Open Access Article
Open Access ArticleFluorescent nucleobase analogue for cellular visualisation and regulation of immunostimulatory CpG oligodeoxynucleotides†
Tatum Melati Andini‡
 ab,
Satoshi Tada‡c,
Tomotaka Kumagaid,
Yuki Takahashi
ab,
Satoshi Tada‡c,
Tomotaka Kumagaid,
Yuki Takahashi c,
Yuriko Higuchic,
Yusuke Kawamoto
c,
Yuriko Higuchic,
Yusuke Kawamoto *c and
Soyoung Park
*c and
Soyoung Park *a
*a
aImmunology Frontier Research Center, Osaka University, Yamadaoka, Suita, Osaka 565-0871, Japan. E-mail: spark@ifrec.osaka-u.ac.jp
bDepartment of Genome Informatics, Research Institute for Microbial Diseases, Graduate School of Medicine, Osaka University, Yamadaoka, Suita, Osaka 565-0871, Japan
cGraduate School of Pharmaceutical Sciences, Kyoto University, 46-29 Yoshida-shimoadachicho, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: y.kawamoto@pharm.kyoto-u.ac.jp
dDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawaoiwakecho, Sakyo-ku, Kyoto 606-8502, Japan
First published on 28th January 2025
Abstract
In this study, we explored the chemical modification of toll-like receptor 9 (TLR9) agonist DNA using a highly fluorescent thymine analogue, ThexT, focusing on its structural and photophysical characteristics. ThexT-labelled CpG oligonucleotides effectively demonstrated intracellular localisation within macrophage cell lines. Notably, immunostimulatory activity varied depending on the site of ThexT incorporation within the TLR9 agonist sequence. The introduction of fluorescent nucleobases offers a useful approach for visualising immunostimulatory oligonucleotides and for modulating immune responses.
Introduction
Nucleic acid sensing plays a crucial role in the detection of and response to pathogens in the innate immune system. Certain toll-like receptors (TLRs), a type of pattern recognition receptor (PRR), are designed to recognise nucleic acids as pathogen-associated molecular patterns (PAMPs) that initiate innate immune responses. This mechanism is an essential component of the first line of defence against invading pathogens such as viruses and bacteria. Toll-like receptor 9 (TLR9) is predominantly expressed in immune cells such as plasmacytoid dendritic cells (pDCs), B cells, and macrophages. TLR9 recognises unmethylated CpG motifs (cytosine–phosphate–guanine), which are common in bacterial and viral DNA but are rarely found in vertebrate methylated DNA. The CpG motif is characterised by the sequence PuPuCGPyPy, where Pu denotes purine (adenine or guanine) and Py denotes pyrimidine (cytosine or thymine). When this motif is detected, TLR9 initiates a potent innate immune response, triggering a cascade of signalling pathways that result in the production of pro-inflammatory cytokines and subsequent activation of the adaptive immune response, allowing the host to effectively combat infection. Several oligonucleotides containing unmethylated CpG motifs have been identified as TLR9 agonists, and these molecules have gained attention for their potential as therapeutic agents capable of modulating immune responses.1–4 Exploiting the DNA-induced activation of innate immunity, chemically synthesised oligodeoxynucleotides (ODNs) containing CpG motifs have been investigated for clinical applications such as vaccine adjuvants, protection against infections, mitigation of allergic reactions, and cancer immunotherapy.5–8 Very recently, Jovasevic et al. demonstrated that TLR9 signalling is required for genomic DNA repair and the formation of learning-induced memories in mice.9 This seminal study suggests a close connection between the nervous and immune systems in cognitive processes. This suggests that TLR9 may be a potential therapeutic target for degenerative neurological diseases and learning disabilities. However, the immunogenic nature of CpG ODNs raises safety concerns regarding therapeutic antisense oligonucleotides, prompting investigations into chemical modifications to avoid innate immune activation.10,11 In this context, various chemical modifications have been explored to improve and regulate the immunostimulatory activity of CpG ODNs.12–14 Amongst these, studies on nucleobase modifications have primarily focused on the CpG dinucleotide recognised by TLR9.15–20 Although structural studies have shown that nucleobases outside PuPuCGPyPy interact with TLR9 residues,21 the effects of chemical modifications of other nucleobases and those beyond the CpG motifs on immunostimulatory activity have not been well studied.22Fluorescent nucleobase analogues are potent tools for detecting nucleic acid molecules and examining biomolecular dynamics, interactions, and reactions associated with nucleic acids.23–29 These modified nucleobase analogues not only possess fluorescent properties but also provide insights into the structure–activity relationships of nucleic acid ligands due to their unique structures that differ from canonical nucleobases.30 Unlike the attachment of fluorescent organic dyes at oligonucleotide termini, fluorescent nucleobase technology allows for site-specific labelling within oligonucleotides, making it suitable for elucidating interactions and reactions of oligonucleotides at the molecular level. However, there are few reports on the visualisation of chemically synthesised oligonucleotides labelled with fluorescent nucleobases in cellular environments,31–33 likely because fluorescence is quenched within polymerised nucleotides,34 resulting in insufficient fluorescence intensity for cellular observation. Agrawal et al. demonstrated that substitution of cytosine in the CpG dinucleotide with a fluorescent bicyclic nucleobase could induce immunostimulatory activity.17,35 Our group found that substitution of guanines outside the CpG motif with a fluorescent thieno[3,4-d]pyrimidine guanine mimic could induce the immunostimulatory activity of CpG DNAs.22,26 Although these fluorescent nucleobases improved the immunostimulatory activity of modified ODNs, cellular observations using their fluorescence have not been reported.
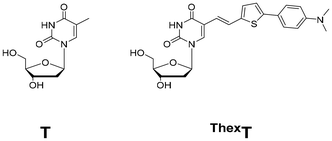
Recently, our group reported a highly emissive molecular rotor-type thymine analogue termed ThexT (Fig. 1).36 This modified pyrimidine nucleoside has thiophene and dimethylaniline moieties connected through an olefinic bond to the 5-position of thymine. In the visible region (470–530 nm), the ThexT nucleoside exhibited significant fluorescence (Φ = 0.61 in DMSO). Oligonucleotides containing ThexT exhibit significant fluorescence changes that are visible to the naked eye in response to structural changes in nucleic acids and binding interactions with target proteins. The π-extended hydrophobic structure and intense fluorescence of ThexT prompted us to investigate the potential cellular applications of oligonucleotides that contain fluorescent nucleobases.
In this study, we incorporated fluorescent nucleobases into oligonucleotides and assessed their fluorescence properties and functionality. Specifically, we investigated the chemical modification of TLR9 agonist DNA with the ThexT nucleobase, focusing on its structural features and emissive properties. Although interactions between TLR9 and thymine have been previously reported,21 few studies have examined the effect of structural alterations in thymine on TLR9 agonist activity. Recognising this gap, we sought to understand the consequences of integrating ThexT at specific sites with TLR9 agonists. Our experiments, conducted using the murine macrophage cell line RAW 264.7, revealed that this fluorescent nucleobase allowed effective visualisation of agonist DNA inside the cells. Furthermore, we found that immunostimulatory activity varied based on the position of the ThexT modification within the TLR9 agonist sequence.
Results and discussion
Synthesis of chemically modified immunostimulatory oligodeoxynucleotides
CpG ODNs can be divided into three distinct classes based on their sequences and structural characteristics. Each class of CpG ODNs exhibits distinct immunostimulatory properties and activates different immune cell types. In this study, we focused on ODN 1668, a prototypical sequence within the CpG-B class (also known as K′-type CpG ODN), as a configurable synthetic TLR9 agonist. To evaluate the effect of substituting thymine nucleobases with the chemically modified fluorescent analogue ThexT, we identified substitution sites as the 1st, 5th, 10th, 11th, 14th, and 17th thymine from the 5′-terminus of ODN 1668 (ODNThexT1 to ODNThexT17, Table 1).| Name | Sequence (5′ to 3′, X = ThexT) |
|---|---|
| a The CpG motif (GACGTT) is underlined.b All phosphate linkages were replaced with phosphorothioates. | |
| ODN 1668 | TCCAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) CCTGATGCT CCTGATGCT |
| ODNThexT1 | XCCAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) CCTGATGCT CCTGATGCT |
| ODNThexT5 | TCCAX![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) CCTGATGCT CCTGATGCT |
| ODNThexT10 | TCCAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[X with combining low line]](https://www.rsc.org/images/entities/b_char_0058_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) CCTGATGCT CCTGATGCT |
| ODNThexT11 | TCCAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[X with combining low line]](https://www.rsc.org/images/entities/b_char_0058_0332.gif) CCTGATGCT CCTGATGCT |
| ODNThexT14 | TCCAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) CCXGATGCT CCXGATGCT |
| ODNThexT17 | TCCAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) CCTGAXGCT CCTGAXGCT |
Additionally, the phosphodiester backbones of ODN 1668 were replaced by nuclease-resistant phosphorothioate backbones. Synthetic oligonucleotides (ODNs) with phosphorothioate modifications enhance nuclease resistance and improve cellular uptake. These chemical modifications have facilitated their widespread application as vaccine adjuvants, enhancing immunogenicity by activating innate immune receptors, such as TLR9, and in cancer therapy, where they are employed to elicit antitumour immune responses. Using solid-phase synthesis and ThexT phosphoramidite prepared as previously outlined (Fig. S1†), a series of CpG ODNs with various ThexT substitution positions were synthesised. The synthesised ODNs were characterised by ESI-MS (Table S1†).
Photophysical property of ThexT-modified immunostimulatory oligodeoxynucleotides
To investigate the fluorescence behaviour of ThexT-modified ODNs, we measured the absorption and emission spectra of ODNThexT1–ODNThexT17 (Fig. S2†). The absorption spectra revealed peaks at approximately 260 and 385 nm, which corresponded to the native nucleobase and ThexT-modified nucleobase, respectively. At the same concentration (2 μM), all ThexT-modified ODNs displayed a comparable absorbance at 260 nm, originating from intrinsic nucleobases. The maximum fluorescence intensity was observed at ∼445 nm. Notably, the fluorescence intensity varied depending on the position of ThexT within ODNs. As a result, ODNThexT14 exhibited relatively low fluorescence intensity, whereas ODNThexT17 showed relatively high fluorescence intensity. This was likely due to the fluorescent feature of ThexT as a molecular rotor, indicating distinct microenvironments within ODNs. While we could not directly examine interactions between ThexT-modified ODN and the TLR9 protein in this study, the microenvironment formed upon binding to the protein could influence fluorescence intensity of ThexT.Cellular uptake of ThexT-modified oligodeoxynucleotides
To investigate the fluorescence behaviour of ThexT-containing CpG ODNs in a cellular context, we examined their cellular uptake and localisation using confocal microscopy. Macrophages exhibit a strong response to bacterial DNA and biologically active CpG ODN owing to the presence of unmethylated CpG motifs, which the immune system recognises as pathogen-associated molecular patterns (PAMPs). These motifs trigger macrophage activation, resulting in the upregulation of MHC class II and co-stimulatory molecules, and the release of pro-inflammatory cytokines such as TNF-α, IL-1, IL-6, and IL-12. Macrophages serve as an appropriate immune cell model for evaluating the biological activity of synthetic CpG ODN given their crucial role in mediating innate immune responses. We selected the murine macrophage cell line RAW 264.7 which can internalise the designed oligonucleotides without the need for transfection reagents. Upon treating live RAW 264.7 cells with 10 μM of ThexT-labelled ODNs, we observed distinct cellular fluorescence of ThexT-labelled ODNThexT1 to ODNThexT17 using confocal microscopy with 405 nm laser excitation (Fig. 2). In contrast, cells treated solely with the medium exhibited much weaker signals, likely owing to autofluorescence. This indicated that the highly emissive nucleobases in ThexT effectively visualised internalised CpG oligonucleotides. Co-staining with the nuclear stain DRAQ5 confirmed that the ThexT-labelled ODNs were localised outside the nucleus (Fig. S3†), consistent with previous reports of immunostimulatory ODNs with fluorescent dyes co-localising with cytoplasmic proteins.37 Furthermore, these results indicate that ThexT is a reliable tool for visualising the cellular localisation of oligonucleotides, providing crucial insights into their internalisation, transport, and function within cellular compartments.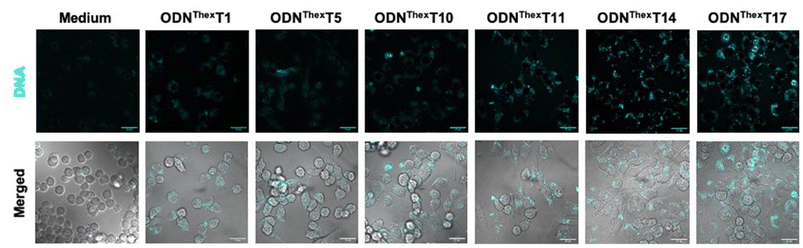 | ||
| Fig. 2 Confocal microscopic images of living RAW 264.7 cells treated with emissive ThexT-modified ODNs for 4 hours. Images of only medium and 10 μM of DNA-treated cells are shown. Scale bars: 20 μm. | ||
This highlights the importance of visualising nucleic acid-based drug delivery and cellular interactions, emphasising their indispensable roles in these processes.
Effect of ThexT-modification on immunostimulatory activity
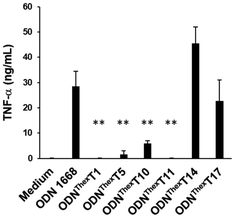
To examine the effect of the substitution of ThexT on the immunostimulatory properties of ODN 1668, we evaluated the production of pro-inflammatory cytokine TNF-α by RAW 264.7 cells following the treatment of modified immunostimulatory ODNs. In this experiment, none of the ODNs contained a 5-methylated CpG motif, as this modification could reduce the immunostimulatory activity of TLR9 agonists. ODN 1668 used in this study contains six thymine sites at the 1st, 5th, 10th, 11th, 14th, and 17th positions from the 5′-end of the oligonucleotide. We generated ThexT-modified ODNs by sequentially replacing each thymine site. Fig. 3 shows the levels of TNF-α release from RAW 264.7 cells in response to unmodified ODN 1668 and the series of ThexT-modified CpG ODNs. Notably, immunostimulatory activity varied significantly depending on the position of ThexT (Fig. 3). ODNThexT14 and ODNThexT17 induced cytokine release comparable to that induced by ODN 1668, whereas treatment with ODNThexT1 to ODNThexT11 resulted in a marked decrease in TNF-α release, with some responses close to the baseline levels observed in untreated cells. Agrawal et al. reported that substituting these thymine bases in the two adjacent nucleotide positions on the 3′-side to the CpG dinucleotide with abasic nucleoside severely impaired immunostimulatory activity.38 Our finding suggests that ODNThexT10 and ODNThexT11 substitutions substantially reduced immunostimulatory activity in RAW 264.7 cells. This implies that the two pyrimidine bases on the 3′-side of CpG dinucleotides are critical for TLR9 recognition. In addition to the absence of pyrimidine moieties,38 the π-extension of nucleobases at these locations can significantly influence TLR9 activation. Regarding the substitution outside core CpG motif (PuPuCGPyPy), ODNThexT1 and ODNThexT5, with alterations in the 5′-flanking sequence of the CpG motif, exhibited significantly weaker immunostimulatory effect. In contrast, both ODNThexT14 and ODNThexT17, with ThexT sites located in the 3′-flanking sequence of the CpG motif, elicited immunostimulatory activity comparable to ODN 1668.This result aligns with our previous finding that chemical modification of guanines in the 3′-flanking sequence of the CpG motif not only maintained the interaction between TLR9 and CpG ODNs but also enhanced immunostimulatory activity.22
To gain structural insight, we performed a molecular modelling study based on the previously published crystal structure of the TLR9-agonistic CpG ODN complex.21 Shimizu et al. previously reported the crystal structure of TLR9 complexed with an agonistic CpG-DNA, which consists of 12 mer sequences derived from ODN 1668.
Structural analysis demonstrated that agonistic DNA formed a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 dimeric complex with TLR9, and thymine bases in the core hexamer CpG motif formed van der Waals interactions with TLR residues.
2 dimeric complex with TLR9, and thymine bases in the core hexamer CpG motif formed van der Waals interactions with TLR residues.
We selected ODNThexT11 and ODNThexT14 for our modelling study. Both variants had thymine bases substituted with ThexT. However, we observed significant differences in their immunostimulatory activities depending on the ThexT incorporation site. Using the PDB structure (3WPC), in which the agonistic ODN 1668_12 mer was bound to TLR9, we constructed a molecular model in which ODNThexT11 and ODNThexT14 were bound to TLR9 (Fig. 4). By comparing these two, we sought to comprehend how ThexT incorporation position affects the binding propensity, anticipating that this would provide valuable insights.
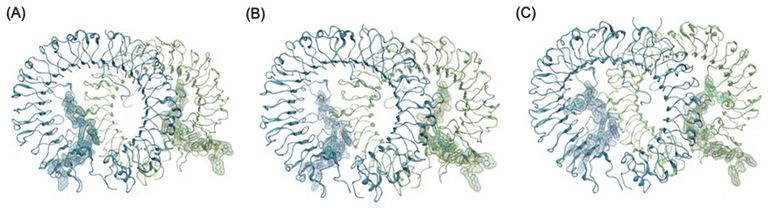 | ||
| Fig. 4 Overall structural models of TLR9-agonistic DNA complexes; (A) crystal structure of TLR9 with unmodified ODN1668_12 mer (PDB 3WPC), (B) plausible energy minimized binding model of TLR9 with (B) ODNThexT11_12 mer (C) ODNThexT14_12 mer based on PDB 3WPC. The DNA molecules are shown in stick representation and semi-transparent surface representation. Sequences of ODN 1668_12 mer, ODNThexT11_12 mer and ODNThexT14_12 mer are 5′-CATGACGTTCCT-3′, 5′-CATGACGTThexTCCT-3′ and 5′-CATGACGTTCCThexT-3′, respectively. | ||
The agonistic ODN 1668-bound TLR9 structure revealed that the core hexamer CpG motif (GACGTT) and the surrounding bases interact with the proximal amino acids in TLR9. For instance, the thymine base at the 11 position forms a hydrogen bond with Ser72 and stacks upon Trp96 (Fig. 5A and C).
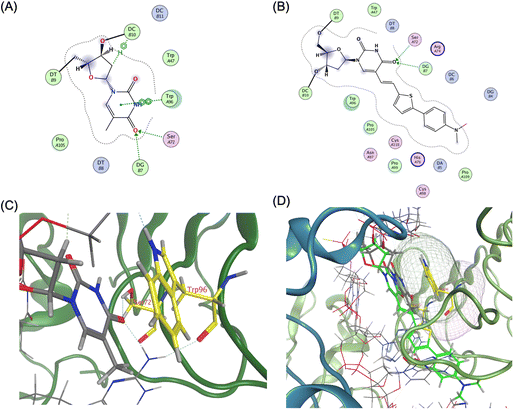 | ||
| Fig. 5 Ligand interaction between TLR9 and agonistic DNA complexes, (A) ligand interaction diagram of native T11 of ODN 1668_12 mer, (B) ligand interaction diagram of ThexT11 of ODNThexT11_12 mer, (C) magnified view of ligand interaction of native T11 of ODN1668_12 mer in TLR9, (D) superposition of ODN 1668_12 mer and ODNThexT11_12 mer by MOE, yielding an root-mean-square deviation (RMSD) value of 0.66 Å. ThexT is depicted as a green stick representation. | ||
Compared with the unmodified ODN 1668-bound TLR9 structure, the energy-minimised model of the ODNThexT11-TLR9 complex appeared to reduce the original interactions. Visualisation of van der Waals interactions revealed the spatial arrangement and contact regions between Trp96 and T11, with the binding mode of the incorporated ThexT appearing to weaken this interaction (Fig. 5B and D). This suggests that the alteration in T11 through ThexT incorporation may decrease the binding affinity, potentially explaining the observed reduction in immunostimulatory activity.
In contrast, the adjacent base T14 not only forms hydrogen bonds with Arg262 and Glu616 but also engages in multiple van der Waals interactions (Fig. 6A and C).
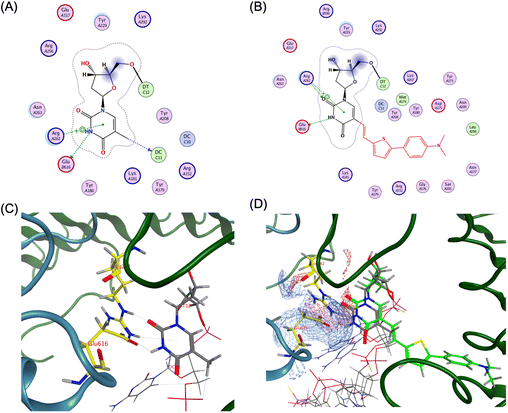 | ||
| Fig. 6 Ligand interaction between TLR9 and agonistic DNA complexes, (A) ligand interaction diagram of native T14 of ODN1668_12 mer, (B) ligand interaction diagram of ThexT ODNThexT14_12 mer, (C) magnified view of ligand interaction of native T14 of ODN 1668_12 mer in TLR9, (D) electrostatic map of the superposition of ODN 1668_12 mer and ODNThexT14_12 mer generated by MOE, yielding a root-mean-square deviation (RMSD) value of 0.64 Å. The Glu616 residue of second TLR9 is represented with asterisk. | ||
Notably, Glu616, a residue located at interface 2, suggested that the thymine base at position 14 contributes to interactions with both interfaces 1 and 2.
The energy-optimised model of the ODNThexT14-TLR9 complex revealed that the original interactions observed in the unmodified ODN 1668-bound TLR9 structure were largely conserved.
An electrostatic model visualisation of the interaction between T14 and the TLR9 binding site demonstrated that incorporated ThexT at 14 position exhibits a well-aligned spatial fit and overlapping contact regions with Arg262 and Glu616, thus reinforcing the proposed binding mode (Fig. 6B and D).
These findings align with the experimental results showing that ODNThexT14 retains immunostimulatory activity comparable to ODN 1668. This suggests that the modification at T14 may allow it to function as an adaptable flanking base. This alteration is expected to contribute to interactions with both interfaces 1 and 2 while maintaining CpG motif binding in the core hexamer (Fig. S4 and S5†).
Conclusions
Fluorescent nucleobases are versatile tools for tagging oligonucleotides and monitoring their interactions and reactions in biological environments. They are also being explored for potential use as nucleic acid surrogates. In this study, we demonstrated that ThexT, a fluorescent thymidine nucleoside with a π-extended structure at the 5th position of thymine, effectively highlights oligonucleotides internalised within cells. Furthermore, ThexT modification of the TLR9 agonist ODN influences immunostimulatory responses in a position-dependent manner. Notably, ThexT inhibited immunostimulatory activity at specific locations without the need for methylated CpG motifs.The incorporation of fluorescent nucleobases permits the visualisation of immunostimulatory oligonucleotides within immune cells and represents a promising strategy to modulate oligonucleotide-mediated immunostimulatory response to infections and other immune-related diseases. These findings provide valuable insight into the structural design of immunostimulatory oligonucleotides. Building on this research, our laboratory is currently developing therapeutic oligonucleotides with reduced immunogenicity and enhanced functional properties, guided by the structural information of the agonistic CpG ODN-TLR9 complex.
Data availability
The data supporting this study are included within the article and its ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Japan Society for the Promotion of Science (Grant-in-Aid for Transformative Research Areas (A) “Biophysical Chemistry for Material Symbiosis” 23H04076 to S. P., 23H04072 to Y. K.), and Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED (JP23ama121025). S. P. would like to acknowledge the “Rising Fellow Program” from Toyota Physical and Chemical Research Institute.References
- A. M. Krieg, A.-K. Yi, S. Matson, T. J. Waldschmidt, G. A. Bishop, R. Teasdale, G. A. Koretzky and D. M. Klinman, Nature, 1995, 374, 546–548 CrossRef CAS PubMed.
- F. Takeshita, C. A. Leifer, I. Gursel, K. J. Ishii, S. Takeshita, M. Gursel and D. M. Klinman, J. Immunol., 2001, 167, 3555–3558 CrossRef CAS PubMed.
- S. Bauer, C. J. Kirschning, H. Häcker, V. Redecke, S. Hausmann, S. Akira, H. Wagner and G. B. Lipford, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 9237–9242 CrossRef CAS PubMed.
- J. Vollmer and A. M. Krieg, Adv. Drug Delivery Rev., 2009, 61, 195–204 CrossRef CAS.
- A. F. Carpentier, G. Auf and J. Y. Delattre, Front. Biosci., 2003, 8, 115–127 CrossRef.
- D. Klinman, Nat. Rev. Immunol., 2004, 4, 248–257 CrossRef.
- M. Roman, E. Martin-Orozco, J. S. Goodman, M.-D. Nguyen, Y. Sato, A. Ronaghy, R. S. Kornbluth, D. D. Richman, D. A. Carson and E. Raz, Nat. Med., 1997, 3, 849–854 CrossRef CAS.
- H. Shirota and D. Klinman, Expert Rev. Vaccines, 2014, 13, 299–312 CrossRef CAS PubMed.
- V. Jovasevic, E. Wood, A. Cicvaric, H. Zhang, Z. Petrovic, A. Carboncino, K. Parker, T. Bassett, M. Moltesen, N. Yamawaki, H. Login, J. Kalucka, F. Sananbenesi, X. Zhang, A. Fischer and J. Radulovic, Nature, 2024, 628, 145–153 CrossRef PubMed.
- D. M. Klinman, Antisense Nucleic Acid Drug Dev., 1998, 8, 181–184 CrossRef.
- T. Yoshida, T. Hagihara, Y. Uchida, Y. Horiuchi, K. Sasaki, T. Yamamoto, T. Yamashita, Y. Goda, Y. Saito, T. Yamaguchi, S. Obika, S. Yamamoto and T. Inoue, Sci. Rep., 2024, 14, 11540 CrossRef.
- E. Kandimalla and S. Agrawal, Nucleic Acid Drugs, 2012, vol. 249, pp. 61–93 Search PubMed.
- Y. Kawamoto, W. Liu, J. H. Yum, S. Park, H. Sugiyama, Y. Takahashi and Y. Takakura, ChemBioChem, 2022, 23, e202100583 CrossRef PubMed.
- M. E. Distler, J. P. Cavaliere, M. H. Teplensky, M. Evangelopoulos and C. A. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2215091120 CrossRef.
- E. Kandimalla, D. Yu, Q. Zhao and S. Agrawal, Bioorg. Med. Chem., 2001, 9, 807–813 CrossRef.
- E. Kandimalla, L. Bhagat, D. Wang, D. Yu, F. Zhu, J. Tang, H. Wang, P. Huang, R. Zhang and S. Agrawal, Nucleic Acids Res., 2003, 31, 2393–2400 CrossRef PubMed.
- E. Kandimalla, L. Bhagat, F. Zhu, D. Yu, Y. Cong, D. Wang, J. Tang, J. Tang, C. Knetter, E. Lien and S. Agrawal, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14303–14308 CrossRef.
- M. Jurk, A. Kritzler, H. Debelak, J. Vollmer, A. Krieg and E. Uhlmann, ChemMedChem, 2006, 1, 1007–1014 CrossRef PubMed.
- M. Putta, F. Zhu, Y. Li, L. Bhagat, Y. Cong, E. Kandimalla and S. Agrawal, Nucleic Acids Res., 2006, 34, 3231–3238 CrossRef PubMed.
- D. Yu, M. Putta, L. Bhagat, Y. Li, F. Zhu, D. Wang, J. Tang, E. Kandimalla and S. Agrawal, J. Med. Chem., 2007, 50, 6411–6418 CrossRef PubMed.
- U. Ohto, T. Shibata, H. Tanji, H. Ishida, E. Krayukhina, S. Uchiyama, K. Miyake and T. Shimizu, Nature, 2015, 520, 702–705 CrossRef.
- Y. Araie, S. Ohtsuki, S. Park, M. Nagaoka, K. Umemura, H. Sugiyama, K. Kusamori, Y. Takahashi, Y. Takakura and M. Nishikawa, Bioorg. Med. Chem., 2021, 29, 115864 CrossRef CAS PubMed.
- W. Xu, K. M. Chan and E. T. Kool, Nat. Chem., 2017, 9, 1043–1055 CrossRef CAS PubMed.
- D. Dziuba, P. Didier, S. Ciaco, A. Barth, C. A. M. Seidel and Y. Mely, Chem. Soc. Rev., 2021, 50, 7062–7107 RSC.
- Y. Tor, Acc. Chem. Res., 2024, 57, 1325–1335 CrossRef CAS PubMed.
- S. Park, H. Otomo, L. Zheng and H. Sugiyama, Chem. Commun., 2014, 50, 1573–1575 RSC.
- I. Okamura, I. S. Park, R. Hiraga, S. Yamamoto and H. Sugiyama, Chem. Lett., 2017, 46, 245–248 CrossRef CAS.
- S. Hirashima, J. H. Han, H. Tsuno, Y. Tanigaki, S. Park and H. Sugiyama, Chem. – Eur. J., 2019, 25, 9913–9919 CrossRef CAS.
- A. W. Wee, J. H. Yum, H. Sugiyama and S. Park, RSC Chem. Biol., 2021, 2, 876–882 RSC.
- Y. Li, A. Fin, A. Rovira, Y. Su, A. Dippel, J. Valderrama, A. Riestra, V. Nizet, M. Hammond and Y. Tor, ChemBioChem, 2020, 21, 2595–2598 CrossRef CAS.
- J. Nilsson, T. Baladi, A. Gallud, D. Bazdarevic, M. Lemurell, E. Esbjörner, L. Wilhelmsson and A. Dahlén, Sci. Rep., 2021, 11, 11365 CrossRef CAS PubMed.
- J. Nilsson, C. Benitez-Martin, H. Sansom, P. Pfeiffer, T. Baladi, H. Le, A. Dahlén, S. Magennis and L. Wilhelmsson, Phys. Chem. Chem. Phys., 2023, 25, 20218–20224 RSC.
- B. Mir, I. Serrano-Chacón, P. Medina, V. Macaluso, M. Terrazas, A. Gandioso, M. Garavís, M. Orozco, N. Escaja and C. González, Nucleic Acids Res., 2024, 52, 3375–3389 CrossRef.
- D. Ward, E. Reich and L. Stryer, J. Biol. Chem., 1969, 244, 1228–1237 CrossRef.
- C. Liu and C. Martin, J. Mol. Biol., 2001, 308, 465–475 CrossRef PubMed.
- T. Kumagai, B. Kinoshita, S. Hirashima, H. Sugiyama and S. Park, ACS Sens., 2023, 8, 923–932 CrossRef PubMed.
- P. Ahmad-Nejad, H. Häcker, M. Rutz, S. Bauer, R. Vabulas and H. Wagner, Eur. J. Immunol., 2002, 32, 1958–1968 CrossRef.
- D. Yu, E. Kandimalla, Q. Zhao, L. Bhagat, Y. Cong and S. Agrawal, Bioorg. Med. Chem., 2003, 11, 459–464 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ob02034k |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |