Open Access Article
Open Access ArticleUltrasound-activated nano-oxygen sensitizer for sonodynamic–radiotherapy of esophageal cancer†
Jiayin Liua,
Manru Shia,
Huijia Zhaob,
Xin Baib,
Quan Lina,
Xin Guana,
Bolin Wu *b and
Mingyan E.*a
*b and
Mingyan E.*a
aDepartment of Radiation Oncology, Harbin Medical University Cancer Hospital, No. 150, Haping Road, Nangang District, Harbin, Heilongjiang Province 150081, China. E-mail: emingyan0996@163.com; Fax: +86 451 86298500; Tel: +86 13936662229
bDepartment of Ultrasound, Harbin Medical University Cancer Hospital, No. 150, Haping Road, Nangang District, Harbin, Heilongjiang Province 150081, China. E-mail: wubolin@hrbmu.edu.cn; Fax: +86 451 85718392; Tel: +86 15663615088
First published on 20th February 2025
Abstract
Background: owing to the intricate nature, variability, and persistent oxygen-deficient environment associated with esophageal cancer (EC) tissues, radiotherapy (RT) sometimes doesn't work as well because some cancer cells can resist the radiation to a certain extent. This can lead to the cancer coming back in the same spot or even making the treatment ineffective. The integration of RT with oxygenation strategies is a common approach in cancer treatment. The advent of oxygen-enhancing sonodynamic therapy (SDT), leveraging the cytotoxic effects of reactive oxygen species (ROS), has garnered significant attention as an innovative approach to inducing cell death. Methods: this study utilized nanobubbles (NBs) containing the acoustic sensitizer indocyanine green (ICG) to create a nanoplatform (ICG@O2 NBs) that incorporates oxygen-enhanced SDT and RT. Besides, NBs are paired with low-frequency ultrasound (LFUS), known as ultrasound-targeted nano-bubble destruction (UTND), for precise drug release and improved safety. Results: experimental findings, including JC-1/DCFH-DA assays, demonstrate that ICG@O2 NBs effectively enhance the performance of both RT and SDT. RNA sequencing (RNA-seq) demonstrated differential expression of mRNA and LncRNA prior to and after co-treatment. KEGG and GO pathway analysis were then conducted for enriching and recognizing target genes and pathways correlated with the sensitivity of RT, which were revealed to be remarkably clustered in RT-associated pathways. Conclusion: in vitro and in vivo investigations have indicated significant efficacy of synergistic treatments, highlighting the potential of combining NBs with SDT and RT for managing EC.
1. Introduction
Radiotherapy (RT), which employs X-rays, is one of the most successful cancer therapies. It reduces cell proliferation, produces highly toxic hydroxyl (˙OH) free radicals, and destroys double-stranded DNA.1,2 For esophageal cancer (EC), it has continuously been a preferred therapeutic therapy strategy.3 However, radioresistance, which results from strong intracellular antioxidant defenses and insufficient X-ray energy deposition, frequently limits its efficacy.4 Recent advancements in nanomedical research have demonstrated that combining radiotherapy with other treatments, e.g. chemotherapy,5,6 photothermal therapy,7,8 photodynamic therapy,9 chemodynamic therapy,10,11 as well as immunotherapy, may improve therapeutic effect. Nevertheless, most documented combination therapies have failed to significantly overcome radioresistant tumors, depending on direct addition of numerous treatments.12,13 Unsatisfactory therapeutic effects and excessive doses of radiation are still prevalent, underscoring the urgent need for synergistic strategies to enhance radiosensitivity and improve radiotherapeutic effectiveness.Sonodynamic therapy (SDT) stands out as a highly promising noninvasive cancer treatment, capable of precisely targeting and eliminating tumors.14 SDT primarily evolved from the principles of photodynamic therapy (PDT). In the process of SDT, sonosensitizers, upon stimulation by ultrasonic energy, can elicit an increased production of ROS within cells.15 High-intensity focused ultrasound is a thermal ablation method that uses a combination of mechanical and thermal factors to cause tissue necrosis and cell death.16 The ultrasonic energy and intensity employed in SDT, in contrast, are substantially lower. SDT utilizes nonthermal mechanisms to preserve the biosafety of normal tissues surrounding the tumor. Unlike PDT light, US can penetrate deeply into tissues safely with minimal attenuation.17,18 However, the hypoxic environment within tumors significantly limits the effectiveness of SDT in combating cancer. Additionally, during the SDT process, substantial oxygen is progressively depleted, further exacerbating the hypoxic tumor microenvironment (TME). Therefore, it is crucial to establish methods for delivering oxygen to tumor sites to enhance therapeutic efficacy.10
Research indicates that combining SDT with RT enhances tumor sensitivity to radiation by increasing blood supply and reducing hypoxic cells, thereby boosting therapeutic efficacy through synergistic ROS production.19–21 Additionally, it can lower the laser exposure intensity in SDT and reduce the X-ray radiation dose in RT, thereby enhancing patient tolerance.22–24 Thus, integrating SDT with RT represents an effective and promising therapeutic approach.
The effectiveness of SDT and RT has been increased by the development of many nanoplatforms that reduce or even reverse hypoxia. NBs have gained widespread attention as carriers in combination with low-frequency ultrasound (LFUS), a method known as UTND, offering advantages over other nanocarriers. NBs can serve as cavitation nuclei, capable of expanding, compressing, and collapsing when stimulated by ultrasound.25 Micro-jets created by the collapse during cavitation provide shear stress on cells, which causes reversible pore creation in cell membranes. Drug delivery into the cells is made easier by this procedure, which momentarily increases the permeability of cell membrane without endangering cell survival.26 Lipid–chitosan core NBs have been shown in several investigations to efficiently reverse tumor hypoxia by inhibiting HIF-1α expression.26,27 NBs are therefore seen as a viable subject for more study.
Next-generation high-throughput RNA-Seq is a pioneer sequencing technology that enables high-throughput quantitative analysis of the entire transcriptome. In contrast to other sequencing methods, RNA-Seq offers a more precise depiction of isoform and transcript levels. Incorporating RNA-Seq technology into our investigations can yield important insights into the processes of tumor treatment and help us better understand how combination therapy enhances the sensitivity of RT and uncover related gene alterations.
In light of the aforementioned considerations, we created shell–core structured oxygen-enriched NBs, which have an oxygen-filled gas core and a lipid shell that contains the sonosensitizer ICG. As far as we know, this is the time that oxygen and ICG have been incorporated into NBs for use in SDT. Furthermore, the application of the UTND technique can enhance the accumulation of ICG in tumor cells. Additionally, ultrasound can break down the NBs, accelerating the release of oxygen to raise the dosage of tumor-specific oxygen. After then, therapeutic ultrasonic irradiation was employed for SDT, which induced cell apoptosis or necrosis, thereby significantly improving the SDT effectiveness. When X-ray irradiation is added, the resultant ˙OH free radicals cause further DNA damage to tumor cells, which effectively inhibits them from proliferating. Meanwhile, ICG@O2 NBs show great potential as a tool for imaging with contrast-enhanced ultrasound (CEUS). Thus, ICG@O2 NBs hold promise for realize the integration of tumor imaging along with treatment. Subsequently, RNA-Seq investigates the underlying mechanisms and determines associated gene alterations, providing a viable approach and possible therapy options for EC.
2. Materials and methods
2.1 Chemicals
1,2-Stearoyl-sn-glycerol-3-phosphoethanolamine-N-(methoxy [polyethylene glycol]-2000) (DSPE-PEG-2000) and 1,2-distearoyl-sn-glycerol-3-phosphocholine (DSPC) were provided by Avanti Polar Lipids (Alabaster, AL). In addition, 2′,7′-dichlorofluorescin diacetate (DCFH-DA) was included, ICG was acquired from Shanghai Yuanye Bio-Technology Co., Ltd (Shanghai, China). CCK-8 and Annexin V-FITC Apoptosis Detection Kit were supplied by BD Pharmingen.2.2 Preparation of ICG@O2 NBs
ICG@O2 NBs were produced via the thin-film hydration ultrasonic technique, as previously reported.28 DSPE-PEG-2000 and DSPC were first combined in a solvent combination of methanol and methylene chloride (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, vol/vol) at a mass ratio of 9
2, vol/vol) at a mass ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The solution is moved to a beaker and evaporated to create a lipid film once it has been thoroughly dissolved and combined. To create a lipid suspension, 5 mL of 100 μg mL−1 ICG in PBS was added to the dried lipid film. Afterwards, via applying a micro-extruder (Avanti Polar Lipids, Alabaster, AL), the suspension was passed through a 100 nm film and extruded 20 times. ICG@C3F8 NBs and ICG@O2 NBs were formed by transferring the extruded solution to a sealed vial, where the syringe was removed and replaced with either perfluoropropane (C3F8) gas or pure oxygen. After 60 seconds of mechanical shaking in a dental mixer (YJT Medical Apparatus and Instruments, Shanghai, China), the mixture was reconstituted in 2 mL of PBS solution and stored at 4 °C. All procedures were carried out in darkness.
1. The solution is moved to a beaker and evaporated to create a lipid film once it has been thoroughly dissolved and combined. To create a lipid suspension, 5 mL of 100 μg mL−1 ICG in PBS was added to the dried lipid film. Afterwards, via applying a micro-extruder (Avanti Polar Lipids, Alabaster, AL), the suspension was passed through a 100 nm film and extruded 20 times. ICG@C3F8 NBs and ICG@O2 NBs were formed by transferring the extruded solution to a sealed vial, where the syringe was removed and replaced with either perfluoropropane (C3F8) gas or pure oxygen. After 60 seconds of mechanical shaking in a dental mixer (YJT Medical Apparatus and Instruments, Shanghai, China), the mixture was reconstituted in 2 mL of PBS solution and stored at 4 °C. All procedures were carried out in darkness.
2.3 Characterization
Examine the morphology and distribution of ICG@O2 NBs utilizing a scanning transmission electron microscope (TEM, Hitachi, Japan). With a dynamic light scattering (DLS) analyzer (ZEN3600, Malvern Instruments), the zeta potentials, stability, and particle sizes of ICG@O2 NBs were examined.2.4 Detection of ˙OH generation
A brilliant green fluorescence with emission and excitation peaks at 515 and 490 nm is produced when aminophenyl fluorescein (APF, ThermoFisher, USA) interacts with ˙OH. In ddH2O with 5 μM APF, ICG@O2 NBs, ICG, and PBS were suspended. The RAD SOURCE RS2000 system (USA) was employed to expose each reagent (100 μL) to X-ray irradiation at dosages of 0, 2, 4, 6, or 8 Gy. An AniView SE system (Guangzhou Biolight Biotechnology Co., Ltd) was applied to promptly measure the fluorescence signals.2.5 Detection of 1O2 generation
Singlet Oxygen Sensor Green (SOSG, Thermo Fisher, USA) reacts and emits a bright green fluorescence with emission and excitation maxima at 525 and 504 nm, separately. In ddH2O with 12.5 μM SOSG, ICG@O2 NBs, ICG, and PBS were suspended for SDT. The AniView SE system (Guangzhou Biolight Biotechnology Co., Ltd) was exploited to promptly measure the fluorescence signals.2.6 Cell culture
KYSE-150 and TE-1, the human esophageal cancer cell lines, were acquired from the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, CAS. The cultivation of cells was conducted at 37 °C with 5% CO2 in RPMI-1640 media with 10% FBS in a humidified incubator.2.7 Cell cytotoxicity assay
Cytotoxicity of ICG@O2 NBs was assessed with a conventional CCK-8 test. In particular, esophageal cancer cells (KYSE-150 and TE-1) were plated in 96-well plates at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well, and were incubated for 24 hours at 37 °C with 5% CO2. Following the treatment of US, the medium was replaced for new medium that contained varying concentrations of ICG@O2 NBs. Following a period of 24 hours incubation, cell viability was determined through the standard methodology of CCK-8 assay.
000 cells per well, and were incubated for 24 hours at 37 °C with 5% CO2. Following the treatment of US, the medium was replaced for new medium that contained varying concentrations of ICG@O2 NBs. Following a period of 24 hours incubation, cell viability was determined through the standard methodology of CCK-8 assay.
In order to assess the RT killing effect of ICG@O2 NBs, KYSE-150 and TE-1 cells weretreated with or without ICG@O2 NBs for six hours. This was followed by X-ray exposure and/or US therapy (1.0 W cm−2, 5 minutes, Focused Ultrasound Therapeutic Device, Model-CZF, Chongqing Haifu Medical Technology Co., Ltd, China). The CCK-8 test was applied to investigate the cell-killing impact following a further 18 hours of incubation, as previously mentioned. KYSE-150 and TE-1 cells (1 × 105 cells per well) were cultured with or without ICG@O2 NBs for 6 hours before being exposed to X-rays or US treatment (1.0 W cm−2, 5 minutes) for the live/dead co-staining experiment. All cells were stained with calcein-AM/propidium iodide and investigated under a fluorescence microscope after an extra 18 hours.
2.8 Apoptosis analysis
Flow cytometry was applied to measure apoptosis. Cells at different stages of apoptosis were counted utilizing the FITC-Annexin V/PI detection kit. After being plated in 6-well plates, KYSE-150 along with TE-1 cells were subjected to DMEM (negative control), SDT + RT, RT, and SDT. Subsequently, 1 × 105 cells was resuspended in 100 μL of 1× binding buffer. The cell suspension was incubated for 15 minutes in darkness following the addition of FITC-Annexin V and PI. Following adding 400 μL of 1× binding buffer to the cells, the samples were prepared for analysis with flow cytometry.2.9 DNA double-strand breaks
KYSE-150 and TE-1 cells in the irradiation groups were subjected to X-rays and incubated for an extra hour following various treatments (RT, SDT, DMEM, and SDT + RT). The cells were then fixed with 4% paraformaldehyde, blocked with 10% goat serum, and permeabilized with 0.5% Triton. The anti-γH2AX antibody was subsequently cultured with the cells at a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 for 12 hours. Following the application of a secondary antibody, a confocal laser scanning microscope (CLSM) was exploited to view the cells.
200 for 12 hours. Following the application of a secondary antibody, a confocal laser scanning microscope (CLSM) was exploited to view the cells.
2.10 ROS detection
To detect ROS within cells, a DCFH-DA fluorescent probe was employed. Following 6 hours of incubation with or without ICG@O2 NBs, KYSE-150 and TE-1 cells were subjected to an X-ray (100 cGy min−1, 8 Gy), and then treatment with US (1.0 W cm−2, 5 min), and incubated for 18 hours. Following incubation with a cell medium containing DCFH-DA, the cells were examined through a CLSM.2.11 Mitochondrial membrane potential detection
Following six hours of incubation with or without ICG@O2 NBs, KYSE-150 along with TE-1 cells were treated by US (1.0 W cm−2, five minutes), exposed to an X-ray (100 cGy min−1, 8 Gy), and then incubated for 18 hours. The same laser excitation at 488 nm was applied to determine JC-1 aggregates and monomers at emission wavelengths of 590 nm (red) and 530 nm (green).2.12 In vivo tumor model
The six-week-old male naked mice were acquired from the Laboratory Animal Center of Harbin Medical University. A cell suspension comprising 5 × 106 KYSE-150 cells was subcutaneously injected into the right flank of mice to establish tumor-bearing mice. This investigation was authorized via the Animal Ethics Committee of Harbin Medical University. All animal experiments, encompassing euthanasia, were conducted in compliance with the regulations of the Institutional Animal Care and Use Committee and Association for Assessment and Accreditation of Laboratory Animal Care.2.13 In vivo fluorescence imaging
Tumor-bearing mice (n = 3) received intravenous administration of ICG@O2 NBs or not. 24 hours following the injection, the mice were observed via the In Vivo Imaging System (Bruker, FX Pro). All of the mice were euthanized at the conclusion of the trial, and the major organs and tumors were gathered for imaging.2.14 In vivo antitumor therapy
First, there were four groups (n = 5) of tumor-bearing nude mice: (1) PBS, (2) SDT, (3) RT, and (4) RT + SDT. Mice in groups (1) and (3) received PBS intravenously, but those in groups (3) and (4) received RT. Mice in groups (2) and (4) were given ICG@O2 NBs. X-ray (8 Gy, 100 cGy min−1) and/or LFUS (1 W cm−2, 5 minutes) were administered to the tumors 24 hours following injection for the groups undergoing X-ray and/or US treatments. Every 4 days of the trial, measurements of tumor volume (tumor volume (mm3) = 1/2 × length × width2.) and body weight were made. All organs and tumors were gathered at the end of the experiment for further pathological examination.The healthy mice (n = 3) were administered ICG@O2 NBs. At designated time points following injection, blood samples were collected for routine blood analysis.
2.15 Construction, sequencing and analysis of whole transcriptome library
The RNA sequencing experiment was conducted with four groups: Group A, TE-1 cells before RT + SDT treatment; Group B, TE-1 cells after RT + SDT treatment; Group C, KYSE-150 cells before RT + SDT treatment; and Group D, KYSE-150 cells after RT + SDT treatment. Extraction of total RNA was conducted from KYSE-150 and TE-1 cells both before and after RT + SDT treatment and subjected to quality control. Deep sequencing and the creation of whole transcriptome library were carried out by Novogene Bioinformatics Technology Co., Ltd (Beijing, China).2.15.2.1 Quality control. The raw data (fastq format) was initially processed with an in-house Perl script. In this step, low-quality reads, poly-N reads, and adapter sequences were eliminated from the raw data to deliver clean data (clean reads). The GC, Q30, and Q20 contents of clean data were also computed. This clear, high-quality data acted as the basis for all further analysis.
2.15.2.2 Coding potential analysis. Without depending on established annotations, CNCI (Coding-Non-Coding-Index) (v2) efficiently separates protein-coding from non-coding sequences via analyzing nearby nucleotide triplets. CNCI was utilized in this investigation using its default parameters.
CPC2 (v0.1) employs four sequence inherent properties that are clearly interpretable and biologically significant. The Fickett score determines the preferred positions of bases within the sequence at the DNA level. The ORF (open reading frame) integrity and length are crucial at the RNA level, as protein-coding transcripts typically have long and intact ORFs. Additionally, CPC2 incorporates several peptide-level features, on the basis of hypothesis that peptides found in non-coding transcripts have diverse chemical characteristics from those encoded by legitimate coding sequences. Ultimately, CPC2 incorporates the isoelectric point (pI) as a key feature in its final SVM model.29
We applied Pfam Scan (v1.3) to determine any known protein family domains from the Pfam database (version 27), combining Pfam A and B, after translating each transcript in all three reading frames.30 Subsequent studies did not include transcripts with a Pfam hit. The default settings of -E 0.001 and --domE 0.001 were utilized for Pfam searches.31
Conservation of evolutionary features specific to coding areas are analyzed by PhyloCSF (phylogenetic codon substitution frequency) (v20121028). It recognizes high-frequency synonymous codon substitutions and conserved amino acid variations, while simultaneously observing low-frequency other missense and nonsense mutations, thereby distinguishing between non-coding and protein-coding transcripts.32 Utilizing the default parameters, we ran PhyloCSF and created multi-species genome sequence alignments.
Transcripts that were exhibited to have coding potential utilizing any of the four aforementioned tools were eliminated, and those that did not were regarded as our candidate lncRNAs.
2.16 Statistical analysis
GraphPad Prism (version 8.0) was employed to statistically analyze the experimental data. The data are represented as mean ± SD after one-way analysis of variance (ANOVA) was applied. P-Values less than 0.05 were regarded as statistically significant.3. Results and discussion
3.1 Characterization of ICG@O2 NBs
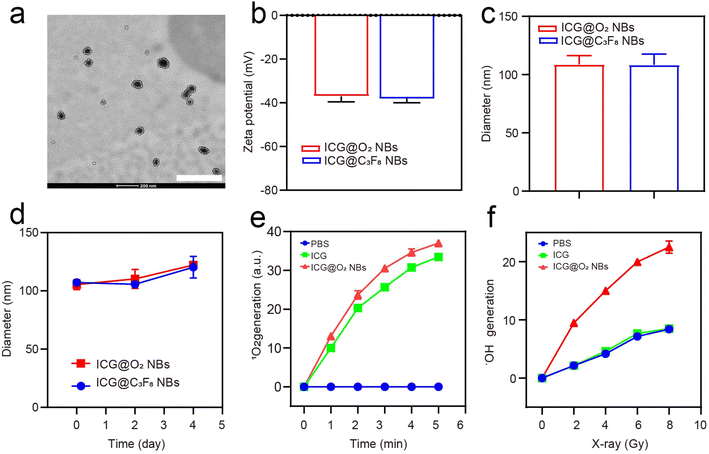
ICG@C3F8 NBs and ICG@O2 NBs were created through a thin-film hydration-sonication approach. In this procedure, hydrophobic ICG was enclosed within the lipid bilayer, with oxygen serving as the core. To increase the stability of NBs, DSPE-PEG-2000 was also conjugated to their surface (Scheme 1). TEM images indicated that the ICG@O2 NBs had a smooth surface and a homogeneous spherical shape, with a diameter of around 100 nm, as exhibited in Fig. 1a. The zeta potential of ICG@O2 NBs were −37.04 ± 2.53 mv (Fig. 1b). The synthesized ICG@O2 NBs had a size distribution of 108.8 ± 7.5 nm (Fig. 1c). And the size of ICG@O2 NBs and ICG@C3F8 NBs did not change in 4 days, which suggests good stability (Fig. 1d).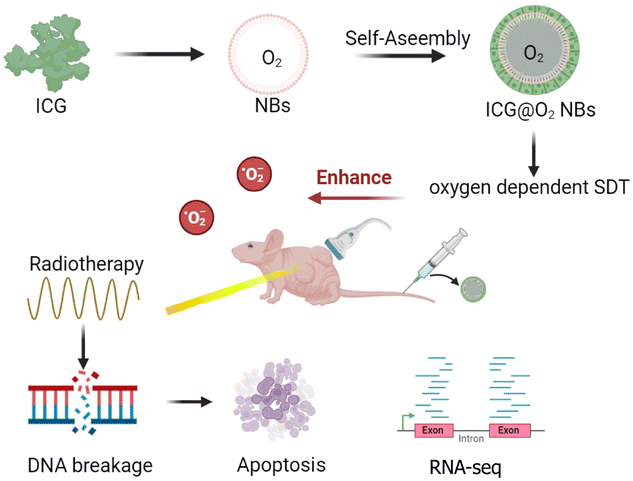 | ||
| Scheme 1 Schematic illustration of ultrasound-inspired ICG@O2 NBs nanotheranostics for the powerful tumor cell invasion synergistic sonodynamic therapy (SDT)–radiotherapy (RT). | ||
3.2 In vitro efficacies assessment of ICG@O2 NBs
The SOSG probe was applied to measure the SDT effectiveness of ICG@O2 NBs in order to identify the generation of singlet oxygen. As exhibited in Fig. 1e, ICG@O2 NBs demonstrated a significant generation of singlet oxygen, with a ∼37-fold increase compared to PBS when exposed to ultrasound at a density of 1 W cm−2. Next, an APF test was exploited to assess the radio-enhancement impact of ICG@O2 NBs. This assay detects the generation of hydroxyl radicals (˙OH) by activating fluorescence upon reaction with ˙OH. As shown in Fig. 1f, ICG@O2 NBs produced significantly more hydroxyl radicals (˙OH) compared to PBS and X-ray treatment alone (8 Gy), with a ∼3-fold increase in relative enhancement upon X-ray irradiation. Additionally, the production of hydroxyl radicals (˙OH) was dose-dependent, indicating that X-ray irradiation was the primary factor responsible for inducing ˙OH generation. These results suggest that ICG@O2 NBs, when combined with X-ray/US, exhibit enhanced SDT and radio-sensitizing efficacy.3.3 In vitro SDT efficacy of ICG@O2 NBs
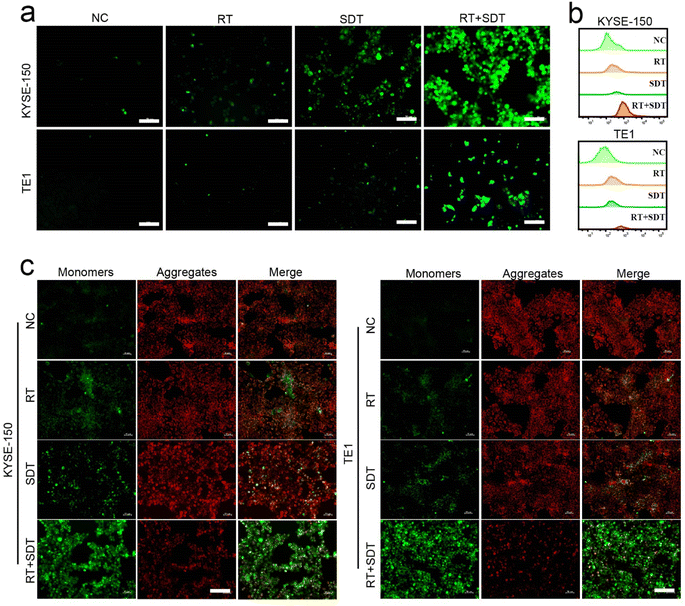
To determine the 1O2 generation efficiency of ICG@O2 NBs, DCFH-DA was utilized to visualize the levels of ROS, which would react with 1O2 and cause DCF to fluoresce green. KYSE-150 and TE-1 cells with RT + SDT treatment exhibited the strongest intracellular DCFH-DA fluorescence with LFUS (US density of 1 W cm−2). In comparison, cells with RT treatment displayed moderate levels of intracellular fluorescence, whereas cells treated with SDT displayed the highest levels of ROS by flow cytometry analysis (Fig. 2a and b), indicating that SDT elevated the production of intracellular 1O2 related to RT.To further confirm the efficiency of the production of single-linear oxygen in cells, 1O2-induced mitochondrial membrane potential variations were evaluated employing JC-1 staining. In the mitochondrial matrix, JC-1 builds up and forms aggregates that emit fluorescence. On the other hand, JC-1 exists as monomers and emits green fluorescence when mitochondrial membrane depolarization stops it from accumulating in mitochondrial matrix. Following RT + SDT therapy, a decreased ratio of red (JC-1 aggregate) to green (JC-1 monomer) fluorescence intensity suggested mitochondrial membrane depolarization, as displayed in Fig. 2c. On the other hand, cells treated with RT alone had lesser green fluorescence intensity and stronger red fluorescence intensity, indicating less mitochondrial damage. These findings indicate that RT + SDT can generate a high level of ROS, leading to significant cytotoxicity.
3.4 In vitro SDT–RT synergistic efficacy
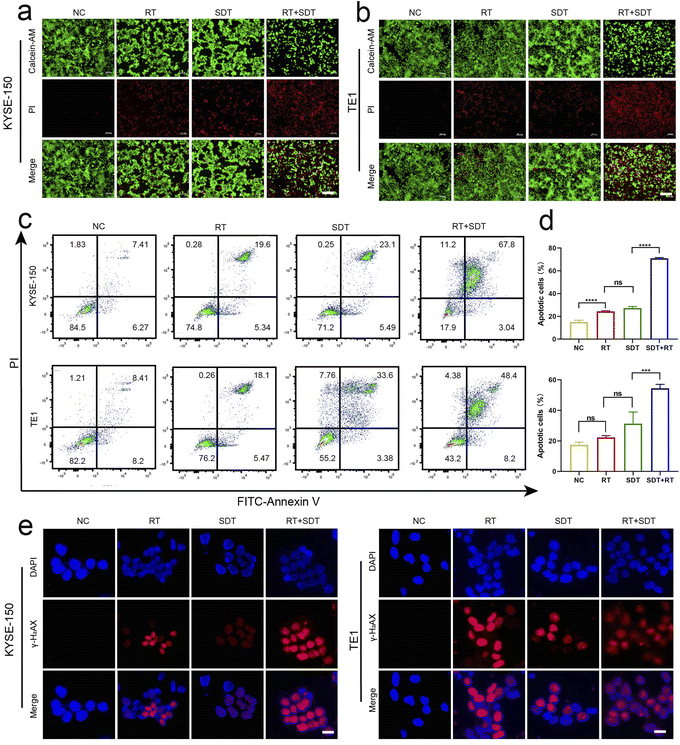
To assess whether SDT and RT synergistically reduced cell viability, CCK-8 assays were conducted (Fig. S1†). After incubating KYSE-150 and TE-1 cells with ICG@O2 NBs nanosystems for 24 hours in darkness, there was a minimal decrease in cell viability, suggesting that ICG@O2 NBs have good biocompatibility. After treatment with X-ray (8 Gy) and/or US (1.0 W cm−2, 5 min), 86.17% of the cells in the RT + SDT group were killed, which was significantly higher than the 46.29% cell death observed in the SDT group and the 21.98% in the RT group. The CCK-8 results were in agreement with the outcomes of the live/dead cell test utilizing calcein-AM/propidium iodide (PI) co-staining (Fig. 3a and b) and the apoptosis analysis with Annexin V-FITC/PI co-staining (Fig. 3c and d). These outcomes provide further evidence that cellular apoptosis is more markedly induced by RT and SDT together than by either treatment alone.3.5 In vitro radiosensitization effect of ICG@O2 NBs
The primary source of radiation-induced cytotoxicity is cellular DNA damage. With confocal microscopy, γ-H2AX foci—a quantifiable indicator of DNA double-strand breaks—were observed in the cell nuclei to confirm the radiosensitizing action of ICG@O2 NBs (Fig. 3e and f). The fluorescence intensity of γ-H2AX foci was considerably greater in the RT + SDT group than in the RT group, indicating that the ICG@O2 NBs nanosystem effectively induced DNA breakage.3.6 In vivo SDT–RT imaging and antitumor effect of ICG@O2 NBs
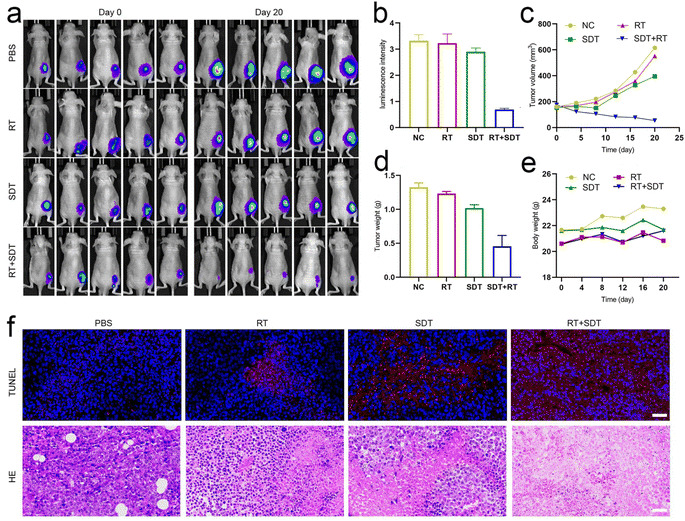
For biodistribution studies, mice bearing the tumor were administered free ICG and ICG@O2 NBs via tail vein injection, and after 24 h, for ex vivo fluorescence imaging, the major organs along with tumor were obtained (Fig. S2†). The ICG@O2 NBs group's fluorescent signal was still detectable in the tumor tissue after 24 h and showed higher fluorescence signal compared to mice treated with ICG. This demonstrates that the generation of ICG@O2 NB produces long-term, intense ICG enrichment at the tumor site.For assessing the anti-tumor effects of ICG@O2 NBs in combination with SDT–RT, we established subcutaneous xenograft models in nude mice. Tumor-bearing mice were allocated randomly to four groups (5 mice per group) when the tumor volume achieved about 100-150 mm3: (1) PBS, (2) RT, (3) SDT, and (4) RT + SDT. The treatments, including US (1 W cm−2) for SDT, X-ray irradiation (8 Gy) for RT, and combined SDT–RT (US, 1 W cm2; X-ray, 8 Gy), were administered 24 hours after injection of ICG@O2 NBs. During the RT procedure, to protect the healthy tissues of mice from radiation exposure, lead plates were applied. Over the course of the 20 days treatment period, digital pictures of the mice along with their tumor tissues were taken from each group (Fig. 4a). Every four days, the tumor volume was detected. As revealed in Fig. 4b and c, tumors in the PBS group exhibited rapid growth. In contrast, the groups treated with either SDT or RT alone exhibited some degree of suppression of tumor growth. Notably, the ICG@O2 NBs in combination with X-ray therapy confirmed a more marked decline of tumor development than the RT-only group, indicating that ICG@O2 NBs might improve the effectiveness of RT. Interestingly, the combination treatment of LFUS and X-ray in the ICG@O2 NBs groups resulted in significant tumor ablation, resulting in the smallest observed tumor volume. The weight of the resected tumor was in line with the tumor growth pattern observed (Fig. 4d), further confirming that the synergistic SDT–RT treatment using ICG@O2 NBs effectively inhibited tumor growth. Besides, during the combination treatment, there was no discernible decline in body weight (Fig. 4e), indicating that the treatment was well tolerated by the mice.
Next, we conducted immunohistochemical staining on tissue sections. TUNEL staining was performed to assess cell apoptosis, as it is a well-established marker for detecting apoptotic cells. The tumors in the RT + SDT group had a noticeably greater proportion of TUNEL-positive cells than those in the other treatment groups, as displayed in Fig. 4f, suggesting improved apoptosis. Hematoxylin and eosin (H&E) staining of tumor tissues displayed significant structural damage in the ICG@O2 NBs + SDT + RT group, with a noticeable decrease in cell density in the tumors. These findings indicate that the ICG@O2 NBs nanosystem, when combined with LFUS and X-ray irradiation, can effectively suppress esophageal cancer growth by inducing apoptosis and suppressing cancer cell proliferation in tumor cells.
3.7 In vivo biosafety analysis of ICG@O2 NBs
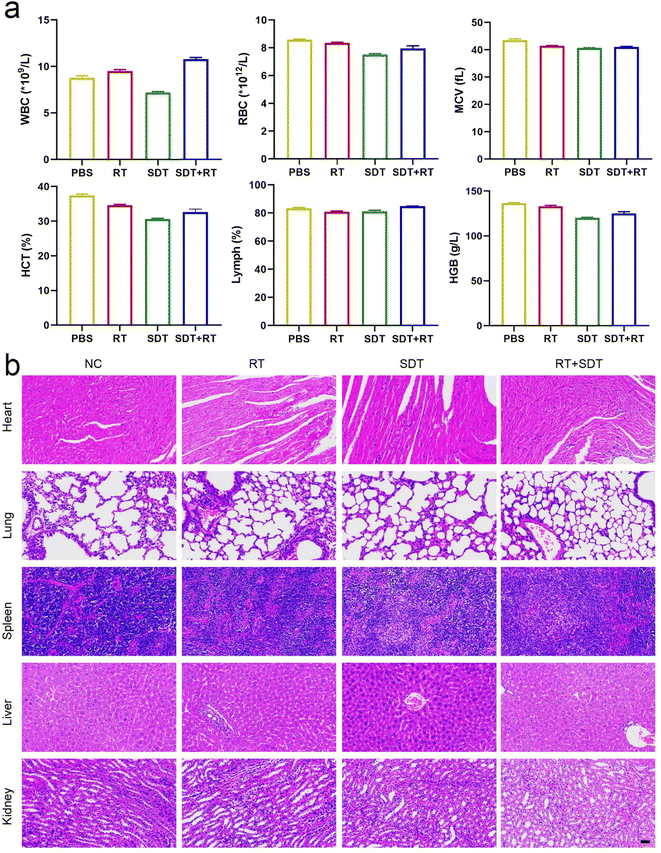
Following 20 days of treatment, blood samples were taken for investigation of blood cell counts in order to evaluate the biosafety of the ICG@O2 NBs nanosystem. The blood cell counts exhibited no discernible abnormalities, encompassing red blood cells (RBC), lymphocytes (LY), white blood cells (WBC), platelets (PLT) and hemoglobin (HGB) (Fig. 5a). These results indicate that the ICG@O2 NBs nanosystem exhibits good biosafety. During the combination therapy, no apparent damage to major organs was found, like the spleen, liver, heart, kidneys and lungs (Fig. 5b). This suggests that ICG@O2 NBs combined with LFUS/X-ray irradiation does not cause significant acute biological toxicity. These findings demonstrate that the combination of ICG@O2 NBs with RT and SDT can effectively inhibit the growth of tumors and decrease the mortality rate caused by tumors without significant long-term side effects.3.8 Differential expression analysis
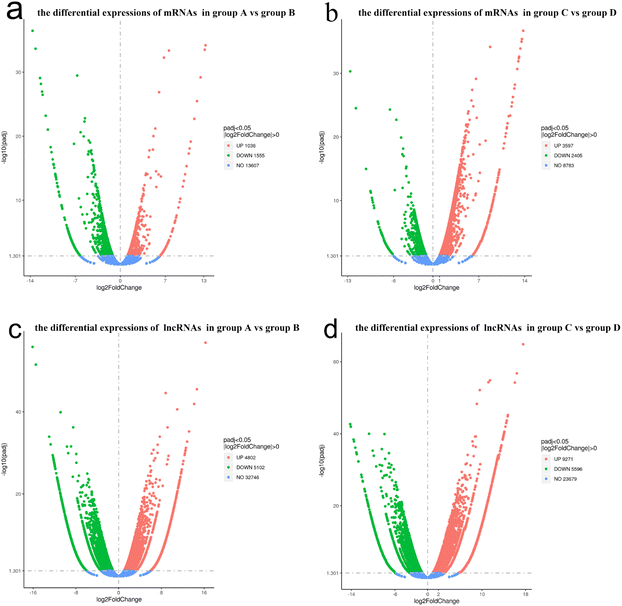
The lncRNA and mRNA transcripts levels were assessed based on their expression levels. As a result, 2591 mRNA transcripts showed differential expression between group A and B, with 1036 transcripts being upregulated and 1555 downregulated (P < 0.05, Fig. 6a). In total, there were 6002 mRNA transcripts that were differentially expressed between group C and D, with 3597 transcripts upregulated and 2405 downregulated (P < 0.05, Fig. 6b).Similarly, lncRNA transcripts also exhibited differential expression. In total, there were 9904 lncRNA transcripts that were differentially expressed between group A and B, of which 5102 were down-regulated and 4802 were up-regulated (P < 0.05, Fig. 6c). There were 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 867 lncRNA transcripts that were differentially expressed between groups C and D, of which 5596 were down-regulated and 9271 were up-regulated (P < 0.05, Fig. 6d).
867 lncRNA transcripts that were differentially expressed between groups C and D, of which 5596 were down-regulated and 9271 were up-regulated (P < 0.05, Fig. 6d).
3.9 Functional enrichment analysis: GO and KEGG
In accordance with GO analysis, 88 differential mRNA GO terms were clearly enriched in groups A versus B (Table S1,† P < 0.01), while 51 terms displayed considerable enrichment in group C vs. D (Table S2,† P < 0.01). At the same time, 31 differential lncRNA target GO terms were significantly enriched in group A vs. B (Table S3,† P < 0.01), while 20 terms showed significant enrichment in group C vs. D (Table S4,† P < 0.01). Table 1 and 2 display the typical enriched terms for differential mRNA along with targets, separately. Fig. S3† displays the comprehensive group classification of GO terms.| GO accession | Description | Term type | P Valuea | P Valueb |
|---|---|---|---|---|
| a Means P value between group A and group B.b Means P value between group C and group D. | ||||
| GO:0006119 | Oxidative phosphorylation | Biological process | 4.45 × 10−7 | 1.09 × 10−5 |
| GO:0042775 | Mitochondrial ATP synthesis coupled electron transport | Biological process | 7.31 × 10−6 | 9.09 × 10−6 |
| GO:0000786 | Nucleosome | Cellular component | 1.51 × 10−14 | 0.000514 |
| GO:0005746 | Mitochondrial respiratory chain | Cellular component | 7.71 × 10−8 | 1.39 × 10−6 |
| GO:0003735 | Structural constituent of ribosome | Molecular function | 0.000241 | 1.48 × 10−12 |
| GO accession | Description | Term type | P Valuea | P Valueb |
|---|---|---|---|---|
| a Means P value between group A and group B.b Means P value between group C and group D. | ||||
| GO:0044391 | Ribosomal subunit | Cellular component | 1.97 × 10−5 | 0.000479 |
| GO:0005840 | Ribosome | Cellular component | 1.98 × 10−5 | 0.00013 |
| GO:0044448 | Cell cortex part | Cellular component | 0.000223 | 0.000253 |
| GO:0005743 | Mitochondrial inner membrane | Cellular component | 0.005852 | 3.26 × 10−7 |
| GO:0098798 | Mitochondrial protein complex | Cellular component | 0.008315 | 0.000997 |
Table 3 and 4 provide the top five pathway enrichments for differential mRNA and lncRNA targets, separately. Additionally, the detailed KEGG pathway enrichment data can be accessed through NCBI's Sequence Read Archive or upon request from the corresponding author. On the other hand, Fig. S4† displays the pathway enrichment results for lncRNA and mRNA targets across several groups.
| Term | Database | ID | P-Valuea | P Valueb |
|---|---|---|---|---|
| a Means P value between group A and group B.b Means P value between group C and group D. | ||||
| Chemical carcinogenesis – reactive oxygen species | KEGG PATHWAY | hsa05208 | 0.000672 | 0.000263855 |
| Ribosome | KEGG PATHWAY | hsa03010 | 3.27 × 10−5 | 3.53 × 10−11 |
| Oxidative phosphorylation | KEGG PATHWAY | hsa00190 | 1.14 × 10−8 | 1.84 × 10−8 |
| Nucleotide excision repair | KEGG PATHWAY | hsa03420 | 0.044381 | 0.00133868 |
| Cytosolic DNA-sensing pathway | KEGG PATHWAY | hsa04623 | 0.00133 | 0.021292642 |
| Term | Database | ID | P-Valuea | P Valueb |
|---|---|---|---|---|
| a Means P value between group A and group B.b Means P value between group C and group D. | ||||
| Ribosome | KEGG PATHWAY | hsa03010 | 0.001127 | 0.002237 |
| Nucleotide excision repair | KEGG PATHWAY | hsa03420 | 0.044381 | 0.001339 |
| Ubiquitin mediated proteolysis | KEGG PATHWAY | hsa04120 | 0.00399 | 0.011191 |
| Necroptosis | KEGG PATHWAY | hsa04217 | 0.000444 | 0.000493 |
| Chemical carcinogenesis – reactive oxygen species | KEGG PATHWAY | hsa05208 | 0.003199 | 0.008812 |
4. Discussion and conclusions
In this work, bionic ICG@O2 NBs have been designed as a nanocarrier encapsulating an ultrasonic sensitizer ICG and oxygen for US/fluorescent image-guided cancer sonodynamic therapy to increase the sensitivity of radiation therapy. This new core–shell structure could provide enough oxygen to diagnose and treat cancer at the same time. With UTND technology, ICG@O2 NBs can achieve directional delivery of sound sensitizer ICG to release oxygen at the tumor site in a controlled manner to reverse the internal oxygen-depleted state.At the same time, the enhanced sonodynamic effect of oxygen can produce more singlet oxygen and improve the sensitivity of radiation therapy. ICG@O2 NBs can effectively induce the death of esophageal cancer cells KYSE-150 and TE-1 by low frequency focused ultrasound and X-ray irradiation, combined with the efficiency of SDT and radiation therapy (Fig. 2 and 3). Real-time monitoring by fluorescence imaging, ICG@O2 NBs radiosensitizer gives priority to long-term accumulation of tumor sites (Fig. 4). In addition, through the synergistic effect of SDT and RT, ICG@O2 NBs showed excellent antitumor properties without systemic toxicity in xenografted nude mouse esophageal cancer models (Fig. 5).
However, the underlying mechanism by which sonodynamic therapy enhances radiation therapy is not known. A state-of-the-art next-generation technique called NA sequencing has enabled the creation of a posttranscriptional regulatory network for pathways and genes associated with SDT–RT in esophageal cancer (EC), offering unprecedented insights into the roles of mRNAs and lncRNAs. Predicting the lncRNAs and mRNAs that control radiation sensitivity in EC was the main goal of this investigation. Based on the results, the lncRNAs and mRNAs may be viable targets for co-chemotherapy for EC in the future.
Initially, the Illumina HiSeq 2500 platform was employed to identify the differences in lncRNA and mRNA expression between groups A and B, as well as between groups C and D. According to the analysis, a total of 8593 dysregulated mRNA transcripts (Fig. 6a and b) and 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 771 apparently dysregulated lncRNA transcripts (Fig. 6c and d) were detected between both parallel experimental groups.
771 apparently dysregulated lncRNA transcripts (Fig. 6c and d) were detected between both parallel experimental groups.
GO enrichment analysis indicated that mRNA transcripts were primarily enriched in terms associated with “nucleosome”, “mitochondrial ATP synthesis coupled electron transport”, “oxidative phosphorylation”, “structural constituent of ribosome” and “mitochondrial respiratory chain”. These enriched terms, which are displayed in Table 1, were associated with ribosomes and mitochondria, suggesting that SDT is an essential part of the therapeutic process. Owing to mitochondria are the primary targets of SDT. Additionally, the cellular component contained five typical differential lncRNA target enrichment terms (Table 2). These terms represent the important structure of tumor cells. Therefore, another factor that enhances the radiosensitivity of SDT may be attributed to the destruction of tumor cell structures.
Next, to provide a clear visualization of the GO enrichment results for other researchers, the top 20 enriched GO terms were selected for display. Due to limitations in figure resolution and manuscript length, the figures depicting differential lncRNA and mRNA target enrichments are presented in Fig. S3.†
Third, another significant finding was the enrichment of pathways. Differential mRNAs were mainly enriched in key pathways, as shown in Table 3, including “chemical carcinogenesis-reactive oxygen species”, “ribosome”, “oxidative phosphorylation”, “nucleotide excision repair” and “cytosolic DNA-sensing pathway”.
The differential lncRNA targets were primarily enriched in key pathways, as listed in Table 4, including terms for instance “ubiquitin-mediated proteolysis”, “nucleotide excision repair”, and “ribosome”. “necroptosis” and “chemical carcinogenesis-reactive oxygen species”. These KEGG pathways were all involved in the principles of radiation therapy and sonodynamic therapy.
All these in vivo and in vitro investigations show that ICG@O2 NBs nanosystems based on bionic engineering strategies have broad application prospects in US/fluorescence image-guided collaborative SDT–RT and further translational studies.
Ethics approval and consent to participate
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Harbin Medical University and approved by the Animal Ethics Committee of Harbin Medical University.Data availability
According to acceptable requirements, all the resources or data utilized in the paper are accessible to Prof. Mingyan E., the leading correspondence.Conflicts of interest
The authors state that no conflict of interest exists.Acknowledgements
This work was funded by the National Natural Science Foundation of China (grant number 82072027 and 82202163), natural Science Foundation of Heilongjiang Province (grant number PL2024H187) and the Haiyan Science Foundation of Harbin Medical University Cancer Hospital (grant number JJMS2021-30 and JJJQ2024-06). We thank Prof. Kuikun Yang (School of Life Science and Technology, Harbin Institute of Technology) for supplying the altruistic guidance on the project.References
- C. Allen, S. Her and D. A. Jaffray, Adv. Drug Deliv. Rev., 2017, 109, 1–2 CrossRef CAS PubMed.
- D. E. Citrin, N. Engl. J. Med., 2017, 377, 1065–1075 CrossRef CAS PubMed.
- G. Petroni, L. C. Cantley, L. Santambrogio, S. C. Formenti and L. Galluzzi, Nat. Rev. Clin. Oncol., 2022, 19, 114–131 CrossRef CAS PubMed.
- M. Diehn, R. W. Cho, N. A. Lobo, T. Kalisky, M. J. Dorie, A. N. Kulp, D. Qian, J. S. Lam, L. E. Ailles, M. Wong, B. Joshua, M. J. Kaplan, I. Wapnir, F. M. Dirbas, G. Somlo, C. Garberoglio, B. Paz, J. Shen, S. K. Lau, S. R. Quake, J. M. Brown, I. L. Weissman and M. F. Clarke, Nature, 2009, 458, 780–783 CrossRef CAS PubMed.
- Y. Kang, X. Yu, X. Fan, Aod. Aodenggerile, S. Zhao, C. Tu, Z. Yan, R. Wang, W. Li and H. Qiu, ACS Nano, 2020, 14, 4336–4351 CrossRef CAS PubMed.
- D. Manoharan, L. C. Chang, L. C. Wang, Y. S. Shan, F. C. Lin, L. C. Wu, H. S. Sheu, W. P. Su and C. S. Yeh, ACS Nano, 2021, 15, 9084–9100 CrossRef CAS PubMed.
- Y. Yang, M. Chen, B. Wang, P. Wang, Y. Liu, Y. Zhao, K. Li, G. Song, X. B. Zhang and W. Tan, Angew Chem. Int. Ed. Engl., 2019, 58, 15069–15075 CrossRef CAS PubMed.
- R. Cai, H. Xiang, D. Yang, K. T. Lin, Y. Wu, R. Zhou, Z. Gu, L. Yan, Y. Zhao and W. Tan, J. Am. Chem. Soc., 2021, 143, 16113–16127 CrossRef CAS PubMed.
- W. Deng, K. J. McKelvey, A. Guller, A. Fayzullin, J. M. Campbell, S. Clement, A. Habibalahi, Z. Wargocka, L. Liang, C. Shen, V. M. Howell, A. F. Engel and E. M. Goldys, ACS Cent. Sci., 2020, 6, 715–726 CrossRef CAS PubMed.
- Y. Li, J. Yang, G. Gu, X. Guo, C. He, J. Sun, H. Zou, H. Wang, S. Liu, X. Li, S. Zhang, K. Wang, L. Yang, Y. Jiang, L. Wu and X. Sun, Nano Lett., 2022, 22, 963–972 CrossRef CAS PubMed.
- Y. K. Hou, Z. J. Zhang, R. T. Li, J. Peng, S. Y. Chen, Y. R. Yue, W. H. Zhang, B. Sun, J. X. Chen and Q. Zhou, ACS Appl. Mater. Interfaces, 2023, 15, 2602–2616 CrossRef CAS PubMed.
- Y. Pan, W. Tang, W. Fan, J. Zhang and X. Chen, Chem. Soc. Rev., 2022, 51, 9759–9830 RSC.
- H. Li, Q. Luo, H. Zhang, X. Ma, Z. Gu, Q. Gong and K. Luo, Chem. Soc. Rev., 2023, 52, 47–96 RSC.
- S. Liang, X. Deng, P. Ma, Z. Cheng and J. Lin, Adv. Mater., 2020, 32, e2003214 CrossRef PubMed.
- A. K. Wood and C. M. Sehgal, Ultrasound Med. Biol., 2015, 41, 905–928 CrossRef PubMed.
- P. Saccomandi, A. Lapergola, F. Longo, E. Schena and G. Quero, Int. J. Hyperther., 2018, 35, 398–418 CrossRef PubMed.
- A. P. McHale, J. F. Callan, N. Nomikou, C. Fowley and B. Callan, Adv. Exp. Med. Biol., 2016, 880, 429–450 CrossRef CAS PubMed.
- K. Wu, H. Zhao, Z. Sun, B. Wang, X. Tang, Y. Dai, M. Li, Q. Shen, H. Zhang, Q. Fan and W. Huang, Theranostics, 2019, 9, 7697–7713 CrossRef CAS PubMed.
- W. Sun, L. Luo, Y. Feng, Y. Qiu, C. Shi, S. Meng, X. Chen and H. Chen, Adv. Mater., 2020, 32, e2000377 CrossRef PubMed.
- Q. Wang, N. Liu, Z. Hou, J. Shi, X. Su and X. Sun, Adv. Healthcare Mater., 2021, 10, e2000802 CrossRef PubMed.
- G. D. Wang, H. T. Nguyen, H. Chen, P. B. Cox, L. Wang, K. Nagata, Z. Hao, A. Wang, Z. Li and J. Xie, Theranostics, 2016, 6, 2295–2305 CrossRef CAS PubMed.
- X. Yu, X. Liu, W. Wu, K. Yang, R. Mao, F. Ahmad, X. Chen and W. Li, Angew Chem. Int. Ed. Engl., 2019, 58, 2017–2022 CrossRef CAS PubMed.
- D. Dinakaran, J. Sengupta, D. Pink, A. Raturi, H. Chen, N. Usmani, P. Kumar, J. D. Lewis, R. Narain and R. B. Moore, Acta Biomater., 2020, 117, 335–348 CrossRef CAS PubMed.
- L. Luo, W. Sun, Y. Feng, R. Qin, J. Zhang, D. Ding, T. Shi, X. Liu, X. Chen and H. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 12591–12599 CrossRef CAS PubMed.
- Y. Tian, Z. Liu, H. Tan, J. Hou, X. Wen, F. Yang and W. Cheng, Int. J. Nanomed., 2020, 15, 401–418 CrossRef CAS PubMed.
- H. Tan, Y. Tian, H. Yang, Z. Liu, X. Liang, B. Li and W. Cheng, J. Biomed. Mater. Res. B Appl. Biomater., 2021, 109, 1796–1806 CrossRef CAS PubMed.
- M. S. Khan, J. Hwang, Y. Seo, K. Shin, K. Lee, C. Park, Y. Choi, J. W. Hong and J. Choi, Artif. Cell Nanomed. Biotechnol., 2018, 46, S318–s327 CrossRef PubMed.
- X. Wang, C. Wang, H. Tian, Y. Chen, B. Wu and W. Cheng, Int. J. Nanomed., 2023, 18, 6199–6212 CrossRef CAS PubMed.
- Y. J. Kang, D. C. Yang, L. Kong, M. Hou, Y. Q. Meng, L. Wei and G. Gao, Nucleic Acids Res., 2017, 45, W12–w16 CrossRef CAS PubMed.
- M. Punta, P. C. Coggill, R. Y. Eberhardt, J. Mistry, J. Tate, C. Boursnell, N. Pang, K. Forslund, G. Ceric, J. Clements, A. Heger, L. Holm, E. L. Sonnhammer, S. R. Eddy, A. Bateman and R. D. Finn, Nucleic Acids Res., 2012, 40, D290–D301 CrossRef CAS PubMed.
- A. Bateman, E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall and E. L. Sonnhammer, Nucleic Acids Res., 2002, 30, 276–280 CrossRef CAS PubMed.
- M. F. Lin, I. Jungreis and M. Kellis, Bioinformatics, 2011, 27, i275–i282 CrossRef CAS PubMed.
- C. Trapnell, B. A. Williams, G. Pertea, A. Mortazavi, G. Kwan, M. J. van Baren, S. L. Salzberg, B. J. Wold and L. Pachter, Nat. Biotechnol., 2010, 28, 511–515 CrossRef CAS PubMed.
- A. C. Frazee, G. Pertea, A. E. Jaffe, B. Langmead, S. L. Salzberg and J. T. Leek, Nat. Biotechnol., 2015, 33, 243–246 CrossRef CAS PubMed.
- M. D. Young, M. J. Wakefield, G. K. Smyth and A. Oshlack, Genome Biol., 2010, 11, R14 CrossRef PubMed.
- M. Kanehisa, M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda, T. Tokimatsu and Y. Yamanishi, Nucleic Acids Res., 2008, 36, D480–D484 CrossRef CAS PubMed.
- X. Mao, T. Cai, J. G. Olyarchuk and L. Wei, Bioinformatics, 2005, 21, 3787–3793 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5na00042d |
| This journal is © The Royal Society of Chemistry 2025 |