Graphene-based electrochemical sensors for antibiotics: sensing theories, synthetic methods, and on-site monitoring applications
Yangguang
Zhu
abd,
Chen
Ye
cd,
Xiao
Xiao
 h,
Zhuang
Sun
ad,
Xiufen
Li
h,
Zhuang
Sun
ad,
Xiufen
Li
 b,
Li
Fu
b,
Li
Fu
 e,
Hassan
Karimi-Maleh
e,
Hassan
Karimi-Maleh
 fg,
Jun
Chen
*h and
Cheng-Te
Lin
fg,
Jun
Chen
*h and
Cheng-Te
Lin
 *acd
*acd
aKey Laboratory of Advanced Marine Materials, Ningbo Institute of Materials Technology and Engineering (NIMTE), Chinese Academy of Sciences, Ningbo 315201, P. R. China. E-mail: linzhengde@nimte.ac.cn
bLaboratory of Environmental Biotechnology, School of Environmental and Civil Engineering, Jiangnan University, Wuxi 214122, P. R. China
cCenter of Materials Science and Optoelectronics Engineering, University of Chinese Academy of Sciences, Beijing 100049, P. R. China
dQianwan Institute, Ningbo Institute of Materials Technology and Engineering (NIMTE), Chinese Academy of Sciences, Ningbo 315201, P. R. China
eCollege of Materials and Environmental Engineering, Hangzhou Dianzi University, Hangzhou 310018, P. R. China
fSchool of Resources and Environment, University of Electronic Science and Technology of China, Chengdu 611731, P. R. China
gSchool of Engineering, Lebanese American University, Byblos 1102-2801, Lebanon
hDepartment of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA. E-mail: jun.chen@ucla.edu
First published on 7th October 2024
Abstract
Owing to the extensive use of antibiotics for treating infectious diseases in livestock and humans, the resulting residual antibiotics are a burden to the ecosystem and human health. Hence, for human health and ecological safety, it is critical to determine the residual antibiotics with accuracy and convenience. Graphene-based electrochemical sensors are an effective tool to detect residual antibiotics owing to their advantages, such as, high sensitivity, simplicity, and time efficiency. In this work, we comprehensively summarize the recent advances in graphene-based electrochemical sensors used for detecting antibiotics, including modifiers for electrode fabrication, theoretical elaboration of electrochemical sensing mechanisms, and practical applications of portable electrochemical platforms for the on-site monitoring of antibiotics. It is anticipated that the current review will be a valuable reference for comprehensively comprehending graphene-based electrochemical sensors and further promoting their applications in the fields of healthcare, environmental protection, and food safety.
Wider impactThe key points discussed in this review include the fabrication of graphene-based electrochemical sensors for antibiotics, the theoretical underpinnings of electrochemical sensing mechanisms, and the evolution of portable electrochemical platforms for on-site monitoring. These advancements are of significant wider interest as they not only enhance our ability to detect and quantify antibiotics in various samples but also contribute to the broader fields of environmental science, public health, and materials engineering. The future is expected to be marked by further refinement of sensor technology, with a focus on improving detection limits, selectivity, and the practicality of on-site monitoring. The insights provided in this review will be instrumental in guiding future research, particularly in understanding the complex interactions at the electrode surface and the redox processes that facilitate the detection of specific antibiotics. By addressing the questions of detection performance improvement, material modification on graphene nanosheets, the underlying sensing mechanisms, and the status of equipment platforms, this review aims to clarify the current state of graphene-based electrochemical sensors for antibiotics and their readiness for real-world applications. The potential for miniaturization and portability is a critical aspect as it will enable more widespread and accessible monitoring of antibiotic residues, thus safeguarding both ecological systems and human health. |
1. Introduction
Antibiotics can be utilized as revolutionary medicines for curing various infectious diseases in livestock and humans.1–3 However, inappropriate use of antibiotics leads to the excessive release of residual antibiotics into the aquaculture industry, medications, and wastewater.4 The accumulation of residual antibiotics in the environment produces harmful effects on nontarget organisms, and causes contamination of food and natural water resources and associated human health threats by bacterial resistance to antibiotics.5–7 In particular, antibiotic consumption has reached over 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ton and 0.7 million deaths per year related to antibiotic resistance is estimated on a global scale.8 According to the declaration of the World Health Organization, numerous antibiotics are identified as potential environmental contaminants and classified as furans, amphenicols, β-lactams, quinolones, macrolides, sulfonamides, tetracyclines and aminoglycosides.9–11 For human health and ecological safety, it is urgent to monitor the residual antibiotics with accuracy and convenience.
000 ton and 0.7 million deaths per year related to antibiotic resistance is estimated on a global scale.8 According to the declaration of the World Health Organization, numerous antibiotics are identified as potential environmental contaminants and classified as furans, amphenicols, β-lactams, quinolones, macrolides, sulfonamides, tetracyclines and aminoglycosides.9–11 For human health and ecological safety, it is urgent to monitor the residual antibiotics with accuracy and convenience.
Conventional analytical methods, such as high-performance liquid chromatography (HPLC), liquid chromatography–mass spectrometry (LC–MS), gas chromatography–mass spectrometry (GC–MS), and electrochemical detection methods, for antibiotic detection have been under development for decades.12–15 Among them, chromatography enables the simultaneous detection of multiple antibiotics and offers advantages such as high sensitivity and strong anti-interference capability.12,13 Yet, it still faces bottlenecks, such as costly equipment, complex sample pretreatments, and high detection costs, making it suitable only for sample analysis in laboratory research. Hence, constructing electrochemical sensors has been preferably accepted due to its high sensitivity, simplicity, and time efficiency.16–18 However, direct electrochemical detection of antibiotics on conventional electrodes like glassy carbon electrodes (GCEs) usually confronts low sensitivity. Thus, adopting chemical modification with various nanomaterials on electrochemical electrodes has become a widely used strategy.19–21
Graphene is a two-dimensional nanomaterial consisting of a single layer of carbon atoms arranged in a hexagonal honeycomb lattice. Its unique atomic structure and electronic properties endow it with exceptional electrical, optical, and chemical characteristics.22 When considering its electrochemical aspects, it has been regarded as a perfect electrode modification material due to its excellent properties, such as high surface-to-volume ratio, good electrical conductivity, favourable biocompatibility and rapid electron transferring rate.18,23,24 Moreover, electrochemical signals can be further enhanced through the functionalization of graphene via covalent and non-covalent modifications.25 Recently, graphene-based electrochemical sensors for antibiotics have been extensively constructed, and the detection performance including selectivity, limit of detection (LOD) and real sample analysis have been greatly improved.26–28
As far as we know, the focus of the reported reviews is on describing the status via bibliometric analysis and highlighting the research on graphene-based nanocomposite-modified electrodes in the context of materials science and their application in determining antibiotics in real samples within a laboratory environment.29–34 However, it is inadequate for comprehensively reviewing graphene-based electrochemical sensors for antibiotics. The specific function of graphene with various materials in electrode fabrication is not elaborated. The antibiotic detection mechanism involved in electrochemical sensing is not clear yet, especially referring to the redox reaction process involving electron transferring and interfacial interactions between the electrode surface and the adsorbed molecules at the atomic level. Furthermore, whether the electrochemical sensors constructed in current stage achieve the requirement for the on-site monitoring of antibiotics in real samples is unknown.
In this review, we focus on three aspects: graphene-based modifiers for electrode fabrication, theoretical analyses of electrochemical sensing mechanisms, and portable electrochemical platforms for the on-site monitoring of antibiotics in real samples. It is anticipated that our review will be a valuable reference for comprehending the whole fabrication process of graphene-based electrochemical sensors, and further promoting the development of the sensors for practical applications in trace level detection of antibiotics.
2. Overview of the construction of graphene-based electrochemical sensors for antibiotics
Since 2D graphene with various forms can be fabricated in mass production, graphene-based electrochemical sensors for antibiotics have been constructed after extensive research.35–37 Antibiotics can be classified as aminoglycosides, tetracyclines, sulfonamides, macrolides, quinolones, beta-lactams, amphenicols and furans. Their general characteristics and types of electrodes are shown in Table 1. A development timeline of graphene-based electrochemical sensors for antibiotics including aspects as antibiotics, modifiers and LOD performance is summarized in Fig. 1a. Modifiers based on graphene can be classified as metal nanoparticles (NPs), carbon nanotubes (CNTs), metal–organic frameworks (MOF), metal oxides, antibodies and aptamers. Based on the data during the past ten decades, the distribution of antibiotic types to be detected is inhomogeneous, wherein amphenicol and aminoglycoside antibiotics occupy the majority. Hence, graphene-based electrochemical sensors for detecting other types of antibiotic molecules need to be enhanced in the future.| Antibiotic class | Type | Chemical formula | Mechanism of action | Side-effects | Electrodes | Ref. |
|---|---|---|---|---|---|---|
| Aminoglycosides | Streptomycin | C21H39N7O12 | Inhibits protein synthesis | Ototoxicity, nephrotoxicity | GO/P(NIPAm-MPTC-GMA) | 38 |
| Kanamycin | C18H36N4O11 | GO/Pt–Cu alloy/aptamer | 39 | |||
| Tetracyclines | Tetracycline | C22H24N2O8 | Inhibits protein synthesis | Endocrine disruption of aquatic species, | rGO/Fe3O4/aptamer | 40 |
| Oxytetracycline | C22H24N2O9 | GO/Au NPs/aptamer | 41 | |||
| Sulfonamides | Sulfamethoxazole | C12H14N4O4S | Inhibits folic acid synthesis | Diarrhea, vomiting | Graphene/ZnO | 42 |
| Sulfadiazine | C11H11N3O2S | GO/COF | 43 | |||
| Macrolides | Azithromycin | C38H72N2O12 | Inhibits protein synthesis | Decreased shelf-life | GO/CNTs | 44 |
| Erythromycin | C37H67NO13 | Graphene/Au–Pt NPs | 45 | |||
| Quinolones | Ciprofloxacin | C17H18FN3O3 | Inhibits DNA replication | Severe hepatic toxicity | rGO/Au NPs/aptamer | 46 |
| Ofloxacin | C18H20FN3O4 | GO/ionic liquid | 47 | |||
| Beta-lactams | Amoxicillin | C16H19N3O5S | Inhibits cell wall synthesis | Rashes, fever | GO/CdTe/Au NPs | 48 |
| Penicillin | C16H18N2O4S | GO/Fe3O4/CNTs/aptamer | 49 | |||
| Amphenicols | Chloramphenicol | C11H12Cl2N2O5 | Inhibits protein synthesis | Inhibition of bone marrow | rGO/Co3O4 | 50 |
| Furans | Nitrofurantoin | C8H6N4O5 | Inhibits protein synthesis | Ecological risks, human health damage | rGO/GdFeO3 | 51 |
| Furazolidone | C8H7N3O5 | GO/GeW | 52 |
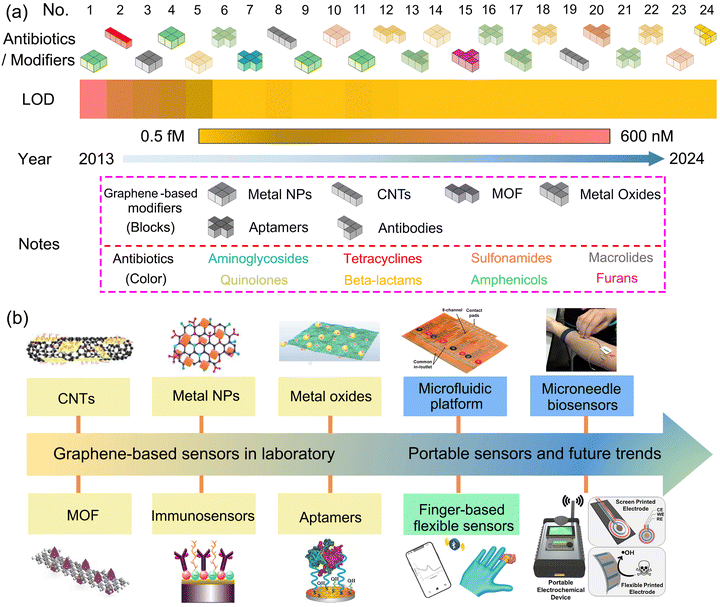 | ||
| Fig. 1 Development timeline of graphene-based electrochemical sensors for antibiotics. (a) Aspects such as antibiotics, modifiers and LOD performance, and all corresponding data originate from ref. 17, 27, 46 and 53–73. (b) Evolution history of equipment platforms.44,62,69,74–78 | ||
Aptamers or immunosensors based on graphene-modified electrodes exhibit superior detection performance towards antibiotics than other types of modifiers with a lower LOD and deserve to be further developed. The equipment platforms constructed in electrochemical sensors is essential to real applications successfully and its evolution history is depicted in Fig. 1b. Graphene-based sensors in laboratory require bulky equipment and multiple processing involved in the conventional assays, while in the contrary, potable sensors being capable of on-site monitoring indicates miniaturized device, simplified processing and wireless transmission technology, and prevails in the future.
Fig. 2a depicts the relationships between three aspects as A, B and C. Suitable modifiers designed for electrode fabrication exhibit superior detection performance, and their upper limit was determined by real-sample analysis in this work. Conversely, the interpretation of sensing mechanism provides theoretical prediction of suitable modifiers for specific antibiotics. The sensing mechanism for antibiotics includes two parts: electrochemical reaction and interfacial reaction at the atomic level, as presented in Fig. 2b. Fig. 2c illustrates the miniaturization of sensors containing an integrated platform and screen-printed electrodes for on-site monitoring in real applications. The three aspects as A, B and C will be comprehensively discussed in the further sections.
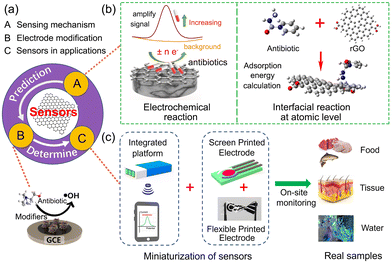 | ||
| Fig. 2 Schematic of key review elements involved in graphene-based electrochemical sensors for antibiotics. (a) Relationship between the above-mentioned three aspects. (b) Electrochemical sensing mechanism in theory. Reproduced with permission.79 Copyright from 2022, American Chemical Society. (c) Miniaturized sensing devices for on-site monitoring in real-time applications. | ||
3. Theoretical analyses of electrochemical sensing mechanism
Electrochemical sensing signal is produced by the electron transfer of analytes via a redox reaction on the modified electrode, which is usually proportional to the analyte concentration in practical detection.29 The interpretation of antibiotic sensing mechanism theoretically is more important for pursuing excellent detection performance by various electrochemical techniques. Here, the antibiotic sensing mechanism includes two parts: the analysis of electrical responses at the electrochemical level, and the redox reaction process involving electron transferring and interfacial interactions between the electrode surface and the adsorbed molecules simulated by density functional theory.80,813.1 Electrochemical reaction mechanism
The principle of electrochemical antibiotic sensors is based on sensing signals generated by the redox reaction of antibiotics on the surface of graphene-based modifiers and converted into identifiable electrical signals proportional to the concentrations of antibiotics.82 The sensing electrical signal was detected using various electroanalytical techniques such as cyclic voltammetry (CV), differential pulse voltammetry (DPV), electrochemical impedance spectroscopy (EIS) and chronoamperometry (i–t). To construct optimal electrochemical sensors for specific antibiotic detection, three key aspects including electrode modification, sensors in real-time applications and sensing mechanism will be fully discussed. The operation modes of electrochemical sensors for antibiotics are schematically illustrated in Fig. 3a.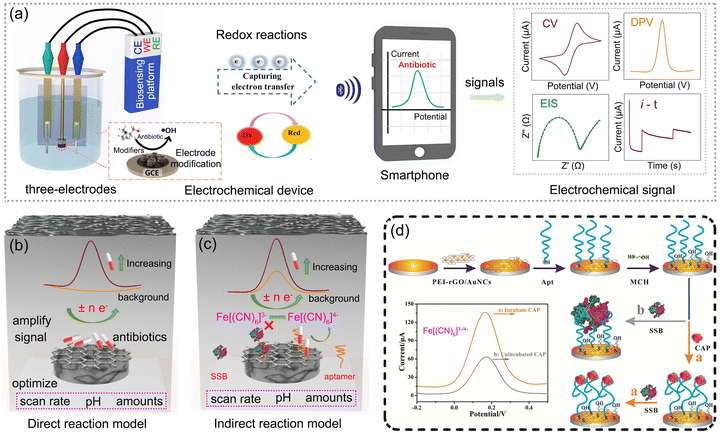 | ||
| Fig. 3 Working mechanism of electrochemical sensors for antibiotics: (a) electrochemical operation modes. (b) and (c) Scheme of direct reaction model and indirect reaction model. (d) Scheme of electrochemical aptasensor for chloramphenicol (CAP) detection. Reproduced with permission.69 Copyright from 2021, Elsevier. | ||
The electrical signals produced from different electrochemical detection models show a large difference due to the different construction designs of the sensing system.51,69,83 Through the review of the above-mentioned electrochemical sensors for antibiotics, two types of electrochemical detection models can be summarized as a direct reaction model and an indirect reaction model, as shown in Fig. 3b and c. Notably, the choice of electrochemical sensors depends on the properties of the target substance, leading to varying applicability of the two principles. For antibiotics containing structural components that readily participate in redox reactions, such as furazolidone and sulfamethoxazole, the direct model is appropriate for the detection. Conversely, for target antibiotics that exhibit difficulty in catalytic redox reactions, such as streptomycin and tetracyclines, the indirect model is the alternative option.
The electrical signals produced in the direct model are based on the direct catalytic reaction of antibiotics and enhanced with the increased concentrations of antibiotics. Through the optimization of parameters such as scan rate, pH, and amounts of modified materials, the electrochemical performance of the electrode modified for antibiotic detection is adjusted to the maximum. Then, the electrochemical redox reaction for antibiotic detection can be concluded as, for example, the irreversible reduction of a nitro group (R-NO2) of nitrofurantoin (NFT) into a phenylhydroxylamine group (R-NHOH) corresponding to an equal number of electrons (4e−) and protons (4H+) transferred.51 The current responses triggered by electrochemically active GdFeO3/rGO nanocomposites were enhanced with the increasing concentrations of NFT. However, the direct catalytic reduction of antibiotics consumes more energy and causes increased difficulty in the fabrication of hybrid materials with higher sensitivity.
To further improve the detection performance of antibiotics, indirect reaction models are developed based on the redox reactions of a pair of Fe[(CN)6]3−/4− and the special design of aptasensors. Ascribed to the easier catalytic reactions of a pair of Fe[(CN)6]3−/4− and the specific binding of aptamers and antibiotics, the electrical signals toward antibiotic detection from the indirect reaction models trigger the higher, and obtain better sensitivity than direct reaction models. Chloramphenicol is taken as a typical antibiotic for illustrating the indirect reaction model, as shown in Fig. 3d, wherein polyethyleneimine-functionalized reduced graphene oxide and gold nanocubes (PEI-rGO/AuNCs) served as the modified electrode to immobilize the aptamers.69 The large size and negative charge of single-stranded DNA-binding protein hinder the redox reactions of Fe[(CN)6]3−/4− and reduce the electrical signal drastically. When chloramphenicol is present in the electrolyte, the aptamer–chloramphenicol complex is formed, which inhibits the combination of single-stranded DNA-binding protein with the aptamer, resulting in a stronger response. Herein, the modification of PEI onto the rGO surface can be achieved via hydrogen bonding and electrostatic attraction, effectively anchoring it to the electrode.
In particular, due to the advantages of large specific surface area, strong interfacial charge transfer capability, good stability, and ease of functionalization, graphene is an ideal material for electrode modification, which triggers intense electrochemical responses in electrochemical sensors.
3.2 Interfacial interaction mechanism by density functional theory
Conventional experimental studies are typically carried out in laboratories, involving repeated practices that are both costly and time-consuming. Computational modelling ahead of the experiments is an alternative solution to afford valuable simulations for materials screening, design and mechanism elaboration.84 Density functional theory (DFT) satisfies the requirement for providing the theoretical prediction of material research, gaining popularity in various domains as energy storage, battery materials, environmental catalysis and sensors.85–88 DFT modelling is carried out using software packages such as Vienna ab initio Simulation Package (VASP), Quantum Espresso, Materials Studio, CP2K and CRYSTAL.89 With regard to electrochemical sensors specifically, it offers valuable atomic-scale insights into the structure, properties, and performance of graphene-based nanocomposites and their interfacial interaction with adsorbed antibiotics at the atomic level.| fk+ = qk(N + 1) − qk(N) | (1) |
| fk− = qk(N) − qk(N − 1) | (2) |
The next is to focus on interfacial interactions between the electrode surface and adsorbed antibiotic molecules. The molecular electrostatic potential (MEP) map depicts the electrostatic potential distribution of positive and negative regions in the electrode-antibiotic system, identifying the sites for nucleophilic and electrophilic attacks.92 Qian et al. used the MEP map of isoniazid and different oxygen contents of rGO to reveal interfacial interaction between graphene and target antibiotics.79 The primary distribution of electron density on the oxygen groups of rGO created repulsive forces that prevented isoniazid from approaching the rGO sheet, resulting in worse detection performance. Notably, MEP offers an intuitive way to estimate the best sites of the system for interfacial interactions. Density of states (DOS) provides details about the chemical bonding between graphene-based nanocomposites and antibiotics, exploring the active sites and interfacial interactions from the perspective of orbital interaction.93 A quantitative tool to evaluate the stability of antibiotics adsorbed on various graphene sensing substrates was performed by adsorption energy (Ead) calculation, where the negative value indicates a spontaneous reaction involved in the adsorption process.79 In particular, the more negative value of Ead presents a stronger bond between the antibiotic and the substrate.94 The Ead value can be calculated using eqn (3), where E(sub/atb), E(sub) and E(atb) depict the total energies of the antibiotic-adsorbed graphene material, sensor substrate and antibiotic, respectively. Ren et al. studied the influence of N-doped graphene on the adsorption of phenol ions using Ead calculations.95 It revealed that N doping can alter the electronic structure of graphene and create new active sites, thereby enhancing the adsorption of target molecules.
To further investigate the type of interfacial interactions between graphene-based nanocomposites and antibiotics, the reduced density gradient (RDG) is applied using eqn (4), where ρ(r) denotes the electron density.92 The various types of interactions are discriminated by the function Sign![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) λ2(r)·ρ(r), where Sign
λ2(r)·ρ(r), where Sign![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) λ2(r) indicates the sign of the electron density Hessian matrix's second eigenvalue. Herein, Sign
λ2(r) indicates the sign of the electron density Hessian matrix's second eigenvalue. Herein, Sign![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) λ2(r)·ρ(r) < 0 implies the strong attractive interactions such as hydrogen bond or halogen bond, while Sign
λ2(r)·ρ(r) < 0 implies the strong attractive interactions such as hydrogen bond or halogen bond, while Sign![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) λ2(r)·ρ(r) ≈ 0 depicts the lesser attractive interactions as van der Waals (vdW) attraction and Sign
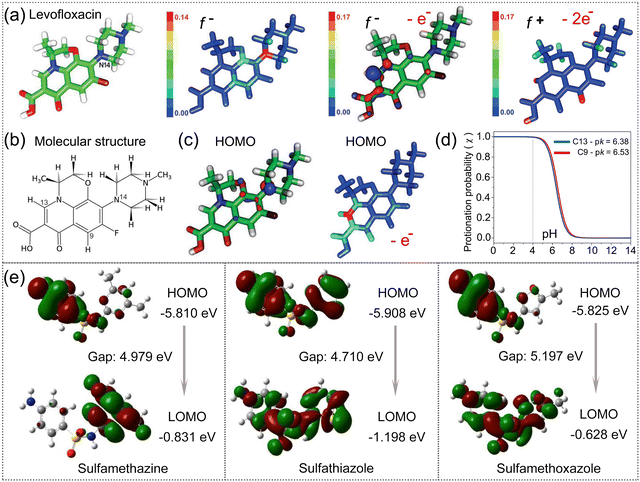
λ2(r)·ρ(r) ≈ 0 depicts the lesser attractive interactions as van der Waals (vdW) attraction and Sign![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) λ2(r)·ρ(r) > 0 presents the strong repulsive interactions as the steric effect. Using eqn (3) and (4), we can interpret the interfacial interactions between graphene-based nanocomposites and antibiotics quantitatively, and predict the suitable electrode modifiers to the specific antibiotic theoretically. Adekoya et al. adopted the RDG map to determine the specific type of interfacial interactions between graphene and antibiotic cephalexin (CEX).92 Hydrogen bonding facilitates the successful binding of CEX on GO/PEG in the [Sign
λ2(r)·ρ(r) > 0 presents the strong repulsive interactions as the steric effect. Using eqn (3) and (4), we can interpret the interfacial interactions between graphene-based nanocomposites and antibiotics quantitatively, and predict the suitable electrode modifiers to the specific antibiotic theoretically. Adekoya et al. adopted the RDG map to determine the specific type of interfacial interactions between graphene and antibiotic cephalexin (CEX).92 Hydrogen bonding facilitates the successful binding of CEX on GO/PEG in the [Sign![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) λ2(r)·ρ(r) < 0] region, confirming a strong interaction between CEX and GO/PEG.
λ2(r)·ρ(r) < 0] region, confirming a strong interaction between CEX and GO/PEG.
| Ead = E(sub/atb) − E(sub) − E(atb) | (3) |
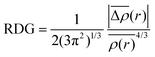 | (4) |
 | ||
| Fig. 4 Identification of active sites and chemical reactivity of antibiotic structures. (a) Illustration of the 3D structure, the lowest energy of conformation, f− indices, (b) molecular structure, (c) HOMO distribution of levofloxacin and (d) protonation probability for C9 and C13 atoms functioned with buffer pH. (a)–(d) Reproduced with permission.90 Copyright from 2018, Wiley-VCH. (e) HOMO, LOMO and the energy gap of sulfamethazine, sulfathiazole and sulfamethoxazole. Reproduced with permission.91 Copyright from 2015, Walter de Gruyter. | ||
In conclusion, to assess the chemical reactivity of the target antibiotic, the HOMO–LUMO energy gap is the suitable evaluation index. A lower value indicates that the target antibiotic is highly polarizable, often exhibiting strong chemical reactivity but low stability. Fukui indexes can be further applied to identify the vulnerable sites of specific antibiotics to redox reactions.
 | ||
| Fig. 5 Interfacial interactions of antibiotics adsorbed on graphene-based nanocomposites. (a) Optimized structure of GO/PEG–CEX. (b) MEP distribution of CEX, GO/PEG and GO/PEG–CEX. The blue regions indicate low electron density, while the red domains depict electron enrichment. RDG isosurface map for determining the type of interfacial interactions in (c) GO, (d) GO/PEG and (e) GO/PEG–CEX. (a)–(e) Reproduced with permission.92 Copyright from 2022, American Chemical Society. | ||
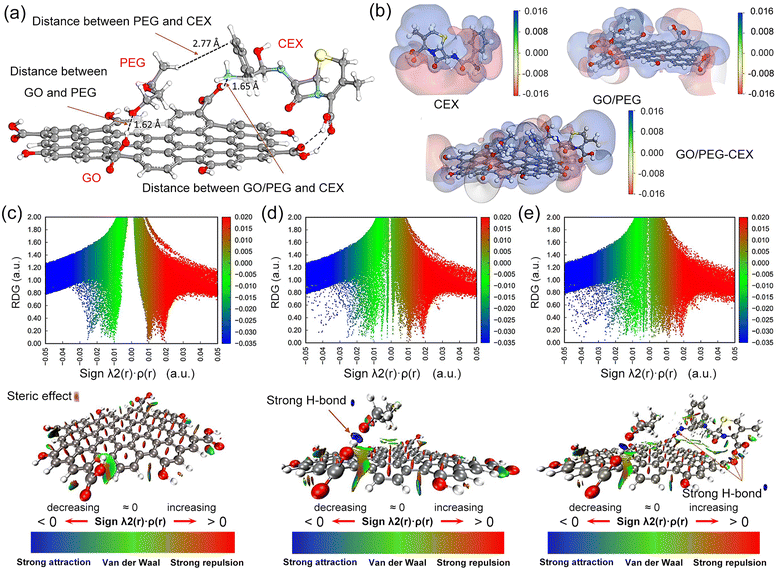
The influences of the electronic structure of graphene regulated by doping with elements such as nitrogen (N), varying oxygen content or hybridization with other materials are vital to elaborate the interfacial interactions of antibiotics adsorbed on graphene-based nanocomposites. Ren et al. investigated the influence of N-doped graphene on the adsorption of phenol ions by DFT modelling.95 As depicted in Fig. 6a–d, the distances between the pristine rGO plane and its next to graphitic N (γ1 and γ2), pyridinic N and O atom of phenol were 2.834, 1.597, 1.508 and 2.426 Å, respectively. Compared to pristine rGO, the shorter C–O distance of N-doped rGO demonstrated that N doping can alter the electronic structure of graphene and create new active sites for enhancing the adsorption of phenol ions. The rGO plane adjacent to graphitic N presents a higher affinity for adsorbing phenol ions than pyridinic N, while pyrrolic N indicates ineffective adsorption capacity. Therefore, N doping enhances the metal-like property of graphene due to the induction of a high positive charge density on the ortho-carbon atoms, promoting its adsorption capacity.
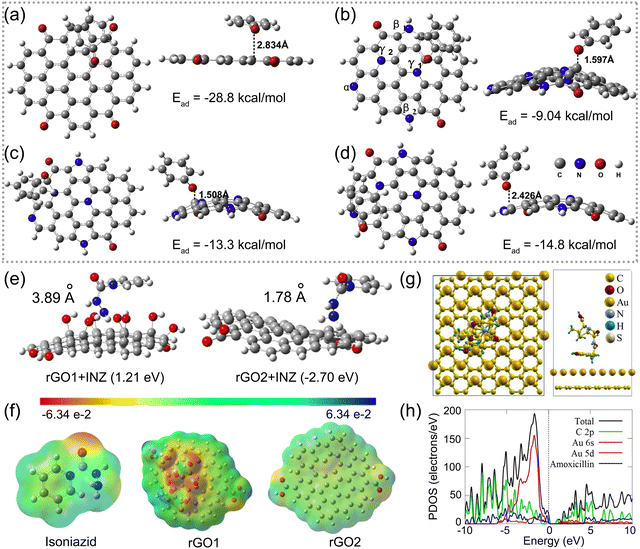 | ||
| Fig. 6 Interfacial interactions of antibiotics adsorbed on graphene-based nanocomposites. Optimal models for the adsorption of a phenol ion on (a) pristine rGO and (b)–(d) various sites of N-doped rGO including three types of N as pyridinic (α), pyrrolic (β) and graphitic (γ). (a)–(d) Reproduced with permission.95 Copyright from 2018, Elsevier. (e) Optimized structures of isoniazid adsorption on rGO1 and rGO2. (f) Molecular electrostatic map of isoniazid, rGO1, and rGO2. (e) and (f) Reproduced with permission.79 Copyright from 2021, American Chemical Society. (g) Illustration of low-energy adsorption of amoxicillin onto the Au/graphene surface. (h) Total density of states (DOS) and partial density of states (PDOS) contributed by the Au/graphene electrode and amoxicillin. (g) and (h) Reproduced with permission.93 Copyright from 2021, Elsevier. | ||
Qian et al. conducted a DFT calculation to reveal the roles of rGO oxygen content in developing graphene-based electrochemical detection of isoniazid with high performance.79 As shown in Fig. 6e, the positive Ead of the rGO1 sensor with a higher oxygen content and isoniazid exhibited repulsive forces that prevented isoniazid from approaching the rGO sheet. Fig. 6f shows the molecular electrostatic potential of isoniazid, rGO1 and rGO2, and indicates the primary distribution of electron density on the oxygen groups to form repulsive forces. The computational results confirmed the experimental data that rGO1 behaved with weaker voltammetric responses. Osikoya et al. investigated the adsorption behaviour of amoxicillin on the Au/graphene electrode by DFT calculation.93 As presented in Fig. 6g, amoxicillin presenting a vertical configuration with a –NH2 group preferred to be adsorbed stably onto the Au/graphene electrode surface, with an Ead value of −2.03 eV. The adsorption behaviour can be attributed to the formation of a strong X–H⋯Au (X = C or N) interaction between the Au atom and the –CH3 or –NH2 group of amoxicillin. Partial density of states results presented in Fig. 6h suggest that amoxicillin has a high affinity towards the Au/graphene nanointerface.
In summary, the adsorption of the target antibiotic preferentially occurred on graphene-based nanocomposites via hydrogen bonding, electrostatic interaction or covalent bonding. Carboxylic, epoxy or hydroxyl functional groups are the common adsorption sites on graphene-based electrodes for targeting antibiotics that contain a –CH3 or –NH2 group. The abundant oxygen functional groups on graphene preferentially facilitate the adsorption of target antibiotic stably through various interfacial interactions.
4. Synthesis of graphene-based modifiers for electrode fabrication
The derivative graphene is widely used as pristine graphene, graphene oxide (GO), and reduced graphene oxide (rGO), exhibiting various attributes with various forms and structures, as shown in Fig. 7a. Pristine graphene comprises carbon atoms arranged in a planar hexagonal lattice, producing excellent electronic and mechanical properties.22 Unfortunately, it still suffers difficulties in bulk production, water dispersion and isolation, hindering its further application in sensing areas.97 The GO surface is loaded with oxygen-containing moieties such as peripheral carboxylic acid (–COOH), in-plane epoxide (–O–) and hydroxyl (–OH) groups, possessing the stronger colloidal stability, hydrophilicity and chemical surface modification.98 However, considerable defect density resulting from oxygen functional groups in GO decreases its electrochemical conductivity up to an electrical insulator, which is weak to sensing applications. rGO, reduced oxygen content from GO, exhibits superior electrical conductivity.30 Yet, removing oxygen groups results in less hydrophilicity and difficulty in water dispersion to fully absorb antibiotics. To develop practical applications of graphene based on its superior physical/chemical properties, the investigation of mass-production methods compatible with the industry standard is highly demanded. The state-of-the-art synthesis approaches include Hummers’ method, mechanical exfoliation in the liquid phase, and chemical vapor deposition (CVD), with the tunability of lateral size, crystallinity, and price for their own applications.99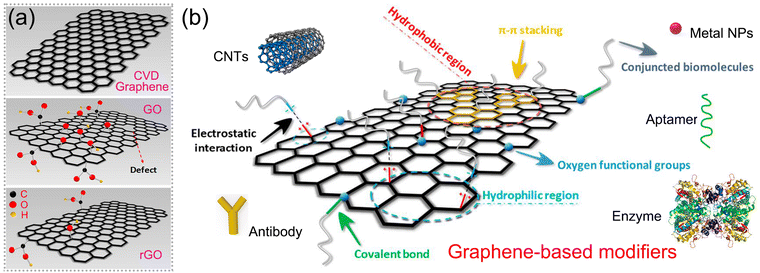 | ||
| Fig. 7 Surface functionalization/decoration of graphene: (a) graphene and its derivatives. (b) Schematic of various modifiers functionalized on graphene. Reproduced with permission.100 Copyright from 2019, Multidisciplinary Digital Publishing Institute. | ||
As graphene's electrocatalytic activity is restricted, the functionalization of graphene is considered the effective way to facilitate its applications further and is classified as covalent and non-covalent modification, as illustrated in Fig. 7b. Covalent modification can be achieved by forming amide and carbamate ester bonds with the carboxyl and hydroxyl groups, and non-covalent modification includes weak van der Waals forces, π–π stacking, and electrostatic interaction.25,101,102 A series of materials such as metal NPs, metal oxides, enzymes, aptamers and others are applied as modifiers to combine with graphene and fabricate electrodes to form electrochemical sensors for antibiotics.64,68,93,103,104 Our reviewing in this part focuses on the prevalent methods to fabricate various graphene-based hybrid structures and the relationship between material structure and sensing performance, providing valuable references for the synthesis of graphene-based modifiers for electrode fabrication.
4.1 Solution blending method
Solution blending is the universal and cost-effective method to prepare graphene-based hybrids based on weak van der Waals forces and π–π stacking. Zhu et al. fabricated Ti3C2Tx/TiO2 NPs by in situ oxidation of Ti3C2Tx/nanosheets in a controllable manner and used a small amount of rGO (<2.5 wt%) as a conductive additive to blend with Ti3C2Tx/TiO2 NPs to form Ti3C2Tx/TiO2 NPs/rGO heterojunction electrodes, as illustrated in Fig. 8a.105 The rGO served as an electrically conductive cross-linked network, optimizing the interfacial structure to facilitate electron transfer. Zhu et al. prepared TiO2 nanosheets (NSs) by in situ oxidation of Ti3C2Tx NSs and blended them with polyethyleneimine (PEI)-grafted rGO to obtain a rGO/PEI/TiO2 hybrid, as shown in Fig. 8b. The branched PEI surface is abundant in positively charged amino groups, serving as binding sites to interact with rGO via hydrogen bonding or electrostatic attraction. The van der Waals and/or hydrogen bonding are the primary forces responsible for forming the hybrid and constructing the novel electrochemical aptasensing platform. By this fabrication method, various components with specific sensing functions can be easily modified on graphene, exhibiting good voltammetric responses. Despite the simplicity and cost-effectiveness of the solution blending method, it still encounters shortcomings such as weak interactions and the inability to precisely control the structure and layers of the hybrid on substrates. Thus, sensing performance cannot be precisely regulated in an optimum state.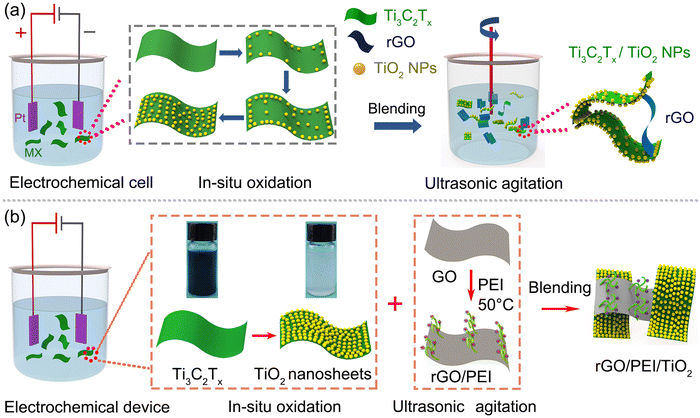 | ||
| Fig. 8 Solution blending method for preparing graphene-based hybrids as (a) Ti3C2Tx/TiO2 NPs/rGO. Reproduced with permission.105 Copyright from 2024, Wiley-VCH. (b) rGO/PEI/TiO2. Reproduced with permission.106 Copyright from 2023, Elsevier. | ||
4.2 Interfacial electrostatic self-assembly method
The interfacial electrostatic self-assembly method not only offers high controllability but also provides an inexpensive and straightforward approach to preparing films with diverse graphene-based hybrid structures on substrates. The electrostatic interaction is the primary driving force involved in the interfacial electrostatic self-assembly, through the reaction between graphene and the opposite charged modifiers.107 Xu et al. fabricated the electroactive rGO/SiO2 nanospheres with huge specific surface area by applying the electrostatic self-assembly method, with Tannic acid (TA) to reduce GO to rGO electrochemically, as illustrated in Fig. 9a.108,109 The hybrid with a 3D structure, as shown in Fig. 9b, not only prevents the aggregation of rGO but also increases the specific surface area, ultimately improving its sensing performance.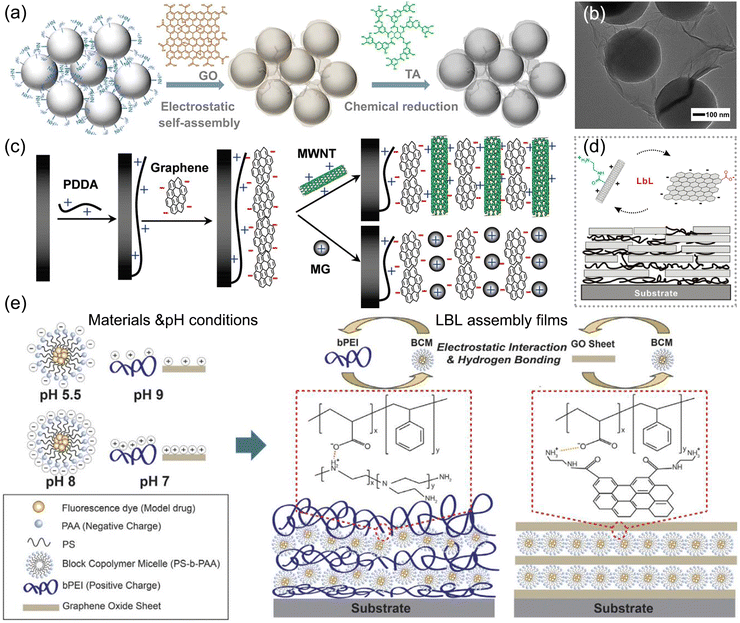 | ||
| Fig. 9 Interfacial electrostatic self-assembly method for preparing films with various graphene-based hybrid structures. (a) Illustrative preparation of rGO/SiO2 nanospheres and (b) their TEM image. (a) and (b) Reproduced with permission.108 Copyright from 2021, American Chemical Society. (c) Controllable formation of electroactive nanostructures by the LBL assembly method. Reproduced with permission.110 Copyright from 2011, American Chemical Society. (d) Multilayered structure of rGO/CNTs by the LBL assembly method. Reproduced with permission.109 Copyright from 2010, American Chemical Society. (e) pH regulation on the structure of LBL-assembled multilayered films. Reproduced with permission.111 Copyright from 2016, Springer Nature. | ||
Furthermore, to precisely regulate the film thickness formed on substrates, the layer-by-layer (LBL) assembly method is commonly used to obtain optimal coating films.112 Multilayered nanostructures composed of graphene and CNTs can be controllably formed on electrodes via the LBL method, using electrostatic and π–π interactions, as illustrated in Fig. 9c and d.110 Typically through the π-stacking interactions, CNTs adhere to GO flakes well and disperse in water stably by the high solubility of GO as a surfactant, facilitating the formation of the films uniformly. The controlled layering process yields optimal performance for sensing applications. The thickness and structure of the LBL-assembled films could be controlled and altered by pH with electrostatic and hydrogen bonding, providing an innovative perspective to fabricate the hybrid film by the LBL method, as shown in Fig. 9e.111
In conclusion, an interfacial electrostatic self-assembly offers an optimal method for fabricating a uniform hybrid structure with an enlarged specific surface area and improved target adsorption, thereby boosting its sensing performance.
4.3 In situ growth method
The intermolecular interactions between graphene and other specific modifiers can be further enhanced by an in situ growth method. The method encompasses two approaches: the in situ growth of graphene on a conductive substrate by atomic deposition and the in situ growth of modifiers on graphene by solvo-hydrothermal synthesis. Fig. 10a presents the formation process of graphene on the conductive substrate at a high temperature in a H2 atmosphere, providing an effective way to form stable electrodes by achieving strong interfacial contact. Typically, the growth of graphene on high-pressure high-temperature (HPHT) diamond occurs by sp3-to-sp2 transformation,113 as illustrated in Fig. 10b. Fig. 10c and d depict the surface morphology before and after treatment, showcasing the transformation from smooth and flat morphology into an island-like structure. The Raman results presented in Fig. 10e and f reveal the formation of few layers of graphene on HPHT diamond. Fig. 10g presents the mechanism of sp3-to-sp2 transformation, showing the precipitation of carbon atoms on molten Cu to form graphene layers at the substrate interface. The transformation results in a strong interfacial bonding between graphene and HPHT, forming stable electrodes of anti-fouling in practical sensing platforms.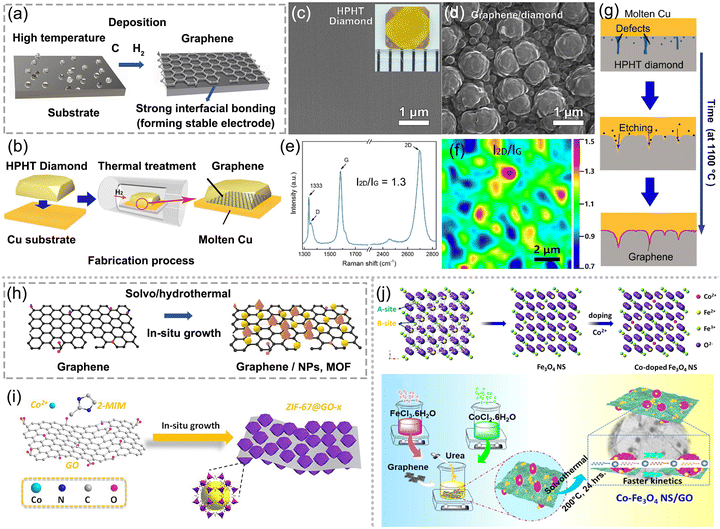 | ||
| Fig. 10 In situ growth method for obtaining strong interfacial contact between graphene and modifiers. (a) Illustration of graphene deposition on the substrate at high temperatures. (b) In situ growth of graphene on HPHT diamond by thermal treatment. SEM images of HPHT diamond (c) before and (d) after treatment. (e) Raman spectrum and (f) its mapping of graphene-diamond. (g) Formation mechanism of graphene-diamond. (b)–(g) Reproduced with permission.113 Copyright from 2018, Elsevier. (h) Solvo-hydrothermal method for the in situ growth of graphene-based modifiers. (i) Electrode modified with zeolitic imidazole framework-derived Co and nitrogen co-doped carbon polyhedrons (ZIF-67C) and GO hybrids. Reproduced with permission.114 Copyright from 2019, Springer. (j) Formation of Co2+ doping and synthesis procedure of Co–Fe3O4 nanospheres/GO. Reproduced with permission. Copyright from 2021, American Chemical Society. | ||
Fig. 10h shows the solvo-hydrothermal method for the in situ growth of graphene-based modifiers. Chen et al. synthesized a homogeneous nanocomposite of zeolitic imidazole framework-derived Co and nitrogen co-doped carbon polyhedrons and rGO by an in situ growth method,114 as depicted in Fig. 10i. The composite material derived from MOF and rGO demonstrates excellent stability, selectivity and sensitivity towards Metronidazole detection, owing to its enhanced mass transfer, abundant electroactive sites, and superior conductivity. Nehru et al. fabricated cobalt-doped Fe3O4 nanospheres deposited on GO via a facile hydrothermal technique,66 as shown in Fig. 10j. The schematic Fe3O4 crystal structure (site A and B) represents a cubic crystal feature. Here, site A has been occupied by Fe3+ and site B has been occupied by an equal number of Fe2+ and Fe3+ ions. During the introduction of Co2+ into the Fe3O4 matrix, Fe3+ was replaced by Co2+ in the site A, while Fe2+ was transformed to Fe3+ in site-B to sustain electric neutrality. The in situ growth of Co-doped Fe3O4 on GO improved the performance shortages of the Fe3O4 system due to agglomeration and less conductivity, and enhanced its operation stability due to the strong intermolecular interactions. Thus, the hybrid boosts the electrochemical kinetics and exhibits a low detection limit of 1.04 nM towards chloramphenicol. In summary, the in situ growth of graphene-based modifiers renders superior mechanical stability, electrical conductivity, abundant electro-active sites, and electrocatalytic activity and boosts the electron-transfer kinetics for antibiotic detection.40,60,115
5. Portable electrochemical platform for the on-site monitoring of antibiotics in real samples
Conventional electrodes and electrochemical apparatus based on the tag/anti-tag system are bulky and time/energy consuming, hindering their potential for the on-site monitoring of environmental, clinical and food samples. Thus, the fabrication of miniaturized electrochemical platforms has attracted attention.116–118 Integrating miniaturized electrodes as screen printed, flexible forms, microfluidic systems and potentiostat into a portable detection platform with automation will facilitate its on-site applications, exhibiting superiorities as portability, cost-efficiency, data availability in less time and manipulation outside the laboratories with facility.119–122 In this section, the state-of-the-art portable electrochemical platform can be summarized into three categories: (a) glove-based wearable sensors for monitoring antibiotics in food samples, (b) epidermal and microneedle-based wearable device for clinical monitoring, and (c) microfluidic electrochemical chip proposed for multianalyte monitoring.5.1 Glove-based wearable sensors for monitoring antibiotics in food samples
When considering antibiotic monitoring in the food industry, the needs for real-time, high-accuracy and on-site analysis are imperative for ensuring the food safety and preservation. Glove-based wearable sensors were developed for electrochemical monitoring antibiotics in food samples,77,123 as depicted in Fig. 11a. Mishra et al. reported a glove-based wearable sensor with a printable electrode system to monitor pesticide-contaminated agricultural products.123,124 Before electrochemical measurements, swipe sampling with finger scan on agricultural products was carried out for collecting analyte residues. The electrochemical cell was formed by joining the scan finger and collection finger to perform measurements. Portable potentiostat was loaded along with electrodes to emit signals through wireless transmission to a smartphone. The fabrication and sensing process of glove-based sensors are exhibited in Fig. 11b–f. The proposed wearable “lab-on-a-glove” platform exhibits potential for future monitoring antibiotics in food supervision.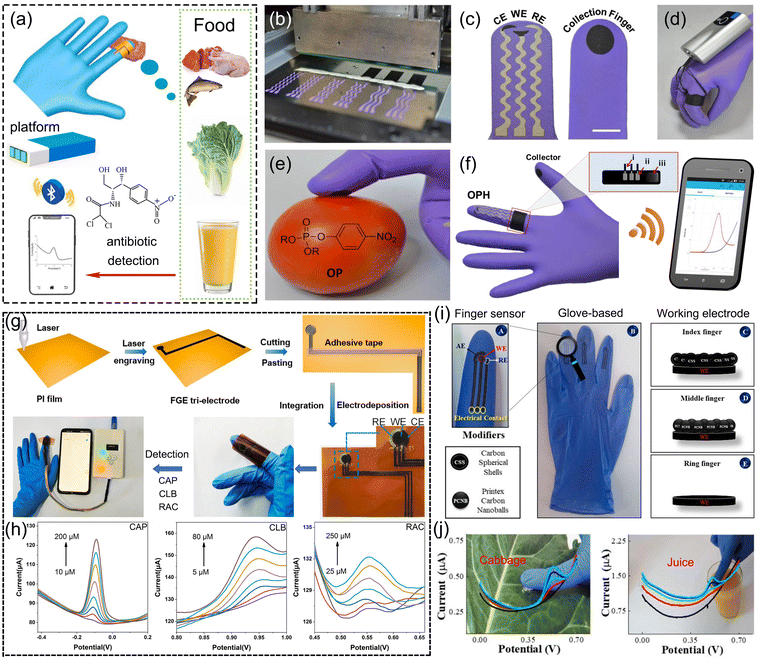 | ||
| Fig. 11 Glove-based wearable sensors for antibiotic detection. (a) Schematic. (b) Configuration of the glove-based stretchable device, with (c) scan finger (left) containing biosensing electrodes and collection finger (right). (d) Photograph of the glove-based sensor, containing a ring bandage connecting the electrodes with a portable potentiostat. (e) and (f) Swipe sampling protocol on the glove to complete the sensing function. (b)–(f) Reproduced with permission.123 Copyright from 2017, American Chemical Society. (g) A laser-induced flexible electrochemical sensing system integrated on the finger. (h) DPV curves of antibiotic detection on flexible graphene electrodes. (g) and (h) Reproduced with permission.77 Copyright from 2022, Elsevier. (i) and (j) Photograph of the glove-based sensor for on-site monitoring in food samples. Reproduced with permission.124 Copyright from 2021, Elsevier. | ||
Li et al. constructed a laser-induced flexible electrochemical sensing system on fingers for rapid real-time on-site identification of chloramphenicol, clenbuterol, and ractopamine in meat.77 As depicted in Fig. 11g, flexible graphene electrodes were facilely patterned and prepared by CO2 laser and integrated with disposable blue nitrile gloves for constructing a finger-based sensing system. Through the connection with a portable electrochemical analyser, electrochemical signals could be received by the directly touching object under DPV tests with fingertips and displayed on mobile phone. The LOD of chloramphenicol and the other two feed additives, namely, clenbuterol and ractopamine was 2.70, 1.29 and 7.81 μM, respectively, enabling the successful application of the sensing platform on the finger for food security, as shown in Fig. 11h. Raymundo-Pereira et al. developed a non-enzymatic sensor system printed on three fingers of a rubber glove for the detection of carbendazim in food samples, as depicted in Fig. 11i.124 In Fig. 11j, the sensor was capable of monitoring carbendazim in a rapid and cost-effective manner by directly touching the samples with the glove, exhibiting 47 nM LOD for carbendazim by the DPV method, and was successfully applied for cabbage and juice samples.
5.2 Epidermal and microneedle-based wearable device for clinical monitoring
In clinical medicine critically ill patients are treated with inadequate antibiotic dosing, resulting in antimicrobial resistance or bad antimicrobial therapy for infections.125 The wide ranging variations in individual pharmacokinetic–pharmacodynamic target attainment request the emergency for accurate antibiotic monitoring to facilitate individualized dose optimization.125 Thus, the promotion of wearable biosensor is necessary for the on-site and non-invasive monitoring of antibiotics in human interstitial fluids such as sweat to diagnose disease at early stage, leading to the development of epidermal and microneedle-based wearable sensors for antibiotic monitoring in clinical treatment. As an invasive biofluid with abundant physiological information, sweat is suitably used to be a target fluid for health monitoring combined with an epidermal sensor. Fig. 12a depicts the epidermal sensor involving the integration of skin multilayer models. Tai et al. fabricated a wearable sweat band (s-band) platform for on-body and non-invasive drug monitoring, as presented in Fig. 12b–d.126 Printed circuit boards and carbon electrodes were integrated into this wearable platform to construct the flexible electrochemical sensor, and DPV measurement was employed for caffeine detection after sweat extraction by the iontophoresis method. This s-band portable platform may achieve continuous and non-invasive recognition of antibiotic levels, showing the potential for future applications in clinical medicine and healthcare-related system.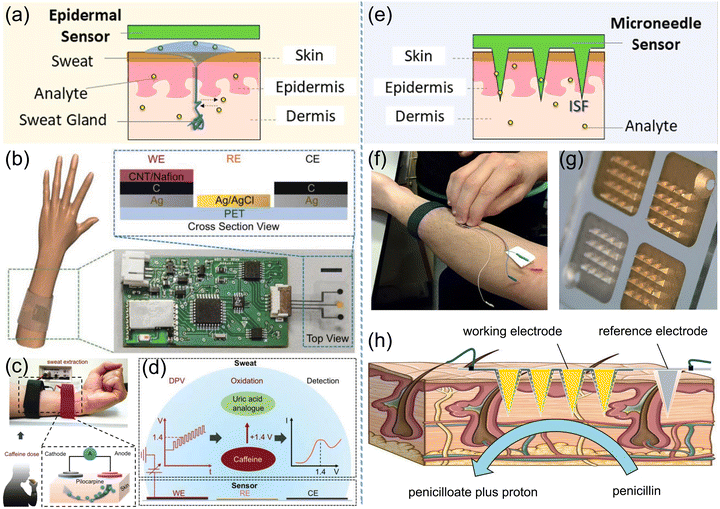 | ||
| Fig. 12 Epidermal and microneedle-based wearable sensors through sweat or interstitial fluid sampling for antibiotic detection in clinical treatment. (a) Scheme of an epidermal sensor involving the integration of skin multilayer models. Reproduced with permission.116 Copyright from 2020, American Chemical Society. (b) A wearable platform integrated into a wristband for noninvasive drug monitoring and the cross-section view of the flexible electrodes. (c) Sweat extraction through iontophoresis method and (d) DPV signals from caffeine detection. (b)–(d) Reproduced with permission.126 Copyright from 2018, Wiley-VCH. (e) Scheme of microneedle transdermal sensing via interstitial fluid access. (f) and (g) Microneedle array biosensor to the forearm with 60 s firm pressure. (h) Diagram of the penetration of microneedle array into the dermal-interstitial space. (f)–(h) Reproduced with permission.75 Copyright from 2019, Elsevier. | ||
To overcome the difficulty of accessing human interstitial fluids, the microneedle array has been developed to substitute hypodermic needles for transdermal drug delivery and sampling human interstitial fluids. Microneedle-based wearable sensors are constructed by the integration of multiple microneedle electrodes on a wearable patch device for real-time diagnostic evaluation of the antibiotic level.75,127Fig. 12e illustrates the microneedle transdermal sensing via interstitial fluid access. Rawson et al. developed a microneedle β-lactamase biosensor for real-time and minimally invasive monitoring of penicillin V in vivo, as shown in Fig. 12f and g.75 The open circuit potential of the working electrode versus the reference electrode was recorded as detection signals, as depicted in Fig. 12h. Penicillin V, as a common antibiotic to cure bacterial infections, diffuses from the extracellular fluid to the hydrogel layer, and hydrolysed to penicilloate and a proton by β-lactamase. Therefore, the increasing concentration of penicillin V in the tissue promoted the protons generated at the sensor surface, inducing the elevations of open circuit potential. The LOD of penicillin V by the microneedle method was estimated as 0.17 mg L−1, prospecting potential antibiotic monitoring for individualized dose optimization.
5.3 Microfluidic electrochemical chip proposed for multianalyte monitoring
Point-of-care (POC) monitoring proposes the requirements for rapid, accurate, and low sample volume detection in a regulated bioanalytical environment. Moreover, simultaneous detection of multi-antibiotics in human biofluids with a very low volume raises the need of a multiplexed biosensor platform. Here, a microfluidic electrochemical chip provides the opportunity to achieve multi-antibiotic POC monitoring with operation simplicity and size miniaturization.74,128,129Kling et al. constructed a microfluidic platform enabling the electrochemical readout of up to eight enzyme-linked assays for the simultaneous detection of two antibiotics, namely, tetracycline and streptogramin in spiked human plasma, as presented in Fig. 13a.74 The microchannel network contained a single electrochemical cell with a three-electrode setup for the amperometric signal detection, and was connected to the substrate solution reservoir via the common to the outlet. The signal amplification of antibiotic detection was obtained using a defined stop-flow measurement technique, and the corresponding function principle is illustrated in Fig. 13b. The bound enzyme glucose oxidase generated limited H2O2 when the glucose substrate was constantly supplied in the microchannel and catalyzed the reaction to more H2O2 when the flow was stopped. When restarting the flow, the accumulated clouds were flushed over the working electrode, and the generated peak signal from H2O2 oxidation was measured amperometrically, as depicted in Fig. 13c and d. The formula of antibiotic detection in the human plasma involved a DNA–protein interaction sensing mechanism.74 The repressor protein showed a conformational change in the antibiotic presence, indicating no binding capability to their designated operator DNA. The higher concentration of antibiotics resulted in lesser proteins bound to operator DNA, demonstrating the decreased electrochemical signal. The platform concluded the LOD of 6.33 and 9.22 ng mL−1 for tetracycline and pristinamycin respectively, as shown in Fig. 13e.
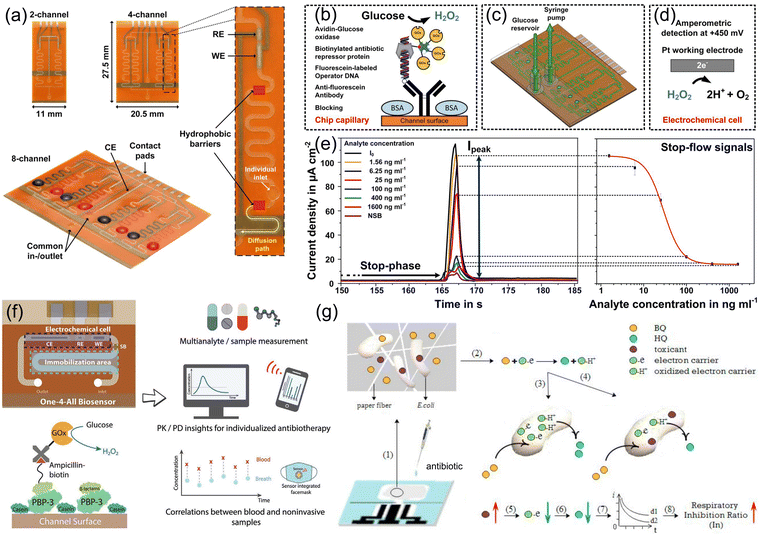 | ||
| Fig. 13 Microfluidic electrochemical chip proposed for the simultaneous detection of multi-antibiotics. Microfluidic biosensors comprising multiple immobilization sections: (a) system configuration; (b) measurement principle; (c) sensing device; (d) reaction mechanism of H2O2 oxidation at the working electrode; (e) amperometric responses and the resulting on-chip calibration curve. (a)–(e) Reproduced with permission.74 Copyright from 2016, American Chemical Society. (f) A microfluidic electrochemical biosensor constructed for multianalyte monitoring based on enzymatic catalysis. Reproduced with permission.128 Copyright from 2022, Wiley-VCH. (g) Paper-based microfluidic device for multianalyte monitoring based on the respiratory inhibition of E. coli. Reproduced with permission.129 Copyright from 2019, Elsevier. | ||
Dincer et al. constructed a microfluidic electrochemical biosensor consisting of an electrochemical cell, immobilized region, and a hydrophobic barrier in prevention of electrode fouling during detection, as depicted in Fig. 13f.128 The measurement signal could be obtained from the catalysis reaction of H2O2 by glucose oxidase, and the platform proved the feasibility for the on-site monitoring of multi-antibiotics such as piperacillin, tazobactam and meropenem in human biofluids. Zhang et al. developed a paper-based microfluidic device for multianalyte monitoring, as displayed in Fig. 13g,129 which was operated based on the respiratory inhibition of E. coli due to the interference of antibiotics in the environment, resulting in the amplification of electrochemical signal. The paper-based microfluidic platform offers a new perspective for the on-site monitoring of multi-antibiotics.
In summary, a portable electrochemical platform for the on-site monitoring of antibiotics depends on specific application requirements, leading to varying applicability of the three wearable scenarios. For the convenient monitoring of antibiotics in food samples, a wearable “lab-on-a-glove” platform shows great potential to achieve the goal. Yet, this platform still faces a bottleneck in the limit of detection of antibiotics.
For the non-invasive monitoring of antibiotics in human interstitial fluids such as sweat and blood, an epidermal and microneedle-based platform is the optimum choice for clinical monitoring. It can achieve trace-level or ultra trace-level detection limits of target antibiotics.
Moreover, the microfluidic electrochemical chip provides an opportunity for the simultaneous detection of multi-antibiotics in human biofluids with accuracy, but it suffers from the limitation of complicated device construction. The selection of graphene-based electrodes for diverse wearable platforms is contingent upon specific application scenarios, whereas graphene facilitates signal amplification during detection periods.
6. Conclusions and future perspectives
The excessive emissions of residual antibiotics pose an increasing threat to the ecosystem and human health due to extensive unreasonable use of antibiotics to cure various infectious diseases in livestock and humans. For human health and ecological safety, it is critical to determine the residual antibiotics with accuracy and convenience. In this regard, electrochemical detection platforms for antibiotics have been developed due to their high sensitivity, simplicity, and time saving. This comprehensive review retrospects the recent advances in graphene-based electrochemical sensors for antibiotics, focusing on the theoretical description of electrochemical sensing mechanisms and practical applications of portable electrochemical platforms for on-site monitoring. Table 2 summarizes the sensor performance, including the detection linear range and LOD. Obviously, graphene-based sensors including immunosensors and aptasensors demonstrate superior detection performance with an LOD several thousands of times lower than that of sensors modified with noble metal NPs, CNTs, metal oxides and MOFs. This superiority is attributed to the specific antigen–antibody reaction and the precise recognition and binding with the target antibiotic.31 Additionally, modifiers such as noble metal NPs, CNTs, metal oxides and MOFs have been extensively applied to sensing electrodes due to their excellent biocompatibility, large surface area, high electron transfer rate, good chemical stability and mechanical strength.50,62,67,130| Graphene-based modifying electrode | Analyte | Technique | Detection range | LOD | Real sample | Ref. | |
|---|---|---|---|---|---|---|---|
| Noble metal NPs | rGO/Ag NPs/NiF | Amikacin | DPV | 0.05–15.0 μM | 38.0 nM | Urine | 131 |
| rGO/Au NPs/Pd NPs | Lomefloxacin | SWV | 4.0–500.0 μM, 30.0–350.0 μM | 38.0 nM | Urine | 56 | |
| Amoxicillin | 9.0 μM | ||||||
| rGO/Au NPs | Chloramphenicol | LSV | 2.0–80.0 μM | 0.59 μM | Eye drops | 53 | |
| rGO/Pd NPs | Chloramphenicol | DPV | 0.05–1.0 μM | 0.05 μM | Honey, tap water | 132 | |
| GO/Au NPs | Clindamycin | SWV | 0.95–140.0 μM | 0.29 μM | Urine, river water | 73 | |
| rGO/Au NPs/polypyrrole | Doxorubicin | CV | 0.02 μM–25.0 mM | 0.02 μM | Drugs | 133 | |
| rGO/Cu NPs | Metronidazole | i–t | 0.002–210.0 μM | 0.6 nM | Drugs | 60 | |
| GO/Ag NPs | Metronidazole | i–t | 0.09–4594.0 μM | 0.07 μM | Drugs | 134 | |
| CNTs | GO/CNTs | Azithromycin | LSV | 0.1–10 μM | 0.07 μM | Urine, capsules | 44 |
| rGO/VS2/CNTs | Azithromycin | DPV | 2.8–300 nM | 0.9 nM | Human serum, urine | 67 | |
| rGO/CNTs | Natamycin | ASV | 0.05–2.5 μM | 0.01 μM | Red wine, beverage | 58 | |
| GO-CNTs | Tetracycline | DPV | 20.0–310.0 μM | 0.36 μM | River water | 54 | |
| MOF | rGO/Z-800 | Chloramphenicol | DPV | 1.0–180.0 μM | 0.25 μM | Milk, honey | 135 |
| rGO/NH2-UiO-66 | Ciprofloxacin | ASV | 0.02–1.0 μM | 6.67 nM | Tap water, lake water | 62 | |
| Metal oxides | rGO/Eu2O3 | Chloramphenicol | i–t | 0.02–800.3 μM | 1.32 nM | Milk, honey | 63 |
| rGO/TiO2 | Furazolidone | DPV | 1.0–150.0 pM | 0.43 pM | Blood serum | 136 | |
| Graphene/MnMoO4 | Ornidazole | i–t | 0.01–0.77 μM | 0.85 nM | River water | 68 | |
| Graphene/ZnO | Sulfamethoxazole | DPV | 1–170 μM | 0.4 μM | Urine, serum, lake water | 42 | |
| Trimethoprim | 1–170 μM | 0.3 μM | |||||
| GO/Fe3O4/Co | Chloramphenicol | DPV | 0.005–152.2 μM | 1.04 nM | Food | 66 | |
| Immunosensors | Graphene/prussian/chitosan | Kanamycin | DPV | 0.04–28.8 nM | 13.0 pM | Food | 137 |
| GO/P(NIPAm-MPTC-GMA) | Streptomycin | DPV | 0.09–170.0 nM | 2.89 pM | Milk | 38 | |
| Graphene/Zn/Ni-ZIF/Au NPs | Monensin | DPV | 0.38–150.0 nM | 0.17 nM | Milk | 138 | |
| rGO/Au NPs/peroxidase | Oxytetracycline | DPV | 1.0 pM–4.0 μM | 1.0 pM | Food | 41 | |
| Aptamers | GO/Ag NPs/aptamer | Chloramphenicol | DPV | 10.0 pM–0.2 μM | 3.3 pM | Milk, honey | 65 |
| rGO/Au NPs/aptamer | Ciprofloxacin | SWV | 0.001–1.0 μM | 1.0 nM | Milk | 46 | |
| Graphene/Pt–Cu alloy/aptamer | Kanamycin | DPV | 1.0 pM–10.3 nM | 0.87 pM | Food | 39 | |
| GO/aptamer | Tetracycline | EIS | 0.1 pM–10.0 μM | 0.03 pM | Blood serum | 139 | |
| GR/Fe3O4/Au NPs/aptamer | Streptomycin | DPV | 0.09–340.0 nM | 0.05 nM | Milk | 57 | |
Although electrochemical sensing methodology has been successfully applied for antibiotic detection, there are still many scientific and technical challenges that need to be overcome in the future. As illustrated in Fig. 14, the current challenges can be listed as follows:
(1) The category distribution of electrochemical sensors for antibiotics is inhomogeneous. Due to the presence of multiple antibiotics in the environment, the current sensors primarily designed for common antibiotics, typically amphenicols and aminoglycosides, are not sufficient and beneficial for the trace determination of antibiotic residues in the environment. Constructing electrochemical sensors for antibiotics covering as many species as possible is necessary.
(2) The electrodes designed on graphene-based modifiers without the selective capability suffer technical difficulty in the selectivity of target antibiotics, especially in the presence of multiple antibiotic species with similar structures in real samples. While for the current immunosensors or aptasensors aimed to the selectivity issue, the enduring preservation and maintenance of bioactive molecules as immune components and aptamers pose a challenge to ensure their consistence performance over time. Thus, constructing electrochemical sensors for antibiotics that possess both high sensitivity and excellent selectivity towards various types of antibiotics is still urgent and meaningful. Particularly, ensuring the long-term stability of graphene-based sensors is essential for the sustained monitoring and practical applications.
(3) The theoretical interpretation of the antibiotic sensing mechanism is vitally important for pursuing excellent detection performance with various electrochemical techniques. Yet, the sensing mechanism of antibiotics reported in current research papers mainly refers to the analysis of electrical responses. The redox reaction process involving electron transfer and interfacial interactions between the electrode surface and the adsorbed molecules at the atomic level should be carried out for in-depth exploration, using theoretical calculation software of the quantum-mechanical method. It not only aggravates the interpretation of the antibiotic sensing mechanism theoretically, but also provides key information about the specific nanomaterials with sensitivity to every antibiotic.
(4) Various forms of antibiotics exist in environment such as in river water, food, human serum, urine, and honey, increasing the difficulty for the development of portable electrochemical platforms for the on-site monitoring of antibiotics. Thus, the platform should be automated, conforming to the scenario requirements of antibiotic detection in various real samples. The optimization of the microneedle, epidermal or microfluidic platform is hopeful for on-site therapeutic antibiotic monitoring in human biofluids towards clinical treatment. Glove-based wearable sensors show potential for the future on-site monitoring of antibiotics in the environment. Moreover, the cost of constructing graphene-based electrochemical sensors to achieve mass production should be reduced to an accepted extent to realize the commercial application.
Overall, the future development of graphene-based electrochemical sensors for antibiotics requires addressing the challenges at the technological, engineering, and application levels. The deep exploration within the scope enables the further utilization of the sensors in the fields of healthcare monitoring, environmental protection, and food safety.
Data availability
The raw/processed data required to reproduce the findings of this study cannot be shared at this time due to legal/ethical reasons.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
C. T. Lin acknowledges the financial support by the National Natural Science Foundation of China (52272053), the National Key R&D Program of China (2022YFA1203100), and Ningbo Key Scientific and Technological Project (2021Z120, 2021Z115, 2022Z084).References
- K. K. Brandt, A. Amezquita, T. Backhaus, A. Boxall, A. Coors, T. Heberer, J. R. Lawrence, J. Lazorchak, J. Schonfeld, J. R. Snape, Y. G. Zhu and E. Topp, Environ. Int., 2015, 85, 189–205 CrossRef CAS PubMed.
- H. K. Allen, J. Donato, H. H. Wang, K. A. Cloud-Hansen, J. Davies and J. Handelsman, Nat. Rev. Microbiol., 2010, 8, 251–259 CrossRef CAS.
- R. F. Yuan, Y. D. Zhu, B. H. Zhou and J. Y. Hu, Chem. Eng. J., 2019, 359, 1527–1536 CrossRef CAS.
- B. J. A. Berendsen, J. Lahr, C. Nibbeling, L. J. M. Jansen, I. E. A. Bongers, E. L. Wipfler and M. G. M. van de Schans, Chemosphere, 2018, 204, 267–276 CrossRef CAS PubMed.
- Y. Luo, D. Mao, M. Rysz, Q. Zhou, H. Zhang, L. Xu and P. J. J. Alvarez, Environ. Sci. Technol., 2010, 44, 7220–7225 CrossRef PubMed.
- D. Tadic, M. J. Bleda Hernandez, F. Cerqueira, V. Matamoros, B. Pina and J. M. Bayona, J. Hazard. Mater., 2021, 401, 123424 CrossRef CAS.
- J. C. Underwood, R. W. Harvey, D. W. Metge, D. A. Repert, L. K. Baumgartner, R. L. Smith, T. M. Roane and L. B. Barber, Environ. Sci. Technol., 2011, 45, 3096–3101 CrossRef CAS PubMed.
- R. Singh, A. P. Singh, S. Kumar, B. S. Giri and K.-H. Kim, J. Cleaner Prod., 2019, 234, 1484–1505 CrossRef CAS.
- M. Kumar, S. Jaiswal, K. K. Sodhi, P. Shree, D. K. Singh, P. K. Agrawal and P. Shukla, Environ. Int., 2019, 124, 448–461 CrossRef CAS PubMed.
- K. Kummerer, Chemosphere, 2009, 75, 417–434 CrossRef PubMed.
- K. Kummerer, Chemosphere, 2009, 75, 435–441 Search PubMed.
- H. Y. Shen and H. L. Jiang, Anal. Chim. Acta, 2005, 535, 33–41 CrossRef CAS.
- H. Y. Liu, S. L. Lin and M. R. Fuh, Talanta, 2016, 150, 233–239 CAS.
- A. Azzouz and E. Ballesteros, Food Chem., 2015, 178, 63–69 CAS.
- J. N. Baby, B. Sriram, S. F. Wang and M. George, J. Hazard. Mater., 2021, 408, 124940 CAS.
- T. Kokulnathan, T. J. Wang, N. Duraisamy, E. A. Kumar and S. An Ni, J. Hazard. Mater., 2021, 412, 125257 CAS.
- T. Kokulnathan and S. M. Chen, J. Hazard. Mater., 2020, 384, 121304 Search PubMed.
- F. Chen, Z. Fan, Y. Zhu, H. Sun, J. Yu, N. Jiang, S. Zhao, G. Lai, A. Yu, C. T. Lin, C. Ye and L. Fu, Materials, 2020, 13, 777 Search PubMed.
- N. A. Shad, S. Z. Bajwa, N. Amin, A. Taj, S. Hameed, Y. Khan, Z. Dai, C. Cao and W. S. Khan, J. Hazard. Mater., 2019, 367, 205–214 CrossRef CAS PubMed.
- Y. Zhang, Z. Liu, Y. Wang, X. Kuang, H. Ma and Q. Wei, J. Hazard. Mater., 2020, 398, 122778 CAS.
- M. Li, T. Zhe, F. Li, R. Li, F. Bai, P. Jia, T. Bu, Z. Xu and L. Wang, J. Hazard. Mater., 2022, 435, 129059 CrossRef CAS PubMed.
- W. Dai, L. Lv, J. Lu, H. Hou, Q. Yan, F. E. Alam, Y. Li, X. Zeng, J. Yu, Q. Wei, X. Xu, J. Wu, N. Jiang, S. Du, R. Sun, J. Xu, C. P. Wong and C. T. Lin, ACS Nano, 2019, 13, 1547–1554 Search PubMed.
- Y. Zhu, Q. Tian, X. Li, L. Wu, A. Yu, G. Lai, L. Fu, Q. Wei, D. Dai, N. Jiang, H. Li, C. Ye and C. T. Lin, Biosensors, 2021, 11, 462 Search PubMed.
- N. Cui, P. Guo, Q. Yuan, C. Ye, M. Yang, M. Yang, K. W. A. Chee, F. Wang, L. Fu, Q. Wei, C. T. Lin and J. Gao, Sensors, 2019, 19, 2979 Search PubMed.
- A. Criado, M. Melchionna, S. Marchesan and M. Prato, Angew. Chem., Int. Ed., 2015, 54, 10734–10750 CAS.
- Y. Yao, C. Jiang and J. Ping, Biosens. Bioelectron., 2019, 123, 178–184 Search PubMed.
- S. Liu, G. Lai, H. Zhang and A. Yu, Microchim. Acta, 2017, 184, 1445–1451 Search PubMed.
- O. Bunkoed, P. Raksawong, R. Chaowana and P. Nurerk, Talanta, 2020, 218, 121168 CrossRef CAS.
- Q. Wang, Q. Xue, T. Chen, J. Li, Y. Liu, X. Shan, F. Liu and J. Jia, Chin. Chem. Lett., 2021, 32, 609–619 CrossRef CAS.
- L. Fu, S. Mao, F. Chen, S. Zhao, W. Su, G. Lai, A. Yu and C. T. Lin, Chemosphere, 2022, 297, 134127 CrossRef CAS.
- A. Joshi and K. H. Kim, Biosens. Bioelectron., 2020, 153, 112046 CrossRef CAS.
- L. Lan, Y. Yao, J. Ping and Y. Ying, Biosens. Bioelectron., 2017, 91, 504–514 CrossRef CAS PubMed.
- L. Qian, S. Durairaj, S. Prins and A. Chen, Biosens. Bioelectron., 2021, 175, 112836 CrossRef CAS.
- T. T. T. Tran, M. N. Do, T. N. H. Dang, Q. H. Tran, V. T. Le, A. Q. Dao and Y. Vasseghian, Environ. Res., 2022, 208, 112744 CrossRef CAS PubMed.
- T. W. Chen, U. Rajaji, S. M. Chen, S. Chinnapaiyan and R. J. Ramalingam, Ultrason. Sonochem., 2019, 56, 430–436 CAS.
- Z. Jiang, G. Li and M. Zhang, Sens. Actuators, B, 2016, 228, 59–65 CAS.
- M. Rouhani and A. Soleymanpour, Microchem. J., 2021, 165, 106160 CrossRef CAS.
- X. He, H. Han, L. Liu, W. Shi, X. Lu, J. Dong, W. Yang and X. Lu, ACS Appl. Mater. Interfaces, 2019, 11, 13676–13684 CAS.
- X. Qin, Y. Yin, H. Yu, W. Guo and M. Pei, Biosens. Bioelectron., 2016, 77, 752–758 CrossRef CAS.
- X. Zhan, G. Hu, T. Wagberg, S. Zhan, H. Xu and P. Zhou, Microchim. Acta, 2015, 183, 723–729 CrossRef.
- S. Liu, Y. Wang, W. Xu, X. Leng, H. Wang, Y. Guo and J. Huang, Biosens. Bioelectron., 2017, 88, 181–187 CrossRef CAS PubMed.
- X. Yue, Z. Li and S. Zhao, Microchem. J., 2020, 159, 105440 CrossRef CAS.
- Y. Sun, J. He, G. I. N. Waterhouse, L. Xu, H. Zhang, X. Qiao and Z. Xu, Sens. Actuators, B, 2019, 300, 126993 CrossRef CAS.
- K. Zhang, L. Lu, Y. Wen, J. Xu, X. Duan, L. Zhang, D. Hu and T. Nie, Anal. Chim. Acta, 2013, 787, 50–56 CrossRef CAS.
- W. Lian, S. Liu, J. Yu, X. Xing, J. Li, M. Cui and J. Huang, Biosens. Bioelectron., 2012, 38, 163–169 CAS.
- M. Mahmoudpour, H. Kholafazad-Kordasht, J. E. Nazhad Dolatabadi, M. Hasanzadeh, A. H. Rad and M. Torbati, Anal. Chim. Acta, 2021, 1174, 338736 CrossRef CAS PubMed.
- A. Wong, T. A. Silva, F. C. Vicentini and O. Fatibello-Filho, Talanta, 2016, 161, 333–341 CrossRef CAS.
- S. Li, X. Ma, C. Pang, H. Li, C. Liu, Z. Xu, J. Luo and Y. Yang, Anal. Chim. Acta, 2020, 1127, 69–78 CrossRef CAS PubMed.
- W. Guo, A. Umar, M. A. Alsaiari, L. Wang and M. Pei, Microchem. J., 2020, 158, 105270 CrossRef CAS.
- M. Yadav, V. Ganesan, R. Gupta, D. K. Yadav and P. K. Sonkar, Microchem. J., 2019, 146, 881–887 CrossRef CAS.
- V. Mariyappan, M. Keerthi, S. M. Chen and T. Jeyapragasam, J. Colloid Interface Sci., 2021, 600, 537–549 CrossRef CAS PubMed.
- K. D. Ramadhass, M. Ganesan, T.-W. Chen, S.-M. Chen, Q. Hao, W. Lei and G. Gopalakrishnan, Colloids Surf., A, 2021, 628, 127275 CrossRef CAS.
- J. Borowiec, R. Wang, L. Zhu and J. Zhang, Electrochim. Acta, 2013, 99, 138–144 CrossRef CAS.
- A. Wong, M. Scontri, E. M. Materon, M. R. V. Lanza and M. D. P. T. Sotomayor, Electroanal. Chem., 2015, 757, 250–257 CrossRef CAS.
- R. Karthik, M. Govindasamy, S.-M. Chen, V. Mani, B.-S. Lou, R. Devasenathipathy, Y.-S. Hou and A. Elangovan, J. Colloid Interface Sci., 2016, 475, 46–56 CrossRef CAS.
- N. Kumar, Rosy and R. N. Goyal, Sens. Actuators, B, 2017, 243, 658–668 CrossRef CAS.
- J. Yin, W. Guo, X. Qin, J. Zhao, M. Pei and F. Ding, Sens. Actuators, B, 2017, 241, 151–159 CrossRef CAS.
- X. Yang, X. Yu, Y. Heng and F. Wang, Electroanal. Chem., 2018, 816, 54–61 CrossRef CAS.
- T. Kokulnathan, T. S. K. Sharma, S.-M. Chen, T.-W. Chen and B. Dinesh, J. Taiwan Inst. Chem. Eng., 2018, 89, 26–38 CrossRef CAS.
- V. Velusamy, S. Palanisamy, T. Kokulnathan, S. W. Chen, T. C. K. Yang, C. E. Banks and S. K. Pramanik, J. Colloid Interface Sci., 2018, 530, 37–45 CrossRef CAS PubMed.
- M. Govindasamy, S. F. Wang, S. Kumaravel, R. J. Ramalingam and H. A. Al-Lohedan, Ultrason. Sonochem., 2019, 52, 382–390 CrossRef CAS PubMed.
- X. Fang, X. Chen, Y. Liu, Q. Li, Z. Zeng, T. Maiyalagan and S. Mao, ACS Appl. Nano Mater., 2019, 2, 2367–2376 CrossRef CAS.
- U. Rajaji, S. Manavalan, S.-M. Chen, M. Govindasamy, T.-W. Chen and T. Maiyalagan, Composites, Part B, 2019, 161, 29–36 CrossRef CAS.
- M. H. Ghanbari, A. Khoshroo, H. Sobati, M. R. Ganjali, M. Rahimi-Nasrabadi and F. Ahmadi, Microchem. J., 2019, 147, 198–206 CrossRef CAS.
- M. Roushani, Z. Rahmati, S. Farokhi, S. J. Hoseini and R. H. Fath, Mater. Sci. Eng., C, 2020, 108, 110388 CrossRef CAS PubMed.
- R. Nehru, C.-D. Dong and C.-W. Chen, ACS Appl. Nano Mater., 2021, 4, 6768–6777 CrossRef CAS.
- H. Mater Mahnashi, A. M. Mahmoud, A. Saad Alkahtani and M. M. El-Wekil, Microchem. J., 2021, 163, 105925 CrossRef CAS.
- K. Venkatesh, R. Rajakumaran, S. M. Chen, C. Karuppiah, C. C. Yang, S. K. Ramaraj, M. A. Ali, F. M. A. Al-Hemaid, M. S. El-Shikh and B. M. A. Almunqedhi, Chemosphere, 2021, 273, 129665 CrossRef CAS.
- M. Lu, C. Cao, F. Wang and G. Liu, Mater. Des., 2021, 199, 109409 CrossRef CAS.
- Y. G. Zhu, X. F. Li, M. F. Wu, M. J. Shi, Q. C. Tian, L. Fu, H. S. Tsai, W. F. Xie, G. S. Lai, G. Wang, N. Jiang, C. Ye and C. T. Lin, Anal. Chim. Acta, 2023, 1275, 341607 CrossRef CAS.
- J. Huang, Z. Qiu, J. Lin, J. Lin, F. Zhu, G. Lai and Y. Li, Microchem. J., 2023, 188, 108444 CrossRef CAS.
- P. Kolhe, A. Roberts and S. Gandhi, Food Chem., 2023, 398, 133846 CrossRef CAS PubMed.
- A. Wong, C. A. Razzino, T. A. Silva and O. Fatibello-Filho, Sens. Actuators, B, 2016, 231, 183–193 CAS.
- A. Kling, C. Chatelle, L. Armbrecht, E. Qelibari, J. Kieninger, C. Dincer, W. Weber and G. Urban, Anal. Chem., 2016, 88, 10036–10043 CAS.
- T. M. Rawson, S. A. N. Gowers, D. M. E. Freeman, R. C. Wilson, S. Sharma, M. Gilchrist, A. MacGowan, A. Lovering, M. Bayliss, M. Kyriakides, P. Georgiou, A. E. G. Cass, D. O'Hare and A. H. Holmes, Lancet Digital Health, 2019, 1, e335–e343 Search PubMed.
- S. Kumar, P. Karfa, K. C. Majhi and R. Madhuri, Mater. Sci. Eng., C, 2020, 111, 110777 CrossRef CAS PubMed.
- J. Li and X. Bo, J. Hazard. Mater., 2022, 423, 127014 CAS.
- R. Umapathi, S. M. Ghoreishian, S. Sonwal, G. M. Rani and Y. S. Huh, Coord. Chem. Rev., 2022, 453, 214305 CrossRef CAS.
- L. Qian, A. R. Thiruppathi, J. van der Zalm and A. Chen, ACS Appl. Nano Mater., 2021, 4, 3696–3706 CrossRef CAS.
- C. A. Donini, M. K. L. da Silva, R. P. Simões and I. Cesarino, Electroanal. Chem., 2018, 809, 67–73 CrossRef CAS.
- C. L. Malonga Matanou, B. R. Malonda-Boungou, M. N'Dollo, M. D. Nkoua Ngavouka, P. S. Moussounda, A. T. Raji and B. M'Passi-Mabiala, J. Phys. Chem. Solids, 2020, 138, 109162 CrossRef CAS.
- A. R. Cardoso, A. C. Marques, L. Santos, A. F. Carvalho, F. M. Costa, R. Martins, M. G. F. Sales and E. Fortunato, Biosens. Bioelectron., 2019, 124–125, 167–175 CrossRef CAS PubMed.
- K. Abnous, N. M. Danesh, M. Ramezani, A. S. Emrani and S. M. Taghdisi, Biosens. Bioelectron., 2016, 78, 80–86 CrossRef CAS.
- L. Gao, Q. Mao, S. Luo, L. Cao, X. Xie, Y. Yang, Y. Deng and Z. Wei, Environ. Pollut., 2020, 259, 113795 CrossRef CAS.
- Q. He, B. Yu, Z. Li and Y. Zhao, Energy Environ. Mater., 2019, 2, 264–279 CrossRef CAS.
- X. Wu, F. Kang, W. Duan and J. Li, Prog. Nat. Sci., 2019, 29, 247–255 CrossRef CAS.
- X. Duan, H. Sun and S. Wang, Acc. Chem. Res., 2018, 51, 678–687 CrossRef CAS PubMed.
- P. Rebelo, J. G. Pacheco, I. V. Voroshylova, A. Melo, M. N. D. S. Cordeiro and C. Delerue-Matos, Sens. Actuators, B, 2021, 329, 129112 CrossRef CAS.
- P. Zhang, Y. Yang, X. Duan, Y. Liu and S. Wang, ACS Catal., 2021, 11, 11129–11159 CrossRef CAS.
- M. K. L. Da Silva, R. Plana Simões and I. Cesarino, Electroanalysis, 2018, 30, 2066–2076 CrossRef CAS.
- J. S. Won, J. Kaewsuk, J. H. Jo, D.-H. Lim and G. T. Seo, J. Adv. Oxid. Technol., 2015, 18, 31–38 CAS.
- O. C. Adekoya, G. J. Adekoya, R. E. Sadiku, Y. Hamam and S. S. Ray, ACS Omega, 2022, 7, 33808–33820 CrossRef CAS PubMed.
- A. O. Osikoya, F. Opoku and P. P. Govender, Chem. Phys. Lett., 2021, 764, 138278 CrossRef CAS.
- M. Ding, D. C. Sorescu and A. Star, J. Am. Chem. Soc., 2013, 135, 9015–9022 CrossRef CAS PubMed.
- X. Ren, H. Guo, J. Feng, P. Si, L. Zhang and L. Ci, Chemosphere, 2018, 191, 389–399 CrossRef CAS PubMed.
- J. Moens, P. Jaque, F. De Proft and P. Geerlings, J. Phys. Chem. A, 2008, 112, 6023–6031 CrossRef CAS.
- C. Ye, F. Zhang, X. Tan, H. F. Sun, W. Dai, K. Yang, M. H. Yang, S. Y. Du, D. Dai, J. H. Yu, N. Jiang, W. T. Su, L. Fu, H. Li, J. Kong and C. T. Lin, Chin. Chem. Lett., 2020, 31, 2507–2511 CrossRef CAS.
- X. Ding, H. Liu and Y. Fan, Adv. Healthcare Mater., 2015, 4, 1451–1468 CrossRef CAS.
- K. S. Novoselov, V. I. Fal'ko, L. Colombo, P. R. Gellert, M. G. Schwab and K. Kim, Nature, 2012, 490, 192–200 CrossRef CAS.
- H. P. Bei, Y. Yang, Q. Zhang, Y. Tian, X. Luo, M. Yang and X. Zhao, Molecules, 2019, 24, 658 CrossRef CAS.
- S. Ayyaru and Y. H. Ahn, J. Membr. Sci., 2017, 525, 210–219 CrossRef CAS.
- N. Golafshan, M. Kharaziha and M. Fathi, Carbon, 2017, 111, 752–763 CrossRef CAS.
- K. Abnous, N. M. Danesh, M. Alibolandi, M. Ramezani, S. M. Taghdisi and A. S. Emrani, Sens. Actuators, B, 2017, 240, 100–106 CrossRef CAS.
- F. Wang, L. H. Zhu and J. D. Zhang, Sens. Actuators, B, 2014, 192, 642–647 CrossRef CAS.
- Y. Zhu, T. H. Liu, W. Zhou, M. Shi, M. Wu, P. Shi, N. Zhao, X. Li, Z. Zhang, D. Zhang, Y. Lv, W. Wu, H. S. Tsai, G. Lai, L. Fu, H. Karimi-Maleh, H. Li, N. Jiang, C. Ye and C. T. Lin, Small Struct., 2024, 5, 2400034 CrossRef CAS.
- Y. Zhu, X. Li, M. Wu, M. Shi, Q. Tian, L. Fu, H.-S. Tsai, W.-F. Xie, G. Lai, G. Wang, N. Jiang, C. Ye and C.-T. Lin, Anal. Chim. Acta, 2023, 1275, 341607 CrossRef CAS.
- X. Hong, B. Zhang, E. Murphy, J. Zou and F. Kim, J. Power Sources, 2017, 343, 60–66 CrossRef CAS.
- M. Xu, Y. Zhu, S. Gao, Z. Zhang, Y. Gu and X. Liu, ACS Appl. Nano Mater., 2021, 4, 12442–12452 CrossRef CAS.
- T.-K. Hong, D. W. Lee, H. J. Choi, H. S. Shin and B.-S. Kim, ACS Nano, 2010, 4, 3861–3868 CrossRef CAS.
- X. Wang, J. Wang, H. Cheng, P. Yu, J. Ye and L. Mao, Langmuir, 2011, 27, 11180–11186 CrossRef CAS.
- U. Han, Y. Seo and J. Hong, Sci. Rep., 2016, 6, 24158 CrossRef CAS.
- F.-X. Xiao, J. Miao and B. Liu, J. Am. Chem. Soc., 2014, 136, 1559–1569 CrossRef CAS.
- Q. Yuan, Y. Liu, C. Ye, H. Sun, D. Dai, Q. Wei, G. Lai, T. Wu, A. Yu, L. Fu, K. W. A. Chee and C.-T. Lin, Biosens. Bioelectron., 2018, 111, 117–123 CrossRef CAS.
- H. Chen, X. Wu, R. Zhao, Z. Zheng, Q. Yuan, Z. Dong and W. Gan, Mikrochim. Acta, 2019, 186, 623 CrossRef PubMed.
- N. Sebastian, W.-C. Yu and D. Balram, Inorg. Chem. Front., 2019, 6, 82–93 RSC.
- H. Teymourian, M. Parrilla, J. R. Sempionatto, N. F. Montiel, A. Barfidokht, R. Van Echelpoel, K. De Wael and J. Wang, ACS Sens., 2020, 5, 2679–2700 CrossRef CAS.
- S. A. N. Gowers, D. M. E. Freeman, T. M. Rawson, M. L. Rogers, R. C. Wilson, A. H. Holmes, A. E. Cass and D. O’Hare, ACS Sens., 2019, 4, 1072–1080 CrossRef CAS.
- J. Tu, R. M. Torrente-Rodríguez, M. Wang and W. Gao, Adv. Funct. Mater., 2019, 30, 1906713 CrossRef.
- S. Eyvazi, B. Baradaran, A. Mokhtarzadeh and M. D. L. Guardia, Trends Food Sci. Technol., 2021, 114, 712–721 CrossRef CAS.
- H. Sohrabi, A. Hemmati, M. R. Majidi, S. Eyvazi, A. Jahanban-Esfahlan, B. Baradaran, R. Adlpour-Azar, A. Mokhtarzadeh and M. de la Guardia, TrAC, Trends Anal. Chem., 2021, 143, 116344 CrossRef CAS.
- H. M. Mohamed, TrAC, Trends Anal. Chem., 2016, 82, 1–11 CrossRef CAS.
- L. Fu, Y. Zheng, P. Zhang, H. Zhang, Y. Xu, J. Zhou, H. Zhang, H. Karimi-Maleh, G. Lai, S. Zhao, W. Su, J. Yu and C. T. Lin, Biosens. Bioelectron., 2020, 159, 112212 CrossRef CAS PubMed.
- R. K. Mishra, L. J. Hubble, A. Martin, R. Kumar, A. Barfidokht, J. Kim, M. M. Musameh, I. L. Kyratzis and J. Wang, ACS Sens., 2017, 2, 553–561 CrossRef CAS PubMed.
- P. A. Raymundo-Pereira, N. O. Gomes, F. M. Shimizu, S. A. S. Machado and O. N. Oliveira, Chem. Eng. J., 2021, 408, 127279 CrossRef CAS.
- J. A. Roberts, M. H. Abdul-Aziz, J. Lipman, J. W. Mouton, A. A. Vinks, T. W. Felton, W. W. Hope, A. Farkas, M. N. Neely, J. J. Schentag, G. Drusano, O. R. Frey, U. Theuretzbacher and J. L. Kuti, Lancet Infect. Dis., 2014, 14, 498–509 CrossRef PubMed.
- L. C. Tai, W. Gao, M. Chao, M. Bariya, Q. P. Ngo, Z. Shahpar, H. Y. Y. Nyein, H. Park, J. Sun, Y. Jung, E. Wu, H. M. Fahad, D. H. Lien, H. Ota, G. Cho and A. Javey, Adv. Mater., 2018, 30, e1707442 CrossRef.
- R. K. Mishra, K. Y. Goud, Z. Li, C. Moonla, M. A. Mohamed, F. Tehrani, H. Teymourian and J. Wang, J. Am. Chem. Soc., 2020, 142, 5991–5995 CrossRef CAS PubMed.
- H. C. Ates, H. Mohsenin, C. Wenzel, R. T. Glatz, H. J. Wagner, R. Bruch, N. Hoefflin, S. Spassov, L. Streicher, S. Lozano-Zahonero, B. Flamm, R. Trittler, M. J. Hug, M. Kohn, J. Schmidt, S. Schumann, G. A. Urban, W. Weber and C. Dincer, Adv. Mater., 2022, 34, e2104555 CrossRef.
- J. Zhang, Z. Yang, Q. Liu and H. Liang, Talanta, 2019, 202, 384–391 CrossRef CAS.
- A. Hrioua, A. Loudiki, A. Farahi, M. Bakasse, S. Lahrich, S. Saqrane and M. A. El Mhammedi, Bioelectrochemistry, 2021, 137, 107687 CrossRef CAS.
- N. Sharma, S. Panneer Selvam and K. Yun, Appl. Surf. Sci., 2020, 512, 145742 CrossRef CAS.
- W. Yi, Z. Li, C. Dong, H.-W. Li and J. Li, Microchem. J., 2019, 148, 774–783 CrossRef CAS.
- M. Behravan, H. Aghaie, M. Giahi and L. Maleknia, Diamond Relat. Mater., 2021, 117, 108478 CrossRef CAS.
- S. Chinnaraj, V. Palani, S. Yadav, M. Arumugam, M. Sivakumar, V. Maluventhen and M. Singh, Sens. Bio-Sens. Res., 2021, 32, 100425 CrossRef.
- Y. Yuan, X. Xu, J. Xia, F. Zhang, Z. Wang and Q. Liu, Mikrochim. Acta, 2019, 186, 191 CrossRef PubMed.
- A. A. Ensafi, M. Sohrabi, M. Jafari-Asl and B. Rezaei, Appl. Surf. Sci., 2015, 356, 301–307 CrossRef CAS.
- B. Y. Zhao, Q. Wei, C. Xu, H. Li, D. Wu, Y. Cai, K. Mao, Z. Cui and B. Du, Sens. Actuators, B, 2011, 155, 618–625 CrossRef.
- M. Hu, X. Hu, Y. Zhang, M. Teng, R. Deng, G. Xing, J. Tao, G. Xu, J. Chen, Y. Zhang and G. Zhang, Sens. Actuators, B, 2019, 288, 571–578 CrossRef CAS.
- A. Benvidi, M. D. Tezerjani, S. M. Moshtaghiun and M. Mazloum-Ardakani, Microchim. Acta, 2016, 183, 1797–1804 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |