Advances in antibacterial agents for Mycobacterium fortuitum†
Carlos
Roberto Tavolari Jortieke
 ,
Angélica
Rocha Joaquim
,
Angélica
Rocha Joaquim
 and
Fernando
Fumagalli
and
Fernando
Fumagalli
 *
*
Department of Pharmacy, Health Sciences Centre, Federal University of Santa Maria, Santa Maria, RS, Brazil. E-mail: fernando.fumagalli@ufsm.br; Tel: +55 (55) 3220 9372
First published on 18th October 2024
Abstract
Mycobacterium fortuitum is an emerging human pathogen, characterized by an increase in prevalence and antibacterial resistance over the years, highlighting the need for the development of new drugs against this rapidly growing nontuberculous mycobacterium (NTM). To support this crusade, this review summarizes findings from the past two decades concerning compounds with antimycobacterial activity against M. fortuitum. It identifies the most promising and effective chemical frameworks to inspire the development of new therapeutic alternatives for infections caused by this microorganism. Most compounds effective against M. fortuitum are synthetic, with macozinone, featuring a 2-piperazine-benzothiazinone framework, standing out as a notable drug candidate. Among natural products, the polyphenolic polyketide clostrubin and the sansanmycin peptide analogs have shown efficacy against this NTM. Some compounds' mechanisms of action on M. fortuitum have been studied, including NITD-916, which acts as an enoyl-acyl carrier protein reductase inhibitor, and TBAJ-5307, which inhibits F-ATP synthase. Moreover, this review discusses the pathogenic molecular mechanisms and potential therapeutic targets within this mycobacterium.
Introduction
Mycobacterium fortuitum, also known as Mycolicibacterium fortuitum,1,2 is a rapidly growing nontuberculous mycobacterium (NTM) associated with opportunistic infections in humans and animals.3 Considered an emerging human pathogen, its infections have been increasingly prevalent in recent years.4–6 The NTM can withstand a wide range of environmental temperatures,7 and its infection incidence varies by country, ranging from 1 to 40 cases per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 population,8 with 3–30% attributable to M. fortuitum.9–12
000 population,8 with 3–30% attributable to M. fortuitum.9–12
Commonly found in the environment, M. fortuitum can cause respiratory, skin, or soft tissue infections in both immunodeficient and immunocompetent individuals.13,14 In terms of microbial susceptibility and therapeutic regimen, infections caused by different NTM present some particularities, despite susceptibility tests being indicated.15 An official guideline for the management of pulmonary diseases related to NTM recommends an oral or parenteral regimen with drugs such as macrolides, clofazimine, linezolid, amikacin, imipenem, cefoxitin, or tigecycline to treat infections by M. abscessus, and macrolides, rifamycin, ethambutol, amikacin, or streptomycin for infections by the M. avium complex.16,17
To date, there is no specific guideline for the therapy of pulmonary infections associated with M. fortuitum. However, susceptibility studies have shown that M. fortuitum clinical isolates are sensitive to amikacin, imipenem, moxifloxacin, ciprofloxacin, and doxycycline.18,19M. fortuitum isolates may be intrinsically resistant to macrolides.20,21 Additionally, some cases of M. fortuitum resistance to quinolones22,23 and omadacycline24 have been reported. M. fortuitum and M. abscessus are the most common NTM isolated in skin infections.25 For skin and soft tissue infections caused by NTM, surgery may be necessary in conjunction with drug therapy.25,26 This may involve a combination of cefoxitin, imipenem, amikacin, and macrolides for M. abscessus skin infections or fluoroquinolones, macrolides, doxycycline, or antifolate for M. fortuitum skin infections.27
In addition to commercial issues,28 challenges in the development of new drugs for NTM stem from the microorganism's characteristics, such as its lipid-rich outer membrane and the ability to adhere to surfaces to form biofilms.29,30 Additionally, the lack of animal models that accurately mimic human infection31–33 and the limited physiological similarities between NTM and M. tuberculosis restrict the applicability of drugs developed for M. tuberculosis to NTM.34 While recent reviews have discussed molecular targets and new antimicrobial agents for M. tuberculosis,35–39 only a few have focused on NTM,40–42 and none exclusively on M. fortuitum.
Additionally, limited information is available on the biochemical pathways of M. fortuitum that can be used as targets for new antimycobacterial compounds.43 Given the urgency of developing new treatments for NTM,44 including M. fortuitum, this review aims to present information from the last two decades on compounds known for their antimicrobial activity against M. fortuitum, analyzing their in vitro activity, uncovering structural and physicochemical insights for the development of new effective agents, and discussing potential targets involved in the pathogenicity molecular mechanism of this microorganism.
In vitro activity against M. fortuitum
The in vitro evaluation of antimicrobial activity is a critical step in the early phases of drug discovery. It provides quick, reproducible, and cost-effective data on how a chemical structure affects biological activity.45,46 Though it has some limitations,47 it serves as a benchmark to determine if a chemical entity will succeed in the drug development process.Drugs aimed at treating M. fortuitum infections show low micromolar values in in vitro tests (Table 1), consistent with other anti-infective drugs.48 This micromolar range activity can serve as a criterion during the drug development process. In this study, we applied these parameters to identify promising compounds from a range of diverse compounds already tested against M. fortuitum. For clarity, we have divided them into compounds derived from natural sources and synthetic ones. The graphs illustrating the relationship between biological activity and physicochemical properties are presented in the ESI.†
| Drug | Class | MICStrain [μM] |
|---|---|---|
| MIC = minimum inhibitory concentration as the lowest concentration at which no bacterial growth was observed. | ||
| Amikacin | Aminoglycoside | MICATCC6841 = 3.41 |
| MICATCC49404 = 5.33 | ||
| Moxifloxacin | Quinolone | MICATCC6841 = 0.15 |
| Clarithromycin | Macrolide | MICATCC6841 = 1.34 |
| Doxycycline | Tetracycline | MICATCC6841 = 0.56 |
| Imipenem | Carbapenem | MICATCC6841 = 13.36 |
| Sulfamethoxazole | Sulfonamide | MICATCC6841 = 126.3 |
| Linezolid | Oxazolidinone | MICATCC6841 = 23.71 |
| Clofazimine | Riminophenazine | MICATCC6841 = 4.22 |
Natural products related compounds
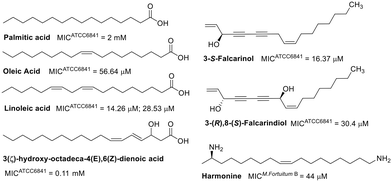
Natural products showing promising activity against M. fortuitum cover a wide range of classes, including fatty acids, alkaloids, steroids, terpenes, phenolic derivatives, peptides, and polyamides. The minimum inhibitory concentrations (MIC) of fatty acids evaluated against M. fortuitum range from 14.26 μM to 2 mM.53–56 The activity of these natural products is influenced by their degree of unsaturation (Fig. 1), as evidenced by linolenic acid being two to four times more active than oleic acid.53,54 A reduction in chain length from eighteen to sixteen carbon atoms, as well as the absence of unsaturation (palmitic acid), significantly decreases the activity.53 The position of unsaturation also plays a crucial role in antimicrobial efficacy, as demonstrated by the lower activity of 3(ζ)-hydroxy-octadeca-4(E),6(Z)-dienoic acid compared to linoleic acid. Moreover, the introduction of a hydroxyl group at C3 has been shown to be unfavorable.55,56Fatty acid derivatives have been explored as well, such as 3-(S)-falcarinol, a derivative of oleic acid.57 The MIC of 3-(S)-falcarinol was approximately threefold lower than that of its precursor,58 highlighting the critical role of unsaturation in the antimycobacterial activity of fatty acids and related compounds. The enantiomer, 3-(R)-falcarinol, exhibited a similar MIC value of 16.4 μM,59 indicating comparable activity. However, the dihydroxylated derivative, 3-(R),8-(S)-falcarindiol, was shown to be less active, suggesting that the addition of a hydroxyl group at C8 is detrimental to its activity.59 The stearic acid derivative, harmonine,60 also displayed comparable activity to that of oleic acid,61 which has a single unsaturation.
Interestingly, the FabI enzyme (enoyl-acyl carrier protein reductase), critical in fatty acid biosynthesis in bacteria, has been identified as a molecular target for linoleic acid and other unsaturated fatty acids in Staphylococcus aureus and Escherichia coli.62 This enzyme is also a target of isoniazid in Mycobacterium tuberculosis.63
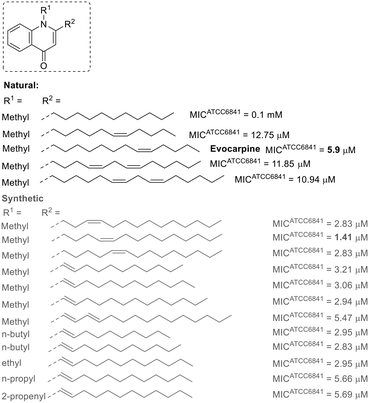
A similar relationship between unsaturation and anti-M. fortuitum activity is observed in some natural 2-alkylquinolones (Fig. 2).64 Various alkyl chains have been examined, with the alkaloid evocarpine (a biosynthetic derivative of myristoleic acid) displaying the best activity in this group, featuring a thirteen-carbon chain and a single unsaturation at the substituent at position 2. Either reducing the length of the alkyl chain or introducing a second unsaturation diminished the activity by approximately half. Furthermore, the absence of unsaturation led to a substantial reduction in activity.64 Synthetic 2-alkylquinolones with unsaturated chains showed encouraging results, with some being more active than evocarpine.65–67 Compounds with double bonds conjugated to the quinolone ring were particularly promising, with their activity appearing to depend more on the substituent at position 1 than on the size of the alkyl chain at position 2. Smaller alkyl chains (containing up to four carbons) at position 1 generally led to more active derivatives.65–67
Alkylquinolones have been identified as inhibitors of MurE ligase (UDP-N-acetylmuramoylalanyl-D-glutamate-2,6-diaminopimelate ligase) in M. tuberculosis.65 This ATP-dependent ligase plays a crucial role in the synthesis of cell wall peptidoglycan and has emerged as a target for new antimicrobial agents.68,69
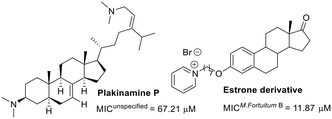
Regarding steroid-related compounds, the limited number of molecules evaluated against M. fortuitum suggests that this area is ripe for exploration in drug discovery (Fig. 3). Adamec and colleagues discovered estrone derivatives exhibiting noteworthy activity. The series substituted with pyridines at C3 showed similar efficacy.70 Exploring other substitutions at C3 might optimize this hit. In another study, plakinamine P, isolated from a sponge, exhibited modest activity against M. fortuitum, despite its significant activity against M. tuberculosis (MIC = 3.81 μM).71
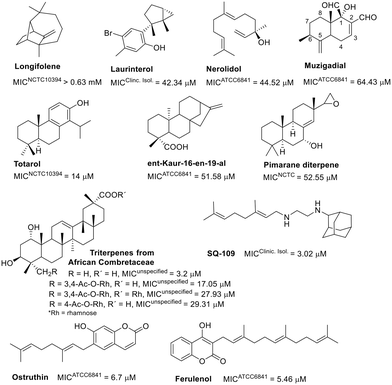
An intriguing correlation was noted when assessing the activity of another class of isoprene derivatives. Lipophilicity seems critical for the anti-M. fortuitum activity of certain evaluated terpenoids (Fig. 4). The hydroxylated sesquiterpenes, laurinterol and nerolidol displayed comparable activities,72,73 as well as clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values (4.18 and 5.40, respectively).74 Muzigadial, the most hydrophilic sesquiterpene in this group (clog
P values (4.18 and 5.40, respectively).74 Muzigadial, the most hydrophilic sesquiterpene in this group (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.16),74 was less active.54 Longifolene, consisting only of carbon and hydrogen atoms, is one exception; despite its high clog
P = 1.16),74 was less active.54 Longifolene, consisting only of carbon and hydrogen atoms, is one exception; despite its high clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value (4.06),74 it showed reduced activity.75
P value (4.06),74 it showed reduced activity.75
The activity of a small selection of diterpenes also demonstrated this correlation. The diterpene totarol (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 5.34)74 was more active than ent-Kaur-16-en-19-al73,75 (clog
P = 5.34)74 was more active than ent-Kaur-16-en-19-al73,75 (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 4.11)74 and a pimarane diterpene76 (clog
P = 4.11)74 and a pimarane diterpene76 (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 3.7).74 Substituting a hydroxyl group for a carbonyl in a pimarane diterpene decreased its activity eightfold.76 A triterpene from African Combretaceae (clog
P = 3.7).74 Substituting a hydroxyl group for a carbonyl in a pimarane diterpene decreased its activity eightfold.76 A triterpene from African Combretaceae (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 5.54)74 exhibited significant activity against M. fortuitum. Incorporating rhamnose into this framework negatively affected the activity,77 emphasizing the importance of lipophilicity. Although natural products with higher lipophilicity showed better in vitro activity, this is unlikely to translate to in vivo models due to low aqueous solubility, high metabolism, and promiscuity-related off-target toxicity.78
P = 5.54)74 exhibited significant activity against M. fortuitum. Incorporating rhamnose into this framework negatively affected the activity,77 emphasizing the importance of lipophilicity. Although natural products with higher lipophilicity showed better in vitro activity, this is unlikely to translate to in vivo models due to low aqueous solubility, high metabolism, and promiscuity-related off-target toxicity.78
Coumarins and alkaloids with isoprene units displayed notable activity, as shown in Fig. 4. Geranyl and farnesyl coumarins achieved similar MIC values.79,80 Ferulenol derivatives, modified at the end of the alkyl chain (with hydroxyl, benzoyloxyl, or acetyl groups), were less effective than the hit compound.79
SQ-109, a diamine with a geranyl group, showed a MIC of 3.02 μM against M. fortuitum,81 comparable to that of amikacin (refer to Table 1), and is under investigation in clinical trials for tuberculosis treatment.82–84 In M. tuberculosis, this compound targets MmpL3 (mycobacterial membrane protein large 3), a crucial protein for transporting cell wall components.85,86 Furthermore, SQ-109 has recently been reported to exhibit immunomodulatory properties.87
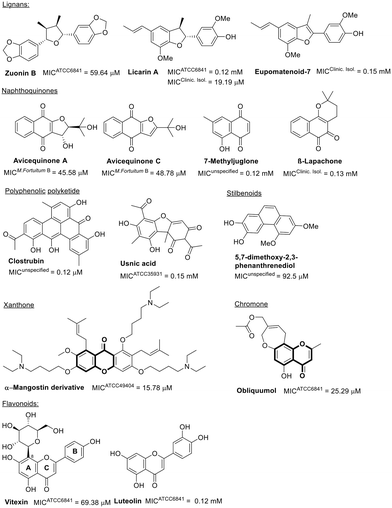
Phenols represent another significant class of natural products with antibacterial activity.88,89 Despite the phenolic compounds' issues of low bioavailability due to high metabolism,90,91 a diverse collection has been evaluated against M. fortuitum, as indicated in Fig. 5.
Among the neolignans, licarin A exhibits superior activity, with MICs against clinical isolates comparable to those of linezolid (Table 1). However, against the standard strain ATCC 6841, licarin A demonstrated poor activity. Enhancing the lipophilicity of derivatives from the licarin A hydroxyl group can potentially improve its antimicrobial efficacy (ESI†).92 Saturation in the furan moiety within the central core of this natural product is crucial for activity, as illustrated by eupomatenoid-7, which has an unsaturated benzofuran, exhibiting even weaker antimicrobial effects.93 Beyond this class of natural products, the lignan zuorin B also modestly impacts M. fortuitum growth.73
Another group of phenolic compounds evaluated biologically against M. fortuitum is the naphthoquinones, all of which displayed modest to weak activity. Notably, the furo-naphthoquinones found in the twigs of Avicennia marina, avicequinone A and C, differ from neolignans in that saturation of the furan moiety is not essential for antimicrobial activity (Fig. 5).94–96
The natural polyketides, considered among the most significant microbial metabolites in human antibiotic medicines (e.g., erythromycin and doxycycline), are noteworthy.97 Clostrubin, a polyphenolic polyketide antibiotic featuring a unique benzo[a]tetraphene scaffold, has shown excellent antimicrobial activity against M. fortuitum (Fig. 5).98
Other polyphenolic natural products, including usnic acid99 and compounds with stilbenoid or flavonoid scaffolds, have demonstrated moderate to poor activity against M. fortuitum (Fig. 5).100 Interestingly, the presence of glucopyranoside in C8 of ring A in the flavonoid vitexin seems beneficial for activity against M. fortuitum,101 a benefit not observed with the disaccharide rutinose bonded to C3 in ring C.102
Xanthone and chromone, two other phenolic compound classes active against M. fortuitum that share a benzopyrone moiety, are also of interest (Fig. 5). A semisynthetic xanthone derivative from alpha-mangostin exhibits good in vitro activity against M. fortuitum and its biofilm formation.103 Obliquumol, a chromone isolated from Ptaeroxylon obliquum common in Southern Africa, is slightly less active.104
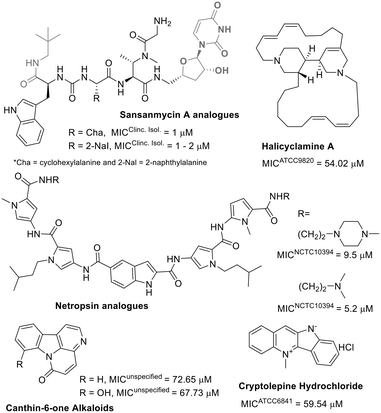
Among alkaloids, polyamides, and peptide natural products, the latter two are highlighted as potent antibacterial agents against M. fortuitum, whereas the former shows moderate activity (Fig. 6).
Sansanmycin A, a known uridylpeptide natural product, has been used as a scaffold inspiration in developing M. tuberculosis translocase inhibitors. These inhibitors prevent the formation of a lipid compound, a key intermediate in mycobacterial peptidoglycan synthesis.105 Recently, the same research group described new analogues of this natural uridylpeptide (Fig. 6) and identified the critical role of the appended neopentylamide moiety in the antimycobacterial activity of these compounds.106 The low micromolar anti-M. fortuitum activity of these sansanmycin A analogues is comparable to some of the most active known drugs (Table 1).
Unlike sansanmycin A, netropsin is a non-peptidic polyamide but has also been used as a scaffold for developing analogues with effective activity against M. fortuitum growth (Fig. 6), possibly due to DNA minor groove binding.107 For these polyamide compounds, replacing the methyl-piperazine with a morpholine ring results in inactive compounds (MIC > 77.7 μM).107
Alkaloids, as a class, do not appear to be effective antibiotics against M. fortuitum (Fig. 6). The indole alkaloids canthin-6-one and cryptolepine exhibited weak anti-M. fortuitum activity.56,108 Not significantly different, halicyclamine A, a marine sponge alkaloid, was the most potent among them against M. fortuitum (Fig. 6).109 Although alkaloids alone seem ineffective against M. fortuitum, evidence suggests that their combination with known antibiotics can potentiate anti-mycobacterial activity.110
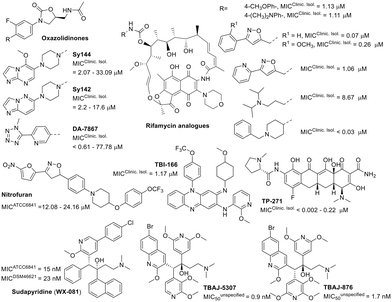
Synthetic compounds
The development of new drugs often employs the “me-too” strategy to produce what are known as “follow-on drugs”. This approach leverages the structure of existing drugs, implementing modifications to enhance activity and/or aspects related to absorption, distribution, metabolism, excretion, and toxicity (ADMET).111,112 This strategy has been used to develop new antimycobacterial agents, evaluating compounds related to known anti-infective medications against M. fortuitum (Fig. 7). One example is DA-7867, an oxazolidinone derivative with a tetrazole moiety, demonstrating greater activity than linezolid against twenty-four clinical isolates of M. fortuitum with MICs ranging from <0.61 to 19.45 μM.113 Oxazolidinones Sy142 and Sy144 also showed greater activity than the control drug linezolid, with MICs of 23.41–189.72 μM.114 These findings suggest that the isosteric substitution of a morpholine ring with a piperazine and the incorporation of fused N-heterocycles, such as imidazo[1,2-a]pyrimidine (Sy142) and imidazo[1,2-b]pyridazine (Sy144), are beneficial modifications.The bedaquiline analogue sudapyridine (WX-081) demonstrated significant anti-M. fortuitum activity, comparable to the control drug, with MIC values in the nanomolar range.115 This compound has been the subject of clinical trials for the treatment of tuberculosis, showing advantages in safety and pharmacokinetics compared to bedaquiline.116,117 The other two bedaquiline analogues, TBAJ-5307 and TBAJ-876, were recently described as F-ATP synthase inhibitors. This enzyme provides the biological energy ATP and maintains ATP homeostasis under hypoxic stress conditions. These two drug candidates showed broad-spectrum anti-NTM activity with excellent in vitro performance against M. fortuitum in the low nanomolar range.118
Another compound displaying noteworthy activity was TP-271, a tetracycline analogue, exhibiting superior activity to doxycycline (Table 1) against M. fortuitum.119 TP-271 has undergone phase I clinical trials as a potential pneumonia treatment.120
Rifamycin analogues featuring a substituted carbamate group at the C25 position exhibited significant activity against M. fortuitum, with MIC values ranging from <0.03 to 8.67 μM.121 Within this series, the compound with a 1-benzyl-4-methylpiperidinyl substituent as the carbamate group was found to be the most active. The presence of aromatic ring substituents in these derivatives appears to enhance activity, although only a single aliphatic substituent was investigated (Fig. 7).121
Other studies have investigated analogues within the nitrofuran and riminophenazine classes, which have demonstrated promising anti-M. fortuitum activity and efficacy in tuberculosis animal models.122,123
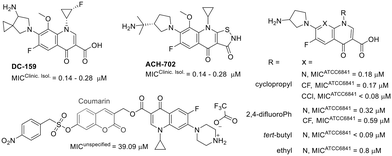
Furthermore, studies on fluoroquinolone-related compounds against M. fortuitum have revealed that derivatives retaining the quinolone pharmacophore (the carboxylic acid at C3) exhibited comparable activity against M. fortuitum (Fig. 8).124–126
Interestingly, compound ACH-702, which contains an isothiazol-3(2H)-one moiety fused to C2/C3, showed activity similar to that of its analogue DC-159, which incorporates the quinolone pharmacophore at C3.124,127 A coumarin–quinolone hybrid exhibited reduced activity, highlighting the importance of the carboxylic acid at C3.128 This compound was designed to function as a prodrug, being activated by nitroreductases.128 Structural modifications at positions 1, 7, and 8 were also explored. The structure–activity relationship studies indicate that bulky substituents at position 1 generally decrease activity. Furthermore, alterations at position 1 and other substituted heterocycles at position 7 appear to be tolerable.125
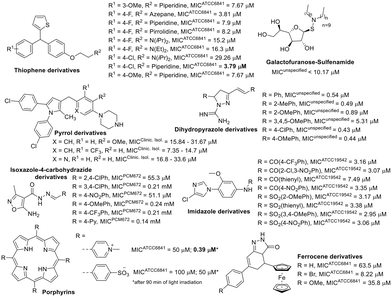
In addition to compounds inspired by known drugs, a diverse range of synthetic compounds was also developed to combat M. fortuitum infection. These compounds, focusing on five-membered heterocycles, showed promising activity against M. fortuitum (Fig. 9).
Among the non-aromatic five-membered rings, tetrahydrofuran in a galactofuranose with a lipophilic sulfenamide demonstrated significant activity against M. fortuitum. A smaller lipophilic side chain in the structure had a detrimental effect on the antimycobacterial activity of these compounds.129 Lipophilicity also seemed to influence the activity of dihydropyrazole derivatives evaluated against M. fortuitum. Compared to the previously mentioned sugar derivative, the dihydropyrazole performed better against M. fortuitum (Fig. 9) despite being less lipophilic. This could be attributed to the free amidine in the structure, which is essential for anti-M. fortuitum activity.130 The lipophilic balance derived from the benzene ring supports various substituents, except for the trimethoxy substitution that reduces the clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of this class of compounds to less than 3, diminishing the activity tenfold (ESI†).
P of this class of compounds to less than 3, diminishing the activity tenfold (ESI†).
Considering compounds based on aromatic five-membered rings, isoxazole derivatives exhibited the lowest anti-M. fortuitum activity (Fig. 9).131 Pyrrole derivatives also showed moderate activity,132 except when part of the porphyrin structure was activated by irradiation.133 The ferrocene derivatives did not stand out as excellent agents against M. fortuitum.134
Generally, the thiophene-containing trisubstituted methane evaluated against M. fortuitum had good activity (Fig. 9) but was not as potent as moxifloxacin (Table 1), a drug used as a positive control.135 However, two thiophene compounds from this set also demonstrated in vivo activity, clearing mice of bacilli in acute infections,135 and one of them was the less active thiophene derivative (Fig. 9). Regarding compounds with an imidazole ring active against M. fortuitum, amide derivatives did not significantly differ from sulphonamide derivatives, except for the thienyl amide, which was less active than its related sulphonamide. Nevertheless, all the imidazole derivatives in Fig. 9 showed good anti-M. fortuitum activity, comparable to rifampicin, another positive control drug.136
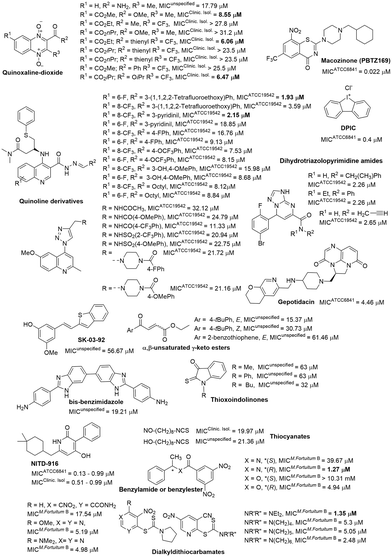
In addition to the five-membered heterocyclic derivatives, compounds centered on 6,6-fused heterocycles also displayed excellent antimycobacterial activity (Fig. 10).
The most extensively investigated core was quinoline, where compounds with a C4 substitution by triazole exhibited only modest anti-M. fortuitum activity.137 Compounds featuring a carbohydrazone moiety at C2 and an amino-phenyl-thio butanamide at C3 achieved low micromolar range activity against M. fortuitum (Fig. 10).138 Quinoxaline derivatives were also evaluated for their anti-M. fortuitum activity, with di-N-oxide quinoxalines that have smaller esters at C7 showing more conducive activity (Fig. 10).139,140
Benzothiazinone represents another 6,6-fused heterocycle used in antimycobacterial agents, notable in the drug candidate macozinone, which is undergoing clinical trials for M. tuberculosis.141 It has demonstrated excellent in vitro142 and in vivo143 activity against M. fortuitum (Fig. 10). The mechanism of action of macozinone against M. tuberculosis involves the inhibition of decaprenyl-phosphoryl-ribose 2′-epimerase (DprE1), a critical enzyme in the biosynthesis of vital cell wall polysaccharides.144
Interestingly, simple frameworks have also shown low micromolar MICs for M. fortuitum, including benzylamide, dihydrotriazolopyrimidine, and thiocarbamate derivatives (Fig. 10).145–147 However, a diverse set of compounds exhibited moderate to poor activity against M. fortuitum, among them benzothiophene SK-03-92, bis-benzimidazole, keto ester, thioxoindolinones, and thiocyanate derivatives.148–153
Compounds featuring tricyclic fused-ring system cores were identified among those with low micromolar MICs for M. fortuitum. Diphenyleneiodonium chloride (DPIC, Fig. 10), discovered during an in vitro screening of the LOPAC®1280 small molecule library, showed potential in vivo activity comparable to the amikacin control.154 Another notable compound, the triazaacenaphthylene antibiotic gepotidacin, which, (Fig. 10) by inhibiting DNA replication through topoisomerase inhibition,155 showed a significant reduction in the bacterial load in various organs at a concentration ten times lower than that of amikacin.156,157
Lastly, the enoyl-ACP reductase (InhA) inhibitor,158 NITD-916 (Fig. 10), with a low micromolar MIC against M. fortuitum, demonstrated anti-M. fortuitum activity in a macrophage model and was highly effective in protecting infected zebrafish larvae during short-duration treatments.159
Pathogenic molecular mechanism and potential therapeutic targets
Although a recent study explored the proteomics for M. fortuitum,43 structural biological data for M. fortuitum remain scarce. To date, the Protein Data Bank (PDB) repository contains only seven 3D structures related to M. fortuitum, of which two (PDB: 5K21 and 7PDA) are enzymes involved in the biotransformation of phenazine-related compounds.160 Most of the remaining structures have not been described in publications.Mycobacterium species have lipid mycolic acids that play essential roles in physiology and virulence.161 In this context, enoyl-acyl carrier protein reductase InhA is a crucial enzyme for fatty acid synthesis; its inhibition halts mycolic acid biosynthesis. As previously mentioned, NITD-916 directly inhibits the InhA of M. fortuitum, and the chemical interactions with the target are elucidated in the resolved crystal structures available (PDB: 7U0O and 7K73), where the pyridone central core is responsible for three hydrogen bonds while the cyclohexyl occupies a hydrophobic pocket.159
Other type II fatty acid components have been evaluated in knockdown studies for M. fortuitum. While the 3-hydroxyacyl-ACP dehydratase (HadC) is not essential for the survival and growth of M. fortuitum,162 the fabG4 gene is crucial for maintaining cell envelope physiology and is required for the formation of the M. fortuitum pellicle and biofilm.163
While much basic mycobacterial research has focused on lipid composition, carbohydrate features are underexplored in drug discovery, with only two enzymes having 3D information available: glycosyl hydrolase (PDB: 4W65) and shikimate 5-dehydrogenase (PDB: 4XIJ). Glycoside hydrolases are important for the persistence, virulence, and general biology of these microorganisms and are promising drug targets.164 Similarly, the shikimate pathway, absent in humans but essential for mycobacteria, is an attractive target for developing new selective antimycobacterial drugs.165
Like other microorganisms, M. fortuitum has shown drug resistance, and the cleavage of β-lactam antibiotics is one of the mechanisms. In mycobacteria, β-lactam resistance is primarily due to the production of class-A β-lactamase,166 which has been described in M. fortuitum (PDB: 2CC1) with structural features that confer more efficient cephalosporinase activity.167 The rRNA methylase gene is another mechanism of resistance against macrolides in M. fortuitum.20 Resistance to isoniazid is related to two unrelated catalase-peroxidases (KatGI and KatGII) in M. fortuitum.168
Few studies discuss the pathogenic mechanisms of M. fortuitum. One mechanism is the ability of M. fortuitum to produce porins to facilitate nutrient uptake through the mycobacterial cell wall, which also increases the colony size of this microorganism.169 Understanding these pathogenic mechanisms is crucial, as is recognizing those not involved in this process. For example, extracellular DNA (eDNA), an essential component of biofilm formation, is scarcely present in the rapidly growing mycobacteria M. fortuitum.170
Conclusions
This review summarizes the major findings of the last two decades on active compounds against Mycobacterium fortuitum. During this period, around two hundred compounds have shown some level of anti-M. fortuitum activity, with a quarter originating from natural sources and the majority from synthetic origins. Among natural products, the polyphenolic polyketide clostrubin and the sansanmycin peptide analogs have emerged as the most promising and effective against M. fortuitum. However, more potent compounds were identified in the synthetic category.In only a few cases there were enough data to establish a structure–activity relationship, and even fewer compounds were also tested in vivo. Notably, analogs of antibiotics approved for other uses demonstrated significant activity against this microorganism. The drug candidates macozinone, gepotidacin, TBAJ-5307, and NIT-916 deserve special mention for their outstanding in vitro and/or in vivo anti-M. fortuitum activity. It is important to note that no strict correlation exists between the in vitro anti-M. fortuitum activity and the physicochemical properties of the evaluated compounds (ESI†).
For most active compounds discussed, there is a lack of information on their mechanism of action on M. fortuitum. Moreover, only recently studies begun to identify potential M. fortuitum targets, which are primarily associated with mycobacterial lipid composition, although carbohydrate-related targets are also being recognized as potentially significant. Despite our promising findings, much remains to be done to develop a new drug. A consideration will be whether a new agent has sufficiently broad NTM spectrum coverage to increase patient recruitment in future clinical trials and help attract pharmaceutical companies' interest.
Data availability
No primary research results, software or code have been included as part of this review. Furthermore, any data supporting this article have been included as part of the ESI.†Author contributions
The manuscript was written through contributions of all authors and all authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors kindly acknowledge FAPERGS [Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul] for the financial support provided (Grant no. 19/2551-0001273-0).References
- R. S. Gupta, B. Lo and J. Son, Front. Microbiol., 2018, 9, 67 CrossRef.
- E. Tortoli, B. A. Brown-Elliott, J. D. Chalmers, D. M. Cirillo, C. L. Daley, S. Emler, R. A. Floto, M. J. Garcia, W. Hoefsloot, W.-J. Koh, C. Lange, M. Loebinger, F. P. Maurer, K. Morimoto, S. Niemann, E. Richter, C. Y. Turenne, R. Vasireddy, S. Vasireddy, D. Wagner, R. J. Wallace, N. Wengenack and J. Van Ingen, Eur. Respir. J., 2019, 54, 1900795 CrossRef PubMed.
- I. Pavlik, V. Ulmann and J. O. Falkinham, Microorganisms, 2022, 10, 1516 CrossRef.
- M. J. Donohue, BMC Infect. Dis., 2021, 21, 258 CrossRef PubMed.
- C. S. Thornton, M. Mellett, J. Jarand, L. Barss, S. K. Field and D. A. Fisher, Eur. Respir. Rev., 2021, 30, 200299 CrossRef.
- P. Suresh, A. Kumar, R. Biswas, D. Vijayakumar, S. Thulasidharan, G. Anjaneyan, A. Kunoor and L. Biswas, Am. J. Trop. Med. Hyg., 2021, 105, 1335–1338 Search PubMed.
- C. N. Ratnatunga, V. P. Lutzky, A. Kupz, D. L. Doolan, D. W. Reid, M. Field, S. C. Bell, R. M. Thomson and J. J. Miles, Front. Immunol., 2020, 11, 303 CrossRef CAS PubMed.
- C. L. Daley and D. E. Griffith, in Murray and Nadel's Textbook of Respiratory Medicine, Elsevier, 2016, pp. 629–645.e6 Search PubMed.
- P. M. Cassidy, K. Hedberg, A. Saulson, E. McNelly and K. L. Winthrop, Clin. Infect. Dis., 2009, 49, e124–e129 CrossRef.
- A. K. Maurya, V. L. Nag, S. Kant, R. A. S. Kushwaha, M. Kumar, A. K. Singh and T. N. Dhole, BioMed Res. Int., 2015, 2015, 1–5 CrossRef PubMed.
- M. Shteinberg, N. Stein, Y. Adir, S. Ken-Dror, D. Shitrit, D. Bendayan, L. Fuks and W. Saliba, Eur. Respir. J., 2018, 51, 1702469 CrossRef PubMed.
- A. Jarchow-MacDonald, M. Smith, A.-L. Seagar, C. D. Russell, P. Claxton, I. F. Laurenson and O.-L. Moncayo-Nieto, Open Forum Infect. Dis., 2022, 10, ofac665 CrossRef PubMed.
- D. Z. Uslan, T. J. Kowalski, N. L. Wengenack, A. Virk and J. W. Wilson, Arch. Dermatol., 2006, 142, 1287–1292 Search PubMed.
- S. Park, G. Y. Suh, M. P. Chung, H. Kim, O. J. Kwon, K. S. Lee, N. Y. Lee and W.-J. Koh, Respir. Med., 2008, 102, 437–442 CrossRef PubMed.
- K. M. Pennington, A. Vu, D. Challener, C. G. Rivera, F. N. U. Shweta, J. D. Zeuli and Z. Temesgen, J. Clin. Tuberc. Other Mycobact. Dis., 2021, 24, 100244 CrossRef CAS.
- C. L. Daley, J. M. Iaccarino, C. Lange, E. Cambau, R. J. Wallace, C. Andrejak, E. C. Böttger, J. Brozek, D. E. Griffith, L. Guglielmetti, G. A. Huitt, S. L. Knight, P. Leitman, T. K. Marras, K. N. Olivier, M. Santin, J. E. Stout, E. Tortoli, J. Van Ingen, D. Wagner and K. L. Winthrop, Eur. Respir. J., 2020, 56, 2000535 CrossRef PubMed.
- K. Kumar, C. L. Daley, D. E. Griffith and M. R. Loebinger, Eur. Respir. Rev., 2022, 31, 210212 CrossRef.
- S. R. Kim, J. Y. Yu, S.-Y. Kim, D. H. Kim and B. W. Jhun, Microbiol. Spectrum, 2023, 11, e02051-23 CrossRef.
- S. Tasaka, T. Urano, M. Mori, K. Yamaguchi and M. Kanazawa, Kekkaku, 1995, 70, 31–35 CAS.
- K. A. Nash, Y. Zhang, B. A. Brown-Elliott and R. J. Wallace, J. Antimicrob. Chemother., 2005, 55, 170–177 CrossRef CAS.
- Y. Shen, X. Wang, J. Jin, J. Wu, X. Zhang, J. Chen and W. Zhang, BioMed Res. Int., 2018, 2018, 1–10 Search PubMed.
- K. Kurokawa, N. Harada, H. Sasano, H. Takagi, S. Takei, A. Nakamura, K. Kamada, A. Yoshida, K. Kikuchi and K. Takahashi, BMC Infect. Dis., 2020, 20, 866 CrossRef PubMed.
- R. J. Wallace, G. Bedsole, G. Sumter, C. V. Sanders, L. C. Steele, B. A. Brown, J. Smith and D. R. Graham, Antimicrob. Agents Chemother., 1990, 34, 65–70 CrossRef CAS.
- B. A. Brown-Elliott and R. J. Wallace, Antimicrob. Agents Chemother., 2021, 65, e01947-20 CrossRef PubMed.
- T. M. Gonzalez-Santiago and L. A. Drage, Dermatol. Clin., 2015, 33, 563–577 CrossRef CAS.
- W. J. Lee, S. M. Kang, H. Sung, C. H. Won, S. E. Chang, M. W. Lee, M. N. Kim, J. H. Choi and K. C. Moon, J. Dermatol., 2010, 37, 965–972 CrossRef.
- C. Franco-Paredes, L. A. Marcos, A. F. Henao-Martínez, A. J. Rodríguez-Morales, W. E. Villamil-Gómez, E. Gotuzzo and A. Bonifaz, Clin. Microbiol. Rev., 2018, 32, e00069-18 CrossRef PubMed.
- G. S. Timmins, Expert Opin. Drug Discovery, 2020, 15, 7–9 CrossRef.
- J. O. Falkinham, Front. Microbiol., 2018, 9, 1613 CrossRef.
- A. C. Pereira, B. Ramos, A. C. Reis and M. V. Cunha, Microorganisms, 2020, 8, 1380 CrossRef CAS.
- M. D. Johansen and L. Kremer, Front. Cell. Infect. Microbiol., 2020, 10, 357 CrossRef CAS.
- M. D. Johansen, H. P. Spaink, S. H. Oehlers and L. Kremer, Trends Microbiol., 2024, 32, 663–677 CrossRef CAS.
- F. M. Entwistle and P. J. Coote, J. Med. Microbiol., 2018, 67, 585–597 CrossRef CAS.
- I. Soni, M. A. De Groote, A. Dasgupta and S. Chopra, J. Med. Microbiol., 2016, 65, 1–8 CrossRef CAS.
- G. S. Shetye, S. G. Franzblau and S. Cho, Transl. Res., 2020, 220, 68–97 CrossRef CAS PubMed.
- R. Vohra, M. Gupta, R. Chaturvedi and Y. Singh, Recent Pat. Anti-Infect. Drug Discovery, 2006, 1, 95–106 CAS.
- H. M. A. Hameed, M. M. Islam, C. Chhotaray, C. Wang, Y. Liu, Y. Tan, X. Li, S. Tan, V. Delorme, W. W. Yew, J. Liu and T. Zhang, Front. Cell. Infect. Microbiol., 2018, 8, 114 CrossRef.
- G. Cazzaniga, M. Mori, L. R. Chiarelli, A. Gelain, F. Meneghetti and S. Villa, Eur. J. Med. Chem., 2021, 224, 113732 CrossRef CAS.
- A. Bahuguna and D. S. Rawat, Med. Res. Rev., 2020, 40, 263–292 CrossRef PubMed.
- Y. Gu, W. Nie, H. Huang and X. Yu, Front. Cell. Infect. Microbiol., 2023, 13, 1243457 CrossRef CAS PubMed.
- C. R. Cantelli, A. Dassonville-Klimpt and P. Sonnet, Future Med. Chem., 2021, 13, 1367–1395 CrossRef CAS.
- W. Addison, M. Frederickson, A. G. Coyne and C. Abell, RSC Med. Chem., 2022, 13, 392–404 RSC.
- A. Sharma, S. Bansal, N. Kumari, J. Vashistt and R. Shrivastava, Appl. Microbiol. Biotechnol., 2023, 107, 6029–6046 CrossRef CAS PubMed.
- S. Hoffner, M. M. Chan, E. D. Chan and D. Ordway, in Drug Discovery Targeting Drug-Resistant Bacteria, Elsevier, 2020, pp. 361–376 Search PubMed.
- A. Astashkina, B. Mann and D. W. Grainger, Pharmacol. Ther., 2012, 134, 82–106 CrossRef CAS.
- J. E. Ekert, J. Deakyne, P. Pribul-Allen, R. Terry, C. Schofield, C. G. Jeong, J. Storey, L. Mohamet, J. Francis, A. Naidoo, A. Amador, J.-L. Klein and W. Rowan, SLAS Discovery, 2020, 25, 1174–1190 CrossRef CAS PubMed.
- S. Saeidnia, A. Manayi and M. Abdollahi, Curr. Drug Discovery Technol., 2015, 12, 218–224 CAS.
- M. B. Fontoura and F. Fumagalli, Lett. Drug Des. Discovery, 2024, 21, 203–208 CrossRef CAS.
- V. D. C. Flores, F. D. S. Siqueira, C. R. Mizdal, P. C. Bonez, V. A. Agertt, S. T. Stefanello, G. G. Rossi and M. M. A. D. Campos, Microb. Pathog., 2016, 99, 229–235 CAS.
- T. T. Aung, W. H. J. Chor, J. K. H. Yam, M. Givskov, L. Yang and R. W. Beuerman, Ocul. Surf., 2017, 15, 770–783 CrossRef PubMed.
- J. Luo, X. Yu, G. Jiang, Y. Fu, F. Huo, Y. Ma, F. Wang, Y. Shang, Q. Liang, Y. Xue and H. Huang, Antimicrob. Agents Chemother., 2018, 62, e00072-18 CrossRef PubMed.
- S. Das, T. Garg, S. Chopra and A. Dasgupta, J. Antimicrob. Chemother., 2019, 74, 1317–1322 CrossRef CAS PubMed.
- M. Stavri, R. Schneider, G. O'Donnell, D. Lechner, F. Bucar and S. Gibbons, Phytother. Res., 2004, 18, 774–776 CrossRef CAS PubMed.
- A. A. Wube, F. Bucar, S. Gibbons and K. Asres, Phytochemistry, 2005, 66, 2309–2315 CrossRef CAS.
- M. Stavri, K. T. Mathew and S. Gibbons, Phytochemistry, 2006, 67, 1530–1533 CrossRef CAS.
- G. O'Donnell and S. Gibbons, Phytother. Res., 2007, 21, 653–657 CrossRef.
- P. Santos, L. Busta, W. C. Yim, E. B. Cahoon and D. K. Kosma, J. Exp. Bot., 2022, 73, 2889–2904 CrossRef CAS.
- M. Stavri and S. Gibbons, Phytother. Res., 2005, 19, 938–941 CrossRef CAS.
- A. Schinkovitz, M. Stavri, S. Gibbons and F. Bucar, Phytother. Res., 2008, 22, 681–684 CrossRef CAS.
- E. Haulotte, P. Laurent and J. Braekman, Eur. J. Org. Chem., 2012, 2012, 1907–1912 CrossRef CAS.
- C. R. Röhrich, C. J. Ngwa, J. Wiesner, H. Schmidtberg, T. Degenkolb, C. Kollewe, R. Fischer, G. Pradel and A. Vilcinskas, Biol. Lett., 2012, 8, 308–311 CrossRef PubMed.
- C. J. Zheng, J.-S. Yoo, T.-G. Lee, H.-Y. Cho, Y.-H. Kim and W.-G. Kim, FEBS Lett., 2005, 579, 5157–5162 CrossRef CAS.
- Y. Zhang, T. Garbe and D. Young, Mol. Microbiol., 1993, 8, 521–524 CrossRef CAS PubMed.
- M. Adams, A. Wube, F. Bucar, R. Bauer, O. Kunert and E. Haslinger, Int. J. Antimicrob. Agents, 2005, 26, 262–264 CrossRef CAS.
- J. D. Guzman, A. Wube, D. Evangelopoulos, A. Gupta, A. Hüfner, C. Basavannacharya, Md. M. Rahman, C. Thomaschitz, R. Bauer, T. D. McHugh, I. Nobeli, J. M. Prieto, S. Gibbons, F. Bucar and S. Bhakta, J. Antimicrob. Chemother., 2011, 66, 1766–1772 CrossRef CAS.
- A. A. Wube, F. Bucar, C. Hochfellner, M. Blunder, R. Bauer and A. Hüfner, Eur. J. Med. Chem., 2011, 46, 2091–2101 CrossRef CAS PubMed.
- A. A. Wube, A. Hüfner, C. Thomaschitz, M. Blunder, M. Kollroser, R. Bauer and F. Bucar, Bioorg. Med. Chem., 2011, 19, 567–579 CrossRef CAS PubMed.
- C. Basavannacharya, G. Robertson, T. Munshi, N. H. Keep and S. Bhakta, Tuberculosis, 2010, 90, 16–24 CrossRef CAS PubMed.
- V. Hervin, V. Roy and L. A. Agrofoglio, Molecules, 2023, 28, 8076 CrossRef CAS PubMed.
- J. Adamec, R. Beckert, D. Weiß, V. Klimešová, K. Waisser, U. Möllmann, J. Kaustová and V. Buchta, Bioorg. Med. Chem., 2007, 15, 2898–2906 CrossRef CAS.
- C. Rodrigues Felix, J. C. Roberts, P. L. Winder, R. Gupta, M. C. Diaz, S. A. Pomponi, A. E. Wright and K. H. Rohde, Mar. Drugs, 2019, 17, 707 Search PubMed.
- S. García-Davis, K. Leal-López, C. A. Molina-Torres, L. Vera-Cabrera, A. R. Díaz-Marrero, J. J. Fernández, P. Carranza-Rosales and E. Viveros-Valdez, Mar. Drugs, 2020, 18, 287 Search PubMed.
- A. O. Pereira, J. M. Avila, G. Do Carmo, F. S. Siqueira, M. M. A. Campos, D. F. Back, A. F. Morel and I. I. Dalcol, Ind. Crops Prod., 2018, 121, 461–467 CAS.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef.
- A. Y. Gordien, A. I. Gray, S. G. Franzblau and V. Seidel, J. Ethnopharmacol., 2009, 126, 500–505 CrossRef CAS PubMed.
- M. Stavri, A. Paton, B. W. Skelton and S. Gibbons, J. Nat. Prod., 2009, 72, 1191–1194 CrossRef CAS.
- D. R. Katerere, A. I. Gray, R. J. Nash and R. D. Waigh, Phytochemistry, 2003, 63, 81–88 CrossRef CAS.
- R. R. Miller, M. Madeira, H. B. Wood, W. M. Geissler, C. E. Raab and I. J. Martin, J. Med. Chem., 2020, 63, 12156–12170 CrossRef CAS.
- G. Appendino, E. Mercalli, N. Fuzzati, L. Arnoldi, M. Stavri, S. Gibbons, M. Ballero and A. Maxia, J. Nat. Prod., 2004, 67, 2108–2110 CAS.
- A. Schinkovitz, S. Gibbons, M. Stavri, M. J. Cocksedge and F. Bucar, Planta Med., 2003, 69, 369–371 CAS.
- M. N. Protopopova, L. Einck, B. Nikonenko and P. Chen, US2009/0192173A1, Sequella, Inc., 2009.
- N. Heinrich, R. Dawson, J. Du Bois, K. Narunsky, G. Horwith, A. J. Phipps, C. A. Nacy, R. E. Aarnoutse, M. J. Boeree, S. H. Gillespie, A. Venter, S. Henne, A. Rachow, P. P. J. Phillips, M. Hoelscher, A. H. Diacon, On behalf of the Pan African Consortium for the Evaluation of Antituberculosis Antibiotics (PanACEA), A. M. Mekota, N. Heinrich, A. Rachow, E. Saathoff, M. Hoelscher, S. Gillespie, A. Colbers, G. P. Van Balen, R. Aarnoutse, M. Boeree, A. Bateson, T. McHugh, K. Singh, R. Hunt, A. Zumla, A. Nunn, P. Phillips, S. Rehal, R. Dawson, K. Narunsky, A. Diacon, J. Du Bois, A. Venter, S. Friedrich, I. Sanne, K. Mellet, G. Churchyard, S. Charalambous, P. Mwaba, N. Elias, C. Mangu, G. Rojas-Ponce, B. Mtafya, L. Maboko, L. T. Minja, M. Sasamalo, K. Reither, L. Jugheli, N. Sam, G. Kibiki, H. Semvua, S. Mpagama, A. Alabi, A. A. Adegnika, E. Amukoye and A. Okwera, J. Antimicrob. Chemother., 2015, 70, 1558–1566 CAS.
- M. J. Boeree, N. Heinrich, R. Aarnoutse, A. H. Diacon, R. Dawson, S. Rehal, G. S. Kibiki, G. Churchyard, I. Sanne, N. E. Ntinginya, L. T. Minja, R. D. Hunt, S. Charalambous, M. Hanekom, H. H. Semvua, S. G. Mpagama, C. Manyama, B. Mtafya, K. Reither, R. S. Wallis, A. Venter, K. Narunsky, A. Mekota, S. Henne, A. Colbers, G. P. Van Balen, S. H. Gillespie, P. P. J. Phillips and M. Hoelscher, Lancet Infect. Dis., 2017, 17, 39–49 CAS.
- S. E. Borisov, E. M. Bogorodskaya, G. V. Volchenkov, E. V. Kulchavenya, A. O. Maryandyshev, S. N. Skornyakov, O. B. Talibov, A. M. Tikhonov and I. A. Vasilyeva, Tuberculosis and Lung Diseases, 2018, 96, 6–18 CrossRef.
- K. Tahlan, R. Wilson, D. B. Kastrinsky, K. Arora, V. Nair, E. Fischer, S. W. Barnes, J. R. Walker, D. Alland, C. E. Barry and H. I. Boshoff, Antimicrob. Agents Chemother., 2012, 56, 1797–1809 CrossRef CAS PubMed.
- K. A. Sacksteder, M. Protopopova, C. E. Barry, K. Andries and C. A. Nacy, Future Microbiol., 2012, 7, 823–837 CrossRef CAS PubMed.
- M. Singh, S. Kumar, B. Singh, P. Jain, A. Kumari, I. Pahuja, S. Chaturvedi, D. V. R. Prasad, V. P. Dwivedi and G. Das, Commun. Biol., 2022, 5, 759 CrossRef CAS PubMed.
- K. Ecevit, A. A. Barros, J. M. Silva and R. L. Reis, Future Pharmacol., 2022, 2, 460–498 CrossRef.
- A. C. Kauffmann and V. S. Castro, Antibiotics, 2023, 12, 645 CrossRef.
- B. R. Albuquerque, S. A. Heleno, M. B. P. P. Oliveira, L. Barros and I. C. F. R. Ferreira, Food Funct., 2021, 12, 14–29 CAS.
- Á. Fernández-Ochoa, M. D. L. L. Cádiz-Gurrea, P. Fernández-Moreno, A. Rojas-García, D. Arráez-Román and A. Segura-Carretero, Molecules, 2022, 27, 777 Search PubMed.
- D. J. Alvarenga, L. M. F. Matias, L. M. Oliveira, L. P. M. O. Leão, J. A. Hawkes, B. V. B. Raimundo, L. D. F. D. Castro, M. M. A. D. Campos, F. D. S. Siqueira, T. D. Santos and D. T. Carvalho, Microb. Pathog., 2020, 144, 104203 Search PubMed.
- R. León-Díaz, M. Meckes, S. Said-Fernández, G. M. Molina-Salinas, J. Vargas-Villarreal, J. Torres, J. Luna-Herrera and A. Jiménez-Arellanes, Mem. Inst. Oswaldo Cruz, 2010, 105, 45–51 Search PubMed.
- L. Han, X. Huang, H.-M. Dahse, U. Moellmann, H. Fu, S. Grabley, I. Sattler and W. Lin, J. Nat. Prod., 2007, 70, 923–927 Search PubMed.
- L. J. McGaw, N. Lall, T. M. Hlokwe, A. L. Michel, J. J. M. Meyer and J. N. Eloff, Biol. Pharm. Bull., 2008, 31, 1429–1433 CAS.
- J. L. D. Silva, A. R. Mesquita and E. A. Ximenes, Mem. Inst. Oswaldo Cruz, 2009, 104, 580–582 Search PubMed.
- K. J. Weissman and P. F. Leadlay, Nat. Rev. Microbiol., 2005, 3, 925–936 CAS.
- S. Pidot, K. Ishida, M. Cyrulies and C. Hertweck, Angew. Chem., Int. Ed., 2014, 53, 7856–7859 CAS.
- D. F. Ramos and P. E. Almeida Da Silva, Pharm. Biol., 2010, 48, 260–263 CAS.
- D. R. Katerere, A. I. Gray, R. J. Nash and R. D. Waigh, Fitoterapia, 2012, 83, 932–940 CrossRef CAS.
- M. A. Aderogba, B. Madikizela and L. J. McGaw, S. Afr. J. Bot., 2019, 126, 371–376 CrossRef CAS.
- D. Lechner, S. Gibbons and F. Bucar, Phytochem. Lett., 2008, 1, 71–75 CrossRef CAS.
- T. T. Aung, J. K. H. Yam, S. Lin, S. M. Salleh, M. Givskov, S. Liu, N. C. Lwin, L. Yang and R. W. Beuerman, Antimicrob. Agents Chemother., 2016, 60, 24–35 CrossRef CAS PubMed.
- T. E. Ramadwa, M. D. Awouafack, M. S. Sonopo and J. N. Eloff, Nat. Prod. Commun., 2019, 14, 1–7 CrossRef.
- A. T. Tran, E. E. Watson, V. Pujari, T. Conroy, L. J. Dowman, A. M. Giltrap, A. Pang, W. R. Wong, R. G. Linington, S. Mahapatra, J. Saunders, S. A. Charman, N. P. West, T. D. H. Bugg, J. Tod, C. G. Dowson, D. I. Roper, D. C. Crick, W. J. Britton and R. J. Payne, Nat. Commun., 2017, 8, 14414 CrossRef.
- W. Tran, A. S. Kusay, P. M. E. Hawkins, C.-Y. Cheung, G. Nagalingam, V. Pujari, D. J. Ford, A. Stoye, J. L. Ochoa, R. E. Audette, E. Hortle, S. H. Oehlers, S. A. Charman, R. G. Linington, E. J. Rubin, C. G. Dowson, D. I. Roper, D. C. Crick, T. Balle, G. M. Cook, W. J. Britton and R. J. Payne, J. Med. Chem., 2021, 64, 17326–17345 CrossRef CAS.
- A. I. Khalaf, A. H. Ebrahimabadi, A. J. Drummond, N. G. Anthony, S. P. Mackay, C. J. Suckling and R. D. Waigh, Org. Biomol. Chem., 2004, 2, 3119 RSC.
- S. Gibbons, F. Fallah and C. W. Wright, Phytother. Res., 2003, 17, 434–436 CAS.
- M. Arai, M. Sobou, C. Vilchéze, A. Baughn, H. Hashizume, P. Pruksakorn, S. Ishida, M. Matsumoto, W. R. Jacobs and M. Kobayashi, Bioorg. Med. Chem., 2008, 16, 6732–6736 CAS.
- K. Puk and L. Guz, Pol. J. Vet. Sci., 2022, 479–481 CAS.
- F. Giordanetto, J. Boström and C. Tyrchan, Drug Discovery Today, 2011, 16, 722–732 Search PubMed.
- M. L. Lanthier, K. W. Kerr and K. L. Miller, Clin. Pharm. Ther., 2019, 106, 1125–1132 Search PubMed.
- L. Vera-Cabrera, B. A. Brown-Elliott, R. J. Wallace, J. Ocampo-Candiani, O. Welsh, S. H. Choi and C. A. Molina-Torres, Antimicrob. Agents Chemother., 2006, 50, 4027–4029 CAS.
- W. Zhao, Y. Jiang, P. Bao, Y. Li, L. Tang, Y. Zhou and Y. Zhao, Jpn. J. Infect. Dis., 2015, 68, 520–522 CAS.
- R. Zhu, Y. Shang, S. Chen, H. Xiao, R. Ren, F. Wang, Y. Xue, L. Li, Y. Li, N. Chu and H. Huang, Microbiol. Spectrum, 2022, 10, e01372-22 CrossRef.
- X. Yang, C. Li, X. Wang, Z. Zheng, P. Sun, C. Xu, L. Chen, J. Jiang, S. Normark, B. Henriques-Normark and X. You, Engineering, 2024, 38, 52–68 CrossRef CAS.
- C. Yu, H. Qian, Q. Wu, Y. Zou, Q. Ding, Y. Cai, L. Liang, J. Xu, L. Li, B. Zan, Y. Li and Y. Liu, Clin. Transl. Sci., 2024, 17, e13718 CrossRef CAS PubMed.
- P. Ragunathan, P. Sae-Lao, C. Hamela, M. Alcaraz, A. Krah, W. H. Poh, C. J. Ern Pee, A. Y. Hou Lim, S. A. Rice, K. Pethe, P. J. Bond, T. Dick, L. Kremer, R. W. Bates and G. Grüber, J. Biol. Chem., 2024, 300, 105618 CrossRef CAS PubMed.
- M. Cynamon, J. Jureller, B. Desai, K. Ramachandran, M. Sklaney and T. H. Grossman, Antimicrob. Agents Chemother., 2012, 56, 3986–3988 CrossRef CAS.
- J. Sellarès-Nadal, J. Burgos, V. Falcó and B. Almirante, J. Exp. Pharmacol., 2020, 12, 529–538 CrossRef.
- K. D. Combrink, A. R. Ramos, S. Spring, S. Schmidl, K. Elizondo, P. Morin, B. De Jesus and F. P. Maurer, Bioorg. Med. Chem. Lett., 2019, 29, 2112–2115 CrossRef CAS PubMed.
- Rakesh, D. F. Bruhn, M. S. Scherman, L. K. Woolhiser, D. B. Madhura, M. M. Maddox, A. P. Singh, R. B. Lee, J. G. Hurdle, M. R. McNeil, A. J. Lenaerts, B. Meibohm and R. E. Lee, PLoS One, 2014, 9, e87909 CrossRef CAS PubMed.
- J. Xu, B. Wang, L. Fu, H. Zhu, S. Guo, H. Huang, D. Yin, Y. Zhang and Y. Lu, Antimicrob. Agents Chemother., 2019, 63, e02155-18 CrossRef.
- A. Disratthakit and N. Doi, Antimicrob. Agents Chemother., 2010, 54, 2684–2686 CrossRef CAS PubMed.
- T. E. Renau, J. W. Gage, J. A. Dever, G. E. Roland, E. T. Joannides, M. A. Shapiro, J. P. Sanchez, S. J. Gracheck, J. M. Domagala, M. R. Jacobs and R. C. Reynolds, Antimicrob. Agents Chemother., 1996, 40, 2363–2368 CrossRef CAS.
- M. C. Bagchi, D. Mills and S. C. Basak, J. Mol. Model., 2006, 13, 111–120 CrossRef PubMed.
- C. A. Molina-Torres, J. Ocampo-Candiani, A. Rendón, M. J. Pucci and L. Vera-Cabrera, Antimicrob. Agents Chemother., 2010, 54, 2188–2190 CrossRef CAS PubMed.
- K. A. Pardeshi, T. A. Kumar, G. Ravikumar, M. Shukla, G. Kaul, S. Chopra and H. Chakrapani, Bioconjugate Chem., 2019, 30, 751–759 CrossRef CAS.
- D. J. Owen, C. B. Davis, R. D. Hartnell, P. D. Madge, R. J. Thomson, A. K. J. Chong, R. L. Coppel and M. V. Itzstein, Bioorg. Med. Chem. Lett., 2007, 17, 2274–2277 CrossRef CAS PubMed.
- D. F. Ramos, G. Fiss, C. P. Frizzo, M. A. P. Martins, H. G. Bonacorso, N. Zanatta and P. E. Almeida Da Silva, Int. J. Antimicrob. Agents, 2014, 43, 481–483 CAS.
- P. Ploszaj, A. Junka, B. Szponar, M. Maczynski, S. Ryng, M. Bartoszewicz and A. Piwowar, Acta Pol. Pharm., 2018, 75, 637–647 CAS.
- S. K. Arora, N. Sinha, S. Jain, R. S. Upadhayaya, G. Jana, S. Ajay and R. K. Sinha, WO2004/026828A1, 2004.
- K. B. Guterres, G. G. Rossi, L. B. Menezes, M. M. Anraku De Campos and B. A. Iglesias, Tuberculosis, 2019, 117, 45–51 CAS.
- H. Parveen, M. Y. Wani, S. Mukhtar and A. Ahmad, J. Mol. Struct., 2024, 1311, 138472 CAS.
- V. Kr. Kashyap, R. Kr. Gupta, R. Shrivastava, B. S. Srivastava, R. Srivastava, M. K. Parai, P. Singh, S. Bera and G. Panda, J. Antimicrob. Chemother., 2012, 67, 1188–1197 CAS.
- P. K. Ranjith, R. Pakkath, K. R. Haridas and S. N. Kumari, Eur. J. Med. Chem., 2014, 71, 354–365 CAS.
- K. D. Thomas, A. V. Adhikari, I. H. Chowdhury, E. Sumesh and N. K. Pal, Eur. J. Med. Chem., 2011, 46, 2503–2512 CrossRef CAS PubMed.
- S. Eswaran, A. V. Adhikari, N. K. Pal and I. H. Chowdhury, Bioorg. Med. Chem. Lett., 2010, 20, 1040–1044 CAS.
- R. G. Glushkov, RU2209067C1, 2003.
- I. Palos, J. Luna-Herrera, E. Lara-Ramírez, A. Loera-Piedra, E. Fernández-Ramírez, Ma. Aguilera-Arreola, A. Paz-González, A. Monge, B. Wan, S. Franzblau and G. Rivera, Molecules, 2018, 23, 1453 Search PubMed.
- V. Makarov and K. Mikušová, Appl. Sci., 2020, 10, 2269 CAS.
- J. Shi, J. Lu, S. Wen, Z. Zong, F. Huo, J. Luo, Q. Liang, Y. Li, H. Huang and Y. Pang, Antimicrob. Agents Chemother., 2018, 62, e01314-18 CrossRef.
- L. Zheng, X. Qi, W. Zhang, H. Wang, L. Fu, B. Wang, X. Chen, X. Chen and Y. Lu, Front. Cell. Infect. Microbiol., 2023, 13, 1115530 CrossRef CAS PubMed.
- M. Imran, S. A. Khan, S. M. B. Asdaq, M. Almehmadi, O. Abdulaziz, M. Kamal, M. K. Alshammari, L. I. Alsubaihi, K. H. Hussain, A. S. Alharbi and A. K. Alzahrani, J. Infect. Public Health, 2022, 15, 1097–1107 CrossRef PubMed.
- B. Kappes, T. Neumann, E. Unger, H.-M. Dahse, U. Möllmann and B. Schlegel, DE10158057A1, 2003.
- V. Makarov, J. Antimicrob. Chemother., 2006, 57, 1134–1138 CrossRef CAS.
- A. Babu, K. Sunil, A. M. Sajith, E. K. Reddy, S. Santra, G. V. Zyryanov, T. Venkatesh, S. Bhadrachari and M. Nibin Joy, Pharmaceuticals, 2024, 17, 548 CrossRef CAS PubMed.
- K. Waisser, L. Heinisch, M. Šlosárek and J. Janota, Folia Microbiol., 2006, 51, 25 CrossRef CAS.
- W. R. Schwan, M. Kallaus, S. Krueger, A. Monte, M. S. Kabir and J. M. Cook, J. Infect. Chemother., 2012, 18, 124–126 CrossRef CAS PubMed.
- J. B. Moreira, J. Mann, S. Neidle, T. D. McHugh and P. W. Taylor, Int. J. Antimicrob. Agents, 2013, 42, 361–366 Search PubMed.
- N. Kurepina, B. N. Kreiswirth and A. Mustaev, J. Appl. Microbiol., 2013, 115, 943–954 CrossRef CAS PubMed.
- V. V. N. P. B. Tiruveedhula, C. M. Witzigmann, R. Verma, M. S. Kabir, M. Rott, W. R. Schwan, S. Medina-Bielski, M. Lane, W. Close, R. L. Polanowski, D. Sherman, A. Monte, J. R. Deschamps and J. M. Cook, Bioorg. Med. Chem., 2013, 21, 7830–7840 Search PubMed.
- A. Mustaev and N. Kurepina, US8859798B2, Rutgers, The State University of New Jersey, 2014.
- A. K. Singh, R. Thakare, P. Karaulia, S. Das, I. Soni, M. Pandey, A. K. Pandey, S. Chopra and A. Dasgupta, J. Antimicrob. Chemother., 2017, 72, 3117–3121 CrossRef CAS PubMed.
- A. S. M. Ali and C. S. Anderson, Lancet, 2024, 403, 702–703 CrossRef CAS.
- M. N. Ahmad, T. Garg, S. Singh, R. Shukla, P. Malik, R. V. Krishnamurthy, P. Kaur, S. Chopra and A. Dasgupta, Antimicrob. Agents Chemother., 2022, 66, e00564-22 CrossRef.
- B. A. Brown-Elliott, G. Bush, M. D. Hughes, E. Rodriguez, C. A. Weikel, S. B. Min and R. J. Wallace, Antimicrob. Agents Chemother., 2024, e01684-23 CrossRef.
- U. H. Manjunatha, S. P. S. Rao, R. R. Kondreddi, C. G. Noble, L. R. Camacho, B. H. Tan, S. H. Ng, P. S. Ng, N. L. Ma, S. B. Lakshminarayana, M. Herve, S. W. Barnes, W. Yu, K. Kuhen, F. Blasco, D. Beer, J. R. Walker, P. J. Tonge, R. Glynne, P. W. Smith and T. T. Diagana, Sci. Transl. Med., 2015, 7, 269ra3 Search PubMed.
- F. Roquet-Banères, M. Alcaraz, C. Hamela, J. Abendroth, T. E. Edwards and L. Kremer, Antimicrob. Agents Chemother., 2023, 67, e01607-22 CrossRef PubMed.
- K. C. Costa, N. R. Glasser, S. J. Conway and D. K. Newman, Science, 2017, 355, 170–173 CrossRef CAS.
- H. Marrakchi, M.-A. Lanéelle and M. Daffé, Chem. Biol., 2014, 21, 67–85 CrossRef CAS PubMed.
- A. Sharma, J. Vashistt and R. Shrivastava, Int. J. Mycobact., 2022, 11, 159 CrossRef CAS PubMed.
- A. Sharma, J. Vashistt and R. Shrivastava, J. Basic Microbiol., 2022, 62, 1504–1513 CrossRef CAS PubMed.
- N. Van Wyk, M. Drancourt, B. Henrissat and L. Kremer, Glycobiology, 2017, 27, 112–122 CrossRef CAS PubMed.
- J. E. S. Nunes, M. A. Duque, T. F. De Freitas, L. Galina, L. F. S. M. Timmers, C. V. Bizarro, P. Machado, L. A. Basso and R. G. Ducati, Molecules, 2020, 25, 1259 CrossRef CAS.
- M. J. Nasiri, M. Haeili, M. Ghazi, H. Goudarzi, A. Pormohammad, A. A. Imani Fooladi and M. M. Feizabadi, Front. Microbiol., 2017, 8, 681 CrossRef PubMed.
- E. Sauvage, E. Fonzé, B. Quinting, M. Galleni, J.-M. Frère and P. Charlier, Antimicrob. Agents Chemother., 2006, 50, 2516–2521 CrossRef CAS.
- M. C. Menéndez, J. A. Ainsa, C. Martín and M. J. García, J. Bacteriol., 1997, 179, 6880–6886 CrossRef.
- S. Sharbati, K. Schramm, S. Rempel, H. Wang, R. Andrich, V. Tykiel, R. Kunisch and A. Lewin, BMC Microbiol., 2009, 9, 31 CrossRef.
- A. Ilinov, A. Nishiyama, H. Namba, Y. Fukushima, H. Takihara, C. Nakajima, A. Savitskaya, G. Gebretsadik, M. Hakamata, Y. Ozeki, Y. Tateishi, S. Okuda, Y. Suzuki, Y. S. Vinnik and S. Matsumoto, Sci. Rep., 2021, 11, 10953 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4md00508b |
| This journal is © The Royal Society of Chemistry 2025 |