Open Access Article
Open Access ArticleA novel DNA sequence-selective, guanine mono-alkylating ADC payload suitable for solid tumour treatment†
Paolo Andriolloa,
Daniella di Masciob,
Paul J. M. Jacksona,
Md. Mahbub Hasan ac,
Ilona Pysz-Hoseya,
George Procopiou
ac,
Ilona Pysz-Hoseya,
George Procopiou a,
Keith R. Fox
a,
Keith R. Fox b,
Khondaker Miraz Rahman
b,
Khondaker Miraz Rahman *a and
David E. Thurston
*a and
David E. Thurston a
a
aSchool of Cancer & Pharmaceutical Sciences, King's College London, WC2R 2LS, UK. E-mail: k.miraz.rahman@kcl.ac.uk; Tel: +44 (0)20 7848 1891
bSchool of Biological Sciences, University of Southampton, SO17 1BJ, UK
cDepartment of Genetic Engineering and Biotechnology, Faculty of Biological Sciences, University of Chittagong, Chattogram, Bangladesh
First published on 11th March 2025
Abstract
Pyridinobenzodiazepines (PDDs) are a new class of DNA mono-alkylating antibody–drug conjugate (ADC) payloads that can be linked through their C9 position to a sequence recognition component, guiding them to specific DNA sequences. Compound 18 is a PDD monomer with a unique sequence-selectivity profile and high cytotoxicity in vitro (e.g., IC50 = 0.30 nM in SW60; 1.6 nM in LIM1215 and 0.142 nM in SW48 cell line, after 96 hours incubation). To evaluate its potential as an ADC payload, an amine functionality was introduced into the terminal phenyl ring, and the modified compound was conjugated to trastuzumab (drug–antibody ratio [DAR] = 1.6). The resulting ADC exhibits significant in vivo efficacy in a pancreatic cancer xenograft model using BALB-c mice transplanted with the CAPAN-1 cell line. Complete tumour regression is observed out to 60 days after a single dose of 2 mg kg−1 comparing favourably to a 10 mg kg−1 dose of trastuzumab deruxtecan (Enhertu®). The novel ADC has a good tolerability profile, with a maximum tolerated dose (MTD) above 15 mg kg−1. The tolerability and efficacy profile of compound 18 in an ADC format suggests that PDDs represent a potentially valuable new class of payloads for the treatment of solid tumours.
Introduction
Antibody–drug conjugates (ADCs) are a rapidly growing class of therapeutic agents used for the treatment of different cancer types. At the time of writing, fifteen ADCs have been approved by the FDA. These include belantamab mafodotin, which was withdrawn in 2022, as well as the two immunotoxins moxetumomab pasudotox (Lumoxiti®) and tagraxofusp (Elzonris®).1–6 Recent approvals have highlighted the expanding role of ADCs in both solid and haematologic malignancies, with several new ADCs expected to reach the market in the coming years as the field continues to advance rapidly. ADCs are designed to couple the efficacy of certain cytotoxic agents in triggering cell death with the high selectivity of monoclonal antibodies and are composed of three parts: a monoclonal antibody (mAb) targeted to a cancer-specific antigen, a cytotoxic small molecule (i.e., a payload), and a linker which joins these two moieties together.7,8 The two primary mechanistic families of payloads used in ADCs are microtubule inhibitors and DNA-interactive agents. Within the latter family, the pyrrolobenzodiazepines (PBDs) are a class of molecules that exert their cytotoxicity through covalent bonding with the C2-NH2 of guanine in the minor groove of DNA. Since the isolation of anthramycin, the first natural product PBD to be discovered (from the Streptomyces and Micrococci bacteria in the early 1960s),9 many other naturally-occurring and synthetic PBD compounds have been developed. These include synthetic PBD dimers that consist of two PBD units joined together via a linker and capable of cross-linking DNA.10,11SGD-1882 and SG3199 are two examples of PBD dimers (Fig. 1) which have been conjugated to a number of antibodies and have been evaluated in several cancer types in the clinic.7 In particular, tesirine is the linker-payload used in loncastuximab tesirine (Zynlonta®), which was approved by the FDA in early 2021 for the treatment of refractory and relapsed diffuse large B-cell lymphoma.12 There are several clinical-stage ADCs that are using pyrrolobenzodiazepines as their payloads.13,14
 | ||
| Fig. 1 Structures of the PBD dimers SGD-1882 (A), SG3199 (B), the C8-conjugate PBD monomers KMR-28-39 (C) and the C9-conjugated PDD monomer 18 (D). | ||
Building on the success of PBDs, a novel class of molecules—pyridinobenzodiazepines (PDDs)—has been developed by expanding the five-membered C-ring of the PBD into a six-membered ring (Fig. 2a).15 This structural modification is thought to enhance the flexibility of the PDD core, while maintaining the snug fitting within the DNA minor groove (Fig. 2b).
The six-membered C-ring also facilitates additional van der Waals interactions with the walls of the DNA minor groove which contributes to enhancing the stability of the drug–DNA complex. These enhancements in DNA-binding affinity and stability suggest that PDDs may offer comparable efficacy to PBDs as ADC payloads.15 The novel chemical structure and functionality of PDDs provide a new scaffold for ADC payloads, offering an alternative to the successful PBD platform to optimise DNA-based ADC payloads.
Over the years, guanine mono-alkylating PBD monomers have been extensively studied. C8-conjugated PBD–polyamide compounds, such as KMR-28-39 (Fig. 1), have demonstrated average cytotoxicity in the low picomolar (pM) range and exhibit a strong preference for specific A/T and G/C base pair combinations.16 These molecules consist of two main components connected by a trimethylene spacer: a PBD core, responsible for DNA alkylation, and a C8-linked heterocyclic polyamide tail, which provides DNA sequence-selectivity and stabilises the adduct through non-covalent interactions with DNA base pairs.17
Polyamide structures based on the minor-groove binding agent distamycin scaffold have been extensively studied, and a large number of these lexitropsins have been obtained using a range of heterocycles.18–21 Such structures are known to bind non-covalently within the minor groove of DNA, and different heterocycles have been observed to produce marked differences in sequence selectivity.22
In this study, the PDD core has been linked through its C9-position (Fig. 1) to a heterocyclic polyamide side-chain consisting of four heterocycle units so that the entire compound 18 (a first generation high potency PDD monomer) spans up to 8–9 base pairs as shown in Fig. 3A and S9.† This was used as a basis to design the PDD monomers used in this study, with a C9-side chain containing heteroatoms potentially capable of recognising and binding non-covalently to mixed regions of DNA.17 From an ADC payload perspective, in a similar way to PBD monomers, the primary mechanism of action driving cell death remains DNA alkylation. However, with the addition of the C9 sequence-selective side chain, this payload also achieves cytotoxicity by inhibiting transcription factors, a secondary mechanism established for C8-linked PBD monomers.23 Here, we report for the first time the synthesis, in vitro cytotoxicity, antibody conjugation, and in vivo efficacy of an ADC containing a first generation high potency PDD-based payload.
 | ||
| Fig. 3 (A) Molecular docking representation of a PDD monomer18 containing an MTP building block and four heterocyclic units that fit snugly within the DNA minor groove, spanning 8–9 base pairs. (B) Interaction of the MTP building block with DNA bases within the DNA minor groove, showing H-bond, π-sigma, and π-sulfur interactions. (C) Structures of compound 18 and (D) compound 19. | ||
Results and discussion
Payload design and chemistry
The design of the C9-side chain was inspired by the C8-side chain in KMR-28-39 (Fig. 1), which demonstrated femtomolar cytotoxicity against a panel of human tumour cell lines and exhibited the ability to inhibit the NFκB transcription factor which contributes to its cytotoxicity in addition to DNA alkylation.16 The GC-targeting biaryl methyl phenyl benzenamine (MPB) building block in KMR-28-39 was replaced with a 2-(4-methylthiophen-2-yl)pyridine (MTP) fragment. 2-(Thiophen-2-yl)pyridine containing structures are known to form an intramolecular sigma-hole bond between the atom of sulfur and the nitrogen of the pyridine. This is a non-covalent interaction between the lone pair on the nitrogen and a region of partial positive charge originating from the sulfur atom along the extension of the sigma bonds. This electrostatic interaction forces nitrogen and sulfur atoms into a cis-orientation.24 The presence of a methyl group on the thiophene would then direct both heteroatoms towards the inside of the minor groove. There, molecular modelling suggested that the hydrogen bond-accepting nitrogen in the MTP fragment may form hydrogen-bond interactions with guanine bases, while the building block may also engage in π-sigma and π-sulfur interactions with DNA bases (Fig. 3B). Unlike guanine, AT bases lack an amino group protruding into the DNA minor groove, resulting in reduced steric hindrance for the bulky sulfur atom to accommodate. This characteristic enables the resulting molecule to target GC-AT mixed transcription factor binding sequences, such as those found in the NFκB transcription factor. A C9-linked PDD monomer containing the MTP fragment, compound 18 (Fig. 3C), was designed to evaluate the cytotoxicity and suitability of this type of PDD monomer as an ADC payload. For this purpose, an amine functionality was introduced (compound 19, Fig. 3D) in the terminal phenyl ring to enable connection to a suitable linker that could be joined to an appropriate antibody to form an ADC.The N-Alloc/O-THP-protected PDD core (9) was synthesised by adapting a previously-described method for the synthesis of structurally-related PBD molecules (Scheme 1).25
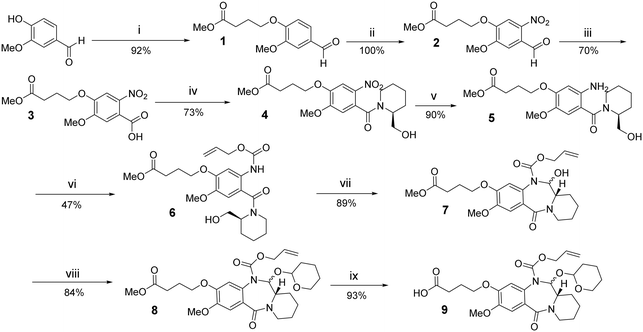
Vanillin was coupled with methyl 4-bromobutyrate (i) in a basic environment to give the ester 1 which was then selectively nitrated using KNO3/TFA to give the nitro benzaldehyde 2. This intermediate was oxidised to the corresponding carboxylic acid 3 with potassium permanganate. The intermediate 4 was obtained by amide coupling of 3 and optically pure (S)-piperidine methanol. Oxalyl chloride was used to activate the carboxylic acid to the corresponding acyl chloride in order to promote coupling in good yield (73%). Palladium(0) on carbon (10%) was used as catalyst in the presence of hydrogen gas (3 atm), to reduce the nitro compound 4 to the corresponding amine, 5. The starting material converted completely to the corresponding aniline that was subsequently Alloc-protected using allyl chloroformate in the presence of pyridine to give 6 after purification by column chromatography. The Alloc-protected PDD ring system 7 was obtained via tandem oxidation/cyclisation, using PIDA/TEMPO. In order to prevent racemisation at the C12a-position of the PDD ring system which might occur under basic conditions, the hydroxide of carbinolamine 7 was protected by introduction of the THP group. This protection was achieved using DHP in the presence of a catalytic amount of pTsOH, with conversion of the starting material to 8 in good yield (84%). The final N-Alloc/O-THP-protected PDD core 9 was obtained by hydrolysis of the methyl ester under basic conditions, using aqueous NaOH (1 M). The overall yield was 14%, and this synthetic sequence was used to prepare up to 20 g of 9 (Scheme 1).
Compound 10 was obtained via N-Boc-protection of the amino group of 5-aminopyridin-2-ol, with a 37% yield. This was then converted into the triflate derivative, 11 using bis(trifluoromethanesulphonyl)aniline as triflating agent. The methyl-thiophene pyridine (MTP) structure (12), comprising of the second and third ring of the payload, was obtained via palladium-catalysed Suzuki cross-coupling between 11 and the boronic acid of methyl thiophene. N-Boc deprotection was achieved by treating compound 12 with HCl in 1,4-dioxane, and the resulting free amine was then reacted with N-methyl pyrrole derivative carboxylic acid, representing the first heterocyclic ring of the C9 side chain. This amide coupling was carried out using EDCI/DMAP and gave 13 in a 71% yield. The amino group of 13 was deprotected in a similar manner and then reacted with the carboxylic acid of the PDD unit 9 via amide coupling to afford 14 as a mixture of C12a diastereomers. Enantiomeric excess was not determined at this stage as the chiral centre in position C12a is lost during the final imine deprotection leading to enantiomerically pure final compounds. The methyl ester of 14 was hydrolysed at room temperature under alkaline conditions to afford the corresponding carboxylic acid, 15 in good yield (96%). A third amide coupling reaction was then carried out between 15 and the terminal aniline. Aniline was reacted with 15 to provide 16 while a benzene-1,4-diamine was used to obtain 17. Finally, the protected compounds, 16 and 17 were subjected to Alloc-deprotection conditions with pyrrolidine and tetrakis(triphenylphosphine)palladium(0) in DCM to afford the final products 18 and 19 with yields of 83% and 75% respectively. The final compound and all intermediates were purified by silica gel column chromatography, and characterised by LC-MS and NMR (Scheme 2).
Compound 19 was coupled to a commercially available MC-Val-Ala-OH linker to facilitate efficient conjugation to trastuzumab. The MC-Val-Ala-OH linker has been previously used to link PBD payloads to an antibody to generate ADCs.26,27 This was accomplished through a standard amide coupling reaction, in dichloromethane and methanol, using EEDQ as the activating agent (Scheme 3). Although the thiosuccinimide linkage obtained in the ADC could undergo retro-Michael elimination and substitution with endogenous thiols such as albumin and glutathione, this maleimide linker technology remains the most widely used among the currently approved ADCs. These include trastuzumab-deruxtecan, to which the newly obtained ADC was compared.28
 | ||
| Scheme 3 Synthesis of compound 20: i) 2-ethoxy-1-ethoxycarbonyl-1,2-dihydroquinoline, methanol, DCM, (6-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)hexanoyl)-L-valyl-L-alanine. | ||
Transcription factor inhibition array assay
Although covalently binding DNA-interactive agents cause cell-death primarily through alkylation of DNA, it is known that the PBD dimers (e.g., SJG-136) and PBD-based monomeric C8-conjugates (e.g., GWL-78 and KMR-28-39) have a secondary mechanism of action and can inhibit the binding of certain transcription factors (TFs) to the DNA helix. This is accomplished through steric interaction caused by preferential binding to certain sequences of DNA base pairs. For example, transcription factors such as NF-γ (in the case of GWL-78) and NF-κB (in the case of KMR-28-39) have been shown to be down-regulated by PBD-based agents.16,29Therefore, the TF-inhibition profile of compound 18 was investigated using a commercially available transcription factor plate array assay in which HeLa cells were treated with 18, and a nuclear extract of the treated cells analysed and compared with that of a non-treated control.30 This assay is luminescence-based and makes use of biotin-labeled probes containing consensus sequences for 96 TFs which are then detected with a streptavidin-HRP conjugate (Fig. 4A).
It was observed that important oncogenic transcription factors such as ROR, HIF, RUNX, PIT1 and Gli-1 were all down-regulated by ≥20%, with a simultaneous up-regulation of a number of transcription factors including p53. Fig. 4B shows all transcription factors up- or down-regulated by more than 20% among those tested. The down-regulation of key transcription factors is expected to make cancer cells more sensitive to chemotherapy by interfering with their ability to adapt, proliferate, and evade therapeutic interventions. For example, HIF-1 binds to hypoxia-response elements (HREs), inducing the expression of numerous hypoxia-responsive genes involved in angiogenesis, metabolic adaptation, cell survival and proliferation, invasion and metastasis.31,32 ROR, Gli-1, PIT1 and RUNX are also known to be over-expressed in some cancer types. For example, ROR seems to play an important role in pancreatic ductal adeno-carcinoma stemness and is associated with aggressive behaviour of the tumour,33 while Gli-1 is often associated with both small- and non-small-cell lung cancers.34 RUNX is known to have an oncosupportive effect in haematological and epithelial malignancies such as skin, oral, breast, ovarian, prostate and neural cancers,35 and PIT1 over-expression has been observed to cause phenotypic changes in proteins involved in the development of breast cancer and its metastasis to liver and lungs.36 Therefore, the down-regulation of HIF suggests a potential use of PDD analogues containing MTP moieties in the treatment of solid tumours, as hypoxia is a condition that most solid tumours need to overcome.
It is also important to note that the observed transcription factor down-regulation profile of compound 18 (Fig. 4B) was found to be different compared to the structurally related C8-linked PBD monomers. For example, KMR-28-39 has been shown to down-regulate the oncogenic transcription factors NF-κB and SMAD.16 Therefore, the transcription factor array data suggests the involvement of other transcription factors compared to the structurally-similar monoalkylating PBDs.23
DNA fluorescence melting
DNA melting assays were undertaken to compare the binding affinity of 18 and 19 to different DNA sequences. In these studies, one DNA strand contained an attached 5′-fluorescein (F), while its complement contained 3′-dabcyl (Q) as a fluorescence quencher. The differences in melting temperature (ΔTm) between the free DNA and drug:DNA adducts were considered to be a representation of the binding affinity of that particular ligand for the sequence of DNA studied.The first DNA fluorescence melting experiments used a sequence (sequence 1) that contained a single central G, to which the ligands should covalently bind, which was flanked by blocks of An.Tn. An established methodology was used for this study,17 and utilized two different ligand concentrations (1 and 10 μM).
The ligands were incubated with the DNA sequence at room temperature overnight (18 h) at concentrations of 0, 1 and 10 μM, and the DNA complexes were then melted by gradual heating, followed by slow annealing as previously described.17 The covalent bond between ligand and DNA is reversible at high temperature and the ligands slowly detach from the DNA at the elevated temperatures. While the DNA double strand re-forms the ligand seems to initially interact non-covalently and to form a covalent bond with guanine only once the re-anneal process has occurred. The annealing temperature for the drug-treated samples are therefore lower than their melting temperatures. The melting process was repeated after leaving the mixtures to reanneal for 5, 30 and 120 min, allowing the ligands to reattach to the duplex. 18 and 19 stabilized the DNA double helix to similar extents at both 10 and 1 μM, and no notable differences were observed with shorter incubation times (Fig. 5). This suggests that both ligands can bind quickly and with high affinity to the DNA sequence used in the study.
A further series of DNA sequences were then designed. In these, the original guanine was retained but a second guanine was introduced with 1–4 adenines between the two Gs, generating sequences with a central 5′-AGAnGA (n = 1–4). 18 and 19 were then incubated with these different sequences so that potential differences in sequence selectivity could be assessed.
While both analogues increased the melting temperature of the An.Tn sequence that contained a single G (sequence 1, Table 1) by over 30 °C, they produced lower stabilization of the sequences that contained a second guanine, with up to three base pairs separating the two alkylation points. When a fourth AT-base pair was added in between the two guanines (sequence 5, Table 1) the stabilisation was similar to that of the sequences with a single central G. This shows that both 18 and 19 have a preference for sequences that contain a guanine flanked by at least four A/T base pairs.
| ΔTM (°C) | |||||
|---|---|---|---|---|---|
| Compound | Sequence 1 | Sequence 2 | Sequence 3 | Sequence 4 | Sequence 5 |
| 18 | 34.2 | 30.1 | 29.4 | 24.9 | 34.0 |
| 19 | 33.0 | 28.8 | 29.5 | 27.5 | 32.0 |
| Key: | |||||
|---|---|---|---|---|---|
| Sequences | |||||
| 1 | 5′-F-A7GA6T3-3′ | ||||
| 3′-Q-T7CT6A3-5′ | |||||
| 2 | 5′-F-A9GAGA5-3′ | ||||
| 3′-Q-T9CTCT5-5′ | |||||
| 3 | 5′-F-A8GAAGA5-3′ | ||||
| 3′-Q-T8CTTCT5-5′ | |||||
| 4 | 5′-F-A7GAAAGA5-3′ | ||||
| 3′-Q-T7CTTTCT5-3′ | |||||
| 5 | 5′-F-A6GAAAAGA5-3′ | ||||
| 3′-Q-T6CTTTTCT5-5′ | |||||
DNase I footprinting
DNase I footprinting was used to determine the sequence selectivity of the synthesized PDD monomers, 18 and 19. This assay is based on a ligand's ability to inhibit deoxyribonuclease (DNase) I cleavage at its binding site. The DNA–ligand complex is briefly digested with DNase I before the enzymatic reaction is stopped. The resulting 32P-labelled DNA fragments are denatured and separated by electrophoresis on a polyacrylamide gel based on their molecular weight. Finally, the gel is dried and exposed to a phosphorus screen, allowing the ligand binding sites to be identified by visual inspection. A GA marker track is included on the gel to indicate the location of purines, enabling sequence determination.This study employed two sequences, HexA and MS1, previously used to assess the DNA-binding potential of DNA-interactive molecules.37,38 For this analysis, these sequences provided preliminary data on the sequence selectivity of 18 and 19. The DNA footprints are presented in Fig. 6. Both analogues showed similar DNA footprints, suggesting that the terminal amine in 19 does not significantly impact its sequence selectivity.
It can also be seen that the two ligands preferentially bind to sequences with fewer guanines, compared to other sites in the sequences. In four out of seven footprints, these sequences are flanked by a series of four A/T base pairs. Given the abundance of G/C-rich regions within the two sequences, this observation is consistent with the DNA melting data. It is important to note that the HexA and MS1 sequences do not contain all possible base-pair combinations spanned by both analogues, so the resulting data offered only an initial indication of sequence selectivity.
Evaluation of cytotoxicity
The in vitro cytotoxicity of compounds 18 and 19 was evaluated in three human colorectal tumour cell lines, i.e., SW48, LIM1215, and SW620. Although these cell lines are known to have relatively low HER2 expression compared to others, such as CAPAN-1, they are routinely employed as a model to evaluate the activity of compounds in solid tumour cell lines, as they have faster replication rates and provide robust reproducible data in MTT assays.39,40 Both compounds exhibited highly potent antitumour activity, with IC50 values in the nanomolar to sub-nanomolar range across all tested cell lines (Table 3). The IC50 for compound 18 in the SW48 cell line was 0.14 nM while it was of 1.07 nM for 19, showing an activity about 7.5-fold lower than 18. However, the activity of the compounds were comparable in the LIM1215 cell line, with 19 and 18 having IC50 values of 1.05 nM and 1.60 nM, respectively. In SW620 cells, compound 18 was again found to be more active with an IC50 of 0.30 nM, whereas 19 had an IC50 of 0.86 nM (Table 2).| Mean IC50 (nM) | ||||||
|---|---|---|---|---|---|---|
| Compound | SW48 | LIM1215 | SW620 | |||
| IC50 | SD | IC50 | SD | IC50 | SD | |
| 18 | 0.14 | ±0.023 | 1.60 | ±0.23 | 0.30 | ±0.04 |
| 19 | 1.07 | ±0.022 | 1.05 | ±0.125 | 0.86 | ±0.106 |
| Mean IC50 (nM) | ||||
|---|---|---|---|---|
| Compound | CAPAN-1 (HER2 +) | MCF7 (HER2 −) | ||
| IC50 | SD | IC50 | SD | |
| 19 | 1.02 | ±0.06 | 1.18 | ±0.13 |
| Trastuzumab–(20) | 1.30 | ±0.04 | 29.7 | ±1.55 |
| Trastuzumab-deruxtecan | 46.0 | ±7.13 | 660.7 | ±50.9 |
The observed differences in cytotoxicity between compounds 18 and 19 may be attributed to the structural variation between the two compounds. Compound 19 includes a terminal amino group on the C9-polycyclic side chain to enable linkage with an antibody, which may affect its interaction with cellular components and permeability across membranes, thus affecting its cytotoxic activity.
Conjugation of compound 19 to trastuzumab
Trastuzumab was chosen as a model antibody for conjugation of the linker-payload (i.e., 20, Fig. 7) to obtain proof of concept data. It is a humanized monoclonal antibody developed to recognise and bind to the extracellular domain of the human epidermal growth factor receptor 2 (HER2).41 This receptor is over-expressed in a number of solid tumours, in particular those of epithelial origin such as bladder, oesophageal and oesophagogastric junction, breast and gall bladder cancers.42 Trastuzumab has been approved for the treatment of breast and gastric cancers and is generally well-tolerated by most patients.41,42 Its targeting capabilities have also been exploited by conversion to an ADC (e.g., Enhertu® which contains the topoisomerase I inhibitor, DXd, as payload and is considered a first line treatment for metastatic breast cancer). Therefore, trastuzumab was considered to be a useful benchmarking antibody to investigate the potential of compound 19 as an ADC payload.A stochastic conjugation of 20 to the reduced disulphide bridges of trastuzumab was carried out to link the antibody and payload. The interchain disulphide bridges were partially reduced with the mild reducing agent tris(2-carboxyethyl)phosphine (TCEP). A total reduction achieved using a stronger reducing agent, such as dithiothreitol (DTT), would have led to higher DARs, which was considered inappropriate for this higher potency payload. Conjugation of the linker-payload to the free thiols of the antibody was carried out in an aqueous medium. After purification using Size-Exclusion Chromatography (SEC), an ADC with an average DAR of 1.6 (74.4% as a monomer and 21.3% as a dimer) was obtained (Fig. S10†). The ADC was then formulated in 30 mM histidine, 200 mM sorbitol at pH 6 to ensure stability, and to limit aggregation. Size Exclusion Chromatography (SEC) and Hydrophobic Interaction Chromatography (HIC) analyses were performed on the ADC to confirm its purity and DAR (Fig. S10†).
Cytotoxicity of the ADCs
The novel ADC was evaluated in the pancreatic cancer HER2-positive cell line CAPAN-1, and in the breast cancer-derived HER2-negative cell line MFC7.30 The cytotoxicities of the unconjugated payload 19 and of trastuzumab-deruxtecan were also evaluated in the same cell lines for comparison purposes. Trastuzumab–(20) exhibited very low nanomolar activity in the HER2-positive CAPAN-1 cell line with an IC50 of 1.30 nM, comparable with the equivalent unconjugated payload (IC50 1.02 nM), while the IC50 value was approximatively 25-fold higher in the HER2-negative MCF7 cell line. However, compound 19 was equally active in both cell lines (Table 3). Although still cytotoxic in MCF-7 cell line, trastuzumab–(20) proved to be about 23-fold more active in the HER2-positive cell line CAPAN-1. This was an increment in cytotoxic activity superior to that observed with trastuzumab-deruxtecan which had IC50 values in MCF7 approximatively 14-fold higher than in CAPAN-1 cells. This observation was consistent with the anticipated behaviour of ADCs, with the payload being released only after the antibody has recognised its antigen and the complex internalised and degraded. As HER2 is expressed by the CAPAN-1 cell line, antigen recognition by the ADC is expected to occur, whereas it is not expected in the case of the MCF7 (HER2-negative) cell line. The cytotoxicity observed in MCF7 cells could be attributable to traces of unconjugated payload or to the presence of the HER-2 antigen which, although expressed in a low copy number, is not completely absent in this cell line.In vivo efficacy and tolerability
In vivo tolerability and efficacy studies were carried out on trastuzumab–(20) in mice. In a maximum tolerated dose (MTD) study, the ADC was administered in a dose-escalation format to three groups of CD1 mice, with bodyweight monitored over a 21 day period. Each group consisted of six mice. Although larger sample sizes are typically used in in vivo studies, this cohort was deemed sufficient for generating preliminary MTD data. The MTD was defined as the lowest dose at which at least one mouse exhibited a bodyweight reduction equal to or exceeding a 15% threshold. The ADC showed good tolerability (Fig. 8), with an MTD estimated at or above 15 mg kg−1. However, two mice in the 15 mg kg−1 group (black line, Fig. 8) displayed bodyweight losses exceeding 10%, suggesting that 15 mg kg−1 was approaching the MTD threshold. It should be noted that, despite having comparable in vitro cytotoxicity, ADCs containing the high-potency PDD payload exhibit much higher tolerability in mice compared to similarly potent PBD dimers, which have a maximum tolerated dose (MTD) in the range of 1 mg kg−1 to 5 mg kg−1.43,44 It is possible that the increased tolerability of PDDs might be linked to their inability to crosslink DNA. This characteristic could potentially make them less likely to trigger double-stranded DNA breaks, which may explain their improved safety profile.The efficacy of the ADC was then evaluated in the CAPAN-1 tumour line in a mouse xenograft model. This cell line was chosen for two main reasons. First, the expression profile of HER2 is high in this tumour type, so it is suitable for investigating a trastuzumab-based ADC. Second, it is a pancreatic tumour line (which are difficult cancers to treat), and one in which other best-in-class trastuzumab-based ADCs (e.g., trastuzumab deruxtecan, DAR ≈ 8) have shown limited efficacy.45
Both trastuzumab–(20) and trastuzumab-deruxtecan induced tumour stasis or regression through day 29 (Fig. 9). However, despite having been administered at a higher dose, significant tumour regrowth was observed at this point in the trastuzumab deruxtecan-treated group (red line, Fig. 9), while trastuzumab–(20) continued to show tumour regression out to day 56, achieving four complete responses (CRs) out of six mice. The lower activity of trastuzumab deruxtecan on this CAPAN-1 derived xenograft model could be due to the lower cytotoxicity of DXd, its payload cytotoxic component, when compared to other toxins used in ADCs.46,47 For example, DXd does not bind covalently to DNA but instead interacts non-covalently with the DNA–topoisomerase I complex, inhibiting re-ligation of the DNA strand.48,49 It is worth noting that, although trastuzumab-deruxtecan was dosed at a higher concentration than trastuzumab–(20), due to its higher tolerability in mice,47 it failed to produce a similar response. The ability of trastuzumab–(20) to prevent tumour recurrence for up to 60 days in this xenograft model, highlights the potential of the C9-linked PDD monomers as an effective payload class for ADC development. This prolonged tumour suppression highlights the suitability of the C9-linked PDD monomers and more broadly the PDD class of DNA minor groove binders as a new class of ADC payload for targeted cancer therapy.
Experimental
Chemistry
Reagents were purchased from standard commercial suppliers. Solvents were purchased from VWR (UK). Anhydrous reactions were carried out in pre-oven-dried glassware under an inert atmosphere of argon. Anhydrous solvents were used as purchased without further drying. Thin Layer Chromatography (TLC) was performed on silica gel aluminium plates (Merck 60, F254), and flash column chromatography was carried out either manually, using silica gel (Merck 9385, 230–400 mesh ASTM, 40–63 μM) (whilst monitoring by thin layer chromatography: UV (254 nm)) or using a Biotage Isolera 1 chromatography system coupled to Dalton mass spectrometer. All NMR spectra were obtained at room temperature using a Bruker DPX400 spectrometer, a Varian Mercury Vx, Agilent 400 Hz spectrometer, or a Bruker DPX600 spectrometer for which chemical shifts are expressed in ppm relative to the solvent and coupling constants are expressed in Hz. Microwave reactions were carried out on a Biotage Initiator+ microwave synthesis reactor. HRMS was performed on a Thermo Scientific-Exactive HCD Orbitrap Mass Spectrometer. Yields refer to isolated material (homogeneous by TLC or NMR) and names are assigned according to IUPAC nomenclature. Liquid Chromatography Mass Spectroscopy (LCMS) (methods A & B) analysis was performed on a Waters Alliance 2695 eluting in gradient. Function type: diode array (535 scans). Column type: monolithic C18 50 × 4.60 mm. Mass spectrometry data were collected using a Waters Micromass ZQ instrument coupled to a Waters 2695 HPLC with a Waters 2996 PDA. Ultra-Performance Liquid Chromatography Mass Spectroscopy (UPLC-MS) (method C) analysis was performed on a Waters Acquity H-class UPLC eluting in gradient. Function type: photo diode array (502.93 nm). Column type: Acquity UPLC BEH C18 1.7 μm 2.1 × 50 mm. Mass spectrometry data were collected using a Waters SQ Detector 2 coupled to a Waters Acquity H Class UPLC with ACQ-PDA.Specific optical rotation ([α]) of final compounds was determined at r.t. using a Bellingham–Stanley ADP 440+ polarimeter. The concentration reported are expressed in g/100 mL. The length of the chamber used was 0.3 dm. Uncorrected melting point and degradation temperature of final compounds were obtained using a Cole-Parmer Electrothermal IA9100 Standard Digital Melting Point Apparatus.
Synthesis of compounds 1–20
1H NMR (400 MHz, CDCl3) δ 9.84 (s, 1H), 7.46–7.37 (m, 2H), 6.98 (d, J = 8.2 Hz, 1H), 4.16 (t, J = 6.3 Hz, 2H), 3.91 (s, 3H), 3.69 (s, 3H), 2.56 (t, J = 7.2 Hz, 2H), 2.20 (quin, J = 6.7 Hz, 2H); 13C NMR (100 MHz, CDCl3) δ 190.9, 173.4, 153.8, 149.9, 130.1, 126.8, 111.6, 109.2, 67.8, 56.0, 51.7, 30.3, 24.2; MS (ES+) m/z = 253 (M + H)+; LCMS (method A): tR = 6.48 min.
1H NMR (400 MHz, CDCl3) δ 10.42 (s, 1H), 7.60 (s, 1H), 7.39 (s, 1H), 4.21 (t, J = 6.3 Hz, 2H), 3.98 (s, 3H), 3.70 (s, 3H), 2.61–2.53 (m, 2H), 2.22 (quin, J = 6.6 Hz, 2H); 13C NMR (100 MHz, CDCl3) δ 187.8, 173.2, 153.5, 151.7, 143.8, 125.5, 109.9, 108.1, 68.6, 56.6, 51.8, 30.2, 24.1; MS (ES+): m/z = 298 (M + H)+; LCMS (method A): tR = 6.97 min.
1H NMR (400 MHz, MeOD) δ 7.47 (s, 1H), 7.25 (s, 1H), 4.13 (t, J = 6.2 Hz, 2H), 3.94 (s, 3H), 3.68 (s, 3H), 2.54 (t, J = 7.2 Hz, 2H), 2.17–2.06 (m, 2H); 13C NMR (100 MHz, MeOD) δ 175.3, 168.6, 153.8, 151.3, 143.1, 122.8, 112.4, 109.2, 69.6, 57.0, 52.2, 31.2, 25.5; MS (ES±): m/z = 314 (M + H)+, 312 (M–H)−; LCMS (method A): tR = 6.22 min.
1H NMR (400 MHz, CDCl3) δ 7.68–7.64 (m, 1H), 6.77–6.70 (m, 1H), 4.16–4.07 (m, 3H), 3.93–3.89 (m, 3H), 3.83 (s, 1H), 3.67 (s, 3H), 3.15 (d, J = 1.4 Hz, 1H), 3.11 (s, 1H), 2.78 (s, 1H), 2.56–2.50 (m, 3H), 2.21–2.12 (m, 4H), 1.74–1.55 (m, 4H); 13C NMR (100 MHz, CDCl3) δ 173.3, 168.1, 154.6, 148.2, 137.4, 127.6, 111.4, 108.3, 68.3, 60.6, 56.7, 53.5, 51.7, 43.3, 38.0, 34.9, 30.3, 24.1, 19.7; MS (ES+): m/z = 411 (M + H)+; LCMS (method A): tR = 6.28 min.
1H NMR (400 MHz, CDCl3) δ 6.69 (s, 1H), 6.27–6.18 (m, 1H), 4.03–3.94 (m, 3H), 3.94–3.82 (m, 3H), 3.81–3.76 (m, 1H), 3.74 (s, 3H), 3.73–3.68 (m, 1H), 3.67–3.65 (m, 3H), 3.56 (d, J = 4.8 Hz, 1H), 3.03 (s, 1H), 2.51 (t, J = 7.2 Hz, 2H), 2.11 (quin, J = 6.7 Hz, 2H), 1.68–1.59 (m, 4H), 1.55–1.40 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 173.6, 171.2, 150.3, 141.8, 141.1, 113.2, 112.3, 102.4, 67.5, 60.8, 60.4, 56.8, 51.6, 30.4, 25.8, 24.3, 21.0, 19.9, 14.2; MS (ES+): m/z = 381 (M + H)+; LCMS (method A): tR = 5.52 min.
1H NMR (600 MHz, CDCl3) δ 8.27 (s, 1H), 7.59 (s, 1H), 6.74 (s, 1H), 5.89 (ddt, J = 17.2, 10.7, 5.5 Hz, 1H), 5.30 (ddd, J = 17.2, 3.0, 1.5 Hz, 1H), 5.17 (ddd, J = 10.5, 2.7, 1.3 Hz, 1H), 5.08–4.62 (m, 1H), 4.63–4.50 (m, 2H), 4.03 (t, J = 6.2 Hz, 2H), 3.84 (s, 1H), 3.76 (s, 3H), 3.64 (s, 4H), 3.57 (s, 1H), 3.33 (s, 1H), 2.99 (s, 1H), 2.49 (t, J = 7.4 Hz, 2H), 2.15–2.08 (m, 2H), 1.60 (br s, J = 12.7 Hz, 4H), 1.50–1.35 (m, 2H); 13C NMR (150 MHz, CDCl3) δ 173.5, 170.5, 153.9, 149.7, 144.9, 132.6, 130.1, 117.6, 117.1, 110.9, 106.2, 67.7, 65.5, 60.6, 56.4, 53.5, 51.6, 30.5, 25.7, 25.3, 24.4, 19.7, 14.8; MS (ES+): m/z = 465 (M + H)+; LCMS (method A): tR = 6.47 min.
1H NMR (600 MHz, CDCl3) δ 7.15 (s, 1H), 6.65 (s, 1H), 5.90 (d, J = 10.1 Hz, 1H), 5.78 (s, 1H), 5.12 (d, J = 12.0 Hz, 2H), 4.66 (dd, J = 13.1, 5.0 Hz, 1H), 4.45 (s, 1H), 4.33 (dd, J = 17.1, 4.3 Hz, 1H), 4.17 (s, 1H), 4.02 (dd, J = 17.6, 4.7 Hz, 2H), 3.88 (s, 3H), 3.66 (s, 3H), 3.46 (dt, J = 10.0, 4.1 Hz, 1H), 3.10–3.02 (m, 1H), 2.51 (t, J = 7.2 Hz, 2H), 2.12 (dd, J = 13.5, 6.9 Hz, 2H), 1.83–1.69 (m, 3H), 1.64–1.59 (m, 3H); 13C NMR (150 MHz, CDCl3) δ 173.5, 169.1, 156.1, 150.1, 149.0, 131.9, 127.4, 125.1, 118.0, 113.6, 110.7, 82.5, 67.9, 66.7, 58.4, 56.1, 55.4, 51.6, 38.7, 30.3, 24.2, 23.1, 23.0, 18.3, 18.1; MS (ES+): m/z = 463 (M + H)+; LCMS (method A): tR = 6.30 min.
1H NMR (400 MHz, CDCl3) δ 7.50 (s, 1H), 6.48 (s, 1H), 6.10–5.85 (m, 1H), 5.70–5.62 (m, 1H), 5.01–4.92 (m, 3H), 4.55–4.20 (m, 2H), 4.18–4.13 (m, 1H), 3.96–3.91 (m, 3H), 3.78 (s, 3H), 3.55 (s, 3H), 3.40–3.34 (m, 2H), 3.00–2.91 (m, 1H), 2.24 (t, J = 7.0 Hz, 2H), 2.05–2.02 (m, 2H), 1.67–1.43 (m, 12H); 13C NMR (150 MHz, DMSO) δ 173.4, 168.5, 150.1, 149.2, 133.2, 127.6, 126.4, 116.9, 114.6, 110.8, 99.8, 94.7, 88.0, 83.9, 68.0, 66.0, 63.3, 62.5, 56.2, 55.4, 51.8, 38.6, 30.6, 30.2, 25.3, 24.3, 23.1, 19.7, 18.3; MS (ES+): m/z = 547 (M + H)+; LCMS (method A): tR = 7.70 min.
1H NMR (400 MHz, (CD3)2SO) δ 12.15 (br. s., 1H), 7.03 (s, 1H), 6.86 (s, 1H), 6.06–5.87 (m, 1H), 5.83–5.69 (m, 1H), 5.11–4.96 (m, 3H), 4.64–4.36 (m, 2H), 4.16–4.02 (m, 1H), 4.00–3.92 (m, 2H), 3.80 (s, 3H), 3.79–3.70 (m, 2H), 3.54–3.46 (m, 1H), 2.89–2.83 (m, 1H), 2.36 (t, J = 7.2 Hz, 2H), 1.96–1.89 (m, 2H), 1.71–1.41 (m, 12H); 13C NMR (100 MHz, (CD3)2SO) δ 174.5, 174.4, 168.5, 168.5, 150.1, 149.1, 133.1, 127.6, 126.3, 114.5, 110.7, 109.1, 99.7, 84.4, 68.0, 67.9, 56.2, 52.9, 38.5, 30.6, 30.3, 30.2, 25.4, 25.3, 23.1, 23.0, 18.3; MS (ES±): m/z = 533 (M + H)+, 531 (M–H)−; LCMS (method A): tR = 6.98 min.
1H NMR (400 MHz, CDCl3) δ 7.58 (s, 1H), 7.45 (dd, J = 9.6, 2.3 Hz, 1H), 6.73 (s, 1H), 6.55 (d, J = 9.7 Hz, 1H), 1.47 (s, 9H); 13C NMR (100 MHz, DCCl3) δ 163.4, 154.5, 126.0, 120.9, 120.1, 85.8, 77.2, 28.4; MS (ES+): m/z = 211 (M + H)+; LCMS (method B): tR = 2.68 min.
1H NMR (400 MHz, CDCl3) δ 7.33 (d, J = 7.0 Hz, 1H), 7.30 (s, 1H), 7.09 (d, J = 9.0 Hz, 1H), 1.50 (s, 9H); 13C NMR (100 MHz, CDCl3) δ 150.3, 136.3, 134.6, 129.4, 126.9, 123.2, 28.1; MS (ES+): m/z = 343 (M + H)+; LCMS (method B): tR = 4.23 min.
1H NMR (600 MHz, (CD3)2SO) δ 9.76 (s, 1H), 8.58 (d, J = 2.4 Hz, 1H), 7.97 (d, J = 7.4 Hz, 1H), 7.88 (d, J = 8.7 Hz, 1H), 7.58 (s, 1H), 3.80 (s, 3H), 2.49 (s, 3H), 1.49 (s, 9H); 13C NMR (150 MHz, (CD3)2SO) δ 162.5, 152.7, 148.1, 146.7, 139.6, 136.2, 129.5, 128.0, 125.6, 122.9, 119.5, 79.9, 51.8, 28.0, 15.9; MS (ES+): m/z = 349 (M + H)+; LCMS (method B): tR = 4.32 min.
1H NMR (600 MHz, (CD3)2SO) δ 10.11 (s, 1H), 9.17 (s, 1H), 8.87 (d, J = 2.4 Hz, 1H), 8.24 (dd, J = 8.7, 2.4 Hz, 1H), 7.93 (d, J = 8.7 Hz, 1H), 7.63 (s, 1H), 7.03 (s, 1H), 6.99 (s, 1H), 3.83 (s, 3H), 3.81 (s, 3H), 2.50 (s, 3H), 1.46 (s, 9H); 13C NMR (150 MHz, (CD3)2SO) δ 162.5, 159.8, 152.9, 148.1, 146.7, 144.9, 141.2, 136.1, 128.2, 127.4, 125.7, 122.7, 122.0, 119.3, 118.5, 105.2, 78.4, 51.8, 36.3, 28.2, 15.9; MS (ES+): m/z = 371 (M + H)+; LCMS (method B): tR = 4.22 min.
1H NMR (400 MHz, CD3OD) δ 8.76 (dd, J = 6.6, 2.2 Hz, 1H), 8.22–8.15 (m, 1H), 7.78 (d, J = 8.7 Hz, 1H), 7.44 (s, 1H), 7.21 (dd, J = 6.8, 1.7 Hz, 1H), 7.14 (d, J = 8.9 Hz, 1H), 7.01–6.83 (m, 2H), 6.18 (d, J = 10.1 Hz, 1H), 6.01–5.65 (m, 1H), 5.10–5.04 (m, 3H), 4.62–4.39 (m, 2H), 4.23–4.02 (m, 3H), 3.90 (d, J = 2.7 Hz, 3H), 3.87 (s, 3H), 3.85 (s, 4H), 3.61–3.51 (m, 1H), 3.45 (dd, J = 9.7, 3.9 Hz, 1H), 3.06 (ddd, J = 11.2, 10.5, 5.6 Hz, 1H), 2.57–2.51 (m, 5H), 2.21–2.15 (m, 2H), 1.85–1.64 (m, 8H), 1.59–1.47 (m, 4H); 13C NMR (100 MHz, CDCl3) δ 169.3, 169.1, 163.2, 156.0, 155.6, 155.1, 150.4, 149.8, 148.0, 146.9, 132.0, 131.9, 130.1, 129.2, 127.8, 127.6, 127.5, 126.6, 126.1, 121.9, 119.1, 118.8, 110.8, 107.9, 100.9, 96.4, 84.0, 81.3, 69.9, 63.0, 55.9, 51.7, 38.9, 36.9, 30.8, 30.5, 25.2, 23.1, 22.9, 19.5, 18.0, 16.2; MS (ES+): m/z = 885 (M + H)+; LCMS (method B): tR = 4.33 min.
1H NMR (400 MHz, CD3OD) δ 8.81–8.73 (m, 1H), 8.19 (ddd, J = 8.6, 6.0, 2.5 Hz, 1H), 7.78 (d, J = 8.7 Hz, 1H), 7.44 (s, 1H), 7.21 (dd, J = 6.8, 1.8 Hz, 1H), 7.13 (d, J = 8.8 Hz, 1H), 7.02–6.84 (m, 2H), 6.19 (d, J = 10.1 Hz, 1H), 6.00–5.67 (m, 1H), 5.11–5.05 (m, 3H), 4.68–4.39 (m, 2H), 4.23–4.03 (m, 3H), 3.90 (d, J = 2.7 Hz, 3H), 3.86 (s, 4H), 3.59–3.51 (m, 1H), 3.45 (dd, J = 9.5, 3.9 Hz, 1H), 3.07 (d, J = 10.2 Hz, 1H), 2.57–2.50 (m, 5H), 2.17 (dt, J = 6.8, 5.8 Hz, 2H), 1.86–1.63 (m, 8H), 1.59–1.44 (m, 4H); 13C NMR (100 MHz, CD3OD) δ 175.1, 171.3, 171.2, 165.8, 161.7, 157.0, 152.0, 151.6, 150.8, 150.6, 148.6, 147.9, 137.0, 133.5, 133.4, 130.4, 129.4, 129.1, 129.0, 123.4, 123.4, 121.6, 120.7, 117.6, 115.7, 115.5, 111.6, 111.3, 102.0, 85.4, 79.2, 69.5, 69.3, 68.1, 56.6, 54.8, 40.1, 40.0, 37.3, 32.1, 31.7, 26.4, 26.3, 24.0, 20.7, 20.5, 19.2, 19.1, 16.4; MS (ES+): m/z = 871 (M + H)+; LCMS (method B): tR = 3.92 min.
1H NMR (600 MHz, CDCl3) δ 8.49–8.33 (m, 1H), 7.67 (s, 1H), 7.64–7.55 (m, 3H), 7.39–7.25 (4H), 7.12 (dd, J = 14.9, 6.7 Hz, 2H), 6.84 (s, 1H), 6.20–6.10 (m, 1H), 6.04–5.88 (m, 1H), 5.83–5.64 (m, 1H), 5.42–525 (m, 1H), 5.15–4.98 (m, 2H), 4.75–4.53 (m, 2H), 4.41–4.25 (m, 2H), 4.15–4.06 (m, 1H), 3.84–3.90 (m, 3H), 3.77–3.56 (m, 6H), 3.18–3.05 (m, 1H), 2.60 (d, J = 8.3 Hz, 3H), 2.44–2.34 (m, 2H), 2.23–2.08 (m, 2H), 1.78–1.55 (m, 12H); 13C NMR (150 MHz, CDCl3) δ 169.7, 169.5, 161.3, 160.7, 160.3, 155.7, 150.4, 145.6, 144.3, 141.8, 138.0, 136.0, 132.1, 131.8, 129.2, 128.3, 128.0, 124.6, 122.0, 121.6, 120.7, 120.3, 119.2, 119.0, 118.8, 118.4, 111.4, 105.5, 100.9, 100.6, 95.2, 88.5, 84.0, 67.2, 64.2, 56.3, 55.9, 39.5, 39.3, 37.0, 33.1, 31.1, 30.7, 25.4, 25.3, 23.2, 23.1, 20.2, 19.7, 18.6, 16.2; MS (ES+): m/z = 946 (M + H)+; LCMS (method B): tR = 4.30 min.
1H NMR (600 MHz, (CD3)2SO) δ 10.17 (s, 1H), 10.02 (d, J = 4.2 Hz, 1H), 9.59 (s, 1H), 8.88 (d, J = 2.0 Hz, 1H), 8.23–8.20 (m, 1H), 7.90 (d, J = 8.7 Hz, 1H), 7.56 (s, 1H), 7.31 (d, J = 8.7 Hz, 2H), 7.25 (dd, J = 3.6, 1.5 Hz, 1H), 7.08 (d, J = 1.8 Hz, 1H), 7.06 (d, J = 3.9 Hz, 1H), 6.82 (s, 1H), 6.55–6.51 (m, 2H), 6.04 (d, J = 10.0 Hz, 1H), 5.86–5.71 (m, 1H), 5.10–4.94 (m, 3H), 4.64–4.34 (m, 4H), 4.15–3.95 (m, 4H), 3.84 (d, J = 2.6 Hz, 3H), 3.81 (t, J = 3.7 Hz, 3H), 3.78–3.72 (m, 1H), 3.58–3.49 (m, 1H), 2.93–2.86 (m, 1H), 2.44 (d, J = 7.8 Hz, 5H), 2.07–2.00 (m, 2H), 1.77–1.34 (m, 12H); 13C NMR (150 MHz, (CD3)2SO) δ 169.0, 168.1, 160.5, 159.9, 148.7, 145.6, 145.3, 144.3, 141.2, 140.8, 135.5, 132.7, 132.5, 127.9, 127.7, 127.6, 122.3, 122.2, 122.0, 119.5, 118.8, 116.5, 114.0, 113.6, 110.4, 105.5, 99.2, 94.2, 83.4, 68.1, 67.8, 65.5, 56.0, 55.7, 54.9, 48.6, 39.5, 38.2, 38.1, 36.3, 31.8, 31.7, 30.2, 24.9, 24.8, 24.5, 22.6, 22.6, 18.5, 17.9, 17.8, 15.5; MS (ES+): m/z = 961 (M + H)+; LCMS (method B): tR = 3.58 min.
[α]D23 = 156.6° (c 0.171, (CH3)2CO); 1H NMR (400 MHz, (CD3)2SO) δ 10.13 (s, 1H), 10.00 (s, 1H), 9.94 (s, 1H), 8.88 (d, J = 2.5 Hz, 1H), 8.22 (dd, J = 8.7, 2.5 Hz, 1H), 8.00 (d, J = 5.7 Hz, 1H), 7.93 (d, J = 8.7 Hz, 1H), 7.71 (d, J = 7.6 Hz, 2H), 7.60 (s, 1H), 7.37–7.31 (m, 2H), 7.27 (s, 1H), 7.25 (d, J = 1.8 Hz, 1H), 7.12–7.07 (m, 2H), 6.80 (s, 1H), 4.16–4.11 (m, 1H), 4.09–3.97 (m, 2H), 3.85 (s, 3H), 3.82 (s, 3H), 3.74–3.63 (m, 1H), 3.16–3.07 (m, 1H), 2.49–2.41 (m, 5H), 2.09–2.01 (m, 3H), 1.92–1.47 (m, 5H); 13C NMR (100 MHz, (CD3)2SO) δ 169.0, 166.3, 164.7, 161.2, 159.8, 150.2, 147.1, 145.4, 144.9, 141.9, 141.2, 139.9, 138.9, 135.7, 131.7, 128.6, 127.8, 127.5, 123.7, 122.3, 122.0, 120.7, 120.3, 119.5, 118.9, 111.4, 109.5, 105.5, 67.8, 55.8, 55.6, 54.2, 49.3, 36.3, 31.9, 24.7, 23.7, 22.6, 17.7, 15.6; MS (ES+): m/z = 760 (M + H)+; LCMS (method B): tR = 3.70 min; HRMS (ESI, m/z): calc. for C41H42N7O6S+ (M + H)+ 760.2912 found 760.2904.
[α]D23 = −92° (c 0.036, (CH3)2CO); 1H NMR (600 MHz, (CD3)2SO) δ 10.11 (s, 1H), 9.93 (s, 1H), 9.57 (s, 1H), 8.85 (d, J = 2.5 Hz, 1H), 8.21 (dd, J = 8.7, 2.5 Hz, 1H), 8.00 (d, J = 5.7 Hz, 1H), 7.90 (d, J = 8.6 Hz, 1H), 7.56 (s, 1H), 7.31 (d, J = 8.7 Hz, 2H), 7.27 (s, 1H), 7.24 (d, J = 1.7 Hz, 1H), 7.07 (d, J = 1.8 Hz, 1H), 6.80 (s, 1H), 6.53 (d, J = 11.7 Hz, 2H), 4.92 (s, 2H), 4.15–4.10 (m, 1H), 4.05–3.94 (m, 2H), 3.85 (s, 3H), 3.82 (s, 3H), 3.70–3.67 (m, 1H), 3.15–3.08 (m, 1H), 2.46–2.43 (m, 5H), 2.07–2.02 (m, 3H), 1.89–1.63 (m, 5H); 13C NMR (150 MHz, (CD3)2SO) δ 169.0, 167.1, 166.3, 164.7, 160.5, 159.8, 150.2, 147.1, 145.6, 145.3, 144.3, 141.1, 140.8, 139.9, 132.5, 127.9, 127.7, 127.6, 122.3, 122.2, 122.0, 120.7, 119.5, 118.8, 113.6, 111.4, 109.5, 105.5, 67.8, 61.2, 55.6, 49.3, 39.5, 36.3, 31.9, 24.7, 23.7, 22.6, 17.7, 15.5; MS (ES+): m/z = 775 (M + H)+; LCMS (method B): tR = 3.00 min; HRMS (ESI, m/z): calc. for C41H43N8O6S+ ([M] + H)+ 775.3021 found 775.3026.
[α]D22 = 39° (c 0.225, (CH3)2SO); 1H NMR (600 MHz, (CD3)2SO) δ 10.12 (s, 1H), 9.94 (s, 2H), 9.87 (s, 1H), 8.87 (d, J = 2.4 Hz, 1H), 8.22 (dd, J = 8.6, 2.3 Hz, 1H), 8.12 (d, J = 7.0 Hz, 1H), 8.00 (d, J = 5.7 Hz, 1H), 7.92 (d, J = 8.7 Hz, 1H), 7.81 (d, J = 8.6 Hz, 1H), 7.63 (d, J = 8.9 Hz, 2H), 7.59 (s, 1H), 7.55 (d, J = 8.9 Hz, 2H), 7.27 (s, 1H), 7.24 (d, J = 1.6 Hz, 1H), 7.07 (d, J = 1.5 Hz, 1H), 6.99 (s, 2H), 6.80 (s, 1H), 4.39 (p, J = 7.0 Hz, 1H), 4.17 (dd, J = 8.5, 6.9 Hz, 1H), 4.14–4.11 (m, 1H), 4.00 (dddd, J = 17.5, 14.0, 9.6, 5.1 Hz, 3H), 3.85 (s, 3H), 3.82 (s, 3H), 3.73–3.66 (m, 2H), 3.37 (t, J = 7.1 Hz, 2H), 3.14–3.09 (m, 1H), 2.47–2.44 (m, 5H), 2.07–2.02 (m, 3H), 1.97 (dd, J = 13.5, 6.8 Hz, 1H), 1.89–1.83 (m, 1H), 1.72 (ddd, J = 16.0, 9.8, 5.2 Hz, 2H), 1.51–1.46 (m, 4H), 1.31 (d, J = 7.1 Hz, 3H), 1.19 (dt, J = 14.8, 7.5 Hz, 4H), 0.85 (dd, J = 23.0, 6.8 Hz, 6H); 13C NMR (150 MHz, (CD3)2SO) δ 181.8, 180.5, 180.5, 180.3, 178.4, 175.8, 174.2, 170.4, 169.3, 159.7, 156.6, 154.9, 154.3, 151.2, 150.6, 149.3, 145.1, 144.3, 143.9, 143.8, 141.2, 137.2, 137.0, 131.8, 131.5, 130.2, 130.1, 129.0, 128.8, 128.4, 120.9, 119.0, 115.0, 67.0, 65.1, 64.4, 58.8, 46.5, 45.8, 44.4, 41.4, 40.1, 39.8, 37.2, 35.3, 34.4, 34.2, 33.2, 32.0, 28.7, 27.6, 27.5, 27.2, 25.1; MS (ES+): m/z = 1139 (M + H)+; LCMS (method A): tR = 6.78 min; HRMS (ESI, m/z): calc. for C59H68N11O11S+ ([M] + H)+ 1138.4815 found 1138.4847.
DNA fluorescence melting assay
For the fluorescence melting experiments a stock solution of the compounds was prepared by dissolving them in DMSO at a concentration of 1 or 10 mM. These were diluted to working concentrations immediately before use.The F- and Q-labelled oligonucleotide pairs were first annealed at a concentration of 0.25 μM in 50 mM sodium phosphate buffer pH 7.4 containing 1 M NaCl. The mixture was heated at 90 °C for 1 min and slowly cooled to room temperature. 4 μL of the ligand was then added to 16 μL of the annealed oligonucleotides to achieve a final ligand concentration of 0, 1 or 10 μM, and the mixture was incubated for 18 hours at room temperature.
Fluorescence melting profiles were measured using a Roche LightCycler using a total reaction volume of 20 μL. Initially, the samples were denatured by heating to 95 °C at a rate of 1 °C min−1. The samples were then maintained at 95 °C for 5 min before cooling to 25 °C at 1 °C min−1. The samples were then held at 25 °C for a further 5 min and re-melted by heating to 95 °C at 1 °C min−1. This process was then repeated holding the samples at 25 °C for 30 and 120 min before re-melting. Annealing steps and melting steps were all recorded and changes in fluorescence were measured at 520 nm. Tm values were obtained from the first derivates of the melting profiles using the Roche LightCycler software.
DNase I footprinting
Footprinting reactions were performed using the DNA fragments HexA, which contains 32 of the 64 possible symmetrical hexanucleotide sequences, and MS1 that contains all possible 134 tetranucleotide sequences. The DNA fragments were obtained by cutting the parent plasmids with HindIII and SacI and were labelled at the 3′-end of the HindIII site with [α-32P]dATP using exo-Klenow fragment. After gel purification, the radiolabelled DNA was dissolved in 10 mM Tris-HCl pH 7.5 containing 0.1 mM EDTA, at a concentration of about 10 c.p.s per μL as determined on a handheld Geiger counter. 1.5 μL of radiolabelled DNA was mixed with 1.5 μL ligand that had been freshly diluted at the desired concentrations in Tris-HCl pH 7.5, containing 10 mM NaCl. The complexes were left to equilibrate for the desired time before digesting with 2 μL DNase I (final concentration about 0.01 units per mL in a solution containing 20 mM NaCl, 2 mM MgCl2 and 2 mM MnCl2). The reactions were stopped after 1 minute by adding 4 μL of formamide containing 10 mM EDTA and bromophenol blue (0.1% w/v). The samples were then heated at 100 °C for 3 minutes before loading onto 8% denaturing polyacrylamide gels containing 8 M urea. Gels were fixed in 10% acetic acid, transferred to 3MM paper, dried and exposed to a phosphor screen overnight, before analysing with a Typhoon phosphorimager.Molecular modelling study
Molecular dynamics simulations were undertaken on the ligand both non-covalently and covalently bound to the relevant sequences used in the study. The DNA sequence was constructed using the AMBER246 module nab. Each ligand was generated using ChemBioOffice and was then docked in the minor groove using AMBER xleap, parm99SB and modified parmbsc0247, and Gaff AMBER force field parameters. Antechamber was used to construct .mol2 files through the addition of Gasteiger charges, and missing parameters were generated using parmchk. A covalent bond was generated between the exocyclic amine groups of selected guanines (guided by molecular mechanics calculations 248). Energy minimisation was then undertaken in a gradient manner by initially placing the DNA under a high force constraint (i.e., 500 kcal mol−1 Å−2), which was then reduced in stages to zero to enable the ligand to find its local energy minimum. This was followed by reduction in force in a periodic manner with a relaxation of restraints. Production simulations in an implicit solvent (GBSA) were run for a period of 10 ns, and atomic coordinates were saved at 1 ps intervals. Analysis of molecular dynamics simulations was undertaken using VMD249, and all models were created using Chimera250.IC50 calculation
Tumor cell lines were maintained in RPMI1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine and 1 mM sodium pyruvate. 1800 cells per well were seeded in a volume of 180 μL in a 96-well flat bottom polystyrene plate. The cells were allowed to adhere overnight at 37 °C in a CO2 incubator. Ligands were initially formulated in dimethyl sulphoxide, and stocks stored at −80 °C. They were then further formulated at 10× concentration in RPMI1640 medium. 20 μL of diluted samples were added into each treatment well. On each plate, blank wells with no cells, and untreated wells containing cells, were included. Plates were then cultured at 37 °C in a CO2 incubator for 96 h. Cytotoxicity was evaluated in triplicate using a tetrazolium salt-based assay, the MTT assay. After 96 h, the supernatant was removed from each well and 200 μL of a sterile filtered 500 μg mL−1 MTT solution in water added to each well. The plates were then incubated at 37 °C in a CO2 incubator for 4 h. The supernatant was then removed and the formazan crystals formed solubilized by adding 150 μL of dimethyl sulphoxide to each well. The plate was then read on a plate reader at 540 nm, and percentage cell survival calculated as follows: ((mean absorbance treated wells at concentration x – mean absorbance blank wells) ÷ (mean absorbance untreated wells at concentration x – mean absorbance blank wells)) × 100. Data were plotted as concentration in nM vs. % cell survival, and IC50 values for inhibition of cell growth were determined with the GraphPad Prism software using non-linear regression.Transcription factor plate array assay
The transcription factor plate array assay kit was obtained from Signosis, Inc (USA). 2 × 106 HeLa cells were treated with 100 nM of compound 18, and incubated for 6 hours before extracting the nuclear protein and carrying out the TF plate array assay according to the manufacturer's protocol.32 In the case of each transcription factor, the RLU value obtained for the cells treated with compound 18 was deducted from the respective values obtained for the cells treated with vehicle control to obtain the differences in TF activation/inhibition.Conjugation methodology and ADC purification
The interchain disulphides of commercially-available trastuzumab formulated at pH 7–8 in 2 mM EDTA were partially reduced with TCEP for 90–180 minutes. The extent of reduction was controlled to achieve a specified drug-to-antibody ratio (DAR) of ∼2. The reduced antibody was then diluted with PBS/2 mM EDTA to 2 mg mL−1. The compounds were dissolved in a mixture of DMA/DMSO, and conjugation of the payloads to the antibody was achieved by addition of an excess of the payload to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of the reduced antibody and propylene glycol, at a final protein concentration of 1 mg mL−1. The antibody/payload mixture was allowed to react for 1 h to form the antibody–drug conjugate (ADC), and the reaction was then quenched with an excess of N-acetyl cysteine to remove unreacted starting material. The mixture was then further diluted with 1
1 mixture of the reduced antibody and propylene glycol, at a final protein concentration of 1 mg mL−1. The antibody/payload mixture was allowed to react for 1 h to form the antibody–drug conjugate (ADC), and the reaction was then quenched with an excess of N-acetyl cysteine to remove unreacted starting material. The mixture was then further diluted with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PBS/3% cyclodextrin and added to a proprietary purifying resin. The resin-bound ADC was washed with PBS/3% cyclodextrin to remove excess small-molecule impurities, and then released from the resin. The ADCs were finally formulated through G25 desalting into the chosen formulation filtered prior to aliquoting and storage at −80 °C.
1 PBS/3% cyclodextrin and added to a proprietary purifying resin. The resin-bound ADC was washed with PBS/3% cyclodextrin to remove excess small-molecule impurities, and then released from the resin. The ADCs were finally formulated through G25 desalting into the chosen formulation filtered prior to aliquoting and storage at −80 °C.
In vivo efficacy study
CAPAN-1 tumours were implanted in the flanks of Balb-c mice and grown to a size of ∼100–150 mm3. The mice were then given a single dose of the payload 19-containing ADC trastuzumab–(20) via tail vein. A dose of 2 mg kg−1 was chosen based on available data for other PDD-containing ADCs.8 The approved HER-2 targeting ADC Enhertu® was also included in the study as a control and was administered as a single dose of 10 mg kg−1.Conclusions
This study provides evidence supporting the suitability of 18, a first-generation high potency pyridinobenzodiazepine monomer containing an MTP building block, as a new ADC payload. A novel ADC, trastuzumab–(20), was developed using this payload, which demonstrated significant efficacy in a human tumour xenograft model of HER2-positive pancreatic cancer. This class of PDD monomer payloads appears to selectively bind DNA sequences with guanine bases flanked by tracts (≥4 base pairs) of AT base pairs, as confirmed by both DNA footprinting and DNA fluorescence melting assays. Payload 18 was found to down-regulate critical oncogenic transcription factors, including HIF-1, suggesting a secondary mechanism of action that could be relevant across various tumour types, including pancreatic cancer as evaluated in this study. The ADC exhibited nanomolar cytotoxicity in in vitro experiments. The trastuzumab–(20) ADC outperformed Enhertu® in human tumour xenograft studies at one-fifth of the dose (2 mg kg−1 versus 10 mg kg−1), achieving complete tumour regression at less than 20% of the maximum tolerated dose (MTD). This suggests that ADCs incorporating payload 18 may represent promising candidates for the treatment of solid tumours in humans. While this study demonstrates the potential of a new class of DNA-interactive payloads in an ADC format, further investigations into cellular uptake, stability, bystander killing, pharmacokinetics, and release profiling will be necessary to fully characterize their pharmacological properties before initiating preclinical studies for ADCs containing this payload class.Ethical statement
All animal procedures were performed in accordance with the guidelines of the Animals (Scientific Procedures) Act 1986. All protocols used in this study were approved by the Animal Welfare and Ethical Review Committee of the CRO (AxisBio, Londonderry, UK).Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Conceptualization and methodology: P. A., D. E. T. and P. J. M. J.; investigation: P. A., I. P. H., D. M., K. R. F. and G. P.; software: P. J. M. J and M. H.; supervision: K. M. R. and D. E. T.; writing – original draft: P. A.; writing – review & editing: K. M. R., visualization: P. A., K. M. R., P. J. M. J. and M. H.; validation: P. J. M. J., K. M. R., D. E. T. and P. A.Conflicts of interest
D. E. T., K. M. R., P. J. M. J. are scientific co-founders and shareholders of Pheon Therapeutics, an ADC Development Company. P. A. and G. P. are shareholders of Pheon Therapeutics.Acknowledgements
The authors are grateful to Pheon Therapeutics Ltd for supporting this study.References
- A. Lee, Drugs, 2021, 81, 1229–1233 CrossRef CAS PubMed.
- U.S. Food and Drug Administration, FDA grants accelerated approval to tisotumab vedotin-tftv for recurrent or metastatic cervical cancer, 2021, available at: https://www.accessdata.fda.gov/scripts/medwatch/index.cfm, accessed [February 04, 2025].
- S. Baah, M. Laws and K. M. Rahman, Molecules, 2021, 26, 2943 CrossRef CAS PubMed.
- U.S. Food and Drug Administration, FDA approves datopotamab deruxtecan-dlnk for unresectable or metastatic HR-positive, HER2-negative breast cancer, 2025, available at: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-datopotamab-deruxtecan-dlnk-unresectable-or-metastatic-hr-positive-her2-negative-breast, accessed [February 04, 2025].
- Y. Y. Syed, Drugs, 2019, 79, 579–583 CrossRef CAS PubMed.
- K. Tsuchikama, Y. Anami, S. Y. Y. Ha and C. M. Yamazaki, Nat. Rev. Clin. Oncol., 2024, 21, 203–223 CrossRef CAS PubMed.
- A. Beck, L. Goetsch, C. Dumontet and N. Corvaïa, Nat. Rev. Drug Discovery, 2017, 16, 315–337 CrossRef CAS PubMed.
- H. Bouchard, C. Viskov and C. Garcia-Echeverria, Bioorg. Med. Chem. Lett., 2014, 24, 5357–5363 CrossRef CAS PubMed.
- J. Mantaj, P. J. Jackson, K. M. Rahman and D. E. Thurston, Angew. Chem., Int. Ed., 2017, 56, 462–488 CrossRef CAS PubMed.
- J. A. Hartley, Expert Opin. Invest. Drugs, 2011, 20, 733–744 CrossRef CAS PubMed.
- E. Grunberg, H. N. Prince, E. Titsworth, G. Beskid and M. D. Tendler, Chemotherapia, 2009, 11, 249–260 CrossRef PubMed.
- B. Xu, Eur. J. Clin. Pharmacol., 2022, 78, 707–719 CrossRef CAS PubMed.
- J. A. Hartley, Expert Opin. Biol. Ther., 2021, 21, 931–943 CrossRef CAS PubMed.
- Y. Kovtun, G. E. Jones, S. Adams, L. Harvey, C. A. Audette and A. Wilhelm, et al., Blood Adv., 2018, 2, 848–858 CrossRef CAS PubMed.
- N. Veillard, F. Cascio, P. J. M. Jackson and D. E. Thurston, Cytotoxic Payloads for Antibody–Drug Conjugates, The Royal Society of Chemistry, 2019, pp. 349–363 Search PubMed.
- K. M. Rahman, P. J. M. Jackson, C. H. James, B. P. Basu, J. A. Hartley and M. De La Fuente, et al., J. Med. Chem., 2013, 56, 2911–2935 CrossRef CAS PubMed.
- M. A. Basher, K. M. Rahman, P. J. M. Jackson, D. E. Thurston and K. R. Fox, Biophys. Chem., 2017, 230, 53–61 CrossRef CAS PubMed.
- B. S. P. Reddy, S. K. Sharma and J. W. Lown, Curr. Med. Chem., 2001, 8, 475–508 CrossRef CAS PubMed.
- D. H. Nguyen, J. W. Szewczyk, E. E. Baird and P. B. Dervan, Bioorg. Med. Chem., 2001, 9, 7–17 CrossRef CAS PubMed.
- M. L. Kopka, D. S. Goodsell, G. W. Han, T. K. Chiu, J. W. Lown and R. E. Dickerson, Structure, 1997, 5, 1033–1046 CrossRef CAS PubMed.
- M. L. Kopka, C. Yoon, D. Goodsell, P. Pjura and R. E. Dickerson, Proc. Natl. Acad. Sci. U. S. A., 1985, 82, 1376–1380 CrossRef CAS PubMed.
- P. G. Baraldi, B. Cacciari, A. Guiotto, R. Romagnoli, A. N. Zaid and G. Spalluto, Il Farmaco, 1999, 54, 15–25 CrossRef CAS PubMed.
- D. B. Corcoran, D. E. Thurston and K. M. Rahman, Small-Molecule Transcription Factor Inhibitors in Oncology, The Royal Society of Chemistry, 2018, pp. 81–124 Search PubMed.
- P. d. S. M. Pinheiro, D. A. Rodrigues, M. A. Alves, L. W. Tinoco, G. B. Ferreira and C. M. R. de Sant'Anna, et al., New J. Chem., 2018, 42, 497–505 RSC.
- D. Antonow and D. E. Thurston, Chem. Rev., 2011, 111, 2815–2864 CrossRef CAS PubMed.
- F. Zammarchi, K. E. Havenith, S. Chivers, P. Hogg, F. Bertelli and P. Tyrer, et al., Mol. Cancer Ther., 2022, 21, 582–593 CrossRef CAS PubMed.
- R. J. Christie, A. C. Tiberghien, Q. Du, B. Bezabeh, R. Fleming and A. Shannon, et al., Antibodies, 2017, 6, 20 CrossRef PubMed.
- Y. Wang, F. Xie, L. Liu, X. Xu, S. Fan and W. Zhong, et al., Drug Delivery, 2022, 29, 754–766 CrossRef CAS PubMed.
- M. Kotecha, J. Kluza, G. Wells, C. C. O'Hare, C. Forni and R. Mantovani, et al., Mol. Cancer Ther., 2008, 7, 1319–1328 CrossRef CAS PubMed.
- G. Procopiou, P. J. Jackson, D. di Mascio, J. L. Auer, C. Pepper and K. M. Rahman, et al., Commun. Biol., 2022, 5, 741 CrossRef CAS PubMed.
- S. Soni and Y. S. Padwad, Acta Oncol., 2017, 56, 503–515 CrossRef CAS PubMed.
- J. C. Jun, A. Rathore, H. Younas, D. Gilkes and V. Y. Polotsky, Curr. Sleep Med. Rep., 2017, 3, 1–9 CrossRef PubMed.
- W. Yao and A. Maitra, Cell, 2019, 177, 516–518 CrossRef CAS PubMed.
- E. Mastrangelo and M. Milani, Lung Cancer: Targets Ther., 2018, 9, 35–43 CAS.
- E. Deltcheva and R. Nimmo, Biochem. J., 2017, 474, 1755–1768 CrossRef CAS PubMed.
- A. Martinez-Ordoñez, S. Seoane, P. Cabezas, N. Eiro, J. Sendon-Lago and M. Macia, et al., Oncogene, 2018, 37, 1430–1444 CrossRef PubMed.
- A. J. Hampshire and K. R. Fox, Anal. Biochem., 2008, 374, 298–303 CrossRef CAS PubMed.
- M. Lavesa and K. R. Fox, Anal. Biochem., 2001, 293, 246–250 CrossRef CAS PubMed.
- A. J. M. Wollman, C. Fournier, I. Llorente-Garcia, O. Harriman, A. L. Payne-Dwyer and S. Shashkova, et al., J. R. Soc., Interface, 2022, 19, 20220088 CrossRef CAS PubMed.
- V. Belli, N. Matrone, S. Napolitano, G. Migliardi, F. Cottino and A. Bertotti, et al., J. Exp. Clin. Cancer Res., 2019, 38, 236 CrossRef PubMed.
- K. McKeage and C. M. Perry, Drugs, 2002, 62, 209–243 CrossRef CAS PubMed.
- M. Yan, M. Schwaederle, D. Arguello, S. Z. Millis, Z. Gatalica and R. Kurzrock, Cancer Metastasis Rev., 2015, 34, 157–164 CrossRef CAS PubMed.
- F. Zammarchi, K. Havenith, S. Chivers, P. W. Hogg, C. Britten and S. Dissanayake, et al., Cancer Res., 2018, 78, 744 CrossRef.
- L. R. Staben, J. Chen, J. d. Cruz-Chuh, G. del Rosario, M. A. Go and J. Guo, et al., J. Med. Chem., 2020, 63, 9603–9622 CrossRef CAS PubMed.
- Y. Aida Ogitani, T. Aida, K. Hagihara, J. Yamaguchi, C. Ishii and N. Harada, et al., Clin. Cancer Res., 2016, 22, 5097–5108 CrossRef PubMed.
- L. Conilh, G. Fournet, E. Fourmaux, A. Murcia, E. L. Matera and B. Joseph, et al., Pharmaceuticals, 2021, 14, 3 CrossRef PubMed.
- W. Li, K. H. Veale, Q. Qiu, K. W. Sinkevicius, E. K. Maloney and J. A. Costoplus, et al., ACS Med. Chem. Lett., 2019, 10, 1386–1392 CrossRef CAS PubMed.
- V. Vidmar, M. Vayssières and V. Lamour, Int. J. Mol. Sci., 2023, 24, 3986 CrossRef CAS PubMed.
- Y. Ogitani, T. Aida, K. Hagihara, J. Yamaguchi, C. Ishii and N. Harada, et al., Clin. Cancer Res., 2016, 22, 5097–5108 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4md01040j |
| This journal is © The Royal Society of Chemistry 2025 |