Open Access Article
Open Access ArticleGraphene quantum dots as potential broad-spectrum antiviral agents†
Younghun
Jung‡
a,
Jaehyeon
Hwang‡
 a,
Hyeonwoo
Cho‡
a,
Hyeonwoo
Cho‡
 bc,
Jeong Hyeon
Yoon
a,
Jong-Hwan
Lee
b,
Jaekwang
Song
bc,
Jeong Hyeon
Yoon
a,
Jong-Hwan
Lee
b,
Jaekwang
Song
 b,
Donghoon
Kim
d,
Minchul
Ahn
*bc,
Byung Hee
Hong
b,
Donghoon
Kim
d,
Minchul
Ahn
*bc,
Byung Hee
Hong
 *bc and
Dae-Hyuk
Kweon
*a
*bc and
Dae-Hyuk
Kweon
*a
aDepartment of Integrative Biotechnology, College of Biotechnology and Bioengineering, Sungkyunkwan University, Suwon, 16419, Republic of Korea
bDepartment of Chemistry, Seoul National University, Seoul, 08826, Republic of Korea
cGraphene Research Center & Graphene Square Chemical Inc., Advanced Institute of Convergence Technology, Suwon, 16229, Republic of Korea
dDepartment of Pharmacology, Peripheral Neuropathy Research Center (PNRC), Dong-A University College of Medicine, Busan, 49201, Republic of Korea
First published on 30th January 2025
Abstract
As pandemic viruses have become a threat to people, various treatments have been developed, including vaccines, neutralizing antibodies, and inhibitors. However, some mutations in the target envelope protein limit the efficiency of these treatments. Therefore, the development of broad-spectrum antiviral agents targeting mutation-free viral membranes is of considerable importance. Herein, we propose graphene quantum dots (GQDs) as broad-spectrum antiviral agents, wherein the amphiphilic properties of GQDs destroy the viral membranes, regardless of the type of viruses, including SARS-CoV-2 and influenza virus. We observed that GQDs suppress both viral infection and replication and demonstrated their low cytotoxicity in a cell line and a mouse model, revealing the potential of GQDs as a universal first-line treatment for various viral diseases.
1. Introduction
According to the World Health Organization (WHO), the coronavirus disease (COVID-19) has killed nearly 7 million people worldwide since its outbreak in 2019. COVID-19 caused by SARS-CoV-2 has caused a widespread threat and disruption worldwide, and it is now marking a chapter of a global pandemic in the history of humanity. Several viral outbreaks in the past 10 decades, including the Spanish flu of 1918, HIV, SARS, MERS, Ebola, ZIKA and the ongoing COVID-19, have been caused by highly fatal enveloped viruses.1,2 In addition to pandemic diseases, such enveloped viruses are involved in human infectious diseases such as hepatitis, herpes, and smallpox.3 Accordingly, efforts to develop vaccines are fast-paced around the world to counter these emerging viruses.However, vaccine development is an expensive and time consuming process because it requires many efficacy and safety studies prior to clinical usage.4,5 Additionally, the lack of understanding about newly emerging viruses often leads to difficulties in vaccine development. Furthermore, the repetitive re-emerging of viral mutants may periodically demand additional vaccine development as in the case of influenza vaccines. For instance, the E484K mutation of COVID-19, which was identified in South Africa and quickly spread in the UK, has become more resistant to vaccines owing to mutations in their spike protein.6 Sole dependence on vaccine development is inadequate for rapid response to viral threats, as evidenced by COVID-19 and applicable to future emerging viral diseases.
Hence, the requirement for universal antiviral drugs have increased, and numerous studies have suggested potential broad-spectrum antiviral drugs based on peptides,7–9 nanoparticles,10–12 and small molecules.13 To overcome the mutational vulnerability of viral proteins, these universal antiviral agents are designed to target the viral envelopes that are similar to cell membranes and disrupt them physically or chemically, thereby irreversibly inactivating the viruses.14 In particular, graphene derivatives, such as modified graphene oxides, have been studied as antiviral agents owing to their high surface area, tunable chemical functionality, and amphiphilicity.10,15–25 Although graphene derivatives show broad-spectrum antiviral activity, their cytotoxicity caused by their large sizes (100 nm to 1 μm) makes their development as actual drugs challenging. Herein, we propose a graphene quantum dot (GQD) with an average size of 5 nm as a novel antiviral agent that is effective against a wide range of viruses. GQDs are the smallest fragments of few-layered graphene nanoflakes that contain hydrophilic edges and hydrophobic basal planes. In recent years, GQDs with amphiphilic characteristics have been utilized to disaggregate protein fibrils like α-synucleins, β-amyloids, and cholesterol that cause degenerative brain diseases.26,27 In addition, GQDs are less toxic than other large graphene derivatives because they are less than 20 nm in size and can be quickly removed by renal clearance.28–35 Moreover, in our previous study, we have confirmed that oral administration of GQDs do not affect the gut microbiota.32 These ideal properties of amphiphilicity and low toxicity of GQDs indicate that they can be used as potential antiviral agents for regulating multiple strains of viruses.
The antiviral properties of GQDs against enveloped viruses have been reported to be enhanced by functionalization and doping.36,37 However, to the best of our knowledge, no studies have been conducted using GQDs against non-enveloped viruses. Additionally, while studies have been conducted on enveloped viruses, there have been no reports optimizing antiviral efficacy through size control of GQDs for these viruses.
Herein, we synthesized GQDs through oxidative exfoliation and investigated their antiviral effects with respect to their sizes. On performing a series of in vitro experiments, we observed that GQDs significantly hindered the infection of several enveloped viruses, including SARS-CoV-2 and influenza virus, as well as the non-enveloped adenovirus. In addition, the virucidal effect by GQDs was visualized using bio-transmission electron microscopy (Bio-TEM). Notably, the complete disruption of the virus inhibited viral infection in vivo, and no symptoms and lethality were observed in mice. Thus, our results indicated that GQDs can potentially inhibit multiple viruses by disrupting the viral membranes in vitro and in vivo. Furthermore, the results from in vitro viral inhibition suggested their potential as a therapeutic and preventing agent.
2. Results and discussion
2.1 Preparation and characterizations of amphiphilic GQDs
To investigate the antiviral effects of GQDs, they were synthesized through the acidic oxidation of carbon black (Super P®), followed by filtration with a disc filter of 20 nm and dialysis with 1 kDa dialysis membrane for 3 days.38,39 The dialyzed solution was lyophilized for 2 days to obtain a brownish-black powder with a yield of about ∼15%. The synthesized GQDs were characterized using TEM and dynamic light scattering (DLS) to confirm their morphological structure and size distribution, respectively. GQDs were observed to be monodispersed graphene fragments, and the average lateral size of GQDs was 2.36 ± 0.69 nm (Fig. 1a and b). Using the fast Fourier transform technique in the TEM image, the lattice space of GQDs was observed to be 0.24 nm, which was close to the d value of the graphene (100) plane.40 The hydrodynamic size of GQDs was 5.08 ± 1.54 nm, and the zeta potential of GQDs was −10.6 ± 4.65 mV (Fig. S1†). We also performed XPS, FT-IR, Raman spectroscopy, and UV-vis spectroscopy to confirm the chemical structures of GQDs.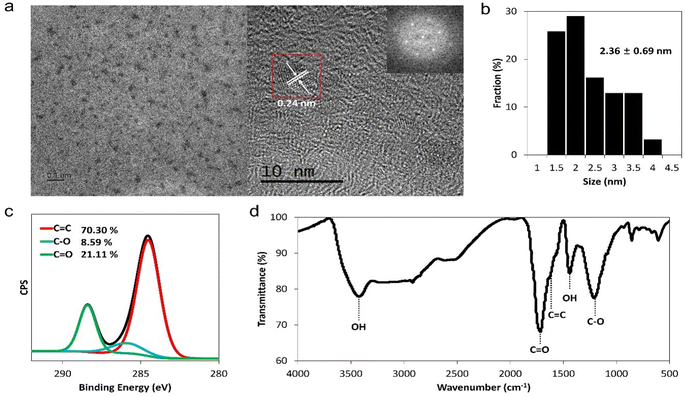 | ||
| Fig. 1 Characteristics of GQDs. (a) TEM images and fast Fourier transform pattern of GQDs (inner box). (b) Size distribution of GQDs. (c) XPS C 1s spectra and (d) FT-IR spectra of GQDs. | ||
In the XPS spectra, we observed that the ratio of the C 1s to O 1s peak was about 0.8, which was similar to the C/O ratio obtained from elemental analysis (Fig. S2, Table S1†). C 1s peaks of GQDs were deconvoluted into three peaks at 284.5 eV, 286 eV, and 288 eV, which corresponded to the sp2 carbon (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), alcohol group (C–O), and carbonyl group (O–C
C), alcohol group (C–O), and carbonyl group (O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), respectively (Fig. 1c). In addition, FT-IR spectra demonstrated that GQDs contain aromatic rings (1650 cm−1 peaks, C
O), respectively (Fig. 1c). In addition, FT-IR spectra demonstrated that GQDs contain aromatic rings (1650 cm−1 peaks, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching band) and diverse functional groups, such as carbonyl (1720 cm−1, C
C stretching band) and diverse functional groups, such as carbonyl (1720 cm−1, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching band) and hydroxyl groups (3400 cm−1, O–H stretching) (Fig. 1d). The D band (1350 cm−1) and G band (1580 cm−1) corresponded to the characteristic Raman peak for graphene (Fig. S3†).41 The UV-visible absorption band at 280 nm, which was related to the π–π* transition of conjugated C
O stretching band) and hydroxyl groups (3400 cm−1, O–H stretching) (Fig. 1d). The D band (1350 cm−1) and G band (1580 cm−1) corresponded to the characteristic Raman peak for graphene (Fig. S3†).41 The UV-visible absorption band at 280 nm, which was related to the π–π* transition of conjugated C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds, indicated that GQD samples had graphitic carbon cores.42 As can be seen from the fluorescence image of GQDs at 365 nm excitation, GQDs exhibited yellow emission and emission peaks at ∼480 nm and ∼560 nm (Fig. S4†). From the above-mentioned results, we confirmed that GQDs were composed of a sp2 carbon core and diverse functional groups, including oxygen, indicating that GQDs are amphiphilic agents.
C bonds, indicated that GQD samples had graphitic carbon cores.42 As can be seen from the fluorescence image of GQDs at 365 nm excitation, GQDs exhibited yellow emission and emission peaks at ∼480 nm and ∼560 nm (Fig. S4†). From the above-mentioned results, we confirmed that GQDs were composed of a sp2 carbon core and diverse functional groups, including oxygen, indicating that GQDs are amphiphilic agents.
2.2 Virucidal mode of action of GQDs on a viral envelope
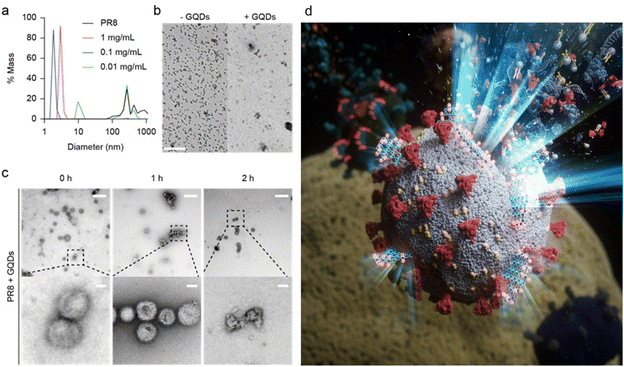
We hypothesised that GQDs would perform a virucidal effect on a viral envelope through the amphipathicity obtained from the hydrophobic plane and hydrophilic edge of the graphene structure. To identify disruption of virions by GQDs, we assessed DLS, which enabled measuring the hydrodynamic size change of viruses on treatment with GQD (Fig. 2a). When treated with 0.1 and 1 mg mL−1 of GQDs, virus peaks in the DLS profile disappeared, indicating the complete disruption of the virus. At a lower concentration (0.01 mg mL−1), intact PR8 viruses were still measured, indicating that a threshold amount of GQDs is required for viral disruption.We performed TEM to verify disruption of the virus membrane by GQDs (Fig. 2b and c). After excess incubation in the presence of 0.05 mg mL−1 GQDs, most virions in the TEM image with a low magnitude disappeared (Fig. 2b). At short incubation time of less than 2 h at room temperature (Fig. 2c), partial disruption of the viral envelope was observed (Fig. 2c, right), whereas no disruption was observed until 1 h of incubation (Fig. 2c, left and center). These results presented the stepwise destruction of the influenza virus from intact virions to completely demolished debris.
2.3 Broad-spectrum antiviral activity against influenza virus and SARS-CoV-2
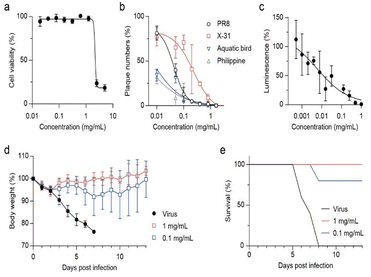
Prior to determining the antiviral activity using a cell-based method, we tested the cytotoxicity of GQDs on Madin–Darby canine kidney (MDCK) cells (Fig. 3a). The cells were exposed to GQDs for 24 h in growth media, stained by crystal violet, and washed to remove dead cells, and absorbance after lysis was measured. Notably, when the amount of GQDs exceeded 2.5 mg mL−1, it induced severe cell death. In contrast, when the amount of GQDs was less than 1.25 mg mL−1, no cytotoxicity was observed. Accordingly, 1.25 mg mL−1 was decided to be the limit concentration for our further cell-based investigation. The CC50 (50% cytotoxic concentration) value was calculated to be 2.18 mg mL−1.As we identified that virucidal mode of-action is independent of strain specificity, plaque forming reduction assay was assessed against various subtypes of influenza A virus, namely, A/Puerto Rico/8/34 (H1N1), A/X-31 (H3N2), A/aquatic bird/Korea/w81/05 (H5N2), and A/Philippine/2/82 (H3N2) (Fig. 3b). GQDs completely inhibited all tested viruses at ≈1 mg mL−1. The IC50 values for PR8 and X31 were 0.043 and 0.231 mg mL−1, respectively, and those for aquatic bird and Philippine were expected to be 0.007 and 0.003 mg mL−1, respectively. Time-of-addition assay results indicated that GQDs inhibited viral infection in the post-treatment scheme, exhibiting an IC50 value of 0.06 mg mL−1 (Fig. S5 and S6†). However, the pre-treatment of GQDs on cells did not suppress viral infection. These results demonstrated the therapeutic potential of GQDs against influenza infection. Moreover, since the disruption of virus was not related to the subtypes of viral proteins, such as hemagglutinin and neuraminidase, GQDs demonstrated great potential as antiviral agents with broad-spectrum antiviral activity.
We further investigated the antiviral potential of GQDs against SARS-CoV-2 pseudoviruses (Fig. 3c and S7†). SARS-CoV-2 pseudoviruses (1.0 × 104 RLU mL−1) were incubated with serially diluted GQDs (2-fold from 1 mg mL−1) for 1 h at RT. HEK293T cells expressing human ACE2 and TMPRSS2 (293T-ACE2/TMPRSS2) were infected with the mixture, and luciferase activity was measured after incubation for 72 h at 37 °C under 5% CO2. GQDs suppressed the infection of pseudoviruses with an IC50 value of 0.006 mg mL−1. Since the cells expressed both ACE2 and TMPRSS2, this result suggested that disruption of virus by GQDs inhibited not only the receptor-mediated endocytosis but also the direct entry through the TMPRSS2-mediated activation of S proteins. In addition, we confirmed that GQDs suppressed the infection of the recombinant human adenovirus type 5 expressing enhanced green fluorescent protein (EGFP) (Fig. S8†).
In three different antiviral experiments against various viruses, including influenza A, adenovirus type 5 and pseudotyped SARS-CoV-2, GQDs exhibited consistent levels of viral inhibition. These results indicate that GQDs can act against a wide range of viruses regardless of their protein types and subtypes. Although their activity is not based on a specific mechanism, our previous research demonstrated the disaggregation of α-syn fibrils through charge interactions between negatively charged GQDs and positively charged part of the fibrils, followed by thermodynamic disassembly.27 In light of these findings, we suggest that negatively charged oxygen-containing functional groups on GQDs are targeted to interact with the positive charge on the surface of viral capsids, resulting in a virucidal effect. For enveloped viruses, it is well known that the amphiphilicity of nanoparticles promotes the interaction with the viral lipid bilayers, thereby exerting a virucidal effect, as shown by graphene derivatives.43–47 In our results, GQDs were found to demonstrate a broad virucidal activity along with viral membrane rupture over time, which is similar to other amphiphilic virucidal peptides. However, unlike large graphene derivatives, GQDs show low cytotoxicity because the cell membrane damage caused by the penetration of GQDs is negligible for lipid bilayers with a large curvature.48–52
We investigated whether the virucidal activity of GQDs can inhibit viral infection in vivo (Fig. 3d and e). When GQD (0, 0.1, or 1.0 mg mL−1)-treated PR8 viruses were intranasally injected in seven-week-old BALB/c female mice, 0.1 mg mL−1 rescued 80% mice from lethal infection, whereas the infected mice without GQDs lost 25% of body weight and were sacrificed at 7 dpi. Furthermore, the loss of body weight rapidly recovered after 3 dpi under the 1.0 mg mL−1 GQD treatment, rescuing all infected mice from lethality. Thus, we confirmed that the antiviral activity of GQDs through virucidal mode of action successfully inhibited the in vivo infection of H1N1 influenza A virus.
2.4 Characterization of well-purified small and large GQD samples
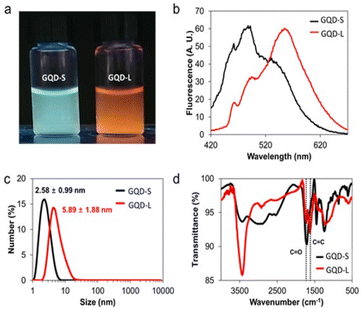
We fractionated synthesized GQDs as GQD-S and GQD-L through dialysis using a dialysis bag of 3 kDa molecular weight for 3 days. The filtered GQDs (<3 kDa) and GQDs remained in the dialysis bag (>3 kDa) were named GQD-S and GQD-L, respectively. As can be seen in Fig. 4a, GQD-S has a greenish-blue fluorescence and GQD-L has an orange fluorescence. GQD-S and GQD-L showed strong fluorescence peaks at 480 nm and 560 nm respectively. These two peaks correspond to the fluorescence peaks observed in the aforementioned GQDs (Fig. 4a, b and S4†). GQD-S was smaller than GQD-L in hydrodynamic size (Fig. 4c). Purified GQD-S and GQD-L were analyzed using FT-IR, XPS, and EA for identifying the functional groups and the carbon to oxygen ratio. In the FT-IR spectra, the ratio of peaks at 1620 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C sp2 carbon bonds) to peaks at 1720 cm−1 (C
C sp2 carbon bonds) to peaks at 1720 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O carboxylic acid) were ∼0.51 and 1.23 in GQD-S and GQD-L, respectively (Fig. 4d). Moreover, through elemental analysis, we found that the C/O ratio of GQD-S was 0.808 and that of GQD-L was 1.317. These results indicate that GQD-S is more hydrophilic than GQD-L (Table S2†). This is due to GQD-S, being smaller than GQD-L, having more edge surface, more functional groups, and higher hydrophilicity.
O carboxylic acid) were ∼0.51 and 1.23 in GQD-S and GQD-L, respectively (Fig. 4d). Moreover, through elemental analysis, we found that the C/O ratio of GQD-S was 0.808 and that of GQD-L was 1.317. These results indicate that GQD-S is more hydrophilic than GQD-L (Table S2†). This is due to GQD-S, being smaller than GQD-L, having more edge surface, more functional groups, and higher hydrophilicity.
2.5 Size-dependent hydrophilicity of GQDs and its influence on antiviral potency against enveloped viruses
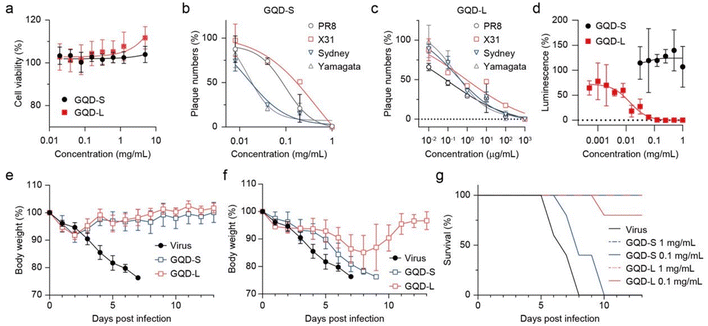
Antiviral activity of the well-refined GQD-S and GQD-L were evaluated in vitro and in vivo (Fig. 5). Both GQDs showed no toxicity on MDCK cells at 5 mg mL−1 (Fig. 5a). Plaque reduction assay showed that GQD-S and GQD-L inhibited viral infection by influenza A and B viruses (Fig. 5b and c). IC50 values of GQD-S against A/PR8, A/X31, and A/Sydney were 0.098, 0.440, and 0.016 mg mL−1, respectively. GQD-L, which was the larger and more hydrophobic GQD, exhibited IC50 values of 0.107, 0.772, and 0.270 μg mL−1 against A/PR8, A/X31, and A/Sydney, respectively. Owing to the virucidal mode of action of GQDs, B/Yamagata viruses were inhibited by these GQDs with size-dependent potency (IC50 values were 0.015 mg mL−1 and 0.270 μg mL−1 for GQD-S and -L, respectively). Interestingly, the inhibition of SARS-CoV-2 pseudoviruses by GQD-S and GQD-L differed significantly (Fig. 5d). At a concentration of 0.0625 mg mL−1, GQD-S failed to inhibit SARS-CoV-2, whereas GQD-L completely suppressed viral infection in vitro (the IC50 value for GQD-L was 0.013 mg mL−1). This gap in activity of GQD-S on influenza and pseudo SARS-CoV-2 viruses indicates that antiviral activity may depend on the membrane composition of the viral envelope.Because influenza viruses are propagated in egg embryo and pseudoviruses are produced in 293T cells, their lipid components are different from each other, and they are expected to showcase varied antiviral potency.53,54 Notably, GQD-L overcame this difference with considerable efficacy through its amphipathicity. This is owing to the large size of GQD-L, which manifests high hydrophobicity and concurrently acts as a more effective surfactant.55,56In vivo results against A/PR8 demonstrated that size-dependent virucidal activity of GQD-S and GQD-L acted effectively in the mouse model (Fig. 5e–g). At a high dose (1 mg mL−1), both GQDs completely removed infectivity in all mice with 100% recovery of body weight (Fig. 5e and g). The antiviral activity was remarkably different at 0.1 mg mL−1 (Fig. 5f and g). Although 40% of the mice in the GQD-S-treated group survived 2 days longer than the negative controls, none of them recovered from infection. In contrast, the GQD-L-treated group showed 80% of survival rate at 13 dpi and fully restored their body weight loss.
3. Method
3.1 Pseudotyped SARS-CoV-2 virus
SARS-CoV-2 pseudoviruses expressing S-glycoproteins and a defective HIV-1 genome encoding luciferase were established. Supernatants containing SARS-CoV-2 pseudovirus were collected 48 h after transfection and utilized in the single-cycle infection of ACE2- and TMPRSS2-transfected 293T cells (293T ACE2/TMPRSS2). Serially diluted GQDs were incubated with 1 × 104 RLU mL−1 of SARS-CoV-2 pseudovirus for 1 h at room temperature prior to cell infection. The medium was replaced with a fresh medium 48 h later and incubated for 24 h. Luciferase activity of infected cells was measured as per the manufacturer's instructions (Promega, Madison, Wisconsin, USA).3.2 Animals
Seven-week-old female BALB/c mice (Koatech, Inc., Pyeongtaek, Korea) were housed at five per cage with access to food and water ad libitum. The mice were intranasally infected with four-fold 50% lethal doses (LD50) of A/PR/8/34 H1N1 virus or GQD-incubated virus, or they were mock-infected. Body weight and survival rate were monitored for 13 days post-infection. When a mouse lost 25% of its initial body weight, it was considered dead and humanely killed. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Sungkyunkwan University, and experiments were approved by the Institutional Animal Care and Use Committee of Sungkyunkwan University (IACUC number: SKKUIACUC2021-05-25-1).4. Conclusions
We investigated the antiviral activity of GQDs to be used as universal agents against viruses, regardless of the viral strain. In this study, we demonstrate that GQDs are capable of inhibiting viral infection in cellular and animal models with broad virucidal effects through direct interaction with viral membranes. Notably, irreversible in vitro inhibition was effective against a wide range of viruses, including enveloped viruses such as influenza and pseudotyped SARS-CoV-2 and even non-enveloped adenovirus.The comparative analysis of GQD-S and GQD-L highlighted the critical role of size and hydrophilicity control in determining antiviral potency. This insight provides valuable guidance for the future design and optimization of GQD-based antiviral therapies. The observed differences in efficacy between GQD-S and GQD-L underscored the importance of fine-tuning these parameters to enhance the antiviral performance.
These results suggest the potential of GQDs to open new pathways to fight against pandemic diseases by offering a rapid and adaptable countermeasure against repeatedly occurring viral diseases. In particular, the unique properties of GQDs, including the size-dependent and hydrophilicity-controlled antiviral activities, would provide alternative strategies to fight the broad-spectrum of viral threats.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Y. J., J. H., H. C. and M. A. conceptualized and conducted the experiments, validated the data, and contributed to writing. J. H. L. and J. S. contributed to materials synthesis and analysis of this work. J. H. Y. and D. K. assisted in antiviral analysis. D. H. K., B. H. H. and M. A. conceived and supervised the project.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea Government (MIST) (No. 2017R1A6A1A03015642, 2020R1A2C2101964, 2022M3E5F1081330, 2022R1A4A5034046 and RS-2021-NF000507).Notes and references
- L. A. Reperant and A. D. M. E. Osterhaus, Vaccine, 2017, 35, 4470–4474 CrossRef PubMed.
- P. R. Saunders-Hastings and D. Krewski, Pathogens, 2016, 5, 66 Search PubMed.
- N. D. Wolfe, C. P. Dunavan and J. Diamond, Nature, 2007, 447, 279–283 Search PubMed.
- B. K. Yoon, W.-Y. Jeon, T. N. Sut, N.-J. Cho and J. A. Jackman, ACS Nano, 2021, 15, 125–148 CrossRef CAS PubMed.
- W. C. Koff, D. R. Burton, P. R. Johnson, B. D. Walker, C. R. King, G. J. Nabel, R. Ahmed, M. K. Bhan and S. A. Plotkin, Science, 2013, 340, 1232910 CrossRef PubMed.
- J. Wise, BMJ, 2021, 372, n359 CrossRef PubMed.
- P. Carravilla, A. Cruz, I. Martin-Ugarte, I. R. Oar-Arteta, J. Torralba, B. Apellaniz, J. Pérez-Gil, J. Requejo-Isidro, N. Huarte and J. L. Nieva, Biophys. J., 2017, 113, 1301–1310 CrossRef CAS PubMed.
- Q. Li, Z. Zhao, D. Zhou, Y. Chen, W. Hong, L. Cao, J. Yang, Y. Zhang, W. Shi, Z. Cao, Y. Wu, H. Yan and W. Li, Peptides, 2011, 32, 1518–1525 CrossRef CAS PubMed.
- Y. Jung, B. Kong, S. Moon, S.-H. Yu, J. Chung, C. Ban, W.-J. Chung, S.-G. Kim and D.-H. Kweon, Biochem. Biophys. Res. Commun., 2019, 517, 507–512 CrossRef CAS PubMed.
- P. Innocenzi and L. Stagi, Chem. Sci., 2020, 11, 6606–6622 RSC.
- V. Cagno, P. Andreozzi, M. D'Alicarnasso, P. Jacob Silva, M. Mueller, M. Galloux, R. Le Goffic, S. T. Jones, M. Vallino, J. Hodek, J. Weber, S. Sen, E.-R. Janeček, A. Bekdemir, B. Sanavio, C. Martinelli, M. Donalisio, M.-A. Rameix Welti, J.-F. Eleouet, Y. Han, L. Kaiser, L. Vukovic, C. Tapparel, P. Král, S. Krol, D. Lembo and F. Stellacci, Nat. Mater., 2018, 17, 195–203 CrossRef CAS PubMed.
- V. Cagno, M. Gasbarri, C. Medaglia, D. Gomes, S. Clement, F. Stellacci and C. Tapparel, Antimicrob. Agents Chemother., 2020, 64, e02001 CrossRef CAS PubMed.
- A. M. Chamoun, K. Chockalingam, M. Bobardt, R. Simeon, J. Chang, P. Gallay and Z. Chen, Antimicrob. Agents Chemother., 2012, 56, 672–681 CrossRef CAS PubMed.
- B. Kong, S. Moon, Y. Kim, P. Heo, Y. Jung, S.-H. Yu, J. Chung, C. Ban, Y. H. Kim, P. Kim, B. J. Hwang, W.-J. Chung, Y.-K. Shin, B. L. Seong and D.-H. Kweon, Nat. Commun., 2019, 10, 185 CrossRef PubMed.
- T. Du, J. Lu, L. Liu, N. Dong, L. Fang, S. Xiao and H. Han, ACS Appl. Bio Mater., 2018, 1, 1286–1293 CrossRef CAS PubMed.
- S. Ye, K. Shao, Z. Li, N. Guo, Y. Zuo, Q. Li, Z. Lu, L. Chen, Q. He and H. Han, ACS Appl. Mater. Interfaces, 2015, 7, 21571–21579 CrossRef CAS PubMed.
- Y.-N. Chen, Y.-H. Hsueh, C.-T. Hsieh, D.-Y. Tzou and P.-L. Chang, Int. J. Environ. Res. Public Health, 2016, 13, 430 CrossRef PubMed.
- T. Du, J. Liang, N. Dong, L. Liu, L. Fang, S. Xiao and H. Han, Carbon, 2016, 110, 278–285 CrossRef CAS.
- A. Łoczechin, K. Séron, A. Barras, E. Giovanelli, S. Belouzard, Y.-T. Chen, N. Metzler-Nolte, R. Boukherroub, J. Dubuisson and S. Szunerits, ACS Appl. Mater. Interfaces, 2019, 11, 42964–42974 CrossRef PubMed.
- D. Iannazzo, A. Pistone, S. Ferro, L. De Luca, A. M. Monforte, R. Romeo, M. R. Buemi and C. Pannecouque, Bioconjugate Chem., 2018, 29, 3084–3093 CrossRef CAS PubMed.
- M. Sametband, I. Kalt, A. Gedanken and R. Sarid, ACS Appl. Mater. Interfaces, 2014, 6, 1228–1235 CrossRef CAS PubMed.
- C. J. Lin, L. Chang, H. W. Chu, H. J. Lin, P. C. Chang, R. Y. L. Wang, B. Unnikrishnan, J. Y. Mao, S. Y. Chen and C. C. Huang, Small, 2019, 15, e1902641 CrossRef PubMed.
- D. Ting, N. Dong, L. Fang, J. Lu, J. Bi, S. Xiao and H. Han, ACS Appl. Nano Mater., 2018, 1, 5451–5459 CrossRef.
- H.-T. Huang, H.-J. Lin, H.-J. Huang, C.-C. Huang, J. H.-Y. Lin and L.-L. Chen, Sci. Rep., 2020, 10, 7343 CrossRef CAS PubMed.
- Z. Song, X. Wang, G. Zhu, Q. Nian, H. Zhou, D. Yang, C. Qin and R. Tang, Small, 2015, 11, 1171–1176 CrossRef CAS PubMed.
- I. Kang, J. M. Yoo, D. Kim, J. Kim, M. K. Cho, S.-E. Lee, D. J. Kim, B.-C. Lee, J. Y. Lee, J.-J. Kim, N. Shin, S. W. Choi, Y.-H. Lee, H. S. Ko, S. Shin, B. H. Hong and K.-S. Kang, Nano Lett., 2021, 21, 2339–2346 CrossRef CAS PubMed.
- D. Kim, J. M. Yoo, H. Hwang, J. Lee, S. H. Lee, S. P. Yun, M. J. Park, M. Lee, S. Choi, S. H. Kwon, S. Lee, S.-H. Kwon, S. Kim, Y. J. Park, M. Kinoshita, Y.-H. Lee, S. Shin, S. R. Paik, S. J. Lee, S. Lee, B. H. Hong and H. S. Ko, Nat. Nanotechnol., 2018, 13, 812–818 CrossRef CAS PubMed.
- Y. Chong, Y. Ma, H. Shen, X. Tu, X. Zhou, J. Xu, J. Dai, S. Fan and Z. Zhang, Biomaterials, 2014, 35, 5041–5048 CrossRef CAS PubMed.
- M. Nurunnabi, Z. Khatun, K. M. Huh, S. Y. Park, D. Y. Lee, K. J. Cho and Y.-k. Lee, ACS Nano, 2013, 7, 6858–6867 CrossRef CAS PubMed.
- J. Li, X. Wang, K.-C. Mei, C. H. Chang, J. Jiang, X. Liu, Q. Liu, L. M. Guiney, M. C. Hersam, Y.-P. Liao, H. Meng and T. Xia, Nano Today, 2021, 37, 101061 CrossRef CAS PubMed.
- B.-C. Lee, J. Y. Lee, J. Kim, J. M. Yoo, I. Kang, J.-J. Kim, N. Shin, D. J. Kim, S. W. Choi, D. Kim, B. H. Hong and K.-S. Kang, Sci. Adv., 2020, 6, eaaz2630 CrossRef CAS PubMed.
- B.-C. Lee, J. Y. Lee, J. Kim, N. Shin, J. M. Yoo, I. Kang, J.-J. Kim, S.-E. Lee, D. Kim, S. W. Choi, B. H. Hong and K.-S. Kang, 2D Mater., 2021, 8, 025036 CrossRef CAS.
- D. Zhang, Z. Zhang, Y. Wu, K. Fu, Y. Chen, W. Li and M. Chu, Biomaterials, 2019, 194, 215–232 CrossRef CAS PubMed.
- S. Wang, I. S. Cole and Q. Li, RSC Adv., 2016, 6, 89867–89878 RSC.
- L. Ou, B. Song, H. Liang, J. Liu, X. Feng, B. Deng, T. Sun and L. Shao, Part. Fibre Toxicol., 2016, 13, 57 CrossRef PubMed.
- C.-T. Hsieh, S. Gu, Y. A. Gandomi, C.-C. Fu, P.-Y. Sung, R.-S. Juang and C.-C. Chen, J. Colloid Interface Sci., 2023, 630, 1–10 CrossRef CAS PubMed.
- D. Iannazzo, S. V. Giofrè, C. Espro and C. Celesti, Expert Opin. Drug Delivery, 2024, 21, 751–766 CrossRef CAS PubMed.
- F. Liu, Y. Sun, Y. Zheng, N. Tang, M. Li, W. Zhong and Y. Du, RSC Adv., 2015, 5, 103428–103432 RSC.
- T. Gao, X. Wang, L.-Y. Yang, H. He, X.-X. Ba, J. Zhao, F.-L. Jiang and Y. Liu, ACS Appl. Mater. Interfaces, 2017, 9, 24846–24856 CrossRef CAS PubMed.
- S. Khan, A. Sharma, S. Ghoshal, S. Jain, M. K. Hazra and C. K. Nandi, Chem. Sci., 2018, 9, 175–180 RSC.
- Y. Li, Y. Hu, Y. Zhao, G. Shi, L. Deng, Y. Hou and L. Qu, Adv. Mater., 2011, 23, 776–780 CrossRef CAS PubMed.
- F. Ehrat, S. Bhattacharyya, J. Schneider, A. Löf, R. Wyrwich, A. L. Rogach, J. K. Stolarczyk, A. S. Urban and J. Feldmann, Nano Lett., 2017, 17, 7710–7716 CrossRef CAS PubMed.
- Y. Li, H. Yuan, A. von dem Bussche, M. Creighton, R. H. Hurt, A. B. Kane and H. Gao, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12295–12300 CrossRef CAS PubMed.
- J. Chen, G. Zhou, L. Chen, Y. Wang, X. Wang and S. Zeng, J. Phys. Chem. C, 2016, 120, 6225–6231 CrossRef CAS.
- X. Zou, L. Zhang, Z. Wang and Y. Luo, J. Am. Chem. Soc., 2016, 138, 2064–2077 CrossRef CAS PubMed.
- X. Zhang, F. Cao, L. Wu and X. Jiang, Langmuir, 2019, 35, 14098–14107 CrossRef CAS PubMed.
- L. Wu, L. Zeng and X. Jiang, J. Am. Chem. Soc., 2015, 137, 10052–10055 CrossRef CAS PubMed.
- L. Liang, Z. Kong, Z. Kang, H. Wang, L. Zhang and J.-W. Shen, ACS Biomater. Sci. Eng., 2016, 2, 1983–1991 CrossRef CAS PubMed.
- J. A. Jackman, G. H. Zan, V. P. Zhdanov and N.-J. Cho, J. Phys. Chem. B, 2013, 117, 16117–16128 CrossRef CAS PubMed.
- J. A. Jackman, R. Saravanan, Y. Zhang, S. R. Tabaei and N.-J. Cho, Small, 2015, 11, 2372–2379 CrossRef CAS PubMed.
- D. Koller and K. Lohner, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 2250–2259 CrossRef CAS PubMed.
- S. R. Tabaei, M. Rabe, V. P. Zhdanov, N.-J. Cho and F. Höök, Nano Lett., 2012, 12, 5719–5725 CrossRef CAS PubMed.
- P. T. Ivanova, D. S. Myers, S. B. Milne, J. L. McClaren, P. G. Thomas and H. A. Brown, ACS Infect. Dis., 2015, 1, 435–442 CrossRef CAS PubMed.
- S. E. Farley, J. E. Kyle, H. C. Leier, L. M. Bramer, J. B. Weinstein, T. A. Bates, J.-Y. Lee, T. O. Metz, C. Schultz and F. G. Tafesse, Nat. Commun., 2022, 13, 3487 CrossRef CAS PubMed.
- M. Simon, M. Veit, K. Osterrieder and M. Gradzielski, Curr. Opin. Colloid Interface Sci., 2021, 55, 101479 CrossRef CAS PubMed.
- P.-Z. Zhang, F.-F. Jiao, Z.-X. Xie, Z. Kong, W. Hu, J.-W. Shen and L.-J. Liang, Mater. Adv., 2022, 3, 6161–6170 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00879k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |