On the design of cell membrane-coated nanoparticles to treat inflammatory conditions
Andreia
Marinho
 ab,
Salette
Reis
ab,
Salette
Reis
 a and
Cláudia
Nunes
a and
Cláudia
Nunes
 *ac
*ac
aLAQV, REQUIMTE, Faculdade de Farmácia, Universidade do Porto, R. Jorge de Viterbo Ferreira 228, 4500-313 Porto, Portugal. E-mail: cdnunes@ff.up.pt
bLAQV, REQUIMTE, Faculdade de Ciências, Universidade do Porto, R. Campo Alegre s/n, 4169-007 Porto, Portugal
cLAQV, REQUIMTE, Instituto de Ciências Biomédicas Abel Salazar, Universidade do Porto, R. Jorge de Viterbo Ferreira 228, 4500-313 Porto, Portugal
First published on 30th October 2024
Abstract
Biomimetic-based drug delivery systems (DDS) attempt to recreate the complex interactions that occur naturally between cells. Cell membrane-coated nanoparticles (CMCNPs) have been one of the main strategies in this area to prevent opsonization and clearance. Moreover, coating nanoparticles with cell membranes allows them to acquire functions and properties inherent to the mother cells. In particular, cells from bloodstream show to have specific advantages depending on the cell type to be used for that application, specifically in cases of chronic inflammation. Thus, this review focuses on the biomimetic strategies that use membranes from blood cells to target and treat inflammatory conditions.
1. Introduction
Clinically characterized by pain, heat, redness, swelling, and dysfunction of the affected tissue,1,2 inflammation represents a protective response of the organism that is triggered by harmful stimuli,3–5 such as pathogens, damaged cells, reactive oxygen species (ROS), and metabolic stress.5 After the harmful stimuli, a complex and coordinated cascade of interactions1,3 between pro- and anti-inflammatory mediators is initiated.1 Normally, this reaction is beneficial – acute inflammation – and ends when the harmful stimuli are eliminated and the tissue is repaired.5 However, if disorders occur that disturb the balance between pro- and anti-inflammatory mediators, acute inflammation can progress to chronic inflammation,1,4,6 contributing to a wide range of chronic inflammatory diseases.4,6In conditions of chronic inflammation, it is necessary to resort to the use of anti-inflammatory drugs that help regulate or suppress inflammation.7 Anti-inflammatory drugs can be divided into several classes, including non-steroidal anti-inflammatory drugs (NSAIDs) and glucocorticoids (GCs), which are prescribed daily at early stages of chronic conditions.7,8 Despite representing the two most prescribed classes of anti-inflammatory drugs worldwide,9 NSAIDs and GCs are associated with several side effects.7–9 Biological drugs, a more recent class, are also being prescribed for the treatment of these pathologies.2,10
The administration of drugs loaded into nanoparticles (NPs) allows overcoming the pharmacokinetic and pharmacodynamic limitations associated with conventional drugs,11–16 which makes them suitable drug delivery vehicles.12 Nanotechnology applied to health – nanomedicine, has achieved remarkable progress, yet understanding the detailed way by which NPs interact with the intrinsic environment in living organisms remains a huge challenge.17–19
Despite increasing the half-life of drugs and allowing greater accumulation at the site of injury, in vivo, NPs face a series of biological barriers6,12,20–22 that, in clinical practice, limit the achievement of adequate therapeutic results.12,20–22 These barriers include opsonization12,20,23–26 and subsequent sequestration by the mononuclear phagocyte system (MPS),12,22,24,25,27,28 nonspecific distribution,12,25 hemorheological flow,12,28 pressure gradient,12 cellular internalization,12,25 leakage from endosomal and lysosomal compartments12 and efflux pumps.12,20 Furthermore, it is essential to recognize that the complexity of these barriers can vary depending on factors like the route of administration, the type of disease, and its stage of progression.12 Additionally, each patient has unique characteristics, which will also contribute to the variability and complexity of the biological barriers that NPs need to face.
To overcome these barriers, the adsorption of hydrophilic polymers, such as polyethylene glycol (PEG), on the surface of NPs can significantly increase their circulation time.20,23,26,29–33 PEGylation involves the grafting of PEG to the surface of NPs, where the ethylene glycol units associate with water molecules, leading to the formation of a hydration layer. This layer, in turn, prevents the adsorption of proteins on the surface of NPs and subsequent clearance by MPS.12,13,23,34,35 As a consequence, the likelihood of passive extravasation into tissue with enhanced permeability and retention (EPR), as in tumor tissues and inflammatory lesions, increases.13,20,22,25,32 The circulation time of PEGylated NPs depends on the length and density of the PEG chains and can range from minutes to several days.26
However, PEG-functionalized NPs have some disadvantages, including: (1) PEG does not confer cell-specific binding capacity on NPs,36,37 and (2) it can induce an “anti-PEG” immune response,13,14,22,25,32,33,35–38 leading to an accelerated clearance rate of PEGylated NPs after multiple doses.22,23,29,32,37,38 There are however some alternatives to PEGylation which include synthetic hydrophilic molecules,39,40 such as zwitterionic units (for example poly(carboxybetaine), poly(sulfobeteine)), hydroxyl group, dendritic polyglycerol sulfate, polyoxazolines39 and other natural polymers.39,40
When designing NPs it is of utmost importance that they demonstrate excellent performance in the complex environments they will encounter in vivo. Hence, considerable effort has been dedicated to investigating techniques aimed at enhancing the interaction of NPs with biological systems.23,41,42 The majority of NPs are artificially created, posing a challenge due to their synthetic nature.21,41 The external composition of NPs can create numerous hurdles, given the human body's expertise in eliminating non-endogenous substances.24,32,41,43 Furthermore, it can be difficult to artificially replicate the biological interactions that occur naturally in the organism.19,41,44
To overcome all these hurdles, there has been a paradigm shift towards cell-based strategies for NPs design. In contrast to the relatively simple components and structures of conventional NPs, cells have a wide variety of tactics to evade attacks by the immune system.21 Escape from the immune system can be achieved by mimicking the physiological mechanisms that regulate cell surface signaling to suppress protein adsorption or selective cell adhesion.20 This can be achieved in several ways, namely by mimicking the physical properties of cells, such as shape and flexibility, or by harnessing materials that naturally derive from cells, such as proteins or antibodies.35
Due to these attractive features, cell-based targeting tactics are very promising for drug delivery due to their high specificity and long-term persistence.21 In this regard, it is of great interest to the scientific community to mimic cellular functions. One of the main strategies for this purpose is the use of liposomes. Structurally, liposomes are composed of at least a phospholipid bilayer and an aqueous core,45–47 thus representing a simplified model of the cell membrane.48,49 Due to their structure, they can trap hydrophilic molecules in the aqueous compartment and hydrophobic molecules in the membrane.47,50,51 As drug delivery systems, they have several advantages, including the possibility of surface functionalization, size adjustment,46,50 modulation of the number of layers, biocompatibility, and biodegradability.46 Despite these characteristics, liposomes remain exogenous to the organism and, therefore, are recognized by MPS and consequently eliminated by the bloodstream,52 in addition to the fact that it is difficult to replicate the complex functionality of the cell membrane50 and the lack of this complexity also contributes to liposomes being more unstable structures, which in itself is a major limitation.49 Thus, they are increasingly being redesigned following a biomimetic approach.
This review provides an overview of the currently used biomimetic techniques, focusing on cell membrane-based biomimetic NPs (Fig. 1). In this sense, the techniques for extracting cell membranes and the methods used for the production of biomimetic NPs are presented, as well as a survey of the most recent studies in the area. In the final section, some examples of non-extracted elements are also highlighted, demonstrating their utilization in emulating biological characteristics.
2. Biomimetics
Biomimetics is an emerging field of science that studies how nature designs, processes and assembles/disassembles molecular building blocks to manufacture high-performance materials and applies these processes to design new molecules and structures with unique properties.20Biomimetic systems attempt to recreate the complex interactions that naturally occur between cells.53 For this, is important to mimic the surface composition, shape and movement of normal cellular physiology.23,54 In other words, biomimetic systems aim to interact directly with the immune system rather than trying to hide from it.30
Normally, surface biofunctionalization of NPs is a bottom-up technique,55 where only some portions are incorporated into the NPs surface through chemical conjugation or non-covalent binding.55,56 This indicates that most of times, only well-known biological interactions are explored.55 But in reality, the efficient biological interface is often multifactorial35,55 and involves mechanisms that are not yet fully understood.55 It also happens that, due to the variety and complexity of proteins on the surface of a cell, it is impractical to synthesize all proteins by conventional chemical methods.13,29,36 Thus, the direct use of the cell membrane (top-down technique) has many advantages, including the ability to prevent various functional moieties in their natural context without the need to resort to complicated methods to identify, synthesize and conjugate individual linkers,25,41,55 and prevent loss of integrity and functionality during the formulation and delivery processes.34
Despite this, the use of bottom-up strategies allows superior physical–chemical control at the end of the process, and chemical conjugation methods remain insufficient to reproduce the membrane complexity. On the other hand, top-down strategies are associated with the difficulty of establishing a standardized protocol.57
Biomimetic nanotechnology takes advantage of the unique biological makeup of cell membranes and combines it with the custom flexibility of synthetic NPs13,29,34,36,38,58–61 and a wide range of payloads to improve targeted delivery.34,36,62 The general concept is to surround NPs with cell-derived membranes that provide complex biological entities found in cell membranes,35,36,63 in a core–shell type design.25,41,63 This biofunctionalization results in evasion-immune properties, reducing the absorption of proteins that lead to opsonization and “self” signals through the surface proteins,29,36,58,59 thus prolonging circulation time in blood,35,39,59 extremely necessary characteristics for the EPR effect.59 On the other hand, biofunctionalization also gives synthetic NPs a natural homing capability,29,58,64 that can be used as an active targeting model.29,38,55,58 Natural lipid components based on cell membranes on the surface of NPs also provide greater colloidal stability and delay drug release (acting as a “molecular fence”).58 On the other hand, the phospholipid bilayer confers biocompatibility to NPs.26
Initially, cell membrane-coated NPs (CMCNPs) were fabricated using a combination of red blood cell membrane and poly lactic-co-glycolic acid (PLGA) through a co-extrusion approach.41,55,59 These particles exhibited significantly enhanced circulation,23,41 which resulted in the preservation of red blood cell self-markers such as CD47 on the surface of NPs.41 Subsequently, a plethora of different platforms have been reported, given the flexibility to choose different membrane materials and different NPs cores.41,65 In the process of designing and developing biomimetic systems, shape, texture and biomimetic movements are factors that must be taken into account.54
Synthetic NPs are, in general, spherical, and rigid in structure, so mimicking the structure (shape and texture) of cells can contribute to increased targeting.20,54 It is reported that elongated filamentous polymer particles have longer circulation times than spherical NPs, probably due to their ability to align with the bloodstream.66 The incorporation of components that adequately respond to the physicochemical characteristics of the target site (such as pH) is also a crucial factor since it can contribute to the adequate release of the drugs they transport.20,54 Conversely, mimicking natural organism movements can offer distinct advantages.20,54 This approach enables the creation of systems characterized by predictable motion, capable of accessing even the deepest tissues.20 Such functionality can be attained through processes like the conversion of adenosine triphosphate to adenosine diphosphate, or similar mechanisms,54 by emulating heartbeats or brain rhythms.20
3. Cell membrane
Cells, one of the fundamental units of life,35,67 are surrounded by highly complex membranes consisting of a lipid bilayer and proteins,68,69 arranged in a highly organized matrix that transmits and receives signals and can manage normal cellular function.68 Functioning as a semipermeable barrier,26,63 the cell membrane separates the inside of the cell from the external environment,26,63,70,71 controlling the traffic of large molecules and ions into and out of the cell.19,26 Thus, lipid integrity is essential for cell function.26The presence of specific biomolecules in the lipid bilayer is critical for the performance of specific functions that ultimately lead to cell–cell interactions, cell signaling and modulation of immunity.44,72,73 Membrane-bound enzymes are responsible for lipid metabolism,19 while membrane-bound proteins are responsible for ion and nutrient transport,19,71 and are also involved in intracellular signaling processes.19 On the other hand, lipids have a structural function, since their amphiphilic nature favors the formation of a bilayer structure.74
Using the cell membrane directly on the surface of NPs faithfully preserves the complexity of the membrane (composition – with all its lipids, proteins, and carbohydrates – and structure).35 This preservation enables NPs coated with the resulting cell membrane to leverage many of the properties exhibited by the parent cell.35 This top-down strategy offers numerous therapeutic opportunities, from cell simulation to multifaceted biointerface properties.3
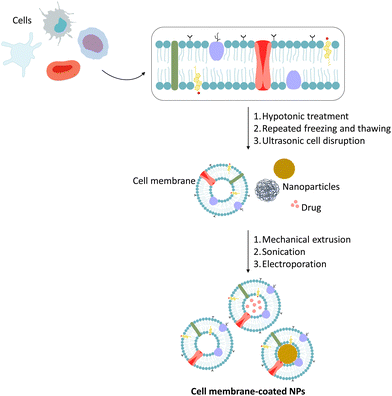
The preparation of CMCNPs is divided into 3 essential steps: membrane extraction, preparation of the NPs and coating of the NPs with membrane.49,67,75 Of these steps, step 3 is the most important, as it will determine the success of the preparation of CMCNPs.49 Different cell types, including red blood cells (RBCs), platelets, leukocytes, stem cells, and cancer cells, have been used as membrane sources for constructing biomimetic systems.64,72,76,77
3.1. Cell membrane extraction
Cell membrane extraction involves three main steps: lysis, membrane collection, and vesicle formation. While procedures may vary, the process typically falls into two categories: cells without a nucleus and cells with a nucleus. For anucleated cells like RBCs and platelets, isolation is achieved via centrifugation, followed by lysis using hypotonic solutions or freeze/thaw cycles.3,67,73,78,79 Then the membrane fragments can be purified by centrifugation and subsequently serially extruded through polycarbonate membranes with decreasing pore size.3,73,78,79 RBCs and platelets can be isolated directly from blood,56 while other cells must first be isolated from tissues and then cultured in vitro.49,56For nucleated cells, membrane extraction is more complex. After isolating cells from cultures, blood, or tissue, a combination of hypotonic lysis, mechanical disruption, and sucrose gradient centrifugation removes the nucleus and cytoplasm.3,64,73,78 DNase and RNase use, help eliminate nuclear components, and reduce concerns due to the administration of genetic material.64 Membranes are then washed with isoionic buffers, followed by additional sonication or extrusion through polycarbonate membranes. The entire extraction step should be performed at 4 °C and as gently as possible. Moreover, protease inhibitors and endotoxin-free solutions are highly recommended for storage of cell membrane-based vesicles at −80 °C to achieve long-term maintenance of membrane proteins function.3,78
Cells isolated from blood must undergo a sterilization process to avoid infections and, in the case of using blood that does not come from the patient, the blood must be screened to guarantee maximum compatibility.15 It should also be noted that membrane extraction approaches are still limited and that they are associated with low yields.64
3.2. NPs coating with membranes
The coating of NPs with the cell membrane must take into account several aspects to successfully obtain CMCNPs, including the surface charge and the size of the NPs.64 Electrostatic interactions between cell membranes and NPs are a key factor in the manufacture of CMCNPs.49,64 Thus, NPs must have a surface charge opposite to the charge on the inner leaflet of the membrane.64After obtaining the membrane and the NPs, these two materials must be fused so that the membrane can cover the surface of the nucleus, producing biomimetic NPs. Currently, there are three coating methods in use: membrane extrusion, sonication, or electroporation.23,63,70 Briefly, in the extrusion process, the cell membrane and synthetic NPs are co-extruded several times through a polycarbonate membrane.49,63,64,70 The sonication process consists of incubating the extracted cell membranes with the synthetic NPs under ultrasonic waves.63,70 In this process, the sonication parameters (such as power, frequency and duration) must be optimized as a way to minimize proteins denaturation and maximize the rate of fusion between the two components.64 Electroporation consists of inducing several pores in the cell membrane through which NPs diffuse into the membranes.63,70 More details about each production's method have already been reviewed in the literature and can be found elsewhere.49,63
Once the CMCNPs are obtained, it becomes necessary to proceed with their characterization. Several techniques have already been reported to assess the stability of these structures. For example, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)56,80,81 and Western blotting to assess the presence of specific proteins,56 scanning and transmission electron microscopy (SEM and TEM, respectively),56,80–82 dynamic light scattering (DLS),56,80,81 lipophilic dye enhanced/advanced fluorescence and/or spectrophotometric techniques for structure evaluation.82
Recently, Liu et al. developed a fluorescence quenching assay to calculate the percentage of fully coated NPs. The work carried out consisted of using synthetic NPs labeled with nitro-2,1,3-benzoxadiazol-4-yl (NBD) coated with membranes extracted from cells and subsequently treated with dithionite (reducing regent that cannot cross membranes). Since dithionite is not able to cross membranes, in NPs completely coated with the membrane, the fluorescence signal of NBD does not change. If the NPs are partially coated or uncoated, the fluorescence intensity changes as dithionite is added. Thus, biomimetic NPs can be distinguished into three classes: uncoated, partially coated, or fully coated NPs.80
4. Cell membrane-derived nanostructures
Due to their bilayer structure, cell membranes allow the attachment of transmembrane proteins without loss of functionality and reliability during the production process of biomimetic NPs. This allows all portions biologically relevant for immune evasion and targeting to be translocated into the NPs.63 With stable and functional compartments, cell membranes preserve cell architecture and support signaling and selective transport mediated by membrane proteins, which is why cell membranes have been used in biomedical applications.83Compared to conventional NPs, biomimetic NPs have more advantages, including: (1) transport specificity;16,54,76 (2) biodegradability; (3) low immunogenicity;54,76 (4) ability to reach hard-to-reach sites, such as the blood–brain barrier, (5) drug release can be driven by means of inactive transport, exocytosis or efflux pumps that can be defined by biological signs,16 and (6) sensitivity and reactivity to the microenvironment.76 Furthermore, CMCNPs coated with different membranes show different characteristics.76,84 For example, RBCs membrane coating of NPs generally prolongs blood circulation time76,84 whereas platelet membrane coating promotes adhesion of NPs to damaged vasculature.76 In the next sections, the different cell membranes that have been used to produce CMCNPs for the treatment of inflammatory conditions will be presented.
4.1. RBCs-derived nanostructures
RBCs, one of the most abundant cell types in the bloodstream,49,64,72 are responsible for transporting oxygen and carbon dioxide to and from tissues.79,85 They are capable of shapeshifting,56 which allows them to leak through capillaries smaller than their diameter.15,73 The presence of immunomodulatory proteins in the membrane increases its circulation time49,72 and contributes to immunoevasive properties.72 An example of this is the transmembrane protein CD47,56,72,73,86 which inhibits phagocytosis72,86,87 and anti-inflammatory responses that reduce the immune behavior of macrophages.72Their membranes have been used for the production of drug transporters due to the inherent characteristics that these cells have: (1) prolonged circulation – about 120 days in humans,16,49,56,77 (2) high capacity for drug loading,77 (3) biocompatibility,16,56,73,76,77,88 (4) biodegradability16,56,73,77 and non-immunogenicity.73,76,77,88 In addition to these advantages, it has also been reported that NPs coated with the RBCs membrane exhibit greater structural rigidity and stability when compared with the uncoated NPs.86 Furthermore, they are cells without a nucleus and mitochondria,16,49,64,76,82 which facilitates membrane extraction and purification processes.49,64,82
Fan et al., used RBCs membranes to coat PLGA nanoparticles into a macroporous alginate scaffold. In fact, the presented results show that in the absence of the RBCs membrane, high levels of IL-12 and tumor necrosis factor α (TNF-α) are produced while with the nanoparticles coated with the RBCs membrane, these levels are significantly reduced.87
In another approach, Safarpour et al., developed RBCs membrane-derived mimetic liposomes for the delivery of curcumin in the healing process of chronic wounds. The results show that liposomes, in addition to being biocompatible, are capable of promoting fibroblasts cell proliferation, which highlights their potential application in wound healing.89
A summary of RBCs-derived nanostructures can be found in Table 1.
| Disease | Core | Payload | Production method | Circulation time | Ref. |
|---|---|---|---|---|---|
| Alzheimer | Carbon quantum dots | Polydopamine | Incubation | 12 h | 90 |
| Polycaprolactone NPs | Curcumin | Sonication and extrusion | — | 91 | |
| Atherosclerosis | Micelles | Simvastatin | Incubation | 26.84 h | 92 |
| PLGA NPs | Probucol | Sonication | 66 h | 93 | |
| Diabetic wounds | Liposomes | Curcumin | Extrusion | — | 94 |
| Lupus erythematosus | PLGA NPs | Cyclosporine A | Sonication and extrusion | — | 95 |
| Liposomes | Mycophenolic acid | Sonication and extrusion | — | 96 | |
| Wound healing | Alginate scaffold | — | Incubation | — | 87 |
| Liposomes | Curcumin | Sonication | — | 89 | |
| Selenium NPs | — | Sonication and extrusion | — | 97 | |
| Stellate ganglion | PLGA microparticles | Curcumin | Sonication | — | 98 |
4.2. Platelets-derived nanostructures
Responsible for the process of blood clotting79,85 and hemostasis,15,64,72,85 platelets are cells without a nucleus,15,49,85 that have a circulation time of 7–10 days.16,49,64,82 When vascular injury occurs, platelets are responsible for repairing ruptured vessels82,88,99 through the release of granules that promote angiogenesis and the recruitment of regenerative cells.82 Unlike RBCs, which are found in the center of blood vessels, platelets are concentrated near blood vessel walls. This characteristic means that platelets are permanently on guard and provide a quick response.85 When compared to other cells, platelets express fewer antigens and are less immunogenic.49 Despite this, they are highly reactive and sensitive, showing the ability to naturally target the sites of injury, properties that are due, in part, to the receptors that they present in the membrane.76Due to its storage capacity and high payload,16 and significant roles in hemostasis, angiogenesis, inflammation, and wound healing,77 the platelet membrane is interesting for use in the development of drug delivery systems.16 It should also be noted that the availability of this type of cell is also a major factor,56 in addition to the fact that specific ligands are expressed on the platelet membrane, such as CD47 and CD55/59,56,73,84 which allow, respectively, immune evasion56,84 and prevent activation of the complement system.56 It is known that the presence of CD47 on the platelet membrane allows its interaction with the signal-regulatory protein α (SIRPα) present in immune cells and, thus, inhibiting the clearance of NPs mediated by the immune system, which causes the NPs coated with the platelet membrane to have a longer circulation time in the bloodstream.100
Based on these properties, Jin et al. developed PLGA NPs coated with platelet membrane loaded with curcumin and resveratrol for the treatment of acute lung injury. The results obtained in vitro showed that biocompatibility increases compared to free drugs. In addition, it was also found that free compounds have some hemolytic activity that is overcome with the use of NPs. In mice with induced acute lung injury, biomimetic NPs, administered intranasally, have been shown to promote accumulation and retention in inflamed lungs. Four hours after administration, the IVIS images showed that the NPs under study accumulate only in the lungs, thus evidencing the specific targeting. It was also verified that the pulmonary vascular permeability and the levels of IL-6, TNF-α, intercellular adhesion molecule 1 (ICAM-1) and inducible nitric oxide synthase (iNOS) decreased 24 h after the administration of the biomimetic NPs. Furthermore, it was found that polarization of macrophages from an M1 state (inflammatory) to an M2 state (anti-inflammatory) occurred, which shows the anti-inflammatory capacity of these biomimetic NPs.101
In another approach, He et al. also used PLGA NPs loaded with FK506 (tacrolimus) and coated with platelet membrane for the treatment of rheumatoid arthritis. Using an in vitro model of the synovial membrane, consisting of collagen IV and synoviocytes, it was found that the NPs under study could adhere to intercellular collagen IV, due to the affinity between collagen and glycoprotein VI present in the platelet membrane. In vivo, fluorescence signals (using the NIRF imaging system) indicate that NPs coated with the platelet membrane selectively accumulate in the inflamed joint, leading to a decrease in inflammation. Blood circulation time also increased and biodistribution studies show a preferential accumulation in inflamed areas, despite also verifying the presence of NPs in the liver and spleen, which are the natural pathways for platelet clearance. In addition, treatment with NPs coated with platelet membrane decreased erosions in bone tissue.102
A summary of platelets-derived nanostructures can be found in Table 2.
| Disease | Core | Payload | Production method | Circulation time | Ref. |
|---|---|---|---|---|---|
| Acute lung injury | PLGA NPs | Curcumin and resveratrol | Extrusion | — | 101 |
| Asthma | PEG/PLGA NPs | Berberine | Incubation and sonication | — | 103 |
| Atherosclerosis | PLGA NPs | Rapamycin | Sonication | — | 104 |
| Exosomes | miRNAs | Sonication and extrusion | — | 105 | |
| PLGA NPs | Glyburide | Extrusion | — | 106 | |
| Bone regeneration | Bioactive glass | — | Sonication | — | 107 |
| Colitis | PEG-PLGA NPs | Patchouli alcohol | Sonication | — | 108 |
| Coronary artery diseases | PLGA NPs | Rapamycin | Sonication | — | 109 |
| Restenosis | Bovine serum albumin- polyethyleneimine | pcDNA3.1-VEGF165 | Incubation | 48 h | 110 |
| Rheumatoid arthritis | PLGA NPs | FK506 | Sonication | — | 102 |
| Ulcerative colitis | PLGA NPs | Platinum/carbon dot nanozymes and piceatannol | Sonication | 2.03 h | 111 |
| Vascular restenosis | PEG NPs | Epigenetic inhibitor (JQ1) | Sonication | — | 112 |
| Wound healing | PLGA NPs | Basic fibroblast growth factor (bFGF) and vascular endothelial growth factor A (VEGFA) | Sonication | — | 113 |
4.3. Leukocytes-derived nanostructures
Leukocytes originate in the bone marrow, where they begin their life cycle by migrating into the bloodstream.25,114 As cells of the immune system, their main function is to protect the body against infectious diseases and foreign invaders,17,23,25,27,30,32,61,115 yet they are heavily involved in inflammatory disorders and cancer.21,61 When body tissues are damaged by infection or injury, generating an inflammatory response, leukocytes are recruited from the bloodstream to sites of inflammation17,25,32,116 to eliminate pathogens through phagocytosis.17 In this physiological process, surface interactions between leukocytes and the endothelium play crucial roles in the recruitment of immune cells into disease-target regions, due to the overexpression of endothelial adhesion molecules (e.g., integrins) to selectively bind with ligands expressed on leukocyte surfaces (e.g., selectins).17,73Generally speaking, leukocyte trafficking can be divided into two phases: movement within the vascular circulation and migration within tissues. The vascular and lymphatic systems are lined by a monolayer of endothelial cells that grow on the basement membrane. These endothelial cells form organized intercellular junctional zones, in this way, the endothelium serves as the main (selectively permeable) barrier between the circulation and the underlying tissues. Each change of environment during this traffic (i.e., movement inwards – intravasation – or outwards – extravasation – from the circulation) requires leukocytes to cross the endothelium (diapedesis).114 They move in an amoeboid fashion, which allows their rapid and effective extravasation from the blood to the tissues,23,25,64,78 making these cells abundant both in the circulation and in extravascular sites.23,25,78
In circulation, leukocytes are divided into five main classes,6,23,61,78,117 based on their morphological and functional characteristics.6,44 Different types of leukocytes are involved in the pathogenesis or prevention of diseases, for example, chronic inflammation is mainly associated with monocytes and lymphocytes while acute inflammation is associated with the activity of granulocyte leukocytes (neutrophils, eosinophils, and basophils).23,61,78 Many of the biological activities of leukocytes are, however, mediated by the molecules they present on the surface of the membrane, which include CD47, lymphocyte function-associated antigen-1 (LFA-1) and macrophage antigen-1 (MAC-1).73
As drug carriers, leukocytes are very attractive. On the one hand, they can be used as drug carriers, thus prolonging the half-lives of drugs, on the other hand, leukocytes have the inherent ability to target inflamed tissues.37 On their surface, leukocytes have several membrane receptors, specific targeting ligands and surface proteins, which makes them exploited for drug delivery, due to their long blood circulation time, easy passage of biological barriers,21,39,49,118 identification of pathologic/inflammation regions and transendothelial migration properties.118
Despite the controllable physical–chemical parameters (e.g., size, component, surface ligands functionalization, homogeneity), it is still highly demanding to reproduce the integrity and complexity of the leukocyte membrane.17 The leukocyte membrane contains a variety of proteins and antigens that retain some of the functions of white blood cells. Their membrane proteins play a crucial role in various cellular processes such as intercellular communication and signal transmission between cells.27
To achieve cell surface functionalization, therapeutics are linked to leukocyte membranes primarily through three techniques: (1) receptor-mediated adhesion – is credible, reproducible, and capable of triggering cell receptor activation and signal transduction (for example, B cells abundantly express the CD44 cell surface receptor which may be useful for attaching hyaluronic acid (HA)-based NPs via the HA-CD44 interaction);65 (2) covalent binding - this binding mode is stronger and stabler than receptor-mediated adhesion. Functional groups on cell surface proteins, such as amino (lysine-NH2) or thiol (cysteine-SH), can be used to modulate binding65 and (3) selectin-mediated adhesion – this is the most effective way to rapidly bind therapeutics to the cell surface of selectin ligands.65
When activated, neutrophils play a crucial role in inflammation.115 Their transmigration to inflamed sites is a highly specific process, so targeting this population of leukocytes may be a novel approach to treating inflammatory diseases.115 Leukocytes have an inherent capacity for chemotaxis, giving them the ability to migrate to a site of infection or inflammation within minutes. Since neutrophils are the first to respond to signals of inflammation, deformable neutrophils can transport cargo through blood vessels and interstitial tissue, following the guidance of chemical signals.116 However, a limitation of this class of leukocytes is their lifetime of less than one week, which is reduced to less than 2 days after activation.117
Neutrophil recruitment is regulated by various adhesion molecules expressed on neutrophils and endothelium,6,21 such as LFA-1 and integrin-1β.21 During inflammation, circulating neutrophils first recognize selectins expressed in the epithelium of blood vessels, then roll and crawl through the blood vessels. This movement of neutrophils facilitates contact with chemokines on the surface of the endothelium, which initiates neutrophil activation. Once activated, neutrophils travel to inflamed sites through cell junctions and transcellular pathways.6,21
Depending on the stimuli they receive, neutrophils can polarize to the N1 or N2 phenotype. The N1 type is anti-tumor and characterized by high levels of TNF-α and ICAM-1. The N2 phenotype is characterized by chemokine upregulation with chemokine ligands 2 and 3 (CCL2 and CCL3, respectively).25
However, other properties of neutrophils make them attractive for the development of nanotherapeutics: (1) while the lifetime of neutrophils is short in the circulation,6,117 the number of neutrophils can be increased by tens of hundreds of hours in a short period of inflammation, which would rapidly increase delivery;6 (2) 50–70% of circulating human leukocytes are neutrophils,6,115,117 therefore, targeting neutrophils may increase therapeutic efficacy and may be translational;6 (3) their migration is regulated by various membrane proteins and the cytokine/chemokine gradient,115 this chemotaxis behavior is a highly promising feature to be translated into nanosystems.72
Due to the properties that neutrophils have in combating inflammation, Wang et al. developed sparfloxacin-loaded NPs coated with the neutrophil membrane for the treatment of pulmonary inflammation (induced by methicillin-resistant staphylococcus aureus – MRSA), which proved to be biocompatible and capable of controlled release of sparfloxacin. In vivo, it appears that 2 hours after administration, NPs begin to accumulate in the lungs. It was also found that after treatment with the developed NPs, the levels of pro-inflammatory cytokines (IL-6, IL-8 and TNF-α) in the blood, inflammatory cells (neutrophils, macrophages, and monocytes) and MRSA in the lungs decreased. Histopathology results show that treatment with NPs prevented alveolar rupture and inflammatory cell infiltration.119
Hassanzadeh et al., developed silk fibroin-based NPs camouflaged with the neutrophils membrane to overcome the low solubility of ferulic acid, as a strategy against acute pancreatitis. The results obtained show that the developed NPs allow controlled release of ferulic acid and a targeted delivery to the inflamed tissues of the pancreas. Furthermore, there is a reduction in the levels of pro-inflammatory cytokines (IL-1β, IL-6 and TNF-α) and even suppression of oxidative stress. In vivo, using this approach to treat induced pancreatitis prevented histopathological changes, such as infiltration of inflammatory cells and disruption of tissue structure.120
In a different approach, Jiang et al. developed neutrophil membrane-camouflaged nanoprobes (Neu-NPs) for the imaging of inflamed high-risk atherosclerotic plaques by interacting with ICAM-1 receptors. Compared with the control probe, the Neu-NPs show more stability in serum and were found to have good targeting for atherosclerotic plaques in mice and rabbit atherosclerosis models. In vivo, after treatment (with rosuvastatin), Neu-NPs can show the reduction of inflammation, which is indicative that Neu-NPs can be used for therapeutic evaluation and drug screening.121
A summary of neutrophil-derived nanostructures can be found in Table 3.
| Membrane | Disease | Core | Payload | Production method | Circulation time | Ref. |
|---|---|---|---|---|---|---|
| Neutrophils | Acute lung injury | Liposomes | Acidic fibroblast growth factor | Extrusion | — | 122 |
| Acute myocardial infarction | PLGA NPs | IL-5 | Sonication | 72 h | 123 | |
| Asthma | Luminol-conjugated α-cyclodextrin NPs | — | Sonication and extrusion | — | 124 | |
| Atherosclerosis | Liposomes | Indocyanine green | Extrusion | — | 121 | |
| Cardiac regeneration | Mesoporous silica NPs | microRNA-10b | Extrusion | 7 days | 125 | |
| Intervertebral disc degeneration | PLGA NPs | Transforming growth factor-beta 1 (TGF-β1) | Extrusion | — | 126 | |
| Lung inflammation | Polycaprolactone-PEG NPs | Sparfloxacin | Sonication | — | 119 | |
| Myocardial ischemia-reperfusion injury | Calixarene NPs | Atorvastatin calcium | Sonication | — | 127 | |
| Pancreatitis | Silk fibroin NPs | Ferulic acid | Sonication | — | 120 | |
| Pneumonia | PLGA NPs | Gentamicin | Sonication | — | 128 | |
| Renal ischemia-reperfusion injury | — | IL-37 | Extrusion | — | 129 | |
| Rheumatoid arthritis | PLGA NPs | — | Sonication | — | 130 | |
| Sepsis | — | — | Sonication | 24 h | 131 | |
| HL-60 neutrophils | Acute lung inflammation/injury | — | Azlocillin and methylprednisolone sodium | Incubation | — | 132 |
| Endothelial inflammation | Liposomes | — | Incubation | — | 133 | |
| Ischemic stroke | Liposomes | Leonurine | Extrusion | 48 h | 134 | |
| Treg | Periodontitis | PLGA NPs | — | Extrusion | — | 135 |
B cells produce antibodies and antigens to establish humoral immunity in the adaptive immune system.6,25 They can specifically recognize antigens even when they are in low concentrations, in addition, they express the major histocompatibility complex (MHC) class II136 and co-stimulatory molecules (such as CD40, CD80, CD86 and OX40L), which allow them to prepare T cells.136,137
The B cell receptor (BCR) is expressed on its membrane, which together with other B cell-specific membrane proteins form a signaling complex, allowing induction of cell-instructive nuclear factor kappa B (NF-kB), phosphoinositide 3-kinase (PI3K), and mitogen-activated protein kinase (MAPK) signalling.138 Among these proteins are CD19, CD22, protein tyrosine phosphatase receptor type C (PTPRC; also known as B220),138 B cell antigen receptor complex-associated protein α chain (Igα; also known as CD79a) and β chain (Igβ; also known as CD79b).138,139 In addition to these proteins, others are also expressed on the surface of B cells, including CD21, CD23, CD24, CD40, CD72.139
NK cells are innate immune cells25,99,140 and secrete perforin to kill pathological cells without antigen stimulation.25,99,140 They regulate the immune response through the release of cytokines, such as TNF-α,140 and promote the maturation of antigen-presenting cells (APCs), which leads to the activation of T cells.99,140 They also have membrane proteins, such as receptor activator of nuclear factor kappa-B ligand (RANKL),140,141 DMAN-1,140 RAB-10, IRGM1, galectin-12 and cannabinoid receptor 1 (CB1)142 capable of interacting with membrane receptors on macrophages and induce its polarization to an M1 state.99,140,142 In addition, they have other receptors that are responsible for tumor recognition and death, including NKp30, NKp44, NKp46, NKG2D.142
The functions of T cells include: (1) attacking foreign invaders directly,6,21,25 (2) secreting cytokines to activate other immune cells, (3) enhancing the response of B cells.6,21 As with macrophages, T cells also divide into subtypes: (1) cytotoxic T cells – which detect and destroy damaged cells; (2) helper T cells – which are involved in activating cytotoxic T cells and regulate the immune response; and (3) memory T cells – can recognize foreign invaders previously encountered and mount a fast immune response.6
Once an invader is identified, T cells can generate specific responses that are customized to maximally eliminate pathogenic or pathogen-infected cells.21 In response to infection by pathogens, some T cells, called helper T cells (Th), generate cytokines that can alter the immune response, while other T cells, called cytotoxic T cells (CTLs), produce toxic granules that contain perforin and lytic enzymes,21 which induce the death of cells infected by pathogens.6,21 Once activated, T cells leave a lasting legacy of the antigens they encountered in the form of memory T cells (Tm). Throughout life, these memory cells will remember each specific pathogen encountered and can mount a strong and rapid response if the same pathogen is detected again,6,21 this is known as acquired immunity.21
T cell membranes are rich in protein, which gives them a variety of functions,82 including: (1) attacking foreign invaders directly; (2) activation of other immune system cells through cytokine release and (3) enhancing B cell response.6 In addition, T cell membrane proteins affect MPS uptake.82
Li et al., describe a biomimetic manipulation strategy for immunosuppression by crosstalk with regulatory T (Treg) cell membrane-coated nanoparticles.135 Treg cells are responsible for providing immunological balance by suppressing effector immune cells or by eliminating aberrant self-attacking clones.135 The results presented by the group show that the nanosystem can inhibit macrophage-osteoclast differentiation due to the inhibition of the macrophage colony stimulating factor (MCSF). Furthermore, maturation of dendritic cells is also inhibited due to the inhibition of dendritic cell costimulatory molecules. In the in vivo model of periodontitis, the results show that the nanosystem is able to suppress the excessive immune response, verifying a reduction in the levels of TNF-α, IL-1β, IL-6, INF-γ, matrix metalloproteinase 8 (MMP-8), monocyte chemoattractant protein-1 (MCP-1) and RANKL.135
Despite their inherent advantageous features, no works have been reported until now, using B and NK cells cell membranes.
A summary of lymphocyte-derived nanostructures can be found in Table 3.
In the bloodstream, monocytes can transmigrate and phagocytose dead cells to prevent the spread of the inflammatory response. However, the long permanence of monocytes (or macrophages) in tissues would cause inflammatory disorders.6 Despite the deleterious roles they play, monocytes are also known to aid in tissue regeneration.15 Due to their biological functions, monocytes have intrinsic targeting to inflamed sites.15
The expression of the CD14 and CD16 receptors on the monocyte membrane is responsible for their classification into three subtypes: (1) classic – CD14++CD16−; (2) intermediates – CD14++CD16+ or (3) non-classical – CD14+CD16++.141,143,144 It is known that each subtype plays divergent roles in different diseases, as well as differing in the ability to secrete cytokines and respond to pathogen-associated molecular patterns (PAMPs).144 Classical monocytes are phagocytic and secrete inflammatory mediators, whereas non-classical monocytes are poorly phagocytic and are suggested to secrete TNF-α in response to some stimuli, but less of other pro-inflammatory molecules. It has already been reported that intermediate monocytes are increased in pathologies such as asthma and rheumatoid arthritis, and that classic monocytes are involved in atherosclerosis.144 Furthermore, classical monocytes express high levels of CCR2, CD62L, CD11b, TRL4 and low levels of CX3CR1, non-classical monocytes express high levels of CX3CR1, CXCR4, HLA-DR and LFA-1 and low levels of CCR2 and CD11b.141
Shen et al. developed ginsenoside and catalase-coloaded porous PLGA nanoparticles, surface modified with monocyte cell membranes. The results obtained show that the developed NPs can eliminate free radicals, do not present cytotoxicity (HUVEC and RAW264.7 cell lines) for the tested concentrations and do not present hemolytic risk. It was also shown that these NPs can target endothelial cells that are highly involved in the atherosclerosis process, while at the same time being able to overcome early uptake by macrophages. In vivo, it was found that the NPs under study accumulate in atherosclerotic plaques and that they manage to delay the progression of the disease since there is a decrease in inflammatory mediators (TNF-α and IL-1β) and in hyperlipidemia. In addition, it was found that the administration of NPs did not significantly change liver parameters, thus demonstrating the biosafety of the nanosystems.145
Similarly, Wang et al. report that PLGA NPs loaded with rapamycin and coated with the monocyte membrane can reduce inflammation and alleviate ischemia/reperfusion injury by preventing monocyte infiltration and suppressing microglia proliferation. In vitro results show that coating NPs with the monocyte membrane significantly reduces phagocytosis, which is indicative of prolonged circulation time. Adhesion of monocytes to the inflamed endothelium was also achieved after treatment with the NPs under study. In vivo, after 6 days it was still possible to detect NPs in circulation. Furthermore, biodistribution studies have shown that NPs preferentially accumulate in the brain, which may reduce side effects and nonspecific toxicity. In vivo anti-ischemic stroke effect shows that NPs can reduce the infarct volume and improve the neurological deficit. In cases of brain injury due to ischemia/reperfusion, proliferation of microglia occurs, which, in turn, induces the recruitment of macrophages. However, after treatment with the developed NPs, the number of microglia and macrophages decreases, which is indicative of a decrease in inflammation.146
A summary of monocyte-derived nanostructures can be found in Table 4.
| Membrane | Disease | Core | Payload | Production method | Circulation time | Ref. |
|---|---|---|---|---|---|---|
| Monocytes | Atherosclerosis | PLGA NPs | Verteporfin | Sonication | — | 147 |
| Atherosclerosis | PLGA NPs | Ginsenoside and catalase | Extrusion | — | 145 | |
| Ischemia/reperfusion injury | PLGA NPs | Rampamycin | Sonication and extrusion | 6 days | 146 | |
| Macrophages | Atherosclerosis | Liposomes | Hydroxysafflower yellow A | Incubation | — | 148 |
| Bone regeneration | Gold nanocages | Resolvin D1 | Extrusion | — | 149 | |
| Colitis | — | E. coli strain Nissle1917 | Incubation | 24 h | 150 | |
| Glomerulonephritis | Nanomicelles | Dexamethasone and HA | Sonication | — | 151 | |
| J774 macrophages | Atherosclerosis | Liposomes | Rapamycin | Microfluidic | — | 152 |
| COVID-19 | Liposomes | Dexamethasone | Microfluidic | — | 153 | |
| Endothelial inflammation | Liposomes | — | Microfluidic | — | 154 | |
| Inflamed vasculature | Liposomes | — | Extrusion | — | 57 | |
| Osteoarthritis | — | — | Extrusion | 7 days | 155 | |
| Rheumatoid arthritis | — | Celastrol | Sonication and extrusion | — | 156 | |
| Sepsis | PLGA NPs | — | Sonication | 17.2 h | 157 | |
| Vascular inflammation | Liposomes | — | Extrusion | — | 22 | |
| Raw 264.7 macrophages | Acute respiratory distress syndrome | Calcium phosphate NPs | Dexamethasone sodium phosphate | Sonication and extrusion | 48 h | 158 |
| Atherosclerosis | ROS responsive NPs | Atorvastatin | Extrusion | 13.32 h | 159 | |
| ROS responsive NPs | Rapamycin | Sonication and extrusion | — | 160 | ||
| PLGA-chitosan NPs | Metronidazole and simvastatin | Sonication | — | 161 | ||
| PLGA NPs | Atorvastatin and metformin | Extrusion | 2.26 h | 162 | ||
| Prussian blue NPs | Rosuvastatin | Sonication and extrusion | 3.5 h | 163 | ||
| Diabetic wounds | PLGA NPs | MLN4924 | Extrusion | — | 164 | |
| Endometriosis | Manganese dioxide nanosheet | — | Extrusion | — | 165 | |
| Gout | Platinum nanozyme | Prussian blue | Extrusion | — | 166 | |
| Hemophagocytic lymphohistiocytosis | PLGA NPs | Ruxolitinib | Sonication | 1.145 h | 167 | |
| Hepatic ischemia-reperfusion injury | PLGA NPs | — | Extrusion | — | 168 | |
| Inflammatory bowel disease | Polydopamine NPs | Cathelicidin-related antimicrobial peptide | Sonication and extrusion | — | 169 | |
| Inflammatory osteolysis | Porous Se@SiO2 nanospheres | — | Sonication and extrusion | — | 170 | |
| Myocardial infarction | PLGA NPs | Vascular endothelial growth factor (VEGF) and N-acetyl-L-cysteine | Sonication | — | 171 | |
| Myocardial ischemia-reperfusion injury | PLGA NPs | peroxynitrite (ONOO−)-triggered CO donor | Sonication and extrusion | — | 172 | |
| Osteoarthritis | Polyethylenimine-HA NPs | Nucleic acids | Extrusion | — | 173 | |
| Osteomyelitis | Magnetic NPs | — | Incubation or extrusion | — | 174 | |
| Periodontitis | Silk fibroin nanoparticles | Minocycline hydrochloride | Extrusion | — | 175 | |
| Peri-prosthetic osteolysis | Exosomes | — | Extrusion | — | 176 | |
| Rheumatoid arthritis | Prussian blue NPs | SiRNAsT/I | Extrusion | — | 177 | |
| PEG NPs | Methotrexate | Sonication | — | 178 | ||
| Sepsis | Antimicrobial peptide NPs | — | Extrusion | — | 179 | |
| Ulcerative colitis | Liposomes | Carbon-dots | Sonication | 158 min | 180 | |
| PLGA NPs | Tasquinimod | Extrusion | — | 181 | ||
| THP-1 macrophages | COVID-19 | PLGA NPs | Lopinavir | Sonication | — | 182 |
As the body's “protective shield”, macrophages are immediately activated and recruited after signs of infection/inflammation or tissue damage are detected. Based on these unique characteristics, macrophage membranes are well suited to coat NPs and thus build biomimetic systems.72 Coated with macrophage membrane, NPs can exhibit a long blood circulation time23,59 similar to NPs coated with RBCs membrane. In addition, NPs camouflaged with macrophage membranes may have the ability to cross vascular barriers and molecular recognition capacity in tumor cells through functional proteins residing in the membranes.59
Due to intracellular degradation, loading drugs directly into macrophages would likely lead to premature drug inactivation leading to uncontrolled release, resulting in limited drug content at the desired sites. Incorporating the drug into the NPs would be useful to reduce drug disintegration within cells. When macrophages are used as a delivery platform, they can be loaded first with the ex vivo nanoparticulate drug delivery system, followed by reinfusion back into the host to deliver their contents to the tissues where the macrophages reside.21
Chengcheng et al., reported biomimetic anti-inflammatory nano-capsules that can block cytokines and promote M2 macrophage polarization, presenting a positive role for bone tissue repair. For this, the membrane of macrophages stimulated with LPS was used enveloping gold nanocage as “cytokine blocker”. Resolvin D1, loaded inside the gold nanocage, was used to promote the polarization of macrophages to an M2 state, and can be released in a controlled manner through the use of NIR irradiation. The studies carried out show that the nanosystem can neutralize the levels of TNF-α and IL-6, which in turn inhibits the activation of neutrophils. In vitro, studies also show that the nanosystem, compared to gold nanocage, is capable of inducing polarization in M2 macrophages, after being irradiated with near-infrared radiation (NIR). In vivo, these results were corroborated, since, after treatment, the levels of inflammatory cells (monocytes, macrophages, and neutrophils) decreased, the polarization of macrophages to an M2 state was verified due to the presence of arginase-1 and the presence of arginase-1 was also verified formation of new bone tissue.149
Other examples report the polarization of macrophages from an M1 state to an M2 state. Zhang et al., report a nanosystem consisting of pH-sensitive nanomicelle, based on dexamethasone and hyaluronic acid, subsequently camouflaged with the macrophage membrane, for the treatment of glomerulonephritis. In this approach HA is responsible for the polarization of macrophages and dexamethasone for mediating the death of mesangial cells. The membrane coating of macrophages endows the nanosystem with active targeting to the inflamed glomerulus. In vitro, it was found that the nanosystem was capable of inhibiting the proliferation of mesangial cells – which proliferate abnormally in cases of glomerulus inflammation – which consequently leads to a decrease in IL-1β and TNF-α levels. The in vivo studies confirmed the results obtained previously, showing again that the nanosystem under study is capable of accumulating in the renal tissue, decreasing the levels of proteinuria, IL-1β and TNF-α, which demonstrates its potential anti-inflammatory applications.151
A summary of macrophage-derived nanostructures can be found in Table 4.
4.4. Mesenchymal stem cells
Mesenchymal stem cells (MSCs) are multipotent progenitor cells, which gives them the ability to differentiate into different types of cells,49,64,73,86 including adipocytes, fibroblasts, and osteoblasts.86 They also have regenerative capacity,64,73,76,185 can be extracted from diverse tissues,49 are easy to isolate185,186 and to cultivate in vitro.49,56,185,186 On the surface they have receptors for circulating ligands, which include receptors for cytokines, growth factors and chemokines, as well as proteins for cell interactions and cell-matrix adhesion.73 Numerous receptors express in the freshly isolated MSCs, including C–X–C chemokine receptor 1 (CXCR1), CXCR4, CXCR5 and CXCR6, C–C chemokine receptor 2 (CCR2), CCR7, CCR9 and CCR10. However, the expression of receptors varies depending on MSC sources, in vitro culturing processes, and cell age.185As a drug delivery system, MSCs are interesting due to the advantages they present: (1) prolonged circulation in the bloodstream;56,73 (2) immune elusion;56 (3) targeting inflamed sites;56,76,82,86,186 (4) tumor affinity;49,56,76,82,86 (5) low immunogenicity73,186 and (6) biocompatibility.49
Zhang et al., used the membrane of bone marrow MSCs, genetically engineered to exhibit high expression of the CXCR4 receptor, to camouflage red fluorescent copolymer nanoparticles loaded with kartogenin for cartilage protection. In vitro, the results show excellent compatibility and that the nanosystem was internalized by chondrocytes (located in the membrane). Furthermore, in the model of inflamed chondrocytes, it was found that the nanosystem under study led to an increase in uptake compared to the control (normal bone marrow MSCs-derived cell membrane camouflaged NPs). In vivo, the nanosystem was shown to lead to a simple cartilage structure, evenly distributed chondrocytes as well as a decrease in pathological scores, compared to the control (normal MSCs-derived cell membrane camouflaged NPs).187
Meng et al., camouflage polydopamine Au–Ag nanoparticles with the membrane of human umbilical MSCs for photothermal acne therapy. In vitro, using cells from the sebaceous gland, it is found that the use of the MSCs membrane leads to a reduction in cytotoxicity, compared with polydopamine Au–Ag nanoparticles. After laser irradiation, these biomimetic nanosystems show a strong antiproliferative effect. In vivo, after 30 days of treatment, it is verified that the biomimetic nanosystem decreases the size of the sebaceous glands inducing a decrease in the release of sebum.188
The study by Chen et al. consisted of modifying bone marrow MSCs with an aptamer targeting the nuclear chromatin protein and loaded with IL-4 mRNA, called cargocyte, to modulate the immune microenvironment and locally treat wounds. The results obtained show a reduction in the production of neutrophil extracellular traps (NETosis) and inhibition of macrophage polarization to the M1 (pro-inflammatory) state, which leads to an improvement in the immune microenvironment of wounds, which in turn leads to reduced inflammation and promotes wound healing.189
A summary of mesenchymal stem cell-derived nanostructures can be found in Table 5.
4.5. Hybrid membranes
Biomimetic hybrid membrane-based nanoparticles (BHMNPs) are another strategy for the production of membrane-based biomimetic NPs, which consists of combining the membrane of different cells on a single surface.73,186,191 This approach raises the level of complexity of the obtained NPs.191 where a membrane is usually used to target the site of action and another membrane aims to increase specific biological functions.192 For example, in BHMNPs produced with tumor cell membrane and platelet membrane, the first membrane targets targeting the nanosystem to tumor sites via homotypic adhesion and, and platelet membranes that target tumor cells via P-selectin.192 It should be noted that the portion of each membrane influences the final function of the BHMNPs, including their circulation time in the body and their distribution in the tissues.192BHMNPs can be produced by two different methods (1) by extraction of each of the membranes individually (as described above) and subsequent fusion, or (2) by fusing the two cells and subsequent extraction of the membrane. In the first method, the two membranes can be fused by stirring at 37 °C and placing them in an ice bath, another alternative is to sonicate the mixture of membranes.192,193 On the other hand, in the second method, membrane fusion is usually achieved by stimulation with polyethylene glycol or electrofusion.192 A disadvantage of this method is unwanted fusion, i.e., fusion between cells of the same type.193
Lin et al. developed Prussian blue nanocomplexes camouflaged with a hybrid membrane of RBCs and M0 macrophages for targeted sinomenine delivery in rheumatoid arthritis. DSPE-PEG2000-HA was added to enhance targeting to inflamed macrophages and synoviocytes in joints. In vitro studies showed that the camouflaged nanocomplex with HA evaded the immune system and was preferentially taken up by inflamed cells. It demonstrated good biocompatibility, with no cytotoxicity in macrophages or liver cells, low hemolytic rate, and reduced platelet aggregation. The nanocomplex inhibited inflamed synoviocyte proliferation, decreased inflammatory mediators (TNF-α, granulocyte colony-stimulating factor (G-CSF) and ROS), and accumulated in arthritic joints in vivo, extending the drug's half-life by 9.36 times.194
In a similar approach, neutrophil-RBC hybrid membranes-coated dexamethasone loaded hollow copper sulfide nanoparticles for targeted and photothermal/anti-inflammatory therapy of osteoarthritis were developed by Xue et al. In vitro, it was shown that the nanosystem, when irradiated with NIR radiation, presents a controlled release profile. The nanosystem has good levels of cytocompatibility in chondrocytes, endotheliocytes, and macrophages. Furthermore, no significant effects on cell proliferation were observed with the combined use of the nanosystem with photothermal therapy. In macrophages, it was verified that the nanosystem was able to reduce the release of TNF-α, IL-1β and IL-6. After applying NIR radiation, the effects of inhibiting the release of TNF-α and IL-6 were more evident due to the release of dexamethasone. Studies with M1 macrophages also show that the cloaked nanosystem is internalized more efficiently than the nanosystem in the absence of the neutrophil-RBC hybrid membrane coating. In vivo, the results show that the nanosystem can target the inflamed joint cavity, with the ability to accumulate in the joint after 1 hour of administration. Plasma levels of TNF-α, IL-1β and IL-6 were also decreased after combined treatment with the nanosystem and photothermal therapy.195
You et al., describe M0 macrophages-RBC hybrid membrane-coated atorvastatin-loaded graphene oxide quantum dots for the therapy of atherosclerosis. As in the previously reported example, the authors functionalized the hybrid membrane with DSPE-PEG2000-HA to increase targeting to inflamed macrophages. In vitro, the results obtained show that the coating of graphene oxide quantum dots with the hybrid membrane leads to a controlled release of atorvastatin at different pH values. It was also shown that the nanosystem has viability rates above 90%, hemolytic rates below 5% and that it manages to be absorbed by macrophages and foam cells. The nanosystem can escape the immune system, but there is a high absorption in activated macrophages. Moreover, the results show that the nanosystem can reduce the production of ROS, nitric oxide (NO) and oxidized low-density lipoprotein ox-LDL. In vivo, it was verified that the circulation half-life of the nanosystem is prolonged with the use of the hybrid membrane. Functionalization with DSPE-PEG2000-HA increases circulation time to 12 h, the targeting of the nanosystem to atherosclerotic plaques and inhibits the degradation of atorvastatin in serum. The secretion of TNF-α, IL-1β, IL-6 and MCP-1 is inhibited, and IL-10 production is stimulated. It was also found that lipid influx was inhibited, which slows the progression of atherosclerosis by regulating the NF-κB/NLRP3 pathway.196
5. Other examples: fully synthetic options towards biomimetic
Non-extracted elements are also being used to mimic biological features and cover the surface of NPs. Some examples of this strategy are shown below.Gao et al., developed bovine serum albumin (BSA)-based biomimetic nanoparticles loaded with retinoic acid and curcumin for anti-inflammatory and neuroprotective in spinal cord injury. In vitro, the results obtained show that the nanosystem can eliminate ROS, can inhibit the inflammatory response by preventing the polarization of macrophages to an M1 state and by increasing the polarization to an M2 state. In addition, inhibition of the NF-κB pathway was also verified. IL-6 and TNF-α levels decreased and IL-4 (anti-inflammatory cytokine) levels increased. The nanosystem is also able to stimulate cell proliferation, neurite outgrowth into neuronal cells, neuronal differentiation of mesenchymal stem cells from bone marrow, and differentiation of neural stem cells into β3-tubulin+ neurons. In vivo, results show that the nanosystem can accumulate in pathological zones bypassing the normal spinal cord. It was also found that the nanosystem was able to restore motor function and promote the formation of scar tissue.197
Chronic inflammation can be one of the impediments to diabetic wound healing. In this sense, Liu et al., report a sprayable methacrylic anhydride-modified gelatin hydrogel combined with bionic neutrophils nanoparticles loaded with glucose oxidase and chloroperoxidase for scar-free wound healing of diabetes mellitus. In vitro, the biomimetic hydrogel was shown to be biocompatible and able to promote fibroblast proliferation. In addition, and since bacterial proliferation delays wound healing, the biomimetic hydrogel was also found to have antibacterial activity against Escherichia coli and Staphylococcus aureus. In vivo, the treatment of wounds with the biomimetic hydrogel showed that after 14 days of treatment, the wound had healed completely, the tissue had a multilayered and connected epithelial structure identical to the healthy state, and after 21 days of treatment, the degree of wound re-epithelialization and collagen expression was high, and on the other hand, hyperglycemia was lower. Treatment with the biomimetic hydrogel also led to a decrease in levels of TNF-α, IL-1β, IL-6.198
Also, for the healing of chronic diabetic wounds, Zhang et al., developed conventional and deformable phosphatidylserine-containing nanoliposomes that mimic apoptotic cells. In vitro, the deformable phosphatidylserine-containing nanoliposomes exhibit obvious resistance to macrophage uptake. At the end of 24 h they were mainly attached to the membrane, probably due to their soft structure that suffers deformation during internalization, which involves the expenditure of energy leading to a longer stay at the membrane of macrophages. In activated macrophages, the results show that the deformable phosphatidylserine-containing nanoliposomes are more effective in inhibiting TNF-α expression and promoting the expression of IL-10 and platelet-derived growth factor-BB (PDGF-BB). In vivo, deformable phosphatidylserine-containing nanoliposomes were found to exhibit a longer residence time in the wound, indicating that they can effectively resist clearance. At the end of 14 days of treatment with deformable phosphatidylserine-containing nanoliposomes led to an almost intact epidermis, with better re-epithelialization due to collagen deposition in the wound bed.199
Yang et al., inspired by the unique “bottlebrush” structure feature of natural proteoglycans, developed a novel injectable brush-like biolubricant was prepared by chemically grafting chondroitin sulfate onto the surface of diclofenac-loaded chitosan NPs. In vitro, it was shown that the nanosystem led to lower coefficients of friction than the corresponding nanosystem without functionalization with chondroitin sulfate, this is because chondroitin sulfate has negatively charged groups that interact with water creating a hydration layer. They also showed the ability of the nanosystem to eliminate reactive oxygen species. In vivo, it was shown that the nanosystem was able to reduce the release of inflammatory mediators (TNF-α, IL-1β, IL-6), decrease erosion and the formation of osteophytes. Furthermore, after treatment it was found that the cartilage was smooth, the joint space uniform and that there was an increase in collagen production.200
6. Conclusion and future outlook
The use of NPs in drug delivery has revolutionized health sciences by overcoming the limitations associated with conventional drugs. Although promising, in vivo, NPs face challenges in traversing complex biological barriers to reach their intended targets. Conventional NPs can be tailored with diverse characteristics based on the therapeutic target, but comprehending the intricate interactions between NPs and organisms remains a challenge, impairing the development of highly efficient systems.In the design process of NPs, it is crucial to consider that the organism can eliminate substances that it does not recognize as endogenous, due to the complex defence mechanism of the immune system, which often prevents the NPs from reaching the target site. The relentless pursuit of alternatives led to the development of biomimetic NPs, a strategy that seeks to mimic the complex interactions that occur naturally in the organism.
This innovative generation facilitates the translocation of properties from the cells of origin to the NPs, particularly relevant for active targeting. Also, it extends circulation time due to more efficient immune escape, enhancing drug delivery efficiency at the target site. The incorporation of hybrid membranes further allows the combination of functions from different cell types, augmenting the biological capabilities of biomimetic NPs.
Despite the promise of this strategy, significant challenges remain before it can be translated into clinical applications. A key obstacle is scaling up production, as current methods are not equipped to handle large volumes and lack standardization, potentially resulting in CMCNPs with inconsistent sizes and surface characteristics. Among the cell types discussed, RBCs stand out due to their abundance in human blood and the availability of transfusions, which provide easy access to blood type-matched cells, enhancing the biocompatibility of the nanosystems.
While membrane extraction methods are simple, they still require optimization to prevent contamination, especially from intracellular components. RBCs are particularly advantageous in this aspect, as their membranes can be easily extracted using simple techniques such as lysis and centrifugation. Their less complex membrane structure also supports better reproducibility in CMCNP production.
Additionally, robust and user-friendly techniques are needed to confirm that synthetic nanoparticles are properly camouflaged by cell membranes during production. Establishing regulatory guidelines to ensure safety is equally crucial for bringing these strategies to market. Nonetheless, using cell membranes directly from patients holds great potential for personalized medicine, minimizing immunogenic reactions and enabling more tailored, effective treatments.
Ethical approval
This article does not include any studies with human participants or animals performed by any of the author.Consent for publication
All authors have given their consent for publication.Author contributions
AM conducted the literature review and drafted the initial version of the manuscript. SR and CN oversaw the process and provided revisions to the manuscript.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research received financial support from PT national funds (FCT/MCTES, Fundação para a Ciência e Tecnologia and Ministério da Ciência, Tecnologia e Ensino Superior) through the projects 10.54499/LA/P/0008/2020, 10.54499/UIDP/50006/2020 and 10.54499/UIDB/50006/2020. AM thanks CCRN (Comissão de Coordenação da Região Norte) for her PhD grant Norte-08-5369-FSE-000050. CN thanks FCT (Fundação para a Ciência e Tecnologia) for funding through the Individual Call to Scientific Employment Stimulus, 10.54499/2022.05608.CEECIND/CP1724/CT0002. The authors also thank Manuela Barros and Vânia Dias for administrative and technical support.References
- K. B. Megha, X. Joseph, V. Akhil and P. V. Mohanan, Phytomedicine, 2021, 91, 153712 CrossRef CAS PubMed.
- D. Placha and J. Jampilek, Pharmaceutics, 2021, 13, 64 CrossRef CAS.
- H. Yan, D. Shao, Y.-H. Lao, M. Li, H. Hu and K. W. Leong, Adv. Sci., 2019, 6, 1900605 CrossRef PubMed.
- L. Chen, H. Deng, H. Cui, J. Fang, Z. Zuo, J. Deng, Y. Li, X. Wang and L. Zhao, Oncotarget, 2018, 9, 7204–7218 CrossRef.
- R. Brusini, M. Varna and P. Couvreur, Adv. Drug Delivery Rev., 2020, 30094–30096, DOI:10.1016/j.addr.2020.07.010.
- X. Dong, D. Chu and Z. Wang, Theranostics, 2017, 7, 751–763 CrossRef CAS PubMed.
- L. P. Alves, K. da Silva Oliveira, J. A. da Paixão Santos, J. M. da Silva Leite, B. P. Rocha, P. de Lucena Nogueira, R. I. de Araújo Rêgo, J. A. Oshiro-Junior and B. P. G. D. L. Damasceno, J. Drug Delivery Sci. Technol., 2020, 60, 102008 CrossRef CAS.
- Z. Deng and S. Liu, Drug Delivery Transl. Res., 2021, 11, 1475–1497 CrossRef CAS.
- A. B. Kovačević, S. M. C. Silva and S. Doktorovová, in Emerging Nanotechnologies in Immunology, ed. R. Shegokar and E. B. Souto, Elsevier, Boston, 2018, pp. 103–133 DOI:10.1016/B978-0-323-40016-9.00005-1.
- I. B. McInnes and E. M. Gravallese, Nat. Rev. Immunol., 2021, 21, 680–686 CrossRef CAS.
- N. Matougui, L. Boge, A.-C. Groo, A. Umerska, L. Ringstad, H. Bysell and P. Saulnier, Int. J. Pharm., 2016, 502, 80–97 CrossRef CAS.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS.
- B. T. Luk and L. Zhang, J. Controlled Release, 2015, 220, 600–607 CrossRef CAS PubMed.
- D. M. Valcourt, J. Harris, R. S. Riley, M. Dang, J. Wang and E. S. Day, Nano Res., 2018, 11, 4999–5016 CrossRef CAS.
- S. T. Yurkin and Z. Wang, Nanomedicine, 2017, 12(16), 2007–2019 CrossRef CAS.
- S. Zargar, D. Hafshejani, A. Eskandarinia, M. Rafienia and A. Kharazi, J. Med. Signals Sens., 2019, 9, 181–189 CrossRef.
- X. Ai, M. Hu, Z. Wang, W. Zhang, J. Li, H. Yang, J. Lin and B. Xing, Bioconjugate Chem., 2018, 29, 838–851 CrossRef CAS PubMed.
- M. A. Dobrovolskaia and S. E. McNeil, Nat. Nanotechnol., 2007, 2, 469–478 CrossRef CAS PubMed.
- T. Penkauskas and G. Preta, Biochimie, 2019, 157, 131–141 CrossRef CAS.
- C. Alvarez-Lorenzo and A. Concheiro, Curr. Opin. Biotechnol, 2013, 24, 1167–1173 CrossRef CAS.
- Y. Huang, X. Gao and J. Chen, Acta Pharm. Sin. B, 2018, 8, 4–13 CrossRef PubMed.
- J. O. Martinez, R. Molinaro, K. A. Hartman, C. Boada, R. Sukhovershin, E. De Rosa, D. Kirui, S. Zhang, M. Evangelopoulos, A. M. Carter, J. A. Bibb, J. P. Cooke and E. Tasciotti, Theranostics, 2018, 8, 1131–1145 CrossRef CAS.
- Y. Liu, J. Luo, X. Chen, W. Liu and T. Chen, Nano-Micro Lett., 2019, 11, 100 CrossRef CAS PubMed.
- A. Parodi, R. Molinaro, M. Sushnitha, M. Evangelopoulos, J. O. Martinez, N. Arrighetti, C. Corbo and E. Tasciotti, Biomaterials, 2017, 147, 155–168 CrossRef CAS PubMed.
- M. Fan and M. Jiang, J. Drug Targeting, 2020, 1–9, DOI:10.1080/1061186x.2020.1757102.
- Y. K. Gong and F. M. Winnik, Nanoscale, 2012, 4, 360–368 RSC.
- A. Li, J. Zhao, J. Fu, J. Cai and P. Zhang, Asian J. Pharm. Sci., 2019, 16, 161–174 CrossRef.
- A. Pasto, F. Giordano, M. Evangelopoulos, A. Amadori and E. Tasciotti, Clin. Transl. Med., 2019, 8, 8 CrossRef PubMed.
- Z. Chai, X. Hu and W. Lu, Sci. China Mater., 2017, 60, 504–510 CrossRef CAS.
- D. Dehaini, R. H. Fang and L. Zhang, Bioeng. Transl. Med., 2016, 1, 30–46 CrossRef PubMed.
- R. M. Samarasinghe, R. K. Kanwar and J. R. Kanwar, Int. J. Mol. Cell. Med., 2012, 1, 133–144 Search PubMed.
- H. Zhang, S. Dong, Z. Li, X. Feng, W. Xu, C. M. S. Tulinao, Y. Jiang and J. Ding, Asian J. Pharm. Sci., 2019, 15, 397–415 CrossRef PubMed.
- S. Y. Fam, C. F. Chee, C. Y. Yong, K. L. Ho, A. R. Mariatulqabtiah and W. S. Tan, Nanomaterials, 2020, 10, 787 CrossRef CAS.
- W. Gao and L. Zhang, J. Drug Targeting, 2015, 23, 619–626 CrossRef CAS.
- R. H. Fang, A. V. Kroll, W. Gao and L. Zhang, Adv. Mater., 2018, 30, 1706759 CrossRef.
- J. C. Harris, M. A. Scully and E. S. Day, Cancers, 2019, 11, 1836 CrossRef CAS PubMed.
- L. Pang, C. Zhang, J. Qin, L. Han, R. Li, C. Hong, H. He and J. Wang, Drug Delivery, 2017, 24, 83–91 CrossRef CAS PubMed.
- Z. Chai, X. Hu and W. Lu, Sci. China Mater., 2017, 60, 504 CrossRef CAS.
- M. Gagliardi, Ther. Delivery, 2017, 8, 289–299 CrossRef CAS PubMed.
- S. Zou, B. Wang, C. Wang, Q. Wang and L. Zhang, Nanomedicine, 2020, 15, 625–641 CrossRef CAS PubMed.
- R. H. Fang, Y. Jiang, J. C. Fang and L. Zhang, Biomaterials, 2017, 128, 69–83 CrossRef CAS.
- M. Sushnitha, M. Evangelopoulos, E. Tasciotti and F. Taraballi, Front. Bioeng. Biotechnol., 2020, 8, 627 CrossRef PubMed.
- S. Li, J. Liu, M. Sun, J. Wang, C. Wang and Y. Sun, Front. Pharmacol., 2020, 11, 24 CrossRef CAS.
- S. Ghosh, K. Girigoswami and A. Girigoswami, Nanomedicine, 2019, 14, 2067–2082 CrossRef CAS PubMed.
- S. Herianto, P.-J. Chien, J.-A. A. Ho and H.-L. Tu, Biomater. Adv., 2022, 142, 213156 CrossRef CAS.
- P. Nakhaei, R. Margiana, D. O. Bokov, W. K. Abdelbasset, M. A. Jadidi Kouhbanani, R. S. Varma, F. Marofi, M. Jarahian and N. Beheshtkhoo, Front. Bioeng. Biotechnol., 2021, 9 Search PubMed.
- J. Wang, M. Zhu and G. Nie, Adv. Drug Delivery Rev., 2021, 178, 113974 CrossRef CAS.
- E. Rideau, R. Dimova, P. Schwille, F. R. Wurm and K. Landfester, Chem. Soc. Rev., 2018, 47, 8572–8610 RSC.
- C.-H. Xu, P.-J. Ye, Y.-C. Zhou, D.-X. He, H. Wei and C.-Y. Yu, Acta Biomater., 2020, 105, 1–14 CrossRef CAS PubMed.
- W. Gao, C.-M. J. Hu, R. H. Fang and L. Zhang, J. Mater. Chem. B, 2013, 1, 6569–6585 RSC.
- A. E. Mona and N. Ayman, in Translational Studies on Inflammation, ed. C. F. N. Ane, IntechOpen, Rijeka, 2019, ch. 9 DOI:10.5772/intechopen.84776.
- A. Akbarzadeh, R. Rezaei-Sadabady, S. Davaran, S. W. Joo, N. Zarghami, Y. Hanifehpour, M. Samiei, M. Kouhi and K. Nejati-Koshki, Nanoscale Res. Lett., 2013, 8, 102 CrossRef.
- S. C. Balmert and S. R. Little, Adv. Mater., 2012, 24, 3757–3778 CrossRef CAS PubMed.
- C. Sabu, C. Rejo, S. Kotta and K. Pramod, J. Controlled Release, 2018, 287, 142–155 CrossRef CAS PubMed.
- A. V. Kroll, R. H. Fang and L. Zhang, Bioconjugate Chem., 2017, 28, 23–32 CrossRef CAS PubMed.
- C. Guido, G. Maiorano, B. Cortese, S. D’Amone and I. Palama, Bioengineering, 2020, 7 Search PubMed.
- R. Molinaro, C. Corbo, J. O. Martinez, F. Taraballi, M. Evangelopoulos, S. Minardi, I. K. Yazdi, P. Zhao, E. De Rosa, M. B. Sherman, A. De Vita, N. E. Toledano Furman, X. Wang, A. Parodi and E. Tasciotti, Nat. Mater., 2016, 15, 1037 CrossRef CAS.
- R. J. C. Bose, S.-H. Lee and H. Park, Drug Discovery Today, 2016, 21, 1303–1312 CrossRef CAS PubMed.
- R. Li, Y. He, S. Zhang, J. Qin and J. Wang, Acta Pharm. Sin. B, 2018, 8, 14–22 CrossRef PubMed.
- J. Jin and Z. M. Bhujwalla, Front. Oncol., 2020, 9 Search PubMed.
- K. Jin, Z. Luo, B. Zhang and Z. Pang, Acta Pharm. Sin. B, 2018, 8, 23–33 CrossRef PubMed.
- P. Zhang, G. Liu and X. Chen, Nano Today, 2017, 13, 7–9 CrossRef CAS.
- V. Vijayan, S. Uthaman and I.-K. Park, Polymers, 2018, 10, 983 CrossRef.
- M. Wu, W. Le, T. Mei, Y. Wang, B. Chen, Z. Liu and C. Xue, Int. J. Nanomed., 2019, 14, 4431–4448 CrossRef CAS.
- Y. Sun, J. Su, G. Liu, J. Chen, X. Zhang, R. Zhang, M. Jiang and M. Qiu, Eur. J. Pharm. Sci., 2017, 96, 115–128 CrossRef CAS.
- A.-M. Chacko, E. D. Hood, B. J. Zern and V. R. Muzykantov, Curr. Opin. Colloid Interface Sci., 2011, 16, 215–227 CrossRef CAS.
- M. Qin, G. Du and X. Sun, Biomater. Sci., 2020, 8, 530–543 RSC.
- R. A. Meyer, J. C. Sunshine and J. J. Green, Trends Biotechnol., 2015, 33, 514–524 CrossRef CAS.
- H. Watson, Essays Biochem., 2015, 59, 43–69 CrossRef PubMed.
- V. Vijayan, S. Uthaman and I. K. Park, Adv. Exp. Med. Biol., 2018, 1064, 45–59 CrossRef CAS PubMed.
- Y.-x Shen, P. O. Saboe, I. T. Sines, M. Erbakan and M. Kumar, J. Membr. Sci., 2014, 454, 359–381 CrossRef CAS.
- M. Xuan, J. Shao and J. Li, Natl. Sci. Rev., 2019, 6, 551–561 CrossRef CAS.
- N. Jan, A. Madni, S. Khan, H. Shah, F. Akram, A. Khan, D. Ertas, M. F. Bostanudin, C. H. Contag, N. Ashammakhi and Y. N. Ertas, Bioeng. Transl. Med., 2023, 8, e10441 CrossRef CAS.
- A. Luchini and G. Vitiello, Biomimetics, 2021, 6, 3 CrossRef CAS.
- Y. Zhai, J. Su, W. Ran, P. Zhang, Q. Yin, Z. Zhang, H. Yu and Y. Li, Theranostics, 2017, 7, 2575–2592 CrossRef CAS.
- Z. Yi, W. Hong-Hui, P. Yuan and G. Jian-Qing, Nanofabrication, 2019, 5, 1–18 Search PubMed.
- M. Y. Thanuja, C. Anupama and S. H. Ranganath, Adv. Drug Delivery Rev., 2018, 132, 57–80 CrossRef CAS.
- L. Yao, L. Jingshan, C. Xiaojia, L. Wei and C. Tongkai, Nano-Micro Lett., 2019, 11, 100 CrossRef.
- H. Wang, Y. Liu, R. He, D. Xu, J. Zang, N. Weeranoppanant, H. Dong and Y. Li, Biomater. Sci., 2020, 8, 552–568 RSC.
- L. Liu, X. Bai, M.-V. Martikainen, A. Kårlund, M. Roponen, W. Xu, G. Hu, E. Tasciotti and V.-P. Lehto, Nat. Commun., 2021, 12, 5726 CrossRef CAS PubMed.
- H.-Y. Chen, J. Deng, Y. Wang, C.-Q. Wu, X. Li and H.-W. Dai, Acta Biomater., 2020, 112, 1–13 CrossRef CAS PubMed.
- S. Yaman, U. Chintapula, E. Rodriguez, H. Ramachandramoorthy and K. Nguyen, Cancer Drug Resist., 2020, 3, 879–911 CAS.
- X. Chen and J. Li, Mater. Chem. Front., 2020, 4, 750–774 RSC.
- X. Zhao and C. Yan, Nanoscale Res. Lett., 2022, 17, 36 CrossRef CAS PubMed.
- A. Sen Gupta, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9, e1464 Search PubMed.
- E. Soprano, E. Polo, B. Pelaz and P. del Pino, J. Nanobiotechnol., 2022, 20, 538 CrossRef CAS.
- Z. Fan, P. Y. Li, J. Deng, S. C. Bady and H. Cheng, Nano Res., 2018, 11, 5573–5583 CrossRef CAS.
- L. Yang, G. Zang, J. Li, X. Li, Y. Li and Y. Zhao, Regener. Biomater., 2020, 7, 349–358 CrossRef CAS PubMed.
- F. Safarpour, M. Kharaziha and R. Emadi, Int. J. Pharm., 2022, 613, 121419 CrossRef CAS PubMed.
- J. Liu, M. Chi, L. Li, Y. Zhang and M. Xie, J. Colloid Interface Sci., 2024, 663, 856–868 CrossRef CAS PubMed.
- J. Gu, C. Yan, S. Yin, H. Wu, C. Liu, A. Xue, X. Lei, N. Zhang and F. Geng, J. Controlled Release, 2024, 366, 448–459 CrossRef CAS.
- M. Shen, H. Li, S. Yao, X. Wu, S. Liu, Q. Yang, Y. Zhang, J. Du, S. Qi and Y. Li, Mater. Sci. Eng., C, 2021, 126, 112164 CrossRef CAS PubMed.
- X. Liang, H. Li, A. Zhang, X. Tian, H. Guo, H. Zhang, J. Yang and Y. Zeng, Nanomedicine, 2022, 41, 102519 CrossRef CAS.
- Q. Tang, M. Dong, Z. Xu, N. Xue, R. Jiang, X. Wei, J. Gu, Y. Li, R. Xin, J. Wang, X. Xiao, X. Zhou, S. Yin, Y. Wang and J. Chen, J. Controlled Release, 2023, 361, 871–884 CrossRef CAS.
- X. Hao, H. Zhang, R. Liu, J. Che, D. Zhang, J. Liang and L. Sun, Mater. Today Adv., 2022, 16, 100294 CrossRef CAS.
- X. Hao, J. Gan, J. Cao, D. Zhang, J. Liang and L. Sun, Mater. Today Bio, 2023, 20, 100625 CrossRef CAS PubMed.
- M. Fang, H. Zhang, Y. Wang, H. Zhang, D. Zhang and P. Xu, Eng. Regener., 2023, 4, 152–160 Search PubMed.
- B. Liu, D. Zhang, H. Tu, O. A. Alimi, Y. Kong, R. Satyanarayana, M. Kuss, Y. Li and B. Duan, Acta Biomater., 2023, 161, 201–212 CrossRef CAS PubMed.
- C. Y. Beh, R. P. Prajnamitra, L. L. Chen and P. C. Hsieh, Molecules, 2021, 26 Search PubMed.
- H. Han, R. Bártolo, J. Li, M.-A. Shahbazi and H. A. Santos, Eur. J. Pharm. Biopharm., 2022, 172, 1–15 CrossRef CAS PubMed.
- H. Jin, R. Luo, J. Li, H. Zhao, S. Ouyang, Y. Yao, D. Chen, Z. Ling, W. Zhu, M. Chen, X. Liao, J. Pi and G. Huang, Front. Pharmacol., 2022, 13 Search PubMed.
- Y. He, R. Li, J. Liang, Y. Zhu, S. Zhang, Z. Zheng, J. Qin, Z. Pang and J. Wang, Nano Res., 2018, 11, 6086–6101 CrossRef CAS.
- H. Jin, J. Li, M. Zhang, R. Luo, P. Lu, W. Zhang, J. Zhang, J. Pi, W. Zheng, Z. Mai, X. Ding, X. Liu, S. Ouyang and G. Huang, Front. Pharmacol., 2021, 12 Search PubMed.
- Y. Song, Z. Huang, X. Liu, Z. Pang, J. Chen, H. Yang, N. Zhang, Z. Cao, M. Liu, J. Cao, C. Li, X. Yang, H. Gong, J. Qian and J. Ge, Nanomedicine, 2019, 15, 13–24 CrossRef CAS.
- Q. Li, Z. Huang, Z. Pang, Q. Wang, J. Gao, J. Chen, Z. Wang, H. Tan, S. Li, F. Xu, J. Chen, M. Liu, X. Weng, H. Yang, Y. Song, J. Qian and J. Ge, Chem. Eng. J., 2023, 452, 138992 CrossRef CAS.
- Z. Karami, M. Akrami, J. Mehrzad, M. Esfandyari-Manesh, I. Haririan and S. Nateghi, Giant, 2023, 16, 100206 CrossRef CAS.
- W. Gao, L. Xiao, Y. Mu and Y. Xiao, iScience, 2022, 25 Search PubMed.
- Y. Song, Y. He, L. Rong, Z. Wang, Y. Ma, N. Zhang and B. Wang, Biomater. Adv., 2023, 148, 213378 CrossRef CAS PubMed.
- Y. Li, B. Zhang, X. Liu, H. Wan, Y. Qin, H. Yan, Y. Wang, Y. An, Y. Yang, Y. Dai, L. Yang and Y. Wang, Biomaterials, 2023, 302, 122288 CrossRef CAS.
- X. Hao, W. Gai, F. Ji, J. Zhao, D. Sun, F. Yang, H. Jiang and Y. Feng, Acta Biomater., 2022, 142, 221–241 CrossRef CAS PubMed.
- X. Yan, J. Song, Y. Zhang, M. Yang, Z. Deng, B. Gao, Y. Zhu, C. Xu, C. Ding, M. Zhang and B. Zhang, Nano Today, 2024, 54, 102139 CrossRef CAS.
- X. Hao, W. Gai, F. Ji, L. Wang, J. Zhao, F. Yang, H. Jiang and Y. Feng, Chem. Eng. J., 2022, 431, 133452 CrossRef CAS.
- B. Wang, J. Chen, C. Zhang, Q. Zhang, Z. Zhu, L. Qiu, J. Yan, Z. Li, X. Zhu, Y. Zhang and Y. Jiang, Int. Immunopharmacol., 2023, 125, 111164 CrossRef CAS.
- C. V. Carman, J. Cell Sci., 2009, 122, 3025–3035 CrossRef CAS PubMed.
- D. Chu, X. Dong, X. Shi, C. Zhang and Z. Wang, Adv. Mater., 2018, 30, e1706245 CrossRef PubMed.
- T. Li, H. Dong, C. Zhang and R. Mo, Nano Res., 2018, 11, 5240–5257 CrossRef CAS.
- S. Ding, C. P. O’Banion, J. G. Welfare and D. S. Lawrence, Cell Chem. Biol., 2018, 25, 648–658 CrossRef CAS.
- A. Narain, S. Asawa, V. Chhabria and Y. Patil-Sen, Nanomedicine, 2017, 12, 2677–2692 CrossRef CAS PubMed.
- K. Wang, Y. Lei, D. Xia, P. Xu, T. Zhu, Z. Jiang and Y. Ma, Colloids Surf., B, 2020, 188, 110755 CrossRef CAS PubMed.
- P. Hassanzadeh, E. Arbabi and F. Rostami, Life Sci., 2021, 270, 119128 CrossRef CAS PubMed.
- Z. Z. Jiang, X. R. Geng, L. L. Su, A. N. Chen, Z. H. Sheng and T. A. Jiang, Mater. Today Chem., 2022, 26, 101062 CrossRef CAS.
- Z. Huang, H. Wang, J. Long, Z. Lu, C. Chun and X. Li, Int. J. Pharm., 2022, 624, 121971 CrossRef CAS.
- D. Han, F. Wang, Z. Qiao, B. Wang, Y. Zhang, Q. Jiang, M. Liu, Y. Zhuang, Q. An, Y. Bai, J. Shangguan, J. Zhang, G. Liang and D. Shen, Bioact. Mater., 2023, 23, 369–382 Search PubMed.
- J. Cai, H. Tao, H. Liu, Y. Hu, S. Han, W. Pu, L. Li, G. Li, C. Li and J. Zhang, Bioact. Mater., 2023, 28, 12–26 Search PubMed.
- Q. Wang, Y. Song, J. Gao, Q. Li, J. Chen, Y. Xie, Z. Wang, H. Tan, H. Yang, N. Zhang, J. Qian, Z. Pang, Z. Huang and J. Ge, Acta Pharm. Sin. B, 2023, 13, 4999–5015 CrossRef CAS PubMed.
- L. Zhou, F. Cai, H. Zhu, Y. Xu, J. Tang, W. Wang, Z. Li, J. Wu, Z. Ding, K. Xi, L. Chen and Y. Gu, Bioact. Mater., 2024, 37, 132–152 CAS.
- X. Hu, F.-Y. Chen, J. Ding, Z. Zhai, H. Yang, L. Shen, Y. Zhu, D.-S. Guo and C. Gao, Chem. Eng. J., 2024, 480, 148138 CrossRef CAS.
- J. Liu, X. Chen, L. Xu, F. Tu, X. Rui, L. Zhang, Z. Yan, Y. Liu and R. Hu, Nanomedicine, 2023, 48, 102640 CrossRef CAS PubMed.
- W. Ma, D. Wu, C. Long, J. Liu, L. Xu, L. Zhou, Q. Dou, Y. Ge, C. Zhou and R. Jia, J. Controlled Release, 2024, 370, 66–81 CrossRef CAS PubMed.
- Q. Zhang, D. Dehaini, Y. Zhang, J. Zhou, X. Chen, L. Zhang, R. H. Fang, W. Gao and L. Zhang, Nat. Nanotechnol., 2018, 13, 1182–1190 CrossRef CAS.
- Y. Xiao, C. Ren, G. Chen, P. Shang, X. Song, G. You, S. Yan, Y. Yao and H. Zhou, Mater. Today Bio, 2022, 14, 100244 CrossRef CAS PubMed.
- J. Gao, Y. Su and Z. Wang, Biomaterials, 2023, 296, 122071 CrossRef CAS PubMed.
- T. Fukuta, S. Yoshimi, T. Tanaka and K. Kogure, Int. J. Pharm., 2019, 563, 314–323 CrossRef CAS PubMed.
- Z. Tang, S. Meng, Z. Song, X. Yang, X. Li, H. Guo, M. Du, J. Chen, Y. Z. Zhu and X. Wang, Mater. Today Bio, 2023, 20, 100674 CrossRef CAS PubMed.
- S. Li, L. Wang, Y. Gu, L. Lin, M. Zhang, M. Jin, C. Mao, J. Zhou, W. Zhang, X. Huang, C. Corbo, W. Tao, E. Lu and J. Liu, Matter, 2021, 4, 3621–3645 CrossRef CAS.
- S. Häusser-Kinzel and M. S. Weber, Front. Immunol., 2019, 10 Search PubMed.
- T. Miyagaki, M. Fujimoto and S. Sato, Int. Immunol., 2015, 27, 495–504 CrossRef CAS PubMed.
- A. P. Sage, D. Tsiantoulas, C. J. Binder and Z. Mallat, Nat. Rev. Cardiol., 2019, 16, 180–196 CrossRef CAS.
- T. W. LeBien and T. F. Tedder, Blood, 2008, 112, 1570–1580 CrossRef CAS PubMed.
- G. Deng, Z. Sun, S. Li, X. Peng, W. Li, L. Zhou, Y. Ma, P. Gong and L. Cai, ACS Nano, 2018, 12, 12096–12108 CrossRef CAS.
- V. S. Wacleche, C. L. Tremblay, J.-P. Routy and P. Ancuta, Viruses, 2018, 10, 65 CrossRef PubMed.
- P. Gong, Y. Wang, P. Zhang, Z. Yang, W. Deng, Z. Sun, M. Yang, X. Li, G. Ma, G. Deng, S. Dong, L. Cai and W. Jiang, Cancers, 2021, 13, 77 CrossRef CAS.
- D. Aldarondo and E. Wayne, Adv. Drug Delivery Rev., 2022, 182, 114116 CrossRef CAS.
- B. J. Ravenhill, L. Soday, J. Houghton, R. Antrobus and M. P. Weekes, Sci. Rep., 2020, 10, 4560 CrossRef CAS.
- J.-W. Shen, C. Li, M.-Y. Yang, J.-F. Lin, M.-D. Yin, J.-J. Zou, P.-Y. Wu, L. Chen, L.-X. Song and J.-W. Shao, Int. J. Pharm., 2022, 611, 121297 CrossRef CAS.
- Y. Wang, Y. Wang, S. Li, Y. Cui, X. Liang, J. Shan, W. Gu, J. Qiu, Y. Li and G. Wang, J. Nanobiotechnol., 2021, 19, 331 CrossRef CAS PubMed.
- H.-C. Huang, T.-Y. Wang, J. Rousseau, M. Orlando, M. Mungaray, C. Michaud, C. Plaisier, Z. B. Chen and K.-C. Wang, Biomaterials, 2024, 306, 122505 CrossRef CAS PubMed.
- Y. Li, A. Yang, Y. Sun, D. Liu, P. You, Y. Zeng, S. Quan, H. Zhang, H. Zhang, S. Ma, Y. Hao, J. Xiong, B. Liu, G. Li and Y. Jiang, Mater. Des., 2023, 227, 111807 CrossRef CAS.
- C. Yin, Q. Zhao, W. Li, Z. Zhao, J. Wang, T. Deng, P. Zhang, K. Shen, Z. Li and Y. Zhang, Acta Biomater., 2020, 102, 416–426 CrossRef CAS PubMed.
- J. Wang, D. Hu, Q. Chen, T. Liu, X. Zhou, Y. Xu, H. Zhou, D. Gu and C. Gao, Mater. Today Bio, 2023, 20, 100679 CrossRef CAS PubMed.
- H. Zhang, Q. He, J. Wang, Y. Wang, X. Xuan, M. Sui, Z. Zhang and L. Hou, Pharmacol. Res., 2022, 181, 106263 CrossRef CAS.
- C. Boada, A. Zinger, C. Tsao, P. Zhao, J. O. Martinez, K. Hartman, T. Naoi, R. Sukhovershin, M. Sushnitha, R. Molinaro, B. Trachtenberg, J. P. Cooke and E. Tasciotti, Circ. Res., 2020, 126, 25–37 CrossRef CAS PubMed.
- R. Molinaro, A. Pasto, F. Taraballi, F. Giordano, J. A. Azzi, E. Tasciotti and C. Corbo, Nanomaterials, 2020, 10, 2301 CrossRef CAS PubMed.
- A. Zinger, M. Sushnitha, T. Naoi, G. Baudo, E. De Rosa, J. Chang, E. Tasciotti and F. Taraballi, ACS Nano, 2021, 15, 6326–6339 CrossRef CAS PubMed.
- C. Mancino, A. Pasto, E. De Rosa, L. Dolcetti, M. Rasponi, P. McCulloch and F. Taraballi, Heliyon, 2023, 9, e16640 CrossRef CAS.
- B. Wang, J. Shen, X. Wang and R. Hou, Int. Immunopharmacol., 2024, 131, 111822 CrossRef CAS PubMed.
- S. Thamphiwatana, P. Angsantikul, T. Escajadillo, Q. Zhang, J. Olson, B. T. Luk, S. Zhang, R. H. Fang, W. Gao, V. Nizet and L. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 11488–11493 CrossRef CAS.
- Q. Qiao, X. Li, X. Ou, X. Liu, C. Fu, Y. Wang, B. Niu, L. Kong, C. Yang and Z. Zhang, J. Controlled Release, 2024, 369, 746–764 CrossRef CAS PubMed.
- C. Gao, Q. Huang, C. Liu, C. H. T. Kwong, L. Yue, J.-B. Wan, S. M. Y. Lee and R. Wang, Nat. Commun., 2020, 11, 2622 CrossRef CAS.
- D. Tang, Y. Wang, A. Wijaya, B. Liu, A. Maruf, J. Wang, J. Xu, X. Liao, W. Wu and G. Wang, Regener. Biomater., 2021, 8 Search PubMed.
- S. Guo, J. Gu, Y. Jiang, W. Cui, J. Chen, L. Li, K. Zheng and Y. Xu, Mater. Des., 2022, 223, 111155 CrossRef CAS.
- P. You, A. Yang, Y. Sun, H. Zhang, Y. Zeng, Y. Hao, J. Xiong, S. Ma, H. Zhang, B. Liu and Y. Jiang, Mater. Des., 2023, 234, 112316 CrossRef CAS.
- D. Liu, A. Yang, Y. Li, Z. Li, P. You, H. Zhang, S. Quan, Y. Sun, Y. Zeng, S. Ma, J. Xiong, Y. Hao, G. Li, B. Liu, H. Zhang and Y. Jiang, J. Pharm. Anal., 2024, 14, 100937 CrossRef PubMed.
- R. Zeng, B. Lv, Z. Lin, X. Chu, Y. Xiong, S. Knoedler, F. Cao, C. Lin, L. Chen, C. Yu, J. Liao, W. Zhou, G. Dai, M.-A. Shahbazi, B. Mi and G. Liu, Bioact. Mater., 2024, 34, 366–380 CAS.
- Q. Sun, J. Chen, M. Yang, X. Ding, H. Zhang, Z. Huang, Q. Huang and Q. Chen, Colloids Surf., B, 2024, 233, 113633 CrossRef CAS PubMed.
- R. Wang, T. Liu, X. Li, E. Lu, Y. Chen, K. Luo, T. Wang, X. Huang, Z. Zhang, S. Du and X. Sha, Asian J. Pharm. Sci., 2024, 100913, DOI:10.1016/j.ajps.2024.100913.
- H. Wang, Y. Wang, H. Liu, X. Li, C. Sun, Z. Pang, B. Zhang and Y. Hu, Int. J. Pharm., 2024, 657, 124127 CrossRef CAS PubMed.
- Z. Ou, H. Zhong, L. Zhang, M. Deng, W. Zhang, J. Wang, H. Feng, J. Gong, C. Miao and Z. Yi, Int. J. Nanomed., 2020, 15, 4125–4138 CrossRef CAS PubMed.
- M. Bao, K. Wang, J. Li, Y. Li, H. Zhu, M. Lu, Y. Zhang, Q. Fan, L. Han, K. Wang, D. Wang, Y. Gao, B. Peng, Z. Ming and W. Liu, Acta Biomater., 2023, 161, 250–264 CrossRef CAS.
- C. Ding, C. Yang, T. Cheng, X. Wang, Q. Wang, R. He, S. Sang, K. Zhu, D. Xu, J. Wang, X. Liu and X. Zhang, J. Nanobiotechnol., 2021, 19, 382 CrossRef CAS.
- K. Zhu, K. Wang, Y. Yao, Y. Zhu, Z. Zhu, W. Wang, R. Qian, Z. Deng, J. Zhao, Y. Shen and F. Shao, Chem. Eng. J., 2024, 151438, DOI:10.1016/j.cej.2024.151438.
- J. Zhang, L. Liu, Z. Dong, X. Lu, W. Hong, J. Liu, X. Zou, J. Gao, H. Jiang, X. Sun, K. Hu, Y. Yang, J. Ge, X. Luo and A. Sun, Bioact. Mater., 2023, 28, 480–494 CAS.
- K. Zhou, C. Yang, K. Shi, Y. Liu, D. Hu, X. He, Y. Yang, B. Chu, J. Peng, Z. Zhou and Z. Qian, Biomaterials, 2023, 295, 122036 CrossRef CAS PubMed.
- M. Shi, K. Shen, B. Yang, P. Zhang, K. Lv, H. Qi, Y. Wang, M. Li, Q. Yuan and Y. Zhang, Theranostics, 2021, 11, 2349–2363 CrossRef CAS.
- Y. Deng, M. Ren, P. He, F. Liu, X. Wang, C. Zhou, Y. Li and S. Yang, Front. Bioeng. Biotechnol., 2023, 11 Search PubMed.
- J. Xie, Y. Hu, H. Li, Y. Wang, X. Fan, W. Lu, R. Liao, H. Wang, Y. Cheng, Y. Yang, J. Wang, S. Liang, T. Ma and W. Su, Acta Biomater., 2023, 160, 297–310 CrossRef CAS PubMed.
- J. Chen, S. Zeng, Q. Xue, Y. Hong, L. Liu, L. Song, C. Fang, H. Zhang, B. Wang, A. C. Sedgwick, P. Zhang, J. L. Sessler, C. Liu and J. Chen, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2213373119 CrossRef CAS.
- X. Hao, W. Gai, Y. Zhang, W. Zhou and Y. Feng, Mater. Des., 2024, 239, 112806 CrossRef CAS.
- Z. Meng, L. Pan, S. Qian, X. Yang, L. Pan, R. Chi, J. Chen, J. Pan and C. Shi, Mater. Des., 2023, 229, 111883 CrossRef CAS.
- Y. Ma, J. Zhao, Z. Deng, B. Gao, C. Xu, X. Yan, M. Yang, Y. Zhang, Q. Xu, M. Zhang and C. Xu, Chem. Eng. J., 2023, 477, 146796 CrossRef CAS.
- Z. Li, X. Zhang, C. Liu, Q. Peng, Y. Wu, Y. Wen, R. Zheng, Q. Yan and J. Ma, J. Innate Immun., 2021, 14, 380–392 CrossRef PubMed.
- Q. Tan, L. He, X. Meng, W. Wang, H. Pan, W. Yin, T. Zhu, X. Huang and H. Shan, J. Nanobiotechnol., 2021, 19, 173 CrossRef CAS.
- W. King, K. Toler and J. Woodell-May, BioMed Res. Int., 2018, 2018, 6510842 Search PubMed.
- Y. Wu, S. Wan, S. Yang, H. Hu, C. Zhang, J. Lai, J. Zhou, W. Chen, X. Tang, J. Luo, X. Zhou, L. Yu, L. Wang, A. Wu, Q. Fan and J. Wu, J. Nanobiotechnol., 2022, 20, 542 CrossRef CAS.
- H.-H. Wu, Y. Zhou, Y. Tabata and J.-Q. Gao, J. Controlled Release, 2019, 294, 102–113 CrossRef CAS PubMed.
- L. Zhu, Y. Zhong, S. Wu, M. Yan, Y. Cao, N. Mou, G. Wang, D. Sun and W. Wu, Mater. Today Bio, 2022, 14, 100228 CrossRef CAS.
- X. Zhang, Y. Sun, W. Chen, J. Yang, J. Chen and S. Chen, Biomater. Adv., 2022, 136, 212802 CrossRef CAS PubMed.
- T. Meng, R. Jiang, S. Wang, J. Li, F. Zhang, J.-H. Lee, J. Jiang and M. Zhu, Colloids Surf., B, 2020, 192, 111145 CrossRef CAS PubMed.
- Z. Chen, Y. Zou, H. Sun, Y. He, K. Ye, Y. Li, L. Qiu, Y. Mai, X. Chen, Z. Mao, C. Yi and W. Wang, Adv. Mater., 2024, 2412253 CrossRef CAS.
- R. J. C. Bose, B. J. Kim, Y. Arai, I.-B. Han, J. J. Moon, R. Paulmurugan, H. Park and S.-H. Lee, Biomaterials, 2018, 185, 360–370 CrossRef CAS.
- R. Rampado, P. Caliceti and M. Agostini, Nanomaterials, 2022, 12, 1543 CrossRef CAS PubMed.
- Y. Liao, Y. Zhang, N. T. Blum, J. Lin and P. Huang, Nanoscale Horiz., 2020, 5, 1293–1302 RSC.
- Y. Zhao, A. Li, L. Jiang, Y. Gu and J. Liu, Biomacromolecules, 2021, 22, 3149–3167 CrossRef CAS.
- Y. Lin, O. Yi, M. Hu, S. Hu, Z. Su, J. Liao, W. Wang, S. Wang, L. Liu, B. Liu and X. Cai, J. Controlled Release, 2022, 348, 42–56 CrossRef CAS.
- X. Xue, H. Liu, S. Wang, Y. Hu, B. Huang, M. Li, J. Gao, X. Wang and J. Su, Composites, Part B, 2022, 237, 109855 CrossRef CAS.
- P. You, A. Mayier, H. Zhou, A. Yang, J. Fan, S. Ma, B. Liu and Y. Jiang, Appl. Mater. Today, 2022, 26, 101386 CrossRef.
- X. Gao, Z. Han, C. Huang, H. Lei, G. Li, L. Chen, D. Feng, Z. Zhou, Q. Shi, L. Cheng and X. Zhou, Bioact. Mater., 2022, 18, 569–582 CAS.
- C. Liu, H. Zeng, Z. Chen, Z. Ge, B. Wang, B. Liu and Z. Fan, Int. J. Biol. Macromol., 2022, 202, 418–430 CrossRef CAS.
- G. Zhang, H. Xue, D. Sun, S. Yang, M. Tu and R. Zeng, Acta Biomater., 2021, 131, 452–463 CrossRef CAS PubMed.
- L. Yang, H. Huang, H. Zeng, X. Zhao, R. Wang, Z. Ma, Z. Fan, Y.-M. Liang, S. Ma and F. Zhou, Carbohydr. Polym., 2023, 304, 120503 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |