Open Access Article
Open Access ArticleDescribing the complex chemistry of benthic seawater: from exometabolite sampling strategies to MS-based metabolomics†
Morgane
Mauduit‡
 ,
Stéphane
Greff
,
Stéphane
Greff
 ,
Marie
Derrien
,
Marie
Derrien
 and
Charlotte
Simmler
and
Charlotte
Simmler
 *
*
IMBE (Institut Méditerranéen de Biodiversité et d'Écologie Marine et Continentale), CNRS 7263, IRD 237, Aix Marseille Université, Avignon Université, Endoume Marine Station, rue de la Batterie des Lions, 13007 Marseille, France. E-mail: charlotte.simmler@imbe.fr
First published on 20th February 2025
Abstract
Covering: 1982 up to the end of 2024
Marine exometabolites (EMs) are small molecules released by marine (micro)organisms into the seawater. Collectively, all of the released EMs contribute to the chemical seascape of a marine ecosystem. Accessing and describing these waterborne molecules are a key focus of various disciplinary fields that aim to study marine biogeochemical cycles, translate the chemical language of the oceans (chemical ecology), or discover new structural entities with biological properties (natural product discovery). Beginning with the semantics of marine exometabolites, this review elucidates the different sampling methods and MS-based metabolomic analyses that are used to describe the chemical composition of seawater of benthic ecosystems. These technical and analytical advances offer promising avenues for describing the structural diversity of marine exometabolites and deciphering their functions in various ecological contexts.
1. Introduction
The oceans represent 70% of the Earth's surface, encompassing more than 95% of the total volume of water. The exploration of such marine environments has expanded since the 1960s with the democratisation of SCUBA (Self-Contained Underwater Breathing Apparatus) and the advances in marine/underwater engineering. By offering possibilities in exploring new horizons, these technological breakthroughs benefited both marine taxonomists, ecologists and natural product chemists.1 Our knowledge of the chemical composition of marine organisms has considerably improved, with more than 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules being reported to date and annually reviewed by A. Carroll's team.2 As such, keystone benthic species in marine ecosystems, such as corals, algae, and sponges, are known to produce a large structural diversity of specialized metabolites. Through their metabolic activities, they also release a part of their metabolites into the surrounding seawater. These exometabolites (EMs) are then mixed with other molecules released by diverse (micro)organisms, contributing to the cycle of nutrients and energy in the benthos. Some EMs can also act as distant chemical cues that affect the life and behaviour of other nearby organisms, thereby structuring the biodiversity.3,4 Describing the chemical composition of seawater is of prime importance in order to understand the driving factors in the functioning of various marine ecosystems, ranging from the shallow tropical coral reefs to the deep sea. Nevertheless, accurately identifying organic molecules in the vast seawater remains more challenging than characterizing the chemical composition of the collected marine organisms. This is notably because marine EMs are highly diluted (≤ picomolar range) in seawater and mixed with a plethora of other molecules from diverse biosynthetic origins, collectively contributing to marine dissolved organic matter (DOM).5,6 Recent advances in sampling and analytical methods, including the ultra-high resolution (UHR) and high-resolution (HR) mass spectrometry (MS) instruments for untargeted metabolomics and the increased use of in silico spectral annotation tools, have collectively enabled marine ecologists and chemists to deepen their understanding of seawater molecular diversity in different ecosystems. This review addresses the key technical and analytical progresses in the characterization of EMs released by benthic organisms. A special focus is given to the evolution of seawater sampling methods and in situ devices from early 1982 up to 2024. Furthermore, previously reviewed7–9 MS instrumentation and spectral annotation tools will be updated with a focus on the comparative methodologies used to describe EM molecular diversity. The final section illustrates how MS-based metabolomic investigations of benthic marine EMs from 2017 to 2024 have enhanced our understanding of the functioning of marine ecosystems.
000 molecules being reported to date and annually reviewed by A. Carroll's team.2 As such, keystone benthic species in marine ecosystems, such as corals, algae, and sponges, are known to produce a large structural diversity of specialized metabolites. Through their metabolic activities, they also release a part of their metabolites into the surrounding seawater. These exometabolites (EMs) are then mixed with other molecules released by diverse (micro)organisms, contributing to the cycle of nutrients and energy in the benthos. Some EMs can also act as distant chemical cues that affect the life and behaviour of other nearby organisms, thereby structuring the biodiversity.3,4 Describing the chemical composition of seawater is of prime importance in order to understand the driving factors in the functioning of various marine ecosystems, ranging from the shallow tropical coral reefs to the deep sea. Nevertheless, accurately identifying organic molecules in the vast seawater remains more challenging than characterizing the chemical composition of the collected marine organisms. This is notably because marine EMs are highly diluted (≤ picomolar range) in seawater and mixed with a plethora of other molecules from diverse biosynthetic origins, collectively contributing to marine dissolved organic matter (DOM).5,6 Recent advances in sampling and analytical methods, including the ultra-high resolution (UHR) and high-resolution (HR) mass spectrometry (MS) instruments for untargeted metabolomics and the increased use of in silico spectral annotation tools, have collectively enabled marine ecologists and chemists to deepen their understanding of seawater molecular diversity in different ecosystems. This review addresses the key technical and analytical progresses in the characterization of EMs released by benthic organisms. A special focus is given to the evolution of seawater sampling methods and in situ devices from early 1982 up to 2024. Furthermore, previously reviewed7–9 MS instrumentation and spectral annotation tools will be updated with a focus on the comparative methodologies used to describe EM molecular diversity. The final section illustrates how MS-based metabolomic investigations of benthic marine EMs from 2017 to 2024 have enhanced our understanding of the functioning of marine ecosystems.
2. Semantics of marine exometabolites (EMs)
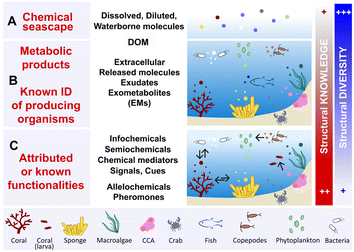
The exometabolome is defined as the set of all metabolites released by an organism, which are therefore termed exometabolites (EMs). The characterization of marine EMs is a central focus across various disciplines, including marine biogeochemistry, chemical ecology, and natural product chemistry. Consequently, the terminologies used to describe these compounds often differ, depending on the scientific field and research questions. This variation in terminology across studies and disciplines can obscure information about the techniques and methodologies that have been developed to investigate marine EMs. This section provides a brief compilation of these different terminologies, as illustrated in Fig. 1.The term marine chemical seascape10,11 (Fig. 1A) echoes the notion of a chemical landscape in terrestrial environments, and refers to the concept of odour landscapes, a recognized phenomenon in chemical ecology.12 Chemical seascapes consist of all molecules released by living organisms, encompassing inorganic molecules and any other synthetic compounds (e.g., anthropogenic, xenobiotic pollutants) present in the environment. The chemical seascape includes dissolved and diluted molecules floating freely or bound to small marine particles (e.g., cells, cellular debris), and integrates with marine DOM.
Marine dissolved organic matter (DOM) is considered one of the most complex mixtures on earth, containing hundreds of thousands of distinct molecules belonging to different structural classes (Fig. 1A). The average litre of seawater contains ≤1 mg of DOM, making its concentration approximately 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times lower than that of inorganic salts.5,6,13 All EMs released by marine primary producers, such as phytoplankton, and through the metabolic activities of marine bacteria contribute to the high molecular diversity of DOM in both pelagic and benthic ecosystems.13–15 The metabolic activities of corals, algae and sponges—abundant sessile species of benthic ecosystems—can lead to the release of EMs, thereby also contributing to the marine DOM pool (Fig. 1B).16–18 Hence, DOM encompasses all marine waterborne dissolved organic molecules, including exhalent,16 released molecules, exometabolites (EMs),17 extracellular metabolites,19 and exudates. Marine biogeochemists categorize DOM into short-lived (labile) DOM, which is rapidly transformed by microbial mineralization, and recalcitrant/refractory DOM, which accumulates and corresponds to the fraction that is most commonly detected and analysed in seawater.13,20–22
000 times lower than that of inorganic salts.5,6,13 All EMs released by marine primary producers, such as phytoplankton, and through the metabolic activities of marine bacteria contribute to the high molecular diversity of DOM in both pelagic and benthic ecosystems.13–15 The metabolic activities of corals, algae and sponges—abundant sessile species of benthic ecosystems—can lead to the release of EMs, thereby also contributing to the marine DOM pool (Fig. 1B).16–18 Hence, DOM encompasses all marine waterborne dissolved organic molecules, including exhalent,16 released molecules, exometabolites (EMs),17 extracellular metabolites,19 and exudates. Marine biogeochemists categorize DOM into short-lived (labile) DOM, which is rapidly transformed by microbial mineralization, and recalcitrant/refractory DOM, which accumulates and corresponds to the fraction that is most commonly detected and analysed in seawater.13,20–22
Another layer of terminology is added when a functionality is attributed to well-identified marine EMs (Fig. 1C). Within the complexity of the chemical seascape, some marine EMs may convey distant information essential to the functioning of ecosystems.4 Molecules with such functionalities, as studied in chemical ecology, include infochemicals, semiochemicals, chemical signals or chemical cues. These chemical mediators can be further defined as allelochemicals when they intervene mainly in interspecific relationships, (e.g., defence, cooperation), or pheromones when they are involved in intraspecific relationships (e.g., reproduction, danger signaling).23–25
In the literature, the term marine DOM tends to be used to describe the global molecular composition of seawater, notably with unidentified biogenic contributors. It is also commonly used in marine biogeochemistry and ecology studies that focus on understanding organic matter cycle and trophic networks (i.e., the exchanges of energy and nutrients) in the oceans.15,18 Conversely, the terms marine EMs,10,17 infochemicals,25 or waterborne allelochemicals26 tend to be used to describe small molecules (<1500 Da) diluted in seawater and whose biosynthetic origins are often known, or those identified in studies dedicated to marine (chemical) ecology and natural product chemistry.
Thus, words pertaining to marine DOM and EMs have different meanings, depending on who uses them in the marine science community. Hence, the choice of term used to design marine EMs is often based on the field of research and objectives of the study. However, it is also based on the degree of chemical knowledge that is involved: known biosynthetic origins, and/or known functionalities in the ecosystem. All these terminologies illustrate that seawater is a highly dynamic environment, where thousands of molecules, that are labile, recalcitrant, biogenic, or xenobiotic intermingle. Some of them are nutrients, other may convey information, and some other may interfere with or alter species interactions.
3. EM sampling methods
Standardized in situ sampling methods are essential prior to any metabolomic analyses designed to reproducibly study the chemical composition of seawater. The majority of the reported methods aim to concentrate marine EMs or DOM by preparing enriched extracts from defined volumes of collected seawater. The techniques employed are principally based on solid phase adsorption/extraction (SPE) principles, which are further detailed below.3.1. Strategic uses of solid phase extraction (SPE)
The most commonly employed adsorbents, as reported in the literature, are silica-based reversed phase (e.g. C18)11,26–28 or polymeric styrene-divinylbenzene (DVB).10,29–31 These adsorbents are used through different supports to concentrate EMs from seawater: cartridges (e.g., Bond Elut PPL [Priority PolLutant], Cation exchange [CX]),30,32,33 membranes, fibers and blades, such as those used for solid phase microextraction (SPME),34 disks10,11,28 and resins (e.g., Diaion® HP20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29,31 Amberlite™ XAD®
29,31 Amberlite™ XAD®![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35–37). The different approaches typically used for EM collection and concentration can be differentiated as either passive or active capture methods (Fig. 2).
35–37). The different approaches typically used for EM collection and concentration can be differentiated as either passive or active capture methods (Fig. 2).
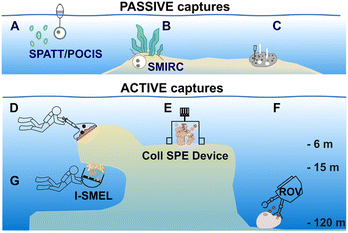 | ||
| Fig. 2 Key examples of in situ devices/methods for capturing marine EMs by solid phase adsorption and extraction (SPE). The small colour-coded dots represent the released EMs. (A–C) Encompass methods using passive processes to capture EMs. (A) SPATT (Solid Phase Adsorption Toxin Tracking)/POCIS (Polar Organic Chemical Integrative Sampler) uses membranes or polymeric resin to capture EMs from the water column. (B) SMIRC (Small Molecule In Situ Resin Capture) uses polymeric resin in a nylon mesh deposited at the surface of marine sediments; here, a seagrass meadow. (C) SPME (Solid Phase Micro Extraction) membranes and blades are inserted in the osculum and ostioles of a sponge to adsorb its inhalant/exhalent. (D–G) Illustration of active processes used to capture EMs. (D) Direct sampling of a defined volume of seawater around a sessile organism. (E) First in situ SPE instrument using a chamber to enclose organisms developed by Coll et al. in 1982.26 (F) Remotely Operated Vehicle (ROV) collecting seawater on SPE cartridges above Geodia barretti. (G) I-SMEL (In Situ Marine moleculE Logger) instrument operated by a SCUBA diver using SPE disks placed above a chamber. | ||
3.2. Devices and techniques using passive capture
During a passive capture, EM adsorption occurs with the flow regardless of the volume of seawater that is processed.31,34,38 Devices that have been developed for passive capture typically involve either membranes (POCIS, SPME) or polymeric resins (SPATT, SMIRC). In the field of aquatic ecotoxicology, Polar Organic Chemical Integrative Samplers (POCIS) are used to capture and facilitate the detection of organic molecules from anthropogenic origins, such as pharmaceuticals, herbicides, and insecticides. Another technique involves the Solid Phase Adsorption Toxin Tracking (SPATT) device, which is deployed to capture toxins that are released during harmful algal blooms. SPATT core components are usually polymeric adsorbent resins that are enclosed in a permeable membrane or mesh, collectively closed and maintained in seawater by a rigid frame. Both POCIS and SPATT devices, along with their applications, have been described in detail in recent reviews (Fig. 2A).38,39 The Small Molecule in situ Resin Capture (SMIRC) device, designed similarly to the SPATT, can be deposited on the surface of marine sediments for 2–8 days. It has been tested in different ecosystems, including seagrass meadows (Fig. 2B). The resin used in the SMIRC can be complemented with agar to favour in situ bacterial growth and their EMs adsorption.31 In a different study, to achieve rapid on-site sampling, a series of membranes and blades with different adsorption phases were placed on the ostioles and in the oscula of the sponge Sarcotragus foetidus to differentiate the incoming dissolved molecules (inhalant) from its exhalent (Fig. 2C).34 Adsorbed EMs on the membranes or blades can be directly analysed after thermal desorption (for GC analysis) or extracted in analytical solvents (for LC analysis).343.3. Devices and techniques using active capture
In active capture, a defined volume of seawater is intentionally pushed towards the stationary phase with a controlled flow rate. Active EM capture methods can be performed either in situ10,16,40,41 to account for the environmental influence on the production of EMs, or in aquaria to target EMs specifically released by collected organisms.11,17,35–37,42 Both approaches, whether in situ or in aquarium, are complementary. The latter approach is generally conducted to concentrate species-specific EMs under controlled conditions and further investigate their ecological functions (e.g., defence, induction of larval settlement).17,35–37,42The most direct and widely deployed strategy is to collect a fixed volume of seawater at the vicinity of a targeted organism (Fig. 2D), and subsequently perform SPE in the laboratory to recover the collected EMs.28,40,41,43 This sampling strategy is reproducible and can be standardized, especially when small volumes of seawater are collected (∼1 L).7,30 For collecting EMs in a targeted ecosystem regardless of the producing organisms, the Niskin bottle allows for the collection of larger volumes of seawater at once (e.g., 8–10 L).19,44 Direct seawater collection is commonly used to study the metabolization of DOM by benthic holobiont species or the comparison of EMs composition across ecosystems.16–19
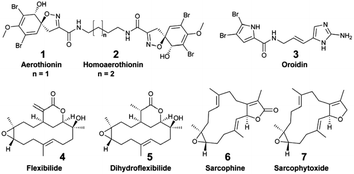
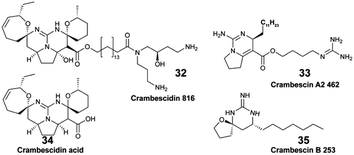
Focusing on the identification of waterborne allelochemicals, chemical ecologists have developed strategies to favour their pre-concentration. For example, Walker et al. used an in situ hermetic chamber that enclosed the sponge Aplysina fistularis in a fixed volume of seawater.41 Following solid phase extraction in the laboratory, they identified and quantified aerothionin (1) and homoaerothionin (2) as EMs. Such method was also deployed to capture bromopyrrole alkaloids released by the sponge Agelas conifera.43 Although this species is known to produce oroidin (3), the authors did not confirm the structural identity of the released brominated EMs.43
Despite their simplicity, such seawater sampling methods have two major drawbacks: (1) the transport of collected seawater back to the laboratory is constrained by the total volumes that can be processed in multiple replicates, (2) bio-transformations of diluted EMs may occur (e.g., metabolization by microorganisms), prior to performing SPE.
Hence, additional sampling strategies that enabled the rapid, on-site adsorption of EMs were developed to eliminate the need for transporting sampled seawater. In aquaria, for example, polymeric resins (XAD-16,36 XAD-7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35,37) were filled in a cylindrical holder or in a nylon mesh, and directly connected to the aquarium pump.30–32 Using these techniques, continuous EM adsorption and accumulation were successfully achieved, while minimizing potential bio-transformations.36,37In situ SPE devices were originally developed forty years ago by marine chemical ecologists aiming to capture waterborne allelochemicals upon their release.26 These devices are derived from benthic chambers, which are used to study the metabolic processes of organisms in shallow waters (e.g., respiration, photosynthesis). They are made of two compartments: (1) a chamber to enclose benthic species in a delimited space, and (2) a pump to direct a fixed volume of seawater from the chamber towards the SPE adsorbents. In 1982, Coll et al. developed the “first totally submersible sampling apparatus” (Fig. 2E) to capture EMs emitted by two alcyonaceans (soft corals), Sinularia flexibilis and Sarcophyton crassocaule.26 After a 30 min seawater filtration at 5 L h−1, the team identified flexibilide (4) and dihydroflexibilide (5), sarcophine (6) and sarcophytoxide (7), which were released from each species respectively. The instrument required an external battery-powered submersible bilge pump with a connection at the surface, and operated at a depth of 5–6 m. A decade later, a modified version of this instrument was designed by Schulte et al. with a submersible battery and an inverted plexiglass funnel. It could be deployed by SCUBA divers, and was tested on the same alcyonacean species at depths down to 13 m.27 The first submersible SPE instrument and the modified one worked both with SEP-PAKs C18 cartridges.26,27 Nevertheless, this apparatus by Schulte et al. required ∼4 hours of operation to accumulate detectable quantities of 4 and 5.27 In 2002, Kubanek et al. tested a manual version of the latter instrument, using a syringe to drive water above the sponges Erylus formosus and Ectyoplasia ferox through a column filled with Diaion® HP-20.29 The authors were looking for triterpene glycosides, but did not detect them as EMs after a 30 min sampling (4.5 L of seawater).29
35,37) were filled in a cylindrical holder or in a nylon mesh, and directly connected to the aquarium pump.30–32 Using these techniques, continuous EM adsorption and accumulation were successfully achieved, while minimizing potential bio-transformations.36,37In situ SPE devices were originally developed forty years ago by marine chemical ecologists aiming to capture waterborne allelochemicals upon their release.26 These devices are derived from benthic chambers, which are used to study the metabolic processes of organisms in shallow waters (e.g., respiration, photosynthesis). They are made of two compartments: (1) a chamber to enclose benthic species in a delimited space, and (2) a pump to direct a fixed volume of seawater from the chamber towards the SPE adsorbents. In 1982, Coll et al. developed the “first totally submersible sampling apparatus” (Fig. 2E) to capture EMs emitted by two alcyonaceans (soft corals), Sinularia flexibilis and Sarcophyton crassocaule.26 After a 30 min seawater filtration at 5 L h−1, the team identified flexibilide (4) and dihydroflexibilide (5), sarcophine (6) and sarcophytoxide (7), which were released from each species respectively. The instrument required an external battery-powered submersible bilge pump with a connection at the surface, and operated at a depth of 5–6 m. A decade later, a modified version of this instrument was designed by Schulte et al. with a submersible battery and an inverted plexiglass funnel. It could be deployed by SCUBA divers, and was tested on the same alcyonacean species at depths down to 13 m.27 The first submersible SPE instrument and the modified one worked both with SEP-PAKs C18 cartridges.26,27 Nevertheless, this apparatus by Schulte et al. required ∼4 hours of operation to accumulate detectable quantities of 4 and 5.27 In 2002, Kubanek et al. tested a manual version of the latter instrument, using a syringe to drive water above the sponges Erylus formosus and Ectyoplasia ferox through a column filled with Diaion® HP-20.29 The authors were looking for triterpene glycosides, but did not detect them as EMs after a 30 min sampling (4.5 L of seawater).29
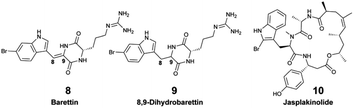
In 2011, to collect the brominated diketopiperazines barettin (8) and 8,9-dihydrobarettin (9) released by the sponge Geodia barretti found at a depth of 120 m, Sjörgren et al. installed C18 SPE cartridges connected to a pump on a Remotely Operated Vehicle (ROV) (Fig. 2F).45 In 2014, the Artificial Marine Sponge, an autonomous underwater instrument, was designed to filter water similarly to sponges to capture marine microorganisms and concentrate their EMs.46 The device comprised a particulate filter, a hollow-fiber bioreactor, and polymeric resin in cartridges. It was operated for 7–14 days on flat benthic surfaces (depth 10–15 m), and afforded <10 μg of jasplakinolide (10) with two other depsipeptide derivatives.
In 2018, the patented Somartex® device (Self Operating MARine Trapping EXtractor) was designed, following the results obtained in aquaria with the encrusting sponge Crambe crambe.36 The shape of the Somartex® closely resembles the device optimized by Schulte et al.,27 but requires a solar panel for continuous operation.36 The Somartex® has not been tested underwater yet. All of these described instruments have been previously used to target specific EMs from identified species. However, despite their technical ingenuity, they have had a limited scope of applications.
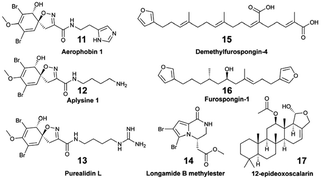
An in situ SPE sampling approach that can be easily and reproducibly performed in different ecosystems is preferred for metabolomic analyses to describe and compare the diversity of marine EMs. The newly developed in situ hand-held SPE instrument called I-SMEL (In Situ Marine moleculE Logger) worked in different configurations (e.g., underwater caves, overhang cliffs), while facilitating the concentration of species-specific EMs on SPE disks (Fig. 2G).10 In a 10 min seawater sampling (∼10 L) at various depths (15–20 m), I-SMEL successfully enriched EMs from three Mediterranean sponges. Bromo-spiroisoxazoline alkaloids aerothionin (1), aerophobin 1 (11), aplysine 1 (12), and purealidin L (13) were all found to be reproducibly released by Aplysina cavernicola. Longamide B methylester (14) was the only brominated alkaloid detected from Agelas oroides, whereas characteristic demethylfurospongin-4 (15), furospongin-1 (16), and 12-epideoxoscalarin (17) were detected in enriched EM extracts from Spongia officinalis. As such, I-SMEL facilitates temporally and spatially informed seawater sampling, offering a rapid and standardized method to enrich EMs in replicate samples required for downstream comparative metabolomics.
3.4. SPE desalting and accessing polar EMs
The presence of large amounts of inorganic sea salts (typically 35–38 g L−1), along with suspended particles like cell debris, poses significant challenges during marine EM adsorption. Such particles may rapidly obstruct the SPE surface, precluding further EM enrichment. For that reason, disks and resins are preferred because they can provide a larger contact area with polymeric adsorbents. They allow the filtering of a larger volume of seawater, favouring the progressive accumulation of highly diluted EMs. Residual sea salts can also alter the MS detection of EMs through ion suppression. Hence, a desalting step with deionized water is most often required prior to eluting EMs from the SPE supports. Nevertheless, this desalting step can also lead to a loss of small polar metabolites. To minimize such losses, standardized SPE protocols proposed by Dittmar and co-workers recommended acidifying the seawater with 0.1% v/v formic acid (pH 2) to protonate small organic molecules, making them less likely to be lost during the desalting step.30 Despite this optimization, the same authors determined that a variable but substantial portion of marine DOM is lost during SPE, with a recovery estimated between 40–60% w/w.30,32 Moreover, such acidification step can protonate nitrogen-containing compounds, thus reducing their retention.32 Consequently, accessing and enriching polar marine EMs remain a continuous endeavour, and various methods have been proposed to address this challenge. Sacks et al. demonstrated that cation exchange (CX)-SPE effectively enriches small, positively charged, and zwitterionic metabolites from seawater, capturing a complementary fraction of dissolved metabolites compared to the standardized protocol on PPL-SPE.33 Other methods proposed the derivatization of polar molecules bearing distinct structural functions (e.g., amine, alcohol) with reagents like benzoyl chloride or dansyl derivatives to improve their retention either on SPE or on reversed phase chromatographic columns.44,47–494. MS-based metabolomic analyses
Over the past ten years, the expansion of untargeted metabolomic analyses applied to the study of marine EMs has closely aligned with advances in ultra-high- and high-resolution MS instruments. This development has been further supported by the widespread availability of annotation methods, facilitating the ability to describe the structural diversity of metabolites involved in biogeochemical cycles, nutrient exchanges, and interactions between organisms and their environment. The MS analysers largely used for the untargeted metabolomic analyses of marine DOM and EMs include Fourier Transform-Ion Cyclotron Resonance Mass Spectrometers (FT-ICR-MS), Orbitrap and quadrupole Time of Flight (qToF).4.1. Ultra-high resolution (UHR) Fourier-transform ion cyclotron resonance mass spectrometry
FT-ICR MS is one of the most powerful tools used to analyse complex mixtures, such as marine DOM or EMs, with minimal sample preparation.5,6 The unparalleled sensitivity of FT-ICR MS, along with its high mass resolution for discriminating close m/z, and high mass accuracy (below 0.1 ppm), allows for the processing of samples through direct infusion via electrospray ionization (ESI). This capability also enables the proposal of molecular formula for each detected mass peak in a sample, while acquiring semi-quantitative information.15,18,50–54 As such, FT-ICR-MS yields thousands of molecular formulas per DOM sample, including molecular signals that would otherwise remain uncharacterized using conventional analytical techniques.14,15,54 These instruments contributed toward better describing the composition and complexity of DOM.54 Nevertheless, some limitations persist through signal suppression and with the difficulty to resolve a high proportion of isomeric compounds, characterizing the complexity of marine DOM and EM mixtures. Chromatographic separation is generally preferred to improve the dynamic range of detected metabolites, while also resolving the adducts and in-source fragments generated in the ESI probe. The coupling of separation techniques with FT-ICR MS is not as straightforward as with other MS analysers, and generally requires a series of adjustments, as explained in the review by Gosset-Erard and co-workers.52 Recently, Lechtenfield et al. proposed a LC-FT-ICR MS method for the analysis of marine DOM without the prior need for solid phase extraction of metabolites from seawater.53 Their method focused on accessing compounds that might not be extractable or recovered through SPE (e.g., polar molecules). In general, FT-ICR MS instruments suffer from their relatively low availability combined with high operational costs that collectively restrict their access to limited laboratories, as opposed to the high-resolution (HR) MS instruments.4.2. Orbitrap and quadrupole-time of flight (qToF)
Orbitrap and quadrupole-Time of Flight (qToF) are HR-MS analysers typically combined with LC instruments. In the last decade, they have been widely used for the untargeted metabolomic analyses of marine EMs. Both Orbitrap and qToF analysers are characterized by lower mass accuracy (<5 ppm) than FT-ICR MS. However, the level of structural information on separated metabolites is improved with the data-dependent acquisition (DDA) of tandem MS spectra (MS2). Workflows for DDA acquisitions and processing using HR-MS and MS2 for the analysis of complex marine DOM have been thoroughly described previously by Petras and co-workers.7–9 New methodologies continue to be regularly proposed by the same team.55–57 Recently, Stincone et al. detailed the key acquisition parameters for Q Exactive Orbitrap metabolomic analysis of complex environmental samples (e.g., marine DOM).56 A 2D chromatographic method was also developed to access to a wider polarity range of marine dissolved organic molecules with cleaner MS2 spectra, favouring their identification.57 DDA selectively fragments high-intensity precursor ions, and are thus biased towards the detection of more ionizable molecules. In contrast, data-independent acquisition (DIA) methods are based on wider isolation windows to capture more comprehensive MS2 data. Both DDA and DIA methods have been used in untargeted MS-based metabolomics, and each has their own distinct advantages.58 Guo and Huan demonstrated that while DIA provides comprehensive MS2 data sampling, DDA produces higher-quality MS2 spectra.58 The latter may be preferred when a higher rate of putative molecular identification is desired, as also confirmed by Patrone et al.59 These authors combined metabolomic outcomes from direct infusion and LC separation using the DIA MS method to gain a deeper understanding of the molecular compositions of different DOM.59 They found out that LC was more efficient for the detection of heteroatomic molecules, such as halogenated ones. However, direct infusion offered a wider number of detected molecules, favouring more polar chemical entities. To that end, the authors concluded that future research should explore the use of other chromatographic methods, such as HILIC (Hydrophilic Interaction Liquid Chromatography), to expand the chemical space of analysed DOM.4.3. Ion mobility and GC-MS
Exometabolomic studies using ion mobility mass spectrometers remain scarce, despite their capacity to unravel isomers of key metabolites in complex matrices.60 Gas chromatography coupled to mass spectrometry (GC-MS) for the untargeted analyses of marine EMs has been rarely employed up to now.34,61,62 For example, non-polar and volatile EMs from the sponge Sarcotragus foetidus were analysed by GC-MS, following their thermo-desorption from polydimethyl siloxane membranes that were initially placed on the sponge surface.34 Sogin et al. proposed the SeaMet GC-MS metabolomic method, which includes a derivatization step, to overcome salts related limitations (e.g., MS ion suppression, see Section 3.4) and access more polar EMs.61 The volatilome of the sponge Ircinia felix in aquarium and in situ was measured by GC-MS through the dynamic headspace extraction of volatile EMs from sampled seawater.624.4. Absolute quantification and targeted analyses
Targeted analyses are required for the absolute quantification and confident identification of key marine EMs using calibration curves or standard addition methods with structurally identical molecules. Such analyses employed HR-MS or triple Quadrupole (QQQ) instruments. The latter apparatuses are low-resolution mass spectrometers, which are principally used for targeted LC-MS analyses with selective reaction monitoring to improve detection limits. The majority of these analyses focused on the quantification of structurally known polar EMs (e.g., amino acids, vitamins, sugars, and nucleosides), which are most often involved in nutrients and energy cycling, and for which commercial standards are available.16,19,63,64 Other methods were proposed to expand the analysis of polar EMs. Targeted HILIC-HR-MS methods were used to either assess the recovery of polar marine EMs from cation exchange CX-SPE,33 or to quantify polar marine biotoxins (e.g., saxitoxins, tetrodotoxin) after SPE enrichment.65 A series of methods employed the chemical derivatization of small polar molecules, diluted in highly saline media, and bearing distinct structural functions (e.g., amine, alcohol, carboxyl functions). Such derivatizations favor their chromatographic retention and subsequent detection.44,47–49 Comparatively, only a few studies reported the quantification of specialized EMs through targeted HR-MS analyses. For that purpose, the purification of specialized metabolites from the organism's crude extracts was necessary to produce the analytical standards.28,375. Describing the chemodiversity of marine EMs
5.1. Van Krevelen & Kendrick mass defect plots
FT-ICR MS data can be processed and further analysed using the open access software (e.g., formularity66 or ICBM-Ocean67). Results from FT-ICR MS analyses of marine DOM are often presented in Van Krevelen18,50,68 and Kendrick Mass Defect (KMD)59 plots generated with raw formulas (Fig. 3A). Venn diagrams are used to assess the EMs richness and visualize the distribution of all identified signals across samples (Fig. 3B).15,50 These three types of graphical representations are among those most frequently used to illustrate the chemodiversity of complex organic mixtures, notably in studies related to marine biogeochemistry. Van Krevelen plots give an approximation of the molecular distribution in structural classes, according to the H/C-to-O/C ratios of their molecular formulas.6 However, overlaps between compound families can occur in such diagrams, as more elements than H, C, and O have to be considered for chemical classification.69 The latter is also determined by structures, rather than molecular formulas alone. Rivas Ubach et al. therefore proposed a multidimensional stoichiometric compound classification (MSCC), integrating more stoichiometric ratios with nitrogen and phosphorus, for the categorization of molecular formulas to compound classes.69 In KMD plots, molecules are distributed according to their nominal Kendrick masses, and a homologous series of chemically related compounds are distributed horizontally. These plots can assist in the discovery of transformation products within complex environmental samples, as recently reviewed.70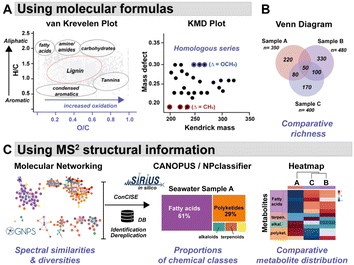 | ||
| Fig. 3 Graphical representation of the EM chemical diversity. (A) Using molecular formulas, both Van Krevelen (adapted from Nebbioso and Piccolo)6 and Kendrick Mass Defect (KMD) plots can be used to interrogate the structural diversity and transformation within a set of samples. (B) Venn diagram illustrating the distribution of molecular signals (n = number of signals/sample) across samples. (C) Chemodiversity of EMs analysed using molecular networking with HR-MS2 datasets. Structural dereplication is performed through automatic databases (DB) interrogation, and additional putative structure identification can be achieved using in silico tools, such as those within the SIRIUS environment. CANOPUS & NPclassifier facilitate the organization of molecules in chemical classes.80,82 The distribution of these chemical classes across samples is illustrated as a heatmap. | ||
5.2. Chemodiversity through tandem MS analyses
Untargeted tandem MS analyses are facilitated by the widespread implementation of open access software (e.g., XCMS,71 MS-DIAL,72 MZmine73). They enable the processing of raw LC-MS2 data for the detection and deconvolution of thousands of molecular signals per sample, generating data matrices that can be used with various chemo-informatics and dereplication tools. Seeking an improved chemical description of marine EMs, chemists and ecologists have benefited from the expansion of tools within the Global Natural Products Social Molecular Networking systems (GNPS, GNPS2), combined with the simplified deployment of multivariate data analyses, such as those proposed in MetaboAnalyst.8,74,75A common practice in untargeted metabolomics for structural dereplication is to compare the MS2 spectrum of an unknown molecule with those available in spectral databases,10,17,63 or with a spectrum of isolated standards.37 Nevertheless, the rate of marine EM identification through database matching remains low (≤10%), notably because marine (exo)metabolites are poorly represented in freely available spectral databases.10,17,57In silico annotation tools, such as those embedded in SIRIUS,76 have been widely adopted by the community. Such tools increase the annotation rates of a marine dataset (up to ∼16%)59 by proposing (i) molecular formulas from HR-MS, and (ii) putative structures through the fragmentation tree interpretation of MS2 spectra with CSI:FingerID.77 Complementary to SIRIUS, other in silico tools, such as MetFrag78 and CFM-ID,79 also guide the putative identification of unknowns, for which different molecular formulas and different structures are equally envisioned. Within the SIRIUS ecosystem, CANOPUS (Class Assignment aNd Ontology Prediction Using mass Spectrometry) predicts the structural class from the MS2 spectrum, even in the absence of structural reference data or MS2 training data.80 Since seawater samples primarily contain unknown molecules, this software provides a more comprehensive description of their chemical composition (Fig. 3C).
Detected and putatively identified metabolites can be hierarchically organized according to their structural entities using ClassyFire,81 and/or to their highest probable natural product classes (e.g., alkaloids, terpenoids) using NPclassifier.82 ConCISE (Consensus Classifications of in silico Elucidations) was recently developed as a standalone software to leverage the in silico CANOPUS annotations with matches obtained through GNPS spectral libraries, thereby increasing the confidence in spectral annotations.83 Within a generated GNPS molecular network, one should keep in mind that ConCISE does not integrate single nodes in its annotation process.
Implemented bioinformatic workflows should also combine the use of rigorous statistical analyses for the selection of significant markers, together with the interrogations of spectral (e.g., MASST)84 and molecular databases (e.g., the LOTUS initiative),85 contributing to a more extensive metabolome annotation. To that end, a recently published step-by-step guide (“Hitchhiker's guide”) for MS2 metabolomic data interpretation aimed to explain all the essential statistical analyses that can be linked to molecular networks to further describe the chemodiversity of complex datasets.74 When publishing metabolomic results with MS2 spectra annotations, the level of confidence in structural identification should be disclosed, as explained by Schymanski et al.86 Similarly, a set of metadata should accompany the results, including key experimental (e.g., retention times) and processing information, as recently reviewed by Alseekh and colleagues.87
5.3. Challenges in improving structural knowledge
Despite their constant improvement, in silico tools are prone to errors, and their accuracy may also depend on the availability of real spectral data in databases. However, marine metabolites are poorly represented in these databases. As a result, the proposed candidate structures from in silico analyses often require further manual inspection (and ideally validation) using authentic standards to confirm the automated results.10,11,37 Manual analysis can reveal important details, such as (1) the presence of unexpected ions (e.g., Mg2+, Fe2+/Fe3+) through the identification of unusual isotopic patterns, (2) co-elutions that lead to overlapping isotopic patterns, and (3) in-source fragmentations. These key points could be missed by researchers when using automatic annotation tools, and without careful analysis of the datasets.11 Additionally, incorporating orthogonal data (such as retention times) and employing retention indexes would help in distinguishing isomers on conventionally used reversed phase columns and exclude erroneous structure proposals.88,89 Halogenated, and particularly brominated, metabolites are commonly found in marine environments. Their putative identification using in silico tools also requires additional attention, as the proposed molecular formulas are erroneous when the monoisotopic signal is not properly selected or fall below the detection limits.10,37In the absence of standards and MS2 spectral matches, even after careful examination of in silico outcomes, the confidence level for identifying unknown marine EMs remains low, typically at level 3 or 4 (e.g., molecular formula according to Schymanski et al.86), reflecting the high degree of structural uncertainty.11,59 Complete and accurate identification of EMs is most often achieved when standards are commercially available, which is generally the case for known metabolites involved in central metabolism (e.g., amino acids, nucleosides). Nevertheless, obtaining standards of marine specialized EMs requires their purification and structure identification. This is a challenging task because these molecules are present in trace amounts, and are often mixed with a plethora of other EMs in the enriched seawater extracts. The latter account for only a few mg of dried mass,10 which is insufficient to purify an EM in enough quantity for its identification by nuclear magnetic resonance (NMR). Hence, the purification of marine benthic EMs was successful only when they were isolated from identified marine organisms,28,37 when the producing organisms were cultivated in laboratory settings (e.g., microalgae),90 or when SPE capturing devices were maintained in place for several days to efficiently accumulate specialized EMs (see Section 6.3).31,36,46
6. Applications of benthic EMs analyses
We further describe recent studies implementing marine MS-based exometabolomics in benthic ecosystems in the contexts of marine ecology, including chemical and microbial ecology, but also natural product discovery.6.1. Describing molecular dynamics in reef ecosystems
Different targeted MS-based metabolomic studies have shown that small polar EMs are released in a species-specific manner by keystone benthic species (e.g., corals, algae and sponges). These EMs play a key role in fuelling the reef food web, thus contributing to the regulation of ecosystem functioning.44,63,64 Unlike targeted analyses, untargeted metabolomics has been modestly implemented to decipher the broad range of chemical signals that sustain marine biodiversity or inform behavioural observations. Most are deployed in a multi-omic strategy to investigate the link between benthic EMs, microbial diversity, and holobiont species assemblages.Untargeted metabolomic analyses were applied by Roach et al. to investigate the metabolome composition at the coral-turf algal interfaces with samples collected from the Caribbean island of Curacao.91 The authors measured a significant difference in both microbial and metabolomic compositions, with ceramide 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/16
1/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
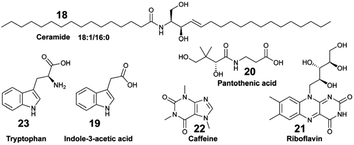
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 (18) being more abundant at the coral–algal interface than in the coral samples. Weber et al. deployed both untargeted and targeted metabolomics on exudates of coral reef species from US Virgin Islands to characterize specific classes of EMs, such as amino acids, nucleosides, vitamins, and indole-based compounds.63 Within the latter chemical family, indole-3-acetic acid (19) was specifically exuded from the octocoral holobiont Gorgonia ventolina, while pantothenic acid (20) and riboflavin (21) were found to be released by both stony corals and octocorals. Caffeine (22) was specifically released by the invasive algae Ramicrusta textilis.
0 (18) being more abundant at the coral–algal interface than in the coral samples. Weber et al. deployed both untargeted and targeted metabolomics on exudates of coral reef species from US Virgin Islands to characterize specific classes of EMs, such as amino acids, nucleosides, vitamins, and indole-based compounds.63 Within the latter chemical family, indole-3-acetic acid (19) was specifically exuded from the octocoral holobiont Gorgonia ventolina, while pantothenic acid (20) and riboflavin (21) were found to be released by both stony corals and octocorals. Caffeine (22) was specifically released by the invasive algae Ramicrusta textilis.
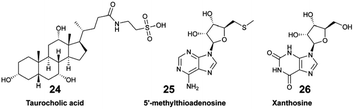
Collectively, the exudate composition was shown to be species-specific, and influenced microbial abundances and community composition in reef waters. The same team developed a derivatization method leading to the detection and quantification of 45 polar EMs from US Virgin Islands' coral reefs.44 The authors demonstrated how variations in targeted polar metabolites were related. For some of them, there was a significant correlation with benthic species composition. For example, the concentration of tryptophan (23) varied significantly across investigated reefs, and was considered as a core EM in coral reefs.44,63 Becker and co-workers deployed a multi-omic strategy to analyse the diversity of microorganisms and dissolved EMs in Florida's coral reef.64 By combining both targeted and untargeted metabolomics, distinct molecular compositions were measured across the investigated reef zones, with two EMs, taurocholic acid (24) and 5′-methylthioadenosine (25), showing significant variation between these zones. The authors suggested that 25 may be a signature metabolite that plays a role in microbial community dynamics within the reef habitat.
In all these studies, the EMs composition varied across different benthic organisms and geographic locations, influencing microbial communities and potentially serving as indicators of reef health (Fig. 4A).44,63,64 Untargeted tandem MS exometabolomic analyses were performed by Kelly et al. on seawater collected from aquaria tanks containing different species of coral reef primary producers from the French Polynesia: three types of algae (macroalgae Dictyota genus, turf, crustose coralline algae-CCA) and two genera of corals (Porites and Pocillopora).17 Feature-based molecular networking combined with deep metabolome annotation using SIRIUS revealed EM chemical class distinctions between corals and algae. Coral EMs were enriched in fatty acyl derivatives, such as oleoyl-taurines and acyl-carnitines. Polyketide macrolactams were detected as EMs released by CCA. Turf algae EMs encompassed nitrogen-containing heterocyclic compounds structurally similar to alkaloids. Both turf algae and CCA EMs were enriched in nitrogen. Furthermore, they had a lower nominal carbon oxidation state compared to coral EMs, thus potentially influencing reef biogeochemistry. The proportion of benthic species-specific EMs in the analysed samples was determined to be between 8–24% of the entire set of detected features. Using the same datasets, Quinlan and co-workers complemented the in silico-based annotations with ConCISE to expand their chemical description, aiming to progressively identify the influencing factors that drive coral reef metabolic shifts.83
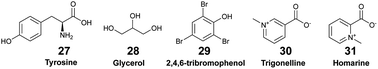
Sponge holobionts, with their diverse microbial communities, and through their impressive filter-feeding activities, significantly impact ecosystem-level nutrient cycling, as reviewed by Pita et al. in 2018.92 It has been demonstrated, through both untargeted and targeted metabolomic analyses, that sponges are primarily responsible for DOM transformation in the oceans. The investigated species from tropical ecosystems were found to release unique DOM profiles,16 which were possibly influenced by the abundance and diversity of their microsymbionts.18,40 Compared to the surrounding seawater, Fiore et al. demonstrated that the released DOM were found to be enriched in nucleosides (e.g., xanthosine 26) and in both tryptophan (23) and tyrosine (27), but were depleted in caffeine (22) and 5′-methylthioadenosine (25).16 Additionally, such released DOM could include sponge-specialized metabolites, such as alkaloids and terpenoids.10,34 Mauduit et al. revealed the presence of specialized metabolites 1, as well as 11–17 in the seawater above each investigated sponge species.10 Collectively, these EMs represented ∼12% of the detected features, which is in agreement with results from Kelly et al.17
6.2. Deciphering distant chemical mediation in the benthos
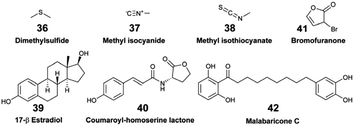
Exhaustive reviews are available on marine chemical ecology in benthic environments3 and planktonic interactions.93,94 Comparative untargeted metabolomic analyses are mainly implemented to study the chemical ecology of marine algae (macro- and micro-algae, phytoplankton), with a particular focus on microbial ecology, and to further understand the factors involved with algal bloom and nutrient cycling in the pelagos.9,93,95 Although it is recognised that untargeted metabolomics is instrumental for identifying yet unknown waterborne chemical cues, its implementation remains scarce in the study of distant chemical mediation in the benthos. Herein, we describe a few recent studies that led to the identification of EMs as allelochemicals in benthic ecosystems.Other applications of MS-based exometabolomics concern the investigations of coral diseases, which are widespread in reef ecosystems due to climate changes and anthropogenic pressures. Ochsenkühn et al. used minimally invasive water sampling above the polyps from two species of coral (genus Acropora and Platygyra), combined with multi-omic analyses to show that both coral surfaces harboured unique bacteria and metabolite profiles.50 Concentration gradients were measured from the mucus to the surrounding seawater. Some of the putatively identified EMs in the mucus were hormones such as 17-β estradiol (39) involved in coral gametogenesis, bacterial quorum regulators such as coumaroyl-homoserine lactone (40) and bromofuranone (41), and antibacterial compounds such as malabaricone C (42). Collectively, these molecules participate in the structuring of the mucus microbial community involved in the defence against pathogens.
6.3. Discovering new molecules as EMs
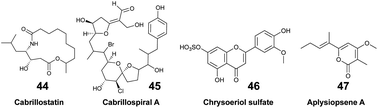
The pool of marine EMs represents a niche of structurally diverse molecules that has attracted the attention of natural product chemists over the past decade. The dual benefit sustaining this interest relies on the fact that (1) exuded molecules that have signal value in nature by analogy can prove to be useful to humans, and (2) new molecules could be discovered with minimal disturbances on marine biodiversity. However, challenges pertaining to the purification and structural description of highly dilute marine EMs tend to overshadow such benefits. The rapidly changing composition of seawater over time and space also complicates the task. Nevertheless, when marine species can be maintained in aquaria, their EMs can be captured on SPE devices and accumulated continuously over a long period of time. By doing so, Vlachou et al. captured and isolated new polycyclic guanidine alkaloids (34, 35) exuded from the sponge Crambe crambe.36 Notwithstanding, the diversity of EMs produced may be lower for cultivated marine organisms that are isolated from neighbouring competing species. Accessing the chemical diversity within marine ecosystems, and thus without collecting any organism, requires optimized in situ sampling methods to reach optimal concentration of new chemical entities. To that end, an artificial sponge was designed to enrich microbial marine EMs from seawater,46 while the SMIRC focused on capturing bacterial EMs from marine sediments.31 Both were deployed several days (2–14 days) on site, and enabled the purification and description of new structures. For example, new structural scaffolds were described from SMIRC enriched extracts.31 The molecules were named cabrillostatin (44) and cabrillospiral (45). The authors performed a MASST84 search with the newly described 44, which was subsequently detected in different shared MS datasets mainly associated with the study of marine DOM. These results therefore contributed to expand the structural lexicon of marine EMs. Other isolated molecules from SMIRC were chrysoeriol sulfate (46), considered to be a seagrass root exudate, and aplysiopsene A (47).7. Challenges & perspectives
Although significant progress has been made in MS-based metabolomics, particularly in data processing and spectral annotation, most of the marine EMs studied to date come from central metabolism with available commercial standards and well-known chemical structures. In contrast, relatively little is known on the proportion, composition, and structural identity of other molecules (including specialized EMs) in the entire DOM pool. Despite their relevance across various scientific fields, from marine biogeochemistry to natural product chemistry, the structural identity and biological functions of the majority of marine EMs remain largely unknown. One key challenge involves increasing the accuracy in their structural description, which will consequently foster a deeper understanding of their biological functions. The optimization of seawater sampling techniques for EM purification, along with the systematic sharing of structural raw data, are interdependent factors that collectively enhance structural knowledge of DOM composition and improve the efficiency of structural annotation tools.7.1. Improving in situ sampling techniques
Since 1982, every decade has seen the development of new in situ devices to capture EMs (often considered as allelochemicals) in a chemical ecology context. Collectively, these devices showcase the technical ingenuity developed for capturing marine molecules, along with the recurring interest of the scientific community. Despite such efforts, the harvested pool of marine-waterborne EMs remains limited in quantity, making their purification, which is essential for both structure elucidation and biological investigations, challenging. To overcome the ‘‘quantity’’ challenge, the sampling methodology should counterbalance the large seawater dilution effect. In line with this first condition, the in situ device should be left alone for several hours per day to accumulate progressively diluted EMs by processing larger quantities of seawater. Secondly, such a device, similar to I-SMEL, should retain the possibility of tracing back the biosynthetic producers of EMs, possibly capturing molecules before any physico-chemical degradation occurs. Knowing the taxonomic identity of EM producers is essential to guide structure identification efforts, as well as studies focused on measuring waterborne chemical exchanges in different ecosystems. Thus, any future improved seawater device should work in different configurations (e.g., overhang cliffs, underwater caves), and favour standardized seawater sampling to aid in mapping the chemical signature of different marine ecosystems.7.2. Limits to chemical space representativeness
It should be noted that the representation of marine chemical space explored through EM captures and MS-based metabolomics is influenced by multiple technical and methodological factors. As with the broader challenge of studying marine ecosystems, every step in the research process, from sample preparation to data processing, impacts the measured chemical diversity. Efforts should be made towards using orthogonal methods combining sampling strategies (e.g., combining SPE supports) and diverse metabolomic analyses (e.g., using different chromatographic analysis or source ionization) to expand the chemical space of the analysed EMs. To that end, various analytical methods have been proposed for the analysis of small polar marine EMs.33,44,47,49,59 Such methods have yet to be widely adopted in marine MS-based exometabolomics. Marine geochemists also usually combine structural information from Nuclear Magnetic Resonance (NMR) and FT-ICR MS for the characterisation of marine DOM.6,54 Nevertheless, such promising combination of orthogonal analytical techniques is not commonly implemented in studies pertaining to marine (chemical) ecology. This is partly due to the molecular complexity of the extracts characterized by a high dynamic range of thousands of EMs. Just as we cannot yet capture the full complexity of ecosystems, no single analytical method or MS-based metabolomic analysis leads to total coverage of the marine EM chemical diversity. This limitation, highlighted by Naman et al.'s photographic snapshot analogy, mirrors the broader difficulties in fully describing the diverse and rapidly changing nature of marine chemical seascapes: “No metabolomics investigation leads to total coverage. At each decision-making juncture, some data are lost knowingly or otherwise”.1017.3. Databases & raw data sharing
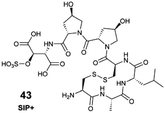
Freely available, raw spectral databases that can be systematically interrogated to rapidly identify metabolites produced by marine organisms remain underdeveloped. In this respect, the marine natural products community lags behind its counterparts in phytochemistry102 and microbiology.103 Nevertheless, researchers' growing commitment to sharing data in public repositories, combined with the community-driven efforts to organize data and metadata, enables global repository-scale analyses with the development of new tools, such as the MS Search Tool (e.g., MASST, microbeMASST).84,103 As such, the discoverability and efficient reuse of acquired spectral information require sharing detailed and ideally standardized metadata, alongside the corresponding raw data. Access to raw and well-documented spectral data from already known marine molecules will provide a critical foundation for improving the description of marine EMs, expanding their structural lexicon. The power of such data-sharing has been nicely illustrated with compound 43 (see Section 6.3).31 The automated interrogation of such shared spectral data (e.g., through molecular networking tools), will also enhance our annotation capabilities via spectral analogy. In addition to what Alseekh et al. proposed,87 incorporating retention indexes in the shared metadata would further enhance inter-laboratory data comparisons, providing an additional level of confidence in MS-based metabolite identification.88,89 The published table of MS2 descriptions could also contain the SMILES (Simplified Molecular Input Line Entry System) notation for putatively identified molecules, or other machine-readable structural information, which will contribute to the growth of molecular databases (e.g., PubChem,104 LOTUS85). Informing the taxonomic origins of identified marine EMs is crucial for accurate downstream dereplication, and to promote the expansion of accurate open-access knowledge. As such, the LOTUS initiative aims to consolidate and share referenced structure-organism pairs through open platforms.85 A series of tools are now available to encourage researchers to take part in the open science movement, including directly contributing to Wikidata, as explained in a recent viewpoint article.105 Additionally, making published and shared data accessible and interconnected will support the development of future Artificial Intelligence (AI) tools. This will advance both our structural and biological knowledge of marine EMs, while benefiting the marine chemistry and ecology communities.7.4. Expanding knowledge on ecosystem functioning
Exploring the molecular composition of seawater at the structural level offers exciting opportunities to compare results across diverse marine ecosystems (e.g., tropical and temperate reefs, underwater caves), and identify both redundant and unique molecular signatures. Such a systematic description will help to better decipher the metabolic contributions of benthic communities, while enabling comparisons of the molecular dynamics governing species interactions. Many of the articles cited herein highlight a consensus among marine ecologists on the need to further investigate the functions of specific EMs in the assemblage of reef holobionts and their role in structuring benthic biodiversity. Up to now, most MS-based marine exometabolomic studies have primarily focused on measuring nutrient exchanges. As a result, interactions in the oceans are often viewed through the lens of trophic relationships, overlooking other functional processes mediated by chemicals, such as homing,11 reproduction,90 and larval recruitment.42 For that purpose, the main challenges that need to be addressed include (1) purifying and identifying waterborne molecules that are highly diluted in seawater, and (2) performing assays either in the laboratory (e.g., in aquaria) or in situ to describe their functions as infochemicals. The second challenge is particularly complex, as not all benthic organisms can be maintained in laboratory settings, and purified compounds may function as cocktails of diluted molecules in unknown proportions rather than as single entities. Marine environmental disturbances, such as ocean acidification and warming, also alter the production and perception of chemical cues by organisms, as explained by Roggatz et al.106 These disruptions can scale up to ecosystem-wide impacts, influencing species distribution and trophic interactions. Therefore, ongoing efforts to develop marine EM metabolomics are crucial for deciphering the chemical mixtures involved in organism behaviours and assessing their resilience to global change over time. Likewise, studying the structural stability of identified marine EMs in seawater could offer valuable insights into their roles as infochemicals and their susceptibility to environmental change.7.5. Funding challenges and societal impacts
Unlike terrestrial ecosystems, where pollination is a well-understood and well-described chemical mediation process, the role of chemical exchanges in marine benthic ecosystems is far less communicated to the public. Understanding the identity of the metabolites produced, released, and perceived by sessile organisms in seawater is essential to unravelling the functioning of diverse marine ecosystems and their resilience to anthropogenic pressures. Nevertheless, research studies on marine benthic EMs and their roles as chemical cues remain underrepresented, partly due to limited funding opportunities and historical difficulties in deciphering seawater chemistry. Moreover, studies focusing on the oceanic world face inherent technical challenges (e.g., hyperbaric pressures), making them naturally expensive. Technological advancements, leading to the development of more efficient in situ devices, will also require appropriate funding to make them more widely accessible to the entire marine science community. Achieving all these objectives requires raising societal awareness about the importance of preserving marine ecosystems, which are increasingly being threatened by global change resulting from human activities. This awareness can be fostered by sharing knowledge about their functions, while emphasizing the services they provide to society, such as food, biotechnologies and medicine.1078. Conclusion
Until the past 20 years, metabolomic investigations of marine EMs had yielded scattered results. The drastic increase in the sensitivity of MS instruments, combined with advances in metabolic and chemometric analyses benefited the fields of marine (chemical) ecology, geochemistry, as well as natural product chemistry. In addition, the development of in silico tools for the annotations of MS2 data towards the categorization of unknown EMs opened new ways to describe the composition of seawater. Applications of marine MS-based exometabolomics are progressively expanding to study the functioning of benthic ecosystems. Most multi-omics studies have focused on examining the relationships between benthic EMs and marine microorganisms, primarily through the lens of trophic exchanges as influential factors that drive biodiversity assemblages. Metabolomic analyses that focus on identifying waterborne chemical mediators and confirming their biological properties within an ecosystem remain limited. However, these studies are crucial to deciphering the functioning of marine ecosystems, and understanding the multiple services they provide. Collectively, these studies also demonstrated the structural diversity of detected and identified marine EMs, from amino acids to steroids, including terpenoid and alkaloid derivatives. As investigations on marine EMs continue to progress, additional efforts should be directed toward their isolation and structural characterization. Moreover, the sharing of raw data from known and new marine molecules is still urgently needed to gain more confidence in the identification process, and promote the accurate description of marine EM chemical space. The challenges faced by marine chemists toward better understanding the chemical composition of the vast seawater have been and will continue to be sources of innovations and biodiscoveries. The availability of new devices to efficiently concentrate molecules from seawater and trace the chemical fingerprints of keystone species offers new opportunities of research. These advancements inspire further efforts toward deciphering the central role of chemical interactions in the functioning of marine ecosystems. Chemists and ecologists alike are encouraged to combine their strengths and expertise to deepen their investigations, either focusing on understanding chemical exchanges and the shaping of marine biodiversity, or concentrating on discovering new bioactive waterborne molecules in the vast seawater. As noted by Eisner and Meinwald, “What remains unknown is of immense potential value, and deserving of protection, lest we be forever impoverished by its loss”.108 The progress made on marine exometabolomics through interdisciplinary research will have a significant impact on the scientific marine community, as well as the society at large.9. Author contributions
M. M., S. G., M. D., C. S.: writing – original draft descriptions, review and editing, visualization. C. S.: supervision.10. Conflicts of interest
There are no conflicts to declare.11. Acknowledgements
The authors acknowledge financial support by the ANR (ANR-20-CE43-0003). The authors are thankful to all members of the NEMO team at the IMBE, in particular Dr Thierry Pérez and Dr Pierre Chevaldonné, for fruitful conversations on the topics of chemical seascape and ecosystem services. The authors also thank the anonymous reviewers for their constructive comments, which improved the quality of our manuscript.12. References
- Research and Discoveries: The Revolution of Science through Scuba, ed. J. R. Pawlik, C. D. Amsler, R. Ritson-Williams, J. B. McClintock, B. J. Baker and V. J. Paul, Smithsonian Institution Scholarly Press, Washington, D.C., 2013, vol. 39 Search PubMed.
- A. R. Carroll, B. R. Copp, T. Grkovic, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2024, 41, 162–207 RSC.
- M. P. Puglisi, J. M. Sneed, R. Ritson-Williams and R. Young, Nat. Prod. Rep., 2019, 36, 410–429 RSC.
- M. E. Hay, Ann. Rev. Mar. Sci., 2009, 1, 193–212 CrossRef PubMed.
- T. S. Catalá, S. Shorte and T. Dittmar, Appl. Microbiol. Biotechnol., 2021, 105, 7225–7239 CrossRef PubMed.
- A. Nebbioso and A. Piccolo, Anal. Bioanal. Chem., 2013, 405, 109–124 CrossRef CAS PubMed.
- D. Petras, I. Koester, R. Da Silva, B. M. Stephens, A. F. Haas, C. E. Nelson, L. W. Kelly, L. I. Aluwihare and P. C. Dorrestein, Front. Mar. Sci., 2017, 4 DOI:10.3389/fmars.2017.00405.
- L. Wegley Kelly, C. E. Nelson, L. I. Aluwihare, M. G. I. Arts, P. C. Dorrestein, I. Koester, S. B. Matsuda, D. Petras, Z. A. Quinlan and A. F. Haas, Front. Mar. Sci., 2021, 8 DOI:10.3389/fmars.2021.630799.
- M. Thukral, A. E. Allen and D. Petras, ISME J., 2023, 17, 2147–2159 CrossRef PubMed.
- M. Mauduit, M. Derrien, M. Grenier, S. Greff, S. Molinari, P. Chevaldonné, C. Simmler and T. Pérez, ACS Cent. Sci., 2023, 9, 2084–2095 CrossRef CAS PubMed.
- M. Derrien, M. Santonja, S. Greff, S. Figueres, C. Simmler, P. Chevaldonné and T. Pérez, Front. Mar. Sci., 2024, 11 DOI:10.3389/fmars.2024.1448616.
- A. Jürgens and M. Bischoff, Funct. Ecol., 2017, 31, 56–64 CrossRef.
- M. Zark, J. Christoffers and T. Dittmar, Mar. Chem., 2017, 191, 9–15 CrossRef CAS.
- S. K. Bercovici, T. Dittmar and J. Niggemann, Limnol. Oceanogr.:Methods, 2022, 20, 350–360 CrossRef CAS.
- B. E. Noriega-Ortega, G. Wienhausen, A. Mentges, T. Dittmar, M. Simon and J. Niggemann, Front. Microbiol., 2019, 10 DOI:10.3389/fmicb.2019.00215.
- C. L. Fiore, C. J. Freeman and E. B. Kujawinski, PeerJ, 2017, 5, e2870 CrossRef PubMed.
- L. Wegley Kelly, C. E. Nelson, D. Petras, I. Koester, Z. A. Quinlan, M. G. I. Arts, L.-F. Nothias, J. Comstock, B. M. White, E. C. Hopmans, F. C. van Duyl, C. A. Carlson, L. I. Aluwihare, P. C. Dorrestein and A. F. Haas, Proc. Natl. Acad. Sci. U.S.A., 2022, 119, e2110283119 CrossRef PubMed.
- T. Hildebrand, H. Osterholz, C. Bunse, H. Grotheer, T. Dittmar and P. J. Schupp, Limnol. Oceanogr., 2022, 67, 2483–2496 CrossRef CAS.
- L. Weber, M. Armenteros, M. Kido Soule, K. Longnecker, E. B. Kujawinski and A. Apprill, Front. Mar. Sci., 2020, 7 DOI:10.3389/fmars.2020.582161.
- M. Zark and T. Dittmar, Nat. Commun., 2018, 9, 3178 CrossRef PubMed.
- D. A. Hansell, Ann. Rev. Mar. Sci., 2013, 5, 421–445 CrossRef PubMed.
- N. Hertkorn, R. Benner, M. Frommberger, P. Schmitt-Kopplin, M. Witt, K. Kaiser, A. Kettrup and J. I. Hedges, Geochim. Cosmochim. Acta, 2006, 70, 2990–3010 CrossRef CAS.
- S. F. Cummins and J. H. Bowie, Nat. Prod. Rep., 2012, 29, 642 RSC.
- J. Meinwald, W. S. Leal and J. Kubanek, ACS Omega, 2018, 3, 4048–4053 CrossRef CAS PubMed.
- M. Saha, E. Berdalet, Y. Carotenuto, P. Fink, T. Harder, U. John, F. Not, G. Pohnert, P. Potin, E. Selander, W. Vyverman, T. Wichard, V. Zupo and M. Steinke, Front. Ecol. Environ., 2019, 17, 530–537 CrossRef.
- J. C. Coll, B. F. Bowden, D. M. Tapiolas and W. C. Dunlap, J. Exp. Mar. Biol. Ecol., 1982, 60, 293–299 CrossRef CAS.
- B. A. Schulte, R. De Nys, G. J. Bakus, P. Crews, C. Eid, S. Naylor and L. V. Manes, J. Chem. Ecol., 1991, 17, 1327–1332 CrossRef CAS PubMed.
- E. Ternon, L. Zarate, S. Chenesseau, J. Croue, R. Dumollard, M. T. Suzuki and O. P. Thomas, Sci. Rep., 2016, 6, 29474 CrossRef CAS PubMed.
- J. Kubanek, K. E. Whalen, S. Engel, S. R. Kelly, T. P. Henkel, W. Fenical and J. R. Pawlik, Oecologia, 2002, 131, 125–136 CrossRef PubMed.
- T. Dittmar, B. Koch, N. Hertkorn and G. Kattner, Limnol. Oceanogr.:Methods, 2008, 6, 230–235 CrossRef CAS.
- A. Bogdanov, M. N. Salib, A. B. Chase, H. Hammerlindl, M. N. Muskat, S. Luedtke, E. B. da Silva, A. J. O'Donoghue, L. F. Wu, S. J. Altschuler, T. F. Molinski and P. R. Jensen, Nat. Commun., 2024, 15, 5230 CrossRef CAS PubMed.
- W. M. Johnson, M. C. Kido Soule and E. B. Kujawinski, Limnol. Oceanogr.:Methods, 2017, 15, 417–428 CrossRef.
- J. S. Sacks, K. R. Heal, A. K. Boysen, L. T. Carlson and A. E. Ingalls, Limnol. Oceanogr.:Methods, 2022, 20, 683–700 CrossRef CAS.
- B. Bojko, B. Onat, E. Boyaci, E. Psillakis, T. Dailianis and J. Pawliszyn, Front. Mar. Sci., 2019, 6 DOI:10.3389/fmars.2019.00632.
- M. F. Mehbub, J. E. Tanner, S. J. Barnett, C. M. M. Franco and W. Zhang, Appl. Microbiol. Biotechnol., 2016, 100, 10609–10626 CrossRef CAS PubMed.
- P. Vlachou, L. Goff, C. Alonso, A. Á. Pedro, J. Gallard, N. Fokialakis and J. Ouazzani, Mar. Drugs, 2018, 16, 0152 CrossRef PubMed.
- M. Mauduit, S. Greff, G. Herbette, J.-V. Naubron, S. Chentouf, T. Huy Ngo, J.-W. Nam, S. Molinari, F. Mabrouki, E. Garayev, B. Baghdikian, T. Pérez and C. Simmler, ACS Omega, 2022, 7, 43068–43083 CrossRef CAS PubMed.
- N. Kamali, F. Abbas, M. Lehane, M. Griew and A. Furey, Molecules, 2022, 27, 7898 CrossRef CAS PubMed.
- M. Roué, H. Darius and M. Chinain, Toxins, 2018, 10, 167 CrossRef PubMed.
- L. K. Olinger, W. K. Strangman, S. E. McMurray and J. R. Pawlik, Front. Mar. Sci., 2021, 8 DOI:10.3389/fmars.2021.665789.
- R. P. Walker, J. E. Thompson and D. J. Faulkner, Mar. Biol., 1985, 88, 27–32 CrossRef CAS.
- Z. A. Quinlan, M.-J. Bennett, M. G. I. Arts, M. Levenstein, D. Flores, H. M. Tholen, L. Tichy, G. Juarez, A. F. Haas, V. F. Chamberland, K. R. W. Latijnhouwers, M. J. A. Vermeij, A. W. Johnson, K. L. Marhaver and L. W. Kelly, Proc. R. Soc. B, 2023, 290, 20231476 CrossRef CAS PubMed.
- E. Richelle-Maurer, M. J. De Kluijver, S. Feio, S. Gaudêncio, H. Gaspar, R. Gomez, R. Tavares, G. Van de Vyver and R. W. M. Van Soest, Biochem. Syst. Ecol., 2003, 31, 1073–1091 CrossRef CAS.
- B. M. Garcia, C. C. Becker, L. Weber, G. J. Swarr, M. C. Kido Soule, A. Apprill and E. B. Kujawinski, J. Proteome Res., 2024, 23, 2041–2053 CrossRef CAS PubMed.
- M. Sjögren, P. R. Jonsson, M. Dahlström, T. Lundälv, R. Burman, U. Göransson and L. Bohlin, J. Nat. Prod., 2011, 74, 449–454 CrossRef PubMed.
- J. J. La Clair, S. T. Loveridge, K. Tenney, M. O'Neil-Johnson, E. Chapman and P. Crews, PLoS One, 2014, 9, e100474 CrossRef PubMed.
- E. Ternon, Y. Wang and K. J. Coyne, Molecules, 2019, 24, 135 CrossRef PubMed.
- C. Xu, S. P. Couvillion, R. L. Sontag, N. G. Isern, Y. Maezato, S. R. Lindemann, T. Roy Chowdhury, R. Zhao, B. R. Morton, R. K. Chu, R. J. Moore, J. K. Jansson, V. L. Bailey, P. J. Mouser, M. F. Romine, J. F. Fredrickson and T. O. Metz, mSystems, 2021, 6(3), e01058 CrossRef CAS PubMed.
- B. Widner, M. C. Kido Soule, F. X. Ferrer-González, M. A. Moran and E. B. Kujawinski, Anal. Chem., 2021, 93, 4809–4817 CrossRef CAS PubMed.
- M. A. Ochsenkühn, P. Schmitt-Kopplin, M. Harir and S. A. Amin, Commun. Biol., 2018, 1, 1–10 CrossRef PubMed.
- R. J. M. Weber, E. Selander, U. Sommer and M. R. Viant, Mar. Drugs, 2013, 11, 4158–4175 CrossRef PubMed.
- C. Gosset-Erard, F. Aubriet, E. Leize-Wagner, Y.-N. François and P. Chaimbault, Talanta, 2023, 257, 124324 CrossRef CAS PubMed.
- O. J. Lechtenfield, J. Kaesler, E. K. Jennings and B. P. Koch, Environ. Sci. Technol., 2024, 58, 4637–4647 CrossRef PubMed.
- M. Seidel, S. P. B. Vemulapalli, D. Mathieu and T. Dittmar, Environ. Sci. Technol., 2022, 56, 3758–3769 CrossRef CAS PubMed.
- D. Petras, J. J. Minich, L. B. Cancelada, R. R. Torres, E. Kunselman, M. Wang, M. E. White, E. E. Allen, K. A. Prather, L. I. Aluwihare and P. C. Dorrestein, Chemosphere, 2021, 271, 129450 CrossRef CAS PubMed.
- P. Stincone, A. K. Pakkir Shah, R. Schmid, L. G. Graves, S. P. Lambidis, R. R. Torres, S.-N. Xia, V. Minda, A. T. Aron, M. Wang, C. C. Hughes and D. Petras, Anal. Chem., 2023, 95, 12673–12682 CrossRef CAS PubMed.
- S. P. Lambidis, T. Schramm, K. Steuer-Lodd, S. Farrell, P. Stincone, R. Schmid, I. Koester, R. Torres, T. Dittmar, L. Aluwihare, C. Simon and D. Petras, Environ. Sci. Technol., 2024, 58, 19289–19304 CrossRef PubMed.
- J. Guo and T. Huan, Anal. Chem., 2020, 92, 8072–8080 CrossRef CAS PubMed.
- J. Patrone, M. Vila-Costa, J. Dachs, S. Papazian, P. Gago-Ferrero and R. Gil-Solsona, Environ. Sci. Technol., 2024, 58, 12454–12466 CrossRef CAS PubMed.
- K. P. Law, W. He, J. Tao and C. Zhang, Front. Microbiol., 2021, 12, 658781 CrossRef PubMed.
- E. M. Sogin, E. Puskás, N. Dubilier and M. Liebeke, mSystems, 2019, 4(6), e00638 CrossRef PubMed.
- C. Duque, A. Bonilla, E. Bautista and S. Zea, Biochem. Syst. Ecol., 2001, 29, 459–467 CrossRef CAS PubMed.
- L. Weber, M. K. Soule, K. Longnecker, C. C. Becker, N. Huntley, E. B. Kujawinski and A. Apprill, ISME Commun., 2022, 2, 1–13 CrossRef PubMed.
- C. C. Becker, L. Weber, B. Zgliczynski, C. Sullivan, S. Sandin, E. Muller, A. S. Clark, M. C. Kido Soule, K. Longnecker, E. B. Kujawinski and A. Apprill, PNAS Nexus, 2023, 2, pgad287 CrossRef PubMed.
- C. Bosch-Orea, J. Sanchís and M. Farré, MethodsX, 2021, 8, 101370 CrossRef CAS PubMed.
- N. Tolić, Y. Liu, A. Liyu, Y. Shen, M. M. Tfaily, E. B. Kujawinski, K. Longnecker, L.-J. Kuo, E. W. Robinson, L. Paša-Tolić and N. J. Hess, Anal. Chem., 2017, 89, 12659–12665 CrossRef PubMed.
- J. Merder, J. A. Freund, U. Feudel, C. T. Hansen, J. A. Hawkes, B. Jacob, K. Klaproth, J. Niggemann, B. E. Noriega-Ortega, H. Osterholz, P. E. Rossel, M. Seidel, G. Singer, A. Stubbins, H. Waska and T. Dittmar, Anal. Chem., 2020, 92, 6832–6838 CrossRef CAS PubMed.
- L. C. Powers, N. Hertkorn, N. McDonald, P. Schmitt-Kopplin, R. Del Vecchio, N. V. Blough and M. Gonsior, Global Biogeochem. Cycles, 2019, 33, 1423–1439 CrossRef CAS.
- A. Rivas-Ubach, Y. Liu, T. S. Bianchi, N. Tolić, C. Jansson and L. Paša-Tolić, Anal. Chem., 2018, 90, 6152–6160 CrossRef CAS PubMed.
- S. Merel, Chemosphere, 2023, 313, 137443 CrossRef CAS PubMed.
- C. A. Smith, E. J. Want, G. O'Maille, R. Abagyan and G. Siuzdak, Anal. Chem., 2006, 78, 779–787 CrossRef CAS PubMed.
- H. Tsugawa, T. Cajka, T. Kind, Y. Ma, B. Higgins, K. Ikeda, M. Kanazawa, J. VanderGheynst, O. Fiehn and M. Arita, Nat. Methods, 2015, 12, 523–526 CrossRef CAS PubMed.
- S. Heuckeroth, T. Damiani, A. Smirnov, O. Mokshyna, C. Brungs, A. Korf, J. D. Smith, P. Stincone, N. Dreolin, L.-F. Nothias, T. Hyötyläinen, M. Oresic, U. Karst, P. C. Dorrestein, D. Petras, X. Du, J. J. J. van der Hooft, R. Schmid and T. Pluskal, Nat. Protoc., 2024, 19, 2597–2641 CrossRef CAS PubMed.
- A. K. Pakkir Shah, A. Walter, F. Ottosson, F. Russo, M. Navarro-Diaz, J. Boldt, J.-C. J. Kalinski, E. E. Kontou, J. Elofson, A. Polyzois, C. González-Marín, S. Farrell, M. R. Aggerbeck, T. Pruksatrakul, N. Chan, Y. Wang, M. Pöchhacker, C. Brungs, B. Cámara, A. M. Caraballo-Rodríguez, A. Cumsille, F. de Oliveira, K. Dührkop, Y. El Abiead, C. Geibel, L. G. Graves, M. Hansen, S. Heuckeroth, S. Knoblauch, A. Kostenko, M. C. M. Kuijpers, K. Mildau, S. Papadopoulos Lambidis, P. W. Portal Gomes, T. Schramm, K. Steuer-Lodd, P. Stincone, S. Tayyab, G. A. Vitale, B. C. Wagner, S. Xing, M. T. Yazzie, S. Zuffa, M. de Kruijff, C. Beemelmanns, H. Link, C. Mayer, J. J. J. van der Hooft, T. Damiani, T. Pluskal, P. Dorrestein, J. Stanstrup, R. Schmid, M. Wang, A. Aron, M. Ernst and D. Petras, Nat. Protoc., 2024, 1–71 Search PubMed.
- Z. Pang, Y. Lu, G. Zhou, F. Hui, L. Xu, C. Viau, A. F. Spigelman, P. E. MacDonald, D. S. Wishart, S. Li and J. Xia, Nucleic Acids Res., 2024, 52, W398–W406 CrossRef PubMed.
- K. Dührkop, M. Fleischauer, M. Ludwig, A. A. Aksenov, A. V. Melnik, M. Meusel, P. C. Dorrestein, J. Rousu and S. Böcker, Nat. Methods, 2019, 16, 299–302 CrossRef PubMed.
- K. Dührkop, H. Shen, M. Meusel, J. Rousu and S. Böcker, Proc. Natl. Acad. Sci. U.S.A., 2015, 112, 12580–12585 CrossRef PubMed.
- C. Ruttkies, S. Neumann and S. Posch, BMC Bioinf., 2019, 20, 376 CrossRef PubMed.
- F. Wang, J. Liigand, S. Tian, D. Arndt, R. Greiner and D. S. Wishart, Anal. Chem., 2021, 93, 11692–11700 CrossRef CAS PubMed.
- K. Dührkop, L.-F. Nothias, M. Fleischauer, R. Reher, M. Ludwig, M. A. Hoffmann, D. Petras, W. H. Gerwick, J. Rousu, P. C. Dorrestein and S. Böcker, Nat. Biotechnol., 2021, 39, 462–471 CrossRef PubMed.
- Y. Djoumbou Feunang, R. Eisner, C. Knox, L. Chepelev, J. Hastings, G. Owen, E. Fahy, C. Steinbeck, S. Subramanian, E. Bolton, R. Greiner and D. S. Wishart, J. Cheminf., 2016, 8, 61 Search PubMed.
- H. W. Kim, M. Wang, C. A. Leber, L.-F. Nothias, R. Reher, K. B. Kang, J. J. J. van der Hooft, P. C. Dorrestein, W. H. Gerwick and G. W. Cottrell, J. Nat. Prod., 2021, 84, 2795–2807 CrossRef CAS PubMed.
- Z. A. Quinlan, I. Koester, A. T. Aron, D. Petras, L. I. Aluwihare, P. C. Dorrestein, C. E. Nelson and L. Wegley Kelly, Metabolites, 2022, 12, 1275 CrossRef CAS PubMed.
- M. Wang, A. K. Jarmusch, F. Vargas, A. A. Aksenov, J. M. Gauglitz, K. Weldon, D. Petras, R. da Silva, R. Quinn, A. V. Melnik, J. J. J. van der Hooft, A. M. Caraballo-Rodríguez, L. F. Nothias, C. M. Aceves, M. Panitchpakdi, E. Brown, F. Di Ottavio, N. Sikora, E. O. Elijah, L. Labarta-Bajo, E. C. Gentry, S. Shalapour, K. E. Kyle, S. P. Puckett, J. D. Watrous, C. S. Carpenter, A. Bouslimani, M. Ernst, A. D. Swafford, E. I. Zúñiga, M. J. Balunas, J. L. Klassen, R. Loomba, R. Knight, N. Bandeira and P. C. Dorrestein, Nat. Biotechnol., 2020, 38, 23–26 CrossRef CAS PubMed.
- A. Rutz, M. Sorokina, J. Galgonek, D. Mietchen, E. Willighagen, A. Gaudry, J. G. Graham, R. Stephan, R. Page, J. Vondrášek, C. Steinbeck, G. F. Pauli, J.-L. Wolfender, J. Bisson and P.-M. Allard, eLife, 2022, 11, e70780 CrossRef CAS PubMed.
- E. L. Schymanski, J. Jeon, R. Gulde, K. Fenner, M. Ruff, H. P. Singer and J. Hollender, Environ. Sci. Technol., 2014, 48, 2097–2098 CrossRef CAS PubMed.
- S. Alseekh, A. Aharoni, Y. Brotman, K. Contrepois, J. D'Auria, J. Ewald, J. C. Ewald, P. D. Fraser, P. Giavalisco, R. D. Hall, M. Heinemann, H. Link, J. Luo, S. Neumann, J. Nielsen, L. Perez de Souza, K. Saito, U. Sauer, F. C. Schroeder, S. Schuster, G. Siuzdak, A. Skirycz, L. W. Sumner, M. P. Snyder, H. Tang, T. Tohge, Y. Wang, W. Wen, S. Wu, G. Xu, N. Zamboni and A. R. Fernie, Nat. Methods, 2021, 18, 747–756 CrossRef CAS PubMed.
- M. Witting and S. Böcker, J. Sep. Sci., 2020, 43, 1746–1754 CrossRef CAS PubMed.
- Q. Chen, Q. Lu, L. Zhang, C. Zhang, J. Zhang, Y. Gu, Q. Huang and H. Tang, Talanta, 2024, 268, 125318 CrossRef CAS PubMed.
- F. A. Klapper, C. Kiel, P. Bellstedt, W. Vyverman and G. Pohnert, Angew. Chem., Int. Ed., 2023, 62, e202307165 CrossRef CAS PubMed.
- T. N. F. Roach, M. Little, M. G. I. Arts, J. Huckeba, A. F. Haas, E. E. George, R. A. Quinn, A. G. Cobián-Güemes, D. S. Naliboff, C. B. Silveira, M. J. A. Vermeij, L. W. Kelly, P. C. Dorrestein and F. Rohwer, Proc. Natl. Acad. Sci. U.S.A., 2020, 117, 13588–13595 CrossRef CAS PubMed.
- L. Pita, L. Rix, B. M. Slaby, A. Franke and U. Hentschel, Microbiome, 2018, 6, 46 CrossRef CAS PubMed.
- C. Kuhlisch, A. Shemi, N. Barak-Gavish, D. Schatz and A. Vardi, Nat. Rev. Microbiol., 2024, 22, 138–154 CrossRef CAS PubMed.
- E. R. Brown, M. R. Cepeda, S. J. Mascuch, K. L. Poulson-Ellestad and J. Kubanek, Nat. Prod. Rep., 2019, 36, 1093–1116 RSC.
- C. Kuhlisch and G. Pohnert, Nat. Prod. Rep., 2015, 32, 937–955 RSC.
- T. Alsufyani, A. Weiss and T. Wichard, Mar. Drugs, 2017, 15, 14 CrossRef PubMed.
- R. X. Poulin, S. Lavoie, K. Siegel, D. A. Gaul, M. J. Weissburg and J. Kubanek, Proc. Natl. Acad. Sci. U.S.A., 2018, 115, 662–667 CrossRef CAS PubMed.
- M. Slattery, M. Hamann, J. McClintock, T. Perry, M. Puglisi and W. Yoshida, Mar. Ecol.: Prog. Ser., 1997, 161, 133–144 CrossRef CAS.
- M. Slattery, in Fouling Organisms of the Indian Ocean, ed. R. Nagabhushanam and M.-F. Thompson, CRC Press, 1st edn, 2020, pp. 135–157 Search PubMed.
- M. Rischer, H. Guo and C. Beemelmanns, Nat. Prod. Rep., 2022, 39, 1833–1855 RSC.
- C. B. Naman, S. T. Puthiyedathu, C. C. Poulin and R. X. Poulin, Anal. Sci. Adv., 2023, 4, 319–323 CrossRef PubMed.
- A. Rutz, M. Dounoue-Kubo, S. Ollivier, J. Bisson, M. Bagheri, T. Saesong, S. N. Ebrahimi, K. Ingkaninan, J.-L. Wolfender and P.-M. Allard, Front. Plant Sci., 2019, 10 DOI:10.3389/fpls.2019.01329.
- S. Zuffa, R. Schmid, A. Bauermeister, P. W. P. Gomes, A. M. Caraballo-Rodriguez, Y. El Abiead, A. T. Aron, E. C. Gentry, J. Zemlin, M. J. Meehan, N. E. Avalon, R. H. Cichewicz, E. Buzun, M. C. Terrazas, C.-Y. Hsu, R. Oles, A. V. Ayala, J. Zhao, H. Chu, M. C. M. Kuijpers, S. L. Jackrel, F. Tugizimana, L. P. Nephali, I. A. Dubery, N. E. Madala, E. A. Moreira, L. V. Costa-Lotufo, N. P. Lopes, P. Rezende-Teixeira, P. C. Jimenez, B. Rimal, A. D. Patterson, M. F. Traxler, R. de C. Pessotti, D. Alvarado-Villalobos, G. Tamayo-Castillo, P. Chaverri, E. Escudero-Leyva, L.-M. Quiros-Guerrero, A. J. Bory, J. Joubert, A. Rutz, J.-L. Wolfender, P.-M. Allard, A. Sichert, S. Pontrelli, B. S. Pullman, N. Bandeira, W. H. Gerwick, K. Gindro, J. Massana-Codina, B. C. Wagner, K. Forchhammer, D. Petras, N. Aiosa, N. Garg, M. Liebeke, P. Bourceau, K. B. Kang, H. Gadhavi, L. P. S. de Carvalho, M. Silva dos Santos, A. I. Pérez-Lorente, C. Molina-Santiago, D. Romero, R. Franke, M. Brönstrup, A. Vera Ponce de León, P. B. Pope, S. L. La Rosa, G. La Barbera, H. M. Roager, M. F. Laursen, F. Hammerle, B. Siewert, U. Peintner, C. Licona-Cassani, L. Rodriguez-Orduña, E. Rampler, F. Hildebrand, G. Koellensperger, H. Schoeny, K. Hohenwallner, L. Panzenboeck, R. Gregor, E. C. O'Neill, E. T. Roxborough, J. Odoi, N. J. Bale, S. Ding, J. S. Sinninghe Damsté, X. L. Guan, J. J. Cui, K.-S. Ju, D. B. Silva, F. M. R. Silva, G. F. da Silva, H. H. F. Koolen, C. Grundmann, J. A. Clement, H. Mohimani, K. Broders, K. L. McPhail, S. E. Ober-Singleton, C. M. Rath, D. McDonald, R. Knight, M. Wang and P. C. Dorrestein, Nat. Microbiol., 2024, 9, 336–345 CrossRef CAS PubMed.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang and E. E. Bolton, Nucleic Acids Res., 2023, 51, D1373–D1380 CrossRef PubMed.
- D. Meijer, M. A. Beniddir, C. W. Coley, Y. M. Mejri, M. Öztürk, J. J. J. van der Hooft, M. H. Medema and A. Skiredj, Nat. Prod. Rep., 2025 10.1039/D4NP00008K.
- C. C. Roggatz, M. Saha, S. Blanchard, P. Schirrmacher, P. Fink, F. Verheggen and J. D. Hardege, Global Change Biol., 2022, 28, 4495–4505 CrossRef CAS PubMed.
- L. E. Fleming, P. J. Landrigan, O. S. Ashford, E. M. Whitman, A. Swift, W. H. Gerwick, J. J. Heymans, C. C. Hicks, K. Morrissey, M. P. White, L. Alcantara-Creencia, K. A. Alexander, T. Astell-Burt, R. G. S. Berlinck, P. J. Cohen, R. Hixson, M. M. Islam, A. Iwasaki, R. A. Praptiwi, H. Raps, J. Y. Remy, G. Sowman, E. Ternon, T. Thiele, S. H. Thilsted, J. Uku, S. Ockenden and P. Kumar, Ann. Glob. Health., 2024, 90, 1–28 CrossRef PubMed.
- T. Eisner and J. Meinwald, Proc. Natl. Acad. Sci. U.S.A., 1995, 92, 1 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Table of described exometabolites, with their taxonomic origins. See DOI: https://doi.org/10.1039/d4np00064a |
| ‡ Is now working at: Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 221, 2800 Kgs, Lyngby, Denmark. |
| This journal is © The Royal Society of Chemistry 2025 |