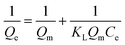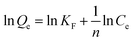Open Access Article
Open Access ArticleSurface imprinted microhelical magnetic polymer nanocomposite fibers for targeted lysozyme separation†
Aakanksha Mohan and
Sutapa Roy Ramanan *
*
Department of Chemical Engineering, BITS Pilani, K K Birla Goa Campus, India. E-mail: sutapa@goa.bits-pilani.ac.in
First published on 17th February 2025
Abstract
Magnetic microhelical structures have recently drawn attention as microswimmers capable of mimicking bacterial propulsion in the low Reynolds number regime. Such structures can be used in microfluidic bioseparation or targeted delivery and their interaction with proteins is extremely important. In this study we fabricated silica coated magnetic microhelices resembling artificial bacterial flagella like structures via electrospinning magnetite nanoparticle incorporated polystyrene nanocomposite solution followed by silica sol coating. Two model proteins, Lysozyme (Lyz) and Bovine Serum Albumin (BSA), were used for protein imprinting along with a polydopamine layer on the magnetic microhelical substrates. The adsorption mechanism of lysozyme on the molecularly imprinted support system was analyzed using adsorption model fitting (Langmuir, Freundlich and Temkin). Adsorption capacity, selective binding and imprinting factor values were calculated for both imprinted (Lyz and BSA) and non-imprinted samples. A significantly higher adsorption capacity was obtained compared to previously reported studies.
1 Introduction
Proteins play a crucial role in multiple industries such as food, biotechnology, pharmaceuticals and cosmetics due to their diverse functional, nutritional and nutraceutical properties. The target applications require high purity proteins necessitating an ongoing demand for low cost and efficient protein purification methods. Although conventional routes provide acceptable standards of homogeneity in the production, the high cost is attributed to the downstream processes involving multiple steps of purification.1 Isolating a specific targeted protein gets complicated for several reasons when the quantity of protein present in a sample is usually very small. Preserving the biological activity and chemical integrity of the protein is yet another challenge as they are sensitive to temperature changes, pH and various other factors.2 Selective protein binding is an important criterion in the protein purification procedure and involves complicated multistep processes. Hence, a considerable amount of research is dedicated to creating a one-step process for isolating a target protein, which demands high selectivity, sensitive detection capabilities and an efficient extraction method. In this regard, several bioinspired frameworks3 and chromatographic membranes4 with high selectivity have been developed for protein purification.Molecular recognition involves the combination of antigen–antibody interaction for specific binding.5 Although it is highly efficient, the high cost involved in this methodology restricts its commercialization. Antibody specificity arises from their self-assembly around a specific antigen. For selective binding of a specific protein, molecular recognition can be mimicked by fabricating a specific template for the given protein.6 This process, referred to as molecular imprinting or molecularly imprinted polymer (MIP), involves polymerizing monomers around a protein template, followed by the removal of the protein to create cavities that mirror the template molecule in size, shape, and functional moieties. It is an extremely cost effective and efficient method that enables selective binding.7
Micro/nanoparticle solid support substrates are popularly used for the MIP study due to their characteristic high surface area.8 Several micro/nano particulate systems such as silica, polystyrene and magnetic particles have been used as MIP supports. The magnetic MIP supports are considered desirable due to their high magnetic susceptibility and easier and faster separation mechanism.9 Geometrical features of the MIP support also play an important role during the adsorption process. Anisotropic geometries such as flower-shapes are known to provide an advantage as they possess multiple facets and planes.10 Core–shell magnetic particles are popularly used as MIP supports where the core material is magnetic in nature and the shell provides a substrate for the protein–polymer interaction.11 Magnetite nanoparticle (MNP) encapsulated polyacrylonitrile nanofibers have also been used as MIP supports.12 Molecular imprinting/surface imprinting technology finds applications in chromatography and sensing applications along with possible biomedical applications such as targeted delivery.13 Recent studies indicated that controlled release and maximum therapeutic benefit were achieved along with selectivity using molecular imprinting technology.14 Protein-based therapeutics are currently being explored in cancers, immune disorders, infections and other diseases due to their high success in clinical trials.15 Developing a magnetic MIP support with molecular recognition capability can be useful in such applications.
Magnetic microhelical structures, resembling bacterial flagella and also referred to as magnetic microswimmers, have recently caught attention due to their unique morphology and their ability to access remote locations precisely compared to the conventional core–shell magnetic particles.16 Lysozyme (Lyz) is widely used as a preservative due to its bactericidal properties17 and its immobilization on the magnetic microswimmer MIP support could be advantageous in applications such as targeted delivery or microfluidic bioseparation.18 Magnetic microswimmers could also be used in the regeneration of membranes by targeted treatment.19
In this study, we synthesized silica coated helical magnetic polystyrene fibers (Si-HMPFs) resembling artificial bacterial flagella (ABF) like structures as MIP solid support substrates. These ABF structures are magnetically active and, hence, can provide guided access to remote locations allowing controlled targeting. Protein immobilization on such structures is expected to provide a better mode for microfluidic bioseparation along with aiding in selective binding, biocompatibility and several other important characteristics for such applications. In the present work, the effectiveness of the synthesized magnetic microhelices as templates was tested using two model proteins (lysozyme and bovine serum albumin) to confirm the compatibility of the approach. Protein adsorption studies on magnetic microhelical structures have not been reported so far to the best of our knowledge.
2 Experimental section
2.1 Materials
Polystyrene (PS, Mw = 360![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), tetraethylorthosilicate (TEOS) and Triton X-100 (TX100) were purchased from Sigma Aldrich. N,N-Dimethylformamide (DMF), ferric chloride anhydrous 98%, ammonium ferrous sulphate, liquor ammonia 30%, nitric acid, 2-butanol, 2-propanol, phosphate buffered saline tablets (pH = 7.4) and Sodium Lauryl Sulphate (SLS) were purchased from Loba Chemie (India). Citric acid (anhydrous) was purchased from Thomas Baker (India). Dopamine hydrochloride (DAH), Tris base, Lysozyme (Lyz) and Bovine Serum Albumin (BSA) were purchased from Himedia (India). Acetic acid (HAc) was purchased from Finar Limited.
000), tetraethylorthosilicate (TEOS) and Triton X-100 (TX100) were purchased from Sigma Aldrich. N,N-Dimethylformamide (DMF), ferric chloride anhydrous 98%, ammonium ferrous sulphate, liquor ammonia 30%, nitric acid, 2-butanol, 2-propanol, phosphate buffered saline tablets (pH = 7.4) and Sodium Lauryl Sulphate (SLS) were purchased from Loba Chemie (India). Citric acid (anhydrous) was purchased from Thomas Baker (India). Dopamine hydrochloride (DAH), Tris base, Lysozyme (Lyz) and Bovine Serum Albumin (BSA) were purchased from Himedia (India). Acetic acid (HAc) was purchased from Finar Limited.
2.2 Electrospinning of helical polystyrene/magnetite nanocomposite fibers
Magnetite nanoparticles (MNPs) were synthesized via the coprecipitation method (ESI†) using ferric chloride and ammonium ferrous sulphate (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) in an aqueous medium followed by subsequent addition of ammonia.20 The as-synthesized MNPs were dispersed in 25% polystyrene solution in a 1
1 ratio) in an aqueous medium followed by subsequent addition of ammonia.20 The as-synthesized MNPs were dispersed in 25% polystyrene solution in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio to prepare polystyrene/magnetite nanocomposite solution. Dimethylformamide solution was used as a solvent and the as-prepared mixture was stirred for 48 hours to obtain good dispersion of magnetite nanoparticles. The solution was then electrospun at 6 kV at a feed rate of 1 mL h−1 with a grounded coagulation collector bath (6 mM SLS aqueous solution), maintained at a 2 cm distance from the spinneret. The helical magnetic polystyrene fiber (HMPF) mats were collected, thoroughly washed multiple times with deionized water (DIW), dried at 40 °C for 24 hours, and then used for further characterization.
2 ratio to prepare polystyrene/magnetite nanocomposite solution. Dimethylformamide solution was used as a solvent and the as-prepared mixture was stirred for 48 hours to obtain good dispersion of magnetite nanoparticles. The solution was then electrospun at 6 kV at a feed rate of 1 mL h−1 with a grounded coagulation collector bath (6 mM SLS aqueous solution), maintained at a 2 cm distance from the spinneret. The helical magnetic polystyrene fiber (HMPF) mats were collected, thoroughly washed multiple times with deionized water (DIW), dried at 40 °C for 24 hours, and then used for further characterization.
2.3 Microhelical structure fabrication via silica sol treatment
Silica sol was synthesized via the sol preparation method detailed in (ESI†) our earlier work.21 The synthesized HMPF mats were immersed in the silica sol and sonicated for 3 hours. The silica-coated fiber mat samples were extracted from the sol and dried at 80 °C for 4 hours in a hot air oven. This was followed by crushing using a mortar and pestle to obtain hydrophilic magnetic microhelical fibers (SiHMPFs).2.4 Preparation of surface imprinted and non-imprinted microhelices
The as-synthesized SiHMPF samples were dispersed in 20 mM Tris buffer solution and sonicated for 30 min. Subsequently, dopamine hydrochloride (DAH) was added to the above solution in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio followed by incubation overnight to modify the SiHMPF surface with a polydopamine (PDA) layer. The samples were then collected and washed with DIW several times and dried at room temperature to obtain the PDA coated SiHMPF (P-SiHMPF). 50 mg of the P-SiHMPF sample was then dispersed into three different sets of 20 mM Tris buffer solution individually and sonicated for 30 min followed by Lysozyme (Lyz) addition. The amount of Lyz was varied as 25 mg, 50 mg and 100 mg for 50 mg of the P-SiHMPF sample to identify the maximum template implantation composition. DAH was further added to the above solution in a 2
1 ratio followed by incubation overnight to modify the SiHMPF surface with a polydopamine (PDA) layer. The samples were then collected and washed with DIW several times and dried at room temperature to obtain the PDA coated SiHMPF (P-SiHMPF). 50 mg of the P-SiHMPF sample was then dispersed into three different sets of 20 mM Tris buffer solution individually and sonicated for 30 min followed by Lysozyme (Lyz) addition. The amount of Lyz was varied as 25 mg, 50 mg and 100 mg for 50 mg of the P-SiHMPF sample to identify the maximum template implantation composition. DAH was further added to the above solution in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (DAH
1 ratio (DAH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Lyz) and the solution was incubated for 24 h to obtain PDA coated Lyz–P-SiHMPF (PLPS) samples. Similarly, replacing Lyz with bovine serum albumin (BSA) resulted in PDA coated BSA–P-SiHMPF (PBPS) samples. The protein imprinted samples were repeatedly washed with 1% SLS/3% HAc solution to remove the imprinted protein followed by DI water and 0.5 M NaCl wash to remove SLS and HAc. The as-synthesized Lyz imprinted (PLPS), BSA imprinted (PBPS) and non-imprinted (P-SiHMPF) samples were used for further studies.
Lyz) and the solution was incubated for 24 h to obtain PDA coated Lyz–P-SiHMPF (PLPS) samples. Similarly, replacing Lyz with bovine serum albumin (BSA) resulted in PDA coated BSA–P-SiHMPF (PBPS) samples. The protein imprinted samples were repeatedly washed with 1% SLS/3% HAc solution to remove the imprinted protein followed by DI water and 0.5 M NaCl wash to remove SLS and HAc. The as-synthesized Lyz imprinted (PLPS), BSA imprinted (PBPS) and non-imprinted (P-SiHMPF) samples were used for further studies.
2.5 Adsorption experiments
The effect of the concentration of Lyz in the precursor solution on the Lyz adsorption capacity of the templated samples was tested by incubating the templated PLPS samples in lysozyme solution. The mass of the PLPS samples used was w = 5 mg, the initial concentration of the Lyz solution used in the adsorption study was Co = 1 mg mL−1 and the volume of the Lyz solution was V = 5 mL. The samples were incubated for 15 minutes and then magnetically decanted to obtain a clear solution to measure the absorbance. The final concentration Cf, was calculated with Cf = (A − yo)/m where A is the absorbance, and yo and m are constants obtained from the calibration curve representing the intercept slope values, respectively. The amount of protein adsorbed on the sample Q was calculated using Q = ((Co − Cf) × V)/w where V is the volume of the solution, w is the mass of the sample, and Co and Cf are the initial and final concentrations of the protein solution before and after incubation. The imprinting factor (IF) was calculated using IF = QPLPS/QPBPS and selective binding (SB) was calculated using QPLPS–QPBPS.Adsorption kinetics was calculated by measuring the amount of Lyz adsorbed on PLPS, Q (mg g−1) with respect to time, with V = 1.5 mL, w = 3 mg and Co = 0.5 mg mL−1. The solution was extracted at a 10 min interval and measured by UV-vis spectrometry. To determine the adsorption equilibrium of the test samples, the concentration of the Lyz solution during the adsorption test was varied from 0.2 mg mL−1 to 1 mg mL−1 with an interval of 0.2 mg mL−1. The samples were incubated for 15 min and magnetically decanted and the supernatant solution was used for analysis. The lysozyme adsorption data for all samples were analyzed using Langmuir, Freundlich, and Temkin models to gain a deeper insight into the adsorption mechanism. The mathematical representations of the models are as follows:
Langmuir model:
Freundlich model:
Temkin model:
Qe = B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln(KTCe) ln(KTCe) |
Selective binding was measured by using non-imprinted and imprinted samples for protein adsorption. The mass of the imprinted and non-imprinted samples used for this study was w = 5 mg, the volume of the Lyz and BSA solutions used was V = 5 mL and the initial concentration of the Lyz and BSA solutions was Co = 0.5 mg mL−1. The as-obtained results of each sample were compared with the opposite protein-imprinted samples to confirm the selectivity.
2.6 Characterization
The surface functionalization of the material was studied by Fourier transform infrared spectroscopy (FTIR) over a range of 400–4000 cm−1 using a PerkinElmer Spectrum Two ATR-FTIR (Attenuated Total Reflectance-FTIR) spectrometer. The surface charge of the as-synthesized samples was determined using a Nanoparticle analyzer (NanoPlus with NanoPlusAT) by measuring the zeta potential value. The morphological characteristics of the HMPF fiber mat and Si-HMPF structures were determined using a field emission scanning electron microscope (Quanta FEG 250). The fiber samples were coated with a 10 nm Au layer using a sputter coater to obtain a conductive surface (LEICA EM ACE 200). The fiber diameter and the coil diameter values of the synthesized HMPFs and SiHMPFs were measured using ImageJ software by calculating the average of 25 data points for the individual sample. A Vibrating Sample Magnetometer (PPMS-VSM) (Quantum Design; PPMS Evercool II) was used to determine the magnetic properties of the citric acid coated magnetite nanoparticles (MNPs) and Si-HMPF samples at 300 K. The hysteresis loop was measured between −10k to +10k Oe at 300 K. 1 mg mL−1 aqueous solution was prepared for the individual studies and sonicated for 20 minutes. 1 mL of this solution was diluted 20 times and used for the zeta potential characterization. The absorption spectra of the protein solutions before and after incubation with surface imprinted and non-imprinted samples were recorded using a Shimadzu (model: UV-1800, Japan) UV-vis spectrophotometer. All the experiments and analyses were performed in triplicate.3 Results and discussion
The chemical configuration of the as-synthesized fibers pre and post functionalization was analyzed using FTIR (Fig. 1a). Helical magnetic polystyrene fibers (HMPFs) showed narrow peaks at 690 cm−1 and 758 cm−1 confirming the presence of benzene rings of polystyrene fibers.22 The peaks at 580 cm−1, 480 cm−1 and 628 cm−1 are attributed to Fe–O vibrations of the magnetite lattice23 The narrow peak observed at 1064 cm−1 corresponded to the symmetric stretching of Si–O–Si bonds24 in the silica coated SiHMPF samples. Dopamine hydrochloride undergoes oxidation and self-polymerizes under alkaline conditions resulting in several functional groups such as quinone, carboxy, amino, imine and catechol structures.25 These groups offer covalent binding to several molecules and are widely used in the binding of divalent metal ions. Polydopamine modification of the surface (P-SiHMPF) was confirmed by the sharp peaks noted at 1630 cm−1 and 1488 cm−1 attributed to the stretching of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N vibrations respectively.26 Minor peaks observed at 1640 cm−1, 1555 cm−1 and 1742 cm−1 could be attributed to the C
N vibrations respectively.26 Minor peaks observed at 1640 cm−1, 1555 cm−1 and 1742 cm−1 could be attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–N and O–C
O, C–N and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds respectively.27
O bonds respectively.27
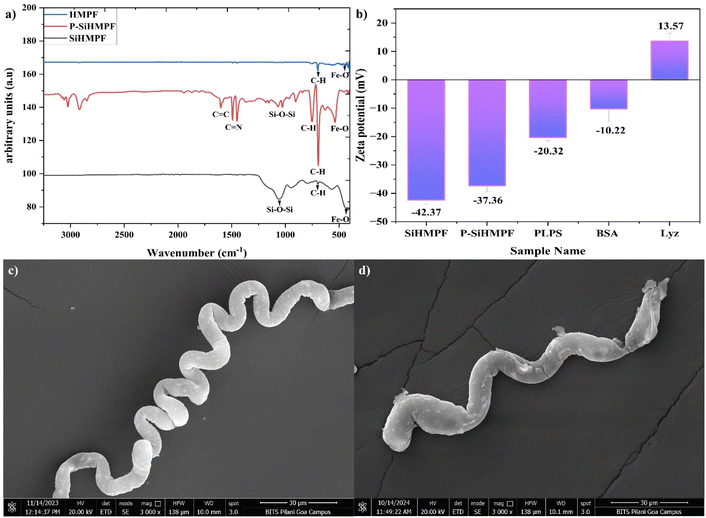 | ||
| Fig. 1 (a) FTIR spectrum of HMPF, SiHMPF and P-SiHMPF; (b) zeta potential values of SiHMPF, P-SiHMPF, PLPS, Bovine Serum Albumin (BSA) and lysozyme (Lyz); FESEM images of (c) HMPF and (d) SiHMPF. | ||
Fig. 1b represents the zeta potential values of the as-synthesized samples (SiHMPF, P-SiHMPF, and PLPS) and the two model proteins lysozyme (Lyz) and bovine serum albumin (BSA). Surface charge plays an important role in the adsorption process as it can lead to electrostatic interactions. Zeta potential values above ±30 mV indicate excellent dispersion which was noted for both the SiHMPF (−42.37 ± 1.15 mV) and P-SiHMPF (−37.36 ± 2.01 mV) samples. Dispersion plays an important role in the adsorption process as it favours the adsorbent–adsorbate interaction due to the increased availability of the active surface area. The zeta potential values for the PLPS samples increased to −20.32 ± 1.18 mV, confirming lysozyme imprinting, since lysozyme is a positively charged molecule with a measured zeta potential of +13.57 ± 2.82 mV (Fig. 1b). Electrostatic interaction can also result in the adsorption of Lyz on non-imprinted samples such as SiHMPF and P-SiHMPF. However, in the absence of molecular recognition capabilities, the efficiency and selectivity of the process will be weak. Surface imprinting would enhance the selectivity of the protein adsorbed onto the support thereby facilitating selective protein separation/purification via the molecular template recognition.
Electron micrographs of a helical magnetic polystyrene fiber (HMPF) and silica coated HMPF (SiHMPF) are shown in Fig. 1c and d respectively. The fiber diameter and coil diameter of these helical fiber samples were measured from a sample size of 25 individual fibers (Fig. S1†). The corresponding average values for HMPF samples were (5.8 ± 1.93 μm) and (24.38 ± 3.9 μm) respectively. The SiHMPF microhelices had an average fiber diameter of 7.27 ± 2.15 μm and a coil diameter of 16.76 ± 4.07 μm respectively. The as-synthesized HMPF samples had a porous surface and high contact angle (hydrophobic surface) whereas post silica modification the porosity and the contact angle of the fiber surface significantly reduced (Fig. S2†). Hydrophilic surfaces provide better dispersion in aqueous medium and hence are an important criterion for protein adsorption studies. The MNPs incorporated in the polystyrene matrix had a uniform distribution throughout the SiHMPF structure (Fig. S3†). The magnetic properties of the synthesized material were determined using a vibrating sample magnetometer (Fig. S4†). Detailed analysis of the contact angle and the magnetic property studies of the HMPF and Si-HMPF structures is included in our earlier published work.21
Several factors such as the surface area of the adsorbent, adsorbent/absorbate mass ratio and surface charge are known to influence the adsorption efficiency. To understand the effect of the mass ratio, the Lyz![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P-SiHMPF ratio was varied as 1
P-SiHMPF ratio was varied as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and labelled 25Lyz, 50Lyz and 100Lyz according to the respective mass of the Lyz used for the surface modification. 5 mg (m) of each of these synthesized samples was used for the adsorption experiment for an initial Lyz concentration of Co = 1 mg mL−1 and volume = 5 mL. Adsorption is a surface phenomenon and the surface area available largely determines the adsorption efficiency. The adsorption capacity of the synthesized samples was determined to be 580.76 mg g−1, 600.89 mg g−1, and 572.62 mg g−1 for 25Lyz, 50Lyz and 100Lyz samples respectively. Although the adsorbed lysozyme quantity for the individual samples was mostly in the same range, the 1
1 and labelled 25Lyz, 50Lyz and 100Lyz according to the respective mass of the Lyz used for the surface modification. 5 mg (m) of each of these synthesized samples was used for the adsorption experiment for an initial Lyz concentration of Co = 1 mg mL−1 and volume = 5 mL. Adsorption is a surface phenomenon and the surface area available largely determines the adsorption efficiency. The adsorption capacity of the synthesized samples was determined to be 580.76 mg g−1, 600.89 mg g−1, and 572.62 mg g−1 for 25Lyz, 50Lyz and 100Lyz samples respectively. Although the adsorbed lysozyme quantity for the individual samples was mostly in the same range, the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mass ratio was chosen for further experiments.
1 mass ratio was chosen for further experiments.
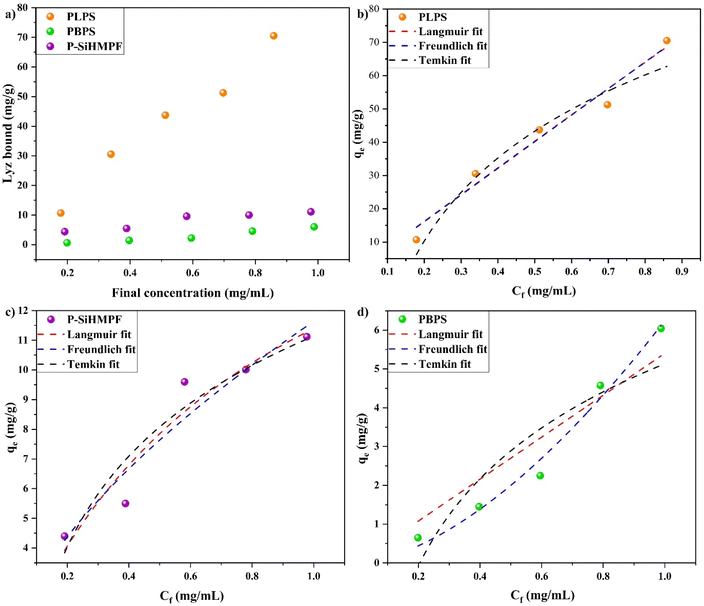
The plot of the adsorption equilibrium experiments for imprinted and non-imprinted samples with various thermodynamic fitting is presented in Fig. 2 where the initial concentration Co of Lyz was varied but the mass of synthesized samples (m = 3 mg) and volume (V = 1.5 mL) of the Lyz solution were maintained constant. The amount of Lyz adsorbed onto the substrates increased with the increase in the given concentration range (Fig. 2a). The adsorption capacity Qe of the Lyz imprinted sample, PLPS (70.53 mg g−1) was 6.34 times higher than that of the non-imprinted sample, P-SiHMPF (11.12 mg g−1) demonstrating the availability of increased active binding sites on the surface. The same study performed using PBPS showed an adsorption capacity of 6.04 mg g−1, confirming the selectivity of Lyz towards the PLPS surface.
To further understand the adsorption behaviour of lysozyme and its interaction with the as-synthesized samples, three different adsorption isotherm models (Langmuir, Freundlich and Temkin) were explored. The adsorption data obtained for all three samples PLPS, P-SiHMPF and PBPS for varying concentrations of lysozyme were fitted individually to the Langmuir, Freundlich and Temkin models (Fig. 2b–d respectively). The fitting parameter data (Table 1) obtained from these models for the individual samples were used to analyze the underlying adsorption mechanism. The Langmuir isotherm model is the most used model in the literature due to its simplicity and it represents the monolayer adsorption mechanism whereas the Freundlich model is an empirical model commonly used in adsorption studies.28 Temkin model accounts for the change in the surface energy level during the adsorption process along with its influence on the adsorption capacity of the synthesized samples.29 These three models are popularly used in the literature to understand the protein adsorption mechanism.17,30,31 To satisfy the Langmuir adsorption model, a surface needs to adhere to the following conditions;32 (a) homogeneous adsorption sites, (b) one protein macromolecule per adsorption site, (c) no interaction between the adsorbed and free proteins in the solution and (d) a dynamic reversible equilibrium process. The Freundlich model, however, describes the monolayer adsorption on the heterogeneous sites where the available binding sites on the surface need not have similar adsorption energy or adsorption rate.29 Even though the Langmuir approach for protein adsorption is misleading as it does not account for several factors such as unfolding or denaturation of the proteins, several studies have reportedly used it to describe the protein adsorption mechanism.33 Both the Langmuir and Freundlich models were applicable for the PLPS (R2 = 0.97) and P-SiHMPF samples, with the curves overlapping for the PLPS surface. However, the Freundlich model could better describe the adsorption mechanism of the synthesized samples as both PDA functional moieties and Lyz templates on the surface could contribute to the Lyz adsorption. Moreover, the shape of the fitted curve obtained for the PLPS and P-SiHMPF samples represents the Freundlich model more closely rather than the Langmuir model (inverted L-shape). The adsorption behavior of the PBPS sample is correlated with the Freundlich model (R2 = 0.98) and it could account for the polydopamine adsorption sites available on the surface. The adsorption capacity Qe of the PLPS samples was significantly larger than the other samples (P-SiHMPF and PBPS) confirming the effectiveness of surface imprinting. Lyz adsorption increased with the increase in concentration. At low concentrations, protein unfolding and denaturation occur at a higher rate and the adsorption of these unfolded proteins occupies a larger surface area compared to the proteins at higher concentration.34 The Temkin model, designed for homogeneous surfaces, was not found to be well suited for either sample and could be attributed to the porous nature of the synthesized surface.29 The above results show that the PLPS and P-SiHMPF structures interact with lysozyme leading to monolayer adsorption due to the surface imprinting of the PLPS sample and the PDA coating on the P-SiHMPF samples.
| Langmuir isotherm | |||||
|---|---|---|---|---|---|
| Sample | Qe | Qmax | KL | Ce | R2 |
| PLPS | 70.53 | 3482.1 | 0.023 | 0.898859393 | 0.97 |
| P-SiHMPF | 11.12 | 20.76 | 1.21 | 0.953328075 | 0.92 |
| PBPS | 6.04 | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 626.3 626.3 |
1.76 | 0.000112077 | 0.89 |
| Freundlich isotherm | |||||
|---|---|---|---|---|---|
| Sample | Qe | KF | Ce | n | R2 |
| PLPS | 70.53 | 80.05 | 0.88156531 | 0.9956 | 0.97 |
| P-SiHMPF | 11.12 | 11.63 | 0.9730167 | 0.61 | 0.92 |
| PBPS | 6.04 | 6.24 | 0.947977113 | 1.64 | 0.98 |
| Temkin isotherm | |||||
|---|---|---|---|---|---|
| Sample | Qe | B | KT | Ce | R2 |
| PLPS | 70.53 | 35.16 | 7.19 | 1.03384155 | 0.96 |
| P-SiHMPF | 11.12 | 4.41 | 12.47 | 0.99821778 | 0.91 |
| PBPS | 6.04 | 3.23 | 4.48 | 1.448235609 | 0.83 |
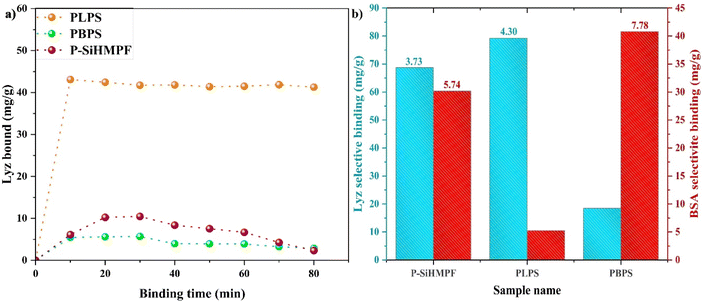
The influence of incubation time on the adsorption capacity of the adsorbents was analyzed using binding kinetic study data (Fig. 3a). All the parameters in the binding kinetic study such as volume (1.5 mL), concentration (0.5 mg mL−1), and mass of PLPS (3 mg) were kept constant and the adsorption/binding time was varied. The amount of protein adsorbed onto the PLPS surface reached a maximum value after 10 min and slightly varied thereafter. This confirms that the surface imprinted PLPS samples provided a quicker adsorption and separation means for protein purification. The earlier reported studies indicate that approximately 30 min is required to attain the maximum adsorption for Lyz.33 The quicker adsorption attained in our study could be a result of the size of the PLPS samples (micron range) which possibly facilitated better binding for the macromolecules such as proteins. The inclusiveness of this approach was further confirmed by imprinting BSA on the P-SiHMPF (PBPS) samples instead of Lyz. The selective binding factor (SB) and imprinting factor (IF) were calculated for both non-imprinted (P-SiHMPF) and protein-imprinted samples (PLPS and PBPS) by considering their respective adsorption capacity values and are mentioned in Table 2. The plot for the selective binding of Lyz and BSA for the individual samples can be seen in Fig. 3b. The imprinting factor and selective binding factor for PLPS and P-SiHMPF were calculated with respect to the PBPS values for Lyz solution and vice versa for BSA solution. The protein adsorption observed in P-SiHMPF samples was clearly not selective and was a result of the favourable functional groups present on the surface. However, a drastic difference in protein adsorption of the imprinted samples was observed indicating specific binding. Fig. 3b confirms that the PLPS and PBPS samples had very high selectivity for Lyz and BSA respectively. Surface imprinting of polymers is known to exhibit increased selectivity for the target protein in both competitive (along with other proteins) and non-competitive environments.35,36 The nature of the binding involved in this study can be confirmed to be non-electrostatic from the zeta potential results (Fig. 1b). Such interactions are known to increase the selectivity of the target protein in a competitive environment along with its adsorption capacity.37 The as-synthesized Lyz-imprinted sample (PLPS) exhibited a high adsorption capacity value along with good selectivity (IF) (Table 2). Several researchers have reported approaches to improve the adsorption capacity of the supports by either selecting a high surface area support material or by enhancing the active binding sites on the surface.10,38 PLPS samples exhibited the highest Qe results when compared to the existing literature data (Table 3). Imprinting factors for the Lyz-imprinted samples in the literature vary in the range of 2.1–6.4.30,33,41 The IF values obtained for the synthesized imprinted samples in this study (PLPS = 4.3; PBPS = 7.78) confirm that the surface imprinting approach for the model protein is efficient and the resulting adsorption is highly selective.
| Sample name | Imprinting factor (IF) | Selective binding factor (SB) |
|---|---|---|
| P-SiHMPF | 3.73 (Lyz); 5.74 (BSA) | 50.27 (Lyz); 24.91 (BSA) |
| PLPS | 4.30 | 60.81 |
| PBPS | 7.78 | 35.57 |
| Sample composition | Qe (mg g−1) | References |
|---|---|---|
| Lyz-imprinted silica NPs | 53.3 | 39 |
| Lyz-imprinted magnetic nanocomposites | 29.0 ± 10.2 | 40 |
| Lyz-imprinted PDA-poly(ethylene glycol) grafting | 116.7 ± 2.1 | 33 |
| Lyz-imprinted graphene oxide | 500 | 38 |
| Lyz-imprinted silica sub microparticles | 90.33 | 30 |
| Lyz-imprinted on MWCNTs-PDA supports | 418 | 19 |
| Lyz-imprinted on MNP-silica core–shell MIPS | 33 | 41 |
| Lyz imprinted on helical magnetic flagella-like structures (this work) | 600.89 | — |
This study confirms that surface imprinting of proteins onto the magnetic microswimmer surfaces offers an effective and efficient method to selectively purify proteins wherein the presence of a magnetic phase significantly reduces the complication of the conventional protein purification methods. The helical morphology of the magnetic microhelices fabricated in this work offered a high surface area and resulted in better adsorption efficiency.
4 Conclusion
Molecularly imprinted polymer (MIP) has been a popular and effective process to surface engineer antigen–antibody like interactions using protein templates. Here, for the first time to the best of our knowledge, we report lysozyme imprinting on magnetic microhelical supports and the study of its adsorption mechanism. The as-synthesized microhelices exhibited high adsorption capacity and selectivity for the imprinted protein (lysozyme or bovine serum albumin). The Freundlich adsorption isotherm model was observed to be a better fit for all the synthesized samples indicating non-homogeneous binding sites on the surface. The results show that magnetic microhelical ABF like structures provide a suitable surface for protein separation and purification via the molecular imprinting method, which can have potential uses in bioseparation.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Aakanksha Mohan – conceptualization, data curation, writing – original draft. Sutapa Roy Ramanan – project administration, supervision, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the Department of Physics, Birla Institute of Technology and Science Pilani K K Birla Goa Campus for the Vibrating Sample Magnetometer facility through the grant no. – SR/FST/PS-1/2017/21, DST, Govt. of India. The CSIF, Birla Institute of Technology and Science Pilani K K Birla Goa Campus, is also acknowledged for providing the FESEM facility.References
- N. E. Labrou, Protein purifi cation: An overview, Methods Mol. Biol., 2014, 1129, 3–10 CrossRef CAS PubMed.
- S. Gräslund, P. Nordlund, J. Weigelt, B. M. Hallberg, J. Bray and O. Gileadi, et al., Protein production and purification, Nat. Methods, 2008, 5, 135–146 CrossRef PubMed.
- J. Wang, Y. Shen, Y. Zhuang, J. Wang and Y. Zhang, Multimodal Affinity-Modulated Efficient Separation of Lysozyme with a Hierarchical MXene@MOF Hybrid Framework, Anal. Chem., 2024, 96(29), 12102–12111 CrossRef CAS PubMed.
- T. Tang, J. Gan, Z. Cao, P. Cheng, Q. Cheng and T. Mei, et al., Ethylene Vinyl Alcohol Copolymer Nanofibrous Cation Exchange Chromatographic Membranes with a Gradient Porous Structure for Lysozyme Separation, Polymers, 2024, 16(8), 1112 CrossRef CAS PubMed.
- S. Ansari and S. Masoum, Molecularly imprinted polymers for capturing and sensing proteins: Current progress and future implications, TrAC, Trends Anal. Chem., 2019, 114, 29–47 Search PubMed.
- H. R. Culver and N. A. Peppas, Protein-Imprinted Polymers: The Shape of Things to Come?, Chem. Mater., 2017, 29(14), 5753–5761 CrossRef CAS PubMed.
- D. Çimen and A. Denizli, Purification of ovalbumin from egg white using molecular imprinted cryogels, Hacettepe J. Biol. Chem., 2022, 50(1), 65–76 CrossRef.
- Y. He and Z. Lin, Recent advances in protein-imprinted polymers: synthesis, applications and challenges, J. Mater. Chem. B, 2022, 10, 6571–6589 RSC.
- T. Jing, H. Du, Q. Dai, H. Xia, J. Niu and Q. Hao, et al., Magnetic molecularly imprinted nanoparticles for recognition of lysozyme, Biosens. Bioelectron., 2010, 26(2), 301–306 Search PubMed.
- E. Roy, S. Patra, S. Saha, D. Kumar, R. Madhuri and P. K. Sharma, Shape effect on the fabrication of imprinted nanoparticles: Comparison between spherical-, rod-, hexagonal-, and flower-shaped nanoparticles, Chem. Eng. J., 2017, 321, 195–206 CrossRef CAS.
- X. Y. Sun, R. T. Ma, J. Chen and Y. P. Shi, Magnetic boronate modified molecularly imprinted polymers on magnetite microspheres modified with porous TiO2 (Fe3O4@pTiO2@MIP) with enhanced adsorption capacity for glycoproteins and with wide operational pH range, Microchim. Acta, 2018, 185(12), 565 CrossRef PubMed.
- D. Li, Y. Gu, X. Xu, Y. Feng, Y. Ma and S. Li, et al., Electrospun polyacrylonitrile fibers with and without magnetic nanoparticles for selective and efficient separation of glycoproteins, Microchim. Acta, 2019, 186(8), 542 CrossRef CAS PubMed.
- S. A. Zaidi, Molecular imprinting: A useful approach for drug delivery, Mater. Sci. Energy Technol., 2020, 3, 72–77 CAS.
- L. Chen, X. Wang, W. Lu, X. Wu and J. Li, Molecular imprinting: Perspectives and applications, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- H. Yavuz, K. Çetin, S. Akgönüllü, A. Denizli and D. Battal, Therapeutic protein and drug imprinted nanostructures as controlled delivery tools, in Design and Development of New Nanocarriers, Elsevier, 2018, pp. 439–473 Search PubMed.
- L. Zhang, K. E. Peyer and B. J. Nelson, Artificial bacterial flagella for micromanipulation, Lab Chip, 2010, 10(17), 2203–2215 Search PubMed.
- Z. Zhang, H. Wang, H. Wang, C. Wu, M. Li and L. Li, Fabrication and evaluation of molecularly imprinted magnetic nanoparticles for selective recognition and magnetic separation of lysozyme in human urine, Analyst, 2018, 143(23), 5849–5856 RSC.
- M. Kiristi, V. V. Singh, B. Esteban-Fernández De Ávila, M. Uygun, F. Soto and D. Aktaş Uygun, et al., Lysozyme-Based Antibacterial Nanomotors, ACS Nano, 2015, 9(9), 9252–9259 CrossRef CAS PubMed.
- X. Xu, P. Guo, Z. Luo, Y. Ge, Y. Zhou and R. Chang, et al., Preparation and characterization of surface molecularly imprinted films coated on multiwall carbon nanotubes for recognition and separation of lysozyme with high binding capacity and selectivity, RSC Adv., 2017, 7(30), 18765–18774 RSC.
- A. Mohan, R. Singhal and S. R. Ramanan, A study on the effect of the collector properties on the fabrication of magnetic polystyrene nanocomposite fibers using the electrospinning technique, J. Appl. Polym. Sci., 2023, 140(6), 4332–4340 CrossRef.
- A. Mohan and S. R. Ramanan, Hydrophobic or hydrophilic microhelices: Crafting surfaces with electrospun magnetic polystyrene fiber and an innovative top-down technique, Polym. Eng. Sci., 2024, 1–10 Search PubMed.
- S. M. Moatmed, M. H. Khedr, S. I. El-Dek, H. Y. Kim and A. G. El-Deen, Highly efficient and reusable superhydrophobic/superoleophilic polystyrene@ Fe3O4 nanofiber membrane for high-performance oil/water separation, J. Environ. Chem. Eng., 2019, 7(6), 103508, DOI:10.1016/j.jece.2019.103508.
- M. A. Dheyab, A. A. Aziz, M. S. Jameel, O. A. Noqta, P. M. Khaniabadi and B. Mehrdel, Simple rapid stabilization method through citric acid modification for magnetite nanoparticles, Sci. Rep., 2020, 10(1), 1–8, DOI:10.1038/s41598-020-67869-8.
- D. Patiño-Ruiz, L. Sanchez-Botero, J. Hinestroza and A. Herrera, Modification of Cotton Fibers with Magnetite and Magnetic Core-Shell Mesoporous Silica Nanoparticles, Phys. Status Solidi A Appl. Mater. Sci., 2018, 215(19), 1–7 Search PubMed.
- Y. Dang, C. M. Xing, M. Quan, Y. B. Wang, S. P. Zhang and S. Q. Shi, et al., Substrate independent coating formation and anti-biofouling performance improvement of mussel inspired polydopamine, J. Mater. Chem. B, 2015, 3(20), 4181–4190 RSC.
- Y. Chen, S. Zhao, M. Chen, W. Zhang, J. Mao and Y. Zhao, et al., Sandwiched polydopamine (PDA) layer for titanium dioxide (TiO2) coating on magnesium to enhance corrosion protection, Corros. Sci., 2015, 96, 67–73 CrossRef CAS.
- L. Zhu, Y. Lu, Y. Wang, L. Zhang and W. Wang, Preparation and characterization of dopamine-decorated hydrophilic carbon black, Appl. Surf. Sci., 2012, 258(14), 5387–5393 CrossRef CAS.
- G. Bayramoglu, V. C. Ozalp, M. Yilmaz, U. Guler, B. Salih and M. Y. Arica, Lysozyme specific aptamer immobilized MCM-41 silicate for single-step purification and quartz crystal microbalance (QCM)-based determination of lysozyme from chicken egg white, Microporous Mesoporous Mater., 2015, 207, 95–104 CrossRef CAS.
- S. Sharma and G. P. Agarwal, Interactions of proteins with immobilized metal ions: A comparative analysis using various isotherm models, Anal. Biochem., 2001, 288(2), 126–140 CrossRef CAS PubMed.
- C. Zhang, Y. Wang, Y. Zhou, J. Guo and Y. Liu, Silica-based surface molecular imprinting for recognition and separation of lysozymes, Anal. Methods, 2014, 6(21), 8584–8591 RSC.
- G. W. Lim, J. K. Lim, A. L. Ahmad and D. J. C. Chan, Molecularly imprinted polymer layers using Navicula sp. frustule as core material for selective recognition of lysozyme, Chem. Eng. Res. Des., 2015, 101, 2–14 CrossRef CAS.
- R. A. Latour, The Langmuir isotherm: A commonly applied but misleading approach for the analysis of protein adsorption behavior, J. Biomed. Mater. Res., Part A, 2015, 103(3), 949–958 CrossRef PubMed.
- W. Han, X. Han, Z. Liu, S. Zhang, Y. Li and J. Lu, et al., Facile modification of protein-imprinted polydopamine coatings over nanoparticles with enhanced binding selectivity, Chem. Eng. J., 2020, 385, 123463 Search PubMed.
- C. F. Wertz and M. M. Santore, Adsorption and relaxation kinetics of albumin and fibrinogen on hydrophobic surfaces: Single-species and competitive behavior, Langmuir, 1999, 15(26), 8884–8894 CrossRef CAS.
- H. R. Culver, S. D. Steichen and N. A. Peppas, A Closer Look at the Impact of Molecular Imprinting on Adsorption Capacity and Selectivity for Protein Templates, Biomacromolecules, 2016, 17(12), 4045–4053 Search PubMed.
- H. Shi and B. D. Ratner, Template recognition of protein-imprinted polymer surfaces, J. Biomed. Mater. Res., 2000, 49(1), 1–11 Search PubMed.
- E. Verheyen, J. P. Schillemans, M. Van Wijk, M. A. Demeniex, W. E. Hennink and C. F. Van Nostrum, Challenges for the effective molecular imprinting of proteins, Biomaterials, 2011, 32, 3008–3020 CrossRef CAS PubMed.
- S. C. Smith, F. Ahmed, K. M. Gutierrez and D. Frigi Rodrigues, A comparative study of lysozyme adsorption with graphene, graphene oxide, and single-walled carbon nanotubes: Potential environmental applications, Chem. Eng. J., 2014, 240, 147–154 CrossRef CAS.
- C. Yang, X. Yan, H. Guo and G. Fu, Synthesis of surface protein-imprinted nanoparticles endowed with reversible physical cross-links, Biosens. Bioelectron., 2016, 75, 129–135 CrossRef CAS PubMed.
- F. Lan, S. Ma, J. Ma, Q. Yang, Q. Yi and Y. Wu, et al., Superparamagnetic nanocomposites based on surface imprinting for biomacromolecular recognition, Mater. Sci. Eng. C, 2017, 70, 1076–1080 CrossRef CAS PubMed.
- Q. Q. Gai, F. Qu, Z. J. Liu, R. J. Dai and Y. K. Zhang, Superparamagnetic lysozyme surface-imprinted polymer prepared by atom transfer radical polymerization and its application for protein separation, J. Chromatogr. A, 2010, 1217(31), 5035–5042 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na01041h |
| This journal is © The Royal Society of Chemistry 2025 |