 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
DynaMate: leveraging AI-agents for customized research workflows
Orlando A.
Mendible-Barreto
 a,
Misael
Díaz-Maldonado
b,
Fernando J.
Carmona Esteva
a,
Misael
Díaz-Maldonado
b,
Fernando J.
Carmona Esteva
 a,
J. Emmanuel
Torres
c,
Ubaldo M.
Córdova-Figueroa
a,
J. Emmanuel
Torres
c,
Ubaldo M.
Córdova-Figueroa
 b and
Yamil J.
Colón
b and
Yamil J.
Colón
 *a
*a
aDepartment of Chemical and Biomolecular Engineering, University of Notre Dame, Notre Dame, IN 46556, USA. E-mail: ycolon@nd.edu
bDepartment of Chemical Engineering, Universidad de Puerto Rico, Mayagüez, PR 00680, USA
cComputer and Data Sciences Department, School of Engineering and Computational Sciences, Merrimack College, North Andover, MA 01845, USA
First published on 22nd May 2025
Abstract
Developments related to large language models (LLMs) have deeply impacted everyday activities and are even more significant in scientific applications. They range from simple chatbots that respond to a prompt to very complex agents that plan, conduct, and analyze experiments. As more models and algorithms continue to be developed at a rapid pace, the complexity involved in building this framework increases. Additionally, editing these algorithms for personalized applications has become increasingly challenging. To this end, we present a modular code template that allows easy implementation of custom Python code functions to enable a multi-agent framework capable of using these functions to perform complex tasks. We used the template to build DynaMate, a complex framework for generating, running, and analyzing molecular simulations. We performed various tests that included the simulation of solvents and metal–organic frameworks, calculation of radial distribution functions, and determination of free energy landscapes. The modularity of these templates allows for easy editing and the addition of custom tools, which enables rapid access to the many tools that can be involved in scientific workflows.
Design, System, ApplicationLarge language models (LLM) are increasingly being adopted to automate complex tasks and scientific workflows. While numerous LLM-based agents have been developed for specific applications, they often lack flexibility for adaptation to tasks beyond their original design. To address this limitation, we developed a modular template for building multi-agent systems designed to simplify the integration of user-defined tools and streamline the automation of repetitive, time-consuming tasks. We demonstrate the utility of this template by constructing a multi-agent framework that automates the setup, execution, and analysis of molecular dynamics simulations. Given a user prompt, the agents coordinate to complete the specified tasks efficiently. The modular design allows new tools and agents to be integrated with minimal changes, typically requiring only three targeted modifications. Any functionality accessible through Python can be incorporated, making the framework highly extensible and usable for various research fields. This approach encourages the development of a growing library of user-contributed scientific tools, and we hope it will foster a collaborative community where researchers share and integrate their tools to collectively build more versatile and robust agent frameworks. |
Introduction
The first half of this decade has been strongly impacted by developments in artificial intelligence (AI) and all its subdivisions. Combined with the unprecedented availability of public and scientific data, it was imminent that various forms of AI made their way into everyday activities. These range from simple algorithms that recommend movies to complex large language models (LLMs) agents that make decisions based on an optimization parameter. The usability of AI tools will continue to increase in everyday activities and the scientific community. For many complex workflows, LLMs and agents will present an opportunity to automate repetitive and time-consuming tasks, allowing researchers to focus their efforts on building new and more complex tools that accelerate the discovery of scientific knowledge.Open-source efforts such as LangChain1 and Huggingface2 have played a significant role in making LLMs and tools publicly available. As a result, more people than ever before can use, modify, and fine-tune various models for specific tasks. Initial efforts evaluating the general chemistry knowledge of GPT-3 and open-source models without fine-tuning showed that, given reasonable prompts, the models could answer expert-level questions with an average accuracy of 72% in topics related to molecular dynamics (MD), cheminformatics, and quantum mechanics.3 Studies like this promoted the interest in the usability of LLMs for more complex tasks, and very rapidly, these models were fine-tuned and modified to predict chemical properties from SMILES strings,4 structures of proteins,5 high-yield products from a forward synthesis process,6 and predict chemical structures from IR spectroscopy.7
Recent developments in LLM agents have improved the capabilities of LLMs by allowing them to interact with tools. In this context, tools are independent algorithms used to solve tasks of interest. For example, a Python function to calculate a value, or a function that computes self-diffusivity from a molecular simulation trajectory. Some initial advancements in this direction were built for retrieval-augmented generation (RAG) tasks. By integrating external data retrieval into the text generation process, the answers given by LLMs are no longer limited to their training context. An external knowledge base can be assigned to an agent as a tool to be used when questions about the specific context included in the external knowledge base are used as prompts.8 This workflow significantly improved the accuracy of LLM-generated answers to domain-specific questions, allowing a new technique to achieve complicated tasks requiring specific documentation. Many tools and RAG frameworks were developed for multiple fields, including biomedical,9 financial,10 regulatory compliance,11 legal question answering,12etc.8
The continued development of more complex tools gave rise to robust frameworks that used RAG capabilities and other tools for autonomous research for catalytic cross-coupling experiments,13 synthesis of organic compounds,14 and other general applications.15,16 These efforts highlight the new ability of LLMs to plan experiments, interact with research hardware to generate data, and analyze this data. The field of molecular simulations has also seen the development of tools and agents that accelerate the process of applying computational methods, validating experimental results with numerical approximations, and analyzing results.17–19 MDCrow,20 for example, is an LLM agentic assistant with over forty expert-designed tools for molecular simulations. It automates complex tasks involved in developing, producing, and analyzing molecular trajectories, effectively reducing the time required for each step. Similarly, ChemCrow21 and ChatMOF22 have demonstrated the capability of automating workflows, designing novel chemical syntheses, and predicting and preparing metal–organic framework materials, respectively. The benefits and performance of these agent frameworks have sparked increasing interest in their usage for custom workflows, but current developments are domain-specific, limiting their transferability between domains.
For this reason, most currently developed frameworks present a steep learning curve in editing or customization for new tools, even more so for entirely new workflows. The LangChain1 community has developed and provided Python libraries that significantly reduce the hardships of building chatbots and agents, but the code structures and modularity of the tools built with this library can be improved. This work focuses on developing a template for a multi-agent framework that can be easily implemented for custom workflows. The modularity of this approach allows users to copy and paste custom tools into a file and rapidly build an LLM assistant for any research workflow. We used the template to build DynaMate, a multi-agent framework that assists with generating, submitting, and analyzing molecular simulations. The usage of this template for this specific workflow highlights its modularity and customizability. The multi-agent framework was created in the context of molecular dynamics, but the developed template can be used to expand its tools in many research areas and applications, including colloidal systems, catalysis, drug delivery, etc. The advantages of the modularity of the template include the automation of time-consuming and repetitive tasks, easy access to in-house tools and workflows through a chatbot, and improved learning processes for researchers starting in a specialized field.
The structure of this paper is organized as follows: first, we describe the code template and discuss how it works and how it can be modified for building multi-agent frameworks for specific tasks of interest. Then, we discuss the agents and tools involved in DynaMate and what they can do. Followed by a detailed discussion of representative examples of complex tasks being performed by the multi-agent framework. The examples present the outcomes of the evaluations, showcasing prompts, outputs, and challenges encountered.
Description of multi-agent-LLM framework code
LLMs can be readily used as chatbots, meaning they can only interpret text and generate responses. Different models exist and they show better and worse capabilities for multiple tests. However, without modifications, LLMs can only interact with the input text. If asked, for example, to generate a plot using Python, it will fail because it does not have the required tools to achieve that goal. Thanks to the teams' efforts at LangChain, we now have access to code that allows the development of frameworks for various tasks using Python code. Now, we can use LangChain packages to give LLMs access to Python functions, for example, and now it can make plots, perform calculations, and run custom functions. This LLM can now be identified as an agent since an Agent is an LLM that has access to tools.LangChain has a series of built-in tools that can be used, but it also offers the ability to use customized tools. This means that tools developed for in-house workflows can be added and used as tools for LLM agents. It is possible to generate several agents for specific tasks and define them as tools for a different agent, referred to as the Scheduler. The scheduler's job is to determine which of the agents it has access to has the correct tool to achieve the goal given in the prompt. In this paper, we present a template for building a multi-agent framework using the tools developed by LangChain and OpenAI. Specifically, we use the framework to build an agent that automates tasks involved in generating and analyzing MD simulations, which we named DynaMate. It is important to mention that the workflow is based on the BaseTool and StructuredTool packages in LangChain. Three main pieces should be carefully constructed when dealing with structured tools for agents. These are the classes for input types, which define the type of inputs and include their description. The Python function that performs the action of the tool being designed, and, finally, the structure tool puts everything together to fully define the tool. A template for these three sections of the code is available in the GitHub repository of this project, and its contents with the three mentioned sections are presented in Fig. 1.
When building agents with structured tools, the first step is to define a Python class using the BaseModel package, which will define the input type (i.e., string, integer, boolean, etc.) and its description, as shown in the red rectangle in Fig. 1. This description must be as clear and specific as possible since it is what the LLM will use when executing the tool. The number of inputs here is dependent on the inputs required by the function shown in the second step (blue rectangle in Fig. 1). The code within this function is the heart of the tool the agent will access, so any Python code used for your workflows can be substituted here to convert it into an agent tool. Once the inputs and the function have been defined, the StructuredTool package is used to combine the class with inputs and the function, as shown in the green rectangle in Fig. 1. The structured tool also has a description, which must be as detailed as possible since the agent will use it to decide which tool to use to solve the task at hand.
Once various tools have been developed, they can be assigned to different LLMs to generate a series of agents with specific abilities. More importantly, we can use these agents as tools for a main agent (i.e., scheduler), which results in a multi-agent framework like the one presented in Fig. 2. The agent that has other agents as tools is the scheduler. As previously mentioned, it oversees interpreting the input and assigning the agent with the correct tools to achieve the goal asked in the input. Once the corresponding agent is selected, it is responsible for choosing which tool to use and generating a response, which is passed back to the scheduler, which generates the final output.
 | ||
| Fig. 2 Schematic of multi-agent workflow architecture. The scheduler receives input and assigns it to the agent with the adequate tools to respond to it. | ||
It is important to mention that obtaining each response becomes less efficient as more tools and agents are included in the framework. This is because of memory requirements, additional steps when passing the input information through the framework, and more loading time for the scheduler to distinguish between the agents. More research is needed in this direction to understand these limitations and how to deal with them appropriately.
Overview of agents and tools
For any workflow, the first step should be defining the task and what precisely we want the agent to achieve. In this case, DynaMate is an LLM agent with multiple capabilities that assist in preparing LAMMPS23 input files and analyzing MD simulations. This workflow has five agents and one scheduler. Table 1 presents the names of the agents, a brief description of the assigned tasks, and the number of tools each agent has access to. In addition, the current version does not include an agent in charge of collecting and organizing metadata from each step or task completed. It is the user's responsibility to maintain a well-organized repository so the agents can identify the paths correctly and use their tools efficiently. Current efforts are focused on developing an administrative agent that will gather and make reports of the generated metadata.| Agent | Name | Description | Number of tools |
|---|---|---|---|
| 1 | System preparation | Generates LAMMPS input and configuration files | 5 |
| 2 | Simulation runner | Runs MD simulation using LAMMPS | 2 |
| 3 | Post-processing | Check the convergence, calculate averages, and thermodynamic properties | 2 |
| 4 | Enhanced sampler | Prepares input for enhanced sampling simulations | 6 |
| 5 | RAG | Extract information from embedded data | 2 |
Agent 1 oversees preparing input files for MD simulations in LAMMPS. These files involve the coordinates of the system to study, force field parameters to describe atomic interactions, and input parameters to define the simulation conditions. This agent also has access to the Moltemplate24 and Packmol25 packages, which aid in generating more complex systems. Moltemplate enables the usage and generation of templates for any system of interest, and Packmol adds molecules to a simulation box while avoiding overlapping atoms. Moltemplate includes the OPLSAA26 and GAFF27 force fields by default, but they offer instructions on including custom force fields. The agent also has access to the lammps_interface28 package, which enables the generation of LAMMPS data files from CIF files. Combining these tools gives this agent a flexible framework to develop systems of interest. Below is a list of this agent's tools and their descriptions.
Tool 1.1: mosdef_tool
• Generate LAMMPS data file for a molecular system using the SMILES string. The inputs are the molecule's name, SMILES string, box size, and number of molecules.∘ Uses MosDEF29 for assigning force fields to molecular systems. Offers the possibility of using custom force fields.
Tool 1.2: rdkit_template_tool
• Generate LAMMPS files for a molecular system starting from the SMILES string of the system of interest. The inputs are the molecule's name, the SMILES string, charge, number of molecules, and box size.∘ Uses RDKit30 to generate molecular systems and Moltemplate for force field values.
Tool 1.3: data_from_cif_tool
• Generate LAMMPS data file using the CIF file of the system of interest. The inputs are the name of the CIF file, force field to be used, and whether to generate a pdb file or not.Tool 1.4: data_to_template_tool
• Generate Moltemplate files from the data file and the input file with force field parameters. The inputs are the name of the molecule, template file, input file, data file, and the name of the molecule in the template.Tool 1.5: packmol_moltemplate_tool
• Generate LAMMPS files for a system with multiple types of molecules. The inputs are a list with the number of molecules of each type, a list with the names of molecules of interest, and the size of the box. Uses lists as inputs.Fig. 3 shows how the template presented in Fig. 1 was modified to create the system preparation agent's mosdef_tool. Similar modifications were performed for all other agents, and the same process can be performed for customized frameworks. First, we define the Python class, which defines the type of inputs and their description. In this case, there are four inputs, three strings, and one integer. By defining what these inputs represent, the LLM can understand them and use them accordingly when used in a prompt. The blue block represents the Python function that performs the task assigned to this tool. In this case, it takes the inputs and generates a LAMMPS data file for a molecular system, given the molecule's name, its SMILES string, the number of molecules in the system, and the size of a cubic box. This function can be replaced by any Python function used in personal workflows and be rapidly implemented into the framework. Finally, everything is put together using the structured tool, where the function and the class of inputs are connected and are ready to assign to an agent. All tools are defined in independent Python scripts, enabling easy modifications to the function and rapid additions or eliminations. The code structure followed in this project defines every agent in an independent folder, which stores all the scripts to that agent's tools. Inside each folder is an additional script called agent_response, which loads all the tool scripts and defines an agent executor, enabling the agent to run. In this way, one can build many specialized agents by simply editing the class of inputs, functions, and structured tools. Once all the directories with the agents are created, the scheduler is created the same way as the other agents but using the agents rather than tools. Therefore, when the user sends a prompt to the scheduler, it reads the structure tool description of the agent to select the appropriate one, then the agent reads the prompt and selects the tool to generate a response. The response is returned to the scheduler, which generates the final output. The modularity of this code enables easy incorporation of multiple specialized agents and the development of very complex workflows.
The tools of Agent 2 enable the preparation of LAMMPS input files, which define the type and parameters of the MD simulation, and the execution of the ‘lmp’ command to start the simulation. The current input file generates the system to undergo energy minimization, then equilibrates its volume using an NPT simulation, and finally, performs an NVT simulation. The temperature and pressure are inputs, but the inputs and type of simulation can be modified by going to the tool's source code. Current efforts aim to increase the flexibility of this tool by allowing an agent to generate the input file from scratch. Below is a list of this agent's tools and their descriptions.
Tool 2.1: lmp_create_tool
• Generate the LAMMPS input file and run the simulation. Inputs are LAMMPS input file name, temperature, pressure, and number of CPUs to use.Tool 2.2: lmp_run_tool
• Run the LAMMPS simulation. Inputs are LAMMPS input file name and number of CPUs to use.Agent 3 can check for the convergence of a simulation's thermodynamical properties. Our in-house code reads properties from a running or finished trajectory to determine if more simulation time is required, given some desired property convergence. This agent also can calculate atom-to-atom radial distribution functions (RDFs) and generate and save plots.
Tool 3.1: RDF_calc_tool
• Calculate RDF from LAMMPS trajectory using the data file, trajectory file, names of the atoms, selections of the atoms, and output file name.Tool 3.2: Ensemble_average_tool
• Computes the ensemble average of a property from a LAMMPS log file. It does this by extracting the trajectory from a LAMMPS log file and then determining convergence based on the change of the cumulative running average across the steps.Agent 4 oversees the generation of PLUMED31 input files for umbrella sampling32 or metadynamics33,34 simulations. The usability of this tool assumes a LAMMPS version compiled with PLUMED. Additional enhanced sampling methods can be implemented by modifying the source code of this agent's tools. Even though unbiasing a simulation is considered a post-processing task, this agent also has the tools to obtain the unbiased free energy surface of the system of interest.
Tool 4.1: UmbSamp_input_gen_tool
• Generate PLUMED input files for umbrella sampling simulations using atomic distance as a collective variable.Tool 4.2: UmbSamp_Multi_run_tool
• Run multiple umbrella sampling simulations in parallel.Tool 4.3: wham_analysis_tool
• Perform weighted histogram analysis method (WHAM) analysis on umbrella sampling simulations using one collective variable.Tool 4.4: UmbSamp_analysis_tool
• Analyze umbrella sampling simulations using one collective variable.Tool 4.5: Prep_Metad_Inps_tool
• Generate PLUMED input files for metadynamics simulations using dihedral angles as collective variables.Tool 4.6: MetaD_analysis_tool
• Analyze the output of a metadynamics simulation in one or two dimensions. The inputs are the path to the metadynamics production run, the dimension of the free energy surface (1 or 2), and the name of the collective variable to be analyzed.Agent 5 can perform retrieval augmented generation (RAG), which is a method in which an LLM can interact with a knowledge base in the form of a vector store of embedded data. This enables the LLM to deliver responses based on the knowledge included in the vector store. By referencing external knowledge, the RAG agent reduces hallucinations and improves the accuracy of the LLM outputs.
Tool 5.1: RAG_DB_gen_tool
• Generate a vector store from a PDF file. First, embed the data and store it for later usage.Tool 5.2: RAG_Retreive_DB_tool
• Retrieve information from a vector store.The framework is complete when all tools are defined and assigned to specific agents, and each agent is designated as a tool for the scheduler agent. When a user submits a prompt, the scheduler analyzes it and forwards it to the agent whose description best matches the prompt. The chosen agent then uses the tool mostly aligned with the prompt's description to complete the task. Users are encouraged to build custom agents and workflows and share them with the community as part of the “Community Agent Framework”, where custom workflows and tools are combined to develop a more robust agent. The code for this framework is available in a GitHub repository: https://github.com/omendibleba/DynaMate.
Representative examples of DynaMate agents
The repository includes a tutorials directory with step-by-step instructions on how each agent's and tool's code functions, providing a useful guide for developing custom tools for specific workflows. This section covers several common examples of each agent's functionality, while comprehensive examples for all agents and tools can be found in the GitHub repository.First, we tested Agent 1 and its tools. The test generates a LAMMPS data file for a molecule, given only the system's SMILES string, number of molecules, and system size. The tool assigns the OPLS-AA force field by default, but this can be customized. Fig. 4 presents the input and the agent's generated response.
The first output that appears is “Entering new AgentExecutor chain…” which refers to the input being processed by the scheduler. In the background, the scheduler decides that it needs to send the task to the system preparation agent (agent 1) since its description best matches the prompt. Then, this latter agent selects which tool it needs to use to complete the task. In this case, the selected tool is the mosdef_tool (i.e., Tool 1.1). Once the tool is invoked, we see that a dictionary is generated, storing the pertinent inputs for the tool. Finally, an output message is generated by agent 1 after performing the task, and this output is passed on to the scheduler. The scheduler, finally, repeats the output message and gives it as an output message to the user. Next to the output is a visualization of the generated data file using VMD.35
The next test evaluates the agent's ability to generate a LAMMPS data file from a CIF file in P1 symmetry, as specified in the lammps_interface documentation. The inputs for this tool are the path of the CIF file and the force field to use. In this case, we use a CIF file for IRMOF-1 and the UFF4MOF force field. Fig. 5 presents the input and response from the agent.
The output shows that the scheduler properly sends the prompt information to the right agent and selects the correct tool for the task. This is confirmed by invoking the ‘lmp_interface_tool’ (i.e., Tool 1.3) and correctly generating the data file of interest. The generated data files are visualized and presented next to the output to confirm the integrity of the files and the structure. Lammps_interface also generates input files defining the type of equations to describe the simulation's interactions. This file should be edited with parameters specific to the type of simulation of interest, and the force fields must be tested and validated before using the files for production runs. The next test involves using the generated data file for IRMOF-1 to generate a Moltemplate file that stores the force field parameters of this system. These templates present a great benefit since they can be later used to generate custom and complex systems involving this or any other molecule of interest. This is achieved by invoking the ‘data_to_template_tool’ (i.e., Tool 1.4) as shown in Fig. 6.
 | ||
| Fig. 6 DynaMate generates template files for Moltemplate using a LAMMPS data file and input file. The files are successfully generated by the agent and are found in the repository. | ||
More complex systems may include multiple types of molecules. Therefore, we are interested in combining multiple templates of MOFs and solvents, for example. The following test combines the generated template for the MOF and the OPLSS force field to describe liquid ethanol interactions. The exact process can be used to generate systems with N types of molecules. Fig. 7 shows the input and output of the agent when asked to generate a template from the given data file.
The agent's output shows that the selected tool to generate the template was packmol_template_tool. The agent successfully called the correct templates and generated the required files for an MD simulation using LAMMPS. The generated input file calls all the required files, but depending on the templates used, it may not be readily usable for simulations and may require manual editing. Next to the agent's output is a visualization of the system generated using the templates for IRMOF-1 and ethanol. The same approach can be used to generate complex systems of interest. DynaMate includes tools for preparing LAMMPS input files with predefined protocols.
The following example shows how to use the framework to generate an input file that includes system minimization, NPT, and NVT equilibrations at the input temperature and pressure for a system of liquid water. This version of DynaMate does not include a tool that allows an agent to generate the input from scratch without a template. Current efforts are focused on this direction and will be discussed in future publications. However, the current version of LLMs may contain sufficient knowledge to generate simple input files that might be sufficient for test simulations. We recognized this as a limitation on the automatization of this workflow, but it represents a step in the right direction. Fig. 8 presents how the developed template was used to generate the LAMMPS input file used in this example. As previously discussed, the class defining the inputs is modified to account for all the inputs of interest and their types. For this example, the inputs are the name for the input file, temperature, pressure, and number of CPUs to run the simulation.
It is important to highlight that the number of inputs can be customized by simply adding them or modifying them in this section. Similarly, the primary function in this example has a general LAMMPS simulation file, which is filled with the values used as inputs. Since this workflow uses Moltemplate, the default name for the data file is ‘system.data’ Specific settings of the simulation may be modified in this file (DynaMate/chatbot/agent_2/A2_tool_1.py). This example demonstrates the framework's modularity and is a clear example of how input files for virtually any simulation engine can be implemented within this multi-agent framework. This template was used to run the simulation and Fig. 9 presents the input prompt and outputs of the tool used to generate a default LAMMPS input file, enabling a simulation to equilibrate a system of interest.
The inputs in the prompt include the temperature (K), pressure (bar), and number of CPUs to run the simulation. Note that the outputs from the function include the path to the directory where the simulation is running. The name of the selected tool is lmp_create_tool (Tool 2.1). Below the agent's output is the simulation output confirming the simulation ran stably for five ns. A future version of this framework will include, among other new tools, an agent capable of generating a LAMMPS input file from scratch with all the required inputs for the simulation of interest. In cases when an input file is readily accessible, DynaMate has a tool called ‘lammps_run_tool’, which takes as input the name of this input file and the number of CPUs to run the simulation. Then, the agent can submit this simulation directly. Fig. 10 shows an example of how to use this tool.
 | ||
| Fig. 10 DynaMate runs a LAMMPS simulation when given the name of the input and the number of CPUs to run the simulation. | ||
Once the output is presented, the simulation will run in the background, and users can open the log files to confirm that it is running properly. Once the simulation is finished, we use the agent for post-processing analysis. DynaMate includes various tools for analyzing results and trajectories for MD simulations, and tutorials for all are presented in the tutorial notebook for agent 3. For this publication, we present an example of using the agent to calculate and plot the atom-to-atom radial distribution function (RDF) of the oxygen–oxygen pair in liquid water. The used prompt and generated output are shown in Fig. 11. The obtained RDF matches the results presented in the literature.36
 | ||
| Fig. 11 DynaMate calculates and plots the radial distribution function of the oxygen–oxygen pair in liquid water. At the left is the agent function, and on the right is the plotted RDF. | ||
The tool being used is called ‘calc_rdf_tool’ (Tool 3.1), and it takes as inputs the name of the data file and trajectory in DCD format, names of the selections, and types of selected atoms. Currently, the user must specify the number of each atomic type of interest. This might present a limitation for inexperienced users, but the agent can be used to read the LAMMPS data file and identify the type of each atom. This information can later be used to ask for the RDF which is currently calculated using PyLAT37 and with MDAnalysis.38,39
Specific types of simulations, such as enhanced sampling simulations, require a LAMMPS executable compiled with additional packages, like PLUMED. DynaMate has tools that can prepare PLUMED input files for umbrella sampling and metadynamics simulations and tools to recover the free energy surface from each of them. Assuming the user has the correct compilation of these packages, the user can use it to prepare an input for a metadynamics simulation. In the following example, DynaMate generates a PLUMED input file for a 2D metadynamics simulation of alanine dipeptide in vacuum. The inputs include the number of atoms that correspond to each collective variable, and biasing parameters such as height, width, and pace of added Gaussians, the bias factor, and temperature. Fig. 12 shows the tested prompt and its outputs.
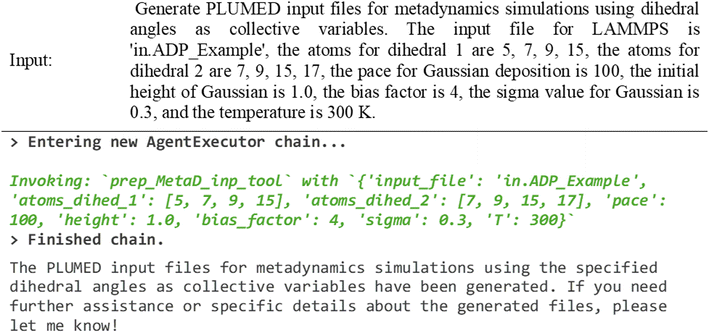 | ||
| Fig. 12 DynaMate prepares a PLUMED input file for a metadynamics simulation of alanine dipeptide in vacuum biasing the phi and psi torsion angles. | ||
The agent calls the correct tool for the task, which is called ‘prep_Metad_inp_tool’ (Tool 4.5), and creates the input dictionary properly from the prompt. Finally, it generates the metadynamics input file, and if all other required files are already accessible, the simulation can be run in the same way presented in Fig. 9. After the simulation finishes, use tools to analyze the sampling on the simulation and recover the unbiased free energy surface. By using the ‘MetaD_analysis_tool’ (Tool 4.6) we can obtain this information using a prompt such as the one presented in Fig. 13.
The default name for the PLUMED output file is “colvar.dat”, but any name should be defined in the input path and loaded properly. The function returns ‘None’ because the structured tool option to return direct is set to True, so after running the function, it returns the function output and not an LLM message. Since this tool generates and shows plots, incorrect LLM messages can appear as if the agent did not properly achieve the goal. Fig. 13, panel A) shows the biased CVs versus time, panel B) shows CV1 vs. CV2, and panel C) is the recovered free energy surface of the system as a function of CV1 and CV2, which agrees with results in the literature.40 This tool also has the capability of recovering the FES of 1 of the two biased variables, and the changes in the analysis will be automatically performed if the PLUMED file has two biased CVs, but the user specifies one dimension in the prompt. More detailed information about this and all other tools is available in the tutorial notebooks.
Agent 5 is currently a typical RAG agent, which enriches its responses from the information in a vector store. For this test case, the knowledge base includes the documentation of LAMMPS, and this agent can be used to answer questions about the inputs and LAMMPS requirements for specific types of simulations. An additional tool enables the agent to generate a new vector store from a given PDF file, which can then be used as a knowledge base. Detailed examples of how to use the agents with its RAG functionalities are available in tutorial notebooks in the project's repository. The retrieval process involves identifying relevant sections from the LAMMPS documentation using semantic similarity calculations, ensuring that the agent accesses the most pertinent information needed to address specific queries about setups. The generation component of the RAG agent then synthesizes this information, presenting answers or suggesting modifications in a coherent and contextually appropriate manner. Augmentation further enhances the response by fine-tuning the information retrieved, ensuring that the suggestions are accurate and aligned with the latest LAMMPS standards.
The presented examples showcase the modularity of DynaMate's agent framework and the relative ease of including custom code related to personal research workflows. Using templates reduces the effort needed to generate tools and enables agent communication. This research represents our first effort in generating an agent framework for generating and analyzing molecular dynamics simulations. Therefore, some tools are currently not at the desired automation level. Future versions of this framework will include newer tools that give the agent more control, allowing for more complex prompts. In addition to the benefit of automation, this modular framework presents a different way of storing and interacting with code from other users.
Outlook
We present a code template that can be used to build multi-agent frameworks that leverage LLMs to automate time consuming tasks involved in complex molecular modeling workflows. Using open-source libraries and models, this modular template enables the facile incorporation of functions in Python code into a semi-automated process. Our tests showed how DynaMate can generate molecular configurations, assign force field parameters, run simulations, and perform various types of post-processing analysis. Moreover, DynaMate provides a modular framework that allows the exchange of tools between researchers and significantly reduces the learning curve related to building tools for non-experts on the task of interest. Due to its versatility, we believe DynaMate will be used to automate repetitive research tasks and aid researchers in developing molecular modeling workflows at a faster pace. Moreover, we believe that the community will continue to build on this framework, which will allow for faster automation and more robust scientific workflows. Users and developers are encouraged to share their tools and agents developed for custom workflows with the objective of building a more robust and interdisciplinary agent framework.Data availability
Data for this article, including source code, instructions, and tutorial material, are available at DynaMate at https://github.com/omendibleba/DynaMate.Conflicts of interest
The authors have no conflict to disclose.Acknowledgements
This work was performed using the computational resources provided by the Notre Dame Center for Research Computing (NDCRC). Y. C. acknowledges funding support from the U.S. Department of Education (award no. P200A210048) and the University of Notre Dame. U. C. acknowledges funding support from the University of Puerto Rico – Mayagüez.References
- V. Mavroudis, Vasilios Mavroudis To Cite This Version: HAL Id: Hal-04817573 LangChain, 2024 Search PubMed.
- S. M. Jain, Hugging Face BT – Introduction to Transformers for NLP: With the Hugging Face Library and Models to Solve Problems, ed. S. M. Jain, Apress, Berkeley, CA, 2022, pp. 51–67, DOI:10.1007/978-1-4842-8844-3_4.
- A. D. White, G. M. Hocky, H. A. Gandhi, M. Ansari, S. Cox, G. P. Wellawatte, S. Sasmal, Z. Yang, K. Liu, Y. Singh and W. J. Peña Ccoa, Assessment of Chemistry Knowledge in Large Language Models That Generate Code, Digital Discovery, 2023, 2(2), 368–376, 10.1039/D2DD00087C.
- K. M. Jablonka, P. Schwaller, A. Ortega-Guerrero and B. Smit, Leveraging Large Language Models for Predictive Chemistry, Nat. Mach. Intell., 2024, 6(2), 161–169, DOI:10.1038/s42256-023-00788-1.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Highly Accurate Protein Structure Prediction with AlphaFold, Nature, 2021, 596(7873), 583–589, DOI:10.1038/s41586-021-03819-2.
- G. Pesciullesi, P. Schwaller, T. Laino and J.-L. Reymond, Transfer Learning Enables the Molecular Transformer to Predict Regio- and Stereoselective Reactions on Carbohydrates, Nat. Commun., 2020, 11(1), 4874, DOI:10.1038/s41467-020-18671-7.
- M. Alberts, T. Laino and A. C. Vaucher, Leveraging Infrared Spectroscopy for Automated Structure Elucidation, Commun. Chem., 2024, 7, 268, DOI:10.1038/s42004-024-01341-w.
- M. Arslan, H. Ghanem, S. Munawar and C. Cruz, A Survey on RAG with LLMs, Procedia Comput. Sci., 2024, 246, 3781–3790, DOI:10.1016/j.procs.2024.09.178.
- G. Xiong, Q. Jin, Z. Lu and A. Zhang, Benchmarking Retrieval-Augmented Generation for Medicine, 2024 Search PubMed.
- A. J. Yepes, Y. You, J. Milczek, S. Laverde and R. Li, Financial Report Chunking for Effective Retrieval Augmented Generation, 2024 Search PubMed.
- J. Kim and M. Min, From RAG to QA-RAG: Integrating Generative AI for Pharmaceutical Regulatory Compliance Process, 2024 Search PubMed.
- J. A. Recio-garcia and M. G. Orozco-del-castillo, Case-Based Reasoning, 2024 Search PubMed.
- D. A. Boiko, R. MacKnight, B. Kline and G. Gomes, Autonomous Chemical Research with Large Language Models, Nature, 2023, 624(7992), 570–578, DOI:10.1038/s41586-023-06792-0.
- C. W. Coley, D. A. Thomas, J. A. M. Lummiss, J. N. Jaworski, C. P. Breen, V. Schultz, T. Hart, J. S. Fishman, L. Rogers, H. Gao, R. W. Hicklin, P. P. Plehiers, J. Byington, J. S. Piotti, W. H. Green, A. J. Hart, T. F. Jamison and K. F. Jensen, A Robotic Platform for Flow Synthesis of Organic Compounds Informed by AI Planning, Science, 2019, 365(6453), eaax1566, DOI:10.1126/science.aax1566.
- Significant-Gravitas, AutoGPT, GitHub repository, GitHub, 2023 Search PubMed.
- yoheinakajima, babyagi, GitHub repository, GitHub, 2023 Search PubMed.
- A. D. McNaughton, G. K. Sankar Ramalaxmi, A. Kruel, C. R. Knutson, R. A. Varikoti and N. Kumar, CACTUS: Chemistry Agent Connecting Tool Usage to Science, ACS Omega, 2024, 9(46), 46563–46573, DOI:10.1021/acsomega.4c08408.
- S. Liu, Y. Lu, S. Chen, X. Hu, J. Zhao, T. Fu and Y. Zhao, DrugAgent: Automating AI-Aided Drug Discovery Programming through LLM Multi-Agent Collaboration, 2024 Search PubMed.
- A. Ghafarollahi and M. J. Buehler, AtomAgents: Alloy Design and Discovery through Physics-Aware Multi-Modal Multi-Agent Artificial Intelligence, 2024 Search PubMed.
- S. Cox, Q. L. Campbell, J. Medina, B. Watterson and A. White, MDCROW: Automating Molecular Dynamics Workflows with Large Language Models, 2025 Search PubMed.
- A. M. Bran, S. Cox, O. Schilter, C. Baldassari, A. D. White and P. Schwaller, Augmenting Large Language Models with Chemistry Tools, Nat. Mach. Intell., 2024, 6(5), 525–535, DOI:10.1038/s42256-024-00832-8.
- Y. Kang and J. Kim, ChatMOF: An Artificial Intelligence System for Predicting and Generating Metal-Organic Frameworks Using Large Language Models, Nat. Commun., 2024, 15(1), 4705, DOI:10.1038/s41467-024-48998-4.
- A. P. Thompson, H. M. Aktulga, R. Berger, D. S. Bolintineanu, W. M. Brown, P. S. Crozier, P. J. in't Veld, A. Kohlmeyer, S. G. Moore, T. D. Nguyen, R. Shan, M. J. Stevens, J. Tranchida, C. Trott and S. J. Plimpton, LAMMPS – A Flexible Simulation Tool for Particle-Based Materials Modeling at the Atomic, Meso, and Continuum Scales, Comput. Phys. Commun., 2022, 271 DOI:10.1016/J.CPC.2021.108171.
- A. I. Jewett, D. Stelter, J. Lambert, S. M. Saladi, O. M. Roscioni, M. Ricci, L. Autin, M. Maritan, S. M. Bashusqeh, T. Keyes, R. T. Dame, J.-E. Shea, G. J. Jensen and D. S. Goodsell, Moltemplate: A Tool for Coarse-Grained Modeling of Complex Biological Matter and Soft Condensed Matter Physics, J. Mol. Biol., 2021, 433(11), 166841, DOI:10.1016/j.jmb.2021.166841.
- L. Martínez, R. Andrade, E. G. Birgin and J. M. Martínez, PACKMOL: A Package for Building Initial Configurations for Molecular Dynamics Simulations, J. Comput. Chem., 2009, 30(13), 2157–2164, DOI:10.1002/jcc.21224.
- W. L. Jorgensen, D. S. Maxwell and J. Tirado-Rives, Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids, J. Am. Chem. Soc., 1996, 118(45), 11225–11236, DOI:10.1021/ja9621760.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, Development and Testing of a General Amber Force Field, J. Comput. Chem., 2004, 25(9), 1157–1174, DOI:10.1002/jcc.20035.
- peteboyd, lammps_interface, GitHub repository, GitHub, 2019 Search PubMed.
- R. S. DeFever, R. A. Matsumoto, A. W. Dowling, P. T. Cummings and E. J. Maginn, MoSDeF Cassandra: A Complete Python Interface for the Cassandra Monte Carlo Software, J. Comput. Chem., 2021, 42(18), 1321–1331, DOI:10.1002/jcc.26544.
- rdkit, Rdkit, GitHub repository, GitHub, 2013 Search PubMed.
- G. A. Tribello, M. Bonomi, D. Branduardi, C. Camilloni and G. Bussi, PLUMED 2: New Feathers for an Old Bird, Comput. Phys. Commun., 2014, 185(2), 604–613, DOI:10.1016/j.cpc.2013.09.018.
- J. Kästner, Umbrella Sampling, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2011, 1(6), 932–942, DOI:10.1002/WCMS.66.
- A. Barducci, M. Bonomi and M. Parrinello, Metadynamics, WIREs Comput. Mol. Sci., 2011, 1(5), 826–843, DOI:10.1002/wcms.31.
- D. Branduardi, F. L. Gervasio and M. Parrinello, From A to B in Free Energy Space, J. Chem. Phys., 2007, 126(5), 54103, DOI:10.1063/1.2432340.
- W. Humphrey, A. Dalke and K. Schulten, VMD – Visual Molecular Dynamics, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- J. Wang, G. Román-Pérez, J. Soler, E. Artacho and M. Fernández-Serra, Density, structure, and dynamics of water: The effect of van der Waals interactions, J. Chem. Phys., 2011, 134(2), 024516, DOI:10.1063/1.3521268.
- M. T. Humbert, Y. Zhang and E. J. Maginn, PyLAT: Python LAMMPS Analysis Tools, J. Chem. Inf. Model., 2019, 59(4), 1301–1305, DOI:10.1021/acs.jcim.9b00066.
- N. Michaud-Agrawal, E. J. Denning, T. B. Woolf and O. Beckstein, MDAnalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations, J. Comput. Chem., 2011, 32(10), 2319–2327, DOI:10.1002/jcc.21787.
- R. J. Gowers, M. Linke, J. Barnoud, T. J. E. Reddy, M. N. Melo, S. L. Seyler, J. Domański, D. L. Dotson, S. Buchoux, I. M. Kenney and O. Beckstein, MD Analysis: A Python Package for the Rapid Analysis of Molecular Dynamics Simulations, in Proceedings of the 15th Python in Science Conference, ed. S. Benthall and S. Rostrup, 2016, pp. 98–105, DOI:10.25080/Majora-629e541a-00e.
- Z. Šućur and V. Spiwok, Sampling Enhancement and Free Energy Prediction by the Flying Gaussian Method, J. Chem. Theory Comput., 2016, 12(9) DOI:10.1021/acs.jctc.6b00551.
| This journal is © The Royal Society of Chemistry 2025 |








