Open Access Article
Open Access ArticleA system for in vitro selection of fully 2′-modified RNA aptamers†
Emily D.
Ziperman‡
 a,
Kate B.
Fitzpatrick‡
a,
Kate B.
Fitzpatrick‡
 a,
Malavika A.
Nair
a,
Alexander W.
Sorum
a,
Malavika A.
Nair
a,
Alexander W.
Sorum
 b,
Linda C.
Hsieh-Wilson
b,
Linda C.
Hsieh-Wilson
 b and
Isaac J.
Krauss
b and
Isaac J.
Krauss
 *a
*a
aDepartment of Chemistry, Brandeis University, Waltham, Massachusetts, USA. E-mail: kraussi@brandeis.edu
bDivision of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, California, USA
First published on 6th February 2025
Abstract
SFM4-3, KOD DGLNK, and Therminator polymerase are investigated for their compatibility with SELection with Modified Aptamers (SELMA), an aptamer discovery method that enables incorporation of large nucleobase modifications such as glycans. We demonstrated that with suitable modifications to the primer design and protocol, these enzymes are compatible with SELMA, enabling 2′-fluoro or 2′-methoxy ribose modifications at all positions. In the case of 2′-fluoro modifications, Therminator exhibits cleaner incorporation of an alkyne-modified nucleobase for click chemistry.
Aptamers are genetically encodable polymers, usually oligonucleotides, that have been selected in vitro from a random library to bind to a protein or molecule of interest.1–4 Oligonucleotide aptamers are typically 20–100 bases in length and can achieve high binding affinities for their targets due to their specific folding, shape complementarity and positioning of functional groups.5 Aptamers possess several advantages compared with antibodies, including straightforward synthesis and quality control, low immunogenicity,6,7 and small size.5,8 Aptamers are typically discovered using SELEX (Systematic Evolution of Ligands by EXponential Enrichment), in which libraries of random DNA or RNA sequences flanked by constant primer regions are incubated with a target of interest and the bound sequences are amplified by PCR. Two aptamer therapeutics have been approved for clinical use. Pegaptanib targets vascular endothelial growth factor (VEGF) for the treatment of age-related macular degeneration.9,10 Very recently in 2023, avacincaptad pegol, an aptamer targeting complement protein C5, was approved for the treatment of geographic atrophy (GA) secondary to age-related macular degeneration.11,12
A barrier to the development of oligonucleotide aptamer therapeutics has been the instability of naked RNA or DNA to nucleases in serum.13,14 To address this, aptamers have been extensively stabilized with modified nucleotides (XNA), such as fluoro-(2′-F-) or methoxy (2′-OMe-) ribose, which confer nuclease resistance.14 Pegaptanib and avacincaptad are illustrative, as they both bear 2′-F- or -OMe at nearly every position. However, these modifications typically must be introduced into natural DNA or RNA aptamers after selection by SELEX, and trial and error is necessary to find substitutions that do not compromise the binding activity of the selected molecule. Incorporation of XNA throughout the selection process would be more desirable and efficient but requires technical innovation, as XNA libraries may not be easily synthesized and/or PCR amplified by polymerases with limited tolerance for modified nucleoside triphosphates (NTPs).15–42
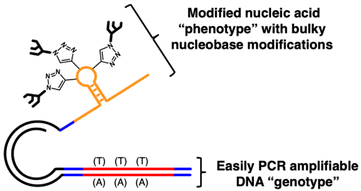
As a solution to this problem, our group has reported SELection of Modified Aptamers (SELMA, Fig. 1),43–48 which allows for extensive modifications, not only of ribose, but of the nucleobase (e.g., oligosaccharide attachment) and includes a variant that can use 2′-F pyrimidine NTPs48 throughout the selection. In SELMA, PCR and/or RT-PCR of modified oligonucleotides is circumvented by tethering an unmodified, PCR-amplifiable DNA copy to each XNA sequence in the library.
However, this technique has not yet been adapted to selections with libraries containing the 2′-F modification on all four nucleotides, which would require an RNA or DNA polymerase capable of incorporating them. Although several polymerases are known to accommodate ribose-modified NTPs, the activity and fidelity of these enzymes remains poorly understood and difficult to assess by traditional sequencing methods. Ultimately, a better understanding of the performance of these enzymes is needed before they can be widely adopted for aptamer selection applications.
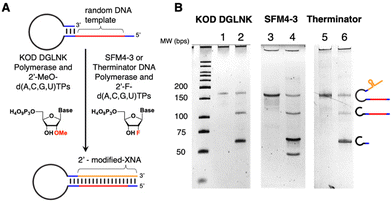
To this end, we have investigated three different polymerases capable of incorporating 2′-modified XNA into the SELMA protocol. Therminator DNA polymerase is a commercially available mutant of 9° N DNA polymerase that is able to incorporate a variety of non-canonical nucleic acids including 2′-F-modified NTPs (2′-F-NTPs).49 SFM4-3, which also incorporates 2′-F-NTPs, is a variant of the Stoffel fragment of TAQ DNA polymerase developed by Chen et al. using directed evolution.50 KOD DGLNK is a mutant KOD polymerase developed by Hoshino et al., that accepts both LNA and 2′-OMe-modified nucleotides.51 We purchased commercially available Therminator and expressed SFM4-3 and KOD DGLNK for generation of the ssXNA “phenotype” strand (G, Fig. 2). Library construction begins with a short ssDNA strand containing the random library template (A), which is annealed to a phosphorylated overhanging “regeneration” primer (B). Bidirectional DNA polymerase extension creates a longer duplex (C) and lambda exonuclease digestion of the phosphorylated strand results in a single stranded hairpin (D). The 3′ end of D is self-priming and can be extended with XNA using one of the three above enzymes to afford double stranded hairpin (E). The XNA strand is then displaced (F) by treatment with a “hairpin poison primer” (HPP) and its extension with BST 2.0 to generate Form G. Compared with the DNA/DNA duplex of our original SELMA method, this relatively long primer is required for the strand displacement of the stronger 2′-F-RNA-DNA interaction.35,52,53 The resulting Form G library contains modified ssXNA that can adopt a sequence-dependent fold and can undergo selection towards a target, while PCR amplification of the tethered dsDNA in the bound library fraction affords a new template in double stranded Form H. The use of the biotinylated primer in PCR allows for strand separation and a return to ssDNA library template (Form A).
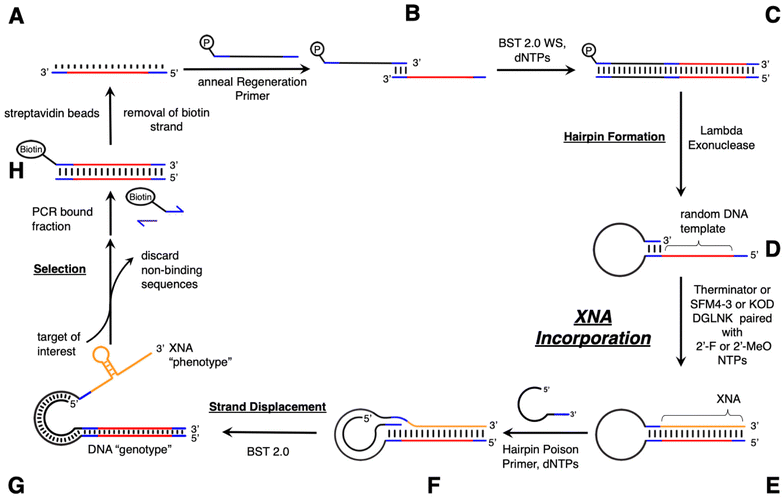 | ||
| Fig. 2 XNA SELMA Library build. Blue = constant primer sites; red = random region; black = constant hairpin region; orange = XNA. | ||
For KOD DGLNK and SFM4-3, some optimization was done to characterize the behavior of each enzyme at various temperatures. We found that, for our applications, increasing the extension temperature from previously reported conditions resulted in more efficient extension and higher yields of Form E (Fig. S3 and S4†).50,51 After determining the ideal extension conditions, each enzyme was used to form double stranded hairpin and subsequently strand displaced as shown in Fig. 3.
To demonstrate compatibility of each enzyme with the SELMA workflow, we performed mock selections in which a known target sequence of 2′-F- or 2′-OMe-RNA was selected from a random library by hybridization to a biotinylated complementary oligonucleotide (Fig. 4A). The target sequence contained an NheI restriction site, which enabled easy detection of the target sequence by the presence of four fragment bands after digestion with NheI (Fig. 4B, Group 4). This sequence was spiked at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 into N25 random libraries generated with 2′-F-RNA using Therminator or SFM4-3, or with 2′-OMe-RNA using KOD DGLNK. Prior to selection, the known sequence was not visible by gel analysis of the spiked libraries following NheI digest (e.g., Fig. 4B, Group 2). Each library was then subjected to one round of selection. Species that bound to the biotinylated complementary oligonucleotide were recovered using streptavidin magnetic beads. Bound material was then PCR amplified and digested with NheI, resulting in the expected four small fragment bands in all three selections (Fig. 4B, Groups 3, 5, 6). Thus, we observed clear enrichment of our target sequence after just one round of selection using libraries generated with each of the enzymes tested here. Control selections containing only random library (Fig. 4B, Group 1) or only NheI (Fig. 4B, Group 4) sequence were also performed in parallel. The successful mock selection demonstrates the capability of the SELMA method to evolve a fully fluorinated or fully methoxy library and generate enrichment without the need for a reverse transcription step.
1000 into N25 random libraries generated with 2′-F-RNA using Therminator or SFM4-3, or with 2′-OMe-RNA using KOD DGLNK. Prior to selection, the known sequence was not visible by gel analysis of the spiked libraries following NheI digest (e.g., Fig. 4B, Group 2). Each library was then subjected to one round of selection. Species that bound to the biotinylated complementary oligonucleotide were recovered using streptavidin magnetic beads. Bound material was then PCR amplified and digested with NheI, resulting in the expected four small fragment bands in all three selections (Fig. 4B, Groups 3, 5, 6). Thus, we observed clear enrichment of our target sequence after just one round of selection using libraries generated with each of the enzymes tested here. Control selections containing only random library (Fig. 4B, Group 1) or only NheI (Fig. 4B, Group 4) sequence were also performed in parallel. The successful mock selection demonstrates the capability of the SELMA method to evolve a fully fluorinated or fully methoxy library and generate enrichment without the need for a reverse transcription step.
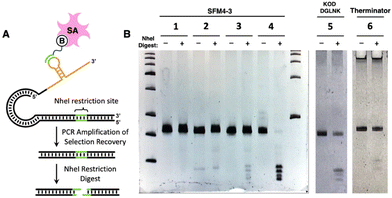 | ||
Fig. 4 (A) NheI selection construct design and enrichment test. B = biotin; SA = streptavidin (B) Gel analysis of NheI-digested templates from selections. NheI-treated samples are indicated with a (+). Group 1 = random library post selection, Group 2 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 NheI template 1000 NheI template![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) random library template pre selection, Group 3 = 1 random library template pre selection, Group 3 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 NheI template 1000 NheI template![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) random library template post selection, Group 4 = NheI template only post selection. Groups 1–4 were prepared with SFM4-3. Groups 5 and 6 are post-selection libraries analogous to Group 3 but made with KOD DGLNK/MeO-NTPs Therminator polymerase/F-NTPs, respectively. Complete gels corresponding to Groups 5 and 6 can be found in Fig. S5.† random library template post selection, Group 4 = NheI template only post selection. Groups 1–4 were prepared with SFM4-3. Groups 5 and 6 are post-selection libraries analogous to Group 3 but made with KOD DGLNK/MeO-NTPs Therminator polymerase/F-NTPs, respectively. Complete gels corresponding to Groups 5 and 6 can be found in Fig. S5.† | ||
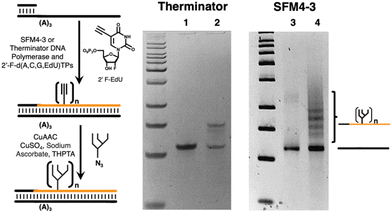
To investigate chemical functionalization of libraries, we focused on 2′-F-RNA and used either Therminator or SFM4-3 to generate an F-RNA strand containing three 5-ethynyluridine (fEdU)48 bases by primer extension against a 56-mer DNA template containing three adenines (Fig. 5). Following CuAAC attachment of oligomannose (Man9) glycan,44 the results were visualized by gel, in which the functionalized F-RNA strand migrates higher due to added glycans. When Therminator was used to generate the F-RNA strand, the corresponding glyco-F-RNA migrated primarily as one band, corresponding to addition of three glycans, with a small amount of a lower band corresponding to incomplete functionalization. However, when SFM4-3 was used to generate the F-RNA, the glyco-F-RNA migrated as a series of bands, corresponding to different numbers of attached glycans in some cases exceeding three. These results suggested substantial misincorporation of fEdU by SFM4-3 and have led us to focus on using Therminator for future libraries.54
Conclusions
In conclusion, we have described several modifications to the SELMA methodology that allow for the incorporation and in vitro selection of fully 2′-modified XNA without the need for reverse transcription or post-SELEX medicinal chemistry optimization to incorporate nuclease resistant modifications. Additionally, we have demonstrated that fully 2′-modified XNA SELMA constructs can be successfully enriched towards a known target. This expansion of SELMA capabilities will simplify discovery of aptamers that could be directly used in the presence of nucleases. We have shown that the 2′F-RNA can be chemically modified by CuAAC, and we have found in the process that Therminator is less prone to misincorporation of fEdU than SFM4-3. Selection applications of SELMA with XNA and CuAAC modifications are currently underway with carbohydrate binding targets. Using this approach, we will develop potent modulators of clinically relevant carbohydrate binding proteins that are directly ready for use in vivo.Data availability
The derived data supporting these findings can be found in the ESI.†Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was supported by NIH Grants R01GM151492 (I. J. K.), R01GM127920 (L. C. H.-W. and I. J. K.) and T32GM139798 (K. B. F.) as well as the NSF Graduate Research Fellowship No. 2139752 (E. D. Z.). I. J. K. acknowledges the NIH Shared Instrumentation grant S10OD034395 for purchase of an NMR spectrometer. We would like to thank Dr Floyd Romesburg for the plasmid of SFM4-3 and Dr Satoshi Obika for the plasmid of KOD DGLNK. Dr Richard Redman and Dr Lizbeth Hedstrom are gratefully acknowledged for helpful discussions.References
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- L. Gold, N. Janjic, T. Jarvis, D. Schneider, J. J. Walker, S. K. Wilcox and D. Zichi, Cold Spring Harbor Perspect. Biol., 2012, 4, a003582 Search PubMed.
- R. Stoltenburg, C. Reinemann and B. Strehlitz, Biomol. Eng., 2007, 24, 381–403 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- V. Thiviyanathan and D. G. Gorenstein, Proteomics: Clin. Appl., 2012, 6, 563–573 CAS.
- Eyetech Study Group, Ophthalmology, 2003, 110, 979–986 CrossRef PubMed.
- Eyetech Study Group, Retina, 2002, 22, 143–152 CrossRef PubMed.
- V. Di Mauro, F. C. Lauta, J. Modica, S. L. Appleton, V. De Franciscis and D. Catalucci, JACC Basic Transl. Sci., 2024, 9, 260–277 CrossRef PubMed.
- E. S. Gragoudas, A. P. Adamis, E. T. Cunningham Jr., M. Feinsod and D. R. Guyer, NEJM, 2004, 351, 2805–2816 CrossRef CAS PubMed.
- J. H. Lee, M. D. Canny, A. De Erkenez, D. Krilleke, Y. S. Ng, D. T. Shima, A. Pardi and F. Jucker, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 18902–18907 CrossRef CAS PubMed.
- G. J. Jaffe, K. Westby, K. G. Csaky, J. Monés, J. A. Pearlman, S. S. Patel, B. C. Joondeph, J. Randolph, H. Masonson and K. A. Rezaei, Ophthalmology, 2021, 128, 576–586 CrossRef PubMed.
- A. Mullard, Nat. Rev. Drug Discovery, 2023, 22, 774 Search PubMed.
- J. M. Healy, S. D. Lewis, M. Kurz, R. M. Boomer, K. M. Thompson, C. Wilson and T. G. McCauley, Pharm. Res., 2004, 21, 2234–2246 CrossRef CAS PubMed.
- C. Kratschmer and M. Levy, Nucleic Acid Ther., 2017, 27, 335–344 CrossRef CAS PubMed.
- Y. Lin, Q. Qiu, S. C. Gill and S. D. Jayasena, Nucleic Acids Res., 1994, 22, 5229–5234 CrossRef CAS PubMed.
- S. Jhaveri, B. Olwin and A. D. Ellington, Bioorg. Med. Chem. Lett., 1998, 8, 2285–2290 CrossRef CAS PubMed.
- P. E. Burmeister, S. D. Lewis, R. F. Silva, J. R. Preiss, L. R. Horwitz, P. S. Pendergrast, T. G. McCauley, J. C. Kurz, D. M. Epstein, C. Wilson and A. D. Keefe, Chem. Biol., 2005, 12, 25–33 CrossRef CAS PubMed.
- J. K. Ichida, K. Zou, A. Horhota, B. Yu, L. W. McLaughlin and J. W. Szostak, J. Am. Chem. Soc., 2005, 127, 2802–2803 CrossRef CAS PubMed.
- N. Minakawa, M. Sanji, Y. Kato and A. Matsuda, Bioorg. Med. Chem., 2008, 16, 9450–9456 CrossRef CAS PubMed.
- V. B. Pinheiro, A. I. Taylor, C. Cozens, M. Abramov, M. Renders, S. Zhang, J. C. Chaput, J. Wengel, S.-Y. Peak-Chew, S. H. McLaughlin, P. Herdewijn and P. Holliger, Science, 2012, 336, 341–344 CrossRef CAS PubMed.
- H. Yu, S. Zhang, M. R. Dunn and J. C. Chaput, J. Am. Chem. Soc., 2013, 135, 3583–3591 CrossRef CAS PubMed.
- M. Kuwahara and S. Obika, Artif. DNA PNA XNA, 2013, 4, 39–48 CrossRef PubMed.
- A. I. Taylor, V. B. Pinheiro, M. J. Smola, A. S. Morgunov, S. Peak-Chew, C. Cozens, K. M. Weeks, P. Herdewijn and P. Holliger, Nature, 2015, 518, 427–430 CrossRef CAS PubMed.
- A. D. Friedman, D. Kim and R. Liu, Biomaterials, 2015, 36, 110–123 CrossRef CAS PubMed.
- A. I. Taylor and P. Holliger, Nat. Protoc., 2015, 10, 1625–1642 CrossRef CAS PubMed.
- I. C. Elle, K. K. Karlsen, M. G. Terp, N. Larsen, R. Nielsen, N. Derbyshire, S. Mandrup, H. J. Ditzel and J. Wengel, Mol. BioSyst., 2015, 11, 1260–1270 RSC.
- I. Alves Ferreira-Bravo, C. Cozens, P. Holliger and J. J. DeStefano, Nucleic Acids Res., 2015, 43, 9587–9599 Search PubMed.
- D. Thirunavukarasu, T. Chen, Z. Liu, N. Hongdilokkul and F. E. Romesberg, J. Am. Chem. Soc., 2017, 139, 2892–2895 CrossRef CAS PubMed.
- Z. Liu, T. Chen and F. E. Romesberg, Chem. Sci., 2017, 8, 8179–8182 RSC.
- A. I. Taylor and P. Holliger, Curr. Protoc. Chem. Biol., 2018, 10, e44 CrossRef PubMed.
- A. E. Rangel, Z. Chen, T. M. Ayele and J. M. Heemstra, Nucleic Acids Res., 2018, 46, 8057–8068 CrossRef CAS PubMed.
- S. Arangundy-Franklin, A. I. Taylor, B. T. Porebski, V. Genna, S. Peak-Chew, A. Vaisman, R. Woodgate, M. Orozco and P. Holliger, Nat. Chem., 2019, 11, 533–542 CrossRef CAS PubMed.
- E. Eremeeva, A. Fikatas, L. Margamuljana, M. Abramov, D. Schols, E. Groaz and P. Herdewijn, Nucleic Acids Res., 2019, 47, 4927–4939 CrossRef CAS PubMed.
- K. M. Rose, I. Alves Ferreira-Bravo, M. Li, R. Craigie, M. A. Ditzler, P. Holliger and J. J. DeStefano, ACS Chem. Biol., 2019, 14, 2166–2175 CAS.
- M. R. Dunn, C. M. McCloskey, P. Buckley, K. Rhea and J. C. Chaput, J. Am. Chem. Soc., 2020, 142, 7721–7724 CrossRef CAS PubMed.
- H. Hoshino, Y. Kasahara, M. Kuwahara and S. Obika, J. Am. Chem. Soc., 2020, 142, 21530–21537 CrossRef CAS PubMed.
- G. Houlihan, S. Arangundy-Franklin, B. T. Porebski, N. Subramanian, A. I. Taylor and P. Holliger, Nat. Chem., 2020, 12, 683–690 CrossRef CAS PubMed.
- L. Zhang and J. C. Chaput, Molecules, 2020, 25, 4194 CrossRef CAS PubMed.
- Q. Shao, T. Chen, K. Sheng, Z. Liu, Z. Zhang and F. E. Romesberg, J. Am. Chem. Soc., 2020, 142, 2125–2128 CrossRef CAS PubMed.
- S. Dey and J. T. Sczepanski, Nucleic Acids Res., 2020, 48, 1669–1680 CrossRef CAS PubMed.
- I. Alves Ferreira-Bravo and J. J. DeStefano, Viruses, 2021, 13, 1983 CrossRef CAS PubMed.
- K. B. Wu, C. J. A. Skrodzki, Q. Su, J. Lin and J. Niu, Chem. Sci., 2022, 13, 6873–6881 RSC.
- I. S. Macpherson, J. S. Temme, S. Habeshian, K. Felczak, K. Pankiewicz, L. Hedstrom and I. J. Krauss, Angew. Chem., Int. Ed., 2011, 50, 11238–11242 Search PubMed.
- J. S. Temme, M. G. Drzyzga, I. S. Macpherson and I. J. Krauss, Chem. – Eur. J., 2013, 19, 17291–17295 CrossRef CAS PubMed.
- J. S. Temme, I. S. Macpherson, J. F. Decourcey and I. J. Krauss, J. Am. Chem. Soc., 2014, 136, 1726–1729 Search PubMed.
- J. S. Temme and I. J. Krauss, Curr. Protoc. Chem. Biol., 2015, 7, 73–92 CrossRef CAS PubMed.
- I. S. Macpherson, J. S. Temme and I. J. Krauss, Chem. Commun., 2017, 53, 2878–2881 RSC.
- R. L. Redman and I. J. Krauss, J. Am. Chem. Soc., 2021, 143, 8565–8571 CrossRef CAS PubMed.
- A. F. Gardner, K. M. Jackson, M. M. Boyle, J. A. Buss, V. Potapov, A. M. Gehring, K. M. Zatopek, I. R. Corrêa Jr., J. L. Ong and W. E. Jack, Front. Mol. Biosci., 2019, 6, 28 CrossRef CAS PubMed.
- T. Chen, N. Hongdilokkul, Z. Liu, R. Adhikary, S. S. Tsuen and F. E. Romesberg, Nat. Chem., 2016, 8, 556–562 CrossRef CAS PubMed.
- H. Hoshino, Y. Kasahara, H. Fujita, M. Kuwahara, K. Morihiro, S.-I. Tsunoda and S. Obika, Bioorg. Med. Chem. Lett., 2016, 26, 530–533 CrossRef CAS PubMed.
- A. Patra, M. Paolillo, K. Charisse, M. Manoharan, E. Rozners and M. Egli, Angew. Chem., Int. Ed., 2012, 51, 11863–11866 CrossRef CAS PubMed.
- N. Martin-Pintado, G. F. Deleavey, G. Portella, R. Campos-Olivas, M. Orozco, M. J. Damha and C. González, Angew. Chem., Int. Ed., 2013, 52, 12065–12068 CrossRef CAS PubMed.
- Error rates for F-RNA synthesis by each of these two enzymes were investigated. However, due to the large error rate of reverse transcription to produce DNA for sequencing, we were unable to discern the specific F-RNA synthesis error rates .
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ob01505c |
| ‡ These authors contributed equally to the content and writing of this manuscript. |
| This journal is © The Royal Society of Chemistry 2025 |