Open Access Article
Open Access ArticleBone-derived nanoparticles (BNPs) enhance osteogenic differentiation via Notch signaling†
Austin
Stellpflug
a,
Justin
Caron
b,
Samantha
Fasciano
b,
Bo
Wang
 *a and
Shue
Wang
*a and
Shue
Wang
 *b
*b
aJoint Department of Biomedical Engineering, Marquette University and the Medical College of Wisconsin, Milwaukee, WI 53226, USA. E-mail: bowang@mcw.edu
bDepartment of Chemistry, Chemical and Biomedical Engineering, University of New Haven, West Haven, CT 06516, USA. E-mail: swang@newhaven.edu
First published on 27th December 2024
Abstract
Mesenchymal stem cell (MSC)-based bone tissue regeneration has gained significant attention due to the excellent differentiation capacity and immunomodulatory activity of MSCs. Enhancing osteogenesis regulation is crucial for improving the therapeutic efficacy of MSC-based regeneration. By utilizing the regenerative capacity of bone ECM and the functionality of nanoparticles, we recently engineered bone-based nanoparticles (BNPs) from decellularized porcine bones. The effects of internalization of BNPs on MSC viability, proliferation, and osteogenic differentiation were first investigated and compared at different time points. The phenotypic behaviors, including cell number, proliferation, and differentiation were characterized and compared. By incorporating a LNA/DNA nanobiosensor and MSC live cell imaging, we monitored and compared Notch ligand delta-like 4 (Dll4) expression dynamics in the cytoplasm and nucleus during osteogenic differentiation. Pharmacological interventions are used to inhibit Notch signaling to examine the mechanisms involved. The results suggest that Notch inhibition mediates the osteogenic process, with reduced expression of early and late stage differentiation markers (ALP and calcium mineralization). The internalization of BNPs led to an increase in Dll4 expression, exhibiting a time-dependent pattern that aligned with enhanced cell proliferation and differentiation. Our findings indicate that the observed changes in BNP-treated cells during osteogenic differentiation could be associated with elevated levels of Dll4 mRNA expression. In summary, this study provides new insights into MSC osteogenic differentiation and the molecular mechanisms through which BNPs stimulate this process. The results indicate that BNPs influence osteogenesis by modulating Notch ligand Dll4 expression, demonstrating a potential link between Notch signaling and the proteins present in BNPs.
Introduction
Bone marrow derived mesenchymal stem cells (MSCs) play a key role in bone repair and regeneration by differentiating into bone-forming osteoblasts and cartilage-forming chondrocytes.1–5 MSCs contribute to bone healing through three different approaches: (a) differentiation and replacement; (b) secretion of cytokines and extracellular vesicles; and (c) immunomodulatory activity.6–8 While identifying the most efficient method for enhancing osteogenic differentiation of MSCs to promote bone regeneration remains challenging, regulating osteogenesis is crucial for improving the therapeutic efficacy. While the differentiation potential of MSCs is well-established, the specific mechanisms governing their plasticity are not fully understood, particularly the processes by which MSCs differentiate into osteoblasts for bone formation. Over the last few decades, unremitting efforts have been devoted to understanding biochemical signals that regulate MSC commitment. Based on these efforts, a number of chemical stimuli (e.g., small bioactive molecules, growth factors, and genetic regulators) have been identified in regulating MSC lineage commitment, including bone morphogenetic protein (BMP), Wnt, and Notch signaling.9–11 Enhancing osteogenesis is crucial for improving the effectiveness of MSC-based therapies in bone tissue engineering and regeneration. Thus, recent research has focused on developing strategies to enhance osteogenesis, which involve biophysical and biochemical stimulation. In recent years, the rapid advancements in nanotechnology and nanomedicine have significantly transformed regenerative medicine, especially in the context of bone diseases.12–14 Nanoparticles (NPs) have emerged as multifunctional tools that integrate diagnostic and therapeutic functions, offering new avenues for treatment.15–19 Moreover, NPs offer precise control over stem cell behavior and enhance drug delivery by overcoming biological barriers due to their small size.20,21NPs can be broadly classified into inorganic particles (such as ceramics, metal, silica, gold, and silver) and organic particles (including synthetic polymers, liposomes, and proteins) based on their chemical structures.22 Although the therapeutic applications of NPs in clinical settings have been extensively studied, several challenges remain in translating these findings into practical treatments. Key issues include their low biocompatibility and the risk of inducing inflammation and tissue damage, which have hindered the wider adoption of NPs in clinical practice, highlighting the need for the development of safer and more effective alternatives.23–25
Our lab has recently developed a novel type of bone-derived nanoparticles (BNPs) from decellularized porcine bones.26 Compared to traditional NPs, BNPs offer excellent biocompatibility due to their natural origin. By crosslinking BNPs, the degradation rate can be controlled for prolonged circulation and retention of therapeutic agents, thereby reducing the side effects associated with traditional methods.26 The utilization of BNPs offers a novel approach to overcoming the limitations of traditional NPs, which will provide a more biocompatible and efficient delivery system for therapeutic agents. Most importantly, these BNPs showed bone regenerative potential both in vitro, by promoting osteogenic differentiation; and in vivo, by repairing bone defects when used as a graft material.26
Despite these findings, the fundamental mechanisms through which BNPs affect the osteogenic differentiation process remain unexplored. Understanding these mechanisms is crucial for optimizing the design and application of BNPs in bone regeneration. Osteogenic differentiation is a dynamic process and involves several significant signaling pathways, including BMP signaling,27 Wnt/b-catenin signaling,28 Hedgehog (HH) signaling,29 and YAP/TAZ (transcriptional coactivator with the PDZ-binding motif),30,31 and Notch signaling.32,33 Our recent studies have shown that Notch signaling is required to modulate shear-stress induced osteogenic differentiation.2 In addition, current studies revealed that internalization of NPs could enhance osteogenic differentiation.34–36 However, the involvement of Notch signaling remains obscure due to the lack of effective tools to detect and monitor gene expression in live cells. Traditional methods for gene detection are limited due to the need for isolation or fixation, which results in the loss of spatial and temporal gene information. Techniques such as RNA in situ hybridization and single cell transcriptomics only apply to fixed cells, thus limiting their utility.37 Although fluorescent protein tagging allows for the monitoring of RNA dynamics in live cells, it is restricted by low transfection efficiency and the requirement for genetic modifications to express engineered transcripts.38 Thus, dynamic monitoring of gene expression in live cells at the single cell level will reveal the fundamental regulatory mechanism of cells during dynamic biological processes, which will eventually open opportunities to develop novel approaches for tissue engineering and regenerative medicine.
Here, we utilized a double-stranded locked nucleic acid/DNA (LNA/DNA) nanobiosensor to investigate the regulatory role of Notch signaling during BNP induced osteogenic differentiation. The effects of internalization of BNPs on MSC viability, proliferation, and osteogenic differentiation were first investigated and compared at different time points. The phenotypic behaviors, including cell number, proliferation, and differentiation were characterized and compared. By incorporating this LNA/DNA nanobiosensor and live cell imaging, we monitored and compared Notch ligand delta-like 4 (Dll4) expression dynamics in the cytoplasm and nucleus during osteogenic differentiation. Pharmacological interventions are used to inhibit Notch signaling to examine the molecular mechanisms involved. The results suggest that Notch inhibition mediates the osteogenic process, with reduced expression of early and late stage differentiation markers (ALP and calcium mineralization). The internalization of BNPs led to an increase in Dll4 expression, exhibiting a time-dependent pattern that aligned with enhanced cell proliferation and differentiation. Our findings indicate that the observed changes in BNP-treated cells could be associated with elevated levels of Dll4 mRNA expression. In summary, this study provides new insights into MSC osteogenic differentiation and the molecular mechanisms through which BNPs stimulate this process. The results indicate that BNPs promote osteogenesis by modulating Notch ligand Dll4 expression, demonstrating a link between Notch signaling and the proteins present in BNPs. Future research will explore the interactions between TGF-β, Notch, and BMP signaling pathways and the impact of BNPs on these interactions during osteogenic differentiation.
Materials and methods
Fabrication of bone-derived nanoparticles
The BNPs were fabricated following our previously published protocol.26 Briefly, fresh porcine tibias were fully demineralized with 0.5 M hydrochloric acid (HCL, Sigma-Aldrich) and decellularized with 0.5% sodium dodecyl sulfate (SDS) and 1% Triton X-100 mixed solution (Sigma-Aldrich). To prepare the digested bone solution, the demineralized and decellularized bones were freeze-dried at −80 °C, milled into a fine powder, and digested in HCL solution (1 gram of extracellular matrix (ECM) powder/100 mL of 1 M HCL) with 15% w/w pepsin under constant stirring at 45 °C until the whole solution became a smooth, uniform liquid without visible solid particles, indicating that the digestion process was complete.26To fabricate the BNPs, the protein concentration of the final digested bone ECM solution was adjusted to a final concentration of 330 μg mL−1 prior to desolvation. Acetone was added dropwise to the ECM solution (the volume ratio (mL) of acetone to ECM solution was 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) under constant stirring at room temperature. 10 minutes after the last drop of the acetone was added to the ECM solution, 50% glutaraldehyde solution was added in a ratio of 33 μL of glutaraldehyde per 1 mL of the initial ECM solution. This solution was left to stir at room temperature for 30 minutes, upon which the pellet of BNPs was collected via centrifugation. The BNPs were then washed with distilled water three times, resuspended again in distilled water, and dispersed via sonication. The BNPs were then freeze-dried at −80 °C and used for experimental characterization or long-term storage.
1) under constant stirring at room temperature. 10 minutes after the last drop of the acetone was added to the ECM solution, 50% glutaraldehyde solution was added in a ratio of 33 μL of glutaraldehyde per 1 mL of the initial ECM solution. This solution was left to stir at room temperature for 30 minutes, upon which the pellet of BNPs was collected via centrifugation. The BNPs were then washed with distilled water three times, resuspended again in distilled water, and dispersed via sonication. The BNPs were then freeze-dried at −80 °C and used for experimental characterization or long-term storage.
Characterization of BNPs
The morphology of the BNPs was characterized using scanning electron microscopy (SEM). BNP samples were resuspended in DI water, sonicated, and 100 μL aliquots were placed on copper mesh grids and left to dry for 24 hours to remove excess moisture. The samples were sputter coated with gold-palladium and observed with a JEOL JSM 35 scanning electron microscope (JEOL USA, Inc., Peabody, MA).Cell culture and reagents
Bone marrow derived MSCs (Lonza) were cultured in MSCBM (Catalog #: PT-3238, Lonza) with GA-1000, L-glutamine, and mesenchymal cell growth factors (Catalog #: PT-4105, Lonza). The MSCs were originally isolated from normal (non-diabetic) adult human bone marrow withdrawn from bilateral punctures of the posterior iliac crests of normal volunteers (based on manufacturer's information). The MSCs used in this paper were from three different purchases. Cells were cultured and maintained in a tissue culture dish at 37 °C and 5% CO2 in a humidified incubator. The medium was replaced every two days and cells were passaged using 0.25% trypsin–EDTA (Invitrogen). Cells were seeded at 0.1 × 106 and 0.05 × 106 cells per well for 12 and 24 well plates, respectively. The newly seeded cells were cultured until they reached 80% confluency before the induction medium was added. Cells from passages 2–6 were used in the experiments. For intracellular uptake of BNPs, BNPs were re-suspended in Opti-MEM (ThermoFisher) and sonicated for 1 minute using a probe sonicator (Brandon). Unless specified, cells were incubated with BNPs at a concentration of 20 μg mL−1 overnight. Osteogenic induction medium (Catalog #: PT-3002, Lonza) was added and replaced every three days. For control groups, cells were maintained in basal medium without induction. To investigate the involvement of Notch signalling, cells were administered with γ-secretase inhibitor DAPT (20 μM) on cell differentiation. It is noted that cells without DAPT treatment were added with DMSO as a control. The differentiation effects were accessed and compared after 7, 14, and 21 days of induction.Design and preparation of the LNA probe
A LNA/DNA nanobiosensor is a complex of a detection probe and a quencher.39 The detection probe is a 20-base nucleotide with alternating LNA/DNA monomers, and its sequence is complementary to the target mRNA sequence. A fluorophore (6-FAM) was labeled at the 5′ of the detection probe for visualization of the mRNA signal in cells. The probe design process has been reported previously, which includes acquiring the target mRNA sequence from GeneBank and choosing a 20-base pair nucleotide sequence.40–42 The selected nucleotide sequence will be characterized and optimized for stability and specificity using the mFold server and NCBI Basic Local Alignment Search Tool (BLAST) database, respectively. The quencher probe consists of a 10-base pair nucleotide sequence, incorporating LNA/DNA monomers, designed to be complementary to the 5′ end of the LNA detection probe. At the 3′ end of this quencher probe, an Iowa Black RQ fluorophore is attached for labeling purposes. The Dll4 LNA detection probe was designed based on target mRNA sequences (5′–3′: +AA +GG +GC +AG +TT +GG +AG +AG +GG +TT). All the probes were synthesized by Integrated DNA Technologies Inc. (IDT).To assemble the LNA/DNA nanobiosensor, the LNA detection probe and quencher probe were first dissolved in 1× Tris EDTA buffer (pH = 8.0) at a concentration of 100 nM. These components were combined in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio and heated to 95 °C for 5 minutes in a dry water bath, and then allowed to cool to room temperature gradually over 2 hours. Once cooled, the mixture can be refrigerated and stored for up to 7 days. For mRNA detection, this nanobiosensor was transfected into cells using Lipofectamine 2000 (ThermoFisher) according to the manufacturer's instructions.
2 ratio and heated to 95 °C for 5 minutes in a dry water bath, and then allowed to cool to room temperature gradually over 2 hours. Once cooled, the mixture can be refrigerated and stored for up to 7 days. For mRNA detection, this nanobiosensor was transfected into cells using Lipofectamine 2000 (ThermoFisher) according to the manufacturer's instructions.
Cell proliferation and viability
The cell proliferation and viability after BNP incubation were evaluated using cell counting kit-8 (cck-8, Sigma Aldrich), following the manufacturers' instructions. Cells were seeded at a concentration of 200 cells per well in 96-tissue culture well plates with a volume of 100 μL culture medium. For the cell toxicity test, cells were treated with BNPs at different concentrations (10, 20, 50, and 100 μg mL−1) after 14 days of incubation. After 14 days, the BNPs were washed, and fresh basal medium was added. For cell proliferation assay, cells were incubated with BNPs overnight, and cell proliferation was evaluated on days 1, 2, 3, 5, and 10, respectively. After incubation, cck-8 was added to the cells and cultured for 4 h. The absorbance was measured at 450 nm for each sample and compared using a fluorescence microplate reader (BioTek, Synergy 2).Live/dead viability staining
The cell viability was further evaluated using live/dead viability assay (ThermoFisher). Cells were stained using propidium iodide (PI, 10 μg mL−1), a fluorescent agent that binds to DNA by intercalating between the bases with little or no sequence preference. Hoechst 33342 was used to stain the cell nucleus at a concentration of 20 μM for 30 minutes. After staining, cells were washed three times with 1× PBS to remove the extra dye. Cells were then imaged using Texas Red (535/617 nm) and DAPI (360/460 nm) filters using an Echo Revolution fluorescence microscope.In situ proliferation assay
For in situ proliferation measurements, a Click-iT EdU Kit (Invitrogen) was used as per the manufacturer's protocol. MSCs were cultured with BNPs at a concentration of 20 μg mL−1 for 6 hours. After incubation, extra BNPs were removed by changing the medium to fresh basal culture medium. Diluted EdU solution (10 μM) was then introduced into all the wells. Before fixation, cells were incubated with EdU solution for 12 h to label newly synthesized DNA. The numbers of total nuclei (stained with Hoechst 33342) and EdU-labeled nuclei were counted manually in the fluorescence images.ALP and Alizarin Red S staining
To evaluate osteogenic differentiation, the expression of the early differentiation marker (alkaline phosphatase, ALP) and late-stage differentiation marker (calcium mineralization) was quantified using an ALP staining kit (Sigma Aldrich) and Alizarin Red S (ARS) staining quantification assay (ScienCell), respectively. For ALP staining, the staining solution was prepared by combining Fast Red Violet solution, Naphthol AS-BI phosphate solution, and water in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. Subsequently, cells were fixed with 4% cold paraformaldehyde (PFA) for 2 minutes to preserve ALP activity. After fixation, the PFA was removed without washing the cells. The staining solution was then added to the fixed cells for 15 minutes at room temperature and protected from light. Following staining, the cells were washed three times with 1× PBS before imaging.
1 ratio. Subsequently, cells were fixed with 4% cold paraformaldehyde (PFA) for 2 minutes to preserve ALP activity. After fixation, the PFA was removed without washing the cells. The staining solution was then added to the fixed cells for 15 minutes at room temperature and protected from light. Following staining, the cells were washed three times with 1× PBS before imaging.
For ARS staining, the culture medium was removed, and the cells were washed with 1× PBS 3 times. The cells were then fixed with 4% PFA for 15 minutes at room temperature. Then the PFA was removed, and the cells were washed 3 times with diH2O. 40 mM ARS solution was added to each well. The wells were shaken gently for 20–30 minutes with the stain. Lastly, the stain solution was removed, and the cells were washed five times with diH2O. Images of the stained cells were then taken.
Imaging and statistical analysis
Images were obtained using an Echo Revolution fluorescence microscope with an integrated digital camera (5MP CMOS Color for bright field, 5 MP sCMOS Mono for fluorescence imaging). To ensure consistency, all images were captured under identical settings, including exposure time and gain. Image analysis and data collection were conducted using NIH ImageJ software. To measure Dll4 mRNA and ALP enzyme activity, the mean fluorescence intensity of each cell was measured, and background noise was subtracted. Cells were quantified within the same field of view, with a minimum of five images analyzed for each condition. Cumulative probability refers to the probability that the fluorescence intensity of cells is lower than or equal to the intensity value on the x-axis. The experiments were conducted at least three times, with over 100 cells quantified per group. The results were analyzed using an independent, two-tailed Student's t-test in Excel (Microsoft). P < 0.05 was considered statistically significant.Results
Fabrication of BNPs and mechanisms of the double-stranded LNA nanobiosensor
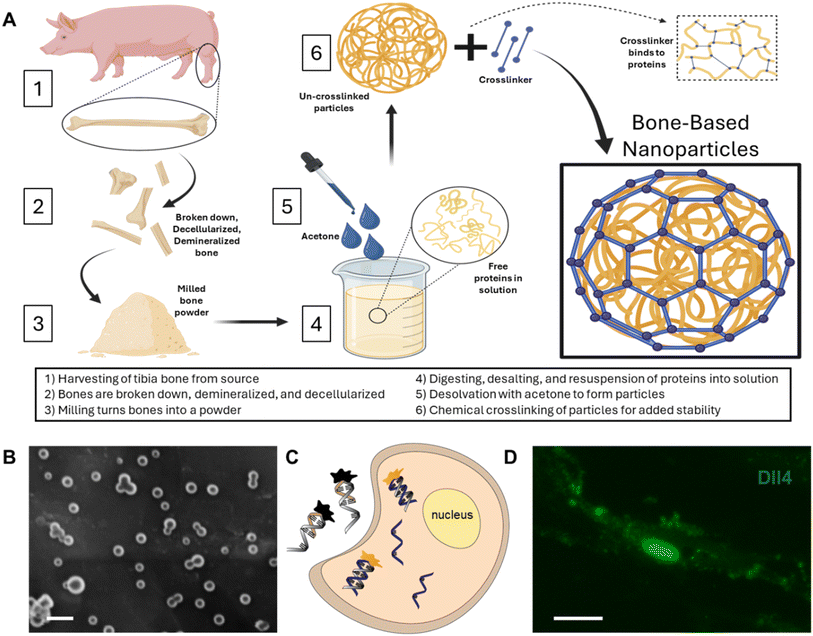
The BNPs fabricated were consistent in morphology with our previous findings.26Fig. 1A shows the fabrication process, including bone decellularization and demineralization, lyophilized bone milling, digesting and desalting bone ECM powder, and final synthesis. The synthesized BNPs have an average size of 79 ± 25.4 nm, Fig. 1B. A double-stranded LNA nanobiosensor was utilized to investigate the involvement of Notch signaling during the dynamic osteogenic differentiation process. The double-stranded DNA/LNA nanobiosensor is a complex of an LNA detection probe and an LNA quenching probe, with lengths of 20 and 10 nucleotides, respectively, Fig. S1.† The LNA detection probe is a single-stranded oligonucleotide sequence with alternating LNA/DNA monomers, which are designed to be complementary to the target mRNA sequence. The LNA nucleotides were chosen due to their higher thermal stability compared to DNA nucleotides, thus enhancing the specificity and sensitivity. To visualize the mRNA expression in live cells, a fluorophore (6-FAM (fluorescein)) was labeled at the 5′ end of the detection probe. The design process of this nanobiosensor has been reported previously.2,4,39 Briefly, the detection LNA probe will spontaneously bind to the quenching probe, forming an LNA–quencher complex. This proximity allows the quencher to quench the fluorescence of the fluorophore at the 5′ end of the LNA probe due to quenching properties. The LNA–quencher complex will be transfected into cells for mRNA detection and visualization. Upon cellular uptake, the LNA detection probe disassociates from the quencher and binds to target mRNA molecules, thus reacquiring a fluorescence signal. This displacement is due to a greater binding free energy difference between the LNA probe and target mRNA compared to that between the LNA probe and quencher. Consequently, the fluorescence intensity within individual cells that contain the LNA/DNA nanobiosensor can quantitatively measure the amount of target mRNA present. In this study, MSCs were transfected with the LNA/DNA nanobiosensor before osteogenic induction.BNPs enhance MSC proliferation
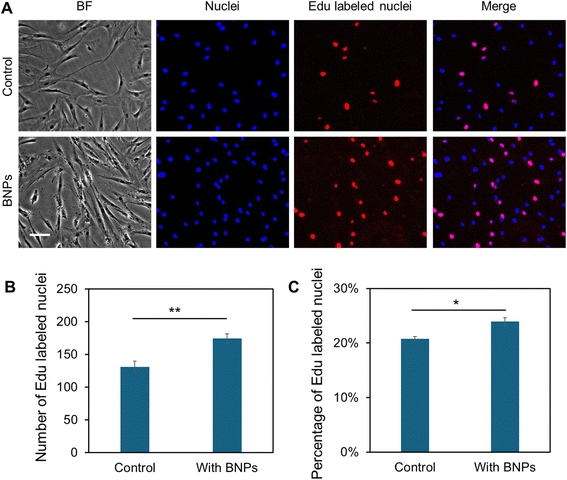
In order to study the effects of different concentrations of BNPs on cell proliferation and viability, MSCs were cultured with BNPs at concentrations of 0, 5, 10, 20, and 50 μg mL−1 for 24 h. The cell viability was first evaluated and compared using cck-8 (Sigma Aldrich) after 14 days of incubation. Fig. S2† shows the comparison of absorbance with and without BNP incubation at different concentrations. Moreover, we evaluated the cell survival using live/dead cell assay after 5 and 7 days, respectively. Fig. S3A† shows the bright field and fluorescence images of MSCs after 5 days of culture in control and BNP co-culture groups, respectively. It was evident that there are no dead cells in both control and BNP co-culture groups (third column of Fig. S3A†), indicating that BNP co-culturing with MSCs did not affect viability and cell survival. To further examine the effects of BNPs on cell proliferation, we quantified and compared the number of cells within different groups. Fig. S3B† shows the comparison of quantified cell numbers with and without BNP co-culturing after 5 and 7 days, respectively. The results showed that the cell number increased by 82.89% and 57.36% after 5 and 7 days, respectively. To characterize cell proliferation, we further performed Click-iT EdU assay to evaluate the newly synthesized DNA nuclei after BNP incubation. Fig. 2A shows representative images of MSCs in control and BNP treated groups. As shown in Fig. 2A and B, the number of EdU-positive nuclei in the BNP treated group was significantly greater compared with the untreated control. The percentage of EdU-positive nuclei relative to the total cell number for the BNP treated group was significantly greater compared to the control (Fig. 2C), indicating that BNP incubation enhanced cell proliferation. Since BNPs were fabricated using the entire ECM of decellularized porcine bone, BNPs contain a variety of ECM proteins.26 These data suggest that the effects of BNPs on cell proliferation occur at an early stage, indicating that the ECM proteins were released quickly after several days of incubation. We further confirmed the effects of BNPs on cell proliferation after 1, 2, 3, 5, and 10 days of co-culturing. Fig. S3C† shows the cumulative absorbance using cck-8 proliferation assay.Internalization of BNPs enhances osteogenic differentiation
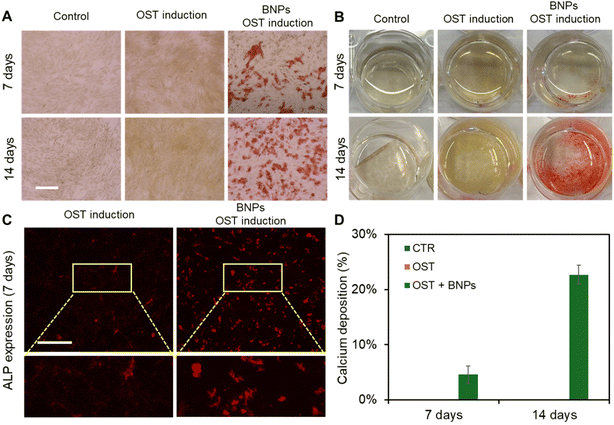
We next evaluated the effects of BNPs on osteogenic differentiation by co-culturing BNPs with MSCs at a concentration of 20 μg mL−1 for 24 h. Briefly, MSCs were seeded in 12-well plates and cultured in the basal medium. Three groups of experiments were conducted, control (CTR group), osteogenic induction (OST group), and BNPs with osteogenic induction (BNPs + OST group). Once the cells reached 70–80% confluency, BNPs were prepared in Opti-MEM and added to the cells in the BNPs + OST group, and the osteogenic induction medium was replaced after 24 h of incubation. After 7 and 14 days of induction, osteogenic differentiation was evaluated using Alizarin Red S (ARS) Staining Quantification Assay (ScienCell). We further assessed calcium mineralization by extracting calcified minerals at low pH and neutralizing them with ammonium hydroxide. The calcium deposition was then quantified using colorimetric detection at 405 nm. Fig. 3A shows the bright field images of MSCs under different treatments after 7 and 14 days of incubation. Fig. 3B shows the images of different wells after ARS staining. The results showed that without osteogenic induction, there is no calcium deposition. With osteogenic induction but without BNP incubation, calcium deposition was not visible after 7 and 14 days of induction, Fig. 3A and B. It is evident that intracellular uptake of BNPs significantly enhanced osteogenic differentiation after 7 and 14 days, with visible calcium deposition. In order to investigate how BNPs accelerate osteogenic differentiation, we evaluated and compared ALP enzyme activity after 7 days of osteogenic induction. Fig. 3C shows representative fluorescence images of MSCs with and without BNP internalization. We further quantified ALP activity by measuring the mean fluorescence intensity of ALP, Fig. S4.† With BNP internalization, the ALP activity increased by 1.5 fold compared to MSCs without BNPs. Fig. 3D shows the quantification and comparison results of calcium mineralization after 7 and 14 days of osteogenic induction, respectively. Without BNP treatments, no calcium mineralization was observed for control (Fig. S5†) and osteogenic groups after 7 and 14 days of induction. BNP internalization significantly enhanced calcium mineralization, with approximately 5% and 23% of cells exhibiting calcium mineralization after 7 and 14 days of induction, respectively.Notch signaling is involved in BNP induced osteogenic differentiation
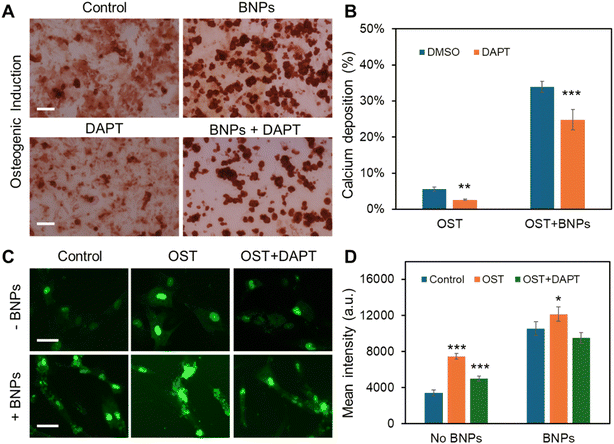
Previous research has demonstrated that Notch signaling plays a role in osteogenic differentiation, influencing ALP activity and the efficiency of osteogenic differentiation.43–45 Additionally, our group has recently identified Dll4 mRNA as a molecular biomarker of osteogenically differentiated MSCs.4 Inhibiting Notch signaling has been found to diminish osteogenic differentiation, accompanied by reduced ALP enzyme activity. However, it is obscure whether Notch signaling is involved in BNP enhanced osteogenic differentiation. To better understand the regulatory role of Notch signaling, we utilized a pharmacological drug, the γ-secretase inhibitor (DAPT) that blocks Notch endoproteolysis, to perturb Notch signaling. MSCs were treated with DAPT (20 μM) before osteogenic induction with or without BNP incubation. The osteogenic differentiation across different treatments was evaluated and compared by quantifying ARS intensity, indicating the amount of calcium deposition. Fig. 4A and S6† show representative ARS staining images of MSCs after 21 days of osteogenic induction under different treatments. We next quantified the percentage of calcium deposition by measuring the average of the ARS-stained region. Fig. 4B shows the comparison results of calcium deposition after 21 days of osteogenic induction. Without BNP incubation, only about 5% of the cells exhibited calcium deposition, and DAPT treatment disturbed osteogenic differentiation, reducing calcium deposition to approximately 2%. Consistent with our earlier findings, treatment with BNPs significantly enhanced osteogenic differentiation, with approximately 34% of cells showing calcium deposition – an increase of about 4.6 times. Additionally, DAPT treatment reduced the effects of BNPs, with about 24% of cells exhibiting calcium deposition. Interestingly, DAPT treatment for the MSCs with BNPs has fewer effects on osteogenic differentiation, suggesting that the intracellular uptake of BNPs counteracted the inhibitory effects of Notch signaling due to pharmaceutical treatment.To further explore the involvement of Notch signaling in the enhancement of osteogenic differentiation by intracellular uptake of BNPs, we examined the Notch 1 ligand, Dll4 mRNA expression of MSCs with or without BNP incubation cultured in basal medium, induction medium, and induction medium with DAPT using an LNA/DNA nanobiosensor. Fig. 4C shows representative fluorescence images of MSCs with and without BNP treatment under different conditions. Dll4 mRNA expression was quantified and compared by measuring the mean fluorescence intensity of each individual cell, Fig. 4D. The results showed that intracellular uptake of BNPs enhanced Dll4 expression in all three groups, control, osteogenic, and DAPT treated osteogenic induction groups.
We also observed an increase in Dll4 expression following osteogenic induction, with a 1.2-fold increase in the control group and a 0.15-fold increase in the BNP-treated group. In the BNP-treated group, DAPT treatment moderated the enhancement of differentiation by BNPs, resulting in Dll4 mRNA expression that showed no significant difference compared to MSCs cultured in basal medium. These results suggest that Notch signaling plays a regulatory role in the osteogenic differentiation of MSCs in BNP incubation. BNP incubation increases Dll4 mRNA expression in MSCs undergoing osteogenic induction, highlighting Notch signaling's role in BNP regulated osteogenic differentiation. Furthermore, inhibiting Notch signaling diminishes the osteogenic differentiation induced by BNPs, resulting in decreased ALP enzyme activity and reduced Dll4 mRNA expression.
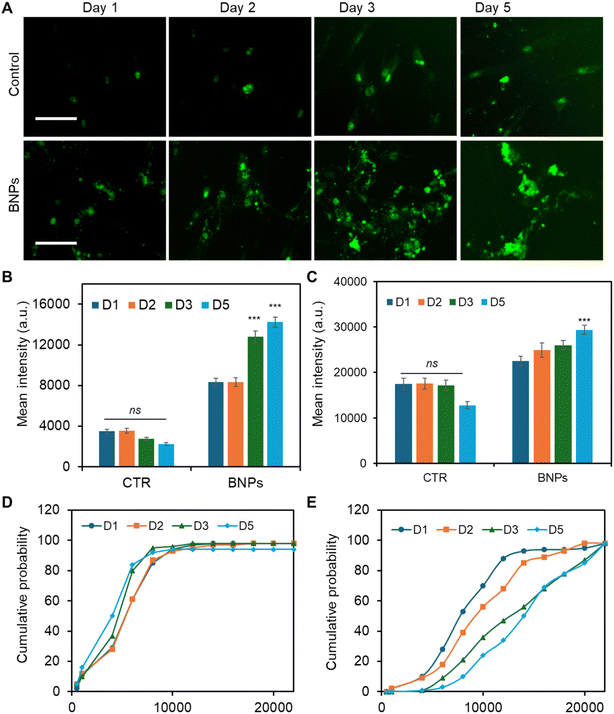
Dynamic monitoring of Dll4 expression during osteogenic differentiation
To better understand and interpret the Notch regulatory mechanisms that contributed to osteogenic differentiation, we monitored Dll4 expression dynamics after osteogenic induction. The effects of BNP incubation were assessed and compared with the control (No BNPs) group. The capability of this LNA/DNA nanobiosensor of live-cell gene detection allowed us to further elucidate the cytoplasmic and nuclear mRNA expression profile. Fig. 5A shows fluorescence images of MSCs expressing Dll4 after 1, 2, 3, and 5 days of osteogenic induction. To further explore and compare the distribution of Dll4 in the cytoplasm and nucleus, we quantified and compared Dll4 expression at different times with different treatments. Examining nuclear Dll4 mRNA expression could potentially reveal transcriptional regulation, including interactions with nuclear factors to influence gene expression and cellular processes. Fig. 5B shows the comparison of cytoplasmic Dll4 expression with and without BNP treatment. For the control group, the cytoplasmic Dll4 expression is relatively low compared to that for the BNP-treated group. Between the different days measured, there is no significant difference. For the BNP-treated group, cytoplasmic Dll4 is significantly higher, particularly on day 3 and day 5, indicating a strong effect of BNPs. The data suggest that BNPs progressively enhance the cytoplasmic Dll4, with the most substantial effects observed on day 3. Fig. 5C shows the comparison of nuclear Dll4 expression over different days with different treatments. For the control group, Dll4 expression across days 1, 2, 3, and 5 showed relatively same levels with no statistically significant changes. In contrast, the BNP-treated group displays a gradual increase, with a significant rise on day 5, indicating the delayed but potent effect of BNPs. These data highlight BNPs' time-dependent efficacy in enhancing osteogenic activity. Nuclear Dll4 expression did not show a significant difference until 5 days of induction, indicating that the nuclear translocation of Dll4 may be a delayed response to signaling or cellular changes, possibly reflecting a time-dependent regulatory mechanism. Furthermore, for the cytoplasmic Dll4, we further conducted cumulative analysis to understand the trend and patterns of Dll4 expression over time, which could potentially reveal insights into the progression and impact of our interventions. Fig. 5D and E show the cumulative probability of Dll4 expression over several days (1, 2, 3, and 5 days) for control and BNP-treated groups, respectively. The cumulative probability curve in Fig. 5D and E reflects the distribution of fluorescence intensities across cells. The x-axis represents fluorescence intensity values, and the y-axis shows the cumulative probability that the fluorescence intensity is less than or equal to a specific value. As shown in Fig. 5D, the steep curve indicates that most cells have similar intensities in the control group, and there is no shift among these curves at different time points, indicating that the fluorescence intensity distribution is similar at different time points (D1, D2, D3, and D5). In contrast, Fig. 5E shows a flatter curve which indicates a wide variation in fluorescence intensities. The shifts among multiple time points (D1, D2, D3, and D5) reveal the differences in the fluorescence intensity distribution. These results confirmed that the effects of BNPs on Dll4 expression are time dependent. These data further support the importance of Notch signaling in regulating BNP enhanced Dll4 expression during osteogenic differentiation, which demonstrates the potent and sustained impact of BNPs, important in fields such as drug delivery or gene therapy.Discussion
In this study, we first investigated the effects of intracellular uptake of BNPs on osteogenic differentiation and examined the regulatory roles of Notch signaling in enhancing this differentiation process, utilizing an LNA/DNA nanobiosensor. This nanobiosensor, unlike conventional approaches for mRNA detection, allows for the monitoring of mRNA expression in live cells at the single-cell level without requiring cell lysis or fixation. This capability permits continuous observation of Dll4 mRNA gene expression dynamics throughout the process of osteogenic differentiation. The specificity and stability of this nanobiosensor have been demonstrated earlier.4 Previous studies have demonstrated that this nanobiosensor can track spatiotemporal RNA dynamics in collective cell migration,46 mice lung cancer,47 wounded corneal tissue repair,48 liver tissue,49 and vasculature formation.42,50 Our group recently reported the application of this nanobiosensor in monitoring Dll4 dynamics during osteogenic differentiation.2,4 It is noted that this nanobiosensor can be utilized to detect other types of RNA detection, i.e., miRNA and long non-coding RNA (lncRNA).39,41,51,52 Moreover, this nanobiosensor is versatile, working effectively across various types and tissue environments. Previous studies have shown that this nanobiosensor can monitor miRNA dynamics during 3D cancer invasion and mRNA and lncRNA dynamics during 3D osteogenic differentiation.41,53 The ability to monitor gene expression in 3D physiological environments opens up possibilities to discover new aspects and mechanisms of cell–cell and cell–matrix interactions, ultimately paving the way for the development of innovative tools in tissue engineering and regenerative medicine.Bone based ECM, derived from decellularized bone tissues, can be used as powder, hydrogel, and electrospun scaffolds in regenerative therapies for bone repair and wound healing.54–57 These scaffolds exhibit robust mechanical properties, inherent osteoinductive and osteoconductive capabilities, and closely mimic natural bone. Although various studies have shown that the decellularized bone matrix promotes bone regeneration, the effects of BNPs on tissue regeneration and its fundamental mechanism remain largely unknown. We recently reported that these BNPs enhance bone repair in vivo.26 Due to their native source, BNPs exhibit unique characteristics, including biocompatibility, bioactivity, osteoinductivity, osteoconductivity, and biodegradability. We have previously reported the degradation of BNPs and the components of BNPs.26
Moreover, these nanoparticles can be engineered to carry drugs, growth factors, or other therapeutic agents directly to the site of bone damage. The targeted drug delivery system can improve the effectiveness of treatments while reducing side effects associated with systemic drug delivery. Importantly, the properties of BNPs can be tailored during their synthesis process to meet specific requirements of different applications, such as varying their size, surface charge, and functionalization with bioactive molecules. These unique features of BNPs harness the natural properties of bone to offer promising solutions in bone tissue engineering, making them a focus of current research in regenerative medicine and related fields.
One of the primary reasons for the unique features of BNPs is the protein content they contain. We have shown that these BNPs contain several major ECM proteins, including TGF-β, fibronectin, and COL1A1.26 It is noted that the enhancement of cell proliferation and osteogenic differentiation can be observed quickly after several days of incubation, Fig. 2 and 3. One reason is that BNPs can gradually release protein over 5–7 days due to the physical connections within the nanoparticles, without the need for crosslinking. The release profile has been studied previously.26 BNPs enhance cell proliferation and osteogenic differentiation, which may be related to the protein contained in the nanoparticles (i.e., TGF-β). Previous studies have shown that TGF-β promotes osteoinduction of osteoblasts and bone marrow stromal cells.58–60 The osteogenic differentiation process is modulated by several growth factors including bone morphogenetic proteins (BMPs), TGF-β, fibroblast growth factor (FGF), and platelet derived growth factor (PDGF). Several signal transduction pathways have been identified during osteogenic differentiation, including BMP signaling, Wnt/b-catenin signaling, and Notch signaling.58–63 Our previous studies have shown that Notch signaling is activated and required for osteogenic differentiation, and inhibition of Notch signaling represses osteogenesis, with reduced ALP activity.2,4 In addition, given the collagen-rich composition of the natural bone microenvironment, the incorporation of collagen in BNPs may facilitate actin clustering, thereby affecting both biophysical and biochemical properties that regulate cellular behavior. It has been demonstrated that densely organized collagen fibers act as bioactive cues, modulating key cellular processes such as migration and differentiation.64–66 Our future work will focus on investigating the mechanisms by which internalized BNPs interact with actin filaments, subsequently activating intracellular biophysical and biochemical signaling pathways that direct cell fate.
Notch signaling is a highly conserved evolutionary pathway that influences cell proliferation, cell fate determination, and stem cell differentiation in both embryonic and adult tissues.67–70 This pathway includes four Notch receptors (Notch1-4) and five distinct ligands (Dll1, Dll3, Dll4, Jag1, and Jag2). Recently, the involvement of Notch signaling in osteogenic differentiation has gained significant attention from researchers, with various studies confirming its activation during this process.43,71 Recently, it has been reported that the Notch ligand Dll4 can promote bone formation in male mice without causing adverse effects in other organs.72 Another study reported that Notch signaling is crucial for regulating the differentiation and function of osteoblasts and osteoclasts, as well as maintaining skeletal homeostasis.73 Cao et al. demonstrated that the Notch receptor Notch1 and Notch ligand Dll1 play roles in osteogenic differentiation.74 Their study showed that inhibiting Notch1 decreased ALP activity during BMP-induced osteogenic differentiation of MSCs in vitro. However, the exact involvement of Notch signaling during BNP enhanced osteogenic differentiation remains largely unexplored, especially the time-dependence of Notch involvement. In this study, we aim to characterize the Notch signaling pathway during spontaneous BNP enhanced osteogenic differentiation during the whole period of cell differentiation. To achieve this aim, we monitored and analyzed Notch ligand Dll4 mRNA expression across different time points. Our results showed that Notch signaling is involved in BNP enhanced osteogenic differentiation. Intracellular uptake of BNPs in MSCs promotes both early (1–7 days) and late stage (8–21 days) differentiation, as indicated by the increase in the production of both early (ALP activity) and late osteogenic markers (calcium mineralization). Interestingly, the analysis of the expression dynamics of Notch ligand Dll4 expression has shown a significant increase of cytoplasmatic Dll4 in BNP treated MSCs during the osteogenic differentiation process, suggesting that intracellular uptake of BNPs could modulate the gene expression profile. To further verify the involvement of Notch signaling, we investigated the effects of γ-secretase inhibitor DAPT on osteogenic differentiation. Without BNPs, disruption of the Notch pathway decreased Dll4 expression and reduced ALP activity and calcium mineralization. The MSCs were treated with BNPs, and the effects of Notch inhibition on osteogenic differentiation were mediated with enhanced Dll4 expression and increased calcium mineralization. Overall, our study suggests that Notch signaling is involved and regulates osteogenic differentiation. Internalization of BNPs enhanced osteogenic differentiation with increased Dll4 expression, indicating that BNPs may activate Notch signaling. To our knowledge, only a few studies have investigated the Dll4 ligand during osteogenic differentiation, and none in our cellular model in live cells.62 Moreover, Dll4 has been reported to be upregulated during endochondral and intramembranous bone regeneration.61
Although it is mentioned in the literature that TGF-β promotes proliferation, early differentiation, and commitment to the osteoblastic lineage through the selective BMP, Wnt, Smad2/3, and Notch signaling,62,63 it is unclear whether the enhancement of osteogenic differentiation is due to single or multiple protein molecules presented in the BNPs. Thus, further mechanistic studies are required to elucidate the molecular and cellular processes that regulate osteogenic differentiation. Specifically, the fundamental regulatory mechanisms of the Notch pathway and its upstream and downstream signaling pathways should be further investigated using loss- and gain-of function experiments. Understanding the fundamental mechanisms of the osteogenic differentiation of MSCs induced by intracellular uptake of BNPs will provide valuable information that can be used for bone regeneration and repair.
Conclusions
In this study, we investigated the effects of intracellular uptake of BNPs on cell proliferation and differentiation and the involvement of Notch signaling during the osteogenic differentiation process. We first showed that BNPs enhance MSC proliferation and osteogenic differentiation, with a significantly increased number of cells, ALP enzyme activity, and calcium deposition. By leveraging an LNA/DNA nanobiosensor, we examined and compared the Notch ligand Dll4 mRNA expression dynamics in the cytoplasm and nucleus during the osteogenic differentiation process. Pharmacological disruption of Notch signaling using γ-secretase inhibitor DAPT mediated the osteogenic process, with reduced expression of early and late stage differentiation markers (ALP and calcium mineralization). The decrease in the expression of osteogenic markers in cells treated with DAPT suggests that the Notch pathway is involved in their regulation. In addition, the expression of Dll4 in BNP treated cells displayed a time-dependent profile, which is consistent with the enhancement of cell proliferation and differentiation. Our results suggest that the changes in BNP treated cells during osteogenic differentiation may be linked to the increased expression in the Dll4 mRNA. In conclusion, this study will add new insights concerning the osteogenic differentiation of MSCs as well as the molecular mechanisms by which BNPs can stimulate the differentiation process. The results showed that BNP induced effects on osteogenesis can be associated with the modulation of Notch signaling which plays an essential role in cell fate and differentiation. Further studies will focus on elucidating the relationship among TGF-β, Notch, and BMP signaling pathways during the osteogenic differentiation process as well as the effects of BNPs.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
B. W. and S. W. conceived the initial idea of the study. A. S., J. C. and S. F. performed the experiments. B. W. and S. W. contributed to the experimental design and data analysis. B. W. and S. W. wrote the manuscript with feedback from all authors.Conflicts of interest
There are no conflicts to declar.Acknowledgements
S. Wang acknowledges the financial support from NSF CAREER (CMMI: 2143151).References
- S. Fasciano, A. Wheba, C. Ddamulira and S. Wang, Biomater. Adv., 2024, 213897 CrossRef CAS PubMed.
- Y. Zhao, K. Richardson, R. Yang, Z. Bousraou, Y. K. Lee, S. Fasciano and S. Wang, Frontiers in Bioengineering and Biotechnology, 2022, 10, 1007430 CrossRef PubMed.
- Y. Zhao, R. Yang, Z. Bousraou and S. Wang, 2021 IEEE 21st International Conference on Nanotechnology (NANO), Montreal, QC, Canada, 2021, pp. 273–276 Search PubMed.
- Y. Zhao, R. Yang, Z. Bousraou, K. Richardson and S. Wang, Sci. Rep., 2022, 12, 10315 CrossRef CAS PubMed.
- P. Charbord, Hum. Gene Ther., 2010, 21, 1045–1056 CrossRef CAS PubMed.
- N. Su, C. Villicana and F. Yang, Biomaterials, 2022, 286, 121604 CrossRef CAS PubMed.
- A. Mohanty, N. Polisetti and G. K. Vemuganti, J. Biosci., 2020, 45, 1–17 CrossRef.
- R. Chen, Z. Hao, Y. Wang, H. Zhu, Y. Hu, T. Chen, P. Zhang and J. Li, Stem Cells Int., 2022, 2022, 7153584 Search PubMed.
- F. Guilak, D. M. Cohen, B. T. Estes, J. M. Gimble, W. Liedtke and C. S. Chen, Cell Stem Cell, 2009, 5, 17–26 CrossRef CAS PubMed.
- Y.-K. Wang, X. Yu, D. M. Cohen, M. A. Wozniak, M. T. Yang, L. Gao, J. Eyckmans and C. S. Chen, Stem Cells Dev., 2012, 21, 1176–1186 CrossRef CAS PubMed.
- S.-J. Heo, T. P. Driscoll, S. D. Thorpe, N. L. Nerurkar, B. M. Baker, M. T. Yang, C. S. Chen, D. A. Lee and R. L. Mauck, Elife, 2016, 5, e18207 CrossRef PubMed.
- K. Qiao, L. Xu, J. Tang, Q. Wang, K. S. Lim, G. Hooper, T. B. Woodfield, G. Liu, K. Tian and W. Zhang, J. Nanobiotechnol., 2022, 20, 141 CrossRef CAS PubMed.
- S. Behzadi, G. A. Luther, M. B. Harris, O. C. Farokhzad and M. Mahmoudi, Biomaterials, 2017, 146, 168–182 CrossRef CAS PubMed.
- J. Zhou, Z. Zhang, J. Joseph, X. Zhang, B. E. Ferdows, D. N. Patel, W. Chen, G. Banfi, R. Molinaro and D. Cosco, Exploration, 2021, 1(2), 20210011 CrossRef PubMed.
- H. Hajiali, L. Ouyang, V. Llopis-Hernandez, O. Dobre and F. R. Rose, Nanoscale, 2021, 13, 10266–10280 RSC.
- J. Scimeca and E. Verron, Mater. Today Adv., 2022, 15, 100260 CrossRef CAS.
- D. Arcos, N. Gómez-Cerezo, M. Saiz-Pardo, D. De Pablo, L. Ortega, S. Enciso, B. Fernández-Tomé, I. Díaz-Güemes, F. Sánchez-Margallo and L. Casarrubios, Acta Biomater., 2022, 151, 501–511 CrossRef CAS PubMed.
- Y. Cui, H. Li, Y. Li and L. Mao, Nanoscale Adv., 2022, 4, 334–352 RSC.
- H. Abu Owida and F. Alnaimat, Adv. Polym. Technol., 2023, 2023, 2934169 Search PubMed.
- L. Ferreira, J. M. Karp, L. Nobre and R. Langer, Cell Stem Cell, 2008, 3, 136–146 CrossRef CAS PubMed.
- L. Accomasso, C. Gallina, V. Turinetto and C. Giachino, Stem Cells Int., 2016, 2016, 7920358 CrossRef PubMed.
- A. Bozorgi, M. Khazaei, M. Soleimani and Z. Jamalpoor, Biomater. Sci., 2021, 9, 4541–4567 RSC.
- A. C. Anselmo and S. Mitragotri, Bioeng. Transl. Med., 2019, 4, e10143 CrossRef PubMed.
- L. Yildirimer, N. T. Thanh, M. Loizidou and A. M. Seifalian, Nano today, 2011, 6, 585–607 CrossRef CAS PubMed.
- A. C. Anselmo and S. Mitragotri, Bioeng. Transl. Med., 2016, 1, 10–29 CrossRef PubMed.
- A. Stellpflug, J. Walls, C. Hansen, A. Joshi and B. Wang, Biomater. Sci., 2024, 12(14), 3633–3648 RSC.
- M. Beederman, J. D. Lamplot, G. Nan, J. Wang, X. Liu, L. Yin, R. Li, W. Shui, H. Zhang and S. H. Kim, J. Biomed. Sci. Eng., 2013, 6, 32 CrossRef PubMed.
- Z. Yuan, Q. Li, S. Luo, Z. Liu, D. Luo, B. Zhang, D. Zhang, P. Rao and J. Xiao, Curr. Stem Cell Res. Ther., 2016, 11, 216–225 CrossRef CAS PubMed.
- T. F. Day and Y. Yang, J. Bone Jt. Surg., Am. Vol., 2008, 90, 19–24 CrossRef PubMed.
- Q. Wei, A. Holle, J. Li, F. Posa, F. Biagioni, O. Croci, A. S. Benk, J. Young, F. Noureddine and J. Deng, Advanced Science, 2020, 7, 1902931 CrossRef CAS PubMed.
- C. Lorthongpanich, K. Thumanu, K. Tangkiettrakul, N. Jiamvoraphong, C. Laowtammathron, N. Damkham, Y. U-pratya and S. Issaragrisil, Stem Cell Res. Ther., 2019, 10, 1–12 CrossRef PubMed.
- C. Lorthongpanich, K. Thumanu, K. Tangkiettrakul, N. Jiamvoraphong, C. Laowtammathron, N. Damkham, U. Yaowalak and S. Issaragrisil, Stem Cell Res. Ther., 2019, 10, 1–12 CrossRef PubMed.
- A. Karystinou, A. J. Roelofs, A. Neve, F. P. Cantatore, H. Wackerhage and C. De Bari, Arthritis Res. Ther., 2015, 17, 147 CrossRef PubMed.
- C. Yi, D. Liu, C.-C. Fong, J. Zhang and M. Yang, ACS Nano, 2010, 4, 6439–6448 CrossRef CAS PubMed.
- Q. Wang, B. Chen, M. Cao, J. Sun, H. Wu, P. Zhao, J. Xing, Y. Yang, X. Zhang and M. Ji, Biomaterials, 2016, 86, 11–20 CrossRef CAS PubMed.
- Z. Xiang, K. Wang, W. Zhang, S. W. Teh, A. Peli, P. L. Mok, A. Higuchi and S. Suresh Kumar, J. Cluster Sci., 2018, 29, 1–7 CrossRef CAS.
- F. Wang, J. Flanagan, N. Su, L.-C. Wang, S. Bui, A. Nielson, X. Wu, H.-T. Vo, X.-J. Ma and Y. Luo, J. Mol. Diagn., 2012, 14, 22–29 CrossRef CAS PubMed.
- J. Yu, J. Xiao, X. Ren, K. Lao and X. S. Xie, Science, 2006, 311, 1600–1603 CrossRef CAS PubMed.
- Y. Zhao and S. Wang, in MicroRNA Detection and Target Identification: Methods and Protocols, Springer, 2023, pp. 75–87 Search PubMed.
- S. Wang, Y. Xiao, D. D. Zhang and P. K. Wong, Biomaterials, 2018, 156, 56–64 CrossRef CAS PubMed.
- S. Fasciano, S. Luo and S. Wang, Analyst, 2023, 148, 6261–6273 RSC.
- S. Wang, J. Sun, D. Zhang and P. Wong, Nanoscale, 2016, 8, 16894–16901 RSC.
- L. Bagheri, A. Pellati, P. Rizzo, G. Aquila, L. Massari, M. De Mattei and A. Ongaro, J. Tissue Eng. Regener. Med., 2018, 12, 304–315 CrossRef CAS PubMed.
- F. Ugarte, M. Ryser, S. Thieme, F. A. Fierro, K. Navratiel, M. Bornhäuser and S. Brenner, Exp. Hematol., 2009, 37, 867–875 CrossRef CAS PubMed.
- Z. Luo, X. Shang, H. Zhang, G. Wang, P. A. Massey, S. R. Barton, C. G. Kevil and Y. Dong, Am. J. Pathol., 2019, 189, 1495–1500 CrossRef CAS PubMed.
- R. Riahi, Z. Dean, T.-H. Wu, M. A. Teitell, P.-Y. Chiou, D. D. Zhang and P. K. Wong, Analyst, 2013, 138, 4777–4785 RSC.
- R. Riahi, S. Wang, M. Long, N. Li, P.-Y. Chiou, D. D. Zhang and P. K. Wong, ACS Nano, 2014, 8, 3597–3605 CrossRef CAS PubMed.
- S. Wang, R. Riahi, N. Li, D. D. Zhang and P. K. Wong, Adv. Mater., 2015, 27, 6034–6038 CrossRef CAS PubMed.
- S. Tao, S. Wang, S. J. Moghaddam, A. Ooi, E. Chapman, P. K. Wong and D. D. Zhang, Cancer Res., 2014, 74, 7430–7441 CrossRef CAS PubMed.
- S. Wang, J. Sun, Y. Xiao, Y. Lu, D. D. Zhang and P. K. Wong, Adv. Biosyst., 2017, 1(1–2), 1600019 CrossRef PubMed.
- Z. S. Dean, R. Riahi and P. K. Wong, Biomaterials, 2015, 37, 156–163 CrossRef CAS PubMed.
- S. Wang, S. Majumder, N. J. Emery and A. P. Liu, Synth. Biol., 2018, 3, ysy005 CrossRef CAS PubMed.
- Z. S. Dean, P. Elias, N. Jamilpour, U. Utzinger and P. K. Wong, Anal. Chem., 2016, 88, 8902–8907 CrossRef CAS PubMed.
- A. Shekaran and A. J. García, J. Biomed. Mater. Res., Part A, 2011, 96, 261–272 CrossRef PubMed.
- A. García-García and I. Martin, Front. Immunol., 2019, 10, 482241 Search PubMed.
- R. E. McMahon, L. Wang, R. Skoracki and A. B. Mathur, J. Biomed. Mater. Res., Part B, 2013, 101, 387–397 CrossRef PubMed.
- A. Mansour, M. A. Mezour, Z. Badran and F. Tamimi, Tissue Eng., Part A, 2017, 23, 1436–1451 CrossRef PubMed.
- G. Chen, C. Deng and Y.-P. Li, Int. J. Biol. Sci., 2012, 8, 272 CrossRef CAS PubMed.
- D. D. Liu, J. C. Zhang, Q. Zhang, S. X. Wang and M. S. Yang, J. Cell. Biochem., 2013, 114, 1105–1114 CrossRef CAS PubMed.
- L. Zhao, S. Jiang and B. M. Hantash, Tissue Eng., Part A, 2010, 16, 725–733 CrossRef CAS PubMed.
- M. I. Dishowitz, S. P. Terkhorn, S. A. Bostic and K. D. Hankenson, J. Orthop. Res., 2012, 30, 296–303 CrossRef CAS PubMed.
- N. Chakravorty, S. Hamlet, A. Jaiprakash, R. Crawford, A. Oloyede, M. Alfarsi, Y. Xiao and S. Ivanovski, Clin. Oral Implants Res., 2014, 25, 475–486 CrossRef PubMed.
- A. Blokzijl, C. Dahlqvist, E. Reissmann, A. Falk, A. Moliner, U. Lendahl and C. F. Ibáñez, J. Cell Biol., 2003, 163, 723–728 CrossRef CAS PubMed.
- R. Y. Nguyen, A. T. Cabral, A. Rossello-Martinez, A. Zulli, X. Gong, Q. Zhang, J. Yan and M. Mak, Adv. Mater., 2023, 35, 2207882 CrossRef CAS PubMed.
- C. Liu, R. Y. Nguyen, G. A. Pizzurro, X. Zhang, X. Gong, A. R. Martinez and M. Mak, Acta Biomater., 2023, 155, 167–181 CrossRef CAS PubMed.
- X. Gong, Z. Wen, Z. Liang, H. Xiao, S. Lee, T. Wright, R. Y. Nguyen, A. Rossello and M. Mak, bioRxiv, 2023, preprint, DOI:10.1101/2023.10.08.561456.
- S. Artavanis-Tsakonas, M. D. Rand and R. J. Lake, Science, 1999, 284, 770–776 CrossRef CAS PubMed.
- K.-i. Mizutani, K. Yoon, L. Dang, A. Tokunaga and N. Gaiano, Nature, 2007, 449, 351–355 CrossRef CAS PubMed.
- B. R. Nelson, B. H. Hartman, S. A. Georgi, M. S. Lan and T. A. Reh, Dev. Biol., 2007, 304, 479–498 CrossRef CAS PubMed.
- C. R. Bjornson, T. H. Cheung, L. Liu, P. V. Tripathi, K. M. Steeper and T. A. Rando, Stem Cells, 2012, 30, 232–242 CrossRef CAS PubMed.
- Y. Wagley, A. Chesi, P. K. Acevedo, S. Lu, A. D. Wells, M. E. Johnson, S. F. Grant and K. D. Hankenson, Stem Cells, 2020, 38, 1332–1347 CrossRef CAS PubMed.
- C. Xu, K. Kruse, H.-W. Jeong, E. C. Watson, S. Adams, F. Berkenfeld, M. Stehling, S. J. Rasouli, R. Fan and R. Chen, Elife, 2022, 11, e60183 CrossRef CAS PubMed.
- J. Yu and E. Canalis, Bone, 2020, 138, 115474 CrossRef CAS PubMed.
- J. Cao, Y. Wei, J. Lian, L. Yang, X. Zhang, J. Xie, Q. Liu, J. Luo, B. He and M. Tang, Int. J. Mol. Med., 2017, 40, 378–388 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00797b |
| This journal is © The Royal Society of Chemistry 2025 |