Open Access Article
Open Access ArticleNanoparticle-protein corona enhances accuracy of Ca-19.9-based pancreatic cancer classification†
Luca
Digiacomo‡
a,
Damiano
Caputo‡
bc,
Roberto
Cammarata
c,
Vincenzo
La Vaccara
c,
Roberto
Coppola
c,
Erica
Quagliarini
 a,
Manuela
Iacobini
a,
Serena
Renzi
a,
Francesca
Giulimondi
a,
Daniela
Pozzi
a,
Manuela
Iacobini
a,
Serena
Renzi
a,
Francesca
Giulimondi
a,
Daniela
Pozzi
 *a,
Giulio
Caracciolo
*a,
Giulio
Caracciolo
 *a and
Heinz
Amenitsch
*a and
Heinz
Amenitsch
 d
d
aNanoDelivery Lab, Department of Molecular Medicine, Sapienza University of Rome, Viale Regina Elena, 291, 00161 Rome, Italy. E-mail: daniela.pozzi@uniroma1.it; giulio.caracciolo@uniroma1.it
bResearch Unit of General Surgery, Department of Medicine and Surgery, University Campus Bio-Medico di Roma, Rome, Italy
cOperative Research Unit of General Surgery, Fondazione Policlinico Universitario Campus Bio-Medico, Roma, Italy
dInstitute of Inorganic Chemistry, Graz University of Technology, 8010 Graz, Austria
First published on 20th January 2025
Abstract
Among the various types of pancreatic cancers, pancreatic ductal adenocarcinoma (PDAC) is the most lethal and aggressive, due to its tendency to metastasize quickly and has a particularly low five-year survival rate. Carbohydrate antigen 19-9 (CA 19-9) is the only biomarker approved by the Food and Drug Administration for PDAC and has been a focal point in diagnostic strategies, but its sensitivity and specificity are not sufficient for early and accurate detection. To address this issue, we introduce a synergistic approach combining CA 19-9 levels with a graphene oxide (GO)-based blood test. This non-invasive technique relies on the analysis of personalized protein corona formed on GO sheets once they are embedded in human plasma. Pairing CA 19-9 values with GO protein patterns from N = 106 donors significantly improved the ability to differentiate between non-oncological and PDAC patients (up to 92%), also boosting the classification of PDAC subjects by 50% compared to CA 19-9 testing alone. Overall, this study sought to bridge the existing gaps in PDAC detection by exploiting the complementary strengths of conventional biomarkers and cutting-edge nanotechnology. Exploration of this combined strategy holds promise for advancing the early detection of PDAC, ultimately contributing to improved patient prognosis and treatment outcomes.
Introduction
Pancreatic cancer is one of the leading causes of cancer-related deaths globally1 and has a very poor five-year survival rate among all types of tumors.2 The poor prognosis of pancreatic cancer, especially pancreatic ductal adenocarcinoma (PDAC), can be attributed primarily to its aggressive nature, which leads to local invasion and distant metastases, coupled with the existing challenges in early-stage detection. While formulating strategies for early detection is deemed crucial for enhancing patient survival, the currently available techniques lack sufficient accuracy. Among these, carbohydrate antigen 19-9 (CA 19-9) is one of the most employed tools in clinical practice. CA 19-9 is a cell surface glycoprotein complex produced by ductal cells in the pancreas, biliary system, and epithelial cells in the stomach, colon, uterus, and salivary glands.3 First described between the late 1970s and the early 1980s,4,5 CA 19-9 is present in small amounts in serum6 and is most commonly associated with PDAC. It is the only marker approved by the Food and Drug Administration (FDA) for the management of pancreatic cancer.7 Although CA 19-9 proved its efficacy in predicting several stages of PDAC,8 recommendations from the American Society of Clinical Oncology advise against using CA 19-9 as a screening or diagnostic test. This caution stems from the potential for the test to yield false-negative results in numerous cases or show abnormally elevated levels in healthy subjects.9 Furthermore, although CA 19-9 has been proved to be able to predict the nodal status in PDAC, the current clinical practice requires also other tools to stage PDAC (e.g., CT scan). The reported median sensitivity and specificity of CA 19-9 for pancreatic cancer are closely related to tumor size, reaching approximately 79% and 82%, respectively.10 Furthermore, the accuracy of CA 19-9 for identification of patients with small resectable cancers is limited, and -in addition- CA 19-9 is frequently elevated in patients with cancers other than pancreatic cancer, and various benign pancreaticobiliary disorders. While its key implication is in PDAC, CA 19-9 is also overexpressed in a wide range of benign and malignant gastrointestinal and extra-gastrointestinal diseases.3 Therefore, CA 19-9 is now considered more useful for patient follow-up than for screening and diagnosis.11 To improve its accuracy in PDAC diagnosis and early detection, CA 19-9 has been tested and employed in tandem with other techniques, such as imaging, biopsy procedures, or in panels with other markers. Unfortunately, despite being promising and having obtained exciting results, these panels are far from being used in daily clinical practice, as they require sophisticated and expensive instrumentation and highly skilled personnel.12In this study, we present a synergistic approach that combines CA 19-9 with a nanotechnology-based test. This test was designed to analyze the protein composition of the layer formed on nanomaterials upon exposure to biological fluids such as blood, serum, or plasma. This biomolecular layer, primarily composed of proteins, is commonly referred to as the protein corona.13,14 The features and composition of the protein corona depend on both the synthetic characteristics of the nanomaterials (e.g., size and surface properties)15 and environmental factors (e.g., protein concentration, temperature, and incubation time).16 Furthermore, and more interestingly, the protein corona has been proven to be personalized and disease-specific, and thus contains information about the health status of individual subjects.17–20 Nanomaterials embedded in biological media can act as “accumulators” of proteins to which they have a distinct affinity.11,21 In other words, proteins present in biological media differentially adsorb to the surface of nanoscale objects, due to specific chemical affinity. Proteins with low dissociation rates from the nanomaterial form a tightly bound layer of biomolecules and the composition of this layer does not merely reflect the human proteome.22 However, under specific and controlled conditions, it may be enriched with disease-specific circulating proteins. Thus, even small differences in the corona obtained from healthy and unhealthy samples can be detected, quantified, and used to develop diagnostic tests.23 It is worth noting that the choice of the nanoplatform has a strong impact on the final test outcomes, as protein-nanomaterial affinity is a driving factor shaping the final composition of the corona. In this regard, graphene oxide (GO) is an optimal candidate.21 GO is a unique material that can be viewed as a single monomolecular layer of graphite with various oxygen-containing functionalities.24 Owing to its large specific area, peculiar physical, chemical, and mechanical properties, and high protein-binding ability, GO has been extensively studied in bio-nanotechnology.25 Furthermore, in some of our previous studies, we demonstrated that the personalized protein corona of GO can be employed to design novel tools for PDAC detection.22,26 These tools rely on electrophoretic analyses (i.e., one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)) of the protein corona patterns associated with single individuals. Thus, owing to the cost-effectiveness and robustness of the technique, along with its high levels of sensitivity, specificity, and accuracy, it fulfilled most of the requirements of the World Health Organization (WHO) for cancer screening and diagnostic procedures (i.e., the RE-ASSURED criteria27). Here, we developed a diagnostic test that utilizes the GO protein corona, validated it on a larger dataset (up to 106 patients), and combined the obtained readouts with CA 19-9 values. This resulted in an enhanced classification ability between non-oncological and PDAC subjects as well as a remarkable reduction in false negatives. Specifically, CA 19-9 testing alone resulted in 8 false negatives, whereas the paired test reduced this to 4, achieving an accuracy of 91.8%. The proposed approach demonstrated the possibility of implementing a noninvasive, affordable, and accurate GO-based tool that may support the diagnosis of PDAC, especially in CA 19-9-negative cases.
Results
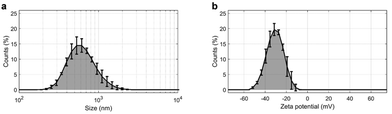
A total number of 48 non-oncological patients (NOP) and 58 subjects suffering from PDAC were included in this study. A complete list of the participants’ demographic and clinical characteristics is provided in Table 1. After blood collection and plasma isolation, samples from each donor were used as protein sources for protein corona formation on the GO sheets. Preliminary characterization of pristine GO in ultrapure water allowed for the determination of the size, polydispersity, and zeta potential of GO sheets. The results are shown in Fig. 1 and summarized in Table 2. Briefly, the measured size of the GO sheets was 602 ± 10 nm, whereas their zeta potential was anionic (i.e. −30.7 ± 0.9 mV) and the solution exhibited a polydispersity index of 0.244 ± 0.009.| Controls (N = 48) | PDAC (N = 58) | |
|---|---|---|
| AJCC indicates American Joint Committee on Cancer; CA 19-9, carbohydrate antigen 19-9. | ||
| Age, median (range) | 54 (23–80) | 71 (43–86) |
| Sex, N (%) | ||
| Male | 24 (50.00%) | 25 (43.10%) |
| Female | 24 (50.00%) | 33 (56.90%) |
| Comorbidity, N (%) | ||
| Cardiac | 1 (2.22%) | 22 (37.92%) |
| Pulmonary | 1 (2.22%) | 15 (25.87%) |
| Diabetes mellitus | 1 (2.22%) | 8 (13.80%) |
| Hypertension | 2 (4.44%) | 13 (22.41%) |
| None | 40 (88.9%) | 0 (0%) |
| Smoking status, N (%) | ||
| Never | 31 (66.67%) | 25 (43.10%) |
| Ex-smoker | 11 (22.22%) | 20 (34.48%) |
| Current | 6 (11.11%) | 13 (22.42%) |
| Tumor stage 8th AJCC edition, N (%) | ||
| I | NA | 18 (31.03) |
| II | NA | 24 (41.38) |
| III | NA | 11 (18.97) |
| IV | NA | 5 (8.62%) |
| CA 19-9 (UI l−1), N (%) | ||
| >37.00 | 0 (0%) | 41 (70.69%) |
| ≤37.00 | 21 (43.75%) | 11 (18.97%) |
| Missing data | 27 (56.25%) | 6 (10.34%) |
| Size (nm) | PdI | Zeta potential (mV) | Zeta deviation (mV) | |
|---|---|---|---|---|
| GO | 602 ± 10 | 0.244 ± 0.09 | −30.7 ± 0.9 | 7.7 ± 0.6 |
These features are fully compatible with previous studies reporting the optimal experimental conditions for PDAC detection by analyzing the personalized protein corona of GO.24
In particular, it has been demonstrated that within 100 and 750 nm, GO lateral size exerts only a marginal influence on the composition of the protein corona.22 Notably, the sizes of GO sheets employed in this study were within this specified range. Thus, GO sheets were exposed to the collected plasma samples (details are provided in the Materials and methods), the resulting protein coronas were isolated by centrifugation (as described in detail in previous studies22,26) and their contents were assessed by 1-dimensional sodium dodecyl sulfate–polyacrylamide gel electrophoresis (1D SDS-PAGE).
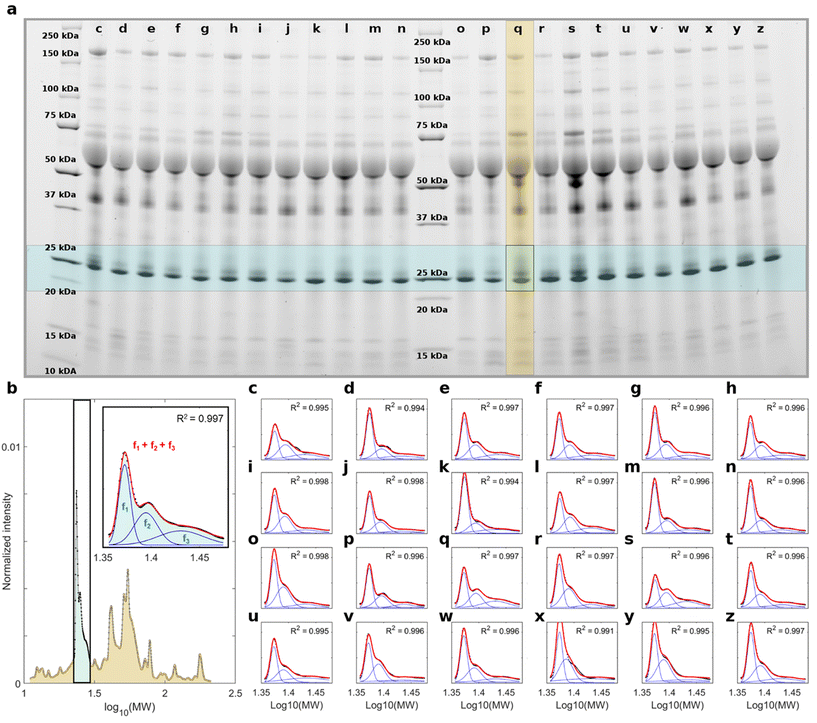
A representative result of the electrophoretic analysis is shown in Fig. 2. Two ladder lanes were used as molecular weight (MW) references, and 24 samples (lanes c–z in Fig. 2a) were loaded onto the gel. Densitometric analysis was performed on each lane, yielding the corresponding protein patterns as normalized intensity distributions, as illustrated in Fig. 2b. Based on previous findings,24,25 we focused on intensity distributions in the MW range of 20–30 kDa. Among the corona proteins adsorbed on GO, those with a MW falling in that specific region have been proven to be the most relevant for PDAC detection by 1D SDS-PAGE.26 The region of interest (ROI) in Fig. 2a is represented by a light blue shaded rectangle and contains a main band at low MW, followed by a secondary band and a weak tertiary component. Consistently, as shown in the inset of Fig. 2b, the intensity profile of the representative lane in the ROI consists of three distinct peaks that partially overlap. Therefore, we aimed to quantify each of the three contributions by fitting the experimental data with a curve equal to the sum of three Gaussian functions, as described in the Materials and methods (eqn (2)). The fitting procedure resulted in a coefficient of determination R2 very close to 1 for all the loaded samples (Fig. 2c–z), specifically 0.991 ≤ R2 ≤ 0.998. This indicates that the computation of the fitting curve and corresponding output parameters were obtained with high reliability. Notably, this achievement was confirmed for all studied samples, which were loaded into five different gels. All gel images can be found in the ESI (Fig. S1†), along with the corresponding densitometric analysis (Fig. S2–S6†).
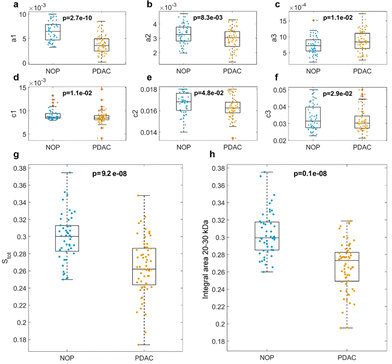
The global NOP and PDAC distributions of the fitting parameters for the three Gaussian components are shown in Fig. 3. In detail, the peak amplitudes are shown in Fig. 3a, b and c, whereas the peak widths are shown in Fig. 3d, e and f, for the first (f1), second (f2), and third (f3) Gaussian functions, respectively.
The peak locations are not shown (but are reported in Fig. S2–S6†), as their values may suffer from undesired but hardly avoidable technical issues. These include (i) an uneven protein band migration for lanes belonging to the same gel (or “smiling effect”, particularly clear for the outer lanes) and (ii) overall differences in protein band migration for lanes belonging to different gels (which are most likely due to fluctuations of applied voltage, temperature, or other environmental factors). Nevertheless, as shown in Fig. 3, the NOP and PDAC distributions exhibited remarkable differences in the amplitudes and widths of the three Gaussian components. Interestingly, when comparing the distributions of amplitudes, p-values from Student's t-test were minimum for a1, which showed larger values for NOP than for PDAC samples. Similarly, when comparing widths, p-values from the Student's t-test were minimal for c1. Globally, these aspects clearly indicate that within the selected ROI, f1 represents the most relevant contribution to the differences between NOP and PDAC samples. Incidentally, among the three Gaussian functions, f1 was also the dominant curve (Fig. 2b and Fig. S2–S6†), and the trend of a1 was reflected by that of Stot (Fig. 3g), that is, the integral areas of the fitting functions within the selected MW range. This is in full agreement with the computed integral areas within 20–30 kDa, as evaluated from the experimental curves (Fig. 3h). In other words, our findings confirmed that the abundance of corona proteins with MW ranging from 20 kDa to 30 kDa were significantly different between NOP and PDAC samples, and in addition, provided a deeper insight by the quantification of the single contributions to the observed global trend.
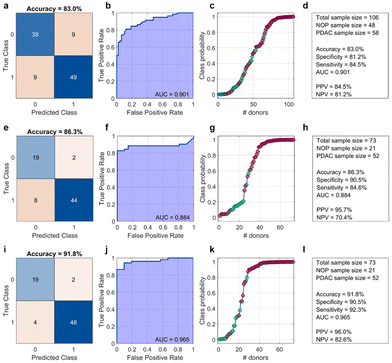
The amplitudes and widths of the primary and secondary bands were used as input variables for binary classification between NOP and PDAC samples by linear discriminant analysis. The obtained outcomes are reported in Fig. 4a–c, in terms of the confusion matrix, Receiver Operating Characteristic (ROC) analysis, and class probability plot, respectively. The corresponding test parameters are listed in Fig. 4d.
As Fig. 4a–d show, a classification test based only on the electrophoretic data reached a global accuracy of 83% (specificity = 81.2%, sensitivity = 84.5%), with an area under the curve (AUC) of the Receiver Operating Characteristic equal to 0.901, positive predictive value (PPV), and negative predictive value (NPV) equal to 84.5% and 81.2%, respectively. Overall, these values are comparable to those obtained by a linear classification that included only CA 19-9 (Fig. 4e–h). Specifically, a CA 19-9-based classification yielded a slightly better accuracy (86.3%) and a lower AUC (0.884), with a remarkable increase in PPV but a significant drop in NPV. Notably, a linear classification analysis that included both electrophoretic data and CA 19-9 values, for patients whose CA-19.9 values were available, resulted in a test with a dramatic improvement in the outcomes, especially global accuracy, sensitivity, and AUC. As the healthy controls were non-oncological patients, CA-19.9 data were only available for those who consented to undergo this assay. The results are shown in Fig. 4(i)–(l), respectively. Among the most interesting aspects, the latter test misclassified only six samples, specifically two NOP and four PDAC samples. In contrast, the CA 19-9-based test misclassified the same number of NOP samples but doubled the number of misclassified PDAC subjects.
The integration of electrophoretic analysis with CA 19-9 values demonstrated a dual benefit: it not only enhanced the overall classification accuracy by providing a more comprehensive diagnostic framework but, more critically, achieved a significant reduction in the number of misclassified PDAC patients.
Discussion
Detecting PDAC in its early stages is crucial in clinical practice and poses significant challenges to the scientific community. PDAC diagnosis typically involves a combination of imaging, endoscopic procedures, and tumor biomarkers. Notably, CA 19-9 is the only biomarker approved by the Food and Drug Administration (FDA) for managing pancreatic cancer.7 However, it exhibits low specificity and sensitivity values.10 Owing to this limitation, potential strategies have been developed to improve the diagnostic accuracy of CA 19-9. For instance, by combining the secretor and Lewis genotypes, the sensitivity of CA19-9 in diagnosing pancreatic cancer was optimized from 80.1% to 88.0%, and the negative predictive value was optimized from 81.2% to 87.1%.10,28 Furthermore, combining CA 19-9 with plasma microRNAs (i.e., miR16 and miR-196a) was found to be more effective than CA 19-9 alone in discriminating pancreatic cancer samples from pancreatitis and normal controls.29 In 2011, researchers at the University of Pittsburgh found that the combination of CA 19-9 with different serum circulating proteins (e.g., Intercellular Adhesion Molecule 1 (ICAM-1), Osteoprotegerin (OPG), and metallopeptidase inhibitor 1 (TIMP1)) obtained promising results in discriminating PDAC from other conditions.30 More recently, Zhang et al. demonstrated that the combination of Mucin-5AC and CA19-9 showed higher accuracy in differentiating PDAC from controls (AUC 0.894; 95% confidence interval (0.844–0.943), sensitivity 0.738, specificity 0.886) than CA19-9 (p = 0.043).31Although these promising approaches require further validation before being adopted in clinical practice, they highlight the potential of combining CA 19-9 with complementary data to significantly enhance the accuracy and utility of CA 19-9 testing. Therefore, in this study, we explored the possibility of employing CA 19-9 in conjunction with a GO-based blood test. GO has been intensively studied in the biomedical field, especially for its large specific surface area, peculiar electrical, chemical, and mechanical properties, and strong protein binding ability.25 This latter aspect is particularly relevant when GO is used as “nanoaccumulator” of biomolecules to detect differences in circulating protein levels, which are specifically associated to PDAC.
With respect to other nanomaterials, GO has been proved to possess an extraordinary ability in adsorbing serum proteins on its surface.25 This is mainly due to the negatively charged oxygenated functional groups at physiological pH, and the hexagonal aromatic graphene structure, which promote hydrogen bonding, electrostatic, hydrophobic, van der Waals, and π–π interactions, thus allowing GO to interact with manifold proteins when it is embedded in biological media.32 As a result, the use of GO in the development of a nanoparticle-based blood test led to improved classification ability26 compared to other nanomaterials, such as gold nanoparticles33 or lipid-based systems.34 In this work, we have consistently used the same GO supplier, and we have further processed the material by sonicating it to reduce its size, which is a standardized step in our procedure. We have also assessed the reproducibility of the entire workflow, from incubation to 1D protein profiling, using both manual and automated approaches. The results showed exceptionally high reproducibility, as reported in one of our previous studies.35
Generally, for diagnostic applications, one of the possible approaches involves exposing nanomaterials to human plasma from non-oncological patients (NOP) and oncological donors, then assessing the compositions of the resulting personalized protein corona, i.e. a disease-specific protein layer that forms on nanomaterials once embedded in biological fluids. Our investigation focused on the 20–30 kDa molecular weight region, as it was identified as the interval exhibiting the most significant differences between PDAC patients and healthy volunteers, guiding our research to this specific range for detailed analysis.26
According to previous mass-spectrometry studies on GO-protein samples from non-oncological and PDAC donors,22 the protein pattern enriching the GO corona in this MW region arises from a delicate, dynamic equilibrium among manifold proteins, including apolipoprotein A1 (which has a high affinity for GO36 and has been recognized as a potential biomarker for PDAC37), immunoglobulin lambda-like polypeptide 5 (IGLL5), and immunoglobulin kappa light chain (IGC).22 Decreased levels of apolipoproteins, including apolipoprotein A1, have been reported in PDAC and proposed as potential biomarkers for this disease. Significant differences in apolipoprotein A1 levels among CA19-9-negative PDAC patients, CA19-9-positive PDAC patients, and healthy controls, with the highest levels observed in the control group. These findings align with the electrophoretic analysis results within the 20–30 kDa range, further supporting the relevance of apolipoprotein A1 in distinguishing disease states. In the present work, three different and partially overlapped bands were recognized within 20–30 kDa (Fig. 2), quantified (Fig. 3), and employed to classify NOP and PDAC samples (Fig. 4). Our results suggest that the amplitude and width of the dominant band are the most relevant parameters for classification. Furthermore, by including the secondary band in the classification analysis, the resulting test exhibited a global accuracy of 83% with an AUC of 0.901. These outcomes are slightly different from those obtained by classification based only on CA 19-9 values (i.e., accuracy = 86%, AUC = 0.884). However, when the electrophoretic data and CA 19-9 values were taken together as input variables, a linear classification yielded largely improved results, reaching an accuracy of 92% and an AUC of 0.965. This represents a noteworthy accomplishment, particularly when considering the absolute numbers of misclassified PDAC subjects, namely four out of 52 for the paired test and eight out of 52 for the CA 19-9-based test. Detecting PDAC at early stages, especially for subjects exhibiting low CA 19-9 values, represents a significant advancement in the accuracy of PDAC diagnosis. Clinically, this improved detection method can lead to earlier and more accurate diagnosis of PDAC, enabling timely treatment and potentially improving patient outcomes. Economically, more accurate early detection can decrease healthcare costs by reducing the need for additional tests and treatments associated with misdiagnosis, and by enabling more effective management of the disease from its onset. From a societal perspective, enhanced diagnostic accuracy reduces the emotional and psychological stress on patients and their families associated with uncertain or incorrect diagnoses. However, the technology still has certain aspects that can be refined and optimized through further research, paving the way for even greater diagnostic precision and clinical applicability.
Table 3 provides a summary of how the proposed test currently meets the World Health Organization's RE-ASSURED criteria for diagnostic tools. So far, the test does not include tools for real-time connection and -due to the need for specific nanomaterials, controlled incubation conditions and instrumentation for protein corona isolation and analysis- it does not fully meet the “equipment-free” requirement. However, the presented GO-based blood test is non-invasive (“Ease of specimen collection”), not expensive (“Affordable”), avoids false negatives (“Sensitive”), avoids false positive (“Specific”), and requires standard laboratory procedures for SDS-PAGE experiment (“User-friendly”). Yet, the time needed for corona formation (1 hour), and isolation (1 hour), SDS-PAGE experiment (1.5 hours), and data processing (30 minutes) is beyond the suggested “rapidity range”, as it is approximately four hours. Therefore, the “Rapidity” requirement has not yet been met by the proposed technology, and it represents an area for potential improvement. Thus, future studies should focus on reducing the total time required for sample treatment, simplifying the necessary equipment to enhance its deliverability to end-users, and exploring its potential for patient follow-up care. This could offer a continuous assessment tool, enabling healthcare providers to tailor treatments more effectively and respond promptly to changes in the patient's condition, thereby optimizing patient outcomes and enhancing personalized care strategies. Finally, we point out that the selection criteria we have identified are the result of a reasoned attempt to eliminate potential biases related to clinical conditions that could influence plasma protein concentrations. However, in choosing these criteria, we also considered that, since the test we propose must be used as a first-level test, it must be applied to subjects who are in a state of “apparent good health” and who therefore would not perform second-level diagnostic tests (e.g. US, CT scan, MRI). A patient with a hematological disease diagnosed by the finding of altered blood tests, such as a patient with a history of neoplasia for example, precisely because of their condition, in common clinical practice would be subjected to second-level tests to reach a diagnosis in the first case and to perform FUP checks in the second.
| RE-ASSURED criteria | Description | Status for GO-based blood test |
|---|---|---|
| Criteria descriptions are reported here as defined in ref. 27. | ||
| R. Real-time connectivity | Tests are connected and/or a reader or mobile phone is used to power the reaction and/or read test results to provide required data to decisionmakers | Not yet met |
| E. Ease of specimen collection | Tests should be designed for use with non-invasive specimens | Met |
| A. Affordable | Tests are affordable to end-users and the health system | Met |
| S. Sensitive | Avoid false negatives | Met |
| S. Specific | Avoid false positives | Met |
| U. User-friendly | Procedure of testing is simple—can be performed in a few steps, requiring minimum training | Met |
| R. Rapid and robust | Results are available to ensure treatment of patient at first visit (typically, this means results within 15 min to 2 hours); the tests can survive the supply chain without requiring additional transport and storage conditions such as refrigeration | Not yet met |
| E. Equipment free or simple | Environmentally friendly Ideally the test does not require any special equipment or can be operated in very simple devices that use solar or battery power. Completed tests are easy to dispose and manufactured from recyclable materials | Unmet |
| D. Deliverable to end-users | Accessible to those who need the tests the most | Not yet met |
Building on the promising results of this technology, we are currently evaluating the most effective strategies for its widespread implementation and distribution. One potential approach involves a centralized model where patient samples are sent to a dedicated laboratory that conducts the test and provides a medical report based on the calculated risk score. Alternatively, a decentralized model could focus on the commercialization of a kit, coupled with user-friendly software for data analysis, potentially operable via a smartphone. In this scenario, a trained operator within the hospital would manage the entire process, including blood collection, test execution, and report generation. Both models aim to seamlessly integrate the GO-based test into clinical workflows, providing clinicians with actionable insights to optimize diagnostic and therapeutic decision-making for individual patients.
Materials and methods
Patients’ enrolment and blood samples collection
A power analysis was performed by using G*power v3.1.9.6 to evaluate the proper sample size for this study. Total effect size was evaluated by considering previous results as a reference26 and read 0.78, alpha error probability was set to 0.01, power equal to 0.95, and allocation ratio NNOP/NPDAC = 1. The power analysis reported a total sample size of approximately 100. Blood samples from 48 healthy donors and 58 PDAC patients were collected and stored according to an established protocol (10/12 ComET CBM and further amendments), which was approved by the Ethical Committee of the University Campus Bio-Medico di Roma. Based on findings from the literature,38 there are observed sex differences in occurrence for PDAC. These sex differences have been considered by including 33 female and 25 male participants, representing approximately 57% and 43% of the study population, respectively. All relevant information regarding the enrolled patients is reported in Table 1. The inclusion criteria for the study were: age >18 years; adequate renal function (creatinine <1.5 mg dL−1, blood urea nitrogen <1.5 × the upper limit); previous personal medical history negative for neoplasticity, renal or liver disease, or blood disorders; no previous chemotherapy or radiotherapy; absence of uncontrolled infections; and written informed consent. All blood samples were collected after the diagnosis of the tumor and before the patient underwent any type of treatment for the disease (e.g., surgical resection, chemotherapy, radiotherapy). Blood plasma was obtained by centrifugation at 37 °C for 15 min at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RCF.
000 RCF.
Preparation of graphene oxide
Graphene oxide was purchased from Graphenea (San Sebastián, Spain). GO water dispersion at 0.25 mg ml−1 was subjected to rapid sonication of 2 min at 125 W (Vibra cell sonicator VC505, Sonics and Materials, United Kingdom) to obtain homogeneous GO solutions.Size and zeta potential experiments
GO was characterized in terms of size and zeta potential using a NanoZetaSizer apparatus (Malvern, United Kingdom) equipped with a 633 nm He–Ne laser and a digital logarithmic correlator. Measurements were performed in ultrapure water using Malvern micro cuvettes (ZEN0040) for size and a Dip Cell Kit (ZEN1002) for zeta potential. The results were reported as the average standard deviation of three independent measurements.Preparation of GO-protein corona complexes
GO-protein mixtures were obtained by incubating 100 μL of GO nanoflakes (0.25 mg ml−1) with 5 μL of 10-fold diluted human plasma from healthy donors and PDAC patients for 60 min at 37 °C, as reported elsewhere.22 After incubation, samples were centrifuged at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620 RCF for 15 min at 4 °C. To remove free proteins, the pellet was washed with ultrapure water. This procedure was repeated three times, and the last pellet, containing GO-protein corona complexes, was used for the next PC analysis through sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The total time required for protein corona formation (incubation) and isolation (centrifugation steps) was approximately 2 hours.
620 RCF for 15 min at 4 °C. To remove free proteins, the pellet was washed with ultrapure water. This procedure was repeated three times, and the last pellet, containing GO-protein corona complexes, was used for the next PC analysis through sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The total time required for protein corona formation (incubation) and isolation (centrifugation steps) was approximately 2 hours.
1D SDS-PAGE experiments
After washing, the pellet containing GO-protein corona complexes was resuspended in 20 μL of Laemmli loading buffer 1×, boiled at 100 °C for 10 min, and centrifuged at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620 RCF for 15 min at 4 °C. Subsequently, 10 μL of supernatant was collected and loaded on a stain-free gradient polyacrylamide gel (4–20% TGX precast gels, BioRad, Hercules, CA, United States), which was run at 150 V for approximately 90 min. Gel images were acquired using a ChemiDocTM imaging system (BioRad, Hercules, CA, United States) and processed using ImageLab Software and custom Matlab (MathWorks, Natick, MA, United States) scripts to evaluate the one-dimensional intensity distribution function of each sample and compute the corresponding one-dimensional molecular weight (MW) distribution. Further details can be found in previous works,22,39 the employed tools for gel image analysis are provided as a supplementary software in ref. 40. Finally, a detailed analysis of the normalized intensity distributions was performed within a 20–30 kDa-range. Specifically, by considering z as an independent variable,
620 RCF for 15 min at 4 °C. Subsequently, 10 μL of supernatant was collected and loaded on a stain-free gradient polyacrylamide gel (4–20% TGX precast gels, BioRad, Hercules, CA, United States), which was run at 150 V for approximately 90 min. Gel images were acquired using a ChemiDocTM imaging system (BioRad, Hercules, CA, United States) and processed using ImageLab Software and custom Matlab (MathWorks, Natick, MA, United States) scripts to evaluate the one-dimensional intensity distribution function of each sample and compute the corresponding one-dimensional molecular weight (MW) distribution. Further details can be found in previous works,22,39 the employed tools for gel image analysis are provided as a supplementary software in ref. 40. Finally, a detailed analysis of the normalized intensity distributions was performed within a 20–30 kDa-range. Specifically, by considering z as an independent variable,| z = log10(MW) | (1) |
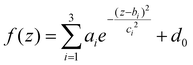 | (2) |
Conclusions
In conclusion, we demonstrated the potential of employing a GO-based blood test in conjunction with CA 19-9 testing. Our approach successfully paired CA 19-9 levels with electrophoretic analysis of the personalized protein corona formed on GO sheets to classify NOP and PDAC subjects. The paired test exhibited high values of accuracy (92%), specificity (91%), sensitivity (92%), and AUC (0.965) and performed significantly better than a classification based only on CA 19-9. In addition, the number of patients with misclassified PDAC was reduced by 50%. Furthermore, it is worth noting that the electrophoretic approach is a noninvasive tool that satisfies most of the WHO guidelines for cancer screening and detection procedures, as it is affordable, sensitive, specific, user-friendly, rapid and robust, simple, and deliverable to end-users. Our laboratory is currently dedicated to developing innovative protocols designed to substantially reduce the exposure time of nanomaterials to human plasma (ideally to as little as 1 minute) and to eliminate the requirement for protein isolation. For these reasons, this study may represent a further step towards the successful integration of GO-based blood tests into clinical practice as supportive diagnostic tools, particularly for CA 19-9-negative PDAC patients. This approach can bridge the existing gaps in diagnostic methodologies, providing a diagnostic tool that is both more sensitive and personalized. This tool has the potential to considerably enhance patient outcomes by facilitating earlier interventions and treatments. Upon validation in a broader cohort, this innovative strategy signifies a significant advancement in the precision of PDAC diagnosis, with extensive implications for patient care, efficiency of healthcare systems, and wider societal and economic dimensions of managing cancer.Author contributions
Conceptualization L. D., D. C., D. P. and G. C.; methodology, L. D., D. C., D. P. and G. C.; software, L. D.; validation, L. D., R. Co, E. Q., S. R., F. G., H. A., and G. C.; formal analysis, L. D., R. Ca., H. A.; investigation, D. C., R. Co., G. C. and D. P.; resources, D. P.; data curation, L. D. and D. C.; writing original draft preparation, L. D., D. C., D. P. and G. C.; writing—review and editing, L. D., D. C., D. P., and G. C.; visualization, L. D. and D. C.; supervision, D. C., G. C. and D. P.; project administration, D. P.; funding acquisition, D. P. All authors have read and agreed to the published version of the manuscript.Ethics approval and consent to participate
The Ethical Committee of the University Campus Bio-Medico di Roma approved this study (Prot. 10/12 ComEt CBM).Consent for publication
All the authors have approved the manuscript for submission.Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This work was supported by the AIRC Foundation under the IG 2020-ID. 24521 project, P.I. Pozzi Daniela.References
- F. Bray, et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- R. L. Siegel, K. D. Miller, H. E. Fuchs and A. Jemal, Cancer statistics, 2021, CA Cancer J. Clin., 2021, 71, 7–33 CrossRef PubMed.
- T. Lee, T. Z. J. Teng and V. G. Shelat, Carbohydrate antigen 19-9—Tumor marker: Past, present, and future, World J. Gastrointest. Surg., 2020, 12, 468 CrossRef PubMed.
- H. Koprowski, et al., Colorectal carcinoma antigens detected by hybridoma antibodies, Somatic Cell Genet., 1979, 5, 957–971 CrossRef CAS PubMed.
- A. Azizian, et al., CA19-9 for detecting recurrence of pancreatic cancer, Sci. Rep., 2020, 10, 1332 CrossRef CAS PubMed.
- S. Scarà, P. Bottoni and R. Scatena, CA 19-9: biochemical and clinical aspects, in Advances in Cancer Biomarkers: From Biochemistry to Clinic for a Critical Revision, 2015, pp. 247–260 Search PubMed.
- B. George, M. Kent, A. Surinach, N. Lamarre and P. Cockrum, The association of real-world CA 19-9 level monitoring patterns and clinical outcomes among patients with metastatic pancreatic ductal adenocarcinoma, Front. Oncol., 2021, 11, 754687 CrossRef PubMed.
- A. Coppola, et al., CA19. 9 serum level predicts lymph-nodes status in resectable pancreatic ductal adenocarcinoma: a retrospective single-center analysis, Front. Oncol., 2021, 11, 690580 CrossRef CAS PubMed.
- A. Dasgupta and A. Wahed, Clinical chemistry, immunology and laboratory quality control: a comprehensive review for board preparation, certification and clinical practice, 2013 Search PubMed.
- G. Luo, et al., Roles of CA19-9 in pancreatic cancer: Biomarker, predictor and promoter, Biochim. Biophys. Acta, Rev. Cancer, 2021, 1875, 188409 CrossRef CAS PubMed.
- L. Digiacomo, et al., Stratifying Risk for Pancreatic Cancer by Multiplexed Blood Test, Cancers, 2023, 15, 2983 CrossRef PubMed.
- R. S. O'Neill and A. Stoita, Biomarkers in the diagnosis of pancreatic cancer: Are we closer to finding the golden ticket?, World J. Gastroenterol., 2021, 27, 4045 CrossRef PubMed.
- T. Kopac, Protein corona, understanding the nanoparticle–protein interactions and future perspectives: A critical review, Int. J. Biol. Macromol., 2021, 169, 290–301 CrossRef CAS PubMed.
- T. Cedervall, et al., Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed.
- M. Lundqvist, et al., Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS PubMed.
- M. Mahmoudi, M. P. Landry, A. Moore and R. Coreas, The protein corona from nanomedicine to environmental science, Nat. Rev. Mater., 2023, 1–17 Search PubMed.
- M. J. Hajipour, S. Laurent, A. Aghaie, F. Rezaee and M. Mahmoudi, Personalized protein coronas: a “key” factor at the nanobiointerface, Biomater. Sci., 2014, 2, 1210–1221 RSC.
- M. J. Hajipour, et al., Personalized disease-specific protein corona influences the therapeutic impact of graphene oxide, Nanoscale, 2015, 7, 8978–8994 Search PubMed.
- E. Quagliarini, L. Digiacomo, S. Renzi, D. Pozzi and G. Caracciolo, A decade of the liposome-protein corona: Lessons learned and future breakthroughs in theranostics, Nano Today, 2022, 47, 101657 CrossRef CAS.
- V. Colapicchioni, et al., Personalized liposome–protein corona in the blood of breast, gastric and pancreatic cancer patients, Int. J. Biochem. Cell Biol., 2016, 75, 180–187 Search PubMed.
- E. Quagliarini, D. Pozzi, F. Cardarelli and G. Caracciolo, The influence of protein corona on Graphene Oxide: implications for biomedical theranostics, J. Nanobiotechnol., 2023, 21, 267 CrossRef CAS PubMed.
- R. Di Santo, et al., Personalized Graphene Oxide-Protein Corona in the Human Plasma of Pancreatic Cancer Patients, Front. Bioeng. Biotechnol., 2020, 8, 491 CrossRef PubMed.
- E. Quagliarini, R. Di Santo, D. Pozzi and G. Caracciolo, Protein corona-enabled serological tests for early stage cancer detection, Sens. Int., 2020, 1, 100025 CrossRef.
- S. C. Ray, Application and uses of graphene oxide and reduced graphene oxide, in Applications of Graphene and Graphene-Oxide based Nanomaterials, 2015, vol. 6, pp. 39–55 Search PubMed.
- S. Kumar and S. H. Parekh, Linking graphene-based material physicochemical properties with molecular adsorption, structure and cell fate, Commun. Chem., 2020, 3, 8 CrossRef CAS PubMed.
- M. Papi, et al., Converting the personalized biomolecular corona of graphene oxide nanoflakes into a high-throughput diagnostic test for early cancer detection, Nanoscale, 2019, 11, 15339–15346 RSC.
- K. J. Land, D. I. Boeras, X.-S. Chen, A. R. Ramsay and R. W. Peeling, REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes, Nat. Microbiol., 2019, 4, 46–54 CrossRef CAS PubMed.
- G. Luo, et al., Optimize CA19-9 in detecting pancreatic cancer by Lewis and Secretor genotyping, Pancreatology, 2016, 16, 1057–1062 CrossRef CAS PubMed.
- J. Liu, et al., Combination of plasma microRNAs with serum CA19–9 for early detection of pancreatic cancer, Int. J. Cancer, 2012, 131, 683–691 CrossRef CAS PubMed.
- F. N. Al-Shaheri, et al., Blood biomarkers for differential diagnosis and early detection of pancreatic cancer, Cancer Treat. Rev., 2021, 96, 102193 CrossRef CAS PubMed.
- J. Zhang, et al., Evaluation of serum MUC5AC in combination with CA19-9 for the diagnosis of pancreatic cancer, World J. Surg. Oncol., 2020, 18, 1–7 CrossRef PubMed.
- M. Sopotnik, et al., Comparative study of serum protein binding to three different carbon-based nanomaterials, Carbon, 2015, 95, 560–572 CrossRef CAS.
- L. Digiacomo, et al., Efficient pancreatic cancer detection through personalized protein corona of gold nanoparticles, Biointerphases, 2021, 16, 011010 CrossRef CAS PubMed.
- D. Caputo, et al., A protein corona-enabled blood test for early cancer detection, Nanoscale, 2017, 9, 349–354 RSC.
- F. Giulimondi, et al., Reproducibility of Biomolecular Corona Experiments: A Primer for Reliable Results, Part. Part. Syst. Charact., 2023, 40, 2200169 CrossRef.
- V. Castagnola, et al., Biological recognition of graphene nanoflakes, Nat. Commun., 2018, 9, 1577 CrossRef CAS PubMed.
- C. Lin, W.-C. Wu, G.-C. Zhao, D.-S. Wang, W.-H. Lou and D.-Y. Jin, ITRAQ-based quantitative proteomics reveals apolipoprotein AI and transferrin as potential serum markers in CA19-9 negative pancreatic ductal adenocarcinoma, Medicine, 2016, 95(31), e4527 CrossRef CAS PubMed.
- R. Nipp, et al., Disparities in cancer outcomes across age, sex, and race/ethnicity among patients with pancreatic cancer, Cancer Med., 2018, 7, 525–535 CrossRef PubMed.
- L. Digiacomo, et al., The biomolecular corona of gold nanoparticles in a controlled microfluidic environment, Lab Chip, 2019, 19, 2557–2567 RSC.
- S. Renzi, et al., Structuring lipid nanoparticles, DNA, and protein corona into stealth bionanoarchitectures for in vivo gene delivery, Nat. Commun., 2024, 15, 9119 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: SDS-PAGE outcomes and data analysis for all the investigated samples (Fig. S1–S11). See DOI: https://doi.org/10.1039/d4nr02435d |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2025 |