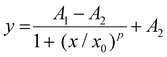Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bioconjugates of photon-upconversion nanoparticles with antibodies for the detection of prostate-specific antigen and p53 in heterogeneous and homogeneous immunoassays†
Ekaterina
Makhneva‡
a,
Pavel
Špaček‡
 a,
Antonín
Hlaváček
*b,
Julie
Weisová
b,
Hans H.
Gorris
a,
Antonín
Hlaváček
*b,
Julie
Weisová
b,
Hans H.
Gorris
 a,
Petr
Skládal
a and
Zdeněk
Farka
a,
Petr
Skládal
a and
Zdeněk
Farka
 *a
*a
aDepartment of Biochemistry, Faculty of Science, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic. E-mail: farka@mail.muni.cz
bInstitute of Analytical Chemistry of the Czech Academy of Sciences, Veveří 967/97, 602 00 Brno, Czech Republic. E-mail: hlavacek@iach.cz
First published on 31st March 2025
Abstract
Sensitive immunoassays for the detection of tumor biomarkers play an important role in the early diagnosis and therapy of cancer. Using luminescent nanomaterials as labels can significantly improve immunoassay performance, especially in terms of sensitivity. Lanthanide-doped photon-upconversion nanoparticles (UCNPs) are nanocrystals capable of converting near-infrared radiation into visible light, and their emission spectra can be tuned by altering the dopant ions. In this study, the bioconjugation between UCNPs and biomolecules was optimized, and different conjugates of Er- and Tm-doped UCNPs (NaYF4:Yb3+,Er3+ and NaYF4:Yb3+,Tm3+) were prepared for detecting prostate-specific antigen (PSA) and tumor protein p53, comparing heterogeneous and homogeneous assay formats. The heterogeneous sandwich immunoassay achieved detection limits of 1.3 pg mL−1 for PSA and 330 pg mL−1 for p53. The homogeneous immunoassays were based on massively parallel spectroscopy (MPS), a novel artificial intelligence-aided single-molecule approach, utilizing conjugates of two different monoclonal antibodies with Er- and Tm-doped UCNPs, respectively. The conjugates provide distinct emission spectra, with only sandwich immunocomplexes of analytes and both types of labels showing signals at the same location. MPS was suitable for observing the immunocomplexes in an aqueous dispersion using only a small sample volume. This innovative method achieved detection limits of 8.2 ng mL−1 and 390 pg mL−1 for PSA and p53, respectively. MPS eliminates the need for time-consuming washing steps required in heterogeneous immunoassays and is amenable to high-throughput applications.
1 Introduction
Immunochemical assays stand out as the predominant and highly sensitive group of methods renowned for their widespread application in clinical diagnostics.1 Since a wide range of clinically relevant molecules can be identified and quantified, immunoassays have become a cornerstone of modern medical laboratories and are indispensable for the detection of various diagnostic markers.2 In particular, the sensitive detection of cancer biomarkers enables early disease diagnosis and the following monitoring of therapy progress. Therefore, several immunoassay formats for cancer biomarker detection have emerged, including enzyme-linked immunosorbent assay (ELISA), radioimmunoassay, fluorescence immunoassay, chemiluminescence immunoassay, and electrochemiluminescence immunoassay.3 Typically, these assays target specific molecular indicators associated with cancer initiation, progression, and response to treatment. Prostate-specific antigen (PSA) and tumor protein p53 are important cancer biomarkers. Serum PSA levels increase in prostate cancer patients; the cut-off values generally range from 2 to 4 ng mL−1.4 Furthermore, PSA levels exceeding 0.2 ng mL−1 in post-radical prostatectomy patients indicate a recurrence of the disease.5 Protein p53 is a crucial tumor suppressor, and its function is deregulated in most human cancers.6 The serum concentration of p53 in healthy individuals is approximately 0.2 ng mL−1 and rises significantly in many malignancies, including several gastrointestinal cancers,7 lung cancer,8 and pancreatic carcinoma.9 In addition, the assessment of p53 levels in cell lysates is also clinically relevant.10ELISA employs bioconjugates of enzymes to produce easily detectable products and, because of its great versatility, is considered the gold standard of immunoassays.11 However, enzymes are prone to denaturation,12 which is associated with their low shelf life,13 and the detection limits (LODs) of ELISA are often insufficient for the detection of low-abundance biomarkers.14 A popular strategy to overcome these drawbacks involves exchanging the conventionally used labels with different nanomaterials, such as noble metal nanoparticles,15,16 quantum dots,17,18 and photon-upconversion nanoparticles (UCNPs).19–21
UCNPs are lanthanide-doped nanocrystals that stand out among the nanoparticle-based labels due to their unique properties, such as excellent photostability and low background interference. UCNPs convert near-infrared radiation into light of a shorter wavelength due to sequential photon absorption, and their emission spectra can be easily tuned by altering the dopant composition.22 Furthermore, core/shell UCNP structures were shown to improve the emission brightness by reducing surface quenching or boosting the upconversion efficiency.23 For their use as immunoassay labels, UCNPs must be conjugated with specific biorecognition molecules, such as streptavidin or antibodies. Before binding the biomolecules, the UCNP surface is usually modified to reduce non-specific interactions, thus decreasing the immunoassay background levels.24 Streptavidin-modified UCNPs allow for applications based on biotinylated detection antibodies,25,26 whereas modification of UCNPs with specific anti-analyte antibodies enables direct binding to the analyte. The utilization of UCNPs enables analog27,28 and digital11,29 readout modes. In the analog readout, the integrated upconversion luminescence intensity is measured. In the digital (single-molecule) readout, the number of UCNP labels is counted, typically via upconversion microscopy, allowing for reaching lower LODs compared to the analog mode.30
A heterogeneous microtiter plate (MTP)-based immunoassay utilizing UCNP labels – upconversion-linked immunosorbent assay (ULISA) – has shown the ability to reach significantly lower LODs compared to the conventional ELISA.14,31 Overall, heterogeneous assays offer high specificity and sensitivity due to separation and washing steps, which, however, are time-consuming. Besides the heterogeneous format,32,33 the use of UCNPs allows for the design of homogeneous immunoassays. Promising examples of homogeneous immunoassays utilizing UCNPs include detection based on Förster resonance energy transfer28 and upconversion cross-correlation spectroscopy (UCCS).34,35 Compared to heterogeneous immunoassays, homogeneous immunoassays are generally less time-consuming but are typically limited by lower specificity and sensitivity.36,37
In this study, heterogeneous (ULISA) and homogeneous (massively parallel spectroscopy, MPS) immunoassays employing antibody-conjugated UCNPs as labels were developed for the detection of two cancer biomarkers, PSA and p53. In the initial optimization steps, human serum albumin (HSA) was used as a model analyte because of the affordability of the respective immunoreagents and its clinical relevance (HSA is a biomarker of several kidney and liver diseases38). The process of bioconjugation of UCNPs with antibodies was systematically studied, comparing different conjugates of UCNPs with polyclonal anti-HSA antibodies in terms of their performance in ULISA. After the optimal bioconjugation conditions were established, the conjugates of UCNPs with polyclonal anti-PSA and anti-p53 antibodies were prepared and employed in ULISA, and their performance was compared with streptavidin-modified UCNPs. Subsequently, UCNP conjugates of different monoclonal anti-PSA and anti-p53 antibodies were synthesized, and their functionality was verified in ULISA for PSA and p53. Finally, the UCNPs modified with monoclonal antibodies were utilized for the first time in a novel homogeneous single-molecule immunoassay format of MPS. This method relies on the simultaneous detection of two distinct UCNP labels bound to a single analyte molecule at the same time. The analyte is incubated with both UCNP labels, a small portion of the mixture is applied onto a glass slide, and the dispersion is recorded via upconversion microscopy. Since a diffraction prism is inserted in front of the detector, the sandwich immunocomplexes appear as double spots, the number of which is evaluated utilizing artificial intelligence. In our previous study,39 a model bioaffinity assay for biotin was designed, and MPS was shown to be feasible. Here, we have advanced the method and developed MPS-based immunoassays for the detection of PSA and p53. This homogeneous format combines shorter analysis time with low sample consumption and shows the potential to evolve into a high-throughput and convenient method for biomarker detection.
2 Materials and methods
2.1 Chemicals and materials
PSA standard (ab78528), monoclonal anti-PSA antibody (ab403), and monoclonal anti-p53 antibody (ab1101) were purchased from Abcam (UK). Polyclonal anti-PSA antibody (AF1344), biotinylated polyclonal anti-PSA antibody (BAF1344), monoclonal anti-PSA antibody (MAB1344), polyclonal anti-p53 antibody (AF1355), biotinylated polyclonal anti-p53 antibody (BAF1355), monoclonal anti-p53 antibody (MAB1355), and p53 protein standard (SP450) were obtained from R&D Systems (USA). HSA, polyclonal anti-HSA antibody (A0433), tris(hydroxymethyl)aminomethane (Tris), copper(II) sulfate pentahydrate, Tween 20, L-ascorbic acid sodium salt, nitrosyl tetrafluoroborate (NOBF4), tris(3-hydroxypropyltriazolylmethyl)amine (THPTA), and a Float-A-Lyzer G2 dialysis device (molecular weight cut-off, MWCO, of 300 kDa) were obtained from Sigma-Aldrich (Germany). Monoclonal anti-HSA antibody (clone AL-01) was purchased from Exbio (Czech Republic). SuperBlock TBS solution was purchased from Thermo Fisher Scientific (USA). All other chemicals were obtained in the highest quality available from Carl Roth (Germany) or Penta (Czech Republic). The 96-well polystyrene transparent MTPs with a flat bottom (Microlon, high-binding) were obtained from Greiner Bio-One (Austria).Buffers included phosphate buffer (PB; 50 mM NaH2PO4/Na2HPO4; pH 7.4), washing buffer (50 mM NaH2PO4/Na2HPO4, 0.05% NaN3, 0.01% Tween 20; pH 7.5), blocking buffer (20% SuperBlock in washing buffer), coating buffer (50 mM NaHCO3/Na2CO3, 0.05% NaN3; pH 9.6), assay buffer (50 mM NaH2PO4/Na2HPO4, 150 mM NaCl, 1 mM KF, 10% SuperBlock, 0.05% NaN3, 0.01% Tween 20; pH 7.5), and dialysis buffer (50 mM Tris, 0.05% NaN3, 1 mM KF; pH 7.5).
The synthesis and characterization of the UCNPs (NaYF4:Yb3+,Er3+ with a diameter of 60 nm and NaYF4:Yb3+,Tm3+ with a diameter of 53 nm) are described in our previous publications.25,39,40 The protocol for bioconjugation of Er-doped UCNPs with streptavidin is also available in our previous publication.27 The protocols for the preparation of azide-modified biomolecules, surface functionalization of the UCNPs, and bioconjugation of Er-doped UCNPs with polyclonal anti-HSA antibody, as well as transmission electron microscopy (TEM) images, emission spectra of the oleic acid-capped UCNPs, and dynamic light scattering (DLS) characterization of the UCNP conjugates and intermediate bioconjugation products, are provided in the ESI.†
2.2 Conjugation of Er- and Tm-doped UCNPs with anti-PSA and anti-p53 antibodies
After the surface functionalization resulting in alkyne-modified UCNPs (described in the ESI†), Cu(I)-catalyzed azide–alkyne cycloaddition was used to conjugate the UCNPs with different antibodies, enabling the detection of PSA and p53. First, 20 μL of Tris-HCl (375 mM in water; pH 7.5), 5 μL of aqueous CuSO4/THPTA solution (25 mM/125 mM), and 2 mg of UCNP-Ner-PEG-alkyne in 1 mM KF were added to the reaction flask. The mixture was purged with Ar for 30 min, and 80 μL of antibody-azide solution (0.5 mg mL−1) in PB was added. After purging the mixture with Ar for another 15 min, 10 μL of an aqueous solution of sodium ascorbate (50 mM) was added to initiate the click reaction. The mixture was kept under Ar purging for 40 min. Finally, the mixture was dialyzed utilizing a Float-A-Lyzer G2 device (300 kDa MWCO) against 5 L of dialysis buffer, which was exchanged nine times. The resulting conjugate was stored at 4 °C.2.3 ULISA for HSA detection
A 96-well high-binding MTP was coated with a monoclonal anti-HSA antibody (AL-01; 1 μg mL−1 in coating buffer, 100 μL per well, 18 h at 4 °C). After the coating, the MTP was washed four times using a HydroFlex washer (Tecan, Switzerland); each washing step included adding 250 μL of washing buffer, followed by liquid aspiration in each well. The MTP was then blocked using the blocking buffer (200 μL per well, 1 h at room temperature, shaking at 200 rpm). Afterward, the MTP was washed four times, and serial HSA dilutions (10−3 to 103 ng mL−1) in assay buffer were added to the wells (100 μL per well) and incubated for 1 h at room temperature at 200 rpm shaking. Then, the plate was washed four times and incubated with 6.5 μg mL−1 of Er-doped UCNPs conjugated with polyclonal anti-HSA antibody. After the last incubation step, the MTP was washed four times and left to dry, followed by measuring upconversion luminescence.2.4 ULISA for PSA and p53 detection
A 96-well high-binding MTP was coated with a monoclonal anti-PSA or anti-p53 antibody (ab403 or MAB1344 in the case of PSA and ab1101 or MAB1355 in the case of p53; 1 μg mL−1 in coating buffer, 100 μL per well, 18 h at 4 °C). After the coating, the MTP was washed four times using the same procedure as in the case of ULISA for HSA. The MTP was then blocked with the blocking buffer (200 μL per well, 1 h at room temperature, shaking at 200 rpm). Afterward, the MTP was washed four times, and serial PSA or p53 dilutions (10−4 to 102 ng mL−1 in the case of PSA and 10−3 to 103 ng mL−1 in the case of p53) in assay buffer were added to the wells (100 μL per well) and incubated for 1 h at room temperature at 200 rpm shaking. After washing four times, the following steps differed due to the use of varying UCNP conjugates: Er-doped UCNPs modified with streptavidin (UCNP–SA) or polyclonal antibodies (UCNP–pAb) and Er- or Tm-doped UCNPs modified with monoclonal antibodies (UCNP–mAb).In the case of UCNP–SA, the analyte binding step was followed by the incubation with biotinylated polyclonal antibody (BAF1344 in the case of PSA and BAF1355 in the case of p53; 0.25 μg mL−1 in assay buffer, 100 μL per well, 1 h at room temperature, shaking at 200 rpm). Then, the MTP was washed four times, and the dispersion of Er-doped UCNP–SA was added (6.5 or 3.3 μg mL−1, 100 μL per well). In the case of UCNP–pAb conjugates, the incubation with the analyte was followed by the addition of the conjugate of Er-doped UCNPs with polyclonal anti-PSA or anti-p53 antibody (concentration of 6.5 or 3.3 μg mL−1). In the case of UCNP–mAb conjugates for PSA detection, the MTP was incubated with Er- or Tm-doped UCNPs modified with MAB1344 or ab403 in a concentration of 6.5 μg mL−1. For p53 protein detection, Er- or Tm-doped UCNPs modified with MAB1355 or ab1101 were used in a concentration of 6.5 μg mL−1. In each case, the MTP was washed four times after the last incubation and left to dry, and the upconversion luminescence was measured.
2.5 Upconversion luminescence scanning
The upconversion luminescence was measured using the UPCON S-Pro reader (Labrox, Finland) equipped with a 980 nm laser excitation source and a 976/30 nm excitation filter. The emission of Er-doped UCNPs was measured utilizing the D800 dichroic mirror and 540/25 nm emission filter. In the case of Tm-doped UCNPs, the emission was measured utilizing the D900 dichroic mirror and 810/40 nm emission filter. In each well, 64 points (8 × 8 square matrix) were raster-scanned using a step size of 100 μm and a signal integration time of 500 ms. The truncated average of luminescence intensities was calculated by excluding the 16 highest and the 16 lowest values, serving as one data point.2.6 Massively parallel spectroscopy (MPS) for PSA and p53 detection
In the MPS-based immunoassay, the analyte was detected by a pair of two monoclonal antibodies, each conjugated to a spectrally distinct type of UCNP (Er- or Tm-doped, respectively). Serial analyte dilutions were prepared in assay buffer, with the concentration range of 10−3 to 105 ng mL−1 for PSA and 0.03 to 100 nM for p53. The analyte solutions were then mixed with the dispersions of Er- and Tm-doped UCNPs modified with corresponding monoclonal antibodies (20 μL of the analyte solution, 10 μL of Er-doped UCNPs, and 10 μL of Tm-doped UCNPs). The mixtures were incubated for 3 h at room temperature on a 3D shaker and then diluted 30× in assay buffer. To sample the dispersion, approximately 1.5 cm × 1.5 cm square of double-sided tape was fixed to a standard microscope glass slide, and a “window” was made by cutting out an approximately 1 cm × 1 cm square. Subsequently, 2 μL of the diluted assay mixture was applied in the “window” and covered with a 170 μm thick glass slide, which adhered to the tape. This assembly resulted in an approximately 80 μm thick layer of the sample dispersion.A laboratory-built epiphoton-upconversion microscope39 was used for MPS analysis. After applying immersion oil onto the cover glass, the microscope objective (60× magnification, 1.40 numerical aperture; Nikon, Japan) was brought into contact with it, and the focal plane was situated ∼10 μm deep in the investigated dispersion. The nanoparticles were excited using a 976 nm laser (Roithner Lasertechnik, Austria); the emission of Er-doped UCNPs was detected at 660 nm, and the emission of Tm-doped UCNPs was detected at 802 nm. The monitored wavelength of Er-doped UCNPs differed from the MTP reader measurements to enable the assessment of the emissions of both types of UCNPs simultaneously using only one emission filter (600 nm long-pass filter). An optical prism was placed in front of the camera sensor, leading to a dispersion of emission. Therefore, if both Er- and Tm-doped UCNPs were bound to the analyte molecule, it appeared as a double spot. For a standard MPS experiment, 1000 images with dimensions of 1024 px × 1024 px (111 μm × 111 μm in the sample plane) using a 10 ms exposure time and 100 ms interval between the images were recorded. To process the images, a convolutional neural network of U-net architecture (Fig. S1†) was trained to detect the double spots as described previously,39 and the number of the sandwich immunocomplexes per experiment was counted.
2.7 Statistical data evaluation
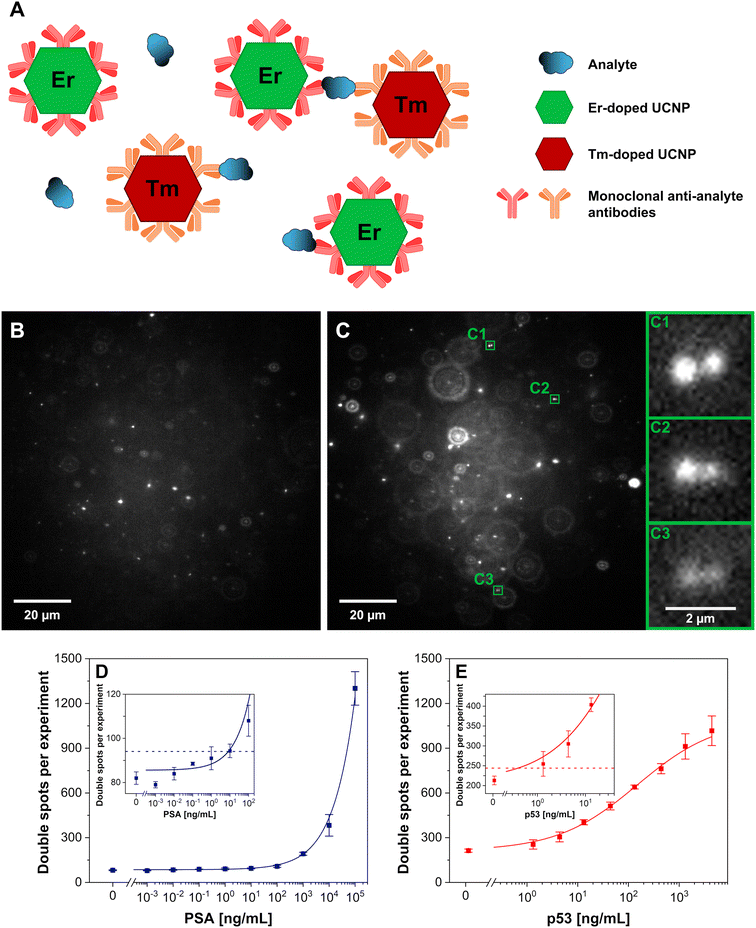
The averages and standard deviations were determined from three independent wells or three independent MPS measurements for ULISA and MPS, respectively. OriginPro 2022 (OriginLab, USA) was used for the regression analysis using a four-parameter logistic function:where y corresponds to upconversion luminescence or the number of MPS double spots per experiment (1000 images), A1 represents the minimum asymptote of the logistic curve, A2 is the maximum asymptote of the curve, x corresponds to the analyte concentration, x0 is the inflection point of the curve, and p is the slope at the inflection point.
The LODs were estimated from the regression curves as concentrations corresponding to the y(LOD) value:
| y(LOD) = A1 + 3 × s0 |
Signal-to-background ratios (S/B) were calculated by dividing the average signal value of the highest concentration by the average signal of the blank. Working ranges were estimated as the intervals between the EC20 and EC80 of the four-parameter logistic curve.
3 Results and discussion
3.1 Optimization of the conjugation of UCNPs with antibodies
To be utilized as immunoassay labels, UCNPs must be conjugated with biorecognition molecules, typically either streptavidin or antibodies. Thus, the first aim of this study was to optimize the bioconjugation process between the UCNPs and antibodies. Before conjugating the UCNPs with antibodies, their size and emission spectra were evaluated (Fig. S2†). Polyclonal anti-HSA antibody was chosen as the model. The bioconjugation procedure consisted of three steps: (1) ligand exchange that replaces oleic acid capping on the UCNP surface by hydrophilic ions, making the UCNPs dispersible in aqueous media, (2) conjugation with a neridronate-PEG-alkyne linker (Ner-PEG-alkyne), neridronate contains a bisphosphonic group that forms strong coordinate covalent bonds with the UCNP surface, resulting in alkyne-modified UCNPs, and (3) Cu(I)-catalyzed cycloaddition between UCNP-Ner-PEG-alkyne and azide-modified biomolecules (Fig. 1A). The resulting conjugates of UCNPs with polyclonal anti-HSA antibody and streptavidin were then tested in ULISA for HSA detection. The ULISA procedure included (1) immobilization of the capture antibody, (2) incubation with serial analyte dilutions, and either (3a) incubation with the UCNP–antibody (UCNP–Ab) conjugate or (3b) incubation with a biotinylated antibody followed by (4) incubation with the UCNP–streptavidin (UCNP–SA) conjugate (Fig. 1B).For the ligand exchange step, HCl41,42 and NOBF4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43,44 are commonly used. Therefore, we decided to compare these ligand exchange agents, preparing two conjugates of UCNPs with polyclonal anti-HSA antibody (UCNP–pAbHSA), with the only difference in the use of either HCl or NOBF4 for the ligand exchange. Each step of the bioconjugation procedure was followed by DLS (Fig. S3†) to ensure correct progress. After either ligand exchange reaction, the UCNPs were prone to aggregation, as indicated by a second DLS peak at ∼2000 nm. The hydrodynamic diameters in the HCl and NOBF4 cases were 54 nm and 55 nm, with polydispersity index (PDI) values of 0.21 and 0.20, respectively. After the PEG-linker binding, the hydrodynamic diameter increased to 91 nm in the case of HCl and to 90 nm in the case of NOBF4. The PDI values notably improved to 0.07 and 0.08, respectively, confirming a monodisperse system in both cases. The binding of the polyclonal anti-HSA antibody to the UCNPs resulted in an increase in the hydrodynamic diameter to 105 nm (with a PDI of 0.12) in the case of HCl and 109 nm (with a PDI of 0.13) in the case of NOBF4. The DLS data show that HCl- and NOBF4-based ligand exchange reactions resulted in very similar conjugates in terms of their hydrodynamic diameters and PDIs. The performance of the resulting UCNP–pAbHSA conjugates in the ULISA for HSA detection also showed no significant differences (Fig. 2A). The conjugate obtained via the HCl ligand exchange procedure achieved a slightly lower LOD of 65 pg mL−1 compared to the NOBF4-based procedure that reached a LOD of 76 pg mL−1. The S/B ratios were 256 and 316 for HCl- and NOBF4-based ligand exchange, respectively. In contrast with the report of Huan Ling et al. that reported better performance of UCNPs modified using NOBF4-based procedure compared to the HCl-based one,45 both ligand exchange procedures in this study resulted in practically identical conjugates. Therefore, HCl was selected as the ligand exchange agent for further bioconjugations as it offers a more convenient procedure and handling.
43,44 are commonly used. Therefore, we decided to compare these ligand exchange agents, preparing two conjugates of UCNPs with polyclonal anti-HSA antibody (UCNP–pAbHSA), with the only difference in the use of either HCl or NOBF4 for the ligand exchange. Each step of the bioconjugation procedure was followed by DLS (Fig. S3†) to ensure correct progress. After either ligand exchange reaction, the UCNPs were prone to aggregation, as indicated by a second DLS peak at ∼2000 nm. The hydrodynamic diameters in the HCl and NOBF4 cases were 54 nm and 55 nm, with polydispersity index (PDI) values of 0.21 and 0.20, respectively. After the PEG-linker binding, the hydrodynamic diameter increased to 91 nm in the case of HCl and to 90 nm in the case of NOBF4. The PDI values notably improved to 0.07 and 0.08, respectively, confirming a monodisperse system in both cases. The binding of the polyclonal anti-HSA antibody to the UCNPs resulted in an increase in the hydrodynamic diameter to 105 nm (with a PDI of 0.12) in the case of HCl and 109 nm (with a PDI of 0.13) in the case of NOBF4. The DLS data show that HCl- and NOBF4-based ligand exchange reactions resulted in very similar conjugates in terms of their hydrodynamic diameters and PDIs. The performance of the resulting UCNP–pAbHSA conjugates in the ULISA for HSA detection also showed no significant differences (Fig. 2A). The conjugate obtained via the HCl ligand exchange procedure achieved a slightly lower LOD of 65 pg mL−1 compared to the NOBF4-based procedure that reached a LOD of 76 pg mL−1. The S/B ratios were 256 and 316 for HCl- and NOBF4-based ligand exchange, respectively. In contrast with the report of Huan Ling et al. that reported better performance of UCNPs modified using NOBF4-based procedure compared to the HCl-based one,45 both ligand exchange procedures in this study resulted in practically identical conjugates. Therefore, HCl was selected as the ligand exchange agent for further bioconjugations as it offers a more convenient procedure and handling.
Afterward, copper(I)-catalyzed cycloaddition between UCNP-Ner-PEG-alkyne and an azide-modified antibody was investigated. The most significant parameter of the catalytical system was found to be the amount of azide-modified antibody. During the typical synthesis of UCNP–SA conjugates developed previously in our group, 10 μg of azide-modified streptavidin per 1 mg of UCNPs is used.46 In this work, different amounts of azide-modified antibody were utilized during the click reaction, and the resulting UCNP–pAbHSA conjugates were compared in terms of their hydrodynamic diameters and PDI values measured by DLS (Fig. S4A†), and performance in the ULISA for the detection of HSA (Fig. 2B). An increase in azide-modified antibody amount during the click reaction caused a slight increase in PDI values of the conjugates and did not show any significant effect on their hydrodynamic diameters, indicating the rising heterogeneity of the conjugates with the increasing amount of azide-modified antibody.
After the DLS characterization, the conjugates were employed in the ULISA for HSA. The summary of the achieved LODs, working ranges, and S/B ratios is provided in Table S1,† showing that the highest S/B ratio of 240 was achieved with 10 μg of azide-modified antibody per 1 mg of UCNPs, and the lowest LOD of 34 pg mL−1 of HSA was achieved with 50 μg of azide-modified antibody per 1 mg of UCNPs. When further increasing the azide-modified antibody amount, the S/B ratio showed a pronounced decrease, and the LODs started to rise, providing an S/B ratio of only 112 and a LOD of 46 pg mL−1 in the case of 100 μg of azide-modified antibody per 1 mg of UCNPs. Both the highest concentration signal and the background signal tended to rise with an increasing amount of azide-modified antibody, indicating an increase in specific and non-specific binding of the conjugates. The resulting analytical parameters of the immunoassay then depended on the extent to which the background and the highest analyte concentration signals increased. The ratio of 20 μg of azide-modified antibody per 1 mg of UCNPs was chosen for the following experiments, representing a compromise between the LOD, S/B, PDI, and antibody consumption.
3.2 Conjugates of UCNPs with polyclonal antibodies and their performance in the ULISA for PSA and p53 detection
After the optimization of the bioconjugation process, the Er-doped UCNPs were conjugated with polyclonal anti-PSA and anti-p53 antibodies, and the conjugates were characterized by DLS (Fig. S4B†). Unexpectedly, the UCNPs conjugated with polyclonal anti-PSA and anti-p53 antibodies showed larger hydrodynamic diameters and higher PDI values compared to the UCNPs conjugated with polyclonal anti-HSA antibody. The anti-PSA and anti-p53 antibodies were produced in goats, whereas the anti-HSA antibody was produced in rabbit. The literature shows that the IgG antibodies from different species differ in their structure,47 which likely influenced the conjugation process. Afterward, the performance of the Er-doped UCNPs conjugated with polyclonal anti-PSA (UCNP–pAbPSA) and anti-p53 (UCNP–pAbp53) antibodies was studied in the ULISA, and the results were compared to UCNP–SA conjugates (Fig. 3).ULISAs for the detection of PSA and p53 were carried out using UCNP label concentrations of 6.5 and 3.3 μg mL−1. The complete summary of the analytical parameters of the assays is provided in Table S2.† When UCNP–pAbPSA was used for PSA detection, LODs and S/B ratios were similar for both label concentrations. Utilizing 6.5 μg mL−1 of UCNP–pAbPSA resulted in better parameters, particularly an LOD of 17 pg mL−1 of PSA and an S/B ratio of 61. As a reference, PSA was detected using the UCNP–SA conjugate. In this case, the label concentration of 6.5 μg mL−1 also provided better results, even though the utilization of the higher concentration caused an increase in the signal of every sample concentration, including the blank. The LOD for PSA detection utilizing 6.5 μg mL−1 of UCNP–SA reached 1.3 pg mL−1, and the S/B ratio was 272. The data show that the UCNP–SA achieved better analytical parameters than the UCNP–pAbPSA conjugate. Nevertheless, the background levels were lower in the case of UCNP–pAbPSA.
For the detection of p53 using the UCNP–pAbp53 conjugate, the lower LOD of 0.84 ng mL−1 and the higher S/B ratio of 50.4 were achieved utilizing the label concentration of 6.5 μg mL−1. In the case of the UCNP–SA conjugate, the LODs were similar with both label concentrations; a slightly better LOD of 0.33 ng mL−1 was obtained utilizing 6.5 μg mL−1 of UCNP–SA. The S/B ratios of the p53 detection using the UCNP–SA conjugate were significantly higher compared to the UCNP–pAbp53 conjugate, and the highest S/B ratio reached 512, achieved using the UCNP–SA concentration of 3.3 μg mL−1.
Overall, the UCNP–pAb conjugates provided lower signal intensities and higher LODs than UCNP–SA. This confirms that utilizing the streptavidin–biotin binding in immunoassays is very beneficial. The bond is very strong, allowing the use of low biotinylated antibody concentrations, and one detection antibody can carry multiple biotin molecules, resulting in stronger signals and, thus, generally better assay performance. However, the ULISA procedure utilizing UCNP–antibody conjugates is one step shorter compared to the ULISA with UCNP–SA labels, and the UCNP–SA conjugates are not suitable for some applications, such as homogeneous immunoassay formats of UCCS and MPS.
3.3 Conjugates of UCNPs with monoclonal antibodies and their performance in the ULISA for PSA and p53 detection
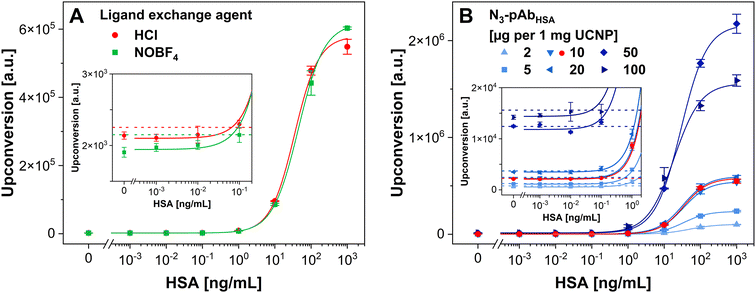
Utilizing UCNPs in MPS requires conjugating two types of UCNPs with different monoclonal antibodies. Therefore, we prepared conjugates of monoclonal anti-PSA (MAB1344 and ab403) and anti-p53 (MAB1355 and ab1101) antibodies with Er- and Tm-doped UCNPs, exhibiting different emission spectra according to the dopants (Fig. S2†). The synthesis utilized the same conditions as in the case of polyclonal anti-PSA and anti-p53 antibodies. The resulting conjugates were analyzed using DLS, revealing results comparable with the UCNP–pAbPSA and UCNP–pAbp53 conjugates, with average hydrodynamic diameters around 210 nm and PDIs ranging from 0.22 to 0.35 (Fig. S5†).Afterward, the Er- and Tm-doped UCNPs conjugated with monoclonal antibodies were tested in ULISAs for PSA and p53 (Fig. 4). The MTP was always coated with a complementary monoclonal antibody, distinct from the one used in the UCNP conjugates. The UCNP conjugates were used at a concentration of 6.5 μg mL−1, which was shown to be the optimal label concentration in the previous assays. The summary of the achieved LODs, working ranges, and S/B ratios is provided in Table S3.† Overall, the Tm-doped UCNPs showed lower signal intensities than the Er-doped ones. Different wavelengths were measured (540 nm in the case of Er-doped UCNPs and 810 nm in the case of Tm-doped UCNPs) utilizing different filters and dichroic mirrors, which influenced the signal intensities. Nevertheless, the LODs and S/B ratios were at a similar level.
The comparison of the performance of Er- and Tm-doped UCNPs conjugated with different monoclonal anti-PSA antibodies showed that the lowest LOD of 16 pg mL−1 of PSA was achieved using the Er-doped UCNP–MAB1344. The other conjugates reached similar LODs, ranging from 32 to 38 pg mL−1 of PSA. The S/B ratios spanned the range from 23 to 42, and the highest S/B of 42 was achieved with the Tm-doped UCNP–ab403. The Er- and Tm-doped UCNPs conjugated with monoclonal anti-p53 antibodies reached LODs ranging from 480 to 780 pg mL−1 of p53. The best LOD of 480 pg mL−1 was achieved utilizing the Tm-doped UCNP–ab1101. The best S/B ratio of 36 was reached with the Er-doped UCNP–MAB1355. Nevertheless, all the obtained S/B ratios were similar, spanning values from 26 to 36.
Compared to the UCNP conjugates with polyclonal anti-PSA antibody, the use of conjugates with monoclonal antibodies for the detection of PSA resulted in increased LODs and lower S/B values. Only the Er-doped UCNP–MAB1344 reached an LOD comparable to the UCNP–pAbPSA. However, in the case of p53, the LODs achieved using UCNP–mAbp53 conjugates were better compared to the UCNP–pAbp53 conjugates, while the S/B ratios were slightly lower. Overall, the UCNP–pAb conjugates showed better performance than the UCNP–mAb ones. The reason may be that polyclonal antibodies can bind to more epitopes, whereas monoclonal antibodies can only bind to one epitope of its target antigen. Nevertheless, the data confirm that conjugates of UCNPs with monoclonal antibodies were successfully synthesized, which was necessary for developing homogeneous MPS-based immunoassays.
Table 1 compares heterogeneous assays using nanomaterials for the detection of PSA and p53. In the case of PSA, the results obtained here correspond with the performance of our previous assays,11,14,27 and the LOD is comparable with the best reports published in the literature. In the case of protein p53, fewer methods are available, as this analyte has not been studied as extensively as PSA. Although several studies report on even lower LODs, the achieved LODs fall within the clinically relevant range.7 Overall, our heterogeneous assays demonstrated high sensitivity and reached low LODs, and the MTP-based procedure allows for the detection of a broad range of analytes.
| Analyte | Method | LOD [pg mL−1] | Working range [ng mL−1] | Ref. |
|---|---|---|---|---|
| PSA | ULISA (UCNP–PEG–SA conjugate) | 0.41 | 0.001–1 | 14 |
| ULISA (UCNP–PEG–SA conjugate) | 0.78 | 0.24–3.1 | 27 | |
| ULISA (UCNP–silica–Ab conjugate) | 20 | 0.1–10 | 11 | |
| UCNP-based LFIA | 0.1 | 0.1–100 | 48 | |
| Fluorescence-linked immunosorbent assay with dual-color quantum dots | 49 | 0.2–800 | 49 | |
| Immunoassay with phosphorescent quantum dots | 17 | 0.05–240 | 50 | |
| Biosensor with luminescent nanorods | 0.64 | 0.001–10 | 51 | |
| SERS aptasensor | 100 | 0.1–20 | 52 | |
| Colorimetric immunosensor with AuNPs | 9 | 0.01–20 | 53 | |
| ULISA (UCNP–PEG–SA conjugate) | 1.3 | 0.3–3.2 | This work | |
| ULISA (UCNP–PEG–Ab conjugate) | 16 | 1.1–15 | This work | |
| p53 | Electrochemiluminescence immunosensor | 1.2 | 0.011–11 | 54 |
| Electrochemical immunosensor | 1.6 | 0.011–0.11 | 55 | |
| Amperometric immunosensor | 1300 | 5–150 | 56 | |
| LFIA with peroxidase-like Pt–Pd NPs | 50 | 0.1–10 | 57 | |
| BioFET immunosensor | 5300 | N/A | 58 | |
| Competitive fluorescence assay | 420 | 2.7–110 | 59 | |
| ULISA (UCNP–SA conjugate) | 330 | 120–370 | This work | |
| ULISA (UCNP–Ab conjugate) | 480 | 30–321 | This work | |
3.4 Conjugates of UCNPs with monoclonal antibodies for MPS-based detection of PSA and p53
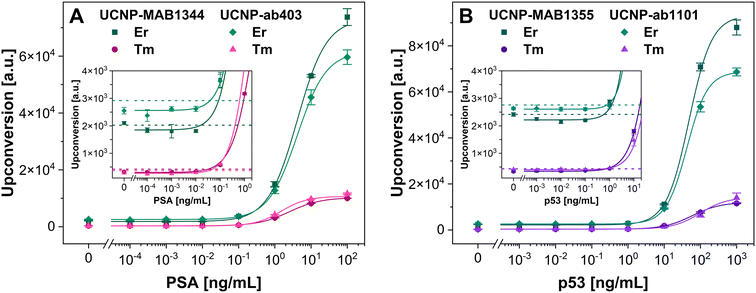
After verifying their performance, the UCNP–mAb conjugates were employed in a homogeneous format of MPS for the detection of PSA and p53. In our previous proof-of-principle study, the MPS of Er-doped streptavidin-modified UCNPs and Tm-doped biotin-modified UCNPs freely diffusing in an aqueous dispersion was described for the first time.39 Streptavidin–biotin interaction is among the strongest known non-covalent interactions, with a dissociation constant of ∼4 × 10−14 M;60 therefore, practically no dissociation of these complexes can be expected. In the current study, we have investigated a more intricate case where the Er- and Tm-doped UCNPs modified with two different monoclonal antibodies were used to detect protein analytes. Each UCNP type was conjugated with one of the monoclonal anti-analyte antibodies; the binding-site specificity of the antibodies was chosen to enable the binding of the respective UCNP types to two different epitopes of the same molecule. Mixing the Er- and Tm-doped UCNP conjugates with the analyte resulted in a formation of sandwich immunocomplexes consisting of an Er-doped UCNP conjugate, an analyte molecule, and a Tm-doped UCNP conjugate (Fig. 5A). Since MPS is a homogeneous method, unbound UCNP conjugates were the predominant component of the dispersion present even in the blanks (Fig. 5B); the total amounts of UCNP conjugates were identical for all the analyte concentrations, and only the number of double spots varied among them. The distinct emission spectra of Er- and Tm-doped UCNPs resulted in the target sandwich immunocomplexes appearing as double spots after the light passed through a diffraction prism (Fig. 5C). For this assay design, it was important to use UCNPs of a similar emission brightness (at 660 and 800 nm for Er- and Tm-doped UCNPs, respectively), to obtain MPS data enabling efficient double spot evaluation. The images were analyzed by the neural network, and the number of sandwich immunocomplexes per experiment was evaluated.First, the most suitable combinations of the Er- and Tm-doped UCNP conjugates were chosen by examining their performance in the analyte detection in terms of S/B levels (Fig. S6†). In the case of PSA, the combination of Er-doped UCNP–ab403 and Tm-doped UCNP–MAB1344 reached an S/B ratio of 3.4, while the Er-doped UCNP–MAB1344 and Tm-doped UCNP–ab403 combination provided an S/B ratio of 4.9. In the case of p53, the combination of Er-doped UCNP–ab1101 and Tm-doped UCNP–MAB1355 reached an S/B ratio of 2.4, and the combination of Er-doped UCNP–MAB1355 and Tm-doped UCNP–ab1101 reached the S/B ratio of 3.8.
The best combinations of Er- and Tm-doped UCNP conjugates were then used for the detection of PSA and p53 with extended analyte ranges, constructing the calibration curves. In the case of MPS detection of PSA (Fig. 5D), the LOD of 8.2 ng mL−1 was achieved, with the S/B of 16. This LOD was 2 to 3 orders of magnitude higher compared to the heterogeneous assays performed in this work. Nevertheless, in the case of p53, the achieved LOD was at a similar level compared to the heterogeneous assays, reaching 390 pg mL−1. However, the S/B of 4.8 was considerably lower compared to the heterogeneous assays. The MPS-based immunoassay was the only case in which the LOD was lower for p53 than for PSA. The increase in signals only at higher PSA concentrations suggests that this was probably caused by the high dissociation constants of the respective immunocomplexes.
Other homogeneous immunoassays using luminescent nanomaterials for the detection of PSA have been reported in the literature. For instance, Craciun et al. utilized the photoluminescence properties of gold nanospheres.61 The authors measured the clustering of anti-PSA antibody-modified gold nanospheres in the presence of PSA by fluorescence correlation spectroscopy and reached an LOD of 4.4 ng mL−1, which is comparable with our results. Homogeneous immunoassays based on time-gated Förster resonance energy transfer between Tb complex donor and quantum dot acceptor have also been used for the detection of PSA, exploiting the long luminescence lifetimes of lanthanide complexes.62–64 These methods reached 1 to 2 orders of magnitude lower LODs compared to MPS. Another homogeneous approach relied on bioluminescence produced by recombinant proteins.65 Two nanobodies were fused with two fragments of nanoluciferase, forming enzymatically active nanoluciferase upon PSA binding, and after the addition of the respective luciferin, bioluminescence was measured. The method reached an LOD of 0.4 ng mL−1 of PSA, which is lower compared to the MPS. However, the nanobodies fused with the luciferase fragments must be tailored for each analyte, which is arguably more complicated than the conjugation of UCNPs with monoclonal antibodies utilized in this work.
As far as we know, there is only one alternative homogeneous immunoassay for tumor protein p53 developed by Heyduk et al.66 This method is based on FRET upon the simultaneous binding of antibody and oligonucleotide to the analyte molecule. However, the work does not provide an LOD or any other analytical parameter of the method. An alternative assay without the need for antibodies by Assah et al.67 utilized p53 DNA response elements as biorecognition molecules. Protein p53 was added to the DNA probe-modified AuNPs, leading to the aggregation of AuNPs, thus changing their absorption spectra and enabling colorimetric detection. This method could distinguish between wild-type p53 and mutant p53; however, the LOD of 5 nM (∼260 ng mL−1) was three orders of magnitude higher compared to MPS.
Heterogeneous assays are generally more sensitive than homogeneous ones due to the phase separation and washing steps. However, MPS stands out in terms of low sample consumption and does not rely on washing steps, which might enable high-throughput analysis of various biomolecules in the future. Many settings of the MPS-based immunoassay can be further optimized, including the incubation time, the amounts of UCNP conjugates, and incubation mixture dilution before the MPS measurements. Therefore, there is a potential to shorten the assay duration from the current time of 3 h. Moreover, artificial intelligence can be further trained to make counting the double spots even more accurate.
4 Conclusions
In this study, the conjugation of UCNPs with antibodies was thoroughly investigated, enabling the preparation of efficient UCNP–antibody conjugates, and the conjugates were employed in heterogeneous and homogeneous immunoassays for the detection of cancer biomarkers: PSA and protein p53. The conjugates with polyclonal antibodies showed better performance in heterogeneous assays than monoclonal antibody conjugates since polyclonal antibodies can bind to multiple epitopes. Compared to heterogeneous immunoassays utilizing UCNP–SA conjugates, those employing polyclonal antibody conjugates demonstrated a 13-fold and 2.5-fold increase in LODs and a 4.5-fold and 8.5-fold decrease in S/B ratios for the detection of PSA and p53, respectively. However, the monoclonal antibody-modified UCNPs enabled the development of a novel homogeneous immunoassay based on MPS. The method involves observing sandwich immunocomplexes between the Er-doped UCNP–monoclonal antibody conjugate, the analyte molecule, and the Tm-doped UCNP–monoclonal antibody conjugate in aqueous medium without solid phase separation and washing steps. This novel AI-implementing method achieved LODs of 8.2 ng mL−1 and 390 pg mL−1 for PSA and p53, respectively. The procedure is fast, and with further optimizations, MPS promises to achieve highly efficient and high-throughput biomolecule detection.Author contributions
E. M.: methodology, investigation, and manuscript writing – original draft; P. Š.: investigation and manuscript writing – review & editing; A. H.: development of instrumentation and software for massively parallel spectroscopy, nanoparticle synthesis, proposing massively parallel spectroscopy for immunochemical assays, and manuscript writing – review & editing; J. W.: nanoparticle synthesis; H. H. G.: manuscript writing – review & editing; P. S.: manuscript writing – review & editing; and Z. F.: supervision, conceptualization, methodology, and manuscript writing – review & editing.Data availability
The data supporting this article have been included as part of the ESI.† Additional data are available upon request.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was funded by grant 21-03156S from the Czech Science Foundation. This publication was produced with support for the long-term conceptual development of the research organizations from the Institute of Analytical Chemistry of the Czech Academy of Sciences (RVO: 68081715). Parts of the graphical abstract were created using BioRender.com. Farka, Z. (2025), https://BioRender.com/m8bov9h and https://BioRender.com/334dd2r.References
- F. Rizzo, Chemosensors, 2022, 10, 326 CrossRef CAS.
- A. Jones, L. Dhanapala, R. N. T. Kankanamage, C. V. Kumar and J. F. Rusling, Anal. Chem., 2020, 92, 345–362 CrossRef CAS PubMed.
- S. Jeong, M.-J. Park, W. Song and H.-S. Kim, Clin. Biochem., 2020, 78, 43–57 CrossRef CAS PubMed.
- Y. Jin, J. H. Jung, W. K. Han, E. C. Hwang, Y. Nho, N. Lee, J. E. Yun, K. S. Lee, S. H. Lee, H. Lee and S.-Y. Yu, Invest. Clin. Urol., 2022, 63, 251 CrossRef PubMed.
- S. Naito, Jpn. J. Clin. Oncol., 2005, 35, 365–374 CrossRef PubMed.
- A. Herrero, E. Rojas, I. Misiewicz-Krzeminska, P. Krzeminski and N. Gutiérrez, Int. J. Mol. Sci., 2016, 17, 2003 CrossRef PubMed.
- A. M. Attallah, M. M. Abdel-Aziz, A. M. El-Sayed and A. A. Tabll, Cancer Detect. Prev., 2003, 27, 127–131 CrossRef CAS PubMed.
- H. Zhang, B. Zhao, Z.-G. Zhai, J.-D. Zheng, Y.-K. Wang and Y.-Y. Zhao, Eur. Rev. Med. Pharmacol. Sci., 2021, 25, 1358–1365 CAS.
- H. Suwa, G. Ohshio, N. Okada, Z. Wang, M. Fukumoto, T. Imamura and M. Imamura, Gut, 1997, 40, 647–653 CrossRef CAS PubMed.
- Y. Han, J. M. Chabu, S. Hu, L. Deng, Y. Liu and S. Guo, Chem. – Eur. J., 2015, 21, 13045–13051 CrossRef CAS PubMed.
- Z. Farka, M. J. Mickert, A. Hlaváček, P. Skládal and H. H. Gorris, Anal. Chem., 2017, 89, 11825–11830 CrossRef CAS PubMed.
- E. Kuah, S. Toh, J. Yee, Q. Ma and Z. Gao, Chem. – Eur. J., 2016, 22, 8404–8430 CrossRef CAS PubMed.
- Z. Farka, V. Čunderlová, V. Horáčková, M. Pastucha, Z. Mikušová, A. Hlaváček and P. Skládal, Anal. Chem., 2018, 90, 2348–2354 CrossRef CAS PubMed.
- M. J. Mickert, Z. Farka, U. Kostiv, A. Hlaváček, D. Horák, P. Skládal and H. H. Gorris, Anal. Chem., 2019, 91, 9435–9441 CrossRef CAS PubMed.
- E. Karakuş, E. Erdemir, N. Demirbilek and L. Liv, Anal. Chim. Acta, 2021, 1182, 338939 CrossRef PubMed.
- Z. Wei, K. Luciano and X. Xia, ACS Nano, 2022, 16, 21609–21617 CrossRef CAS PubMed.
- X. Tang, W. Xia, H. Han, Y. Wang, B. Wang, S. Gao and P. Zhang, ACS Appl. Bio Mater., 2024, 7, 7659–7665 CrossRef CAS PubMed.
- X. Min, S. Huang and C. Yuan, Anal. Chim. Acta, 2022, 1204, 339704 CrossRef CAS PubMed.
- Z. Farka, J. C. Brandmeier, M. J. Mickert, M. Pastucha, K. Lacina, P. Skládal, T. Soukka and H. H. Gorris, Adv. Mater., 2024, 36, 2307653 CrossRef CAS PubMed.
- H. You, X. Hua, L. Feng, N. Sun, Q. Rui, L. Wang and M. Wang, Microchim. Acta, 2017, 184, 1085–1092 CrossRef CAS.
- W. He, M. You, Z. Li, L. Cao, F. Xu, F. Li and A. Li, Sens. Actuators, B, 2021, 334, 129673 CrossRef CAS.
- S. Wen, J. Zhou, K. Zheng, A. Bednarkiewicz, X. Liu and D. Jin, Nat. Commun., 2018, 9, 2415 CrossRef PubMed.
- Y. Zhang, R. Wen, J. Hu, D. Guan, X. Qiu, Y. Zhang, D. S. Kohane and Q. Liu, Nat. Commun., 2022, 13, 5927 CrossRef CAS PubMed.
- U. Kostiv, Z. Farka, M. J. Mickert, H. H. Gorris, N. Velychkivska, O. Pop-Georgievski, M. Pastucha, E. Odstrčilíková, P. Skládal and D. Horák, Biomacromolecules, 2020, 21, 4502–4513 CrossRef CAS PubMed.
- A. Hlaváček, Z. Farka, M. J. Mickert, U. Kostiv, J. C. Brandmeier, D. Horák, P. Skládal, F. Foret and H. H. Gorris, Nat. Protoc., 2022, 17, 1028–1072 CrossRef PubMed.
- M. Pastucha, E. Odstrčilíková, A. Hlaváček, J. C. Brandmeier, V. Vykoukal, J. Weisová, H. H. Gorris, P. Skládal and Z. Farka, IEEE J. Sel. Top. Quantum Electron., 2021, 27, 1–11 Search PubMed.
- E. Makhneva, D. Sklenárová, J. C. Brandmeier, A. Hlaváček, H. H. Gorris, P. Skládal and Z. Farka, Anal. Chem., 2022, 94, 16376–16383 CrossRef CAS PubMed.
- D. Kang, S. Lee, H. Shin, J. Pyun and J. Lee, Biosens. Bioelectron., 2020, 150, 111921 CrossRef CAS PubMed.
- X. Li, L. Wei, L. Pan, Z. Yi, X. Wang, Z. Ye, L. Xiao, H.-W. Li and J. Wang, Anal. Chem., 2018, 90, 4807–4814 CrossRef CAS PubMed.
- J. C. Brandmeier, N. Jurga, T. Grzyb, A. Hlaváček, R. Obořilová, P. Skládal, Z. Farka and H. H. Gorris, Anal. Chem., 2023, 95, 4753–4759 CrossRef CAS PubMed.
- R. Peltomaa, Z. Farka, M. J. Mickert, J. C. Brandmeier, M. Pastucha, A. Hlaváček, M. Martínez-Orts, Á. Canales, P. Skládal, E. Benito-Peña, M. C. Moreno-Bondi and H. H. Gorris, Biosens. Bioelectron., 2020, 170, 112683 CrossRef CAS PubMed.
- Y. Wang, X. Zhao, M. Zhang, X. Sun, J. Bai, Y. Peng, S. Li, D. Han, S. Ren, J. Wang, T. Han, Y. Gao, B. Ning and Z. Gao, J. Hazard. Mater., 2021, 406, 124703 CrossRef CAS PubMed.
- D. Sklenárová, A. Hlaváček, J. Křivánková, J. C. Brandmeier, J. Weisová, M. Řiháček, H. H. Gorris, P. Skládal and Z. Farka, Lab Chip, 2024, 24, 3536–3545 RSC.
- S. Lahtinen, S. Krause, R. Arppe, T. Soukka and T. Vosch, Chem. – Eur. J., 2018, 24, 9229–9233 CrossRef CAS PubMed.
- S. Lahtinen, M. B. Liisberg, K. Raiko, S. Krause, T. Soukka and T. Vosch, ACS Appl. Nano Mater., 2021, 4, 432–440 CrossRef CAS.
- R. J. Dinis-Oliveira, Bioanalysis, 2014, 6, 2877–2896 CrossRef CAS PubMed.
- S. Aydin, Peptides, 2015, 72, 4–15 CrossRef CAS PubMed.
- W. R. Mohamed, N. Mahmoud, F. Abdel Samad, E. Ahmed, M. R. Hamblin and T. Mohamed, Spectrochim. Acta, Part A, 2022, 268, 120646 CrossRef CAS PubMed.
- A. Hlaváček, K. Uhrová, J. Weisová and J. Křivánková, Anal. Chem., 2023, 95, 12256–12263 CrossRef PubMed.
- A. Hlaváček, J. Křivánková, J. Přikryl and F. Foret, Anal. Chem., 2019, 91, 12630–12635 CrossRef PubMed.
- U. Kostiv, Z. Farka, M. J. Mickert, H. H. Gorris, N. Velychkivska, O. Pop-Georgievski, M. Pastucha, E. Odstrčilíková, P. Skládal and D. Horák, Biomacromolecules, 2020, 21, 4502–4513 CrossRef CAS PubMed.
- E. Andresen, C. Würth, C. Prinz, M. Michaelis and U. Resch-Genger, Nanoscale, 2020, 12, 12589–12601 RSC.
- S. Shanwar, L. Liang, A. V. Nechaev, D. K. Bausheva, I. V. Balalaeva, V. A. Vodeneev, I. Roy, A. V. Zvyagin and E. L. Guryev, Materials, 2021, 14, 1657 CrossRef CAS PubMed.
- K. Raiko, A. Lyytikäinen, M. Ekman, A. Nokelainen, S. Lahtinen and T. Soukka, Clin. Chim. Acta, 2021, 523, 380–385 CrossRef CAS PubMed.
- H. Ling, D. Guan, R. Wen, J. Hu, Y. Zhang, F. Zhao, Y. Zhang and Q. Liu, Small, 2024, 20, 2309035 CrossRef CAS PubMed.
- J. C. Brandmeier, K. Raiko, Z. Farka, R. Peltomaa, M. J. Mickert, A. Hlaváček, P. Skládal, T. Soukka and H. H. Gorris, Adv. Healthcare Mater., 2021, 10, 2100506 CrossRef CAS PubMed.
- S. Kabir, Immunol. Invest., 2002, 31, 263–278 CrossRef CAS PubMed.
- X. Hu, J. Liao, H. Shan, H. He, Z. Du, M. Guan, J. Hu, J. Li and B. Gu, Methods, 2023, 215, 10–16 CrossRef CAS PubMed.
- J. Fan, N. Li, F. Wang, Y. Lv, Q. Jin, M. Zhao, Y. Shi, R. Wu, H. Shen and L. S. Li, Sens. Actuators, B, 2023, 375, 132888 CrossRef CAS.
- M. García-Cortés, M. T. Fernández-Argüelles, J. M. Costa-Fernández and A. Sanz-Medel, Anal. Chim. Acta, 2017, 987, 118–126 CrossRef PubMed.
- Z. Yin, L. Zhu, Z. Lv, M. Li and D. Tang, Talanta, 2021, 233, 122563 CrossRef CAS PubMed.
- A. Ouhibi, A. Raouafi, N. Lorrain, M. Guendouz, N. Raouafi and A. Moadhen, Sens. Actuators, B, 2021, 330, 129352 CrossRef CAS.
- P. Karami, H. Khoshsafar, M. Johari-Ahar, F. Arduini, A. Afkhami and H. Bagheri, Spectrochim. Acta, Part A, 2019, 222, 117218 CrossRef CAS PubMed.
- H. Afsharan, F. Navaeipour, B. Khalilzadeh, H. Tajalli, M. Mollabashi, M. J. Ahar and M.-R. Rashidi, Biosens. Bioelectron., 2016, 80, 146–153 CrossRef CAS PubMed.
- H. Afsharan, B. Khalilzadeh, H. Tajalli, M. Mollabashi, F. Navaeipour and M.-R. Rashidi, Electrochim. Acta, 2016, 188, 153–164 CrossRef CAS.
- M. Pedrero, F. Manuel de Villena, C. Muñoz-San Martín, S. Campuzano, M. Garranzo-Asensio, R. Barderas and J. Pingarrón, Biosensors, 2016, 6, 56 CrossRef PubMed.
- T. Jiang, Y. Song, D. Du, X. Liu and Y. Lin, ACS Sens., 2016, 1, 717–724 CrossRef CAS.
- C. Baldacchini, A. F. Montanarella, L. Francioso, M. A. Signore, S. Cannistraro and A. R. Bizzarri, Sensors, 2020, 20, 6364 CrossRef CAS.
- Q. Xu, K. Liang, R.-Y. Liu, L. Deng, M. Zhang, L. Shen and Y.-N. Liu, Talanta, 2018, 187, 142–147 CrossRef CAS PubMed.
- A. Holmberg, A. Blomstergren, O. Nord, M. Lukacs, J. Lundeberg and M. Uhlén, Electrophoresis, 2005, 26, 501–510 CrossRef CAS PubMed.
- A.-M. Craciun, S. Suarasan, M. Focsan and S. Astilean, Talanta, 2021, 228, 122242 CrossRef CAS PubMed.
- S. Bhuckory, L. Mattera, K. D. Wegner, X. Qiu, Y.-T. Wu, L. J. Charbonnière, P. Reiss and N. Hildebrandt, Chem. Commun., 2016, 52, 14423–14425 RSC.
- L. Mattera, S. Bhuckory, K. D. Wegner, X. Qiu, F. Agnese, C. Lincheneau, T. Senden, D. Djurado, L. J. Charbonnière, N. Hildebrandt and P. Reiss, Nanoscale, 2016, 8, 11275–11283 RSC.
- S. Bhuckory, K. D. Wegner, X. Qiu, Y.-T. Wu, T. L. Jennings, A. Incamps and N. Hildebrandt, Molecules, 2020, 25, 3679 CrossRef CAS PubMed.
- M. E. Baghdadi, R. Emamzadeh, M. Nazari and E. Michelini, Enzyme Microb. Technol., 2024, 180, 110474 CrossRef CAS PubMed.
- E. Heyduk, B. Dummit, Y.-H. Chang and T. Heyduk, Anal. Chem., 2008, 80, 5152–5159 CrossRef CAS PubMed.
- E. Assah, W. Goh, X. T. Zheng, T. X. Lim, J. Li, D. Lane, F. Ghadessy and Y. N. Tan, Colloids Surf., B, 2018, 169, 214–221 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5nr00176e |
| ‡ The authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |