3D-printed ultrasensitive SERS substrates via photocrosslinked Pluronic F127 micellar hydrogel with citrate-reduced metallic nanoparticles†
Billy D.
Chinchin-Piñan
 ,
Mateus P.
Bomediano
,
Marcelo G.
de Oliveira
,
Mateus P.
Bomediano
,
Marcelo G.
de Oliveira
 and
Javier E. L.
Villa
and
Javier E. L.
Villa
 *
*
Institute of Chemistry, University of Campinas (UNICAMP), Campinas, SP, Brazil. E-mail: jelv@unicamp.br
First published on 2nd May 2025
Abstract
In this study, we present a rapid and reproducible approach to fabricating flexible surface-enhanced Raman spectroscopy (SERS) substrates using 3D printing technology. We developed 3D-printed ultrasensitive substrates containing metallic nanoparticles (Au or Ag), synthesized by citrate reduction and stabilized by a 3D-printed micellar hydrogel matrix formed by photocrosslinked pluronic F127. The SERS signal can be fine-tuned by adjusting water content within the printed hydrogel, and an analytical enhancement factor of up to 3 × 106 was achieved with excellent repeatability (RSD < 7.0%). These SERS substrates offer robust sensing capabilities, and this versatile fabrication strategy has significant potential for creating a wide range of plasmonic platforms.
Surface-enhanced Raman spectroscopy (SERS) is a sensitive analytical technique able to detect single molecules under favourable conditions. In addition to high sensitivity, the use of a minimal amount of sample for rapid and non-destructive analyses has led SERS to a wide variety of applications.1 For instance, in biomedicine for disease diagnosis,2 in agriculture for pesticide detection,3 in food quality assurance,4 and in pollutant detection in water samples.5 However, one of the most significant challenges lies in improving the low reproducibility that strongly depends on the homogeneity of the metallic nanostructures.6,7 Although lithography-based nanofabrication allows for producing highly regular SERS substrates,8 they are time-consuming and not cost-effective for large-scale production. Therefore, there is a need for rapid alternative methods to fabricate low-cost, sensitive, and reproducible SERS substrates.
An emerging approach for fabricating SERS substrates consists of the use of polymeric matrices to disperse and stabilize metallic nanoparticles by electrostatic, steric, or depletion effects,9,10 thus creating flexible plasmonic structures. Some examples of these polymers include polyethylene glycol, pluronic family, cetyltrimethylammonium bromide (CTAB), and polyvinylpyrrolidone (PVP), which effectively stabilize gold and silver nanoparticles.11–14 However, some of them need to self-assemble onto the nanoparticles to stabilize them (e.g., CTAB and PVP), which may hinder further adsorption of analytes for SERS applications. Pluronic F127 is a synthetic copolymer composed of two hydrophilic poly(ethylene oxide) (PEO) blocks and a hydrophobic poly(propylene oxide) (PPO) core block arranged in a triblock structure ([PEO]99–[PPO]65–[PEO]99). Above its critical micellar concentration, pluronic F127 self-assembles into micelles with hydrophobic cores and hydrophilic coronas, which may further form hexagonal, cubic, or lamellar structures, yielding a thermoreversible hydrogel.10 Pluronic family has demonstrated effective depletion stabilization of gold nanoparticles in aqueous environments.15,16 Furthermore, the methacrylation of pluronic F127's terminal hydroxyl groups, achieved through rapid microwave-assisted reactions, produces pluronic dimethacrylate (pluronic-DM) ink suitable for extrusion-based 3D printing.17 3D printing technology is evolving to meet user, material, and application-specific needs, with hydrogel extrusion in polymer printing showing significant promise in fields such as biomedicine18 (organs and tissues), biotechnology11 (cell culture), and advanced materials development.12 However, the direct fabrication of SERS substrates by 3D printing technology is still in the early stages of exploration within the field.15–18 Based on this background, we hypothesized that photocrosslinkable pluronic-DM can effectively stabilize nanoparticles, facilitating the reproducible 3D printing of homogeneous hydrogel nanocomposites without passivating nanoparticle surfaces, thereby preserving their suitability for subsequent SERS sensing applications.
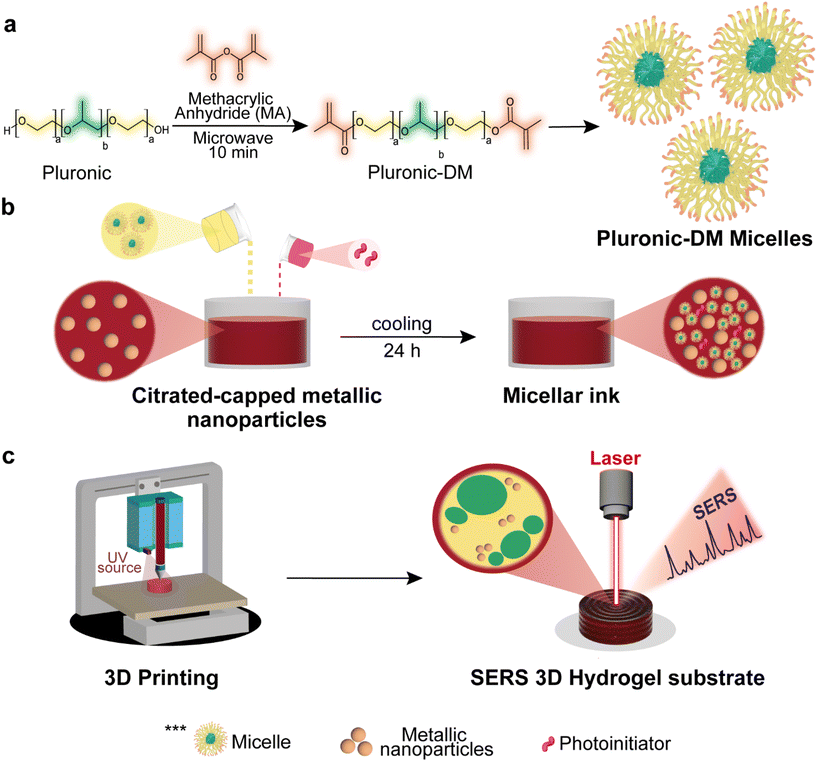
Fig. 1a shows the process for obtaining pluronic F127 dimethacrylated and the subsequent micelle self-assembly. Briefly, a fast esterification reaction with methacrylic anhydride was performed by heating for 10 min in a microwave oven, according to the protocol published elsewhere.19 Next, gold (or silver) metal nanoparticles synthesized by the citrate reduction method (Fig. S1 and S2†) were mixed with pluronic F127-DM. All reagents used were purchased from Sigma-Aldrich. The mixture was left under refrigeration (6–8 °C) for 24 h to promote the solubilization of the polymer, and LAP (lithium phenyl-2,4,6-trimethylbenzoylphosphinate) photoinitiator was then added. After solubilization at low temperature, the solution undergoes phase transition into a gel at room temperature due to micellar ordering, which is expected to stabilize the nanoparticles,10 as shown in Fig. 1b. As a final step, Fig. 1c describes the light-assisted 3D printing process of the micellar inks, in which an extrusion syringe bioprinter with custom temperature control set to print at 20 °C was used. Cylinders of 8 mm in diameter and 3 mm in height were used as computer-aided design models, and the photocrosslinking process was assisted by a UV light source (365–405 nm) to obtain gold (or silver) nanoparticle 3D SERS substrates (see Fig. S3†).
Fig. 2 shows the protocol for analyte detection using the 3D-printed SERS substrate, which was immersed in 5 mL of a crystal violet solution at different concentration levels to promote its adsorption onto the surface of the Au (or Ag) nanoparticles. After three hours, a steady state of maximum swelling was achieved. It is also worth highlighting that this material allowed for efficient analyte diffusion to the surface of the metal nanoparticles, which are covered by easily replaceable citrate molecules compared to CTAB or PVP surfactants (challenging to remove and might hinder SERS measurements). All SERS spectra were acquired using a Raman spectrometer model BWS465-785S iRaman Plus (B&W Tek) in the range of 400 to 1700 cm−1, with a resolution of 4.8 cm−1 and a 785 nm laser coupled to an fiber optic probe (400 mW laser power at the source). All measurements were performed using 30-second accumulation of exposure and 100% laser excitation power. The SERS spectra were recorded at maximum substrate swelling after drying it in a desiccator.
Fig. 3a and b display the SERS spectra of the blank and crystal violet on anhydrous and hydrated 3D-printed hydrogel substrates based on AuNPs and AgNPs. The spectra clearly show the characteristic bands of crystal violet: 420–438 cm−1 (phenyl-C-phenyl asymmetric out-of-plane bending), 526 cm−1 (C–N–C asymmetric bending), 720 cm−1 (C–N–C symmetric stretching), 798 cm−1 (phenyl-H asymmetric out-of-plane bending), 911 cm−1 (out-of-plane deformation of C in the phenyl ring), 1171 cm−1 (C-phenyl, C–H in-plane asymmetric stretching), 1386 cm−1 (C–N, phenyl-C-phenyl asymmetric stretching), 1582 cm−1 (C-phenyl asymmetric stretching), and 1614 cm−1 (C-phenyl in-plane asymmetric stretching).20 The spectrum in the anhydrous state is approximately three times more intense than in the hydrated state, which implies a tunable SERS sensitivity by controlling the water content. The intensity difference probably arises from the increase in hot spots per area as water loss reduces the substrate's volume (i.e., preconcentration of hot spots and analyte). The analytical enhancement factor (EF)21 was calculated as:  where ISERS is the signal obtained from the analyte in the SERS substrate, IRAMAN is the signal obtained from the analyte in the substrate without nanoparticles, CSERS represents the concentration of the analyte detected by the SERS substrate, and CRAMAN is the analyte concentration detected by the substrate without nanoparticles. The average analytical EF estimated for the SERS substrates based on AgNPs (2.51 × 105) and AuNPs (2.96 × 106) showed to be excellent and in good agreement with highly sensitive SERS substrates of AgNPs20,22,23 and AuNPs24,25 previously reported. All the spectra used for these calculations are presented in Fig. S4.† In addition to the high sensitivity of the fabricated substrates, they also displayed excellent repeatability, demonstrated by the relative standard deviations (RSD) of 6.1% and 6,4% for AuNPs and AgNPs-based SERS substrates recorded at 1171 cm−1, respectively. Furthermore, the intensity difference between the anhydrous and hydrated states is less pronounced for AgNPs than AuNPs, probably because of the differences in the shift of the nanoparticle surface plasmons and the localised surface plasmon resonance condition with the Raman laser at 785 nm (see Fig. S1 and S2†). Additional concentration dependence experiments are presented in Fig. S5.†
where ISERS is the signal obtained from the analyte in the SERS substrate, IRAMAN is the signal obtained from the analyte in the substrate without nanoparticles, CSERS represents the concentration of the analyte detected by the SERS substrate, and CRAMAN is the analyte concentration detected by the substrate without nanoparticles. The average analytical EF estimated for the SERS substrates based on AgNPs (2.51 × 105) and AuNPs (2.96 × 106) showed to be excellent and in good agreement with highly sensitive SERS substrates of AgNPs20,22,23 and AuNPs24,25 previously reported. All the spectra used for these calculations are presented in Fig. S4.† In addition to the high sensitivity of the fabricated substrates, they also displayed excellent repeatability, demonstrated by the relative standard deviations (RSD) of 6.1% and 6,4% for AuNPs and AgNPs-based SERS substrates recorded at 1171 cm−1, respectively. Furthermore, the intensity difference between the anhydrous and hydrated states is less pronounced for AgNPs than AuNPs, probably because of the differences in the shift of the nanoparticle surface plasmons and the localised surface plasmon resonance condition with the Raman laser at 785 nm (see Fig. S1 and S2†). Additional concentration dependence experiments are presented in Fig. S5.†
Further investigation of the drying process and the impact on the SERS signal was performed, and the results are shown in Fig. 3c and d. It was performed by monitoring the water loss and the SERS signal at 1171 cm−1 starting from the maximum swelling to the anhydrous state. Fig. 3c illustrates the water loss of the AuNPs substrate alongside the SERS signal; the initial mass at t = 0 was 0.1881 ± 0.0008 g, with an intensity of 8,532 a.u., decreasing to a final mass of 0.03617 ± 0.0031 g after 22 hours of drying at room temperature, with an intensity of 36,600 a.u. According to these data, the gain in sensitivity by drying the 3D-printed AuNPs substrate is about 4.3 times. The consistency of the mass measurements at maximum swelling at each subsequent hour demonstrates the gradual water loss from the hydrogels, with these values serving as a control in the 3D printing process. Once the substrate is completely dry and its mass remains constant, the SERS signal exhibits minimal standard deviation. For the AgNPs-based substrate, the gain in sensitivity was about 2.8 times after the drying step. Therefore, both AuNPs- and AgNPs-based substrates exhibited similar trends in their dehydration curves and corresponding increases in the tunable SERS signal.
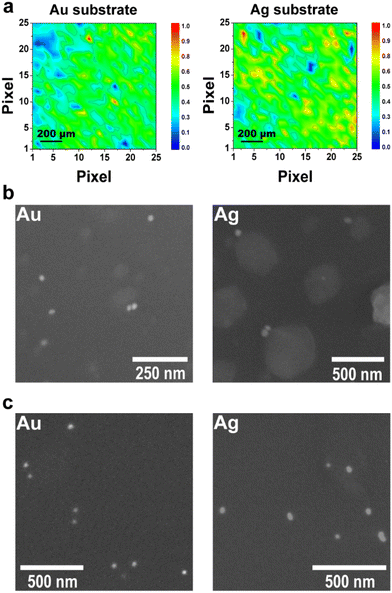
To complement the previous experiments, SERS maps were recorded from the anhydrous plasmonic substrates, and the results are displayed in Fig. 4a and b. These results indicate a homogeneous distribution for both substrates, thereby demonstrating that the proposed 3D-printed plasmonic materials not only provide high sensitivity but also excellent reproducibility. While AgNPs-based SERS substrate produces intense SERS spectra, the sensitivity can be further improved by adjusting the laser wavelength to better match the plasmon resonance or by increasing the recording time. Fig. 4b shows micrographs of the hydrated 3D-printed hydrogel SERS substrates with AuNPs and AgNPs, whereas the anhydrous states are displayed in Fig. 4c. Bright circular dots representing individual or small clusters of metal nanoparticles are visible, thereby confirming that the SERS effect stems from hot spots formed within the fully hydrated 3D-printed hydrogel SERS substrate. Additionally, larger irregular structures in the bulk suggest the formation of polymeric aggregates, potentially comprising crosslinked micelles and disordered non-micellar material (see Fig. S6† for additional micrographs). Moreover, the difference in size of the polymeric aggregates is probably related to the differences in the citrate concentrations remaining from the synthesis of AuNPs and AgNPs. A larger number of nanoparticles per area was observed for both anhydrous substrates than their fully hydrated counterparts. Therefore, the microscopy images corroborated that the increase in hot spots covered by the laser spot and the analyte preconcentration effect caused by the drying step led to an enhanced and tunable SERS signal over time.
We have also demonstrated the potential for biosensing applications by detecting biomolecules such as nicotine, uric acid, and adenine with detection limits down to 500 nmol L−1, 100 nmol L−1, and 10 nmol L−1, respectively (the SERS spectra can be found in Fig. S7a, b, and c†). In addition to detecting these analytes below clinically and biologically relevant concentrations, the fabricated substrates demonstrated applicability in the analysis of urine simulant (Sigma-Aldrich), a sample with a complex matrix of macromolecules, salts, and small biomolecules, thereby unveiling key spectroscopic signal from bioanalytes presented in the sample (Fig. S7d†). To highlight the advantages of our plasmonic 3D-printed micellar hydrogel over previously reported materials, a detailed comparison of features is presented in Table S1.† According to it, our approach provides a facile and rapid preparation of stable plasmonic inks with only three components (pluronic-DM, nanoparticles, and photoinitiator), a straightforward and reproducible photocrosslinking process, and a precise control over substrate shape and size via 3D printing technology that might be extended to medium- and large-scale manufacturing. In addition to the portability of the proposed method, no surface passivation was required, which is a key advantage for further SERS sensing and real-world applications.
Conclusions
We successfully developed an aqueous, plasmonic, and photocrosslinkable ink using citrate-reduced gold and silver nanoparticles, pluronic F127-DM, and a photoinitiator LAP. Colloidal gold and silver were stabilized by the polymer in the ink by the depletion stabilization effect, and, by means of extrusion-based 3D printing, we fabricated ultrasensitive and reproducible SERS hydrogel substrates. By optimizing the SERS sensing protocol, we demonstrated tunable sensitivity from hydrated to anhydrous states, confirmed by large enhancement factors, particularly for AuNPs. This protocol shows promise for a wide range of quantitative and qualitative applications (including the detection of bioanalytes such as uric acid, adenine, and nicotine), and this technology paves the way for further exploration of automated fabrication of ultrasensitive SERS substrates using 3D printing. Additionally, our formulation of a photocrosslinkable micellar ink with citrate-reduced nanoparticles avoids the need for toxic or challenging-to-remove surfactants (e.g., CTAB and PVP) for subsequent SERS measurements.Data availability
To support the validation of data to maintain high standards of research reproducibility, the data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
This work was supported by the São Paulo Research Foundation (FAPESP) (grants 2024/09412-4 – J. E. L. V., 2022/14645-2 – M. G. O., and 2022/13352-1 – M. P. B.) and the National Council for Scientific and Technological Development (CNPq) (grants 406607/2022-2 and 380969/2023-8).References
- S. Schlücker, Angew. Chem., Int. Ed., 2014, 53, 4756–4795 CrossRef.
- N. Choi, H. Dang, A. Das, M. S. Sim, I. Y. Chung and J. Choo, Biosens. Bioelectron., 2020, 164, 112326 CrossRef CAS PubMed.
- Z. Guo, P. Chen, N. Yosri, Q. Chen, H. R. Elseedi, X. Zou and H. Yang, Food Rev. Int., 2023, 39, 1440–1461 CrossRef CAS.
- L. Jiang, M. M. Hassan, S. Ali, H. Li, R. Sheng and Q. Chen, Trends Food Sci. Technol., 2021, 112, 225–240 CrossRef CAS.
- C. W. Yang, X. Zhang, L. Yuan, Y. K. Wang and G. P. Sheng, Water Res., 2023, 232, 119668 CrossRef CAS.
- C. Zong, M. Xu, L.-J. Xu, T. Wei, X. Ma, X.-S. Zheng, R. Hu and B. Ren, Chem. Rev., 2018, 118, 4946–4980 CrossRef CAS PubMed.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. García de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Käll, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J.-F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J.-M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L.-L. Tay, K. G. Thomas, Z.-Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzán, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- M. Chirumamilla, A. Toma, A. Gopalakrishnan, G. Das, R. P. Zaccaria, R. Krahne, E. Rondanina, M. Leoncini, C. Liberale, F. De Angelis and E. Di Fabrizio, Adv. Mater., 2014, 26, 2353–2358 CrossRef CAS PubMed.
- A. B. R. Mayer, Polym. Adv. Technol., 2001, 12, 96–106 CrossRef CAS.
- X. Zhang, M. R. Servos and J. Liu, J. Am. Chem. Soc., 2012, 134, 9910–9913 CrossRef CAS PubMed.
- T. Sakai and P. Alexandridis, Langmuir, 2004, 20, 8426–8430 CrossRef CAS.
- T. Sakai, Y. Horiuchi, P. Alexandridis, T. Okada and S. Mishima, J. Colloid Interface Sci., 2013, 394, 124–131 CrossRef CAS PubMed.
- K. H. Bae, S. H. Choi, S. Y. Park, Y. Lee and T. G. Park, Langmuir, 2006, 22, 6380–6384 CrossRef CAS.
- P. Alexandridis and M. Tsianou, Eur. Polym. J., 2011, 47, 569–583 CrossRef CAS.
- H. Kang, J. T. Buchman, R. S. Rodriguez, H. L. Ring, J. He, K. C. Bantz and C. L. Haynes, Chem. Rev., 2019, 119, 664–699 CrossRef CAS PubMed.
- K. Wang, Z. Yue, X. Fang, H. Lin, L. Wang, L. Cao, J. Sui and L. Ju, Sci. Total Environ., 2023, 856, 159108 CrossRef CAS PubMed.
- W. Wang, Y. Chen, C. Xiao, S. Xiao, C. Wang, Q. Nie, P. Xu, J. Chen, R. You, G. Zhang and Y. Lu, Chem. Eng. J., 2023, 474, 145953 CrossRef CAS.
- G. A. Vinnacombe-Willson, C. García-Astrain, L. Troncoso-Afonso, M. Wagner, J. Langer, P. González-Callejo, D. Di Silvio and L. M. Liz-Marzán, Chem. Mater., 2024, 36, 5192–5203 CrossRef CAS.
- M. P. Bomediano, M. I. Santos, M. V. Lorevice, G. B. Romano, L. C. E. da Silva, A. G. S. Junior and M. G. de Oliveira, MRS Commun., 2024, 14, 1070–1077 CrossRef CAS.
- H.-Y. Chen, M.-H. Lin, C.-Y. Wang, Y.-M. Chang and S. Gwo, J. Am. Chem. Soc., 2015, 137, 13698–13705 CrossRef CAS.
- E. C. Le Ru, E. Blackie, M. Meyer and P. G. Etchegoin, J. Phys. Chem. C, 2007, 111, 13794–13803 CrossRef CAS.
- W. Meng, F. Hu, L.-Y. Zhang, X.-H. Jiang, L.-D. Lu and X. Wang, J. Mol. Struct., 2013, 1035, 326–331 CrossRef CAS.
- F. A. Harraz, A. A. Ismail, H. Bouzid, S. A. Al-Sayari, A. Al-Hajry and M. S. Al-Assiri, Appl. Surf. Sci., 2015, 331, 241–247 CrossRef CAS.
- S. L. Kleinman, B. Sharma, M. G. Blaber, A.-I. Henry, N. Valley, R. G. Freeman, M. J. Natan, G. C. Schatz and R. P. Van Duyne, J. Am. Chem. Soc., 2013, 135, 301–308 CrossRef CAS.
- V. Joseph, A. Matschulat, J. Polte, S. Rolf, F. Emmerling and J. Kneipp, J. Raman Spectrosc., 2011, 42, 1736–1742 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5nr01139f |
| This journal is © The Royal Society of Chemistry 2025 |