, J. Zheng, R. Jacquet, W. J. Landis and M. L. Becker, Concentration-Dependent hMSC Differentiation on Orthogonal Concentration Gradients of GRGDS and BMP-2 Peptides, Biomacromolecules, 2016, 17(4), 1486–1495, DOI:10.1021/acs.biomac.6b00088.
- I. Bilem, P. Chevallier, L. Plawinski, E. D. Sone, M.-C. Durrieu and G. Laroche, Interplay of Geometric Cues and RGD/BMP-2 Crosstalk in Directing Stem Cell Fate, ACS Biomater. Sci. Eng., 2017, 3(10), 2514–2523, DOI:10.1021/acsbiomaterials.7b00279.
- L. Padiolleau, C. Chanseau, S. Durrieu, C. Ayela, G. Laroche and M. Durrieu, Directing hMSCs Fate through Geometrical Cues and Mimetics Peptides, J. Biomed. Mater. Res., 2020, 108(2), 201–211, DOI:10.1002/jbm.a.36804.
- Y. Zhang, M. Remy, T. Leste-Lasserre and M.-C. Durrieu, Manipulating Stem Cell Fate with Disordered Bioactive Cues on Surfaces: The Role of Bioactive Ligand Selection, ACS Appl. Mater. Interfaces, 2024, 16(15), 18474–18489, DOI:10.1021/acsami.4c00262.
- Q. Wei, A. Holle, J. Li, F. Posa, F. Biagioni, O. Croci, A. S. Benk, J. Young, F. Noureddine, J. Deng, M. Zhang, G. J. Inman, J. P. Spatz, S. Campaner and E. A. Cavalcanti-Adam, BMP-2 Signaling and Mechanotransduction Synergize to Drive Osteogenic Differentiation via YAP/TAZ, Adv. Sci., 2020, 7(15), 1902931, DOI:10.1002/advs.201902931.
- S. Vermeulen, Z. Tahmasebi Birgani and P. Habibovic, Biomaterial-Induced Pathway Modulation for Bone Regeneration, Biomaterials, 2022, 283, 121431, DOI:10.1016/j.biomaterials.2022.121431.
- C. López-Serrano, Y. Côté-Paradis, B. Habenstein, A. Loquet, C. Le Coz, J. Ruel, G. Laroche and M.-C. Durrieu, Integrating Mechanics and Bioactivity: A Detailed Assessment of Elasticity and Viscoelasticity at Different Scales in 2D Biofunctionalized PEGDA Hydrogels for Targeted Bone Regeneration, ACS Appl. Mater. Interfaces, 2024, 16(30), 39165–39180, DOI:10.1021/acsami.4c10755.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Fiji: An Open-Source Platform for Biological-Image Analysis, Nat. Methods, 2012, 9(7), 676–682, DOI:10.1038/nmeth.2019.
- D. L. Hern and J. A. Hubbell, Incorporation of Adhesion Peptides into Nonadhesive Hydrogels Useful for Tissue Resurfacing, J. Biomed. Mater. Res., 1998, 39(2), 266–276, DOI:10.1002/(SICI)1097-4636(199802)39:2<266::AID-JBM14>3.0.CO;2-B.
- E. Migliorini, A. Valat, C. Picart and E. A. Cavalcanti-Adam, Tuning Cellular Responses to BMP-2 with Material Surfaces, Cytokine Growth Factor Rev., 2016, 27, 43–54, DOI:10.1016/j.cytogfr.2015.11.008.
- C. A. Hoesli, A. Garnier, P.-M. Juneau, P. Chevallier, C. Duchesne and G. Laroche, A Fluorophore-Tagged RGD Peptide to Control Endothelial Cell Adhesion to Micropatterned Surfaces, Biomaterials, 2014, 35(3), 879–890, DOI:10.1016/j.biomaterials.2013.09.076.
- M.-C. Durrieu and O. Zouani, Substituts osseux greffes par des peptides mimetiques de la proteine humaine bmp-2, EP2968660A1, 2016 Search PubMed.
- D. Lopes, C. Martins-Cruz, M. B. Oliveira and J. F. Mano, Bone Physiology as Inspiration for Tissue Regenerative Therapies, Biomaterials, 2018, 185, 240–275, DOI:10.1016/j.biomaterials.2018.09.028.
- J. Skottke, M. Gelinsky and A. Bernhardt, In Vitro Co-Culture Model of Primary Human Osteoblasts and Osteocytes in Collagen Gels, Int. J. Mol. Sci., 2019, 20(8), 1998, DOI:10.3390/ijms20081998.
- S. W. Lane, D. A. Williams and F. M. Watt, Modulating the Stem Cell Niche for Tissue Regeneration, Nat. Biotechnol., 2014, 32(8), 795–803, DOI:10.1038/nbt.2978.
- X. Wan, Z. Liu and L. Li, Manipulation of Stem Cells Fates: The Master and Multifaceted Roles of Biophysical Cues of Biomaterials, Adv. Funct. Mater., 2021, 31(23), 2010626, DOI:10.1002/adfm.202010626.
- A. Saraswathibhatla, D. Indana and O. Chaudhuri, Cell–Extracellular Matrix Mechanotransduction in 3D, Nat. Rev. Mol. Cell Biol., 2023, 24(7), 495–516, DOI:10.1038/s41580-023-00583-1.
- S. R. Caliari and J. A. Burdick, A Practical Guide to Hydrogels for Cell Culture, Nat. Methods, 2016, 13(5), 405–414, DOI:10.1038/nmeth.3839.
- S. C. Neves, R. F. Pereira, M. Araújo and C. C. Barrias, Bioengineered Peptide-Functionalized Hydrogels for Tissue Regeneration and Repair, Peptides and Proteins as Biomaterials for Tissue Regeneration and Repair, Elsevier, 2018, pp. 101–125 DOI:10.1016/B978-0-08-100803-4.00004-8.
- M. Levin, Y. Tang, C. D. Eisenbach, M. T. Valentine and N. Cohen, Understanding the Response of Poly(Ethylene Glycol) Diacrylate (PEGDA) Hydrogel Networks: A Statistical Mechanics-Based Framework, Macromolecules, 2024, 57(15), 7074–7086, DOI:10.1021/acs.macromol.3c02635.
- N. Rekowska, D. Arbeiter, H. Seitz, R. Mau, A. Riess, T. Eickner, N. Grabow and M. Teske, The Influence of PEGDA's Molecular Weight on Its Mechanical Properties in the Context of Biomedical Applications, Curr. Dir. Biomed. Eng., 2022, 8(2), 181–184, DOI:10.1515/cdbme-2022-1047.
- S. Lin-Gibson, R. L. Jones, N. R. Washburn and F. Horkay, Structure Property Relationships of Photopolymerizable Poly(Ethylene Glycol) Dimethacrylate Hydrogels, Macromolecules, 2005, 38(7), 2897–2902, DOI:10.1021/ma0487002.
- F. Oesterhelt, M. Rief and H. E. Gaub, Single Molecule Force Spectroscopy by AFM Indicates Helical Structure of Poly(Ethylene-Glycol) in Water, New J. Phys., 1999, 1, 6, DOI:10.1088/1367-2630/1/1/006.
- S. Kalakkunnath, D. S. Kalika, H. Lin and B. D. Freeman, Viscoelastic Characteristics of UV Polymerized Poly(Ethylene Glycol) Diacrylate Networks with Varying Extents of Crosslinking, J. Polym. Sci., Part B: Polym. Phys., 2006, 44(15), 2058–2070, DOI:10.1002/polb.20873.
- J. A. Beamish, J. Zhu, K. Kottke-Marchant and R. E. Marchant, The Effects of Monoacrylated Poly(Ethylene Glycol) on the Properties of Poly(Ethylene Glycol) Diacrylate Hydrogels Used for Tissue Engineering, J. Biomed. Mater. Res., Part A, 2010, 92A(2), 441–450, DOI:10.1002/jbm.a.32353.
- Y. Ma, T. Han, Q. Yang, J. Wang, B. Feng, Y. Jia, Z. Wei and F. Xu, Viscoelastic Cell Microenvironment: Hydrogel-Based Strategy for Recapitulating Dynamic ECM Mechanics, Adv. Funct. Mater., 2021, 31(24), 2100848, DOI:10.1002/adfm.202100848.
- H. De Belly, E. K. Paluch and K. J. Chalut, Interplay between Mechanics and Signalling in Regulating Cell Fate, Nat. Rev. Mol. Cell Biol., 2022, 23(7), 465–480, DOI:10.1038/s41580-022-00472-z.
- L. Oliver-Cervelló, H. Martín-Gómez and C. Mas-Moruno, New Trends in the Development of Multifunctional Peptides to Functionalize Biomaterials, J. Pept. Sci., 2022, 28(1), e3335 CrossRef PubMed.
- J. S. Lee, J. S. Lee, A. Wagoner-Johnson and W. L. Murphy, Modular Peptide Growth Factors for Substrate-Mediated Stem Cell Differentiation, Angew. Chem., Int. Ed., 2009, 48(34), 6266–6269, DOI:10.1002/anie.200901618.
- B. Clarke, Normal Bone Anatomy and Physiology, Clin. J. Am. Soc. Nephrol., 2008, 3(Supplement 3), S131–S139, DOI:10.2215/CJN.04151206.
- J. P. Rodríguez, M. González, S. Ríos and V. Cambiazo, Cytoskeletal Organization of Human Mesenchymal Stem Cells (MSC) Changes during Their Osteogenic Differentiation, J. Cell. Biochem., 2004, 93(4), 721–731, DOI:10.1002/jcb.20234.
- L. F. Bonewald, Osteocytes, Marcus and Feldman's Osteoporosis, Elsevier, 2021, pp. 135–163 DOI:10.1016/B978-0-12-813073-5.00007-1.
- E. N. Holland, M. A. Fernández-Yagüe, D. W. Zhou, E. B. O’Neill, A. U. Woodfolk, A. Mora-Boza, J. Fu, D. D. Schlaepfer and A. J. García, FAK, Vinculin, and Talin Control Mechanosensitive YAP Nuclear Localization, Biomaterials, 2024, 308, 122542, DOI:10.1016/j.biomaterials.2024.122542.
- R. McBeath, D. M. Pirone, C. M. Nelson, K. Bhadriraju and C. S. Chen, Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment, Dev. Cell, 2004, 6(4), 483–495, DOI:10.1016/S1534-5807(04)00075-9.
- C. Pizzolitto, F. Scognamiglio, P. Sacco, S. Lipari, M. Romano, I. Donati and E. Marsich, Immediate Stress Dissipation in Dual Cross-Link Hydrogels Controls Osteogenic Commitment of Mesenchymal Stem Cells, Carbohydr. Polym., 2023, 302, 120369, DOI:10.1016/j.carbpol.2022.120369.
- M. Darnell, S. Young, L. Gu, N. Shah, E. Lippens, J. Weaver, G. Duda and D. Mooney, Substrate Stress-Relaxation Regulates Scaffold Remodeling and Bone Formation in Vivo, Adv. Healthcare Mater., 2017, 6(1), 1601185, DOI:10.1002/adhm.201601185.
- M. Walker, E. W. Pringle, G. Ciccone, L. Oliver-Cervelló, M. Tassieri, D. Gourdon and M. Cantini, Mind the Viscous Modulus: The Mechanotransductive Response to the Viscous Nature of Isoelastic Matrices Regulates Stem Cell Chondrogenesis, Adv. Healthcare Mater., 2023, 13(9), 2302571, DOI:10.1002/adhm.202302571.
- Z. Gong, S. E. Szczesny, S. R. Caliari, E. E. Charrier, O. Chaudhuri, X. Cao, Y. Lin, R. L. Mauck, P. A. Janmey, J. A. Burdick and V. B. Shenoy, Matching Material and Cellular Timescales Maximizes Cell Spreading on Viscoelastic Substrates, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(12), E2686–E2695, DOI:10.1073/pnas.1716620115.
- S. J. I. Blackford, T. T. L. Yu, M. D. A. Norman, A. M. Syanda, M. Manolakakis, D. Lachowski, Z. Yan, Y. Guo, E. Garitta, F. Riccio, G. M. Jowett, S. S. Ng, S. Vernia, A. E. del Río Hernández, E. Gentleman and S. T. Rashid, RGD Density along with Substrate Stiffness Regulate hPSC Hepatocyte Functionality through YAP Signalling, Biomaterials, 2023, 293, 121982, DOI:10.1016/j.biomaterials.2022.121982.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ma00165j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |