Chemical diversity of cyanobacterial natural products†
Márcio B.
Weiss
 a,
Ricardo M.
Borges
a,
Ricardo M.
Borges
 b,
Peter
Sullivan
b,
Peter
Sullivan
 c,
João P. B.
Domingues
c,
João P. B.
Domingues
 a,
Francisco H. S.
da Silva
a,
Francisco H. S.
da Silva
 a,
Victória G. S.
Trindade
a,
Victória G. S.
Trindade
 b,
Shangwen
Luo
b,
Shangwen
Luo
 d,
Jimmy
Orjala
d,
Jimmy
Orjala
 e and
Camila M.
Crnkovic
e and
Camila M.
Crnkovic
 *a
*a
aFaculdade de Ciências Farmacêuticas, Universidade de São Paulo, CEP 05508-000, São Paulo, SP, Brazil. E-mail: camilavic@usp.br
bInstituto de Pesquisas de Produtos Naturais Walter Mors, Universidade Federal do Rio de Janeiro, CEP 21941-599, Rio de Janeiro, RJ, Brazil
cHelmholtz Institute for Pharmaceutical Research Saarland, Saarland University, 66123, Saarbrücken, Germany
dState Key Laboratory of Applied Organic Chemistry, College of Chemistry and Chemical Engineering, Lanzhou University, Lanzhou 730000, People's Republic of China
eCollege of Pharmacy, University of Illinois at Chicago, 60612, Chicago, IL, USA
First published on 14th November 2024
Abstract
Covering: 2010 to 2023
Cyanobacterial natural products are a diverse group of molecules with promising biotechnological applications. This review examines the chemical diversity of 995 cyanobacterial metabolites reported from 2010 to 2023. A computational analysis using similarity networking was applied to visualize the chemical space and to compare the diversity of cyanobacterial metabolites among taxonomic orders and environmental sources. Key examples are highlighted, detailing their sources, biological activities, and discovery processes.
1 Introduction
Cyanobacteria are chemically prolific microorganisms that harbor refined biosynthetic machinery for the production of natural products. Capable of performing oxygenic photosynthesis, cyanobacteria contributed for the increase of oxygen in the early atmosphere, allowing for the development of higher forms of life.1,2 Cyanobacteria help sustain the biosphere homeostasis as primary fixers of carbon and nitrogen.3 Over time, they have developed a diversified secondary metabolism which is studied for its ecological implications and biotechnological applications.Cyanobacterial natural products play crucial ecological roles, including feeding deterrence, resource competition, allelopathy, signaling, and UV-protection.4,5 Cyanobacterial toxins are the focus of extensive ecotoxicological studies, due to their association with toxic events and diseases.6 Released during cyanobacterial harmful algal blooms (CyanoHABs), cyanotoxins can induce hazardous effects to exposed individuals, including liver and kidney damage, gastrointestinal disorders, neurological impairment, and dermatitis.7 From a biotechnological perspective, cyanobacteria are a promising source of biofuels,8 food additives,9 supplements,10 and bioactive compounds with pharmaceutical applications.11 Dolastatin 10, for instance, is a cyanobacterial metabolite with potent antiproliferative activity that inspired the synthetic payload of brentuximab vedotin, polatuzumab vedotin, belantamab mafodotin, and enfortumab vedotin which are antibody–drug conjugates currently in use for anticancer treatment.12–17 In addition to cytotoxicity, relevant bioactivities associated with cyanobacterial metabolites include, but are not limited to, antimicrobial, antiviral, protease inhibition, and anti-inflammatory activities.11
The traditional workflow for the discovery of cyanobacterial natural products begins with sample collection, followed by extraction, bioassay or chemical profiling, and bioassay-guided fractionation, leading to the isolation and structure elucidation of the compounds of interest. Over the years, this has been enhanced by the development of more sensitive instruments,18 platforms that allow for the analysis of trace amounts of material,19 and dereplication strategies.20 Omics techniques have expanded possibilities in cyanobacterial natural product research.21,22
By applying a combination of these tools and strategies, research groups continue to discover molecules from marine, freshwater, brackish, and terrestrial cyanobacterial strains. Research programs often focus efforts on investigating cyanobacterial strains from a specific environment type using a few assays for bioactivity screening. Consequently, data from newly reported cyanobacterial metabolites is highly dependent on the research scope of groups focused on identification of cyanobacterial small molecules. Collectively, these efforts have resulted in a group of unique scaffolds, whose chemical diversity is the focus of this paper.
Previous reviews have covered various aspects of cyanobacterial natural products. Baunach et al. (2024) have reviewed new research methods, focusing on genome mining and strategies for high-titer production of cyanobacterial metabolites.21 D'Agostino (2023) has reviewed natural products from symbiotic cyanobacteria and their biosynthesis.23 Kleigrewe et al. (2016)24 highlighted the biosynthetic pathways involved in the secondary metabolism of marine cyanobacteria and Salvador-Reyes and Luesch (2015)25 reviewed the mechanisms of action of marine cyanobacterial metabolites. Dittmann et al. (2015)26 highlighted the biosynthetic diversity of cyanobacterial metabolites. Leão et al. (2012)5 reviewed chemical ecology aspects related to cyanobacterial molecules. Chlipala et al. (2011)27 and Tan (2007)28 have reviewed bioactive compounds from freshwater and marine cyanobacteria, respectively. In a broader perspective, in 2010 Tidgewell et al. have discussed chemical aspects of cyanobacterial molecules found in MarinLit and Scifinder databases along with other literature sources.29 This review adds an assessment of the chemical diversity of all cyanobacterial metabolites published between 2010 and 2023.
The systematic review consisted of ScienceDirect and Web of Science searches for the terms “cyanobacterium” OR “cyanobacteria” AND “natural products”, from January 2010 to December 2023. The search also covered bibliographic references of the included studies. Inclusion criteria were: (1) cyanobacterial origin, (2) first report of the compound or analog in the time period, and (3) unambiguous structure elucidation. This search yielded 365 articles reporting 995 compounds from cyanobacteria collected in different environments (marine, freshwater/terrestrial, and brackish water) and varying geographic locations (Fig. 1). The number of published articles has fluctuated over time with an average of 26 articles per year with slightly more covering on marine cyanobacteria small molecule discovery than strains from the other sources (ESI, Fig. S1†). All 995 compounds are listed in the ESI, Tables S1–S5.† Their metadata were deposited in Zenodo (https://doi.org/10.5281/zenodo.12627504).
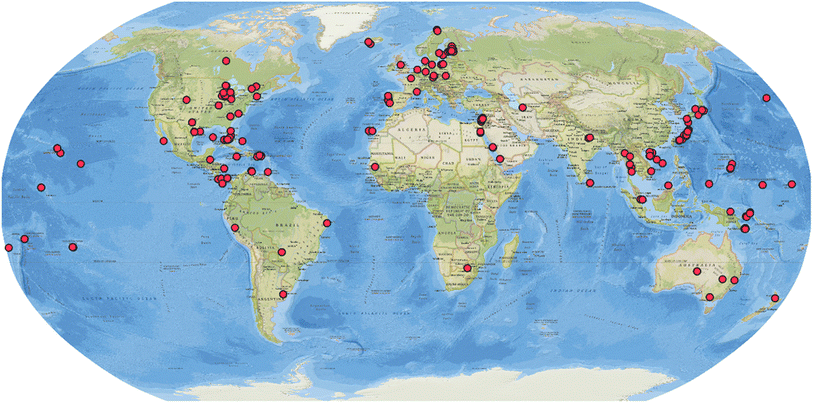 | ||
| Fig. 1 Geographic distribution of the source collection of cyanobacterial strains that are producers of natural products reported between 2010 and 2023. | ||
The similarity networking analysis is detailed in the next section. Selected compounds are described in the following sections, being grouped according to their structural features. Notably, some compounds, mainly those of hybrid biosynthesis, could fit into more than one section of this review. For instance, acyl amides, despite having a hybrid PKS–NRPS biosynthesis, were grouped with fatty acid derivatives due to their structural similarities. In this review, cyanobacterial metabolites were grouped as follows: (a) peptides and lipopeptides, (b) macrolides, cyclophanes, and related compounds, (c) acyl amides and fatty acid derivatives, (d) alkaloids, and (e) miscellaneous compounds (Fig. 2).
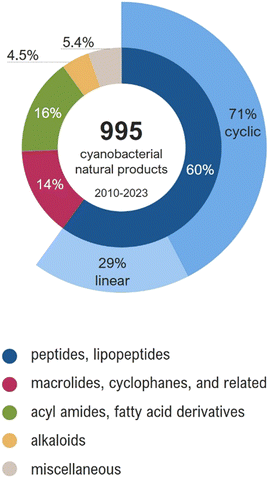 | ||
| Fig. 2 Structural classes of the cyanobacterial metabolites reported between 2010 and 2023 (total of 995 molecules). | ||
Cyanobacterial natural products reported from 2010 to 2023 were produced by strains classified under 57 different genera, with half of them being produced by Microcystis (16%), Nostoc (14%), Lyngbya (11%), and Moorea (9%) (Fig. 3). It is important to note that taxonomic classifications may be based, in some cases, solely on morphology, leading to misrepresentation of genus diversity. These percentages may change as phylogenetic analyses become more widely used and are combined to additional diagnostic criteria in the polyphasic approach for the taxonomic classification of cyanobacteria.30 The environmental sources of the cyanobacterial strains were distributed as follows: marine (52%), freshwater or terrestrial (46%), brackish (2%), and non-reported (1%).
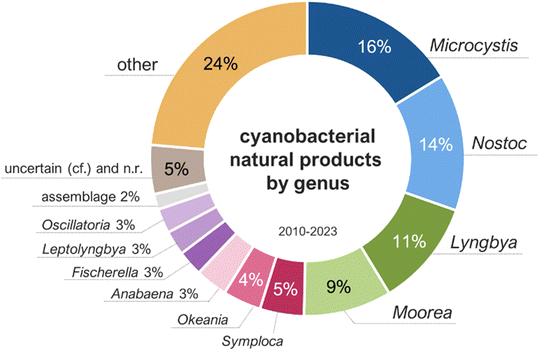 | ||
| Fig. 3 Genus of the cyanobacteria producing metabolites reported between 2010 and 2023 (total of 995 molecules). | ||
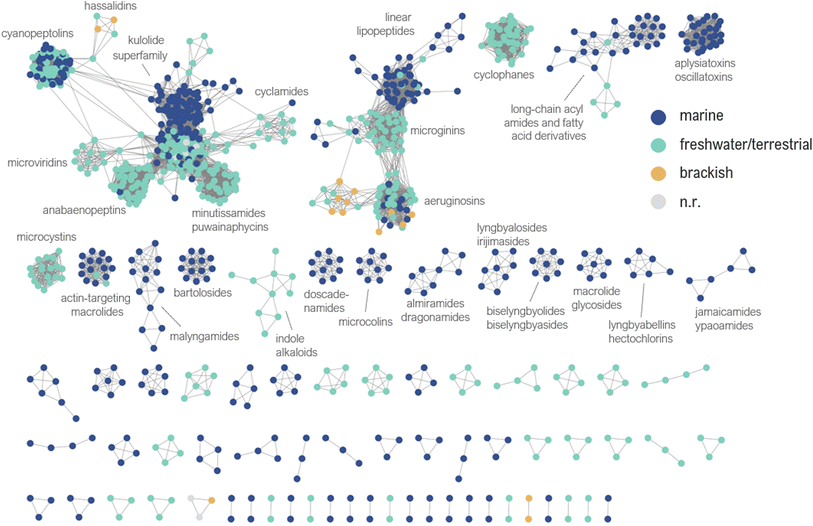
2 Similarity networking
All compounds in this database were analyzed using the DBsimilarity chemoinformatics method to generate similarity networks based on a combination of Morgan fingerprints and Tanimoto similarity coefficients (Fig. 4).31 Similarity values above 0.85 were considered for the identification and formation of distinct clusters according to their ‘main’ structure classification. The two largest clusters in the similarity network are composed of cyclic and linear peptides or lipopeptides, formed by 378 and 141 nodes, respectively.Compounds with low similarity which would typically be represented as isolated nodes are not shown in the network. Instead, they are indicated in the main text and ESI† with an asterisk (*). A total of 63 compounds were found as isolated nodes in the similarity networking analysis, representing 6% of the cyanobacterial natural products reported from 2010 to 2023. Most of them are classified in this review as acyl amides or fatty acid derivatives (27%), followed by alkaloids (22%), miscellaneous (22%), linear peptides or lipopeptides (16%), macrolide or related (11%), and cyclic peptides or lipopeptides (2%).
Both the database and the similarity network have been deposited in Zenodo (https://doi.org/10.5281/zenodo.12627504). Embedded metadata can be used to create visual filters in the similarity network, aiding in inferences and hypothesis development about the chemical diversity of cyanobacterial natural products.
Most often compound families were found to be exclusively produced by cyanobacteria from specific environments, although in some cases structurally similar compounds were discovered from strains collected from different environments (Fig. 4). A similarity network containing all node labels is provided (ESI, Fig. S2†).
With respect to cyanobacterial taxonomy (orders classified according to Komárek et al. 2014),30 it was interesting to identify groups of closely related compounds isolated from cyanobacteria classified under different orders (e.g. actin-targeting macrolides from Oscillatoriales, Nostocales, and Synechococcales) (ESI, Fig. S3†). We invite the readers to explore the similarity network according to their own research interests. It may also be used to assist dereplication or provide valuable insights in cyanobacterial natural product discovery.
3 Peptides and lipopeptides
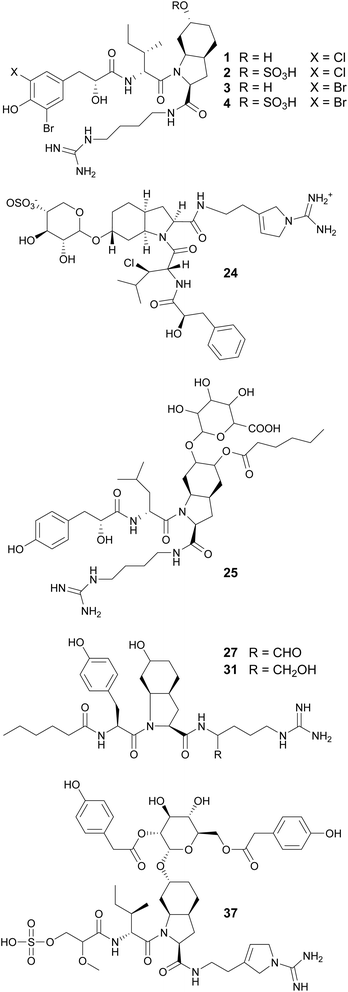
Peptides and lipopeptides account for 60% of the cyanobacterial compounds in this review (Fig. 2). Lipopeptides refer to metabolites of peptidic nature that are prenylated or have lipophilic extensions comprised of polyketide (PK) or fatty acid units. Collectively, these linear and cyclic molecules range from smaller tripeptides to large glycosylated lipopeptides. Other features that greatly increase the chemical diversity of cyanobacterial peptides are the incorporation of D-amino acids, homo variants, and other nonstandard amino acids. Additionally, N-methylation, O-methylation, hydroxylation, reduction, and halogenation are common modifications of cyanobacterial peptides.32 These compounds display biological activities such as protease inhibition, cytotoxicity, protein kinase inhibition, antimicrobial, antifungal, and antiparasitic. About half of the peptides and lipopeptides in this review fall into one of the following classes: aeruginosins, microginins, anabaenopeptins, cyanopeptolins, microcystins, cyclamides, microviridins, and cyanobactins.32Aeruginosins are usually composed of the following residues: a derivative of N-terminal hydroxy-phenyl lactic acid (Hpla), a hydrophobic amino acid, 2-carboxy-6-hydroxyoctahydroindole (Choi), and an arginine derivative, commonly agmatine (Agm). Most aeruginosins are reported from bloom-forming Microcystis and Nodularia, but variants have also been found in Planktothrix and Nostoc strains. Four Ile-containing aeruginosins and one Leu-containing aeruginosin were reported from a freshwater Microcystis aeruginosa bloom Collected in Kibbutz Geva, Israel (1–5).33 They are halogenated variants that were shown to inhibit serine proteases; the hydroxylated derivatives displayed more potent activity than the correspondent sulfated derivatives. Additional aeruginosins (6–23) were isolated from Microcystis.34–39
Aeruginosins 828A (24)40 and 865 (25)41 contain glycosylated Choi units. They were isolated from cultured freshwater Planktothrix sp. and Nostoc sp., respectively. Aeruginosin 828A (24) showed toxicity to the crustacean Thamnocephalus platyurus (LC50 of 22.4 mM) and potent inhibitory activity to thrombin (IC50 of 21.8 nM) and trypsin (IC50 of 112 nM).40 Aeruginosin 865 (25) showed anti-inflammatory activity by inhibiting the translocation of NF-κB (nuclear factor kappa B) to the nucleus.41 From marine strains of Nodularia spumigena, eleven aeruginosin derivatives were found to contain argininal (NAL 1–4, 26–29) or argininol (NOL 1–7, 30–36) instead of Agm on the C-terminus. These compounds possess N-terminal fatty acid moieties of varying chain lengths (C2–C10).42 Varlaxins (37 and 38), from a Nostoc sp. strain, contain a 2-O-methylglyceric acid 3-O-sulfate moiety on the N-terminus and a rare α-D-glucopyranose moiety derivatized with two 4-hydroxyphenylacetic acid residues decorating the Choi unit.4337 showed potent inhibitory activity against human trypsin isoenzymes (low nanomolar IC50 values).
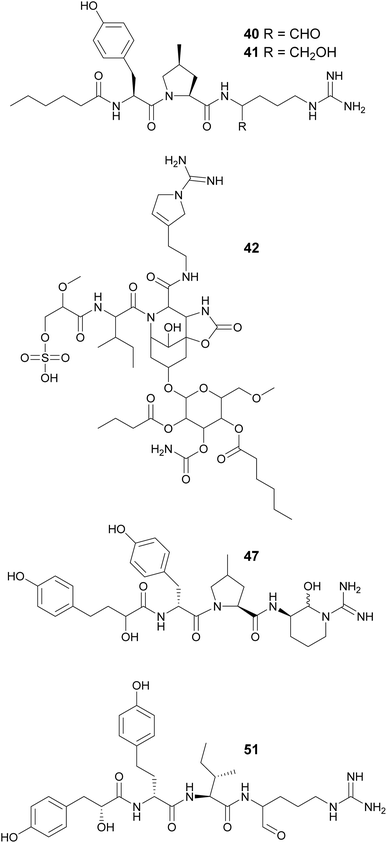
Aeruginosin variants without the Choi unit include pseudoaeruginosins, suomilides, spumigins, and pseudospumigins. Pseudoaeruginosin KT554 (39), from a Microcystis aeruginosa bloom, has a Phe residue in place of Choi.34 Pseudoaeruginosins NS1 (19, 40) and NS2 (20, 41), from Nodularia spumigena, have a fatty acid (hexanoic acid) on the N-terminus and Me-Pro in place of Choi.44 Compound 40 showed potent trypsin inhibitory activity. Evidence suggests that pseudoaeruginosins derive from a hybrid aeruginosin/spumigin pathway.44 Suomilides B–E (42–45) were isolated from a Nostoc sp. strain collected in Kvaløya island, Northern Norway.45 Their unique azabicyclononane core is proposed to originate from prephenate and the putative suomilide biosynthetic gene cluster (BGC) contains genes also present in the aeruginosin and saxitoxin BGCs.45
Spumigins are aeruginosin variants where the Choi residue has been replaced by Pro or Me-Pro. Spumigin J (27,46) was isolated from an extract of the cultured freshwater Anabaena compacta and was shown to inhibit thrombin and cathepsin B, proteases associated with oncological conditions.46 Spumigins K–N (47–50), isolated from a Sphaerospermopsis torques-reginae strain collected in Brazil, contain a 2-hydroxy-4-(4-hydroxyphenyl)butanoic acid residue in the N-terminus.47 Pseudospumigins A–F (51–56) are a new peptide family discovered from a Nostoc sp. strains collected in Brazil, containing Ile, Leu, or Val in place of the Pro or Me-Pro unit found in spumigins. A homology analysis indicated that the BGC encoding for spumigins share similarities with the putative nostosin G (57) BGC.48 Nostosins are tripeptide trypsin inhibitors. Nostosins A and B (58 and 59)49 were isolated from Nostoc sp. and nostosin G (57) from a Dolichospermum sp. strain, along with spiroidesin B (60).50 Microguanidines (61*, 62*, and 63*) are commonly reported from strains that are aeruginosin producers.34,35,51
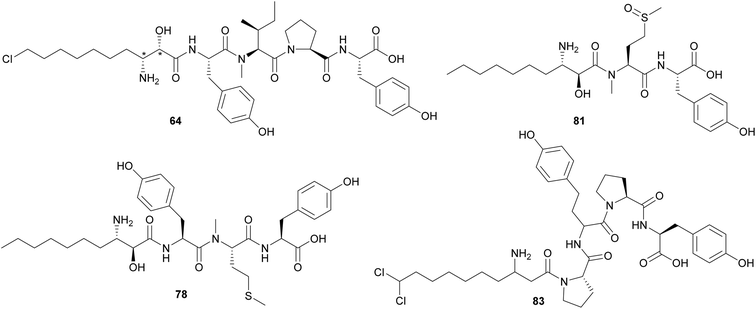
Microginins are linear peptides characterized by the presence of a decanoic acid derivative at the N-terminus, often 3-amino-2-hydroxy-decanoic acid (Ahda), and tyrosine units on the C-terminus. Microginins (64–74) with different absolute configurations and varying substituents in the Ahda moiety were isolated from freshwater Microcystis sp. blooms collected in Israel.52,53 Microginin GH787 (64) showed inhibitory activity against bovine aminopeptidase N and microginins 65–74 showed inhibitory activity against bovine aminopeptidase M with low-micromolar IC50 values.52,53 Three 3-amino-2-hydroxy-octanoic acid (Ahoa)-containing microginins (75–77) were isolated from a cultured Microcystis aeruginosa.54 Using a metabolomics approach, five new microginin variants (78–82) were discovered from a Microcystis aeruginosa in culture.55,56 Microginins 527 (81) and 511 (82) are the first described tripeptide analogues. Heterologous expression of a hybrid polyketide synthase/non-ribosomal peptide synthetase (NRPS–PKS) BGC from a Microcystis aeruginosa into a Escherichia coli led to successful production of microginins by the host, providing experimental proof that the BGC is associated with the biosynthesis of microginins.57 This work also described new variants (83–91) containing 3-aminodecanoic acid fatty acyl units and homophenylalanine and homotyrosine residues, which are unusual in microginins.57
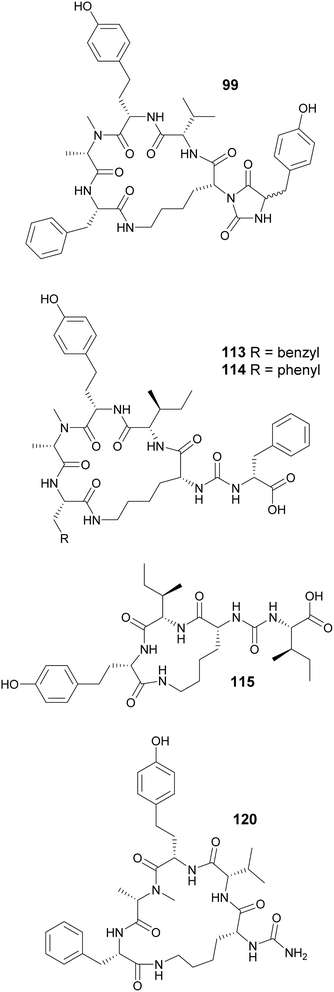
Macrocyclization in cyanobacterial peptides occur in various ways: through regular peptide bonds, ester bonds (depsipeptides), side-chain bridges between two amino acids, or through linkage between the side chain of one residue and the carboxyl group of another, such as in case of the anabaenopeptins. Cyclization of anabaenopeptins occurs between the side chain of a Lys residue and the carboxy group of the C-terminal amino acid residue. Anabaenopeptins (92–98) were isolated from freshwater blooms of Microcystis.35,38,58,59 Anabaenopeptin MM913 (94) contains a rare N-methyl-homoaromatic amino acid at the second position.58 Unique hydantoin derivatives of anabaenopeptins were found in Microcystis biomass collected in Israel (99–103).59 Anabaenopeptin variants were also isolated from Nodularia (104–106),60Brasilonema (107–110),61Woronichinia (111),62 and Nostoc (112)63 strains, highlighting the broad taxonomic distribution of this compound family. Lyngbyaureidamides A (113) and B (114), isolated from a freshwater Lyngbya sp., are the first examples anabaenopeptins containing a D-amino acid as an exocyclic residue.64 Namalides B and C (115 and 116) are anabaenopeptin-like compound, with a tripeptide core, discovered from a Sphaerospermopsis torques-reginae strain.65
A strategy combining high-density cultivation with trascriptomic and metabolomics studies led to the discovery of anabaenopeptin variants (117–119) from terrestrial Nostoc strains.66 Anabaenopeptin KVJ811 (119) was shown to display allelopathic effects against another Nostoc strain.66 The carboxypeptidase-inhibiting anabaenopeptin 679 (120) was reported in as minor compound in the biomass filtered from eight million liters of freshwater lake water (Grand Lake-Saint Marys, Ohio), obtained by countercurrent filtration.67 Anabaenopeptin 120 has a truncated structure, lacking the exocyclic amino acid. Thirteen anabaenopeptins (121–133) were isolated from Planktothrix or Nostoc strains and their structure–activity relationship were investigated with respect to inhibition of TAFIa (activated thrombin activatable fibrinolysis inhibitor).68 Results suggested that the exocyclic amino acid residue plays a critical role in TAFIa inhibition.
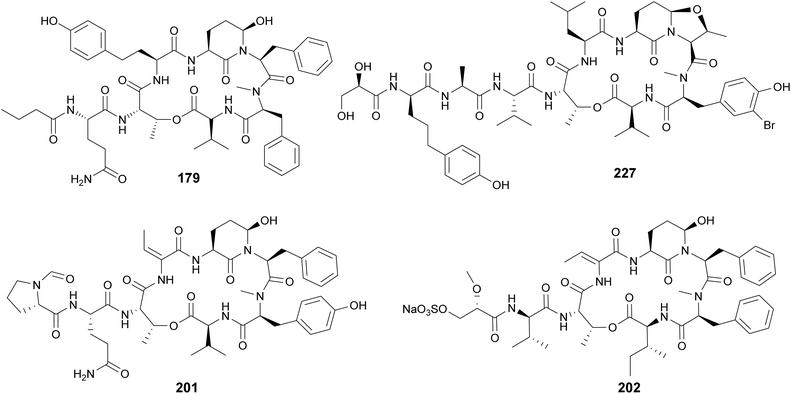
Cyanopeptolins, which include the micropeptins, form another large group of cyclic peptides. They are characterized by the presence of a 3-amino-6-hydroxy-2-piperidone (Ahp) residue and macrocyclization in an ester bond between the C-terminal amino acid and the β-hydroxy group of Thr. They often display protease inhibiting properties. Several micropeptins were isolated from freshwater Microcystis bloom material (134–181), many of them with serine protease inhibitory activity.34,38,39,52,58,69–73 Micropeptin 996 (179) was discovered using an untargeted metabolomics approach to investigate a cultured Microcystis aeruginosa strain.74 Micropeptins 982 (180) and 957 (181) were discovered by a combination of prefractionation, LC-MS/MS molecular networking, and bioassay analyses after processing cyanobacteria-containing biomass collected from a holding lagoon in Michigan, USA.75 Micropeptins 180 and 181 were found to display anti-neuroinflammatory activity in a BV-2 mouse microglial cell assay. Cyanopeptolin variants were isolated from freshwater Microcystis aeruginosa (182)76 and Woronichinia naegeliana (183)62 in culture, as well from a marine Nostoc edaphicum strain (184–200).77,78 Cyanopeptolin 1020 (182) was shown to be a highly potent trypsin inhibitor, with an IC50 of 0.67 nM. It also inhibited factor XIa and kallikrein with IC50 values of 3.9 nM and 4.5 nM, respectively.76 Stigonemapeptin (201) is a cyanopeptolin with elastase and chymotrypsin inhibitory activities discovered in a bloom of freshwater Stigonema sp.79 It features the nonstandard amino acids 2-amino-2-butenoic acid (Abu) and a N-formylated Pro.
Abu residues are also found in marine cyanopeptolins, such as symplostatins 5–10 (202–207),80 tutuilamides A–C (208–210),81 bouillomides (211 and 212),82 and molassamides (213 and 214).83,84 Symplostatins 5–10 (202–207) are potent elastase inhibitors isolated from the marine cyanobacterium Symploca sp. collected in Guam. Elastase has been shown to play a role in diseases associated with chronic inflammation (e.g. cystic fibrosis, asthma, chronic obstructive pulmonary disease, among others). To evaluate the observed activity of symplostatins in a more medically relevant assay, 202 was tested in a bronchial epithelial model system. It was shown to alleviate the cellular effects (viability, detachment, apoptosis) induced by elastase.80 Molassamide (213) was discovered in field collections of a marine Dichothrix utahensis, representing the first peptide from this genus.83 Molassamide B (214) along with rivulariapeptolides (215–218) were identified in an environmental Rivularia sp. biofilm using a native metabolomics screen for protease inhibitors.84 Rivulariapeptolide 1155 (219), from a marine Rivularia sp., was discovered by a combination of untargeted chemical profiling and the use of CANOPUS for dereplication.85 Additional cyanopeptolins from marine cyanobacteria include jizanpeptins A–E (220–224) from Symploca sp.,86 kempopeptin C (225) from Lyngbya confervoides,87 kyanamide (226) from Caldora penicillata,88 and largamide D oxazolidine (227) from Lyngbya cf. confervoides.89 Largamide D oxazolidine (227) has an intriguing intramolecular condensation between Ahp and Thr, which was suggested to modulate protease inhibitory activity.
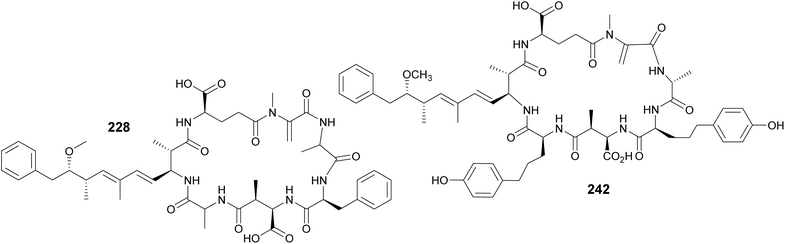
Microcystins are a widely studied group of cyanobacterial cyclic peptides often associated with CyanoHABs. When blooms contaminate drinking water systems, microcystins pose a major public health issue, as they can cause liver damage to exposed humans and animals.90 Their chemical structures include a characteristic (2S,3S,8S,9S)-3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid (Adda) residue, as well as aspartate and glutamate derivatives. Another uncommon amino acid often present in microcystins is N-methyldehydroalanine (Mdha). Four Mdha-containing microcystins (228–231) have been reported from a Microcystis sp. collected in Lake Hakanoa, New Zealand.91,92 From a Microcystis aeruginosa strain collected in Pila, Argentina, seven microcystin congeners were discovered (232–238).93 Highlighting the ubiquitous distribution of these metabolites, three microcystin variants (239–241) were detected from Microcystis strains collected in South Africa and Australia.94 Microcystin variants (242–245) were reported in a biomass collected from a holding lagoon in Michigan, U.S.A, which was concentrated using a filter belt press.95 Microcystins 242–244 contain 2-amino-5-(4-hydroxyphenyl)pentanoic acid (Ahppa) units.
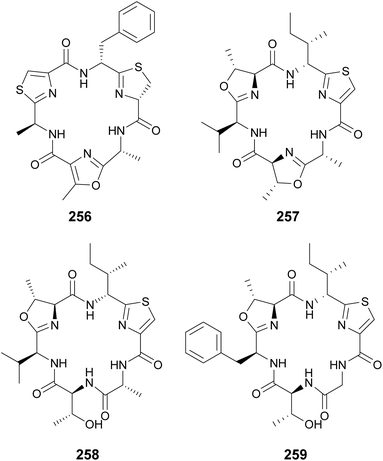
Cyclamides are cyclic hexapeptides characterized by oxazole and thiazole moieties intercalated between amino acids residues. Oxazole and thiazole heterocycles are derived from Thr, Ser, and Cys amino acids.96 Microcyclamides (246–255) were isolated from Microcystis sp. bloom material collected in Israel.97,98 Hapalocyclamide (256) is a phytotoxic cyclamide isolated from a terrestrial Hapalosiphon sp. collected in Thailand.99 It was shown to inhibit lettuce seedling growth in micromolar concentrations. Often, cyclamide-like compounds are identified with non-cyclized rings, i.e. containing unmodified Thr, Ser, and Cys instead of the heterocycles, such as in the balgacyclamides. Balgacyclamides A–C (257–259) were isolated from a Microcystis aeruginosa strain.100 While balgacyclamide A (257) has the typical cyclamide structure with heterocycles in positions 1, 3, and 5, balgacyclamides B (258) and C (259) have non-cyclized Thr residues in position 5. Cyclamides 257 and 258 showed antimalarial activity with IC50 values of 9.0 and 8.2 μM, respectively.
Cyclamides are considered part of the cyanobactin super-family of compounds. Cyanobactins include molecules formed exclusively by proteinogenic amino acids, often cyclized and sometimes prenylated.96 Genomic and metabolomic approaches have been increasingly used to search for new cyanobactin variants. Genome mining for cyanobactin prenyltransferases led to the discovery of tolypamide (260) from a freshwater Tolypothrix sp.101 Anacyclamides (261–271) were identified in Anabaena strains using a combination of genome mining, 34S stable isotope labeling and LC-MS analyses.102 Heterologous expression in Escherichia coli was accomplished for compound 261. Genome mining, stable isotope labeling, and heterologous expression were applied for investigating an inactive gene cluster from Microcystis aeruginosa, leading to the discovery of piricyclamides (272 and 273).103 Genome mining also enabled the discovery of prenylagaramide C (274) from Planktothrix agardhii,104 arthrospiramides (275–278) from Arthrospira platensis (“Spirulina”) supplements,104 and cyanothecamides (279–281) from Cyanothece sp.105 Interestingly, the latter were only detected when cultures were subjected to heat shock (37 °C, 24 h).
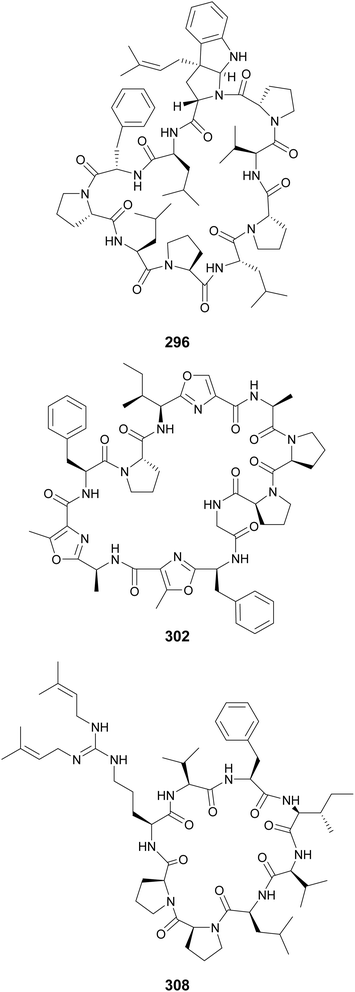
Additional cyclic cyanobactins include aestuaramides (282–293) from the marine Lyngbya aestuarii,106 autumnalamides (294 and 295) from a marine Phormidium autumnale strain,107,108 trikoramides from a marine Symploca hydnoides (296–299),109,110 microphycin KB921 (300) from a freshwater Microcystis spp. bloom,38 microseiramide (301) from a cultured strain of freshwater Microseira sp.,111 wewakazole B (302) from a marine Moorea producens,112 aeruginazoles (303–306) from Microcystis,113,114 sphaerocyclamide (307) from a freshwater Sphaerospermopsis sp.,115 and argicyclamides (308–311) from Microcystis aeruginosa.116,117 Wewakazole B (302) was shown to have cytotoxic activity against MCF-7 breast cancer cells (IC50 value of 0.58 μM) and H460 lung cancer cells (IC50 value of 1.0 μM).112 Compounds 294, 308, 309, and 311 contain unique guanidine prenylations. A C-prenylated cyclotryptophan is found in trikoramides 296–299.
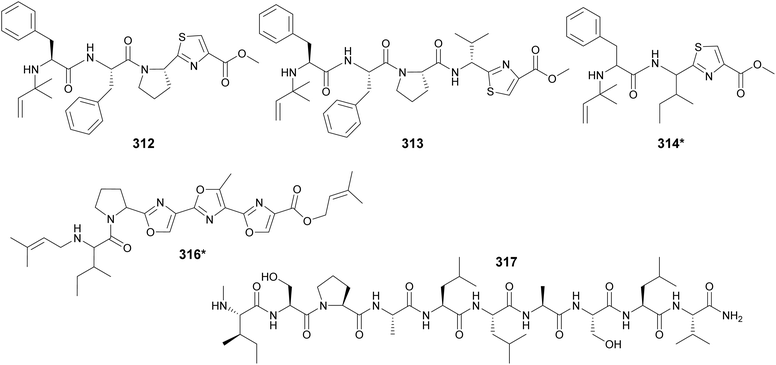
Cyanobactins were traditionally described as cyclic peptides, but a study of 126 cyanobacterial genomes led to the identification of three linear compounds produced by the cyanobactin pathway. Aeruginosamides B and C (312 and 313) were isolated from Microcystis aeruginosa and viridisamide A (314*) from Oscillatoria nigro-viridis.118 They are short peptides (3–5 amino acids) that have O-methyl groups and N-prenyl groups protecting their C- and N-termini, respectively. Aeruginosamide variants (315) were also described from Limnoraphis sp., isolated from Baltic Sea brackish water.119 Muscoride B (316*) is a linear cyanobactin with three contiguous oxazole units and prenylations on both termini.120 Scytodecamide (317) is the longest known linear cyanobactin.121 It was discovered during a metabolomics study investigating the effect of culture conditions on a freshwater Scytonema sp. strain.122
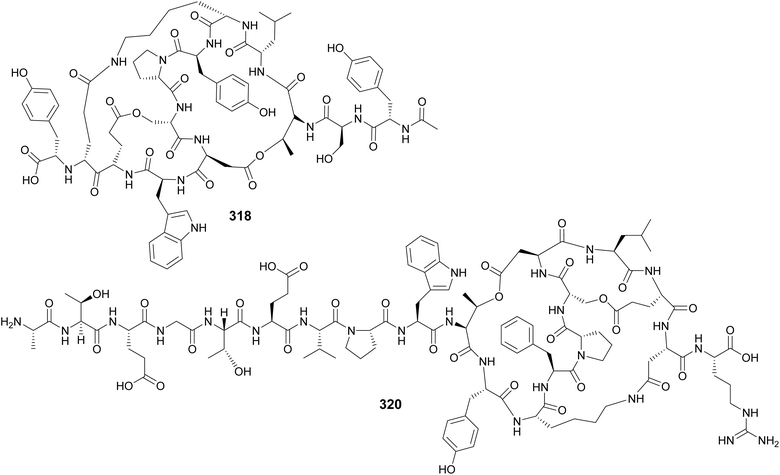
Microviridins are large ribosomally synthesized and post-translationally modified peptides (RiPPs), members of the graspetide family, characterized by a multicyclic structure formed by side chain bridges.123 Microviridin LH1667 (318), from a freshwater bloom of Microcystis spp., is comprised of four macrocycles and 13 amino acid residues. It was found to inhibit chymotrypsin and elastase with IC50 values of 2.8 μM and 20 nM, respectively.37 Microviridin 1777 (319), from a cultured M. aeruginosa, was discovered by a combination of bioactivity screening, genome mining, and metabolite profiling by UHPLC-HRMS/HRMS.124 Genomics, transcriptomics, and high-density cultivation led to the discovery of microviridin variants with unprecedented N-terminal extensions (320–326) from a Nostoc punctiforme strain.125 A study on the chemical ecology of mat-forming Nostoc and its epibionts showed the influence of quorum-sensing signals on the production of microviridins (327–329), suggesting the potential role of microviridins in epibiont–cyanobacterium interactions.126
The aforementioned chemical groups cover 55% of the peptides and lipopeptides included in this review. The following peptidic compounds are chemically diverse and were grouped according to common (or uncommon) molecular features.
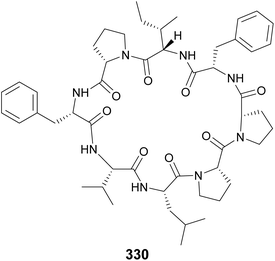
Samoamide A (330) was discovered using a combination of MS/MS molecular networking and bioassay-guided fractionation from a marine sample of cf. Symploca sp. obtained at the American Samoa.127 It showed cytotoxicity against several cancer cells lines, with an IC50 of 1.1 μM against H460 human non-small-cell lung cancer cells being most potent. It contains only proteinogenic amino acids, like the cyanobactins. However, its ribosomal origin cannot be assumed given the presence of a two consecutive Pro residue unit previously shown to inhibit ribosomal translation. Consecutive Pro residues are also found in croissamide (331)128 from a marine Symploca sp. and motobamide (332)129 from a marine Leptolyngbya sp.
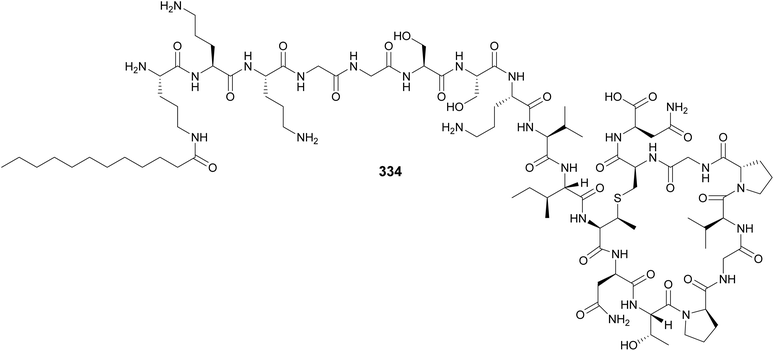
Lingaoamide (333) from a freshwater Oscillatoria sp. is a cyclic heptapeptide composed of proteinogenic amino acids and N-methylations.130 Kamptornamide (334) from a freshwater Kamptonema sp. was discovered by a genome-guided approach aimed to identify ribosomally derived peptides acylated with fatty acid units, followed by heterologous expression in E. coli.131
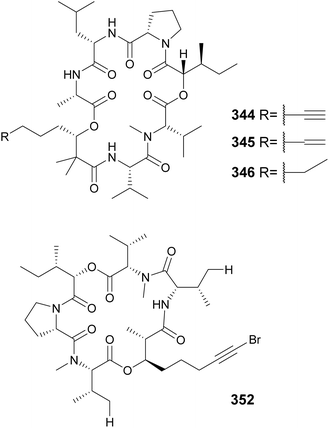
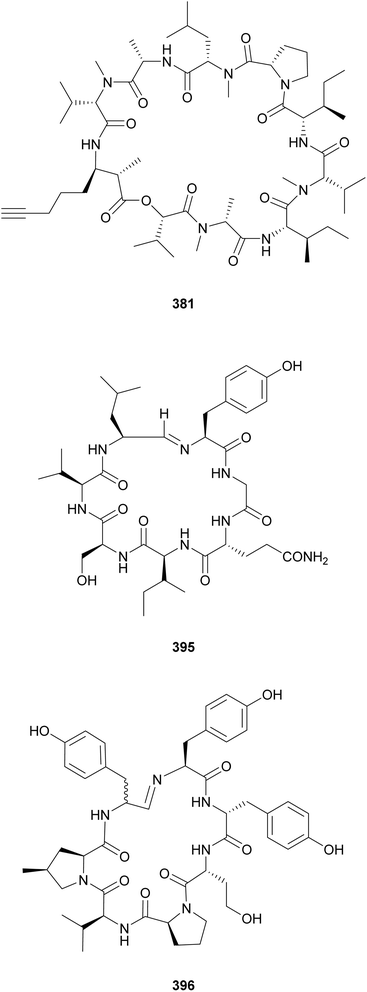
The “kulolide superfamily” of compounds include cyclic depsipeptides, NRPS–PKS hybrids, containing a 2,2-dimethyl-3-hydroxy-7-octynoic acid (Dhoya) or a 3-hydroxy-2-methyl-7-octynoic acid (Hmoya) moiety, or their alkene and alkane analogs 2,2-dimethyl-3-hydroxyoctanoic acid (Dhoea), 2,2-dimethyl-3-hydroxyoctanoic acid (Dhoaa), 3-hydroxy-2-methyl-7-octenoic acid (Hmoea), and 3-hydroxy-2-methyl-7-octanoic acid (Hmoaa). Kulolide-type metabolites containing Dhoya/Dhoea/Dhoaa include pitipeptolides C–F (335–338)132 and benderamide A (339)133 from marine Lyngbya strains, viequeamides A–D (340–343) from a marine Rivularia sp.,134 and kohamamides A–C (344–346) from a marine Okeania sp.135 Also from marine strains, kulolide-type compounds with Hmoya/Hmoea/Hmoaa units include hatupeptins B and C (347 and 348)136 and tiahuramides A–C (349–351)137 from Lyngbya, veraguamides (352–361) from Symploca cf. hydnoides138 and cf. Oscillatoria margaritifera,139 odobromoamide (362) from Okeania sp.,140 and trikoveramides A–C (363–365) from a Symploca hydnoides collected in Indonesia.141 Veraguamide A (352) displayed potent cytotoxic activity against H460 human lung cancer cells with an LD50 of 141 nM. From the same collection of cf. Oscillatoria margaritifera in Panama, the linear veraguamides K and L (366 and 367) were isolated.139
Cyanobacterial peptides containing Dhoya/Hmoya or their reduced derivatives that do not fit into the kulolide group include the cocosamides (368–369),142 floridamide (370),143 pemukainalides (371 and 372),144 yuvalamides (373 and 374),145 guineamide G (375),146 and dudawalamides (376–379).147 Yuvalamide A (373), from a Lyngbya strain collected in Panama, was discovered in a metabolomics analysis of marine algae and cyanobacteria collected around the globe. Guineamide G (375), from Lyngbya, was cytotoxic to mouse neuroblastoma cells with an LC50 of 2.7 μM. The same study also described the discovery of wewakamide A (380).146 Dudawalamides 376–379 were shown to display antiparasitic activity in leishmaniasis, malaria, and Chagas disease assays, with no significant cytotoxicity detected against H-460 human lung carcinoma cells.147
The β-amino acid Amoya (3-amino-2-methyl-7-octynoicacid) shares structural similarities with the hydroxy acid Hmoya. Amoya-containing peptides include the companeramides (381 and 382) from an assemblage of marine filamentous cyanobacteria,148 portobelamides (383 and 384), and caciqueamide (385*) from a marine Caldora sp.149 Other rare β-amino/hydroxy acids found in cyanobacterial peptides are Dmhha (2,2-dimethyl-3-hydroxyhexanoic acid unit) in palmyramide A (386)150 and pitiprolamide (387);151 Amha (3-amino-2-methylhexanoic acid) in precarriebowmide (388),152 medusamide A (389),153 and kakeromamide A (390);154 Amoa (3-amino-2-methyloctanoic acid), in kakeromamide B (391)155 and porpoisamides (392 and 393);156 and Hoa (3-hydroxyoctanoic acid), in scytonemide B (394) from a cultured freshwater Scytonema hofmannii strain.157 From the same strain, scytonemide A (395), a cyclic peptide closed by an unusual imino bond, was also isolated.157
An imino bond is also found in nostocyclopeptide M1 (396), which is composed of several nonproteinogenic amino acids, including 4-methylproline (4-mPro).158 Nostocyclopeptide M1 (396), isolated from a Baltic Sea Nostoc strain, was shown to act as an antitoxin, blocking the uptake of microcystin and nodularin by hepatocytes.159 Nostocyclopeptide Ncp-A2-L (397), from another Baltic Sea Nostoc, is a linear variant with a terminal aldehyde.160 A screening of 116 cyanobacterial strains for 4-mPro biosynthetic genes by PCR led to the discovery of nostoweipeptins W1–W7 (398–404) and nostopeptolides L1–L4 (405–408) from Nostoc sp.161 Nostopeptolides L1–L4 (405–408) contain the fatty acid derivative 2-methylhex-2-enoic acid (Mhea) as an extension of their peptidic macrocycle.
Grassypeptolides are cyclic depsipeptides containing a bis-thiazoline moiety and the β-amino acid 2-methyl-3-aminobutyric acid (Maba). They were isolated from marine Lyngbya confervoides (409 and 410),162Leptolyngbya sp. (411 and 412),163 and Lyngbya majuscula (413 and 414)164 strains. Grassypeptolides demonstrate cytotoxicity to different cancer cells, with grassypeptolides C–E (410–412) being most potent (nanomolar range IC50 values). Along with grassypeptolides D and E (411 and 412), Ibu-epidemethoxylyngbyastatin 3 (415) was also reported.163 It contains a distinct β-keto amino acid 4-amino-2,2-dimethyl-3-oxopentanoic acid (Ibu). Additional β-amino acids with short chains found in marine cyanobacteria include 2-methyl-3-aminopentanoic acid (Map), such as in urumamide (416) from Okeania sp.,165 and 2,4-dimethyl-3-aminopentanoic acid (dolamethylleucine, Dml), in triproamide (417) from Symploca hydnoides.166
Muscotoxins (418 and 419) are 3-amino-2,5-dihydroxydecanoic acid (Ahdoa)-containing lipopeptides isolated from a soil Desmonostoc muscorum strain.167 Muscotoxin A (418) was shown to disrupt mammalian cell membranes by reducing surface fluidity. Desmamides A–C (420–422), from a Desmonostoc muscorum strain isolated from a cycad (Cycas revoluta) root, contain a 3,5-dihydroxy-2-methyldecanoic acid (Dhmda) unit and a glycosylated tyrosine.168 Genome mining and substrate incubation experiments were conducted to described the NRPS–PKS BGC proposed to encode for the production of desmamides.168
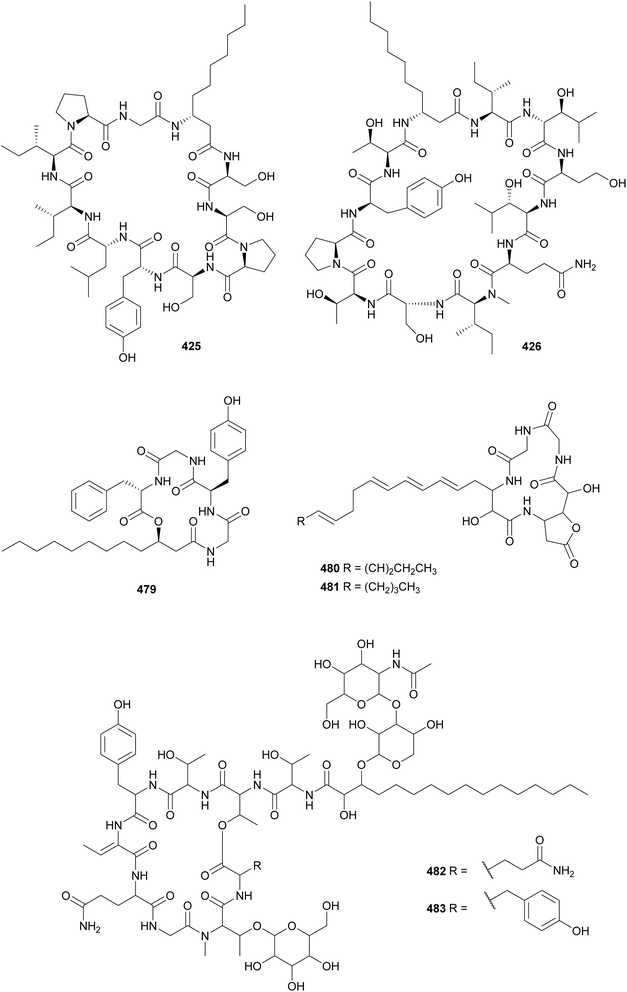
Laxaphycins are cyclic lipopeptides found in both marine and freshwater cyanobacteria that harbor a β-aminodecanoic or β-aminooctanoic acid residue, as well as a few D-amino acids in their structure. They are subclassified as type-A and type-B, containing 11 or 12 amino acid residues, respectively. Laxaphycins reported in recent years include lyngbyacyclamides A and B (423 and 424) from a marine Lyngbya sp.,169 trichormamides A–D (425–428) from freshwater Trichormus sp.170 and cf. Oscillatoria sp. strains,171 laxaphycins B4 (429) and A2 (430) from a marine Hormothamnion enteromorphoides,172 laxaphycins B5 and B6 (431 and 432) from a freshwater Phormidium sp.,173 heinamides (433–440) from a freshwater Nostoc sp.,174 and several laxaphycin analogs including acyclic variants (441–447) from collections of marine Anabaena torulosa.175,176 Acyclic laxaphycins were proposed to be biosynthetic precursors.176 Moderate antimicrobial activity and antiproliferative activity against cancer cell lines were associated with the herein described laxaphycin-type compounds. Evidence shows that type-A and type-B may act synergistically in biological assays.172,174
β-Amino fatty acids with 10–18 carbon chains have been found in lipopeptides from freshwater cyanobacteria such as the minutissamides (448–459)177,178 and puwainaphycins (460–478).179–183 Compounds 448–478 have a decapeptide macrocycle containing a β-amino fatty acid residue with a side chain commonly modified by methylation, chlorination, hydroxylation, or oxidation. Their peptidic macrocycle also contains one or two dehydrobutyrine (Dhb) residues. Minutissamides A–L (448–459) were isolated from Anabaena strains.177,178 They showed biological activity against cancer cell lines in the low micromolar range. Puwainaphycins (460–473) were isolated from Cylindrospermum alatosporum179–182 and puwainaphycins H–L (474–478) were isolated from Nodularia harveyana.184 Genome mining helped identify putative BGCs encoding for puwainaphycin production in C. alatosporum and N. harveyana.180,184 Their biosynthesis starts with a fatty acyl-AMP ligase activating the fatty acid residue, which is elongated by PKS modules, followed by NRPS modules that incorporate the remaining amino acid residues.179
Unnarmicin D (479) is a 5-residue lipopeptide isolated from a marine Trichodesmium thiebautii.185 It features the unusual β-amino fatty acid 3-hydroxydodecanoic acid (3-Hdda), two Gly, D-Tyr, and L-Phe. The nomenclature was established due to the similarity of 479 with the unnarmicins from Photobacterium sp. Unnarmicin D (479) showed no antibacterial or cytotoxic activities. Anabaenolysins A (480) and B (481) are cyclic lipopeptides isolated from two benthic marine Anabaena samples.186 Anabaenolysin A 480 contains a 3-amino-2-hydroxyoctadeca-5,7,9,13,15-pentaenoic acid (Ahopa) while anabaenolysin B 481 contains a 3-amino-2-hydroxyoctadeca-5,7,9,15-tetraenoic acid (Ahota) unsaturated β-amino fatty acids. Both compounds displayed cytotoxic activity against different mammalian cell lines in the low micromolar range (LC50 values ranging from 3 to 17 μM).
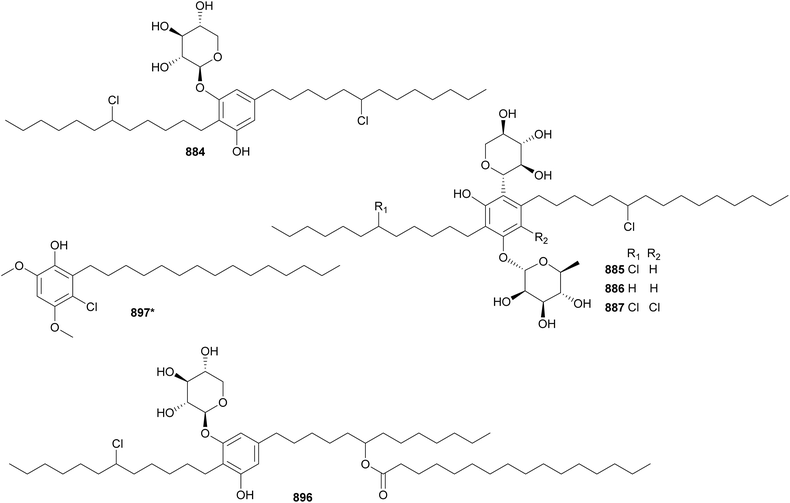
Hassalidins and balticidins are glycolipopeptides with antifungal activity. Hassallidins C (482) and D (483) were discovered during a screening of 99 strains from the genus Anabaena for the presence of the hasV gene, one of the genes involved in the biosynthesis of hassallidins.187 The serendipitous discovery of the hassalidin BGC led to the discovery of hassallidin E (484) in a Planktothrix serta strain.188 Balticidins A–D (485–488) were isolated from an Anabaena cylindrica collected from the Baltic Sea, using a bioassay-guided fractionation approach based on antifungal activity.189
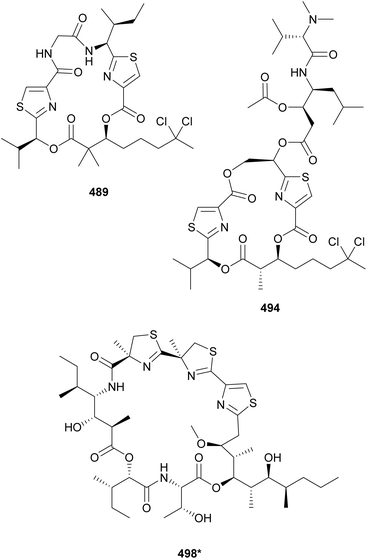
Lyngbyabellins are unique lipopeptides that include a chlorinated 2-(di)methyloctanoate unit and thiazole rings. This family of compounds was expanded with the discovery of 27-deoxylyngbyabellin A (489) and lyngbyabellin J (490) in a marine Lyngbya bouillonii strain from Guam.190 Both compounds displayed cytotoxic activity, with 489 being the most potent (IC50 values of 12 and 7.3 nM against HT-29 and HeLa cells, respectively). Lyngbyabellins K–N (491–494) as well as 7-epi-lyngbyabellin L (495) were reported from marine Moorea bouillonii samples.191 Lyngbyabellin N (494) has a particularly complex structure that includes the lyngbyabellin core and a side chain with a terminal diMe-Val, resembling the structure of the dolastatin 10 family of compounds. The authors suggest that lyngbyabellin 494 might be the result of the biosynthetic hybridization of the two natural product classes. Lyngbyabellin 494 showed strong cytotoxicity against H-460 and HCT116 colorectal carcinoma cells with IC50 values of 4.8 nM–1.8 μM (high variability reported) and 40.9 nM, respectively. Lyngbyabellins O and P (496 and 497) were isolated from a marine Okeania sp. They showed antifouling activity in an Amphibalanus amphitrite larvae assay with EC50 values of 0.38 μM for 496 and of 0.73 μM for 497.192
Hoiamides are cyanobacterial lipopeptides containing two consecutive thiazolines and one thiazole connected to a polyketide unit. Hoiamide B (498*) was obtained from an assemblage of Symploca sp. and cf. Oscillatoria sp. and Hoiamides C (499) and D (500) were obtained from Symploca sp. samples.193,194 Hoiamide B (498*) was shown to influence sodium concentrations in mouse neocortical neurons.193 Hoiamide D (500) was shown to inhibit the interaction between the ubiquitin ligase MDM2 and the transcription factor p53, which represents a target for cancer therapy.194
Bouillonamide (501) is a cyclic depsipeptide containing two PK-derived moieties, a 2-methyl-6-methylamino-hex-5-enoic acid (Mmaha) residue and a 3-methyl-5-hydroxy-heptanoic acid (Mhha) unit.195 It was isolated from a marine Moorea bouillonii. Bouillonamide (501) was cytotoxic to mouse neuroblastoma cells with an IC50 value of 6.0 μM.195 Alotamide B (502), isolated from an assembly of marine cyanobacteria containing Moorea sp., consists of a cyclic depsipeptide closed by a polyketide unit.196
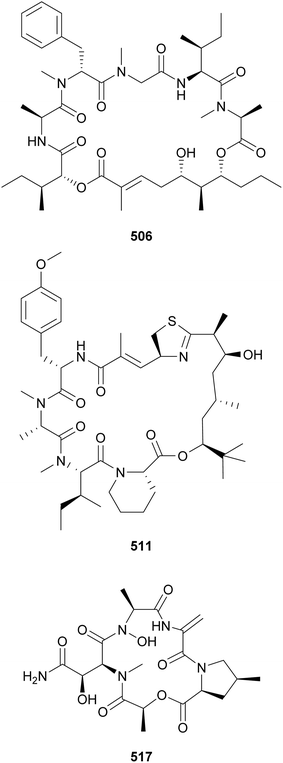
Lagunamides are a recently described family of lipopeptides containing dihydroxy acid polyketide moieties. They may be classified under the “aurilide superfamily” of compounds. Lagunamides A–C (503–505) were isolated from marine Lyngbya majuscula and were shown to display biological activity against Plasmodium falciparum (IC50 values in the low micromolar range) and cancer cells (IC50 values in the nanomolar range). Lagunamides D (506) and D′ (507) were isolated from an assemblage containing mostly Dichothrix sp. and Lyngbya sp.197 Lagunamide D (506) induced human lung adenocarcinoma cells apoptosis and, thus, was cytotoxic with an IC50 of 7.1 nM.198 Odoamide (508), from Okeania sp., represents another member of the aurilide superfamily. It showed potent cytotoxicity against human cervical cancer cells (IC50 = 26.3 nM).199
Apratoxins 509–512 are PK–NRP hybrids that feature a 3,7-dihydroxy-2,5,8,8-tetramethylnonanoic acid (Dtena) residue in their cyclic structures.200,201 Isolated from a marine sample of Lyngbya bouillonii, 509 and 510 displayed potent inhibitory activity against lung cancer cells with IC50 of 2 and 14 nM, respectively. Apratoxin H (511) and apratoxin A sulfoxide (512) were isolated from a cultured Moorea producens.201 Apratoxin H (511) contains a pipecolic acid unit in its structure. It displayed cytotoxicity to lung cancer cells with an IC50 of 3.4 nM.201 Janadolide (513) was isolated from a marine Okeania sp. sample from Janado, Japan.202 It contains a 7-hydroxy-2,5,8,8-tetramethylnon-5-enoic acid-derived moiety as PK-derived unit. It showed potent activity against Trypanosoma brucei brucei (IC50 of 47 nM), with no cytotoxic activity observed against HL-60, MRC-5, or HeLa cells. Additional lipopeptides containing tert-butyl groups in their structures include norbisebromoamide (514)203 from a marine Lyngbya sp. and ypaoamides B and C (515 and 516) from an Okeania sp.204
Gatorbulins (517 and 518) were discovered in an extract of marine Lyngbya cf. confervoides active against isogenic HCT116 colorectal cancer cells with downstream experimentation confirming the gatorbulins as the responsible components.205 Evidence suggested the gatorbulin-1 (517) HCT116 antiproliferative mechanism of action to be microtubule destabilization via tubulin intradimer interface binding.205 They are cyclic depsipeptides composed of modified amino acids and a lactic acid (Lac) moiety.
Aetokthonostatin (519) and two analogs (520 and 521) were discovered from the epiphytic cyanobacterium Aetokthonos hydrillicola, known to be a producer of aetokthonotoxin (see Section 6).206 Aetokthonostatins 519–521 are dolastatin analogs featuring a dolaproline (Dap) and a dolaisoleuine (Dil), common to this family of compounds, and an unusual 3-methoxy-1-phenylbutan-2-amine residue. Aetokthonostatin (519) showed potent cytotoxicity against cancer cells, comparable to that of monomethylauristatin E, the payload of brentuximab vedotin.206
Grassystatins D–F (522–524) are potent cathepsin inhibitors isolated from a marine cyanobacterial collection from Guam.207 These modified peptides are formed by standard, O- and N-methylated amino acids, hydroxy acids, and the statin unit, 4-amino-3-hydroxy-6-methylheptanoic acid (Sta). They also feature a characteristic C-terminus composed of N-Me-Phe and O-Me-Pro. Grassystatin F (524) inhibited cathepsins D and E with IC50 values of 50 and 0.5 nM, respectively.207 Additional Sta-containing peptides with protease inhibitory activity include symplocin A (525) from Symploca sp.,208 maedamide (526) from a marine assemblage of Lyngbya sp.,209 and izenamides A–C (527–529) from a cf. Lyngbya. Tasiamides C–F (530–533), from marine Symploca sp. and Lyngbya sp. collections, are related compounds that lack the Sta unit.210,211 Tasiamide F (533) features a 4-amino-3-hydroxy-5-phenylpentanoic acid, (Ahppa) instead. Phormidepistatin (534), from a freshwater cf. Phormidium sp., contains an epi-Sta unit and a terminal 3,9-dihydroxy-2-methyl-10-phenyldecanoic acid (DMPD) moiety.212 It showed limited protease inhibitory activity due to the epi-Sta unit's 3R,4S configuration, a diastereomer of the more potent 3S,4S-Sta. A phylogenetic analysis showed that the production of secondary metabolites containing Sta or Sta-like moieties is taxonomically widespread across the phylum Cyanobacteria.212
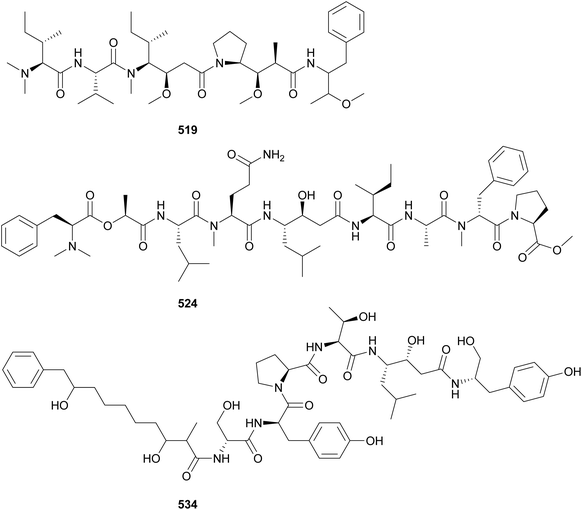
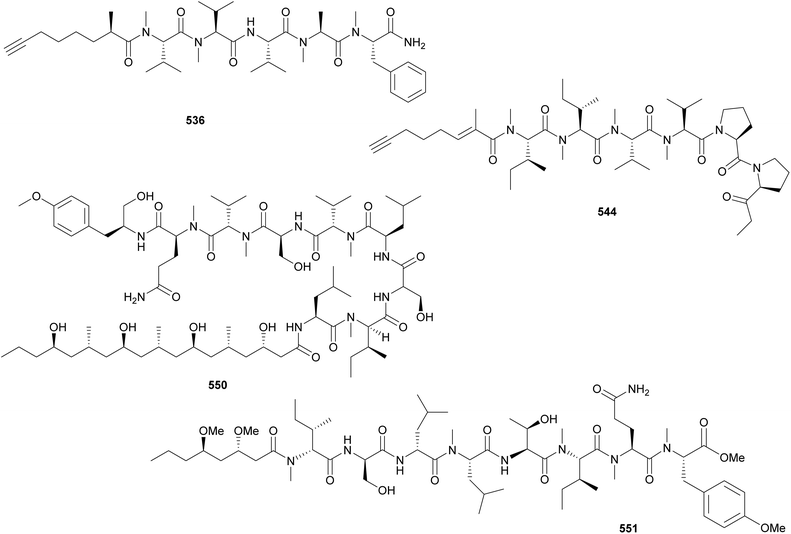
Almiramides (535–542) are linear lipopeptides containing several N-methylated amino acid residues and a terminal fatty acid moiety comprised of a 2-methyloct-7-yonic acid (Moya) unit or derivatives.213,214 Almiramides A–C (535–537) were isolated from a marine Lyngbya majuscula collected in Panama213 and almiramides D–H (538–542) were identified in an Oscillatoria nigroviridis collected in Colombia.214 Compounds 536 and 537 showed potent antiparasitic activity against Leishmania donovani with IC50 values of 2.4 and 1.9 μM, respectively. Compound 535 was inactive, suggesting a role of the terminal unsaturation for bioactivity.
Dragonamide E (543) was discovered in a marine cyanobacterial material collected in Panama.215 It features a 2-methyloct-2-en-7-ynoic acid (Moeya) unit. Dragonamide E (543) displayed activity against Leishmania donovani with an IC50 value of 5.1 μM. Kurahynes A (544) and B (545) also contain Moeya as fatty acid residue, followed by a peptidic sequence ending in 2-(1-oxo-propyl)pyrrolidine (Opp).216,217 Kurahyne 544 was isolated from a marine cyanobacterial assemblage predominantly composed of Lyngbya sp. and kurahyne 545 was found in a marine sample of Okeania sp. Both kurahyne producers were collected near Japan. Jahanyne (546), from a marine Lyngbya sp., and odookeanyne A (547), from an Okeania sp., feature a 2,4-dimethyldec-9-ynoic acid moiety on the N-terminus.218,219 The C-4 demethyl fatty acid was identified in odookeanyne B (548).219 Kurahynes and jahanyne (544–546) displayed cytotoxic activity against HeLa and HL-60 cells in the micromolar range.217
The combination of metabolomics and bioassay-guided fractionation led to the discovery of wenchangamide A (549) from a marine assemblage composed of cf. Neolyngbya sp.220 It contains a fatty acid moiety comprised of repeating hydroxy and methyl groups, similar to minnamide A (550) from a Okeania hirsuta.221 Minnamide A (550) was shown to be a potent inhibitor of HeLa cells (IC50 = 0.17 μM). Ikoamide (551), from a marine Okeania sp., is a linear lipopeptide containing several D-form and N-methylated amino acids, as well as 3,5-dimethoxyoctanoic acid moiety (Dmo).222 It displayed anti-Plasmodium falciparum activity with an IC50 of 0.14 μM.
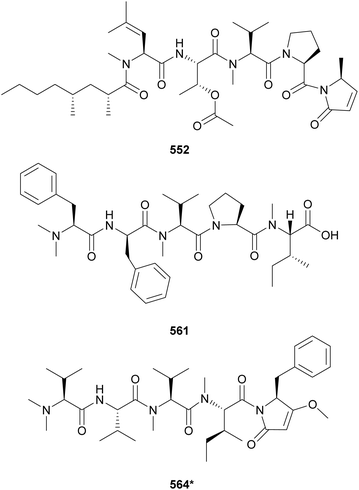
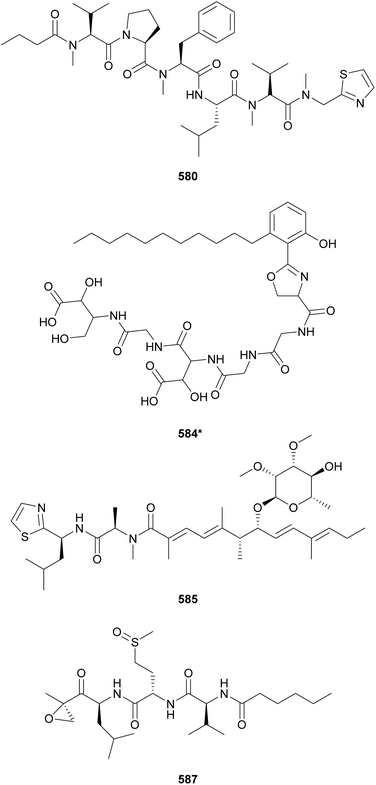
Microcolins E–M (552–560) possess a characteristic 2,4-dimethyloctanoic acid (Dma) unit on the N-terminus.223 Microcolins E–L (552–559) also feature a 5-methyl-1,5-dihydro-2H-pyrrol-2-one (Mdp) moiety on the C-terminus, which was shown to be important for biological activity against cancer cells.223 Terminal pyrrolinone derivatives are also found in iheyamide A, caldoramide, and odopenicillatamide. Iheyamides A–C (561–563) were isolated from a marine Dapis sp.224 Iheyamide A (561), the only analog containing the isopropyl-O-Me-pyrrolinone moiety, displayed an IC50 of 1.5 μM against Trypanosoma brucei with limited cytotoxicity to human cells.224 Caldoramide (564*) is a pentapeptide isolated from a marine Caldora penicillate composed of three sequential Val residues (two of them N-methylated), an N-Me-Ile, and a terminal Bn-O-Me-pyrrolinone. The presence of sequential N-methylvaline residues is an intriguing structural feature that is also found in heptavalinamide A (565) from Symploca sp.225 Odopenicillatamide (566) contains the Bn-O-Me-pyrrolinone unit and was isolated from a Caldora penicillata along with komesuamide (567). Lipopeptide 567 contains an N-terminal 2-methylhexanoic acid (Mha) and a C-terminal Pro-O-Me neighboring an N-Me-Phe. Both 566 and 567 were shown to stimulated glucose uptake without causing cytotoxicity.
Additional linear (lipo)peptides containing a C-terminal Pro-O-Me neighboring an N-Me-Phe residue include tasiamide F (533),211 kinenzoline (568),226 hoshinoamides A–C (569–571),227,228 brintonamides A–E (572–576),229 amantamides A and B (577 and 578),230,231 and mabuniamide (579).232 They were isolated from marine strains classified under the Oscillatoriales order, except for kinenzoline (568) produced by a Salileptolyngbya sp. (Synechococcales). Hoshinoamides A–C (569–571) possess an unusual long chain amino acid moiety connected to 4-(4-hydroxyphenyl)-butanoic acid (Hba). They displayed antiplasmodial activity with IC50 values in the low micromolar range with no cytotoxicity against HeLa cells.227,228 Mabuniamide (579) displayed antiplasmodial activity (IC50 = 1.4 μM) and was shown to stimulate glucose uptake in cultured rat L6 myotubes.232
Biseokeaniamides A–C (580–582), isolated from a marine an Okeania sp., are linear lipopeptides containing an N-terminal butanoic acid unit and a C-terminal N-methyl-2-thiazolemethaneamine (Thz-N-Me-Gly).233 They displayed inhibitory activity in a sterol O-acyltransferase (SOAT) in vitro assay. SOAT is related to intracellular cholesterol storage that may lead to hypercholesterolemia. Lyngbyapeptin D (583), isolated along with lyngbyabellins 489 and 490 from Lyngbya bouillonii, represents another thiazole-containing linear lipopeptide.190 It features N-methylated amino acids, a 3-methoxy-2-butenoic acid (Mba) moiety and a C-terminal Pro-Thz.
Mining cyanobacterial genomes for NRPS–PKS clusters putatively coding for β-hydroxyaspartate (β-OH-Asp), an amino acid often associated with iron chelation, led to the discovery of cyanochelin A (584*).234 Two of the cyanobacterial BGCs putatively associated with the production of β-OH-Asp-containing peptides presented vicinal genes encoding siderophore transporters, further supporting the study hypothesis. Cyanochelin A (584*) was isolated from a Rivularia sp. strain cultured under iron deprivation to strategically induce production.234 It features a distinct hydrophobic tail connected to phenolate and a 2-oxazoline moiety, followed by a peptidic sequence containing β-OH-Asp, ending on a 2,4-dihydroxy, 3-amino butyric acid (Dhaba) unit.
Iezoside (585) is a glycosylated lipopeptide isolated from a marine Leptochromothrix valpauliae collected in Japan.235 It was shown to be a potent inhibitor of the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA), a target associated with human cancers. Iezoside B (586) is the C-31 O-demethyl analogue isolated from a marine assemblage from Florida, USA.236 Both compounds were shown to influence intracellular calcium concentrations and to be cytotoxic to cancer cells.236 Carmaphycins A and B (587 and 588) were discovered from a Symploca sp. collected in Curaçao.237 Featuring a unique α,β-epoxyketone warhead and a methionine sulfoxide/sulfone unit, they were shown to be potent proteasome inhibitors and to display antiproliferative activity against human lung adenocarcinoma and colon cancer cells with IC50 values in the nanomolar range.237
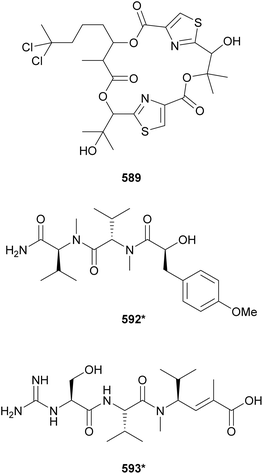
The marine strain Moorea producens JHB collected in Jamaica is known to be a prolific source of secondary metabolites. From this strain, hectochlorin (lipopeptide) and jamaicamides (fatty acid amides) were discovered by structure-guided and bioassay-guided fractionation, respectively.238,239 More recently, its biosynthetic potential was revisited by orthogonal approaches.240 MS/MS molecular networking guided the discovery of hectochlorins B–D (589–591) as well as jamaicamide analogs discussed in Section 5. Complementarily, genome mining revealed several biosynthetic gene clusters encoding short peptides. Investigation of the polar fractions obtained by vacuum liquid chromatography of the cell extract led to the isolation of hectoramide (592*), a linear tripeptide formed by two N-Me-Val residues and 3-(4-methoxyphenyl)lactic acid (Mpla).240 Using the so called “maldiisotopic approach”, strain Moorea producens JHB was analyzed by MALDI-MS after cultivation in normal media and medium containing 15N-sodium nitrate.241 Comparison of the molecular ions and sodium adducts between the two MALDI-MS analyses revealed the number nitrogen atoms present in each detected metabolite. The intriguingly high nitrogen content of a metabolite of m/z 400 led to the discovery of cryptomaldamide (593*), a hybrid tripeptide containing an amidino-serine and a ketide-extended valine.241 Previous reviews have covered the use of metabolomics and biotechnological approached for the discovery of cyanobacterial natural products.21,22
A metagenomic study of the marine sponge Discodermia calyx led to the identification of a NRPS–PKS BGC associated with the production of kasumigamide, a natural product originally discovered from a free-living Microcystis aeruginosa strain.242 The symbiont ‘Entotheonella’ sp. was shown be the producer of kasumigamide found in the sponge extract.242 A search for kas gene homologs revealed it to be widespread in different bacteria, suggesting a potential role in microbial ecology. These findings motivated the reinvestigation of the Microcystis aeruginosa extract, leading to the structure revision of kasumigamide and discovery of deoxykasumigamide (594).243
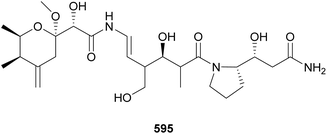
The majority of natural products covered in this review were found in free-living cyanobacteria, with a few examples of metabolites from lichen-associated cyanobacteria. Samples of the terrestrial lichen Peltigera membranacea collected in Iceland were analyzed by whole genome sequencing followed by genome mining for presence of PKS gene clusters.244 Closer investigation into one of the gene clusters, called nsp, revealed it to be derived from the Nostoc symbiont. It also showed closer homology of nsp with the gene clusters responsible for the biosynthesis of pederin, produced by bacteria living in rove beetles, and onnamides, isolated from Theonella swinhoei sponges. Stable isotope labeling allowed for LC-MS identification of nosperin (595) in crude extracts and NMR characterization of 30 μg of the pure compound. Nosperin (595) highlights the importance of symbiotic cyanobacteria as a source for novel secondary metabolites. A review of natural products from symbiotic cyanobacteria was recently published.23 Cusperins A and B (596 and 597) are pederin-like compounds isolated from a freshwater, free-living Cuspidothrix issatschenkoi strain, highlighting potential overlap in secondary metabolism between symbionts and non-symbionts.245
4 Macrolides, cyclophanes, and related compounds
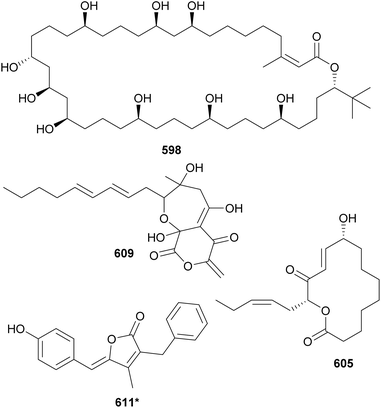
Cyanobacterial polyketides are the second largest category of natural products included in this review. They are constructed as an assembly of basic units, such as acetate, malonate, and/or propionate. A series of reduction and dehydration reactions produce the common moieties observed in polyketides (e.g. hydroxyls and α,β-unsaturations). Their structures are often decorated with halogens, carbamates, and sugar moieties. Hybrid NRP–PK may be found in Sections 3–5 according to structural features. This section is dedicated to cyanobacterial polyketides that form macrocycles, herein subdivided into macrolides and cyclophanes, and related compounds.Bastimolide A (598) is a polyketide characterized by a 40-membered polyhydroxy macrolactone ring containing an α,β-unsaturated carboxyl group and a tert-butyl substituent.246 It was isolated from a marine Okeania hirsuta by 1H NMR-guided fractionation. Bastimolide A (598) showed potent antimalarial activity against four resistant Plasmodium falciparum strains with IC50 values at the nanomolar range. 598 showed some activity against Trypanosoma cruzi, Leishmania donovani, MCF-7 breast cancer cells, and Vero cells at the micromolar range.246 Additional 40-membered polyhydroxy macrolactones with a tert-butyl moiety discovered in the past decade include nuiapolide (599),247 amantelides A and B (600 and 601),248 and palstimolide A (602),249 the latter isolated from a marine Leptolyngbya sp. collection. Bastimolide B (603), isolated from the same Okeania hirsuta collection shown to contain 598, has a 24-membered macrolactone ring extended by an aliphatic chain ending on a terminal tert-butyl group.250 It also displayed antiplasmodial activity. Caylobolide B (604*), from a Phormidium spp. collection, is a 36-membered polyhydroxy macrolactone shown to be moderately cytotoxic to cancer cells.251
Sacrolide A (605) was isolated from the freshwater cyanobacterium Aphanothece sacrum, a noted delicacy in Japanese cuisine.252 The cyanobacterial sample was obtained from a commercial aquaculture pond in Kumamoto, Japan. Sacrolide A (605) is a 14-membered macrolide. It displayed both moderate antimicrobial and cytotoxic activities, raising concerns regarding the safety of the raw ingredient. The same study found no traces of 605 in processed food samples. Further investigation of Aphanothece sacrum led to the discovery of 9-epi-sacrolide A (606) and 15,16-dihydrosacrolide A (607*).253 From a marine Lyngbya sp., a 14-membered macrolide was isolated and named koshikalide (608*).254 An unusual δ-valerolactone ring is found in nostovalerolactone (609), isolated from a Nostoc punctiforme strain along with 9-dehydronostovalerolactone (610) and nostoclides N1 and N2 (611* and 612*).255 Compounds 609–612 were discovered using genome mining and high-density cultivation in a study focused on chemical mediators.
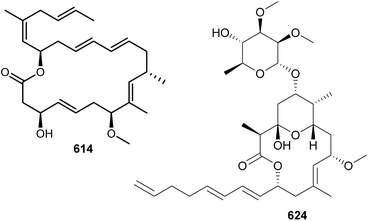
Biselyngbyolides and biselyngbyasides are highly unsaturated cyanobacterial macrolides. After the discovery of biselyngbyaside A in 2009 from a marine Lyngbya sp. collected in Japan,256 a sequence of aglycone and glycoside variants were identified, namely biselyngbyolides A–C (613–615) and biselyngbyasides B–F (616–620).257–260 All of them were identified in Lyngbya sp. from Japanese waters. Biselyngbyolide B (614) displayed potent cytotoxic activity against HeLa S3 and HL-60 cells with IC50 values of 28 and 2.7 nM, respectively. This study also showed that 614 induced the transcription of endoplasmic reticulum (ER) stress-related genes, released calcium from ER, and induced apoptosis.
Lyngbyalosides and irijimasides are macrolide glycosides containing an embedded tetrahydropyran ring and a characteristic unsaturated carbon chain. Three bromine-containing lyngbyalosides (621–623) were isolated from a marine Lyngbya bouillonii collected in Guam, along with lyngbyabellins 489 and 490.190 Low cytotoxicity was reported for 621–623. Irijimasides A–E (624–628), from an Okeania sp., were shown to inhibit the differentiation of mouse macrophage cells into osteoclast-like cells in the presence of RANKL.261 This assay is used as a preliminary in vitro test aiming at the discovery of scaffolds to be further developed for the treatment of periodontitis and osteoporosis.
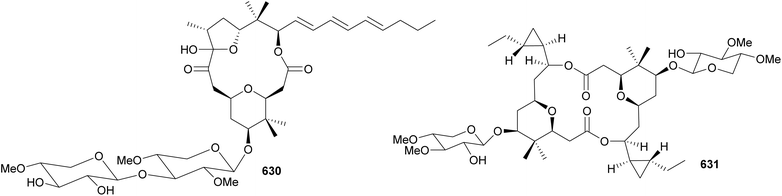
Macrolide glycosides containing a tetrahydropyran ring substituted by a gem-dimethyl group include cyanolide A (629),262 polycavernoside D (630),263 cocosolide (631),264 and akunolides A–D (632–635).265 Polycavernoside D (630), from a marine sample of Okeania sp., is composed of both a tetrahydropyran ring and a tetrahydrofuran ring.263 Polycavernosides are potent toxins originally associated with human death following the ingestion of the edible red algae Polycavernosa tsudai. Later efforts failed in reisolating the polycavernosides from the red algae, and it was suggested that polycavernosides instead derived from associated microorganisms, such as cyanobacteria. Despite their acute toxicity to humans, polycavernosides were shown only to be moderate cytotoxic to cancer cell lines.263 Cocosolide (631), isolated from a marine Symploca sp., has a dimeric structure featuring two cyclopropyl groups.264 It displayed immunosuppressive activity, inhibiting IL-2 production and proliferation of anti-CD3-stimulated T-cells. No cytotoxic activity was observed for 631 against HCT116, RAW macrophage cells, or Jurkat cells.
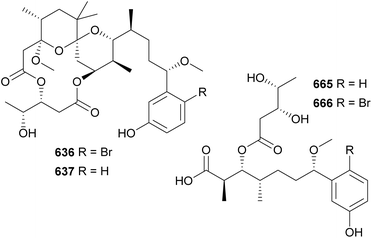
Aplysiatoxins and oscillatoxins are macrolides containing a fused-ring system and a characteristic hydroxy acid unit. A series of aplysiatoxins, oscillatoxins, and derivatives were isolated between 2010–2023 from marine strains under the Oscillatoriales order. They include 3-methoxyaplysiatoxin (636),266 3-methoxydebromoaplysiatoxin (637),266 neo-aplysiatoxin A (638),267 neo-debromoaplysiatoxin A–J (639–648),268–273 oscillatoxins E–M (649–657),270,274–276 and other variants (658–664).274,277 3-Methoxyaplysiatoxin (636) along with the non-brominated variant, 3-methoxydebromoaplysiatoxin (637), were isolated from a marine Trichodesmium erythraeum collected in Singapore.266 They contain the characteristic tetrahydropyran–tetrahydropyran spiro system. Compound 637 displayed anti-Chikungunya virus activity with an IC50 of 2.7 μM. At this concentration, minimal cytotoxicity was observed. The brominated metabolite 636 was inactive at 10 μM.266
Following the identification of aplysiatoxin derivatives in cyanobacterial collections, detailed investigations led to the discovery of nhatrangins A and B (665 and 666) from a Lyngbya majuscula from Vietnam278 and aplysiadione (667) and aplysiaenal (668) from a Moorea producens from Japan.279 These truncated aplysiatoxin variants lack the core macrocycle and are thought to have some biosynthetic homology to the compound family or could be putative biosynthetic intermediates in the pathway. However, it should be noted the aplysiatoxin gene cluster has thus far yet to be elucidated.
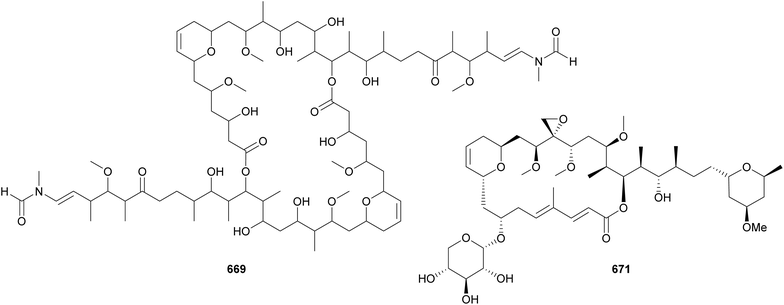
Actin-targeting macrolides, such as the dimeric swinholide-type and the “monomeric” scytophycin-type compounds, have been found in diverse organisms, such as sponges, mollusks, corals, and algae, suggesting that microorganisms may play a role in their biosynthesis.280 Among them, cyanobacteria have been shown to be producers of such macrolides. Luminaolide B (669) was discovered by genome mining in a search for actin inhibitors.280 It was isolated from a cultured freshwater Planktothrix paucivesiculata strain. Its dimeric structure resembles that of luminaolide, which was first discovered from a marine alga.280 7-OMe-scytophycin B (670) was identified in the extract of a freshwater Anabaena sp. that showed antifungal activity.281 From marine cyanobacteria, leptolyngbyolides A–D (671–674) were isolated from Leptolyngbya sp.,282 samholides A–I (675–683) were isolated from cf. Phormidium sp.,283 and symplocolide A (684) was isolated from Symploca sp.284 Symplocolide A (684) was discovered by a combination of bioactivity screening, the NMR-based machine learning tool SMART 2.0, and MS/MS molecular networking.285
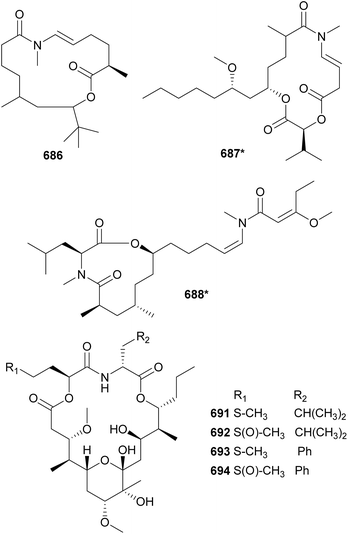
In 2010, two macrocyclic lactones containing an N-enamide and a tert-butyl group were discovered from marine cyanobacteria: laingolide B (685) from Lyngbya bouillonii190 and palmyrolide A (686) from an assemblage of cf. Leptolyngbya and Oscillatoria.286 Palmyrolide A (686) was shown to inhibit the influx of calcium in murine cerebrocortical neurons (IC50 = 3.7 μM). It was also shown to block sodium channels in Neuro-2a cells with no significant cytotoxicity.286 Also featuring an N-methyl enamide, sanctolide (687*) was isolated from a terrestrial Oscillatoria sancta.287 It contains a 2-hydroxyisovaleric acid incorporated into the 14-membered macrolide structure. The mixture of amino and hydroxy acids is found in the macrocyclic structures of kanamienamide (688*) from a marine Moorea bouillonii,288 tricholides A and B (689 and 690) from a marine Trichodesmium thiebautii,185 and looekeyolides A–D (691–694) from Roseofilum.289,290 Looekeyolides 691–694 were isolated from the cyanobacteria associated with black band disease, a lethal coral disease.291,292
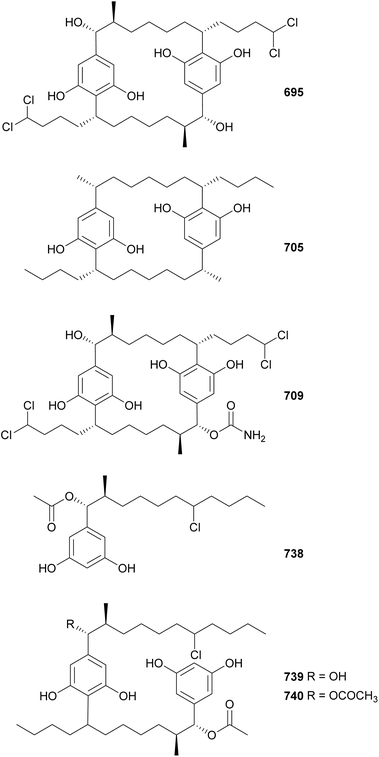
Cyanobacterial cyclophanes have a core [7.7]paracyclophane scaffold often decorated with halogens, carbamates, methyl, and/or hydroxyl groups, which characterize the different subfamilies, including cylindrocyclophanes (695–704),293 merocyclophanes (705–708),294,295 carbamidocyclophanes (709–725),296–299 ribocyclophanes (727–731),300 and nostocyclophanes (732–737).301 Nine cylindrocyclophanes (695–703) were discovered from a terrestrial Nostoc sp. strain.293 They have varying chlorination and hydroxylation patterns, but consistent β-methyl substituents, a key characteristic of the cylindrocyclophane subfamily. Cultivation of the same strain in bromide enriched media led to the discovery of 704. Cylindrocyclophanes 695–703 showed 20S proteasome inhibitory activity and cytotoxicity against HT-29 cancer cells.293
Merocyclophanes A–D (705–708), isolated from terrestrial Nostoc sp. strains, were shown to be cytotoxic to various cancer cells with IC50 values in the low micromolar range.294,295 Merocyclophanes contain characteristic α-branching methyl groups. The putative merocyclophane BGC includes both type I and type III PKS modules.295 Carbamidocyclophanes F–V (709–725) were discovered from freshwater Nostoc and Cylindrospermum strains.296–298,302 As the name suggests, they contain carbamido moieties appended to the paracyclophane core. Carbamidocyclophane F (709) displayed significant activity against Mycobacterium tuberculosis in two assays, MABA (Alamar blue assay) and LORA (low-oxygen recovery assay), with MIC values of 0.8 and 5.4 μM, respectively. Carbamidocyclophanes F and G (709 and 710) were bioactive against Staphylococcus aureus and Enterococcus faecalis, as well as against MDA-MB-435 and HT-29 cancer cells. Carbamidocyclophanes H–U (711–724) showed antibacterial activity against MRSA and other pathogens.297,298 The brominated carbamidocyclophanes M–U (716–724) were isolated after the strain was cultured in KBr-enriched media.298 The same study also revealed the presence of a carbamoyltransferase gene within the gene cluster putatively responsible for the biosynthesis of carbamidocyclophanes.298 Carbamidocyclophane V (725) was isolated from a Cylindrospermum stagnale strain along with carbamidocylindrofridin A (726).302
Cylindrofridins A–C (738–740) were isolated from a freshwater Cylindrospermum stagnale strain using an optimized extraction, enrichment, and separation procedure targeting [7.7]paracyclophanes.303 Cylindrofridin A (738) is a monomeric cyclophane derivative containing an α-acetate, a β-methyl group, and a chlorine on the side chain at the expected site of cyclization. Cylindrofridins B (739) and C (740) are dimeric, uncyclized cyclophane derivatives. Cylindrofridins 738–740 showed no cytotoxicity to HaCaT cells. Only 738 showed weak inhibitory activity against MRSA and S. pneumoniae (IC50 values of 8.6 and 17 μM, respectively). These intriguing structures provided insights into the macrocyclization of cyanobacterial cyclophanes.303
5 Acyl amides and fatty acid derivatives
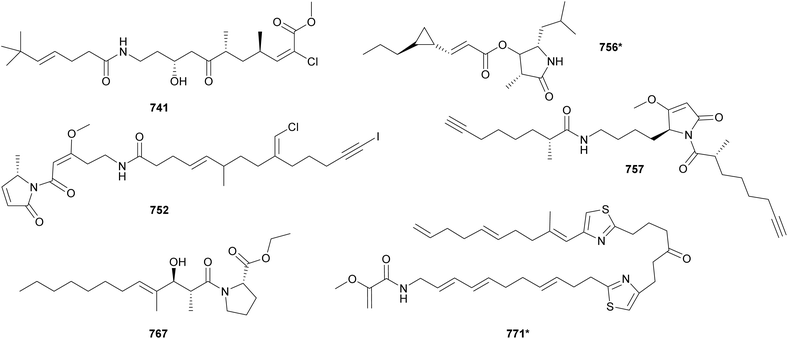
Acyl amides (also called fatty acid amides or lipoamides) are hybrid compounds that contain NRP- and PK-derived moieties, including the characteristic long aliphatic chains. As cyanobacteria have the distinct capability of incorporating fatty acids into their secondary metabolites, this section was dedicated to them. Here we describe acyl amides, fatty acid derivatives and related compounds isolated from cyanobacteria between 2010 and 2023.Luquilloamides (741–746) are a series of highly modified acyl amides isolated from a marine Oscillatoria sp.304 Luquilloamides 741–746 contain a characteristic terminal tert-butyl group. Each luquilloamide has specific modifications, such as the presence of vinyl chloride groups, a cyclohexanone unit, or a lactam head. From the same strain, luquilloamide B (747*) was isolated, which was proposed to be either the product of a truncated biosynthesis or the result of the hydrolysis of luquilloamide C (742).304 A vinyl chloride and a tert-butyl group are also found in the structure of beru'amide (748), an acyl amide isolated in minute amounts from an Okeania sp.305 Its limited quantities hampered degradation and derivatization reactions for absolute configuration determination, which were accomplished by computational methods and chemical synthesis. Synthetic beru'amide (748) displayed biological activity against Trypanosoma brucei rhodesiense with an IC50 value of 1.2 μM.305
A lactam ring is also present in the structures of palmyrrolinone (749*),306 jamaicamides D–F (750–752),240 smenamides C–E (753–755),307 hoshinolactam (756*),308 and doscadenamides (757–766).309,310 Jamaicamides D–F (750–752) were identified by MS/MS molecular networking in the extract of the marine strain Moorea producens JHB, in the same study that reported the discovery of hectochlorins B–D (589–591) (see Section 3).240 To allow for the isolation and characterization of the minor iodinated analog (752), the strain was cultivated in media enriched with sodium iodide. Jamaicamide F (752) was shown to inhibit calcium and sodium influx in neocortical neurons with IC50 values in the micromolar range.240 Smenamides C and E (753 and 755), from a Trichodesmium bloom, also showed cytotoxicity to neuro-2A cells.307 Hoshinolactam (756*) features a γ-lactam ring and a cyclopropane ring within the side chain. It displayed significant biological activity against Trypanosoma brucei brucei (IC50 = 3.9 nM).308 Dosdecamide A (757) is an acyl amide with two Moya-derived side chains. It was isolated from Moorena bouillonii collections from Saipan (USA) and Fingers Reef (Guam).
Ethyl tumonoate A (767) was discovered by a phylogeny-guided approach.311 Marine cyanobacterial samples originally assigned as cf. Oscillatoria margaritifera were shown to be phylogenetically related to a Blennothrix cantharidosmum strain known to produce tumonoic acid. Interestingly, the B. cantharidosmum was initially collected in Papua New Guinea, while the cf. O. margaritifera was from the coast of Curaçao. Their evolutionary relatedness and their significant geographic distance inspired the chemical analysis of the cf. O. margaritifera samples, leading to the discovery of 767. Ethyl tumonoate A (767) features an ethyl ester substituted proline linked to a functionalized fatty acid chain. It showed anti-inflammatory activity in a cell-based nitric oxide assay on murine macrophage (IC50 = 9.8 μM) and inhibitory activity of Ca2+ oscillations at 10 μM in neocortical neurons. A substituted proline is also observed in the structures of okeaniamides A and B (768 and 769), isolated from a marine Okeania sp. Okeaniamides (768 and 769) induced the differentiation of mouse preadipocytes in the presence of insulin, a bioassay employed in the search for new therapeutics for type 2 diabetes mellitus patients.312
Thiazole-containing acyl amides include laucysteinamide A (770*)313 and caldorazole (771*)314 from marine Caldora strains, fischerazole A–C (772–774)315 and aranazoles A–D (775–778)316 from freshwater Fischerella strains, and trichothiazole A (779*)317 from Trichodesmium bloom material. Caldorazole (771*) contains two thiazole rings embedded in a linear structure composed of several olefinic methines and a terminal O-methylenolpyruvamide unit. It showed strong growth-inhibitory activity against HeLa cell lines through the inhibition of the mitochondrial respiratory chain.314 Comparative genomics followed by mass spectrometry analysis guided the discovery of aranazoles A–D (775–778), unusual acyl amides that are extensively chlorinated and contain a dioxane ring.316 Trichothiazole A (779*) is also the product of a mass spectrometry-guided process aiming at the identification of chlorinated metabolites from bloom-forming Trichodesmium strains. It features a terminal alkyne and two vinyl chlorides, in addition to the thiazole ring.317
The first cytotoxic metabolite discovered from a Trichodesmium was trichotoxin A (780), reported in 2011.318 It is a chlorinated polyketide containing a phenyl moiety. Trichotoxin A (780) was cytotoxic to rat pituitary cells and mouse neuroblastoma cells. Additional metabolites isolated from Trichodesmium blooms are trichotoxin B (781),319 the alkyne-containing analog of 780, trichophycins A–I (782, 783*–797*, 788–790),320–322 isotrichophycin C (791),322 and tricholactone (792*).321 Trichophycin A (782) was shown to be the most cytotoxic Trichodesmium metabolite against neuro-2a cells.322
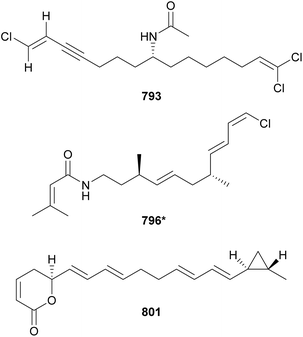
Taveuniamides L–N (793–795) are chlorinated enyne acyl amides isolated by NMR and MS-based metabolomics from a marine Symploca collection from Guam.323 This study also reported the isolation of taveuniamide F, a previously known compound, that was shown to selectively bind to the cannabinoid receptor CNR1.323 The observation of similar 1H NMR profiles led to the discovery of janthielamide A (796*) from a cyanobacterial collection made in Curaçao and kimbeamides A–C (797–799) and kimbelactone A (800*) from a collection made in Papua New Guinea.324 Janthielamide A (796*) and kimbeamide A (797) were shown to block sodium channels in neuro-2a cells. Janthielamide A (796*) was also shown to antagonize sodium influx in cerebrocortical neurons (IC50 = 5.2 μM).324 Containing pyranone rings and polyunsaturated chains, coibacins A–D (801–804) were discovered in marine samples of cf. Oscillatoria sp. collected in Panama.325 Coibacin A (801) displayed most potent activity against Leishmania donovani (IC50 value of 2.4 μM). Coibacins also displayed anti-inflammatory activity, inhibiting nitric oxide (NO) and production of pro-inflammatory proteins.325
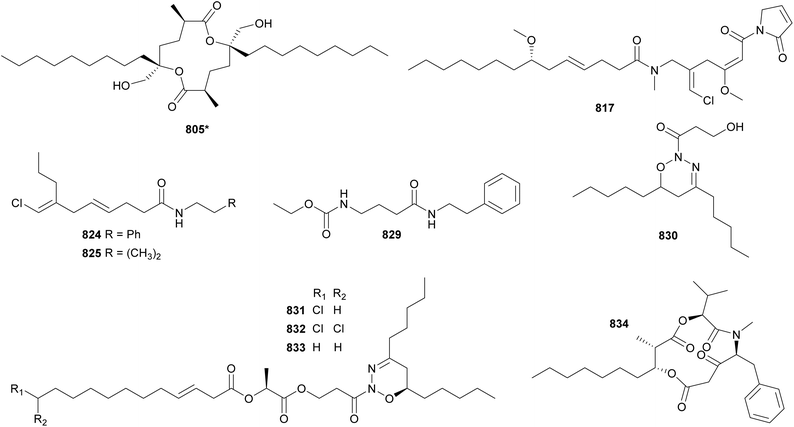
Malyngolide is a fatty acid derivative containing a hydroxymethyl and characteristic six-membered lactone ring. It has been extensively explored in synthetic studies and for potential biological activities.326 From a Lyngbya majuscula collection, a malyngolide dimer (805*) was discovered.326 It showed mild activity against chloroquine-resistant Plasmodium falciparum and human lung cells. The discovery of 805* adds to the list of dimeric polyketides, such as the swinholides (see Section 4).
Honaucins A–C (806*–808) are 4-chlorocrotonic acid derivatives isolated from marine samples of Leptolyngbya crossbyana collected in Hawaii (USA).327 They showed anti-inflammatory activity in murine macrophages (inhibition of nitric oxide production and downregulation of proinflammatory cytokines) and inhibition of quorum sensing signaling in Vibrio harveyi. This activity profile highlights the complexity of intra- and interkingdom chemical interactions and the value of microbial compounds with complementary activities for drug discovery and development.327
Related compounds containing an α-methoxy-β, β′-dimethyl-γ-pyrone were isolated from independent collections of Leptolyngbya sp. They include yoshinones (809–811) from a cyanobacterium collected in Japan328 and kalkipyrone B (812) from a collection made in the American Samoa.329 Yoshinone A (809) was shown to inhibit the differentiation of 3T3-L1 cells into adipocytes (EC50 = 420 nM).
The malyngamides encompass a large family of bioactive fatty acid amides. Their structures are characterized by an aliphatic chain often derived from lyngbic acid [(4E,7S)-7-methoxy-4-tetradecenoic acid]. This chain is often connected by an amide bond to PKS- and/or NRPS-derived moieties and may feature vinyl chloride groups. Twelve malyngamide variants (813–823)142,143,330–336 were discovered between 2010 and 2023. 8-epi-Malyngamide C (813) was found in a marine collection of Lyngbya majuscula.330813 contains an epoxy-ketone ring linked to a vinyl chloride forming the molecule head. 813 was shown to have mild cytotoxic activity against HT-29 cells with an IC50 value of 15.4 μM. Due to the overall structural similarity of these fatty acid amides to acyl homoserine lactones, 813 was tested in a quorum sensing reporter assay, where it displayed inhibitory activity.
Malyngamide 4 (819) was discovered in a marine Moorea producens collection from the Red Sea in Saudi Arabia.335 It showed weak cytotoxic activity against MDA-MB-231 breast adenocarcinoma, HT-29 colorectal carcinoma, and A549 lung carcinoma cell lines. While the majority of malyngamides have a C-14 aliphatic chain, malyngamide Y (820) contains a shorter chain with 12 carbons.143 Forming the molecule head, 820 features a methylated cyclohexanone ring connected to the characteristic 2E vinyl chloride group. It showed cytotoxic activity against NCI-H460 and neuro-2a cancer cells.
Vinyl chloride groups are also found in the structures of credneramides A and B (824 and 825) from a cf. Trichodesmium sp.,337 pitiamides (826 and 827) from Hydrocoleum majus,338 and caracolamide A (828) from a cf. Symploca.339 Credneramides A and B (824 and 825) were shown to modulate intracellular levels of calcium in murine cerebrocortical neurons in the low micromolar range.337 Caracolamide A (828) is composed by a phenethylamide moiety and a rare gem-dichlorovinylidene unit.339 It was shown to affect calcium oscillations in subnanomolar concentrations, while not being cytotoxic to cancer cells.339 A phenethylamide is also observed in santacruzamate A (829), isolated from a cf. Symploca.340 It was isolated by bioassay-guided fractionation based on activity against the malaria parasite and MCF-7 cancer cells. However, structural similarities with suberoylanilide hydroxamic acid (SAHA) inspired the evaluation of 829 for histone deacetylase (HDAC) inhibition. Santacruzamate A (829) was shown to be a potent inhibitor of HDAC2 (IC50 = 0.119 nM), with relatively low inhibition of HDAC4 or HDAC6.340
Nocuolin A (830) is the first reported natural oxadiazine, adding a new family of compounds to the cyanobacterial fatty acid derivatives.341 It was isolated from an active extract of a freshwater Nostoc sp. strain that was screened for apoptotic activity. Genomic analysis led to the identification of a putative biosynthetic gene cluster for 830 (noc), which contained genes for fatty acid binding and modification, aromatic modification, as well as genes encoding for PKS and NRPS modules. Nocuolin A (830) was shown to induce apoptosis in HeLa cells in a caspase-dependent manner. It also showed cytotoxic activity against nine human cancer cell lines (IC50 values from 0.7 to 4.5 μM), with lowest IC50 values recorded in the p53-mutated cell lines.341 Cultivation of the Nostoc sp. strain with fatty acid supplementation led to the discovery of lactylate-containing 830 variants, named nocuolactylates A–C (831–833).342 The same strategy applied to a freshwater Fischerella sp. strain was used for the discovery of hapalosin C–G (834–838).342
Parguerene (839*) was isolated from a marine sample of Moorea producens collected in La Parguera, Puerto Rico.152 It features an alaninol moiety connected to an α,β-unsaturated ketone methylated at the α position, and a terminal monosubstituted phenyl ring. The alaninol residue is also observed in calothrixamides A and B (840 and 841), the first acyl amides from a Calothrix strain and one of the few modified long-chain fatty acid amides reported from freshwater cyanobacteria.343 Calothrixamides 840 and 841 were identified using an in situ sampling probe connected to an LC-UV-MS system that assisted with sample prioritization.343 In addition to the alaninol moiety, 840 features a methyl diene, a carbamate ester, and a terminal alkene, the latter being absent in 841. Structurally similar to 840 and 841, the amino glycerol head-containing mooreamide A (842*) was isolated from a Moorea bouillonii strain.344 It showed cannabinomimetic activity, with potent affinity for the CB1 cannabinoid receptor with a Ki value of 0.47 μM, and selectivity greater than 50-fold over the CB2 receptor.344
Columbamides A–C (843–845) were discovered by a genomic-metabolomic approach applied to three marine cyanobacterial species of the genus Moorea.345 Columbamides 843 and 844 showed sub-micromolar binding affinity to both cannabinoid receptors, with higher affinity for CB1. Additional columbamide analogs (846–855) were isolated from Moorea bouillonii samples collected in Papua New Guinea or Malaysia or by heterologous expression.346–348 Columbamides I–M (851–855) were discovered when the columbamide BGC was expressed in Anabaena PCC 7120 and the engineered Anabaena was grown under different culture conditions.348
Serinolamides (856–859) contain a serinol-derived residue.192,349,350 Serinolamides A and B (856 and 857) were isolated from Lyngbya majuscula349,350 and serinolamides C and D (858 and 859) from an Okeania sp.192 Serinolamide A (856) was found to be a CB1 cannabinoid receptor agonist (Ki = 1.3 μM) with some selectivity over CB2 (>5-fold).349 The same study also reported the isolation of propenediester (860) from an Oscillatoria sp., which showed similar 1H NMR resonances in comparison to 856.349 Additional functionalized fatty esters reported between 2010 and 2023 include 2-hydroxyethyl-11-hydroxyhexadec-9-enoate (861) from a marine Leptolyngbya sp.,351 nostochopcerol (862) from a freshwater edible Nostochopsis lobatus,352 and benderadiene (863) from a marine cf. Lyngbya sp. Benderadiene (863) features a brominated diene moiety connected to a lyngbyoic acid (864)-derived tail.353 The presence of lyngbyoic acid (864) was verified in the cf. Lyngbya sp. sample collected in Singapore by comparison with the experimental data from the first report of the compound from a Lyngbya cf. majuscula sample collected in Florida (USA).353,354 Both 863 and 864 were shown to inhibit Pseudomonas aeruginosa gene expression regulated by quorum-sensing.353,354
Pitinoic acid A (865) was also shown to interfere with P. aeruginosa quorum sensing.355 It was isolated from a marine sample of Lyngbya sp. from Guam, along with pitinoic acid B (866), which is an ester of 865 with a short-chain chlorinated fatty acid. The chlorinated fatty acid, pitinoic acid C, was detected in the crude fraction, but was unstable upon isolation. Pitinoic acid A (865) inhibited expression and production of pyocyanin and elastase in P. aeruginosa, while not interfering with cellular growth. Pitinoic acid B (866), in turn, was shown to reduce transcription of several pro-inflammatory cytokine genes. A crude fraction containing pitinoic acid C showed a similar anti-inflammatory effect as 866, suggesting that this activity might be associated with the chlorinated fatty acid portion of the compound. The authors suggest that 866 may act as a dual-action pro-drug with two pharmacophores eliciting complementary activities.
The fatty acids 7(E)-9-keto-hexadec-7-enoic acid (867)356 and dysidazirine carboxylic acid (868)357 were shown to have anti-inflamatory potential in vitro. Similar to 867, 11-oxopalmitelaidic acid (869) was isolated from a Leibleinia gracilis.358 Puna'auic acid (870) contains a rare allenic moiety.359 It was isolated from a Pseudanabaena sp. from the French Polynesia that grows in shallow waters and interferes with corals and the marine ecosystem.359 Chlorosphaerolactylates A–D (871–874) were isolated from a freshwater Sphaerospermopsis sp.360 They are composed esters of lactic acid and lauric acid with various chlorination patterns. A new cerebroside named mooreaside A (875*) was isolated from a Moorea producens collected in the Red Sea, along with nucleoside derivatives (see Section 6).361
A series of alkylphenols (876, 877*, and 878)362 and alkyl salicylates (879–883)363,364 were discovered from Hormoscilla sp. collected in the Anae Island, Guam. Anaephenes A–C (876–878) represent a new family of cyanobacterial compounds with long alkyl chains featuring different levels and patterns of unsaturation.362 Anaenamides A and B (879 and 880) are also unique structures, originally described as depsipeptides.363 That are composed of hydroxy acid residues and a chlorinated α,β-unsaturated ester in addition to the alkyl salicylic moiety. The same study reported the isolation of anaenoic acid (881), which lacks the halogeneted amino ester unit at the C-terminus, shown to be critical for biological activity.363 Anaenamides C and D (882 and 883) are analogs featuring a primary amide in place of the methyl ester elucidated in 879–881, respectively.364 Anaenamides A and B 879 and 880 were cytotoxic to HCT116 cells in low micromolar levels, while anaenamides C and D 882 and 883 were inactive. Only 882 and 883 showed potential for Nrf2 (nuclear factor erythroid 2-related factor 2) activation at non-cytotoxic concentrations.364
Bartolosides are composed of a dialkylresorcinol structure decorated with halogens and sugar moieties. Bartoloside A (884) was isolated by NMR-guided fractionation of an extract from a marine Nodosilinea sp. collected at S. Bartolomeu do Mar beach, Portugal.365 Concurrently, analogous compounds (bartolosides B–D, 885–887) were isolated from an extract of a marine Synechocystis salina strain collected at the same beach, suggesting a horizontal transfer of the biosynthetic genes. The monoglicosilated analogs, bartolosides E–K (888–894), were isolated from a different Synechocystis salina strain, which was also shown to produce bartoloside A (884).366 Putative brt gene clusters were identified for both Synechocystis strains.365,366In vitro analyses provided biochemical evidence regarding the function of BrtD as ketosynthase and BrtC as aromatase.365 While exploring the function of BrtB as an O-alkylating enzyme, bartoloside A palmitates (895 and 896) were discovered.367 Hierridin C (897*) is a long-chain monoalkylresorcinol isolated from a Cyanobium sp.368 featuring a chlorine on the resorcinol ring. It showed antiplasmodial activity with IC50 values of 1.5 μM against Plasmodium falciparum 3D7 and 2.3 μM against the chloroquine-resistant Plasmodium falciparum Dd2 strain.368
6 Alkaloids
Cyanobacterial alkaloids are a heterogenous group of molecules often biosynthesized by hybrid pathways. It is worth noting that some alkaloids with major peptidic and lipidic skeletons were included in previous sections of this review.Hapalindole X (898) and deschloro hapalindole I (899) were isolated from a terrestrial Westiellopsis sp. from Thailand.369 The same study also reported the discovery of 13-hydroxy dechlorofontonamide (900*) from a freshwater Fischerella muscicola. 900* features an oxidized indole moiety characteristic of the fontonamides. Hapalindole X (898) exhibited growth inhibition against Mycobacterium tuberculosis, Mycobacterium smegmatis, Candida albicans, and Staphylococcus aureus with MIC values of 2.5, 78.8, 2.5, and 9.1 μM, respectively.369 Two formamide-containing hapalindoles, namely hapalindole A-formamide (901) and hapalindole J-formamide (902), were isolated from a freshwater Hapalosiphon sp. strain.370 With a different ring-fusion patten in comparison to hapalindoles, four nitrile-bearing fischerindole derivatives (903–906) were isolated from a Fischerella sp. from Iceland.371 Produced by a Fischerella ambigua, four hapalindole-related alkaloids (907–910) were discovered. While fischambiguine A (907) was inactive against M. tuberculosis, the epoxy-containing fischambiguine B (908) showed an MIC of 2 μM against the pathogen.
Additional indole alkaloids were isolated from a hydrophilic fraction of a marine Moorea producens (911–915),372 from a freshwater Fischerella ambigua (916–918),373 and from a freshwater Aetokthonos hydrillicola (919*).374 A chemistry-guided approach targeting halogenated metabolites led to the discovery of the bisindole tjipanazoles K–M (916–918).373 Tjipanazole M (918) was shown to inhibit the ABCG2 transporter (IC50 = 11 μM), which is implicated in cancer cell drug resistance.373
The cyanobacterial alkaloid aetokthonotoxin (919*) was discovered as the cause of bald eagle mass death events though a complex combination of environmental interplay.374 The aquatic plant Hydrilla verticillate, an invasive species in the southeastern USA, was shown to be colonized by Aetokthonos hydrillicola. In the laboratory, when supplemented with bromide, A. hydrillicola produced 919*, a pentabrominated biindole neurotoxin known to cause vacuolar myelinopathy. The study suggested that, in the environment, bromide may be derived from anthropogenic sources. With H. verticillate colonized by A. hydrillicola as the source, it was proposed 919* was transferred through the food chain leading to bioaccumulation and bald eagle exposure to toxic concentrations of the metabolite. Of note, aetokthonotoxin (919*) showed potent toxicity to Caenorhabditis elegans and zebrafish (LC50 values in the nanomolar range) and was shown to cause brain lesions characteristic of vacuolar myelinopathy in leghorn chickens.374
Four analogs of lyngbyatoxin A (920–923) were isolated from marine samples of Moorea producens.375–377 12-epi-Lyngbyatoxin A (920) was shown to have the same planar structure as lyngbyatoxin A upon HRMS and NMR analyses. However, CD spectra indicated stereochemical differences between the two. 12-epi-Lyngbyatoxin A (920) was toxic to Palaemon paucidens (shrimp) at similar levels of lyngbyatoxin A (LD100 values of 7.5 and 5 mg kg−1, respectively). 920 also caused paralysis in P. paucidens at 0.9 mg kg−1, which was not observed for lyngbyatoxin A. When tested for protein kinase Cδ (PKCδ) affinity, 920 showed significantly lower activity in comparison to lyngbyatoxin A (Ki values of 17 and 0.11 nM, respectively). The oxygenated analogs 2-oxo-3(R)-hydroxy-lyngbyatoxin A (921) and 2-oxo-3(R)-hydroxy-13-N-desmethyl-lyngbyatoxin A (922) displayed significantly less toxicity to P. paucidens and had lower affinity to PKCδ. 2,3-Seco-2,3-dioxo-lyngbyatoxin A (923) was not cytotoxic to HeLa cells, while lyngbyatoxin A displayed an IC50 of 9.2 nM, suggesting the importance of the intact indole ring for this biological activity.377
Two cylindrospermopsin analogs (924 and 925) were reported from a Cylindrospermopsis raciborskii378 and one (926*) was identified in a Raphidiopsis raciborskii.379 The presence of cylindrospermopsin in CyanoHABs carried severe human and animal health risks in addition to potential for triggering economic complications, making the chemical analysis of water sources of critical importance. Despite extensive studies, 924–926 are the only cylindrospermopsin analogs reported in the past two decades, with only two other analogs described prior to that. The basic structures of 7-deoxy-desulfo-cylindrospermopsin (924) and 7-deoxy-desulfo-12-acetylcylindrospermopsin (925) consist of a tricyclic guanidine core connected to an uracil-derived moiety. The discovery of 926 and labeling studies supported that the arginine guanidino group originates the uracil ring in cylindrospermopsin.379In vitro studies also suggest that 924 is a biosynthetic intermediate.379
Three saxitoxin analogs (927*, 928*, and 929*)380,381 and two anatoxin-a analogs (930* and 931*)382 were recently reported from cyanobacteria. 12β-Deoxygonyautoxin 3 (927*), 12β-deoxygonyautoxin 5 (928*), and 12β-deoxysaxitoxin (929*) were identified in a Dolichospermum circinale strain (previously reported as Anabaena circinalis).380,381 Carboxy-homoanatoxin-a (930*) was detected in a Oscillatoria sp. and carboxy-dihydroanatoxin-a (931*) in a Cylindrospermum stagnale.382 The discovery of these compounds sheds light into the biosynthesis of saxitoxin and anatoxin-a, neurotoxic alkaloids of ecotoxicological importance. Formerly known as anatoxin-a(s), the biosynthesis of guanitoxin was investigated in a Sphaerospermopsis torques-reginae, leading to the discovery of pre-guanitoxin (932*).383 This biosynthetic precursor is the product of a N-methyltransferase that dimethylates a primary amine.383
Along with mooreaside A (875*), the nucleoside derivatives 3-acetyl-21-deoxyuridine (933*) and 3-phenylethyl-21-deoxyuridine (934*) were discovered in a marine sample of Moorea producens from Saudi Arabia.361 They showed weak cytotoxic activity against MCF-7 (IC50 values of 18.2 μM for 933* and 22.8 μM for 934*) and no activity against HCT-116 colorectal carcinoma and HepG2 hepatocellular carcinoma cells. Microcystbiopterins A–E (935–939) were isolated from two freshwater blooms of Microcystis spp. collected in Israel.384 Their core structure is composed of fused pyrazine and pyrimidine rings, derived from guanosine triphosphate. In biopterins, the pyrazine ring is further substituted with a dihydroxypropyl moiety at C-6. Even though pterins can be found in eukaryotes and prokaryotes, glycosylated derivatives are mostly isolated from microorganisms, cyanobacteria in particular. Microcystbiopterins are biopterin glycosides with varying O-methylation pattern on the sugar moiety. It is suggested that biopterin glycosides play a role in UV-A radiation protection in cyanobacteria.384
Carriebowlinol (940*) was discovered in an assemblage of two cyanobacteria collected from coral reefs in Belize.385 The chemical investigation of this sample was motivated by results of a phylogenetic analysis that suggested a unique biodiversity for these strains. Antifungal activity-guided fractionation led to the isolation of 940*, a chlorinated tetrahydroquinolinol. Carriebowlinol (940*) showed antifungal activity in the sub-micromolar range against the marine fungi Dendryphiella salina, Lindra thalassiae, and Fusarium sp.385
Aulosirazoles B and C (941* and 942*), featuring a isothiazolonaphthoquinone scaffold, were discovered from a freshwater Nostoc sp. strain.386 They showed biological activity against high-grade serous ovarian cancer cells (OVCAR3) with IC50 values of 0.6 and 3 μM, respectively. A previously known aulosirazole A was isolated from the same strains in higher quantities, allowing for further biological assay testing. Aulosirazole A was shown to activate the nuclear transcription factor FOXO3a, which is associated with better outcomes in ovarian cancer patients.386
7 Miscellaneous compounds
Eucapsitrione (943*) is an anthraquinone derivative isolated from the freshwater Eucapsis sp. strain.387 The crude extract showed potent activity against Mycobacterium tuberculosis guiding the isolation of 943*. Pure 943* showed a MIC value of 3.1 μM in a microplate Alamar blue assay (MABA) against rapidly growing M. tuberculosis and a MIC value of 6.4 μM in a low-oxygen-recovery assay (LORA) with nonreplicating persistent M. tuberculosis.387 Chemical synthesis of 943* yielded a product with spectroscopic properties that differed from those of the natural compound. The structure reassignment has not been reported.388Nostotrebin 6 (944) was isolated from a terrestrial Nostoc sp. based on its potent activity in an acetylcholinesterase inhibition assay.389 The structure of 944 consists of two interconnected cyclopentenedione rings substituted with phenolic moieties. Nostotrebin 6 (944) was shown to inhibit acetylcholinesterase and butyrylcholinesterase with IC50 values ranging from 5.5–7.5 μM.389 Related compounds (945–948) were isolated from the medium extract of a different Nostoc sp. strain.390 Due to their bioactivity profile, they were suggested to be pan-assay interference compounds (PAINs).390
Crossbyanols A–D (949–952) are brominated aryl ethers discovered in marine samples of Leptolyngbya crossbyana.391 This cyanobacterium is known to bloom on Hawaiian reefs, being associated with coral bleaching. 1H NMR-guided fractionation of the cyanobacterial extract led to the isolation of 949–952. Crossbyanol B (950) displayed antibacterial activity against MRSA (MIC of 1.7–3.3 μM) and was toxic to brine shrimp (IC50 of 2.8 ppm).391 Bromoiesol sulfates A and B (953 and 954) were reported from a marine Salileptolyngbya sp. collected in Japan.392 Their hydrolysates bromoiesol A and B were also isolated, but were found to be artifacts of the exposure of 953 and 954 to strong acid conditions. Bromoiesols displayed significant growth inhibition activity against Trypanosoma brucei rhodesience (IC50 values in the low micromolar range), with the hydrolysates being more active than the sulfate analogs.392 Ambigols D and E (955 and 956) are chlorinated aryl ethers isolated from a freshwater Fischerella ambigua.393 They were found to induce prodigiosin production in Serratia spp.393
9-Ethyliminomethyl-12-(morpholin-4-ylmethoxy)-5,8,13,16-tetraaza-hexacene-2,3 dicarboxylicacid (EMTAHDCA, 957*) was isolated from a freshwater Nostoc sp. strain collected in India.394 EMTAHDCA (957*) showed antibacterial activity against Escherichia coli, Proteus vulgaris, and Pseudomonas aeruginosa in a disc diffusion assay. In silico docking studies revealed binding affinity of 957* towards the 30S ribosomal fragment and OmpF porin protein, which are common targets of commercial antibiotics.394
Tolyporphins L–R (958–964) were discovered from a Brasilonema sp. strain collected in Micronesia.395 They feature a tetrapyrrole core modified by oxidation, glycosylation, and/or acetylation. X-ray crystallography assisted with absolute configuration determination, revealing intriguing variations in stereochemistry within this metabolite family.395 Bioassay-guided fractionation for compounds with lipid-reducing activity led to the isolation of a chlorophyll-derivative named 132-hydroxy-pheofarnesin a (965*) from a marine Nodosilinea sp.396 While 965* displayed lipid-reducing activity in the zebrafish Nile red fat metabolism assay (IC50 = 15.5 μM), neither chlorophyll a or b showed any bioactivity in the same assay.396
Glycosylated mycosporine-like amino acids (MAA) were discovered from Nostoc commune (966* and 967)397 and Nostoc sphaericum (968)398 strains. 13-O-(β-Galactosyl)-porphyra-334 was (968) was shown to protect human keratinocytes in culture from UV damage.398 A comparative analysis of the chemical profile of four Nostoc commune strains led to the isolation of a 756-Da MAA, named nostoc-756 (969*). It is the aglycone of 967, which features three chromophores.399 756-Da MAA (969*) was also isolated from a lichen symbiont Nostoc sp. along with aplysiapalythine E (970).400 Genome sequencing and mining identified BGCs associated with MAA production in three locations of the genome of Nostoc sp.400 A bioinformatics analysis supported the high MAA BGC plasticity.400
Dragocins A–D (971–974*) are structurally unique metabolites isolated from a marine cyanobacterium that shows Symploca-like morphology, yet this classification was not supported by 16S rDNA phylogenetic analysis.401 These hybrid metabolites feature a ribose-derived moiety embedded in a nine-membered macrocycle containing a dihydroxypyrrolidine and a OMe-benzoyl units likely derived from a NRPS–PKS pathway.401 Dragocin A (971) was cytotoxic to H-460 human lung cancer cells.401
Leptazolines A–D (975–978) were isolated from a Leptolyngbya sp. culture media extract that showed biological activity against pancreatic adenocarcinoma cells.402 While cyanobacterial compounds from cell extracts are predominant in this review, 975–978 are among the few water-soluble natural products isolated from culture media in recent years. Their chemical diversity further supports the relevance of (re)evaluating hydrophilic fractions for natural product discovery.403
Thiopalmyrone (979*) was isolated along with palmyrrolinone (749*) from an environmental assemblage of cf. Oscillatoria and Hormoscilla spp. collected in Palmyra Atoll.306 It features a α,β-unsaturated thioester group composing a six-membered ring core. Thiopalmyrone (979*) was toxic to Biomphalaria glabrata (LC50 value of 8.3 μM), the intermediate snail host for Schistosoma mansoni. A subtle enhancement in biological activity was observed when 979* and 749* were evaluated as a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture.306
1 mixture.306
Hennaminal (980*) and hennamide (981*) were isolated from a marine Rivularia sp.404 Hennaminal (980*) features a rare β,β-diamino unsaturated ketone and hennamide (981*) has a N-acyl pyrrolinone group. During structural elucidation, 981* dimerized due to the reactivity of the pyrrolinone ring. Compounds 980*-981* were shown to inhibit Trypanosoma brucei rhodesiense with IC50 values of 11 and 9.7 μM, respectively.404
Caldorin (982*) was discovered from a marine Caldora penicillata collected in Japan.405 It has a core structure composed by a cis-fused decalin ring connected to an acylated amide. The decalin ring was proposed to be biosynthesized via an intramolecular Diels–Alder reaction on a polyketide chain.405 Caldorin (982*) was not cytotoxic to cancer cells and was shown to inhibit osteoblast differentiation (IC50 = 6.2 μM).
Decalin rings are also observed in eudesmane-type sesquiterpenoids isolated from a marine cyanobacterial assemblage mostly composed of Neolyngbya sp. (983*)406 and from a freshwater Scytonema sp. (984–986).407 Eudesmacarbonate (983*) was isolated from a cyanobacterial biomass associated with dolphin intoxication events. It features a cyclic carbonate, representing the first report of this moiety in cyanobacterial eudesmane terpenoids. Eudesmacarbonate (983*) showed neurotoxic effects on zebrafish embryos.406 Stigolone (984) and two dihydroxy derivatives (985–986) were isolated from media extracts of a Scytonema sp. in a search for polar compounds.407 The same study also reported the discovery of spirovetivane-type terpenoids, namely spironostoic acid (987), 11,12-didehydrospironostoic acid (988), and 12-hydroxy-2-oxo-11-epi-hinesol (989) from a Calothrix sp. medium extract.407
Cybastacines A and B (990* and 991*) are among the few sesterterpenes reported from cyanobacteria.408 The other known cyanobacterial sesterterpene is scytoscalarol from a Scytonema sp., which features a guanidine moiety similar to 990* and 991*.409 Cybastacines A and B (990* and 991*) were discovered from a Nostoc sp. from the Canary Islands.408 They exhibited antibiotic activity against multiple bacterial strains, with 991* showing stronger effects.408
Four analogs of the meroterpenoid tolypodiol (992–995) were isolated from extracts of a Brasilonema sp. strain.410,411 11-Hydroxytolypodiol (993) displayed significant anti-inflammatory activity by inhibiting the generation of proinflammatory thromboxane B2, being even more potent than aspirin.410 The tolypodiol BGC was identified and successfully expressed in Anabaena sp. UTEX 2576. MS/MS molecular networking confirmed tolypodiol variants (994 and 995) in extracts of the engineered Anabaena sp.411
8 Concluding remarks
Cyanobacteria continue to yield chemically diverse natural products with biotechnological potential. This review highlighted compounds discovered through the collaborative efforts of researchers globally, exploring various environmental sources and innovative discovery methods. The molecules discussed are derived from both field-collected material and cultured cyanobacteria, spanning marine, terrestrial, freshwater, and brackish water environments. The discovery approaches include traditional methods as well as cutting-edge omics-based techniques.The similarity networking analysis assisted with data visualization by displaying chemical families and their sources. This may be valuable information for driving decision-making in cyanobacterial natural product discovery programs. This analysis also revealed structurally unique compounds that share little similarity with other structures in the database.
9 Data availability
Data for this article, including compound database, metadata, source data for figures, and similarity network are available at Zenodo DOI: https://doi.org/10.5281/zenodo.12627504.10 Conflicts of interest
Authors declare no conflict of interest.11 Acknowledgements
We thank Maria Gabriela Silva Bueno and Leonardo de Faria Figueiredo for their assistance with data curation. This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil, Financial code 001; CAPES/Science without Borders BEX 13055-13-5; the National Natural Science Foundation of China (No. 21907047); and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grant numbers 2019/17721-9, 2020/07710-7, 2021/14754-3, and 2022/02872-4.12 References
- B. E. Schirrmeister, J. M. de Vos, A. Antonelli and H. C. Bagheri, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 1791–1796 CrossRef CAS.
- R. E. Blankenship, Plant Physiol., 2010, 154, 434–438 CrossRef CAS PubMed.
- J. F. Kasting and J. L. Siefert, Science, 2002, 296, 1066–1068 CrossRef CAS.
- D. G. Nagle and V. J. Paul, J. Phycol., 1999, 35, 1412–1421 CrossRef CAS.
- P. N. Leão, N. Engene, A. Antunes, W. H. Gerwick and V. Vasconcelos, Nat. Prod. Rep., 2012, 29, 372–391 RSC.
- C. Wiegand and S. Pflugmacher, Toxicol. Appl. Pharmacol., 2005, 203, 201–218 CrossRef CAS PubMed.
- L. Bláha, P. Babica and B. Maršálek, Interdiscip. Toxicol., 2009, 2, 36–41 Search PubMed.
- N. E. Nozzi, J. W. K. Oliver and S. Atsumi, Front. Bioeng. Biotechnol., 2013, 1, 1–6 Search PubMed.
- R. M. M. Abed, S. Dobretsov and K. Sudesh, J. Appl. Microbiol., 2009, 106, 1–12 CrossRef CAS.
- A. Udayan, M. Arumugam and A. Pandey, in Algal Green Chemistry, Elsevier, 2017, pp. 65–89 Search PubMed.
- R. K. Singh, S. P. Tiwari, A. K. Rai and T. M. Mohapatra, J. Antibiot., 2011, 64, 401–412 CrossRef CAS.
- G. R. Pettit, Y. Kamano, C. L. Herald, A. a Tuinman, F. E. Boettner, H. Kizu, J. M. Schmidt, L. Baczynskyj, K. B. Tomer and R. J. Bontems, J. Am. Chem. Soc., 1987, 109, 6883–6885 CrossRef CAS.
- K. Miyazaki, M. Kobayashi, T. Natsume, M. Gondo, T. Mikami, K. Sakakibara and S. Tsukagoshi, Chem. Pharm. Bull., 1995, 43, 1706–1718 CrossRef CAS.
- S. O. Doronina, B. E. Toki, M. Y. Torgov, B. A. Mendelsohn, C. G. Cerveny, D. F. Chace, R. L. DeBlanc, R. P. Gearing, T. D. Bovee, C. B. Siegall, J. A. Francisco, A. F. Wahl, D. L. Meyer and P. D. Senter, Nat. Biotech., 2003, 21, 778–784 CrossRef CAS.
- H. Luesch, R. E. Moore, V. J. Paul, S. L. Mooberry and T. H. Corbett, J. Nat. Prod., 2001, 64, 907–910 CrossRef CAS.
- C. H. Moskowitz, A. Nademanee, T. Masszi, E. Agura, J. Holowiecki, M. H. Abidi, A. I. Chen, P. Stiff, A. M. Gianni, A. Carella, D. Osmanov, V. Bachanova, J. Sweetenham, A. Sureda, D. Huebner, E. L. Sievers, A. Chi, E. K. Larsen, N. N. Hunder and J. Walewski, Lancet, 2015, 385, 1853–1862 CrossRef CAS.
- N. Papon, B. R. Copp and V. Courdavault, Biotechnol. Adv., 2022, 54, 107871 CrossRef CAS PubMed.
- T. F. Molinski, Nat. Prod. Rep., 2010, 27, 321–329 RSC.
- Y. Lin, S. Schiavo, J. Orjala, P. Vouros and R. Kautz, Anal. Chem., 2008, 80, 8045–8054 CrossRef CAS PubMed.
- S. P. Gaudêncio and F. Pereira, Nat. Prod. Rep., 2015, 32, 779–810 RSC.
- M. Baunach, A. Guljamow, M. Miguel-Gordo and E. Dittmann, Nat. Prod. Rep., 2024, 41, 347–369 RSC.
- M. B. Weiss, R. V. Médice, F. R. Jacinavicius, E. Pinto and C. M. Crnkovic, in Microbial Natural Products Chemistry: A Metabolomics Approach, ed. T. Pacheco Fill, Springer International Publishing, Cham, 2023, pp. 21–49 Search PubMed.
- P. M. D'Agostino, Nat. Prod. Rep., 2023, 40, 1701–1717 RSC.
- K. Kleigrewe, L. Gerwick, D. H. Sherman and W. H. Gerwick, Nat. Prod. Rep., 2016, 33, 348–364 RSC.
- L. A. Salvador-Reyes and H. Luesch, Nat. Prod. Rep., 2015, 32, 478–503 RSC.
- E. Dittmann, M. Gugger, K. Sivonen and D. P. Fewer, Trends Microbiol., 2015, 23, 642–652 CrossRef CAS.
- G. E. Chlipala, S. Mo and J. Orjala, Curr. Drug Targets, 2011, 12, 1654–1673 Search PubMed.
- L. T. Tan, Phytochemistry, 2007, 68, 954–979 CrossRef CAS.
- K. Tidgewell, B. R. Clark and W. H. Gerwick, in Comprehensive Natural Products II, ed. H.-W. (Ben) Liu and L. Mander, Elsevier, Oxford, 2010, pp. 141–188 Search PubMed.
- J. Komárek, J. Kaštovský, J. Mareš and J. R. Johansen, Preslia, 2014, 86, 295–335 Search PubMed.
- R. M. Borges, G. de Assis Ferreira, M. M. Campos, A. M. Teixeira, F. das Neves Costa and F. O. Chagas, Phytochem. Anal., 2024, 35, 93–101 CrossRef CAS PubMed.
- M. Welker and H. Von Döhren, FEMS Microbiol. Rev., 2006, 30, 530–563 CrossRef CAS PubMed.
- S. Elkobi-Peer, R. Faigenbaum and S. Carmeli, J. Nat. Prod., 2012, 75, 2144–2151 CrossRef CAS PubMed.
- M. Lifshits and S. Carmeli, J. Nat. Prod., 2012, 75, 209–219 CrossRef CAS PubMed.
- S. Adiv and S. Carmeli, J. Nat. Prod., 2013, 76, 2307–2315 CrossRef CAS PubMed.
- S. Elkobi-Peer, R. K. Singh, T. M. Mohapatra, S. P. Tiwari and S. Carmeli, J. Nat. Prod., 2013, 76, 1187–1190 CrossRef CAS.
- M. Vegman and S. Carmeli, Tetrahedron, 2014, 70, 6817–6824 CrossRef CAS.
- S. Elkobi-Peer and S. Carmeli, Mar. Drugs, 2015, 13, 2347–2375 Search PubMed.
- R. Hasan-Amer and S. Carmeli, Mar. Drugs, 2017, 15, 371 CrossRef PubMed.
- E. Kohler, V. Grundler, D. Häussinger, R. Kurmayer, K. Gademann, J. Pernthaler and J. F. Blom, Harmful Algae, 2014, 39, 154–160 CAS.
- A. Kapuścik, P. Hrouzek, M. Kuzma, S. Bártová, P. Novák, J. Jokela, M. Pflüger, A. Eger, H. Hundsberger and J. Kopecký, ChemBioChem, 2013, 14, 2329–2337 Search PubMed.
- D. P. Fewer, J. Jokela, E. Paukku, J. Österholm, M. Wahlsten, P. Permi, O. Aitio, L. Rouhiainen, G. V. Gomez-Saez and K. Sivonen, PLoS One, 2013, 8, e73618 Search PubMed.
- L. M. P. Heinilä, J. Jokela, M. N. Ahmed, M. Wahlsten, S. Kumar, P. Hrouzek, P. Permi, H. Koistinen, D. P. Fewer and K. Sivonen, Org. Biomol. Chem., 2022, 20, 2681–2692 Search PubMed.
- L. Liu, A. Budnjo, J. Jokela, B. E. Haug, D. P. Fewer, M. Wahlsten, L. Rouhiainen, P. Permi, T. Fossen and K. Sivonen, ACS Chem. Biol., 2015, 10, 725–733 CAS.
- Y. K. H. Schneider, A. Liaimer, J. Isaksson, O. S. B. Wilhelmsen, J. H. Andersen, K. Ø. Hansen and E. H. Hansen, Front. Microbiol., 2023, 14, 1–13 Search PubMed.
- A. R. J. Anas, T. Kisugi, T. Umezawa, F. Matsuda, M. R. Campitelli, R. J. Quinn and T. Okino, J. Nat. Prod., 2012, 75, 1546–1552 Search PubMed.
- M. Sanz, R. K. Salinas and E. Pinto, J. Nat. Prod., 2017, 80, 2492–2501 CAS.
- C.-S. Phan, J. J. Mehjabin, A. R. J. Anas, M. Hayasaka, R. Onoki, J. Wang, T. Umezawa, K. Washio, M. Morikawa and T. Okino, J. Nat. Prod., 2022, 85, 2000–2005 Search PubMed.
- L. Liu, J. Jokela, M. Wahlsten, B. Nowruzi, P. Permi, Y. Z. Zhang, H. Xhaard, D. P. Fewer and K. Sivonen, J. Nat. Prod., 2014, 77, 1784–1790 CrossRef CAS.
- C.-S. Phan, J. J. Mehjabin, A. R. J. Anas, M. Hayasaka, R. Onoki, J. Wang, T. Umezawa, K. Washio, M. Morikawa and T. Okino, J. Nat. Prod., 2022, 85, 2000–2005 CrossRef CAS.
- C. Pancrace, K. Ishida, E. Briand, D. G. Pichi, A. R. Weiz, A. Guljamow, T. Scalvenzi, N. Sassoon, C. Hertweck, E. Dittmann and M. Gugger, ACS Chem. Biol., 2019, 14, 67–75 CrossRef CAS PubMed.
- M. Lifshits, E. Zafrir-Ilan, A. Raveh and S. Carmeli, Tetrahedron, 2011, 67, 4017–4024 CrossRef CAS.
- A. Lodin-Friedman and S. Carmeli, Mar. Drugs, 2018, 16, 78 CrossRef.
- W. K. Strangman and J. L. C. Wright, Tetrahedron Lett., 2016, 57, 1801–1803 CrossRef CAS.
- A. K. Stewart, R. Ravindra, R. M. Van Wagoner and J. L. C. Wright, J. Nat. Prod., 2018, 81, 349–355 CrossRef CAS.
- K. Calabro, G. Genta-Jouve and O. P. Thomas, J. Nat. Prod., 2019, 82, 1040–1044 CrossRef CAS PubMed.
- N. Eusébio, R. Castelo-Branco, D. Sousa, M. Preto, P. D'Agostino, T. A. M. Gulder and P. N. Leão, ACS Synth. Biol., 2022, 11, 3493–3503 CrossRef.
- E. Zafrir-Ilan and S. Carmeli, Tetrahedron, 2010, 66, 9194–9202 CrossRef CAS.
- S. Weisthal Algor, A. Sukenik and S. Carmeli, Mar. Drugs, 2023, 21, 401 CrossRef CAS.
- M. Schumacher, N. Wilson, J. N. Tabudravu, C. Edwards, L. A. Lawton, C. Motti, A. D. Wright, M. Diederich and M. Jaspars, Tetrahedron, 2012, 68, 1622–1628 CrossRef CAS.
- S. Saha, G. Esposito, P. Urajová, J. Mareš, D. Ewe, A. Caso, M. Macho, K. Delawská, A. Kust, P. Hrouzek, J. Juráň, V. Costantino and K. Saurav, Molecules, 2020, 25, 3786 CrossRef CAS.
- B. Bober, E. Chrapusta-Srebrny and J. Bialczyk, Eur. J. Phycol., 2021, 56, 244–254 CrossRef CAS.
- R. Konkel, M. Grabski, M. Cegłowska, E. Wieczerzak, G. Węgrzyn and H. Mazur-Marzec, Int. J. Environ. Res. Public Health, 2022, 19, 12346 CrossRef CAS.
- J. Zi, D. D. Lantvit, S. M. Swanson and J. Orjala, Phytochemistry, 2012, 74, 173–177 CrossRef CAS PubMed.
- M. Sanz, R. K. Salinas and E. Pinto, J. Nat. Prod., 2017, 80, 2492–2501 CrossRef CAS.
- A. Guljamow, M. Kreische, K. Ishida, A. Liaimer, B. Altermark, L. Bähr, C. Hertweck, R. Ehwald and E. Dittmann, Appl. Environ. Microbiol., 2017, 83, e01510–e01517 CrossRef PubMed.
- H. Harms, K. L. Kurita, L. Pan, P. G. Wahome, H. He, A. D. Kinghorn, G. T. Carter and R. G. Linington, Bioorg. Med. Chem. Lett., 2016, 26, 4960–4965 CrossRef CAS PubMed.
- H. Schreuder, A. Liesum, P. Lönze, H. Stump, H. Hoffmann, M. Schiell, M. Kurz, L. Toti, A. Bauer, C. Kallus, C. Klemke-Jahn, J. Czech, D. Kramer, H. Enke, T. H. J. Niedermeyer, V. Morrison, V. Kumar and M. Brönstrup, Sci. Rep., 2016, 6, 32958 CrossRef CAS PubMed.
- E. Zafrir and S. Carmeli, J. Nat. Prod., 2010, 73, 352–358 CrossRef CAS.
- J. D. Isaacs, W. K. Strangman, A. E. Barbera, M. A. Mallin, M. R. McIver and J. L. C. Wright, Harmful Algae, 2014, 31, 82–86 CrossRef CAS PubMed.
- T. Thorskov Bladt, S. Kalifa-Aviv, T. Ostenfeld Larsen and S. Carmeli, Tetrahedron, 2014, 70, 936–943 CrossRef CAS.
- W. K. Strangman, A. K. Stewart, M. C. Herring and J. L. C. Wright, Tetrahedron Lett., 2018, 59, 934–937 CrossRef CAS.
- R. D. Kirk, H. He, P. G. Wahome, S. Wu, G. T. Carter and M. J. Bertin, ACS Omega, 2021, 6, 15472–15478 CrossRef CAS PubMed.
- W. K. Strangman, A. K. Stewart, M. C. Herring and J. L. C. Wright, Tetrahedron Lett., 2018, 59, 934–937 CrossRef CAS.
- R. D. Kirk, H. He, P. G. Wahome, S. Wu, G. T. Carter and M. J. Bertin, ACS Omega, 2021, 6, 15472–15478 CrossRef CAS.
- K. Gademann, C. Portmann, J. F. Blom, M. Zeder and F. Jüttner, J. Nat. Prod., 2010, 73, 980–984 CrossRef CAS.
- H. Mazur-Marzec, A. Fidor, M. Cegłowska, E. Wieczerzak, M. Kropidłowska, M. Goua, J. Macaskill and C. Edwards, Mar. Drugs, 2018, 16, 220 CrossRef PubMed.
- R. Konkel, M. Cegłowska, K. Szubert, E. Wieczerzak, S. Iliakopoulou, T. Kaloudis and H. Mazur-Marzec, Mar. Drugs, 2023, 21, 508 CrossRef CAS PubMed.
- H.-S. Kang, A. Krunic and J. Orjala, J. Nat. Prod., 2012, 75, 807–811 CrossRef CAS.
- L. A. Salvador, K. Taori, J. S. Biggs, J. Jakoncic, D. A. Ostrov, V. J. Paul and H. Luesch, J. Med. Chem., 2013, 56, 1276–1290 CrossRef CAS PubMed.
- L. Keller, K. M. Canuto, C. Liu, B. M. Suzuki, J. Almaliti, A. Sikandar, C. B. Naman, E. Glukhov, D. Luo, B. M. Duggan, H. Luesch, J. Koehnke, A. J. O'Donoghue and W. H. Gerwick, ACS Chem. Biol., 2020, 15, 751–757 CrossRef CAS.
- B. K. Rubio, S. M. Parrish, W. Yoshida, P. J. Schupp, T. Schils and P. G. Williams, Tetrahedron Lett., 2010, 51, 6718–6721 CrossRef CAS.
- S. P. Gunasekera, M. W. Miller, J. C. Kwan, H. Luesch and V. J. Paul, J. Nat. Prod., 2010, 73, 459–462 CrossRef CAS PubMed.
- R. Reher, A. T. Aron, P. Fajtová, P. Stincone, B. Wagner, A. I. Pérez-Lorente, C. Liu, I. Y. Ben Shalom, W. Bittremieux, M. Wang, K. Jeong, M. L. Matos-Hernandez, K. L. Alexander, E. J. Caro-Diaz, C. B. Naman, J. H. W. Scanlan, P. M. M. Hochban, W. E. Diederich, C. Molina-Santiago, D. Romero, K. A. Selim, P. Sass, H. Brötz-Oesterhelt, C. C. Hughes, P. C. Dorrestein, A. J. O'Donoghue, W. H. Gerwick and D. Petras, Nat. Commun., 2022, 13, 4619 CrossRef CAS PubMed.
- K. Dührkop, L.-F. Nothias, M. Fleischauer, R. Reher, M. Ludwig, M. A. Hoffmann, D. Petras, W. H. Gerwick, J. Rousu, P. C. Dorrestein and S. Böcker, Nat. Biotechnol., 2021, 39, 462–471 CrossRef.
- D. A. Gallegos, J. Saurí, R. D. Cohen, X. Wan, P. Videau, A. O. Vallota-Eastman, L. A. Shaala, D. T. A. Youssef, R. T. Williamson, G. E. Martin, B. Philmus, A. E. Sikora, J. E. Ishmael and K. L. McPhail, J. Nat. Prod., 2018, 81, 1417–1425 CrossRef CAS.
- F. Al-Awadhi, L. Salvador, B. Law, V. Paul and H. Luesch, Mar. Drugs, 2017, 15, 290 CrossRef.
- K. Ozaki, A. Iwasaki, K. Suenaga and T. Teruya, Tetrahedron, 2019, 75, 3382–3386 CrossRef CAS.
- S. Matthew, R. Ratnayake, M. A. Becerro, R. Ritson-Williams, V. J. Paul and H. Luesch, Mar. Drugs, 2010, 8, 1803–1816 CrossRef CAS PubMed.
- C. Zhou, H. Chen, H. Zhao and Q. Wang, Algal Res., 2021, 55, 102277 CrossRef.
- J. Puddick, M. R. Prinsep, S. A. Wood, S. C. Cary, D. P. Hamilton and A. L. Wilkins, Phytochem. Lett., 2013, 6, 575–581 CrossRef CAS.
- J. Puddick, M. R. Prinsep, S. A. Wood, C. O. Miles, F. Rise, S. C. Cary, D. P. Hamilton and A. L. Wilkins, Mar. Drugs, 2013, 11, 3025–3045 CrossRef PubMed.
- Y. Qi, L. Rosso, D. Sedan, L. Giannuzzi, D. Andrinolo and D. A. Volmer, Rapid Commun. Mass Spectrom., 2015, 29, 220–224 CrossRef CAS.
- F. F. del Campo and Y. Ouahid, Environ. Pollut., 2010, 158, 2906–2914 CrossRef CAS PubMed.
- H. He, S. Wu, P. G. Wahome, M. J. Bertin, A. C. Pedone, K. R. Beauchesne, P. D. R. Moeller and G. T. Carter, J. Nat. Prod., 2018, 81, 1368–1375 CrossRef CAS PubMed.
- K. Sivonen, N. Leikoski, D. P. Fewer and J. Jokela, Appl. Microbiol. Biotechnol., 2010, 86, 1213–1225 CrossRef CAS.
- A. Raveh, S. Moshe, Z. Evron, E. Flescher and S. Carmeli, Tetrahedron, 2010, 66, 2705–2712 CrossRef CAS.
- E. Zafrir-Ilan and S. Carmeli, Tetrahedron Lett., 2010, 51, 6602–6604 CrossRef CAS.
- I. Koodkaew, S. Matsuyama, Y. Sunohara and H. Matsumoto, Tetrahedron Lett., 2012, 53, 977–979 CrossRef CAS.
- C. Portmann, S. Sieber, S. Wirthensohn, J. F. Blom, L. Da Silva, E. Baudat, M. Kaiser, R. Brun and K. Gademann, J. Nat. Prod., 2014, 77, 557–562 CrossRef CAS PubMed.
- M. Purushothaman, S. Sarkar, M. Morita, M. Gugger, E. W. Schmidt and B. I. Morinaka, Angew. Chem., Int. Ed., 2021, 60, 8460–8465 CrossRef CAS.
- N. Leikoski, D. P. Fewer, J. Jokela, M. Wahlsten, L. Rouhiainen and K. Sivonen, Appl. Environ. Microbiol., 2010, 76, 701–709 CrossRef CAS PubMed.
- N. Leikoski, D. P. Fewer, J. Jokela, P. Alakoski, M. Wahlsten and K. Sivonen, PLoS One, 2012, 7, e43002 CrossRef CAS.
- M. S. Donia and E. W. Schmidt, Chem. Biol., 2011, 18, 508–519 CrossRef CAS PubMed.
- W. E. Houssen, J. Koehnke, D. Zollman, J. Vendome, A. Raab, M. C. M. Smith, J. H. Naismith and M. Jaspars, ChemBioChem, 2012, 13, 2683–2689 CrossRef CAS.
- J. A. McIntosh, Z. Lin, M. D. B. Tianero and E. W. Schmidt, ACS Chem. Biol., 2013, 8, 877–883 CrossRef CAS.
- C. Audoin, J. A. Sánchez, G. Genta-Jouve, A. Alfonso, L. Rios, C. Vale, O. P. Thomas and L. M. Botana, J. Nat. Prod., 2014, 77, 2196–2205 CrossRef CAS.
- C. Clemente, N. Johnson, X. Ouyang, R. V. Popin, S. Dall'Angelo, M. Wahlsten, J. Jokela, A. Colombano, B. Nardone, D. P. Fewer and W. E. Houssen, Chem. Commun., 2022, 58, 12054–12057 RSC.
- M. Y. Phyo, C. Y. G. Ding, H. C. Goh, J. X. Goh, J. F. M. Ong, S. H. Chan, P. Y. M. Yung, H. Candra and L. T. Tan, J. Nat. Prod., 2019, 82, 3482–3488 CrossRef CAS PubMed.
- M. Y. Phyo, T. M. Ben Goh, J. X. Goh and L. T. Tan, Mar. Drugs, 2021, 19, 548 CrossRef CAS.
- S. Luo, A. Krunic, G. E. Chlipala and J. Orjala, Phytochem. Lett., 2015, 13, 47–52 CrossRef CAS PubMed.
- J. A. V. Lopez, S. S. Al-Lihaibi, W. M. Alarif, A. Abdel-Lateff, Y. Nogata, K. Washio, M. Morikawa and T. Okino, J. Nat. Prod., 2016, 79, 1213–1218 CrossRef CAS.
- A. Raveh and S. Carmeli, Org. Lett., 2010, 12, 3536–3539 CrossRef CAS PubMed.
- S. Adiv, R. Ahronov-Nadborny and S. Carmeli, Tetrahedron, 2012, 68, 1376–1383 CrossRef CAS.
- J. Martins, N. Leikoski, M. Wahlsten, J. Azevedo, J. Antunes, J. Jokela, K. Sivonen, V. Vasconcelos, D. P. Fewer and P. N. Leão, Sci. Rep., 2018, 8, 14537 CrossRef PubMed.
- C.-S. Phan, K. Matsuda, N. Balloo, K. Fujita, T. Wakimoto and T. Okino, J. Am. Chem. Soc., 2021, 143, 10083–10087 CrossRef CAS.
- N. Balloo, J. J. Mehjabin, C. Phan and T. Okino, Phycol. Res., 2023, 71, 200–208 CrossRef CAS.
- N. Leikoski, L. Liu, J. Jokela, M. Wahlsten, M. Gugger, A. Calteau, P. Permi, C. A. Kerfeld, K. Sivonen and D. P. Fewer, Chem. Biol., 2013, 20, 1033–1043 CrossRef CAS PubMed.
- M. Cegłowska, K. Szubert, E. Wieczerzak, A. Kosakowska and H. Mazur-Marzec, Mar. Drugs, 2020, 18, 446 CrossRef.
- A. Mattila, R.-M. Andsten, M. Jumppanen, M. Assante, J. Jokela, M. Wahlsten, K. M. Mikula, C. Sigindere, D. H. Kwak, M. Gugger, H. Koskela, K. Sivonen, X. Liu, J. Yli-Kauhaluoma, H. Iwaï and D. P. Fewer, ACS Chem. Biol., 2019, 14, 2683–2690 CrossRef CAS.
- C. M. Crnkovic, J. Braesel, A. Krunic, A. S. Eustáquio and J. Orjala, ChemBioChem, 2020, 21, 845–852 CrossRef CAS.
- C. M. Crnkovic, D. S. May and J. Orjala, J. Appl. Phycol., 2018, 30, 375–384 CrossRef CAS.
- M. Montalbán-López, T. A. Scott, S. Ramesh, I. R. Rahman, A. J. van Heel, J. H. Viel, V. Bandarian, E. Dittmann, O. Genilloud, Y. Goto, M. J. Grande Burgos, C. Hill, S. Kim, J. Koehnke, J. A. Latham, A. J. Link, B. Martínez, S. K. Nair, Y. Nicolet, S. Rebuffat, H.-G. Sahl, D. Sareen, E. W. Schmidt, L. Schmitt, K. Severinov, R. D. Süssmuth, A. W. Truman, H. Wang, J.-K. Weng, G. P. van Wezel, Q. Zhang, J. Zhong, J. Piel, D. A. Mitchell, O. P. Kuipers and W. A. van der Donk, Nat. Prod. Rep., 2021, 38, 130–239 RSC.
- S. Sieber, S. M. Grendelmeier, L. A. Harris, D. A. Mitchell and K. Gademann, J. Nat. Prod., 2020, 83, 438–446 CrossRef CAS.
- D. Dehm, J. Krumbholz, M. Baunach, V. Wiebach, K. Hinrichs, A. Guljamow, T. Tabuchi, H. Jenke-Kodama, R. D. Süssmuth and E. Dittmann, ACS Chem. Biol., 2019, 14, 1271–1279 CrossRef CAS.
- S. Saha, P.-A. Bulzu, P. Urajová, J. Mareš, G. Konert, J. Câmara Manoel, M. Macho, D. Ewe, P. Hrouzek, J. Masojídek, R. Ghai and K. Saurav, mSphere, 2021, 6, e0056221 CrossRef PubMed.
- C. B. Naman, R. Rattan, S. E. Nikoulina, J. Lee, B. W. Miller, N. A. Moss, L. Armstrong, P. D. Boudreau, H. M. Debonsi, F. A. Valeriote, P. C. Dorrestein and W. H. Gerwick, J. Nat. Prod., 2017, 80, 625–633 CrossRef CAS.
- K. Iwasaki, A. Iwasaki, S. Sumimoto, T. Sano, Y. Hitomi, O. Ohno and K. Suenaga, Tetrahedron Lett., 2018, 59, 3806–3809 CrossRef CAS.
- H. Takahashi, A. Iwasaki, N. Kurisawa, R. Suzuki, G. Jeelani, T. Matsubara, T. Sato, T. Nozaki and K. Suenaga, J. Nat. Prod., 2021, 84, 1649–1655 CrossRef CAS.
- A. Iwasaki, N. Kurisawa, T. Wang, X. Li, H. Luo, C. Zhu, G. Patial, X. Yan, S. He, T. Luzzatto-Knaan, F. Tian, C. B. Naman and K. Suenaga, Tetrahedron Lett., 2021, 75, 153214 CrossRef CAS.
- F. Hubrich, N. M. Bösch, C. Chepkirui, B. I. Morinaka, M. Rust, M. Gugger, S. L. Robinson, A. L. Vagstad and J. Piel, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2113120119 CrossRef CAS.
- R. Montaser, V. J. Paul and H. Luesch, Phytochemistry, 2011, 72, 2068–2074 CrossRef CAS PubMed.
- C. Y. G. Ding, J. F. M. Ong, H. C. Goh, C. R. Coffill and L. T. Tan, Mar. Drugs, 2018, 16, 409 CrossRef CAS PubMed.
- P. D. Boudreau, T. Byrum, W. T. Liu, P. C. Dorrestein and W. H. Gerwick, J. Nat. Prod., 2012, 75, 1560–1570 CrossRef CAS.
- A. Iwasaki, I. Shiota, S. Sumimoto, T. Matsubara, T. Sato and K. Suenaga, J. Nat. Prod., 2017, 80, 1948–1952 CrossRef CAS.
- A. Tripathi, J. Puddick, M. R. Prinsep, P. P. F. Lee and L. T. Tan, Phytochemistry, 2010, 71, 307–311 CrossRef CAS PubMed.
- A. Levert, R. Alvariño, L. Bornancin, E. Abou Mansour, A. M. Burja, A. M. Genevière, I. Bonnard, E. Alonso, L. Botana and B. Banaigs, J. Nat. Prod., 2018, 81, 1301–1310 CrossRef CAS PubMed.
- L. A. Salvador, J. S. Biggs, V. J. Paul and H. Luesch, J. Nat. Prod., 2011, 74, 917–927 CrossRef CAS.
- E. Mevers, W. T. Liu, N. Engene, H. Mohimani, T. Byrum, P. A. Pevzner, P. C. Dorrestein, C. Spadafora and W. H. Gerwick, J. Nat. Prod., 2011, 74, 928–936 CrossRef CAS PubMed.
- K. Sueyoshi, T. Kudo, A. Yamano, S. Sumimoto, A. Iwasaki, K. Suenaga and T. Teruya, Bull. Chem. Soc. Jpn., 2017, 90, 436–440 CrossRef CAS.
- M. Y. Phyo, N. P. Katermeran, J. X. Goh and L. T. Tan, Phytochemistry, 2021, 190, 112879 CrossRef CAS.
- S. P. Gunasekera, C. S. Owle, R. Montaser, H. Luesch and V. J. Paul, J. Nat. Prod., 2011, 74, 871–876 CrossRef CAS.
- O. M. Sabry, D. E. Goeger and W. H. Gerwick, Nat. Prod. Res., 2017, 31, 555–561 CrossRef CAS PubMed.
- M. Y. Phyo, J. X. Goh and L. T. Tan, J. Nat. Prod., 2022, 85, 485–492 CrossRef CAS.
- T. Luzzatto-Knaan, N. Garg, M. Wang, E. Glukhov, Y. Peng, G. Ackermann, A. Amir, B. M. Duggan, S. Ryazanov, L. Gerwick, R. Knight, T. Alexandrov, N. Bandeira, W. H. Gerwick and P. C. Dorrestein, Elife, 2017, 6, 1–20 CrossRef.
- B. Han, J. Microbiol. Biotechnol., 2011, 21, 930–936 CrossRef CAS PubMed.
- J. Almaliti, K. L. Malloy, E. Glukhov, C. Spadafora, M. Gutiérrez and W. H. Gerwick, J. Nat. Prod., 2017, 80, 1827–1836 CrossRef CAS.
- O. B. Vining, R. A. Medina, E. A. Mitchell, P. Videau, D. Li, J. D. Serrill, J. X. Kelly, W. H. Gerwick, P. J. Proteau, J. E. Ishmael and K. L. McPhail, J. Nat. Prod., 2015, 78, 413–420 CrossRef CAS.
- O. Demirkiran, J. Almaliti, T. Leão, G. Navarro, T. Byrum, F. A. Valeriote, L. Gerwick and W. H. Gerwick, J. Nat. Prod., 2021, 84, 2081–2093 CrossRef CAS.
- M. Taniguchi, J. K. Nunnery, N. Engene, E. Esquenazi, T. Byrum, P. C. Dorrestein and W. H. Gerwick, J. Nat. Prod., 2010, 73, 393–398 CrossRef CAS.
- R. Montaser, K. A. Abboud, V. J. Paul and H. Luesch, J. Nat. Prod., 2011, 74, 109–112 CrossRef CAS.
- E. Mevers, T. Byrum and W. H. Gerwick, J. Nat. Prod., 2013, 76, 1810–1814 CrossRef CAS.
- A. M. Fenner, N. Engene, C. Spadafora, W. H. Gerwick and M. J. Balunas, Org. Lett., 2016, 18, 352–355 CrossRef CAS.
- F. Nakamura, H. Maejima, M. Kawamura, D. Arai, T. Okino, M. Zhao, T. Ye, J. Lee, Y.-T. Chang, N. Fusetani and Y. Nakao, Bioorg. Med. Chem. Lett., 2018, 28, 2206–2209 CrossRef CAS PubMed.
- A. M. Sweeney-Jones, K. Gagaring, J. Antonova-Koch, H. Zhou, N. Mojib, K. Soapi, J. Skolnick, C. W. McNamara and J. Kubanek, Mar. Drugs, 2020, 18, 167 CrossRef CAS.
- T. Meickle, S. P. Gunasekera, Y. Liu, H. Luesch and V. J. Paul, Bioorg. Med. Chem., 2011, 19, 6576–6580 CrossRef CAS.
- A. Krunic, A. Vallat, S. Mo, D. D. Lantvit, S. M. Swanson and J. Orjala, J. Nat. Prod., 2010, 73, 1927–1932 CrossRef CAS.
- J. Jokela, L. Herfindal, M. Wahlsten, P. Permi, F. Selheim, V. Vasconcelos, S. O. Døskeland and K. Sivonen, ChemBioChem, 2010, 11, 1594–1599 CrossRef CAS.
- L. Herfindal, L. Myhren, R. Kleppe, C. Krakstad, F. Selheim, J. Jokela, K. Sivonen and S. O. Døskeland, Mol. Pharm., 2011, 8, 360–367 CrossRef CAS.
- A. Fidor, K. Cekała, E. Wieczerzak, M. Cegłowska, F. Kasprzykowski, C. Edwards and H. Mazur-Marzec, Biomolecules, 2021, 11, 1483 CrossRef CAS PubMed.
- L. Liu, J. Jokela, L. Herfindal, M. Wahlsten, J. Sinkkonen, P. Permi, D. P. Fewer, S. O. Døskeland and K. Sivonen, ACS Chem. Biol., 2014, 9, 2646–2655 CrossRef CAS.
- J. C. Kwan, R. Ratnayake, K. A. Abboud, V. J. Paul and H. Luesch, J. Org. Chem., 2010, 75, 8012–8023 CrossRef CAS.
- C. C. Thornburg, M. Thimmaiah, L. A. Shaala, A. M. Hau, J. M. Malmo, J. E. Ishmael, D. T. A. Youssef and K. L. McPhail, J. Nat. Prod., 2011, 74, 1677–1685 CrossRef CAS PubMed.
- W. L. Popplewell, R. Ratnayake, J. A. Wilson, J. A. Beutler, N. H. Colburn, C. J. Henrich, J. B. McMahon and T. C. McKee, J. Nat. Prod., 2011, 74, 1686–1691 CrossRef CAS.
- Y. Kanamori, A. Iwasaki, S. Sumimoto and K. Suenaga, Tetrahedron Lett., 2016, 57, 4213–4216 CrossRef CAS.
- M. Y. Phyo, J. X. Goh and L. T. Tan, J. Nat. Prod., 2022, 85, 485–492 CrossRef CAS.
- P. Tomek, P. Hrouzek, M. Kuzma, J. Sýkora, R. Fišer, J. Černý, P. Novák, S. Bártová, P. Šimek, M. Hof, D. Kavan and J. Kopecký, Chem. Res. Toxicol., 2015, 28, 216–224 Search PubMed.
- S. Freitas, R. Castelo-Branco, A. Wenzel-Storjohann, V. M. Vasconcelos, D. Tasdemir and P. N. Leão, J. Nat. Prod., 2022, 85, 1704–1714 CrossRef CAS.
- N. Maru, O. Ohno and D. Uemura, Tetrahedron Lett., 2010, 51, 6384–6387 CrossRef CAS.
- S. Luo, A. Krunic, H.-S. Kang, W.-L. Chen, J. L. Woodard, J. R. Fuchs, S. M. Swanson and J. Orjala, J. Nat. Prod., 2014, 77, 1871–1880 CrossRef CAS.
- S. Luo, H.-S. Kang, A. Krunic, W.-L. Chen, J. Yang, J. L. Woodard, J. R. Fuchs, S. Hyun Cho, S. G. Franzblau, S. M. Swanson and J. Orjala, Bioorg. Med. Chem., 2015, 23, 3153–3162 CrossRef CAS PubMed.
- W. Cai, S. Matthew, Q.-Y. Chen, V. J. Paul and H. Luesch, Bioorg. Med. Chem., 2018, 26, 2310–2319 CrossRef CAS PubMed.
- P. Sullivan, A. Krunic, J. E. Burdette and J. Orjala, J. Antibiot., 2020, 73, 526–533 CrossRef CAS PubMed.
- L. M. P. Heinilä, D. P. Fewer, J. K. Jokela, M. Wahlsten, X. Ouyang, P. Permi, A. Jortikka and K. Sivonen, Org. Biomol. Chem., 2021, 19, 5577–5588 RSC.
- L. Bornancin, F. Boyaud, Z. Mahiout, I. Bonnard, S. Mills, B. Banaigs and N. Inguimbert, Mar. Drugs, 2015, 13, 7285–7300 CrossRef CAS.
- L. Bornancin, E. Alonso, R. Alvariño, N. Inguimbert, I. Bonnard, L. M. Botana and B. Banaigs, Bioorg. Med. Chem., 2019, 27, 1966–1980 CrossRef CAS PubMed.
- H. S. Kang, A. Krunic, Q. Shen, S. M. Swanson and J. Orjala, J. Nat. Prod., 2011, 74, 1597–1605 CrossRef CAS.
- H. S. Kang, M. Sturdy, A. Krunic, H. Kim, Q. Shen, S. M. Swanson and J. Orjala, Bioorg. Med. Chem., 2012, 20, 6134–6143 CrossRef CAS PubMed.
- P. Hrouzek, M. Kuzma, J. Černyý, P. Novák, R. Fisšer, P. Sšimek, A. Lukesšová and J. Kopeckyý, Chem. Res. Toxicol., 2012, 25, 1203–1211 Search PubMed.
- J. Mareš, J. Hájek, P. Urajová, J. Kopecký and P. Hrouzek, PLoS One, 2014, 9, e111904 CrossRef PubMed.
- P. Urajová, J. Hájek, M. Wahlsten, J. Jokela, T. Galica, D. P. Fewer, A. Kust, E. Zapomělová-Kozlíková, K. Delawská, K. Sivonen, J. Kopecký and P. Hrouzek, J. Chromatogr. A, 2016, 1438, 76–83 CrossRef PubMed.
- J. Cheel, P. Urajová, J. Hájek, P. Hrouzek, M. Kuzma, E. Bouju, K. Faure and J. Kopecký, Anal. Bioanal. Chem., 2017, 409, 917–930 CrossRef CAS PubMed.
- K. Saurav, A. Caso, P. Urajová, P. Hrouzek, G. Esposito, K. Delawská, M. Macho, J. Hájek, J. Cheel, S. Saha, P. Divoká, S. Arsin, K. Sivonen, D. P. Fewer and V. Costantino, ACS Omega, 2022, 7, 11818–11828 CrossRef CAS PubMed.
- K. Saurav, A. Caso, P. Urajová, P. Hrouzek, G. Esposito, K. Delawská, M. Macho, J. Hájek, J. Cheel, S. Saha, P. Divoká, S. Arsin, K. Sivonen, D. P. Fewer and V. Costantino, ACS Omega, 2022, 7, 11818–11828 CrossRef CAS PubMed.
- M. Bertin, A. Roduit, J. Sun, G. Alves, C. Via, M. Gonzalez, P. Zimba and P. Moeller, Mar. Drugs, 2017, 15, 206 CrossRef PubMed.
- J. Jokela, L. Oftedal, L. Herfindal, P. Permi, M. Wahlsten, S. O. Døskeland and K. Sivonen, PLoS One, 2012, 7, e41222 CrossRef CAS.
- J. Vestola, T. K. Shishido, J. Jokela, D. P. Fewer, O. Aitio, P. Permi, M. Wahlsten, H. Wang, L. Rouhiainen and K. Sivonen, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E1909–E1917 CrossRef CAS.
- C. Pancrace, J. Jokela, N. Sassoon, C. Ganneau, M. Desnos-Ollivier, M. Wahlsten, A. Humisto, A. Calteau, S. Bay, D. P. Fewer, K. Sivonen and M. Gugger, ACS Chem. Biol., 2017, 12, 1796–1804 CrossRef CAS PubMed.
- T. H. Bui, V. Wray, M. Nimtz, T. Fossen, M. Preisitsch, G. Schröder, K. Wende, S. E. Heiden and S. Mundt, J. Nat. Prod., 2014, 77, 1287–1296 CrossRef CAS.
- S. Matthew, L. A. Salvador, P. J. Schupp, V. J. Paul and H. Luesch, J. Nat. Prod., 2010, 73, 1544–1552 CrossRef CAS.
- H. Choi, E. Mevers, T. Byrum, F. A. Valeriote and W. H. Gerwick, Eur. J. Org. Chem., 2012, 5141–5150 CrossRef CAS PubMed.
- J. G. Petitbois, L. O. Casalme, J. A. V. Lopez, W. M. Alarif, A. Abdel-Lateff, S. S. Al-Lihaibi, E. Yoshimura, Y. Nogata, T. Umezawa, F. Matsuda and T. Okino, J. Nat. Prod., 2017, 80, 2708–2715 CrossRef CAS PubMed.
- H. Choi, A. R. Pereira, Z. Cao, C. F. Shuman, N. Engene, T. Byrum, T. Matainaho, T. F. Murray, A. Mangoni and W. H. Gerwick, J. Nat. Prod., 2010, 73, 1411–1421 CrossRef CAS.
- K. L. Malloy, H. Choi, C. Fiorilla, F. A. Valeriote, T. Matainaho and W. H. Gerwick, Bioorg. Med. Chem. Lett., 2012, 22, 683–688 CrossRef CAS PubMed.
- L. T. Tan, T. Okino and W. H. Gerwick, Mar. Drugs, 2013, 11, 3015–3024 CrossRef.
- K. Aihara, F. Nakamura and Y. Nakao, Chem. Lett., 2023, 52, 270–272 CrossRef CAS.
- D. Luo, M. Y. Putra, T. Ye, V. J. Paul and H. Luesch, Mar. Drugs, 2019, 17, 83 CrossRef CAS.
- D. Luo, M. Y. Putra, T. Ye, V. J. Paul and H. Luesch, Mar. Drugs, 2019, 17, 83 CrossRef CAS PubMed.
- K. Sueyoshi, M. Kaneda, S. Sumimoto, S. Oishi, N. Fujii, K. Suenaga and T. Teruya, Tetrahedron, 2016, 72, 5472–5478 CrossRef CAS.
- K. Tidgewell, N. Engene, T. Byrum, J. Media, T. Doi, F. A. Valeriote and W. H. Gerwick, ChemBioChem, 2010, 11, 1458–1466 CrossRef CAS PubMed.
- C. C. Thornburg, E. S. Cowley, J. Sikorska, L. A. Shaala, J. E. Ishmael, D. T. A. Youssef and K. L. McPhail, J. Nat. Prod., 2013, 76, 1781–1788 CrossRef CAS PubMed.
- H. Ogawa, A. Iwasaki, S. Sumimoto, Y. Kanamori, O. Ohno, M. Iwatsuki, A. Ishiyama, R. Hokari, K. Otoguro, S. Omura and K. Suenaga, J. Nat. Prod., 2016, 79, 1862–1866 CrossRef CAS PubMed.
- H. Sasaki, T. Teruya, H. Fukazawa and K. Suenaga, Tetrahedron, 2011, 67, 990–994 CrossRef CAS.
- K. Sueyoshi, M. Yamada, A. Yamano, K. Ozaki, S. Sumimoto, A. Iwasaki, K. Suenaga and T. Teruya, J. Nat. Prod., 2018, 81, 1103–1107 CrossRef CAS.
- S. Matthew, Q.-Y. Chen, R. Ratnayake, C. S. Fermaintt, D. Lucena-Agell, F. Bonato, A. E. Prota, S. T. Lim, X. Wang, J. F. Díaz, A. L. Risinger, V. J. Paul, M. Á. Oliva and H. Luesch, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2021847118 CrossRef CAS.
- M. Schwark, J. A. Martínez Yerena, K. Röhrborn, P. Hrouzek, P. Divoká, L. Štenclová, K. Delawská, H. Enke, C. Vorreiter, F. Wiley, W. Sippl, R. Sobotka, S. Saha, S. B. Wilde, J. Mareš and T. H. J. Niedermeyer, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2219230120 CrossRef CAS PubMed.
- F. H. Al-Awadhi, B. K. Law, V. J. Paul and H. Luesch, J. Nat. Prod., 2017, 80, 2969–2986 CrossRef CAS.
- T. F. Molinski, K. A. Reynolds and B. I. Morinaka, J. Nat. Prod., 2012, 75, 425–431 CrossRef CAS.
- A. Iwasaki, O. Ohno, S. Sumimoto, S. Suda and K. Suenaga, Tetrahedron Lett., 2014, 55, 4126–4128 CrossRef CAS.
- E. Mevers, F. P. J. Haeckl, P. D. Boudreau, T. Byrum, P. C. Dorrestein, F. A. Valeriote and W. H. Gerwick, J. Nat. Prod., 2014, 77, 969–975 CrossRef CAS.
- F. H. Al-Awadhi, R. Ratnayake, V. J. Paul and H. Luesch, Bioorg. Med. Chem., 2016, 24, 3276–3282 CrossRef CAS PubMed.
- P. Sullivan, A. Krunic, L. J. Davis, H. S. Kim, J. E. Burdette and J. Orjala, J. Nat. Prod., 2021, 84, 2256–2264 CrossRef CAS PubMed.
- L. M. Sanchez, D. Lopez, B. A. Vesely, G. Della Togna, W. H. Gerwick, D. E. Kyle and R. G. Linington, J. Med. Chem., 2010, 53, 4187–4197 CrossRef CAS.
- J. Quintana, L. M. Bayona, L. Castellanos, M. Puyana, P. Camargo, F. Aristizábal, C. Edwards, J. N. Tabudravu, M. Jaspars and F. A. Ramos, Bioorg. Med. Chem., 2014, 22, 6789–6795 CrossRef CAS PubMed.
- M. J. Balunas, R. G. Linington, K. Tidgewell, A. M. Fenner, L. D. Ureña, G. Della Togna, D. E. Kyle and W. H. Gerwick, J. Nat. Prod., 2010, 73, 60–66 CrossRef CAS.
- A. Iwasaki, O. Ohno, S. Sumimoto, S. Suda and K. Suenaga, RSC Adv., 2014, 4, 12840–12843 RSC.
- S. Okamoto, A. Iwasaki, O. Ohno and K. Suenaga, J. Nat. Prod., 2015, 78, 2719–2725 CrossRef CAS.
- A. Iwasaki, O. Ohno, S. Sumimoto, H. Ogawa, K. A. Nguyen and K. Suenaga, Org. Lett., 2015, 17, 652–655 CrossRef CAS PubMed.
- A. Yamano, Y. Asato, N. Natsume, A. Iwasaki, K. Suenaga and T. Teruya, J. Nat. Prod., 2022, 85, 169–175 CrossRef CAS PubMed.
- L. Ding, R. Bar-Shalom, D. Aharonovich, N. Kurisawa, G. Patial, S. Li, S. He, X. Yan, A. Iwasaki, K. Suenaga, C. Zhu, H. Luo, F. Tian, F. Fares, C. B. Naman and T. Luzzatto-Knaan, Mar. Drugs, 2021, 19, 397 CrossRef CAS.
- S. Sumimoto, M. Kobayashi, R. Sato, S. Shinomiya, A. Iwasaki, S. Suda, T. Teruya, T. Inuzuka, O. Ohno and K. Suenaga, Org. Lett., 2019, 21, 1187–1190 CrossRef CAS PubMed.
- K. Iwasaki, A. Iwasaki, S. Sumimoto, T. Matsubara, T. Sato, T. Nozaki, Y. Saito-Nakano and K. Suenaga, J. Nat. Prod., 2020, 83, 481–488 CrossRef CAS.
- H. B. Yu, E. Glukhov, Y. Li, A. Iwasaki, L. Gerwick, P. C. Dorrestein, B. H. Jiao and W. H. Gerwick, J. Nat. Prod., 2019, 82, 2608–2619 CrossRef CAS PubMed.
- N. Kurisawa, A. Iwasaki, G. Jeelani, T. Nozaki and K. Suenaga, J. Nat. Prod., 2020, 83, 1684–1690 CrossRef CAS.
- R. Suo, R. Watanabe, K. Takada, T. Suzuki, H. Oikawa, S. Itoi, H. Sugita and S. Matsunaga, Org. Lett., 2020, 22, 1254–1258 CrossRef CAS PubMed.
- N. Kurisawa, K. Otomo, A. Iwasaki, G. Jeelani, T. Nozaki and K. Suenaga, J. Org. Chem., 2021, 86, 12528–12536 CrossRef CAS.
- A. Iwasaki, T. Tadenuma, S. Sumimoto, I. Shiota, T. Matsubara, Y. Saito-Nakano, T. Nozaki, T. Sato and K. Suenaga, J. Nat. Prod., 2018, 81, 2545–2552 CrossRef CAS.
- A. Iwasaki, K. Ohtomo, N. Kurisawa, I. Shiota, Y. Rahmawati, G. Jeelani, T. Nozaki and K. Suenaga, J. Nat. Prod., 2021, 84, 126–135 CrossRef CAS.
- F. H. Al-Awadhi, B. Gao, M. A. Rezaei, J. C. Kwan, C. Li, T. Ye, V. J. Paul and H. Luesch, J. Med. Chem., 2018, 61, 6364–6378 CrossRef CAS PubMed.
- X. Liang, D. Luo, J.-L. Yan, M. A. Rezaei, L. A. Salvador-Reyes, S. P. Gunasekera, C. Li, T. Ye, V. J. Paul and H. Luesch, Org. Lett., 2019, 21, 1622–1626 CrossRef CAS.
- T. Li, C. Xi, Y. Yu, N. Wang, X. Wang, A. Iwasaki, F. Fang, L. Ding, S. Li, W. Zhang, Y. Yuan, T. Wang, X. Yan, S. He, Z. Cao and C. B. Naman, J. Nat. Prod., 2022, 85, 493–500 CrossRef CAS.
- K. Ozaki, A. Iwasaki, D. Sezawa, H. Fujimura, T. Nozaki, Y. Saito-Nakano, K. Suenaga and T. Teruya, J. Nat. Prod., 2019, 82, 2907–2915 CrossRef CAS PubMed.
- A. Iwasaki, T. Tadenuma, S. Sumimoto, T. Ohshiro, K. Ozaki, K. Kobayashi, T. Teruya, H. Tomoda and K. Suenaga, J. Nat. Prod., 2017, 80, 1161–1166 CrossRef CAS.
- T. Galica, N. Borbone, J. Mareš, A. Kust, A. Caso, G. Esposito, K. Saurav, J. Hájek, K. Řeháková, P. Urajová, V. Costantino and P. Hrouzek, Appl. Environ. Microbiol., 2021, 87, e03128 CrossRef CAS.
- N. Kurisawa, A. Iwasaki, K. Teranuma, S. Dan, C. Toyoshima, M. Hashimoto and K. Suenaga, J. Am. Chem. Soc., 2022, 144, 11019–11032 CrossRef CAS PubMed.
- S. Kokkaliari, D. Luo, V. J. Paul and H. Luesch, Mar. Drugs, 2023, 21, 378 CrossRef CAS.
- A. R. Pereira, A. J. Kale, A. T. Fenley, T. Byrum, H. M. Debonsi, M. K. Gilson, F. A. Valeriote, B. S. Moore and W. H. Gerwick, ChemBioChem, 2012, 13, 810–817 CrossRef CAS.
- B. L. Marquez, K. S. Watts, A. Yokochi, M. A. Roberts, P. Verdier-Pinard, J. I. Jimenez, E. Hamel, P. J. Scheuer and W. H. Gerwick, J. Nat. Prod., 2002, 65, 866–871 CrossRef CAS.
- D. J. Edwards, B. L. Marquez, L. M. Nogle, K. McPhail, D. E. Goeger, M. A. Roberts and W. H. Gerwick, Chem. Biol., 2004, 11, 817–833 CrossRef CAS.
- P. D. Boudreau, E. A. Monroe, S. Mehrotra, S. Desfor, A. Korobeynikov, D. H. Sherman, T. F. Murray, L. Gerwick, P. C. Dorrestein and W. H. Gerwick, PLoS One, 2015, 10, e0133297 CrossRef.
- R. B. Kinnel, E. Esquenazi, T. Leao, N. Moss, E. Mevers, A. R. Pereira, E. A. Monroe, A. Korobeynikov, T. F. Murray, D. Sherman, L. Gerwick, P. C. Dorrestein and W. H. Gerwick, J. Nat. Prod., 2017, 80, 1514–1521 CrossRef CAS.
- Y. Nakashima, Y. Egami, M. Kimura, T. Wakimoto and I. Abe, PLoS One, 2016, 11, e0164468 CrossRef.
- T. Kuranaga, K. Matsuda, M. Takaoka, C. Tachikawa, A. Sano, K. Itoh, A. Enomoto, K. Fujita, I. Abe and T. Wakimoto, ChemBioChem, 2020, 21, 3329–3332 CrossRef CAS.
- A. Kampa, A. N. Gagunashvili, T. A. M. Gulder, B. I. Morinaka, C. Daolio, M. Godejohann, V. P. W. Miao, J. Piel and O. S. Andresson, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E3129–E3137 CrossRef CAS PubMed.
- A. Kust, J. Mareš, J. Jokela, P. Urajová, J. Hájek, K. Saurav, K. Voráčová, D. P. Fewer, E. Haapaniemi, P. Permi, K. Řeháková, K. Sivonen and P. Hrouzek, ACS Chem. Biol., 2018, 13, 1123–1129 CrossRef CAS PubMed.
- C. L. Shao, R. G. Linington, M. J. Balunas, A. Centeno, P. Boudreau, C. Zhang, N. Engene, C. Spadafora, T. S. Mutka, D. E. Kyle, L. Gerwick, C. Y. Wang and W. H. Gerwick, J. Org. Chem., 2015, 80, 7849–7855 CrossRef CAS.
- S. Mori, H. Williams, D. Cagle, K. Karanovich, F. Horgen, R. III and C. Watanabe, Mar. Drugs, 2015, 13, 6274–6290 CrossRef CAS.
- L. A. Salvador-Reyes, J. Sneed, V. J. Paul and H. Luesch, J. Nat. Prod., 2015, 78, 1957–1962 CrossRef CAS.
- L. Keller, J. L. Siqueira-Neto, J. M. Souza, K. Eribez, G. M. LaMonte, J. E. Smith and W. H. Gerwick, Molecules, 2020, 25, 1604 CrossRef CAS.
- C. L. Shao, X. F. Mou, F. Cao, C. Spadafora, E. Glukhov, L. Gerwick, C. Y. Wang and W. H. Gerwick, J. Nat. Prod., 2018, 81, 211–215 CrossRef CAS PubMed.
- L. A. Salvador, V. J. Paul and H. Luesch, J. Nat. Prod., 2010, 73, 1606–1609 CrossRef CAS PubMed.
- N. Oku, M. Matsumoto, K. Yonejima, K. Tansei and Y. Igarashi, Beilstein J. Org. Chem., 2014, 10, 1808–1816 CrossRef.
- N. Oku, S. Hana, M. Matsumoto, K. Yonejima, K. Tansei, Y. Isogai and Y. Igarashi, J. Antibiot., 2017, 70, 708–709 CrossRef CAS.
- A. Iwasaki, T. Teruya and K. Suenaga, Tetrahedron Lett., 2010, 51, 959–960 CrossRef CAS.
- J. Krumbholz, K. Ishida, M. Baunach, J. E. Teikari, M. M. Rose, S. Sasso, C. Hertweck and E. Dittmann, Angew. Chem., Int. Ed., 2022, 61, e202204545 CrossRef CAS PubMed.
- T. Teruya, H. Sasaki, K. Kitamura, T. Nakayama and K. Suenaga, Org. Lett., 2009, 11, 2421–2424 CrossRef CAS.
- M. Morita, O. Ohno and K. Suenaga, Chem. Lett., 2012, 41, 165–167 CrossRef CAS.
- O. Osamu, W. Ayane, M. Maho and S. Kiyotake, Chem. Lett., 2014, 43, 287–289 CrossRef.
- A. Watanabe, O. Ohno, M. Morita, T. Inuzuka and K. Suenaga, Bull. Chem. Soc. Jpn., 2015, 88, 1256–1264 CrossRef CAS.
- M. Morita, O. Ohno, T. Teruya, T. Yamori, T. Inuzuka and K. Suenaga, Tetrahedron, 2012, 68, 5984–5990 CrossRef CAS.
- A. Yamano, N. Natsume, M. Yamada, S. Sumimoto, A. Iwasaki, K. Suenaga and T. Teruya, J. Nat. Prod., 2020, 83, 1585–1591 CrossRef CAS PubMed.
- A. R. Pereira, C. F. McCue and W. H. Gerwick, J. Nat. Prod., 2010, 73, 217–220 CrossRef CAS.
- G. Navarro, S. Cummings, J. Lee, N. Moss, E. Glukhov, F. A. Valeriote, L. Gerwick and W. H. Gerwick, Environ. Sci. Technol. Lett., 2015, 2, 166–170 CrossRef CAS.
- S. P. Gunasekera, Y. Li, R. Ratnayake, D. Luo, J. Lo, J. H. Reibenspies, Z. Xu, M. J. Clare-Salzler, T. Ye, V. J. Paul and H. Luesch, Chem.–Eur. J., 2016, 22, 8158–8166 CrossRef CAS.
- K. Umeda, A. Iwasaki, R. Taguchi, N. Kurisawa, G. Jeelani, T. Nozaki and K. Suenaga, J. Nat. Prod., 2023, 86, 2529–2538 CrossRef CAS.
- D. K. Gupta, P. Kaur, S. T. Leong, L. T. Tan, M. R. Prinsep and J. J. H. Chu, Mar. Drugs, 2014, 12, 115–127 CrossRef PubMed.
- M. Kawaguchi, M. Satake, B.-T. Zhang, Y.-Y. Xiao, M. Fukuoka, H. Uchida and H. Nagai, Molecules, 2020, 25, 457 CrossRef CAS.
- B.-N. Han, T.-T. Liang, L. J. Keen, T.-T. Fan, X.-D. Zhang, L. Xu, Q. Zhao, S.-P. Wang and H.-W. Lin, Org. Lett., 2018, 20, 578–581 CrossRef CAS PubMed.
- Y.-H. Tang, T.-T. Liang, T.-T. Fan, L. J. Keen, X.-D. Zhang, L. Xu, Q. Zhao, R. Zeng and B.-N. Han, Nat. Prod. Res., 2020, 34, 2151–2156 CrossRef CAS PubMed.
- Y.-H. Tang, J. Wu, T.-T. Fan, H.-H. Zhang, X.-X. Gong, Z.-Y. Cao, J. Zhang, H.-W. Lin and B.-N. Han, RSC Adv., 2019, 9, 7594–7600 RSC.
- T.-T. Fan, H.-H. Zhang, Y.-H. Tang, F.-Z. Zhang and B.-N. Han, Mar. Drugs, 2019, 17, 652 CrossRef CAS.
- H.-H. Zhang, X.-K. Zhang, R.-R. Si, S.-C. Shen, T.-T. Liang, T.-T. Fan, W. Chen, L.-H. Xu and B.-N. Han, Toxins, 2020, 12, 733 CrossRef CAS.
- Z. Chen, N. Chen, P. Fu, W. Wang, S. Bian, H. Zhang, S. Shen and B. Han, Molecules, 2023, 28, 2786 CrossRef CAS PubMed.
- H. Nagai, M. Watanabe, S. Sato, M. Kawaguchi, Y.-Y. Xiao, K. Hayashi, R. Watanabe, H. Uchida and M. Satake, Tetrahedron, 2019, 75, 2486–2494 CrossRef CAS.
- H. Nagai, S. Sato, K. Iida, K. Hayashi, M. Kawaguchi, H. Uchida and M. Satake, Toxins, 2019, 11, 366 CrossRef CAS PubMed.
- S. Shen, W. Wang, Z. Chen, H. Zhang, Y. Yang, X. Wang, P. Fu and B. Han, Mar. Drugs, 2021, 19, 630 CrossRef CAS PubMed.
- N. Kanda, B.-T. Zhang, A. Shinjo, M. Kamiya, H. Nagai, H. Uchida, Y. Araki, T. Nishikawa and M. Satake, Nat. Prod. Commun., 2023, 18, 1934578X2311737 CrossRef.
- G. E. Chlipala, P. H. Tri, N. Van Hung, A. Krunic, S. H. Shim, D. D. Soejarto and J. Orjala, J. Nat. Prod., 2010, 73, 784–787 CrossRef CAS PubMed.
- M. Satake, K. Iguchi, R. Watanabe, H. Uchida and H. Nagai, Results Chem., 2021, 3, 100206 CrossRef CAS.
- R. Ueoka, A. R. Uria, S. Reiter, T. Mori, P. Karbaum, E. E. Peters, E. J. N. Helfrich, B. I. Morinaka, M. Gugger, H. Takeyama, S. Matsunaga and J. Piel, Nat. Chem. Biol., 2015, 11, 705–712 CrossRef CAS PubMed.
- T. Shishido, A. Humisto, J. Jokela, L. Liu, M. Wahlsten, A. Tamrakar, D. Fewer, P. Permi, A. Andreote, M. Fiore and K. Sivonen, Mar. Drugs, 2015, 13, 2124–2140 CrossRef CAS PubMed.
- J. Cui, M. Morita, O. Ohno, T. Kimura, T. Teruya, T. Watanabe, K. Suenaga and M. Shibasaki, Chem.–Eur. J., 2017, 23, 8500–8509 CrossRef CAS.
- Y. Tao, P. Li, D. Zhang, E. Glukhov, L. Gerwick, C. Zhang, T. F. Murray and W. H. Gerwick, J. Org. Chem., 2018, 83, 3034–3046 CrossRef CAS PubMed.
- R. Reher, H. W. Kim, C. Zhang, H. H. Mao, M. Wang, L.-F. Nothias, A. M. Caraballo-Rodriguez, E. Glukhov, B. Teke, T. Leao, K. L. Alexander, B. M. Duggan, E. L. Van Everbroeck, P. C. Dorrestein, G. W. Cottrell and W. H. Gerwick, J. Am. Chem. Soc., 2020, 142, 4114–4120 CrossRef CAS.
- R. Reher, H. W. Kim, C. Zhang, H. H. Mao, M. Wang, L.-F. Nothias, A. M. Caraballo-Rodriguez, E. Glukhov, B. Teke, T. Leao, K. L. Alexander, B. M. Duggan, E. L. Van Everbroeck, P. C. Dorrestein, G. W. Cottrell and W. H. Gerwick, J. Am. Chem. Soc., 2020, 142, 4114–4120 CrossRef CAS.
- A. R. Pereira, Z. Cao, N. Engene, I. E. Soria-Mercado, T. F. Murray and W. H. Gerwick, Org. Lett., 2010, 12, 4490–4493 CrossRef CAS.
- H.-S. Kang, A. Krunic and J. Orjala, Tetrahedron Lett., 2012, 53, 3563–3567 CrossRef CAS.
- S. Sumimoto, A. Iwasaki, O. Ohno, K. Sueyoshi, T. Teruya and K. Suenaga, Org. Lett., 2016, 18, 4884–4887 CrossRef CAS PubMed.
- S. P. Gunasekera, J. L. Meyer, Y. Ding, K. A. Abboud, D. Luo, J. E. Campbell, A. Angerhofer, J. L. Goodsell, L. J. Raymundo, J. Liu, T. Ye, H. Luesch, M. Teplitski and V. J. Paul, J. Nat. Prod., 2019, 82, 111–121 CrossRef CAS.
- J. L. Meyer, S. P. Gunasekera, A. L. Brown, Y. Ding, S. Miller, M. Teplitski and V. J. Paul, Mar. Drugs, 2023, 21, 76 CrossRef CAS PubMed.
- S. P. Gunasekera, J. L. Meyer, Y. Ding, K. A. Abboud, D. Luo, J. E. Campbell, A. Angerhofer, J. L. Goodsell, L. J. Raymundo, J. Liu, T. Ye, H. Luesch, M. Teplitski and V. J. Paul, J. Nat. Prod., 2019, 82, 111–121 CrossRef CAS.
- J. L. Meyer, S. P. Gunasekera, A. L. Brown, Y. Ding, S. Miller, M. Teplitski and V. J. Paul, Mar. Drugs, 2023, 21, 76 CrossRef CAS PubMed.
- G. E. Chlipala, M. Sturdy, A. Krunic, D. D. Lantvit, Q. Shen, K. Porter, S. M. Swanson and J. Orjala, J. Nat. Prod., 2010, 73, 1529–1537 CrossRef CAS.
- H.-S. Kang, B. D. Santarsiero, H. Kim, A. Krunic, Q. Shen, S. M. Swanson, H. Chai, A. D. Kinghorn and J. Orjala, Phytochemistry, 2012, 79, 109–115 CrossRef CAS.
- D. S. May, W. L. Chen, D. D. Lantvit, X. Zhang, A. Krunic, J. E. Burdette, A. Eustaquio and J. Orjala, J. Nat. Prod., 2017, 80, 1073–1080 CrossRef CAS PubMed.
- S. Luo, H. S. Kang, A. Krunic, G. E. Chlipala, G. Cai, W. L. Chen, S. G. Franzblau, S. M. Swanson and J. Orjala, Tetrahedron Lett., 2014, 55, 686–689 CrossRef CAS.
- M. Preisitsch, K. Harmrolfs, H. T. L. Pham, S. E. Heiden, A. Fussel, C. Wiesner, A. Pretsch, M. Swiatecka-Hagenbruch, T. H. J. Niedermeyer, R. Muller and S. Mundt, J. Antibiot., 2015, 68, 165–177 CrossRef CAS PubMed.
- M. Preisitsch, S. Heiden, M. Beerbaum, T. Niedermeyer, M. Schneefeld, J. Herrmann, J. Kumpfmüller, A. Thürmer, I. Neidhardt, C. Wiesner, R. Daniel, R. Müller, F.-C. Bange, P. Schmieder, T. Schweder and S. Mundt, Mar. Drugs, 2016, 14, 21 CrossRef PubMed.
- V. T. Pérez, L. A. Ticona, A. H. Cabanillas, S. M. Corral, J. Perles, D. F. R. Valencia, A. M. Quintana, M. O. Domenech and Á. R. Sánchez, Phytochemistry, 2020, 180, 112529 CrossRef.
- D. S. May, H.-S. Kang, B. D. Santarsiero, A. Krunic, Q. Shen, J. E. Burdette, S. M. Swanson and J. Orjala, J. Nat. Prod., 2018, 81, 572–578 CrossRef CAS.
- J. Dai, C. S. Philbin, C. Wakano, W. Y. Yoshida and P. G. Williams, Mar. Drugs, 2023, 21, 101 CrossRef CAS PubMed.
- V. T. Pérez, L. A. Ticona, A. H. Cabanillas, S. M. Corral, J. Perles, D. F. R. Valencia, A. M. Quintana, M. O. Domenech and Á. R. Sánchez, Phytochemistry, 2020, 180, 112529 CrossRef.
- M. Preisitsch, T. H. J. Niedermeyer, S. E. Heiden, I. Neidhardt, J. Kumpfmüller, M. Wurster, K. Harmrolfs, C. Wiesner, H. Enke, R. Müller and S. Mundt, J. Nat. Prod., 2016, 79, 106–115 CrossRef CAS.
- G. J. Kim, S. J. Mascuch, E. Mevers, P. D. Boudreau, W. H. Gerwick and H. Choi, J. Org. Chem., 2022, 87, 1043–1055 CrossRef CAS PubMed.
- R. Taguchi, A. Iwasaki, A. Ebihara, G. Jeelani, T. Nozaki and K. Suenaga, Org. Lett., 2022, 24, 4710–4714 CrossRef CAS PubMed.
- A. R. Pereira, L. Etzbach, N. Engene, R. Müller and W. H. Gerwick, J. Nat. Prod., 2011, 74, 1175–1181 CrossRef CAS.
- C. W. Via, E. Glukhov, S. Costa, P. V. Zimba, P. D. R. Moeller, W. H. Gerwick and M. J. Bertin, Front. Chem., 2018, 6, 1–9 CrossRef.
- H. Ogawa, A. Iwasaki, S. Sumimoto, M. Iwatsuki, A. Ishiyama, R. Hokari, K. Otoguro, S. Ōmura and K. Suenaga, Org. Lett., 2017, 19, 890–893 CrossRef CAS PubMed.
- C. A. Leber, C. B. Naman, L. Keller, J. Almaliti, E. J. E. Caro-Diaz, E. Glukhov, V. Joseph, T. P. Sajeevan, A. J. Reyes, J. S. Biggs, T. Li, Y. Yuan, S. He, X. Yan and W. H. Gerwick, Mar. Drugs, 2020, 18, 515 CrossRef CAS PubMed.
- X. Liang, S. Matthew, Q.-Y. Chen, J. C. Kwan, V. J. Paul and H. Luesch, Org. Lett., 2019, 21, 7274–7278 CrossRef CAS.
- N. Engene, H. Choi, E. Esquenazi, T. Byrum, F. A. Villa, Z. Cao, T. F. Murray, P. C. Dorrestein, L. Gerwick and W. H. Gerwick, J. Nat. Prod., 2011, 74, 1737–1743 CrossRef CAS PubMed.
- K. Ozaki, Y. Asato, N. Natsume, S. Tojo, S. Sumimoto, A. Iwasaki, K. Suenaga and T. Teruya, J. Nat. Prod., 2023, 86, 1564–1570 CrossRef CAS PubMed.
- C. Zhang, C. Naman, N. Engene and W. Gerwick, Mar. Drugs, 2017, 15, 121–131 CrossRef PubMed.
- O. Ohno, A. Iwasaki, K. Same, C. Kudo, E. Aida, K. Sugiura, S. Sumimoto, T. Teruya, E. Tashiro, S. Simizu, K. Matsuno, M. Imoto and K. Suenaga, Org. Lett., 2022, 24, 4547–4551 CrossRef CAS.
- S. A. C. Figueiredo, K. Abt, T. P. Martins, I. Lacomba, A. M. Forero, C. Jiménez, J. Rodríguez and P. N. Leão, ChemRxiv, 2022, pp. 1–9, DOI:10.26434/chemrxiv-2022-v8zlp.
- P. Moosmann, R. Ueoka, M. Gugger and J. Piel, Org. Lett., 2018, 20, 5238–5241 CrossRef CAS.
- R. S. Belisle, C. W. Via, T. B. Schock, T. A. Villareal, P. V. Zimba, K. R. Beauchesne, P. D. R. Moeller and M. J. Bertin, Tetrahedron Lett., 2017, 58, 4066–4068 CrossRef CAS PubMed.
- T. B. Schock, K. Huncik, K. R. Beauchesne, T. A. Villareal and P. D. R. Moeller, Environ. Sci. Technol., 2011, 45, 7503–7509 CrossRef CAS PubMed.
- M. J. Bertin, P. V. Zimba, H. He and P. D. R. Moeller, Tetrahedron Lett., 2016, 57, 5864–5867 CrossRef CAS PubMed.
- M. Bertin, P. Wahome, P. Zimba, H. He and P. Moeller, Mar. Drugs, 2017, 15, 10 CrossRef PubMed.
- M. J. Bertin, J. Saurí, Y. Liu, C. W. Via, A. F. Roduit and R. T. Williamson, J. Org. Chem., 2018, 83, 13256–13266 CrossRef CAS PubMed.
- K. M. McManus, R. D. Kirk, C. W. Via, J. S. Lotti, A. F. Roduit, R. Teta, S. Scarpato, A. Mangoni and M. J. Bertin, J. Nat. Prod., 2020, 83(9), 2664–2671 CrossRef CAS PubMed.
- L. A. Elsadek, E. K. Ellis, G. Seabra, V. J. Paul and H. Luesch, Mar. Drugs, 2023, 22, 28 CrossRef.
- J. K. Nunnery, N. Engene, T. Byrum, Z. Cao, S. V. Jabba, A. R. Pereira, T. Matainaho, T. F. Murray and W. H. Gerwick, J. Org. Chem., 2012, 77, 4198–4208 CrossRef CAS.
- M. J. Balunas, M. F. Grosso, F. A. Villa, N. Engene, K. L. McPhail, K. Tidgewell, L. M. Pineda, L. Gerwick, C. Spadafora, D. E. Kyle and W. H. Gerwick, Org. Lett., 2012, 14, 3878–3881 CrossRef CAS PubMed.
- M. Gutiérrez, K. Tidgewell, T. L. Capson, N. Engene, A. Almanza, J. Schemies, M. Jung and W. H. Gerwick, J. Nat. Prod., 2010, 73, 709–711 Search PubMed.
- H. Choi, S. J. Mascuch, F. A. Villa, T. Byrum, M. E. Teasdale, J. E. Smith, L. B. Preskitt, D. C. Rowley, L. Gerwick and W. H. Gerwick, Chem. Biol., 2012, 19, 589–598 CrossRef CAS PubMed.
- T. Inuzuka, K. Yamamoto, A. Iwasaki, O. Ohno, K. Suenaga, Y. Kawazoe and D. Uemura, Tetrahedron Lett., 2014, 55, 6711–6714 CrossRef CAS.
- M. J. Bertin, O. Demirkiran, G. Navarro, N. A. Moss, J. Lee, G. M. Goldgof, E. Vigil, E. A. Winzeler, F. A. Valeriote and W. H. Gerwick, Phytochemistry, 2016, 122, 113–118 CrossRef CAS PubMed.
- J. C. Kwan, M. Teplitski, S. P. Gunasekera, V. J. Paul and H. Luesch, J. Nat. Prod., 2010, 73, 463–466 CrossRef CAS.
- H. Gross, K. L. McPhail, D. E. Goeger, F. A. Valeriote and W. H. Gerwick, Phytochemistry, 2010, 71, 1729–1735 CrossRef CAS PubMed.
- K. L. Malloy, F. A. Villa, N. Engene, T. Matainaho, L. Gerwick and W. H. Gerwick, J. Nat. Prod., 2011, 74, 95–98 CrossRef CAS PubMed.
- B. Han, U. M. Reinscheid, W. H. Gerwick and H. Gross, J. Mol. Struct., 2011, 989, 109–113 Search PubMed.
- T. T. Chang, S. V. More, I.-H. Lu, J.-C. Hsu, T.-J. Chen, Y. C. Jen, C.-K. Lu and W.-S. Li, Eur. J. Med. Chem., 2011, 46, 3810–3819 Search PubMed.
- L. A. Shaala, D. T. A. Youssef, K. L. McPhail and M. Elbandy, Phytochem. Lett., 2013, 6, 183–188 CAS.
- K. Sueyoshi, A. Yamano, K. Ozaki, S. Sumimoto, A. Iwasaki, K. Suenaga and T. Teruya, Mar. Drugs, 2017, 15, 367 Search PubMed.
- K. L. Malloy, T. L. Suyama, N. Engene, H. Debonsi, Z. Cao, T. Matainaho, C. Spadafora, T. F. Murray and W. H. Gerwick, J. Nat. Prod., 2012, 75, 60–66 Search PubMed.
- W. Cai, J. H. Matthews, V. J. Paul and H. Luesch, Planta Med., 2016, 897–902 Search PubMed.
- C. B. Naman, J. Almaliti, L. Armstrong, E. J. Caro-Díaz, M. L. Pierce, E. Glukhov, A. Fenner, C. Spadafora, H. M. Debonsi, P. C. Dorrestein, T. F. Murray and W. H. Gerwick, J. Nat. Prod., 2017, 80, 2328–2334 Search PubMed.
- C. M. Pavlik, C. Y. B. Wong, S. Ononye, D. D. Lopez, N. Engene, K. L. McPhail, W. H. Gerwick and M. J. Balunas, J. Nat. Prod., 2013, 76, 2026–2033 CAS.
- K. Voráčová, J. Hájek, J. Mareš, P. Urajová, M. Kuzma, J. Cheel, A. Villunger, A. Kapuscik, M. Bally, P. Novák, M. Kabeláč, G. Krumschnabel, M. Lukeš, L. Voloshko, J. Kopecký and P. Hrouzek, PLoS One, 2017, 12, e0172850 Search PubMed.
- S. A. C. Figueiredo, M. Preto, G. Moreira, T. P. Martins, K. Abt, A. Melo, V. M. Vasconcelos and P. N. Leão, Angew. Chem., Int. Ed., 2021, 60, 10064–10072 CAS.
- C. M. Crnkovic, A. Krunic, D. S. May, T. A. Wilson, D. Kao, J. E. Burdette, J. R. Fuchs, N. H. Oberlies and J. Orjala, J. Nat. Prod., 2018, 81, 2083–2090 CrossRef CAS.
- E. Mevers, T. Matainaho, M. Allara’, V. Di Marzo and W. H. Gerwick, Lipids, 2014, 49, 1127–1132 CrossRef CAS.
- K. Kleigrewe, J. Almaliti, I. Y. Tian, R. B. Kinnel, A. Korobeynikov, E. A. Monroe, B. M. Duggan, V. Di Marzo, D. H. Sherman, P. C. Dorrestein, L. Gerwick and W. H. Gerwick, J. Nat. Prod., 2015, 78, 1671–1682 CrossRef CAS.
- J. A. V. Lopez, J. G. Petitbois, C. S. Vairappan, T. Umezawa, F. Matsuda and T. Okino, Org. Lett., 2017, 19, 4231–4234 CrossRef CAS.
- J. J. Mehjabin, L. Wei, J. G. Petitbois, T. Umezawa, F. Matsuda, C. S. Vairappan, M. Morikawa and T. Okino, J. Nat. Prod., 2020, 83, 1925–1930 CrossRef CAS PubMed.
- A. Taton, S. Rohrer, B. Diaz, R. Reher, A. M. Caraballo Rodriguez, M. L. Pierce, P. C. Dorrestein, L. Gerwick, W. H. Gerwick and J. W. Golden, ACS Chem. Biol., 2022, 17, 1910–1923 CrossRef CAS PubMed.
- M. Gutiérrez, A. R. Pereira, H. M. Debonsi, A. Ligresti, V. Di Marzo and W. H. Gerwick, J. Nat. Prod., 2011, 74, 2313–2317 CrossRef.
- R. Montaser, V. J. Paul and H. Luesch, ChemBioChem, 2012, 13, 2676–2681 CrossRef CAS PubMed.
- N. Maneechote, B. Yingyongnarongkul, A. Suksamran and S. Lumyong, Aquacult. Res., 2017, 48, 2088–2095 CrossRef CAS.
- N. Oku, S. Hayashi, Y. Yamaguchi, H. Takenaka and Y. Igarashi, Beilstein J. Org. Chem., 2023, 19, 133–138 CrossRef CAS PubMed.
- N. F. Salleh, J. Wang, B. Kundukad, E. T. Oluwabusola, D. X. Y. Goh, M. Y. Phyo, J. J. L. Tong, S. Kjelleberg and L. T. Tan, Molecules, 2023, 28, 3965 CrossRef CAS.
- J. C. Kwan, T. Meickle, D. Ladwa, M. Teplitski, V. Paul and H. Luesch, Mol. BioSyst., 2011, 7, 1205–1216 CAS.
- R. Montaser, V. J. Paul and H. Luesch, Org. Lett., 2013, 15, 4050–4053 CrossRef CAS PubMed.
- F. H. Al-Awadhi, E. F. Simon, N. Liu, R. Ratnayake, V. J. Paul and H. Luesch, Mar. Drugs, 2023, 21, 553 CrossRef CAS PubMed.
- S. P. Gunasekera, S. Kokkaliari, R. Ratnayake, T. Sauvage, L. A. H. dos Santos, H. Luesch and V. J. Paul, Molecules, 2022, 27, 1717 CAS.
- H. Solanki, M. Pierdet, O. P. Thomas and M. Zubia, Metabolites, 2020, 10, 215 CAS.
- E. Roulland, H. Solanki, K. Calabro, M. Zubia, G. Genta-Jouve and O. P. Thomas, Org. Lett., 2018, 20, 2311–2314 CAS.
- I. Gutiérrez-Del-Río, N. Brugerolle De Fraissinette, R. Castelo-Branco, F. Oliveira, J. Morais, S. Redondo-Blanco, C. J. Villar, M. J. Iglesias, R. Soengas, V. Cepas, Y. L. Cubillos, G. Sampietro, L. Rodolfi, F. Lombó, S. M. S. González, F. López Ortiz, V. Vasconcelos and M. A. Reis, J. Nat. Prod., 2020, 1–6 Search PubMed.
- D. Youssef, S. Ibrahim, L. Shaala, G. Mohamed and Z. Banjar, Molecules, 2016, 21, 324 CrossRef.
- D. Brumley, K. A. Spencer, S. P. Gunasekera, T. Sauvage, J. Biggs, V. J. Paul and H. Luesch, J. Nat. Prod., 2018, 81, 2716–2721 CrossRef CAS.
- D. A. Brumley, S. P. Gunasekera, Q.-Y. Chen, V. J. Paul and H. Luesch, Org. Lett., 2020, 22, 4235–4239 CrossRef CAS PubMed.
- D. A. Brumley, S. P. Gunasekera, T. Sauvage, L. A. H. dos Santos, Q.-Y. Chen, V. J. Paul and H. Luesch, J. Nat. Prod., 2022, 85, 581–589 CrossRef CAS.
- P. N. Leão, H. Nakamura, M. Costa, A. R. Pereira, R. Martins, V. Vasconcelos, W. H. Gerwick and E. P. Balskus, Angew. Chem., Int. Ed., 2015, 54, 11063–11067 CrossRef.
- T. B. Afonso, M. S. Costa, R. Rezende De Castro, S. Freitas, A. Silva, M. P. C. Schneider, R. Martins and P. N. Leão, J. Nat. Prod., 2016, 79, 2504–2513 CrossRef CAS.
- J. P. A. Reis, S. A. C. Figueiredo, M. L. Sousa and P. N. Leão, Nat. Commun., 2020, 11, 1458 CrossRef CAS PubMed.
- M. Costa, I. E. Sampaio-Dias, R. Castelo-Branco, H. Scharfenstein, R. Rezende De Castro, A. Silva, M. P. C. Schneider, M. J. Araújo, R. Martins, V. F. Domingues, F. Nogueira, V. Camoes, V. M. Vasconcelos and P. N. Leao, J. Nat. Prod., 2019, 82, 393–402 CrossRef CAS.
- H. Kim, D. Lantvit, C. H. Hwang, D. J. Kroll, S. M. Swanson, S. G. Franzblau and J. Orjala, Bioorg. Med. Chem., 2012, 20, 5290–5295 CrossRef CAS PubMed.
- T. Chilczuk, C. Steinborn, S. Breinlinger, A. M. Zimmermann-Klemd, R. Huber, H. Enke, D. Enke, T. H. J. Niedermeyer and C. Gründemann, Planta Med., 2020, 86, 96–103 CrossRef CAS PubMed.
- H. Kim, A. Krunic, D. Lantvit, Q. Shen, D. J. Kroll, S. M. Swanson and J. Orjala, Tetrahedron, 2012, 68, 3205–3209 CrossRef CAS PubMed.
- W. Jiang, Y. Bu, M. Kawaguchi, H. Osada, M. Fukuoka, H. Uchida, R. Watanabe, T. Suzuki and H. Nagai, Phytochem. Lett., 2017, 22, 163–166 CrossRef CAS.
- T. Chilczuk, T. F. Schäberle, S. Vahdati, U. Mettal, M. El Omari, H. Enke, M. Wiese, G. M. König and T. H. J. Niedermeyer, ChemBioChem, 2020, 21, 2170–2177 Search PubMed.
- S. Breinlinger, T. J. Phillips, B. N. Haram, J. Mareš, J. A. Martínez Yerena, P. Hrouzek, R. Sobotka, W. M. Henderson, P. Schmieder, S. M. Williams, J. D. Lauderdale, H. D. Wilde, W. Gerrin, A. Kust, J. W. Washington, C. Wagner, B. Geier, M. Liebeke, H. Enke, T. H. J. Niedermeyer and S. B. Wilde, Science, 2021, 371, eaax9050 Search PubMed.
- W. Jiang, W. Zhou, H. Uchida, M. Kikumori, K. Irie, R. Watanabe, T. Suzuki, B. Sakamoto, M. Kamio and H. Nagai, Mar. Drugs, 2014, 12, 2748–2759 Search PubMed.
- W. Jiang, S. Tan, Y. Hanaki, K. Irie, H. Uchida, R. Watanabe, T. Suzuki, B. Sakamoto, M. Kamio and H. Nagai, Mar. Drugs, 2014, 12, 5788–5800 CrossRef CAS PubMed.
- D. T. A. Youssef, L. A. Shaala, G. A. Mohamed, S. R. M. Ibrahim, Z. M. Banjar, J. M. Badr, K. L. McPhail, A. L. Risinger and S. L. Mooberry, Nat. Prod. Res., 2015, 29, 703–709 Search PubMed.
- K. M. Wimmer, W. K. Strangman and J. L. C. Wright, Harmful Algae, 2014, 37, 203–206 CAS.
- A. Méjean, O. Lequin and O. Ploux, J. Am. Chem. Soc., 2022, 144, 14627–14637 CrossRef PubMed.
- T. Minowa, Y. Cho, Y. Oshima, K. Konoki and M. Yotsu-Yamashita, Toxins, 2019, 11, 539 CrossRef CAS PubMed.
- M. Akamatsu, R. Hirozumi, Y. Cho, Y. Kudo, K. Konoki, Y. Oshima and M. Yotsu-Yamashita, Mar. Drugs, 2022, 20, 166 CrossRef CAS.
- A. Kust, A. Méjean and O. Ploux, J. Nat. Prod., 2020, 83, 142–151 CrossRef CAS.
- S. T. Lima, T. R. Fallon, J. L. Cordoza, J. R. Chekan, E. Delbaje, A. R. Hopiavuori, D. O. Alvarenga, S. M. Wood, H. Luhavaya, J. T. Baumgartner, F. A. Dörr, A. Etchegaray, E. Pinto, S. M. K. McKinnie, M. F. Fiore and B. S. Moore, J. Am. Chem. Soc., 2022, 144, 9372–9379 CrossRef CAS PubMed.
- M. Lifshits, D. Kovalerchik and S. Carmeli, Phytochemistry, 2016, 123, 69–74 CrossRef CAS PubMed.
- A. R. Soares, N. Engene, S. P. Gunasekera, J. M. Sneed and V. J. Paul, J. Nat. Prod., 2015, 78, 534–538 CrossRef CAS PubMed.
- L. J. Davis, A. C. Maldonado, M. Khin, A. Krunic, J. E. Burdette and J. Orjala, J. Nat. Prod., 2022, 85, 540–546 CrossRef CAS PubMed.
- M. Sturdy, A. Krunic, S. Cho, S. Franzblau and J. Orjala, J. Nat. Prod., 2010, 73, 1441–1443 CAS.
- G. A. Pullella, D. A. Wild, G. L. Nealon, M. Elyashberg and M. J. Piggott, J. Org. Chem., 2017, 82, 7287–7299 CrossRef CAS PubMed.
- P. Zelík, A. Lukesová, J. Cejka, M. Budesínský, V. Havlícek, A. Cegan and J. Kopecký, J. Enzyme Inhib. Med. Chem., 2010, 25, 414–420 Search PubMed.
- R. Kossack, S. Breinlinger, T. Nguyen, J. Moschny, J. Straetener, A. Berscheid, H. Brötz-Oesterhelt, H. Enke, T. Schirmeister and T. H. J. Niedermeyer, J. Nat. Prod., 2020, 83, 392–400 CAS.
- H. Choi, N. Engene, J. E. Smith, L. B. Preskitt and W. H. Gerwick, J. Nat. Prod., 2010, 73, 517–522 CrossRef CAS.
- A. Ebihara, A. Iwasaki, Y. Miura, G. Jeelani, T. Nozaki and K. Suenaga, J. Org. Chem., 2021, 86, 11763–11770 CrossRef CAS.
- T. Chilczuk, R. Monson, P. Schmieder, V. Christov, H. Enke, G. Salmond and T. H. J. Niedermeyer, ACS Chem. Biol., 2020, 15, 2929–2936 CrossRef CAS.
- Niveshika, E. Verma, A. K. Mishra, A. K. Singh and V. K. Singh, Front. Microbiol., 2016, 7, 1–15 Search PubMed.
- J. R. Gurr, J. Dai, C. S. Philbin, H. T. Sartain, T. J. O'Donnell, W. Y. Yoshida, A. L. Rheingold and P. G. Williams, J. Org. Chem., 2020, 85, 318–326 CrossRef CAS PubMed.
- S. Freitas, N. G. Silva, M. L. Sousa, T. Ribeiro, F. Rosa, P. N. Leão, V. Vasconcelos, M. A. Reis and R. Urbatzka, Mar. Drugs, 2019, 17, 229 CrossRef CAS PubMed.
- K. Matsui, E. Nazifi, S. Kunita, N. Wada, S. Matsugo and T. Sakamoto, J. Photochem. Photobiol., B, 2011, 105, 81–89 Search PubMed.
- K. Ishihara, R. Watanabe, H. Uchida, T. Suzuki, M. Yamashita, H. Takenaka, E. Nazifi, S. Matsugo, M. Yamaba and T. Sakamoto, J. Photochem. Photobiol., B, 2017, 172, 102–108 CrossRef CAS.
- T. Sakamoto, A. Hashimoto, M. Yamaba, N. Wada, T. Yoshida, K. Inoue-Sakamoto, T. Nishiuchi and S. Matsugo, Phycol. Res., 2019, 67, 3–11 CrossRef CAS.
- S. Arsın, E. Delbaje, J. Jokela, M. Wahlsten, Z. M. Farrar, P. Permi and D. Fewer, ACS Chem. Biol., 2023, 18, 1959–1967 CrossRef.
- H. Choi, N. Engene, T. Byrum, S. Hwang, D.-C. Oh and W. H. Gerwick, Org. Lett., 2019, 21, 266–270 CrossRef CAS PubMed.
- J. Bhandari Neupane, R. P. Neupane, Y. Luo, W. Y. Yoshida, R. Sun and P. G. Williams, Org. Lett., 2019, 21, 8449–8453 CrossRef CAS PubMed.
- R. G. S. Berlinck, C. M. Crnkovic, J. R. Gubiani, D. I. Bernardi, L. P. Ióca and J. I. Quintana-Bulla, Nat. Prod. Rep., 2022, 39, 596–669 Search PubMed.
- H. Takahashi, A. Iwasaki, A. Ebihara, R. Taguchi, G. Jeelani, T. Nozaki and K. Suenaga, Org. Lett., 2023, 25, 2400–2404 CrossRef CAS PubMed.
- I. Shiota, A. Iwasaki, S. Sumimoto, H. Tomoda and K. Suenaga, Tetrahedron Lett., 2018, 59, 1261–1263 CrossRef CAS.
- C. A. Lydon, L. Mathivathanan, J. Sanchez, L. A. H. dos Santos, T. Sauvage, S. P. Gunasekera, V. J. Paul and J. P. Berry, J. Nat. Prod., 2020, 83, 2030–2035 CrossRef CAS PubMed.
- T. J. O'Donnell, Y. Luo, W. Y. Yoshida, S. Suzuki, R. Sun and P. G. Williams, J. Nat. Prod., 2022, 85, 415–425 CrossRef PubMed.
- A. H. Cabanillas, V. Tena Pérez, S. Maderuelo Corral, D. F. Rosero Valencia, A. Martel Quintana, M. Ortega Doménech and Á. Rumbero Sánchez, J. Nat. Prod., 2018, 81, 410–413 CrossRef CAS.
- S. Mo, A. Krunic, S. D. Pegan, S. G. Franzblau and J. Orjala, J. Nat. Prod., 2009, 72, 2043–2045 CrossRef CAS.
- J. R. Gurr, T. J. O'Donnell, Y. Luo, W. Y. Yoshida, M. L. Hall, A. M. S. Mayer, R. Sun and P. G. Williams, J. Nat. Prod., 2020, 83, 1691–1695 CrossRef CAS.
- D. Back, T. J. O. Donnell, K. K. Axt, J. R. Gurr, J. M. Vanegas, P. G. Williams and B. Philmus, ACS Chem. Biol., 2023, 18, 1797–1807 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4np00040d |
| This journal is © The Royal Society of Chemistry 2025 |