Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
AlphaFold 3 modeling of DNA nanomotifs: is it reliable?†
Mauricio
Cortes‡
Jr
,
Xindi
Sun‡
,
Anusha
,
Emile Joseph
Batchelder-Schwab
 ,
Jinyue
Li
,
Naseem
Siraj
,
Rishab
Jampana
,
Yuchen
Zhang
,
Yuntian
Bai
and
Chengde
Mao
,
Jinyue
Li
,
Naseem
Siraj
,
Rishab
Jampana
,
Yuchen
Zhang
,
Yuntian
Bai
and
Chengde
Mao
 *
*
Department of Chemistry, Purdue University, West Lafayette, IN 47907, USA. E-mail: cortes5@purdue.edu; sun1208@purdue.edu; aanusha@purdue.edu; ebatchel@purdue.edu; li4195@purdue.edu; nkoomull@purdue.edu; rjampana@purdue.edu; zhangyuchen0620@gmail.com; bai121@purdue.edu; mao@purdue.edu
First published on 22nd May 2025
Abstract
Being able to accurately predict structures is highly desirable for nanoengineering with DNA and other biomolecules. The newly launched AlphaFold 3 (AF3) provides a potential platform for this purpose. In this work, we have used AF3 to model a list of commonly used DNA nanomotifs and compared the AF3 structures with the experimentally observed structures reported in the literature. For asymmetric motifs, AF3 structures are consistent with the experimental observations; but for symmetric motifs, AF3 structures are often substantially different from experimental observations. However, the fails can be rescued if the symmetric motifs are converted into corresponding asymmetric motifs by breaking DNA sequence symmetry while maintaining the backbone symmetry. This study suggests that while AF3 is immensely helpful, we as experimentalists should use it (as it currently stands) with caution. In addition, AF3 needs further development to incorporate the existing experimental data in the training dataset for AF3. At the current stage, a hybrid approach might be beneficial: theoretical modeling softwares calculate the detailed, 3D DNA structures based on secondary DNA structures inspired by experimental observations.
New conceptsProgrammable self-assembly of biomolecules, e.g., structural DNA nanotechnology, provides a superb approach for nanoconstruction. Its potential success critically depends on the structural prediction/design of the biomolecules. Such capabilities are generally missing. The machine-learning-based Alphafold (AF) algorithm has demonstrated excellent capabilities for structural prediction/design for proteins. AF3, the newest version of AF, extends its capability to all major biomolecules (protein, DNA, and RNA) and could be an integrated algorithm for molecular design for all biomolecules. This manuscript provides a systematic evaluation of the application of AF3 to structural DNA nanotechnology. Based on this study, we provide our suggestions for the further development of such a modeling tool for structural DNA nanotechnology. |
Introduction
DNA nanotechnology has rapidly evolved in the last 40 years and has demonstrated great ability for a wide range of nanostructures.1–11 By being limited to Watson–Crick base pairing, the interactions (A–T and G–C) and structures of DNA molecules can be reliably predicted. A wide array of DNA nanostructures can thus be readily constructed. For all DNA nanostructures, the main component is the conventional B-form duplex proposed by Watson and Crick in 1953.12 However, there are some essential elements in DNA nanostructures that distinguish them from the generally perceived, linear, long polymers.1 Introduction of new elements into DNA nanotechnology has expanded the toolbox for the field and brought a new set of nanostructures, e.g. the 4-arm junction and its derivatives,13–20 double crossovers (DX),21,22 and triple crossovers (TX).23 While it is highly desirable, the introduction of new structural elements is very challenging due to the difficulty in making reliable structural predictions. Indeed, structural prediction of biomacromolecules in general is a well-recognized problem. In the last few years, AlphaFold (AF), based on a machine learning algorithm, has emerged as a powerful tool for protein structural prediction. In May 2024, the newly launched AF3 further expanded its capability and became a versatile platform to predict structures of all major biomacromolecules, including proteins, DNAs, and RNAs.24 In addition, AF3 is extremely easy to use for experimentalists with no or minimal knowledge in structural modeling. This new tool will potentially greatly improve the development of DNA nanotechnology. The first step along this direction is to examine whether AF3 can accurately model the commonly used DNA motifs whose structures are known. Herein we have compared AF3-predicted DNA nanostructures with experimentally observed structures. Some of the AF3-predicted structures are consistent with the experimentally observed structures, yet some are not. This study suggests that AF3 is a useful tool for DNA structural nanotechnology, though users should proceed with caution.Results and discussion
We have used AF3 to predict structures for 30 DNA nanomotifs (Fig. S1–S31, ESI†) including single multi-arm branched junctions (Fig. S1–S5, ESI†),13–20 parallelly aligned multi-crossover motifs (Fig. S6–S12, ESI†),21–23,25–28 double crossover-like (DXL) motifs (Fig. S13–S15, ESI†),29–31 a parallelogram (Fig. S16, ESI†),32 a tensegrity triangle (Fig. S17, ESI†),33 point-star motifs (Fig. S18–S22, ESI†),34–38 a T-junction (Fig. S23, ESI†),39 a branched kissing loop (Fig. S24),40 and polyhedra (Fig. S25 and S26, ESI†).41,42 In addition, we have also extended the list to some DNA–RNA hybrid motifs (Fig. S27–S29, ESI†)43 and RNA only motifs (Fig. S20 and S31, ESI†).44,45 All these motifs have been reported in the literature and their structures have been studied by various experimental methods, including polyacrylamide gel electrophoresis (PAGE), fluorescence resonance energy transfer (FRET), atomic force microscopy (AFM), transmission electron microscopy (TEM), nuclear magnetic resonance spectroscopy (NMR), cryogenic electron microscopy (cryoEM), and X-ray crystallography. Except for very few DNA nanomotifs,18,29,33 most DNA nanomotifs mentioned above have low resolution at the nanometer scale. Consequently, the current study examines the accuracy of the AF3 structures at only the nanometer scale instead of angstrom-level accuracy, particularly for the secondary structures and the topologies of the component strands.Fig. 1 shows the workflow for the evaluation of AF3 structural predictions to DNA nanomotifs, using a DNA 4-arm junction (4aJ) as an example.16–18 For the target DNA motif, a set of DNA strands with specific sequences were designed. With these DNA sequences as inputs, AF3 can predict the tertiary (3D) structures from which their corresponding secondary (2nd) structures can be drawn out. Note that the motifs used in this study have all been experimentally studied in the literature. Thus, the AF3-predicted structure can be directly compared with the structure observed in experiments for the same DNA motif. It is worth pointing out two additional issues in this study. (1) In this workflow, potential sequence-specific effects have not been considered. (2) In experiments, the buffers for assembly of DNA nanostructures generally include divalent cations, particularly Mg2+, to screen the strong electrostatic repulsions among negatively charged DNA backbones.16 Thus, we have included 10 Mg2+ and 10 Na+ in modeling to reflect the experimental conditions, though it is not clear the exact role and treatment of cations in the AF3 algorithm. The complete sets of data are provided in the ESI,† as Fig. S1–S31.
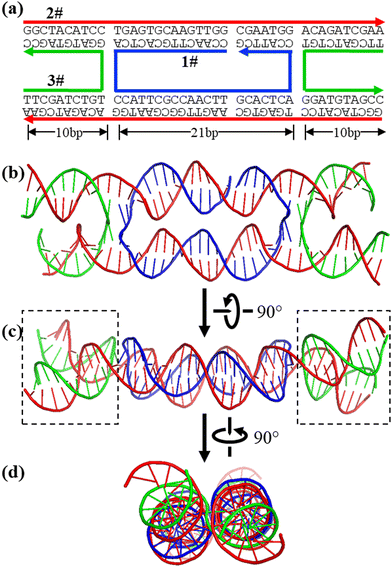
The AF3 results generally agree with the experimental observed structures. For example, DAE is a classic DNA nanomotif and has been extensively used for various DNA nanoconstructions (Fig. 2).21,22 However, there is no direct structural study on this motif by conventional structural biology methods, such as X-ray diffraction, NMR, or cryoEM. AF3 provides a plausible, detailed structural model, which is consistent with all experimental observations so far and our basic knowledge about DNA biophysics and structure. In DAE, all bases are in the correct base pairs and are organized into two duplexes that were roughly anti-parallel and linked by strand crossovers. On the outside, four helical domains (boxed areas in Fig. 2c) significantly deviate from being parallel to minimize electrostatic repulsion of the negatively charged DNA backbone. Such structural features are consistent with numerous AFM observations of DAE-based DNA 2D arrays.22
 | ||
| Fig. 2 AF3 prediction of an antiparallel double crossover motif (DAE). (a) The motif design. (b)–(d) Three orthogonal views of the AF3-predicted structure. | ||
The successful predictions include the following:
(i) Single multi-arm branched junctions: a 3-arm junction (3aJ, Fig. S1, ESI†),13–15 a 4-arm junction (4aJ, Fig. S2, ESI†),16–18 a 6-arm junction (6aJ, Fig. S3, ESI†),19 an 8-arm junction (8aJ, Fig. S4, ESI†),20 and a 12-arm junction (12aJ, Fig. S5, ESI†).20
(ii) Parallelly aligned multi-crossover motifs: an antiparallel double crossover with separation of an even number of half-turns (DAE, Fig. S6, ESI†),21,22 antiparallel double crossover with separation of an odd number of half-turns (DAO, Fig. S7, ESI†),21,22 a triplex crossover (TX, Fig. S8, ESI†),23 a double 6-arm Junction (D6aJ, Fig. S9, ESI†),25 a double 8-arm Junction (D8aJ, Fig. S10, ESI†),25 a paranemic crossover (PX, Fig. S11, ESI†),26,27 and a 6-helix bundle (6HB, Fig. S12, ESI†).28
(iii) Double crossover-like (DXL) motifs, a symmetric DXL with 6-base pair (bp) separation (DXL-6, Fig. S13, ESI†),29 a symmetric DXL with 16-bp separation (DX-16, Fig. S14, ESI†),30 and an asymmetric DXL with 14/18-bp separation (DX-14/18, Fig. S15, ESI†).31
(iv) A parallelogram (Fig. S16, ESI†).32
(v) A symmetric 4-point star (s4PS) motif (Fig. S19, ESI†).25,36
(vi) A T-junction (Fig. S22, ESI†).39
(vii) A branched kissing loop (Fig. S23, ESI†).40
(viii) Polyhedra: a tetrahedron (Fig. S24, ESI†), and a cube (Fig. S25, ESI†).41,42
In addition to confirming the structures of well-established DNA nanomotifs, AF3 can make reasonable structural predictions for less characterized DNA nanostructures. Besides the common 3aJ and 4aJ, other multiple-arm branched DNA junctions (6aJ, 8aJ, and 12aJ) have been designed and assembled.19,20 However, their 3D structures have never been experimentally studied. Here, we used AF3 to predict the 3D structures of a 6aJ (Fig. 3 and Fig. S3, ESI†). A 6aJ has six arms that pairwise stack with each other and are organized into three pseudo-continuous duplexes. Any two adjacent pseudo duplexes are rotated from each other by 60°. The minor groove of one duplex always fits into the major groove of the other duplex to minimize electrostatic repulsion. Most importantly, there is no open space at the center of the junction. All base pairs at the center are stacked with other base pairs and are not solvent accessible. Motifs 8aJ (Fig. S4, ESI†) and 12aJ (Fig. S5, ESI†) adopt similar conformations and all resemble the structural feature of 4aJ (Fig. 1). Such conformation for the multi-arm branched junctions has been speculated before; however, this AF3 modeling supports these speculations for the first time.
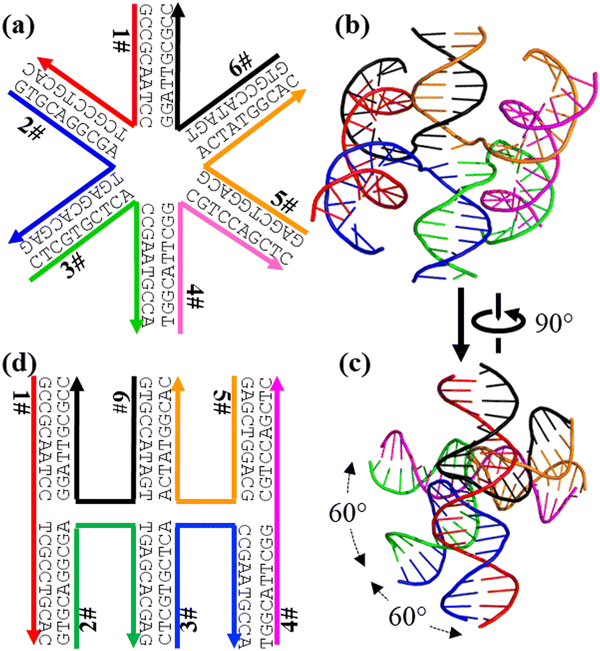 | ||
| Fig. 3 AF3 prediction of a 6-arm junction motif (6aJ). (a) The motif design. (b), (c) Two orthogonal views of the AF3-predicted structure. (d) The corresponding secondary structure of the 6aJ. | ||
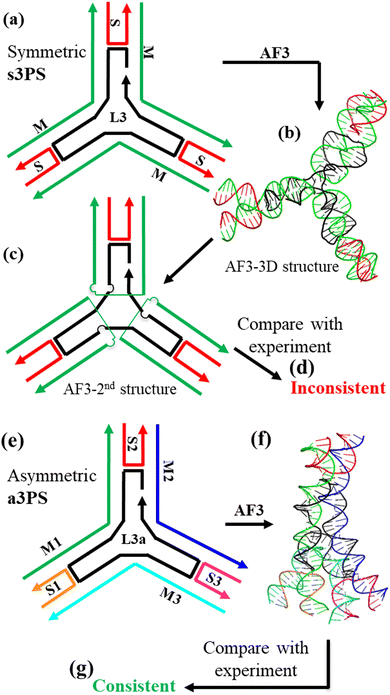
For some DNA motifs, AF3 predictions are substantially different from the structures observed in experiments and against our biophysical knowledge about DNA molecules. One example is the symmetric DNA tensegrity triangle (Fig. 4 and Fig. S17, ESI†). The symmetric DNA tensegrity triangle is composed of three DNA duplexes that are connected through strand crossovers at three points, corresponding to the three vertices of the triangle. It contains one central strand, three identical red strands, and three identical, outer blue strands. With inclusion of complementary sticky ends, the triangles can then further associate with each other in three orthogonal directions to form 3D crystals. Experimentally, tensegrity triangle crystals have been extensively studied by X-ray crystallography and the triangle structures have been solved.33 However, the AF3 structure is vastly different from the experimental results. The red strands crossover from one duplex to another duplex in the AF3 structure; in contrast, the experimental observation shows the red running continuously along one duplex. The structural difference between the AF3 structure and the experimental data is quite dramatic as the secondary structure and DNA topology have drastically changed. The AF3 structure also does not make sense from the point view of DNA biophysics. Each strand crossover costs extra energy compared to continuous DNA duplexes. Such inconsistencies have also been observed for other DNA motifs, including: the symmetric 3PS motif (Fig. S18, ESI†),34 the symmetric 5PS motif (Fig. S21, ESI†),37 and the symmetric 6PS motif (Fig. S22, ESI†).38 We have attributed these failures to inadequate data training. The AF3 is trained with high-resolution experimental data from X-ray crystallography, NMR, and cryoEM. Unfortunately, there is very little such experimental data available for engineered DNA nanostructures. Instead, most available high-resolution data are for DNA/RNA structures with simple topologies such as duplexes or one-stranded folding. Thus, multi-stranded DNA nanomotifs are challenging for AF3.
What structural features of a DNA nanomotif impact the performance of AF3? From all AF3 modeling in this study, there is a general pattern in AF3 performance. All asymmetric DNA motifs are correctly modeled by AF3. All the inconsistent modeling is associated with symmetric DNA motifs (in terms of both backbones and sequences). For example, AF3 gives an incorrect structural model for the symmetric 3PS (s3PS) motif (Fig. 5a–d and Fig. S18, ESI†). An s3PS motif contains a 3-fold rotational axis at the motif center; thus, the three branches are identical to each other. It is assembled from three unique strands: one black L strand with 3-fold repeating sequence, three copies of green strands, and three copies of red strands. Each green strand is supposed to cross from one branch to another at the motif center (Fig. 5a). However, due to the 3-fold rotational symmetry, the green strand could, alternatively and wrongly, make a U-turn at the motif center and stay on the same branch (Fig. 5c). In both situations, all strands are (almost) fully base paired. The current version of AF3 gets confused at this point under such complicated topology and predicts the wrong structure.
The above analysis prompts a hypothesis that AF3 will give correct models if a symmetric motif is converted into an asymmetric motif by breaking the sequence symmetry. We have tested this hypothesis, and the result has proved this hypothesis (Fig. 5e–g and Fig. S19, ESI†). Thus, a strategy is found to help the AF3 algorithm to overcome the symmetry problem in structural modeling. Please note that this strategy follows a general assumption: the exact sequence composition does not significantly change the 3D structures of DNA motifs as long as conventional Watson–Crick base pairs form.
The necessity of including sodium (Na+) and magnesium (Mg2+) in AF3 modeling of DNA nanomotifs is not clear. For most DNA nanomotifs that we have modeled using AF3, the modeling results are the same with or without 10 Na+/10 Mg2+. This observation is likely because AF3 is trained with data from experiments that already include such cations. Some exceptions exist, e.g. 12aJ (Fig. 6). Under both conditions, the 12 arms of the 12aJ pair-wisely stack onto each other to form 6 pseudo-continuous duplexes, akin to the 4aJ. There is no open space at the center. However, the AF3 structure has Na+/Mg2+ packed more densely (Fig. S5, ESI†) than the one without the extra Na+/Mg2+ (Fig. 6), consistent with the notion that cations screen out electrostatic repulsion and allow negatively charged backbones of DNA molecules to come close to each other.
In structural modeling, one important concern is reproducibility: will AF3 give the same structure for the same sequences in multiple, different trials? To address this question, we have used AF3 to model the DAE multiple times as it's the most commonly used DNA nanomotif. The results show that AF3 models are highly reproducible (Fig. 7). Models from multiple rounds of AF3 prediction can be well superimposed for each motif and the calculated root-mean-square deviations (RMSD) are in the range of 0.55–1.91 Å. The two helical domains between the two crossover points are nearly identical for all of the AF3 models. The variation mostly comes from the four helical domains beyond the crossover points.
 | ||
| Fig. 7 Good superimposition of AF3 models from eight rounds for a DAE motif. Every model is coded with a distinct color. (a)–(c) Three orthogonal views of the models. | ||
AF3 is a universal modeling platform for all major biomacromolecules, including DNA and RNA. It allows modeling of DNA–RNA hybrid nanomotifs and RNA-only motifs (Fig. 8 and Fig. S27–S30, ESI†). To evaluate this feature, we used AF3 to model several such motifs. Fig. 8 shows the modeling of a DNA–RNA hybrid DAE motif and an RNA-only DAE motif. They all have been experimentally used for nanoconstruction. Both DAE motifs are symmetric and each of them contains five strands: one long, central strand (L), two copies of outside short strands (S), and two copies of medium continuous strands (M). In the DNA–RNA hybrid DAE, the M strands are RNA and both L and S strands are DNA. Thus, each helical domain is composed of one DNA strand and one other strand and is expected to adopt the A-form duplex conformation (Fig. 8a). The AF3 predicted structure is consistent with the experimental results. Equally successfully, AF3 has produced a structure that is consistent with the experimental data for an RNA-only DAE motif (Fig. 8b).
Conclusion
We have semi-systematically examined the structural prediction power of AF3 for DNA nanomotifs. It is exciting to see that AF3 predictions are mostly consistent with experimental results, but we have also found that some AF3 predictions do not reflect the experimental observations. To DNA nanotechnologists, this study suggests that we should use AF3 with caution. To AF3 developers (or the modeling community in general), this study suggests that further development of AF3 is needed. In particular, some low-resolution structural data (e.g. PAGE, AFM) might be worth being including in the training data set to generate structural constraints for accurate predictions. At the current stage, it would be a great hybrid approach if AF3 could calculate the detailed 3D structures with inputs from human-assigned, secondary DNA structures.Data availability
All data are presented in the manuscript and ESI.†Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was financially supported by NSF (CCF-2107393).References
- N. C. Seeman and H. F. Sleiman, DNA nanotechnology, Nat. Rev. Mater., 2018, 3, 17068 Search PubMed.
- M. R. Jones, N. C. Seeman and C. A. Mirkin, Programmable materials and the nature of the DNA bond, Science, 2015, 347, 840 CrossRef CAS PubMed.
- F. Hong, F. Zhang, Y. Liu and H. Yan, DNA Origami: Scaffolds for creating higher order structures, Chem. Rev., 2017, 117, 12584–12640 CrossRef CAS PubMed.
- A. V. Pinheiro, D. Han, W. M. Shih and H. Yan, Challenges and opportunities for structural DNA nanotechnology, Nat. Nanotechnol., 2011, 6, 763–772 CrossRef CAS PubMed.
- H. Ramezani and H. Dietz, Building machines with DNA molecules, Nat. Rev. Genet., 2020, 21, 5–26 CrossRef CAS PubMed.
- Y. Dong, C. Yao, Y. Zhu, L. Yang, D. Luo and D. Yang, DNA functional materials assembled from branched DNA: design, synthesis, and applications, Chem. Rev., 2020, 120, 9420–9481 CrossRef CAS PubMed.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- T. Zhang, X. Qian, W. Zeng and B. Wei, Custom folding of double-stranded DNA directed by triplex formation, Chemistry, 2023, 9, 1505–1517 CrossRef CAS.
- C. Ng, A. Samanta, O. A. Mandrup, E. Tsang, S. Youssef, L. H. Klausen, M. Dong, M. A. D. Nijenhuis and K. V. Gothelf, Folding double-Stranded DNA into designed shapes with triplex-forming oligonucleotides, Adv. Mater., 2023, 35, 2302497 CrossRef CAS PubMed.
- G. Posnjak, X. Yin, P. Butler, O. Bienek, M. Dass, S. Lee, I. D. Sharp and T. Liedl, Diamond-lattice photonic crystals assembled from DNA origami, Science, 2024, 384, 781–785 CrossRef CAS PubMed.
- H. Liu, M. Matthies, J. Russo, L. Rovigatti, R. P. Narayanan, T. Diep, D. McKeen, O. Gang, N. Stephanopoulos, F. Sciortino, H. Yan, F. Romano and P. Šulc, Inverse design of a pyrochlore lattice of DNA origami through model-driven experiments, Science, 2024, 384, 776–781 CrossRef CAS PubMed.
- J. D. Watson and F. H. C. Crick, Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid, Nature, 1953, 171, 737–738 CrossRef CAS PubMed.
- D. R. Duckett and D. M. Lilley, The three-way DNA junction is a Y-shaped molecule in which there is no helix-helix stacking, EMBO J., 1990, 9, 1659–1664 CrossRef CAS PubMed.
- R. Assenberg, A. Weston, D. L. N. Cardy and K. R. Fox, Sequence-dependent folding of DNA three-way junctions, Nucleic Acids Res., 2002, 30, 5142–5150 CrossRef CAS PubMed; R.-I. Ma, N. R. Kallenbach, R. D. Sheardy, M. L. Petrillo and N. C. Seeman, Three-arm nucleic acid junctions are flexible, Nucleic Acids Res., 1986, 14, 9745–9753 CrossRef PubMed.
- J. B. Welch, D. R. Duckett and D. M. Lilley, Structures of bulged three-way DNA junctions, Nucleic Acids Res., 1993, 21, 4548–4555 CrossRef CAS PubMed.
- N. R. Kallenbach, R.-I. Ma and N. C. Seeman, An immobile nucleic acid junction constructed from oligonucleotides, Nature, 1983, 305, 829–831 CrossRef CAS.
- P. S. Eis and D. P. Millar, Conformational distributions of a four-way DNA junction revealed by time-resolved fluorescence resonance energy transfer, Biochemistry, 1993, 32, 13852–13860 CrossRef CAS PubMed.
- B. F. Eichman, J. M. Vargason, B. H. M. Mooers and P. Shing Ho, The holliday junction in an inverted repeat DNA sequence: Sequence effects on the structure of four-way junctions, Proc. Natl. Acad. Sci. U. S. A., 2000, 97(8), 3971–3976 CrossRef CAS.
- Y. Wang, J. E. Mueller, B. Kemper and N. C. Seeman., Assembly and characterization of five-arm and six-arm DNA branched junctions, Biochemistry, 1991, 30, 5667–5674 CrossRef CAS PubMed.
- X. Wang and N. C. Seeman, Assembly and characterization of 8-arm and 12-arm DNA branched junctions, J. Amer. Chem. Soc., 2007, 129, 8169–8176 CrossRef CAS PubMed.
- T.-J. Fu and N. C. Seeman, DNA Double-Crossover Molecules, Biochemistry, 1993, 32, 3211–3220 CrossRef CAS PubMed.
- E. Winfree, F. Liu, L. A. Wenzler and N. C. Seeman, Design and self-assembly of two-dimensional DNA crystals, Nature, 1998, 394, 539–544 CrossRef CAS PubMed.
- T. H. LaBean, H. Yan, J. Kopatsch, F. Liu, E. Winfree, J. H. Reif and N. C. Seeman, Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes, J. Amer. Chem. Soc., 2000, 122, 1848–1860 CrossRef CAS.
- J. Abramson, J. Adler, J. Dunger, R. Evans, T. Green, A. Pritze, O. Ronneberger, L. Willmore, A. J. Ballard, J. Bambrick, S. W. Bodenstein, D. A. Evans, C.-C. Hung, M. O’Neill, D. Reiman, K. Tunyasuvunakool, Z. Wu, A. Žemgulytė, E. Arvaniti, C. Beattie, O. Bertolli, A. Bridgland, A. Cherepanov, M. Congreve, A. I. Cowen-Rivers, A. Cowie, M. Figurnov, F. B. Fuchs, H. Gladman, R. Jain, Y. A. Khan, C. M. R. Low, K. Perlin, A. Potapenko, P. Savy, S. Singh, A. Stecula, A. Thillaisundaram, C. Tong, S. Yakneen, E. D. Zhong, M. Zielinski, A. Žídek, V. Bapst, P. Kohli, M. Jaderberg, D. Hassabis and J. M. Jumper, Accurate structure prediction of biomolecular interactions with AF3, Nature, 2024, 630, 493–500 CrossRef CAS PubMed.
- M. Li, J. Yu, J. Li, E. Ben Wang, G. Wang and C. Mao, Self-assembly of DNA double multi-arm junctions (DMaJs), RSC Adv., 2016, 6, 76355–76359, 10.1039/c6ra15145k.
- Z. Shen, H. Yan, T. Wang and N. C. Seeman, Paranemic Crossover DNA:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) A generalized holliday structure with applications in nanotechnology, J. Amer. Chem. Soc., 2004, 126, 1666–1674 CrossRef CAS PubMed.
A generalized holliday structure with applications in nanotechnology, J. Amer. Chem. Soc., 2004, 126, 1666–1674 CrossRef CAS PubMed. - X. Wang, A. R. Chandrasekaran, Z. Shen, Y. P. Ohayon, T. Wang, M. E. Kizer, R. Sha, C. Mao, H. Yan, X. Zhang, S. Liao, B. Ding, B. Chakraborty, N. Jonoska, D. Niu, H. Gu, J. Chao, X. Gao, Y. Li, T. Ciengshin and N. C. Seeman, Paranemic Crossover DNA: There and Back Again, Chem. Rev., 2019, 119, 6273–6289 CrossRef CAS PubMed.
- F. Mathieu, S. Liao, J. Kopatsch, T. Wang, C. Mao and N. C. Seeman, Six-helix bundles designed from DNA, Nano Lett., 2005, 5, 661–665 CrossRef CAS PubMed.
- C. Zhang, J. Zhao, B. Lu, N. C. Seeman, R. Sha, N. Noinaj and C. Mao, Engineering DNA crystals toward studying DNA–guest molecule interactions, JACS., 2023, 145, 4853–4859 CrossRef CAS PubMed.
- H. Liu, Y. Chen, Y. He, A. E. Ribbe and C. Mao, Design and self-assembly of two-dimensional DNA crystals, Angew. Chem., 2006, 45, 1942–1945 Search PubMed.
- M. Zheng, Q. Li, V. E. Paluzzi, J. H. Choi and C. Mao, Engineering the nanoscaled morphologies of linear DNA homopolymers, Macromol. Rapid Commun., 2021, 42, 210–217 Search PubMed.
- C. Mao, W. Sun and N. C. Seeman, Designed two-dimensional DNA holliday junction arrays visualized by atomic force microscopy, J. Am. Chem. Soc., 1999, 121, 5437–5443 CrossRef CAS.
- J. Zheng, J. J. Birktoft, Y. Chen, T. Wang, R. Sha, P. E. Constantinou, S. L. Ginell, C. Mao and N. C. Seeman, From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal, Nature, 2009, 461, 74–77 CrossRef CAS PubMed.
- Y. He, Y. Chen, H. Liu, A. E. Ribbe and C. Mao, Self-assembly of hexagonal DNA two-dimensional (2D) arrays, J. Am. Chem. Soc., 2005, 127, 12202–12203 CrossRef CAS PubMed.
- H. Yan, S. H. Park, G. Finkelstein, J. H. Reif and T. H. Labean, DNA-templated self-assembly of protein arrays and highly conductive nanowires, Science, 2003, 301, 1882–1884 Search PubMed.
- Y. He, Y. Tian, Y. Chen, Z. Deng, A. E. Ribbe and C. Mao. Sequence symmetry as a tool for designing DNA nanostructures. Angew. Chem., Intl. Ed., 44, 6694–6696 ( 2005) Search PubMed.
- C. Zhang, M. Su, Y. He, X. Zhao, P. Fang, A. E. Ribbe, W. Jiang and C. Mao, Conformational flexibility facilitates self-assembly of complex DNA nanostructures, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 10665–10669 CrossRef CAS PubMed.
- Y. He, Y. Tian, A. E. Ribbe and C. Mao, Highly Connected Two-Dimensional Crystals of DNA six-point-stars, J. Am. Chem. Soc., 2006, 128, 15978–15979 CrossRef CAS PubMed.
- S. Hamada and S. Murata, Substrate-assisted assembly of interconnected single-duplex DNA nanostructures, Angew. Chem., Int. Ed., 2009, 48, 6820–6823 CrossRef CAS PubMed.
- D. Liu, C. W. Geary, G. Chen, Y. Shao, M. Li, C. Mao, E. S. Andersen, J. A. Piccirilli, P. W. K. Rothemund and Y. Weizmann, Branched kissing loops for the construction of diverse RNA homooligomeric nanostructures, Nat. Chem., 2020, 12, 249–259 CrossRef CAS PubMed.
- R. P. Goodman, I. A. T. Schaap, C. F. Tardin, C. M. Erben, R. M. Berry, C. F. Schmidt and A. J. Turberfield, Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication, Science, 2005, 310, 1661–1665 CrossRef CAS PubMed.
- J. Chen and N. C. Seeman, Synthesis from DNA of a molecule with the connectivity of a cube, Nature, 1991, 350, 631–633 CrossRef CAS PubMed.
- S. H. Ko, M. Su, C. Zhang, A. E. Ribbe, W. Jiang and C. Mao, Synergistic self-assembly of RNA and DNA molecules, Nat. Chem., 2010, 2, 1050–1055 CrossRef CAS PubMed.
- J. M. Stewart, H. K. K. Subramanian and E. Franco, Self-assembly of multi-stranded RNA motifs into lattices and tubular structures, Nucl. Acids Res., 2017, 45, 5628 CrossRef PubMed.
- D. Liu, C. W. Geary, G. Chen, Y. Shao, M. Li, C. Mao, E. S. Andersen, J. A. Piccirilli, P. W. K. Rothemund and Y. Weizmann, Branched kissing loops for the construction of diverse RNA homooligomeric nanostructures, Nat. Chem., 2020, 12, 249–259 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Materials and detailed experimental methods; and a figure for additional AF3 structural predictions. See DOI: https://doi.org/10.1039/d5nh00059a |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |