 Open Access Article
Open Access ArticleSynthesis and preliminary evaluation of cardiac imaging with [68Ga]Ga-NOTA-CTP in normal and infarcted CD1 mice†
Surendra Reddy Gundama,
Manasa Kethamreddya,
Andy González Riveraa,
Aditya Bansala,
Viktoria Krola,
Daniella A. Sahagun b,
Joanna E. Kusmirekc,
Derek R. Johnson
b,
Joanna E. Kusmirekc,
Derek R. Johnson a,
Maliha Zahidb,
Val J. Lowea and
Mukesh K. Pandey
a,
Maliha Zahidb,
Val J. Lowea and
Mukesh K. Pandey *ade
*ade
aDivision of Nuclear Medicine, Department of Radiology, Mayo Clinic, Rochester, MN 55905, USA. E-mail: pandey.mukesh@mayo.edu
bDivision of Comprehensive Cardiology, Department of Cardiovascular Medicine, Mayo Clinic, Rochester, MN 55905, USA
cDivision of Cardiovascular Radiology, Department of Radiology, Mayo Clinic, Rochester, MN 55905, USA
dDepartment of Pharmacology, Mayo Clinic, Rochester, MN 55905, USA
eMayo Clinic Comprehensive Cancer Center, Mayo Clinic, Rochester, MN 55905, USA
First published on 29th May 2025
Abstract
Cell-penetrating peptide-based probes for positron emission tomography (PET) are currently being developed for cardiac imaging. Herein, we have conjugated a synthetic 12 amino acids (NH2-APWHLSSQYSRT-COOH) cardiac targeting peptide (CTP) with a NOTA chelator for 68Ga labeling. The [68Ga]Ga-NOTA-CTP was synthesized with a decay-corrected radiochemical yield of 68.9 ± 12.8% (n = 13) and molar activity (Am) of 1.3 ± 0.5 GBq per μmol (n = 13). The tracer was evaluated in healthy and diseased CD1 mice with myocardial infarction following ligation of the left anterior descending artery. PET/CT imaging and ex vivo biodistribution revealed rapid (within 30 min) clearance of [68Ga]Ga-NOTA-CTP from the blood through renal and hepatobiliary excretion pathways in both healthy and infarcted animals. The uptake of [68Ga]Ga-NOTA-CTP in the heart of healthy and infarcted animals did not show any statistically significant difference for up to 120 min post-injection, but regional differences within healthy and infarcted hearts were detected with [68Ga]Ga-NOTA-CTP by PET/CT imaging at early time points post-injection. Within a healthy heart, the left ventricle standardized uptake value (SUV) was lower than the right ventricle SUV at 10–30 min post-injection. This regional difference between the left and right ventricles was absent in the infarcted heart, likely due to post-ligation changes.
Introduction
Cardiovascular diseases (CVDs) are the leading cause of mortality in the world.1–4 Globally, 523 million CVD cases and associated 18.6 million deaths were reported in 2019.5 Numerous novel radiotracers in positron emission tomography (PET) and single-photon emission computed tomography (SPECT) are being explored in assessing myocardial perfusion, cardiac autonomic dysfunction, atherosclerotic plaques, cardiac metabolism, and viability.6–16 Out of the two modalities, PET is preferred over SPECT due to its higher sensitivity, greater interpretive certainty, lower patient dosimetry, and shorter imaging protocol. Importantly, in the case of CVDs, PET can assess microvascular function by quantitatively measuring the myocardial blood flow at rest and peak stress.17,18In clinical practice, PET imaging of cardiac perfusion allows the detection of cardiac abnormalities. The perfusion imaging is commonly performed with 13N-ammonia (T1/2 = 9.9 min), the potassium analog 82Rb (T1/2 = 76 s), or 15O-water (T1/2 = 2.05 min).18–26 These perfusion markers have their strengths and weaknesses. Administered 13N-ammonia is rapidly cleared from the blood with a first-pass extraction fraction of ∼80% and is taken up by the heart, brain, liver, and kidneys.19,20 The uptake of 13N-ammonia is proportional to coronary blood flow; once taken up, the 13N-ammonia is metabolically trapped in the cardiac cells as 13N-glutamine.19,20 In the case of 82Rb, after administration, 82Rb is rapidly cleared from the body with a first-pass extraction fraction of ∼65% and is taken up primarily in the heart, lungs, and kidneys.23,24 The uptake of 82Rb in the myocardium is also dependent on blood flow, and once taken up, 82Rb is actively transported into the cardiac cells through the Na+/K ATPase pump.23,24 As compared to 13N-ammonia, images from 82Rb scans are inferior in resolution and image quality due to the longer positron range of 82Rubidium (8.6 mm) than 13N-ammonia (2.53–5.4 mm).18–22,25 The third perfusion marker, 15O-water is taken up by the heart by passive diffusion with highest first-pass extraction fraction (∼100%).26 The positron range of 15O-water (4.14 mm) is between the positron range of 13N-ammonia and 82Rb and consequently the imaging quality is better than 82Rb scans but inferior to 13N-ammonia scans.21
Besides imaging quality, other factors need to be considered when choosing a cardiac perfusion tracer. An onsite cyclotron is required for producing 13N-ammonia and 15O-water due to their shorter half-life, whereas the shorter half-life of 82Rb necessitates an onsite 82Sr/82Rb generator. Installing and maintaining an onsite cyclotron or generator can be prohibitive for many hospital sites.18–22 For ease of use and ready availability of perfusion tracers, comparatively longer half-life radiotracers are desirable for broader adoption of PET-based myocardial perfusion imaging in imaging centers lacking onsite cyclotrons or generators. In this context, 18F, with a half-life of 109.7 min and a positron range of 1.03 mm, and 68Ga, with a half-life of 67.71 min and a positron range of 3.5 mm, are possible choices for PET radionuclides for developing perfusion markers.21,27–30
More recently, a promising myocardial perfusion marker, [18F]F-flurpiridaz or BMS-747158-02, is emerging as an option in cardiac imaging.31–33 It is based on the insecticide pyridaben, an inhibitor of the mitochondrial respiratory complex I, a complex of the electron transport chain in the mitochondria.31 It has a first-pass extraction fraction of 94% with primary uptake in the liver, brain, heart, and kidneys.32,33 It has shown encouraging results in phase 3 clinical trials as a perfusion marker for detecting coronary artery disease.33 There are two other 18F labeled perfusion markers, fluorodihydrorotenone ([18F]FDHR)34 derived from insecticide and p-fluorobenzyl triphenylphosphonium cation ([18F]FBnTP)35,36 derived from fungicide that are under investigation at the preclinical stage for their performance as perfusion marker. After administration, these are rapidly cleared from the blood, taken up in the heart in a perfusion-dependent manner, and trapped in the mitochondria of cardiomyocytes. Meanwhile the manuscript is being written [18F]F-flurpiridaz has received food and drug administration's approval in the United States for evaluation of myocardial ischemia and infarction.
Zahid et al. reported a synthetic 12-amino acid cardiac targeting peptide (CTP), NH2-APWHLSSQYSRT-COOH, specifically targeting cardiomyocytes.37–41 The fluorophore-conjugated CTP could accumulate in the heart of Balb/C mice following retro-orbital intravenous injection.37–39 In a different study, accumulation of Cy5.5 labeled CTP was observed in the heart in an isolated rat heart after perfusion in a Langendorff perfusion system.40,41 Following encouraging results in vivo and ex vivo murine models, Zahid et al. tested CTP conjugated-amiodarone (antiarrhythmic drug) in guinea pig42,43 and CTP-conjugated miRNA106a for treating hypertrophic human cardiomyocytes.44 Authors delivered amiodarone to the heart of guinea pigs after intraperitoneal administration of the CTP-amiodarone complex with a functional response.42,43 The ability of CTP to specifically enter cardiomyocytes has been replicated in multiple vertebrate species by at least four independent researchers around the world.45–48
The exact mechanism by which CTP peptide gets internalized in the Cardiomyocytes is not well understood, however we hypothesize that since it is a positively charged peptide at physiological pH, it could cross the cytoplasmic membrane of cardiomyocytes passively like other cell penetrating peptides and charged molecules.49,50 It is known that lipophilic cations are capable of passive diffusion into the cytoplasm and mitochondria of cardiomyocytes in response to a large negative plasma- and mitochondrial- membrane potentials.50,51
Encouraged by previous studies using CTP, an attempt was made to assess if radiolabeled CTPs can be used to image the heart. In this pursuit, the biodistribution of 99mTc radiolabeled CTP as a [99mTc]Tc-HYNIC-CTP was compared to the known myocardial perfusion marker [99mTc]Tc-Sestamibi in CD1 mice using SPECT/CT imaging.40 Among the two, [99mTc]Tc-HYNIC-CTP showed less extra-cardiac uptake than the [99mTc]Tc-Sestamibi. The administered [99mTc]Tc-Sestamibi showed cardiac uptake and significant uptake in the liver, gut, and kidneys.40 On the other hand, [99mTc]Tc-HYNIC-CTP showed a more targeted heart uptake and no uptake in the liver and gut. There was [99mTc]Tc-HYNIC-CTP uptake in the heart at early time point post-injection and renal clearance with radioactivity primarily present in the kidneys and bladder.40 This suggested that [99mTc]Tc-HYNIC-CTP could be used for cardiac imaging with SPECT. The next logical step was to develop a PET radioisotope labeled CTP probe for PET imaging of cardiac abnormalities. Hence, we synthesized [68Ga]Ga-NOTA-CTP with the PET radionuclide 68Ga and tested its PET imaging potential and ex vivo biodistribution in healthy and diseased CD1 mice with myocardial infarction.
Materials and methods
General consideration
All chemicals and solvents were purchased from various commercial manufacturers – Sigma-Aldrich, St Louis, MO, Fisher Scientific, Waltham, MA, and Alfa Aesar, Haverhill, MA and used as received without further purification unless otherwise stated. The NOTA-CTP was custom synthesized by the Peptide & Peptoid Synthesis Facility at the University of Pittsburgh, Pittsburgh, PA, with >97% purity. The mass spectrum, HPLC/UV trace of NOTA-CTP is provided in ESI Fig. S1–S3.† For the radiolabeling, 3.0 M NaOAc (pH 8.4–8.8) buffer was prepared by dissolving 24.6 g of NaOAc in 100 mL of distilled water. Oasis® Millex-GV 0.22 μm (Part No. 186008083) was purchased from Merck, Rahway, NJ. rad-TLC analysis of the reaction mixtures was performed on glass microfiber chromatography paper impregnated with silica gel in 0.1 M sodium citrate solution (pH 5.0) and analyzed using an AR-2000 rad-TLC imaging scanner (Eckert & Ziegler, Valencia, CA). The 0.1 M sodium citrate buffer was prepared by dissolving 0.294 g of sodium citrate dihydrate in 10 mL of distilled water, and the pH was adjusted to 5.0 with 80 μL of concentrated HCl. The final analytical rad-HPLC determined the final purity and identity of compounds performed on a Phenomenex 5 μm, C18(2), 100 Å, LC Column 250 × 4.6 mm (Phenomenex, Torrance, CA), at a UV detector wavelength of 214 nm. Analytical rad-HPLC was performed using H2O + 0.1% TFA (solvent A) and CH3CN + 0.1% TFA (solvent B) with a flow rate of 0.5 mL min−1 by applying a gradient method (0–5 min = 24% B to 32.2% B, 5–20 min = 32.2% B to 35% B). [68Ga]Ga-NOTA-CTP was identified between 8.7–9.3 min retention time.Synthesis of [natGa]-NOTA-CTP
To a solution of 150 μg of NOTA-CTP peptide (1.0 μg μL−1) in Milli-Q water, 10 μL of 3 M sodium acetate (NaOAc) buffer was added and pH was adjusted between 4.0 and 4.5 using 1 M HCl. Immediately afterward, 120 μg of GaCl3 solution (12 μg μL−1) was added, and the reaction mixture was incubated at room temperature for 1 hour. UV-HPLC analysis confirmed the formation of [natGa]-NOTA-CTP with a 90% yield and a retention time of 8.62 minutes (ESI Fig. S5†).Radiolabeling
All experiments were performed manually in a sterile hot cell environment. The peptide precursor solution was prepared by dissolving 1.0 mg of the respective peptide (NOTA-CTP) in 1.0 mL distilled water. For the synthesis of [68Ga]Ga-NOTA-CTP, 1.0 mL of [68Ga]GaCl3 (0.2 ± 0.1 GBq) (n = 13) eluted from the 68Ge/68Ga generator (Eckert & Ziegler, Valencia, CA) was transferred into a 5.0 mL reaction vial. The pH of the reaction mixture was adjusted to 4.5–5.0 with 70 μL of 3 M NaOAc (pH 8.4–8.8) before the addition of 150 μg at a concentration of 1.0 μg μL−1 of NOTA-CTP solution. Then, the reaction mixture was allowed to stir for up to 10 min at RT. The radiolabeling yield was analyzed by rad-TLC using 0.1 M sodium citrate solution (pH 5.0) as the mobile phase. The final pH of the reaction mixture was adjusted to 6.1–6.5 with an additional 170 μL of 3 M NaOAc (pH 8.4–8.8) and passed through a Millex-GV 0.22 μm sterile filter unit. If the radiochemical purity calculated by rad-HPLC was less than 96%, the reaction mixture was purified from a reverse phase rad-HPLC, and final product was concentrated using a C-18 SepPak cartridge and eluted with a solution of 25% EtOH in NaOAc (1 M) followed by addition of a saline solution (0.9%) reaching a final formulation of 10/30/60 v/v/v ethanol/1.0 M sodium acetate/saline. [68Ga]Ga-NOTA-CTP was obtained in 68.9 ± 12.8 (n = 13) decay corrected radiochemical yield, 1.3 ± 0.5 GBq per μmol (n = 13) of molar activity (Am) and >97.8% radiochemical purity after HPLC semipreparative purification confirmed by analytical rad-HPLC at the end of the synthesis (ESI Fig. S4†). Total synthesis and formulation time was 30 min without purification and 60 min with HPLC purification from when [68Ga]GaCl3 was transferred into the reaction vial. The radiochemical purity of the synthesized [68Ga]Ga-NOTA-CTP was analyzed by analytical rad-HPLC having a retention time of 8.76 min (ESI Fig. S4†). Molar activity (Am) was measured by dividing the radioactivity (GBq) present at the end of the synthesis with the amount of NOTA-CTP (μmol) present in the final formulation.Stability analysis
The stability of [68Ga]Ga-NOTA-CTP was assessed in the final formulation (10/30/60 v/v/v ethanol/1.0 M sodium acetate/saline) incubated at room temperature up to 120 min with 20 min intervals using analytical rad-HPLC. The stability in human and mouse sera was assessed by rad-TLC using 0.1 M Sodium citrate buffer (pH = 5.0) as a mobile phase at 0 min, 30 min, 60 min, and 120 min post-incubation. For the serum stability studies, 100 μL of [68Ga]Ga-NOTA-CTP (∼37.0 MBq) was added to 100 μL of human serum or mouse serum in an Eppendorf tube. The mixture was then incubated at 37 °C and stirred at 1500 rpm in a thermomixer for up to 120 min. A small aliquot (∼0.5 μL) was taken out at different time points for rad-TLC.Animals
Healthy CD1 mice (n = 9, body weight 35.45 ± 6.54 g) and CD1 mice with myocardial infarction (n = 7, body weight 31.35 ± 6.32 g) were obtained from Envigo RMS LLC, Indianapolis, IN. These animals were received and housed at the Department of Comparative Medicine facility at the Mayo Clinic, Rochester, MN. Table 1 shows the age and sex of mice used in the study. The myocardial infarction was induced by the ligation of the left anterior descending artery following thoracotomy under surgical anesthesia, leading to ischemia of the anterior and anteroseptal myocardium at Envigo RMS LLC, Indianapolis, IN. Postoperative care was provided for 3 days at Envigo RMS LLC, Indianapolis, IN, to mitigate the complications associated with infarction and anesthesia before being shipped to Mayo Clinic Rochester. At Mayo Clinic, the animals were nourished with a regular diet and water ad libitum and were maintained at optimal conditions of 40% relative humidity and 22 °C temperature with 12-hour light–dark cycles.| Healthy CD1 mice (n = 9) | Age (week) average ± SD | Body weight (g) average ± SD |
|---|---|---|
| Males (n = 5) | 10.69 ± 2.44 | 39.80 ± 4.27 |
| Females (n = 4) | 10.36 ± 2.69 | 30.03 ± 4.36 |
| CD1 mice with myocardial infarction (n = 8) | Age (week) average ± SD | Body weight (g) average ± SD |
|---|---|---|
| Males (n = 4) | 9.79 ± 1.74 | 35.24 ± 5.54 |
| Females (n = 4) | 8.96 ± 1.75 | 27.03 ± 2.24 |
PET/CT imaging
Animals were injected with 2.29 ± 0.83 MBq (62.00 ± 22.31 μCi) of [68Ga]Ga-NOTA-CTP (n = 16) through the tail vein under 1–2.5% isoflurane anesthesia. Animals underwent whole-body dynamic PET-CT using a small animal microPET/CT imaging system, Inveon (Siemens Medical Solutions USA, Malvern, PA), immediately after injection for 30 min with a framing sequence of 4 × 15 s, 8 × 30 s, 5 × 60 s, 4 × 300 s. A CT scan for 7 minutes followed each PET scan. The PET/CT images were reconstructed using Siemens Inveon Micro PET-CT proprietary software. For analysis of PET/CT images, the PET and CT images were overlayed using MIM-7.2.7 software (MIM Software Inc., Cleveland, OH). ROIs were drawn in the heart in the overlayed CT images to delineate the whole heart, left atrium (LA), right atrium (RA), left ventricle (LV), and right ventricle (RV), as shown in ESI Fig. S6.† The mean concentration of the PET signal was quantified in the overlayed PET image in kBq per cc in the ROI for whole heart, left and right ventricle, which was converted to μCi per cc. The SUV was calculated using decay-corrected activity values in the following formula:Ex vivo biodistribution
The animals were transferred to a heating pad under a 1–2.5% isoflurane maintenance dose after the final imaging scan. A midline incision was made, and all the organs were harvested, including the heart. The heart was then placed on a Petri dish and perfused with 10.0 mL of phosphate-buffered saline (PBS) using a 10 mL syringe through the left ventricular apex, clearing the blood in all the chambers. The dissected organs were transferred to labeled and preweighed vials and counted in 2480 Wizard 2 automatic gamma counter (PerkinElmer, Waltham, MA) for decay-corrected 68Ga counts. SUV was then calculated using the following formula:Statistical analysis
The uptake data was analyzed using the Excel spreadsheet program (Microsoft Corporation, Redmond, WA). Uptake was compared multifold between different heart regions and other major organs using unpaired Student t-test analysis. P-Values less than 0.05 were considered statistically significant.Ethical standards
All animal studies were performed after approval and under the Mayo Clinic's Institutional Animal Care and Use Committee (IACUC) guidelines and regulations.Results and discussion
Chemistry
The NOTA-CTP was custom synthesized at the Peptide & Peptoid Synthesis Facility at the University of Pittsburgh, Pittsburgh, PA, with >97% purity (ESI Fig. S1–S3†).Radiochemistry
68Ga-labeling of NOTA-CTP was performed following our previous standardized protocol52 for radiolabeling peptides (Fig. 1). [68Ga]GaCl3 obtained from a 68Ge/68Ga generator was used for the radiolabeling. The labeling was carried out at pH 4.5–5.0 with 150 μg of precursor (optimized concentration, Table 2) by incubating for up to 10 min at room temperature (RT). Radiolabeling yield was confirmed by rad-TLC using 0.1 M sodium citrate solution at pH 5.0 as the mobile phase (ESI Fig. S7†). [68Ga]Ga-NOTA-CTP was obtained with a decay-corrected radiochemical yield of 68.9 ± 12.8% (n = 13) and molar activity (Am) of 1.3 ± 0.5 GBq per μmol (n = 13) with >97.8% radiochemical purity, calculated by rad-HPLC, after HPLC semipreparative purification at the end of the synthesis (ESI Fig. S4†).| S. No | Concentration of NOTA-CTP (μg mL−1) | Reaction time (min) | Reaction conversion (%) based on rad-TLC |
|---|---|---|---|
| Reaction conditions: pH 4.5–5.0, 70 μL of 3 M NaOAc (pH 8.4–8.8), RT.a Reaction conversion was measured by rad-TLC. | |||
| 1 | 69.57 | 10 | 76.00a |
| 2 | 69.57 | 20 | 81.00a |
| 3 | 69.57 | 30 | 87.00a |
| 4 | 100.84 | 10 | 98.00a |
| 5 | 122.95 | 10 | 100.00a |
Stability assessment in formulation and serum
The radiochemical purity of [68Ga]Ga-NOTA-CTP decreased from 98.78 ± 1.08% (n = 4) at 0 min post-synthesis to 92.52 ± 3.66% (n = 4) at 2 h post-synthesis in the formulation (Fig. 2A and ESI Table S1†). Although we saw decreased radiochemical purity by rad-HPLC based on [68Ga]Ga-NOTA-CTP peak, we did not observe free 68Ga leaching from the molecule up to 120 min in the rad-HPLC chromatogram at the retention time of ∼3.35 min (ESI Fig. S7 and S8†). Thus, the impurities contributing to decreased radiochemical purity resulted from the peptide degradation, likely forming labeled fragments with a lower Rf and not related to the stability of the 68Ga-radiolabeling.The stability of [68Ga]Ga-NOTA-CTP was assessed in human and mouse sera incubated at 37 °C in a thermomixer with 1500 rpm for up to 120 min by rad-TLC at 0 min, 30 min, 60 min, and 120 min post-incubation. The results are summarized in Fig. 2B, and representative chromatograms are presented in ESI Fig. S9 and S10,† followed by the tabulated values in ESI Table S2.† Interestingly, [68Ga]Ga-NOTA-CTP maintained stability above 96.33 ± 1.96% in mouse serum (n = 5) and 97.96 ± 0.37% in human serum (n = 4) during the first hour of incubation, with its stability decreasing to 93.55 ± 1.62% in mouse serum (n = 5) and 93.98 ± 0.98% in human serum (n = 4) after 2 hours of incubation at 37 °C. Although it seems [68Ga]Ga-NOTA-CTP was more stable in human serum than in formulation, but it should be noted that stability in formulation was assessed by rad-HPLC and stability in serum was assessed by rad-TLC. The rad-TLC does not have sufficient resolution to identify peptide degradation. It is likely that the labeling is equally stable in formulation and sera with similar peptide degradation but was not visible on the rad-TLC. Considering the lower amount of radioactivity (∼185 MBq) used as 68Ga for the synthesis of [68Ga]-NOTA-CTP, this study does not assess any potential radiolysis during large scale (high radioactivity) clinical production of [68Ga]-NOTA-CTP.
Imaging and biodistribution
Based on the stability profile, the developed radiotracer showed sufficient stability and purity (>98% by rad-TLC) at the time of animal injection.In cardiac studies, the use of larger species, such as canine or porcine ligation models, offers physiology and behavior like humans. However, the disadvantages of larger species include the high cost, the need for dedicated facilities for these species, and an increase in the complexity of handling. According to the International Atomic Energy Agency's recommendations for the preclinical development of radiopharmaceuticals, the use of small animals such as mice and rats has several advantages over large animals.53 The advantages include the possibility of performing whole-body dynamic studies in conventional micro imaging equipment, low cost and easy handling. The infarct model used in this study is well established in mice and commercially available. Our laboratory has a micro-PET scanner (used for this work) that allows us to obtain PET images in mice, which was one of the reasons for selecting the murine model.
The biodistribution of [68Ga]Ga-NOTA-CTP was assessed by PET/CT imaging at several time points post-injection in healthy and diseased CD1 mice with myocardial infarction. In this diseased model, infarction was induced in the heart of CD1 mice by ligating the left anterior descending artery (Fig. 3). Following ligation, the mice were allowed to recuperate for up to 6 weeks with intact ligation in the heart, mimicking chronic myocardial infarction. Prior studies evaluated cardiac changes during the first 8 weeks post ligation of the left anterior descending artery and observed increase in the end-diastolic volume (EDV) and end-systolic volume (ESV) and decrease in the ejection fraction compared to a healthy heart.54–56 This is because the ligation causes rerouting of blood supply with regional decreased to absent perfusion with subsequent cardiac remodeling.54–56 An imaging tracer that could detect these changes at the initial stages would allow the identification of subjects at risk for adverse remodeling, leading to early interventions. Therefore, in our study, [68Ga]Ga-NOTA-CTP was administered within 6 weeks of the surgery, and its distribution was assessed and compared with healthy animals.
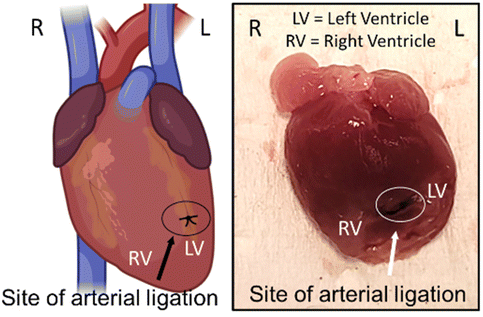 | ||
| Fig. 3 Schematic diagram showing ligation site in the left anterior descending artery of an infarcted heart. | ||
Generally, the administered peptides are primarily cleared from the blood through renal clearance without reabsorption, leading to high urinary bladder activity.57 Serial PET/CT imaging of the healthy animals post-administration of [68Ga]Ga-NOTA-CTP showed that the biodistribution profile of [68Ga]Ga-NOTA-CTP was typical for peptides. The administered [68Ga]Ga-NOTA-CTP had high bladder activity as early as 0–5 min post-injection, with continued urinary clearance until 120 min post-injection (Table 3 and Fig. 4). The major organs taking up [68Ga]Ga-NOTA-CTP were the urinary bladder, kidneys, heart, and liver until 30 min post-injection (Table 3). No or low signal was detected in the heart, kidneys, and liver at 60 min and 120 min post-injection, with the majority of signal present in the urinary bladder (Table 3).
| Timepoint (min) | SUV heart (average ± SD, n = 9) | SUV liver (average ± SD, n = 9) | SUV muscle (average ± SD, n = 9) | SUV kidneys (average ± SD, n = 9) | SUV urinary bladder (average ± SD, n = 9) |
|---|---|---|---|---|---|
| 0–5 | 0.91 ± 0.15 | 1.42 ± 0.49 | 0.36 ± 0.12 | 2.64 ± 0.89 | 7.51 ± 3.49 |
| 6–10 | 0.59 ± 0.09 | 1.66 ± 0.58 | 0.28 ± 0.15 | 2.15 ± 1.21 | 10.73 ± 5.16 |
| 11–20 | 0.46 ± 0.08 | 1.79 ± 0.55 | 0.25 ± 0.15 | 1.91 ± 1.29 | 12.46 ± 5.86 |
| 21–30 | 0.33 ± 0.07 | 1.83 ± 0.54 | 0.21 ± 0.16 | 1.71 ± 1.37 | 14.31 ± 6.88 |
| 60 | 0.04 ± 0.04 (n = 6) | 0.69 ± 0.32 (n = 3) | 0.18 ± 0.13 (n = 7) | 0.92 ± 0.68 (n = 3) | 10.64 ± 6.10 |
| 120 | 0.02 ± 0.01 (n = 6) | 0.14 (n = 1) | 0.19 ± 0.18 (n = 5) | 0.1 (n = 1) | 9.02 ± 8.59 (n = 6) |
An ideal cardiac perfusion marker should show high initial uptake in the heart and rapid clearance from the blood pool.58 Indicative of being a promising perfusion marker, we observed a high uptake of [68Ga]Ga-NOTA-CTP (SUV > 1) in the healthy heart immediately after the intravenous administration, followed by a rapid clearance within 30 min of administration (Table 3 and Fig. 5, ESI Table S3A†). Noticing a faster clearance, [68Ga]Ga-NOTA-CTP uptake was evaluated more closely within 30 min post-injection at several time points (Fig. 5 and 6, ESI Tables S3A and S3B†). The CT images were used to draw regions of interest in the whole heart, left, and right ventricles to assess the uptake of [68Ga]Ga-NOTA-CTP in these regions in the PET scans (Fig. 4 and ESI Fig. S6†).
Regarding the whole heart uptake of [68Ga]Ga-NOTA-CTP, we observed no statistical difference in the uptake in the infarcted heart as compared to the healthy heart at all time points within 30 min post-injection (Fig. 5 and ESI Table S3A†). An infarcted heart is known to undergo remodeling and rerouting of blood supply that could impact uptake of perfusion markers.54–56 Regional differences in [68Ga]Ga-NOTA-CTP uptake in the left ventricle (LV) or right ventricle (RV) in the whole heart (WH) in healthy and infarcted animals were compared (Fig. 5 and 6, and ESI Tables S3A and S3B†). Within a healthy heart, the SUV in LV was lower than the RV at the 10–30 min post-injection time point (Fig. 5 and ESI Table S3A†). This difference between uptake in LV and RV was absent in the infarcted heart (Fig. 5 and ESI Table S3A†). It is possible that dilation of LV and increased EDV in the infarcted heart contributed to higher retention of [68Ga]Ga-NOTA-CTP in LV in the infarcted heart.54–56,59 Additionally, we also compared the ratio of uptake in LV and RV normalized to the entire heart (Fig. 6 and ESI Table S3B†). In both infarcted and healthy hearts, the LV/whole heart SUV ratio was lower than the RV/whole heart SUV ratio at 4–30 min post-injection time point (Fig. 6 and ESI Table S3B†).
In the context of adverse remodeling, after the acute phase of an infarction, the heart undergoes a process of scarring and remodeling, which leads to changes in its geometry, dimensions, and function.60 Adverse remodeling of the left ventricle involves molecular, cellular, and interstitial changes within the tissue.61 This process often occurs following an ST-Segment Elevation Myocardial Infarction (STEMI), which results from the blockage of one or more coronary arteries.61 In such cases, a scarred area typically forms, and the patient may develop HF with either a reduced or mildly reduced ejection fraction (EF). EF is a key measure of the heart's ability to pump blood out the left ventricle. In our experiments, we observed a trend of higher uptake of the radiotracer in the left ventricle of the infarcted model compared to the healthy group (Fig. 5). This observed increase in blood retention of [68Ga]Ga-NOTA-CTP in the infarcted left ventricle implies a lower ejection fraction and potential adverse remodeling of the left ventricle in the infarcted model.
The animals with infarcted hearts showed biodistribution profiles in all organs/tissues similar to healthy animals at 120 min post-injection, as assessed by ex vivo biodistribution except the heart (Table 4). Overall, the uptake of [68Ga]Ga-NOTA-CTP was low in heart; however, the infarcted heart showed statistically higher uptake of [68Ga]Ga-NOTA-CTP (SUV = 0.03 ± 0.02, n = 7) as compared to the healthy heart (SUV = 0.02 ± 0.006, n = 9). Most of the radioactivity was found in organs involved in excretion, like the gallbladder, urinary bladder, small intestine, and cecum at 120 min post injection (Table 4). High urinary and gallbladder uptake also confirmed renal and hepatobiliary excretion of [68Ga]Ga-NOTA-CTP (Table 4).
| Organ | Healthy mice | Infarcted mice |
|---|---|---|
| SUV (average ± SD, n = 9) | SUV (average ± SD, n = 7) | |
| *P value < 0.05 healthy mice vs. infarcted mice | ||
| Brain | 0.01 ± 0.004 | 0.005 ± 0.003 |
| Heart | 0.02 ± 0.006 | 0.03 ± 0.02* |
| Lung | 0.04 ± 0.02 | 0.04 ± 0.04 |
| Liver | 0.22 ± 0.16 | 0.15 ± 0.04 |
| Kidneys | 0.85 ± 0.28 | 0.63 ± 0.21 |
| Spleen | 0.04 ± 0.03 | 0.05 ± 0.04 |
| Small intestine | 2.74 ± 1.29 | 3.38 ± 2.37 |
| Large intestine | 0.94 ± 1.30 | 1.21 ± 1.26 |
| Pancreas | 0.05 ± 0.06 | 0.05 ± 0.04 |
| Muscle | 0.03 ± 0.03 | 0.02 ± 0.01 |
| Urine | 52.68 ± 27.34 | 39.68 ± 37.93 |
| Bone | 0.83 ± 2.35 | 0.03 ± 0.02 |
| Bladder | 5.11 ± 5.63 | 6.53 ± 13.43 |
| Skin | 0.02 ± 0.01 | 0.03 ± 0.03 |
| Adipose | 0.06 ± 0.08 | 0.05 ± 0.05 |
| Stomach | 0.06 ± 0.07 | 0.08 ± 0.10 |
| Cecum | 2.36 ± 2.24 | 2.28 ± 3.64 |
| Blood | 0.05 ± 0.05 | 0.02 ± 0.01 |
| Feces | 5.43 ± 8.97 | 7.00 ± 7.72 |
| Gall bladder | 23.48 ± 19.28 (n = 5) | 12.17 ± 13.13 (n = 3) |
In an earlier study, [68Ga]Ga-DOTA was tested as a cardiac perfusion marker at 24 ± 4 h post-coronary ligation.59 In this myocardial infarction rat model, higher uptake of [68Ga]Ga-DOTA was observed at the infarct site compared to a remote site within the heart at 30 min post-administration of [68Ga]Ga-DOTA.59 This suggests that remodeling and rerouting of the heart's blood supply could be underway as early as 24 ± 4 h, contributing to delayed washout and increased retention of [68Ga]Ga-DOTA at the infarct site aligning with our findings.59 Additionally, in the present study [68Ga]Ga-NOTA-CTP was assessed in the chronically infarcted heart model but there were previous studies where other radiotracers were assessed in acutely infarcted heart model. In an acutely infarcted heart model, the ligation of the left anterior descending coronary artery has occurred within 3–24 h of imaging. Hence, the heart is still in shock with severe perfusion changes.62–65 Perfusion markers can demonstrate profound changes in cardiac perfusion in the acute infarction model. There are many preclinical studies done with acute infarction heart rodent models showing decreased perfusion utilizing [99mTc]Tc-MIBI, 13N-ammonia, [18F]F-flurpiridaz, and 82Rb at the infarct site.62–65 The chronically infarcted heart rodent models use [18F]FDG to assess other parameters such as left ventricular metabolic volume (LVMV), defect/infarct area, and cardiac function.63,64 The infarction model used in this study was generated by Envigo RMS LLC, Indianapolis, IN, followed by a 3-day observation period at their site. Therefore, testing [68Ga]Ga-NOTA-CTP within ∼24 hours post-surgery was impossible. We did not find PET imaging-based perfusion marker studies in a chronically infarcted heart rodent model. Therefore, this opportunity to assess if [68Ga]Ga-NOTA-CTP can detect perfusion differences in a remodeled chronically infarcted heart was reasonable and informative.
Conclusions
After administration, the radiolabeled cardiac targeting peptide, [68Ga]Ga-NOTA-CTP, was rapidly cleared from the body via the renal and hepatobiliary routes. The uptake of [68Ga]Ga-NOTA-CTP in the whole heart was similar in chronically infarcted and healthy hearts, but [68Ga]Ga-NOTA-CTP could identify regional differences in the left ventricular vs. right ventricular uptake between the healthy and diseased hearts. This ability to detect regional differences indicates the potential of [68Ga]Ga-NOTA-CTP as a perfusion marker.Safety and hazards
This study includes use of radioactive materials. Proper institutional guidelines and precautions should be taken while working with radioactive materials.Author contributions
S.R.G.: investigation, validation, curation, and analysis of data, and writing the manuscript. M.K.: investigation, validation, curation, and analysis of data, and writing the manuscript. A.G.R.: investigation, validation, curation, and analysis of data, and writing the manuscript. A.B.: investigation, validation, curation, and analysis of data, and writing the manuscript. V.K.: investigation, validation, curation, and analysis of data, and writing the manuscript. D.A.S.: animal ordering, reviewing, and editing the manuscript. J.E.K.: reviewing and editing the manuscript. D.R.J.: reviewing and editing the manuscript. M.Z.: resources, reviewing and editing the manuscript. V.J.L.: resources, reviewing and editing the manuscript. M.K.P.: conceptualization, fund acquisition, methodology, project administration, resources, supervision, software, and writing, reviewing, and editing the manuscript.Data availability
The entire data generated and analyzed during this study are included in the main manuscript and its ESI.†Conflicts of interest
All the authors declare no competing conflict of interest. M.Z. is the inventor of CTP.Acknowledgements
This study was funded by the Department of Radiology and Department of Cardiovascular Medicine, Mayo Clinic, Rochester, MN USA. MZ is supported by an R01 grant HL153407 from NHLBI.References
- J. P. Duggan, A. S. Peters, G. D. Trachiotis and J. L. Antevil, Surg. Clin. North. Am., 2022, 102, 499–516 CrossRef PubMed.
- K. Wang, X. Shi, Z. Zhu, X. Hao, L. Chen, S. Cheng, R. S. Y. Foo and C. Wang, Genome Med., 2022, 14, 63 CrossRef CAS PubMed.
- J. S. Lawton, J. E. Tamis-Holland, S. Bangalore, E. R. Bates, T. M. Beckie, J. M. Bischoff, J. A. Bittl, M. G. Cohen, J. M. DiMaio, C. W. Don, S. E. Fremes, M. F. Gaudino, Z. D. Goldberger, M. C. Grant, J. B. Jaswal, P. A. Kurlansky, R. Mehran, T. S. Metkus, Jr., L. C. Nnacheta, S. V. Rao, F. W. Sellke, G. Sharma, C. M. Yong and B. A. Zwischenberger, Circulation, 2022, 145, e4–e17 CrossRef PubMed.
- S. S. Virani, A. Alonso, H. J. Aparicio, E. J. Benjamin, M. S. Bittencourt, C. W. Callaway, A. P. Carson, A. M. Chamberlain, S. Cheng, F. N. Delling, M. S. V. Elkind, K. R. Evenson, J. F. Ferguson, D. K. Gupta, S. S. Khan, B. M. Kissela, K. L. Knutson, C. D. Lee, T. T. Lewis, J. Liu, M. S. Loop, P. L. Lutsey, J. Ma, J. Mackey, S. S. Martin, D. B. Matchar, M. E. Mussolino, S. D. Navaneethan, A. M. Perak, G. A. Roth, Z. Samad, G. M. Satou, E. B. Schroeder, S. H. Shah, C. M. Shay, A. Stokes, L. B. VanWagner, N. Y. Wang and C. W. Tsao, Circulation, 2021, 143, e254–e743 CrossRef PubMed.
- G. A. Roth, G. A. Mensah, C. O. Johnson, G. Addolorato, E. Ammirati, L. M. Baddour, N. C. Barengo, A. Z. Beaton, E. J. Benjamin, C. P. Benziger, A. Bonny, M. Brauer, M. Brodmann, T. J. Cahill, J. Carapetis, A. L. Catapano, S. S. Chugh, L. T. Cooper, J. Coresh, M. Criqui, N. DeCleene, K. A. Eagle, S. Emmons-Bell, V. L. Feigin, J. Fernández-Solà, G. Fowkes, E. Gakidou, S. M. Grundy, F. J. He, G. Howard, F. Hu, L. Inker, G. Karthikeyan, N. Kassebaum, W. Koroshetz, C. Lavie, D. Lloyd-Jones, H. S. Lu, A. Mirijello, A. M. Temesgen, A. Mokdad, A. E. Moran, P. Muntner, J. Narula, B. Neal, M. Ntsekhe, G. Moraes de Oliveira, C. Otto, M. Owolabi, M. Pratt, S. Rajagopalan, M. Reitsma, A. L. P. Ribeiro, N. Rigotti, A. Rodgers, C. Sable, S. Shakil, K. Sliwa-Hahnle, B. Stark, J. Sundström, P. Timpel, I. M. Tleyjeh, M. Valgimigli, T. Vos, P. K. Whelton, M. Yacoub, L. Zuhlke, C. Murray and V. Fuster, J. Am. Coll. Cardiol., 2020, 76, 2982–3021 CrossRef PubMed.
- S. T. Dahlberg, J. Nucl. Cardiol., 2009, 16, 493–496 CrossRef PubMed.
- O. O. Sogbein, M. Pelletier-Galarneau, T. H. Schindler, L. Wei, R. G. Wells and T. D. Ruddy, BioMed Res. Int., 2014, 2014, 942960 Search PubMed.
- R. Mikołajczak and P. Garnuszek, Nucl. Med. Rev. Cent. East. Eur., 2012, 15, 39–45 CrossRef.
- D.-Y. Kim, H.-J. Kim, K.-H. Yu and J.-J. Min, Bioconjugate Chem., 2012, 23, 431–437 CrossRef CAS PubMed.
- V. Sharma, J. Sivapackiam, S. E. Harpstrite, J. L. Prior, H. Gu, N. P. Rath and D. Piwnica-Worms, PLoS One, 2014, 9, e109361 CrossRef PubMed.
- A. Y. Ji, Q. M. Jin, D. J. Zhang, H. Zhu, C. Su, X. H. Duan, L. Bian, Z. P. Sun, Y. C. Ni, J. Zhang, Z. Yang and Z. Q. Yin, ACS Med. Chem. Lett., 2017, 8, 191–195 CrossRef CAS PubMed.
- M. K. Pandey, A. P. Belanger, S. Wang and T. R. DeGrado, J. Med. Chem., 2012, 55, 10674–10684 CrossRef CAS PubMed.
- T. R. DeGrado, M. K. Pandey, A. P. Belanger, F. Basuli, A. Bansal and S. Wang, Am. J. Physiol. Endocrinol. Metab., 2019, 316, E251–E259 CrossRef CAS PubMed.
- T. R. DeGrado, F. Bhattacharyya, M. K. Pandey, A. P. Belanger and S. Wang, J. Nucl. Med., 2010, 51, 1310–1317 CrossRef CAS PubMed.
- M. Dietz, C. H. Kamani, V. Dunet, S. Fournier, V. Rubimbura, N. Testart Dardel, A. Schaefer, M. Jreige, S. Boughdad, M. Nicod Lalonde, N. Schaefer, N. Mewton, J. O. Prior and G. Treglia, Front. Med., 2022, 9, 887508 CrossRef PubMed.
- M. K. Pandey, F. Bhattacharyya, A. P. Belanger, S. Wang and T. R. DeGrado, Circulation, 2010, 122, A12657–A12657 Search PubMed.
- T. M. Bateman, J. Nucl. Cardiol., 2012, 19(Suppl 1), S3–11 CrossRef PubMed.
- R. S. Driessen, P. G. Raijmakers, W. J. Stuijfzand and P. Knaapen, Int. J. Cardiovasc. Imaging, 2017, 33, 1021–1031 CrossRef PubMed.
- I. Carvajal-Juarez, A. Monroy-Gonzalez, N. Espinola-Zavaleta, A. Meave-Gonzalez and E. Alexanderson-Rosas, Ann. Nucl. Cardiol., 2019, 5, 63–68 CrossRef.
- H. R. Schelbert, M. E. Phelps, S. C. Huang, N. S. MacDonald, H. Hansen, C. Selin and D. E. Kuhl, Circulation, 1981, 63, 1259–1272 CrossRef CAS PubMed.
- J. Maddahi and R. R. Packard, Semin. Nucl. Med., 2014, 44, 333–343 CrossRef PubMed.
- M. F. Di Carli, S. Dorbala, J. Meserve, G. El Fakhri, A. Sitek and S. C. Moore, J. Nucl. Med., 2007, 48, 783–793 CrossRef PubMed.
- S. Senthamizhchelvan, P. E. Bravo, C. Esaias, M. A. Lodge, J. Merrill, R. F. Hobbs, G. Sgouros and F. M. Bengel, J. Nucl. Med., 2010, 51, 1592–1599 CrossRef PubMed.
- P. Arumugam, D. Tout and C. Tonge, Br. Med. Bull., 2013, 107, 87–100 CrossRef CAS PubMed.
- H. A. Ziessman, J. P. O'Malley, J. H. Thrall and F. H. Fahey, in Nuclear Medicine (Fourth Edition), ed. H. A. Ziessman, J. P. O'Malley and J. H. Thrall, W.B. Saunders, Philadelphia, 2014, p. ix, DOI:10.1016/B978-0-323-08299-0.05001-X.
- H. Iida, I. Kanno, A. Takahashi, S. Miura, M. Murakami, K. Takahashi, Y. Ono, F. Shishido, A. Inugami and N. Tomura and, et al., Circulation, 1988, 78, 104–115 CrossRef CAS PubMed.
- C. C. Yang, Diagnostics, 2021, 11, 2275 CrossRef CAS PubMed.
- J. T. Thackeray, J. P. Bankstahl, Y. Wang, M. Korf-Klingebiel, A. Walte, A. Wittneben, K. C. Wollert and F. M. Bengel, Eur. J. Nucl. Med. Mol. Imaging, 2015, 42, 317–327 CrossRef CAS PubMed.
- E. A. McCutchan, Nucl. Data Sheets, 2012, 113, 1735–1870 CrossRef CAS.
- M. K. Pandey, J. F. Byrne, H. Jiang, A. B. Packard and T. R. DeGrado, Am. J. Nucl. Med. Mol. Imaging, 2014, 4, 303–310 Search PubMed.
- H. M. Sherif, A. Saraste, E. Weidl, A. W. Weber, T. Higuchi, S. Reder, T. Poethko, G. Henriksen, D. Casebier, S. Robinson, H. J. Wester, S. G. Nekolla and M. Schwaiger, Circ. Cardiovasc. Imaging, 2009, 2, 77–84 CrossRef PubMed.
- J. Maddahi, J. Czernin, J. Lazewatsky, S. C. Huang, M. Dahlbom, H. Schelbert, R. Sparks, A. Ehlgen, P. Crane, Q. Zhu, M. Devine and M. Phelps, J. Nucl. Med., 2011, 52, 1490–1498 CrossRef CAS PubMed.
- N. Matsumoto, Ann. Nucl. Cardiol., 2023, 9, 91–93 CrossRef PubMed.
- R. C. Marshall, P. Powers-Risius, B. W. Reutter, J. P. O'Neil, M. La Belle, R. H. Huesman and H. F. VanBrocklin, J. Nucl. Med., 2004, 45, 1950–1959 CAS.
- I. Madar, H. T. Ravert, Y. Du, J. Hilton, L. Volokh, R. F. Dannals, J. J. Frost and J. M. Hare, J. Nucl. Med., 2006, 47, 1359–1366 CAS.
- D. Y. Kim, H. S. Kim, H. Y. Jang, J. H. Kim, H. S. Bom and J. J. Min, ACS Med. Chem. Lett., 2014, 5, 1124–1128 CrossRef CAS PubMed.
- M. Zahid and P. D. Robbins, Methods Mol. Biol., 2011, 683, 277–289 CrossRef CAS PubMed.
- M. Zahid, B. E. Phillips, S. M. Albers, N. Giannoukakis, S. C. Watkins and P. D. Robbins, PLoS One, 2010, 5, e12252 CrossRef PubMed.
- D. Sahagun and M. Zahid, Biomolecules, 2023, 13, 1690 CrossRef CAS PubMed.
- M. Zahid, K. S. Feldman, G. Garcia-Borrero, T. N. Feinstein, N. Pogodzinski, X. Xu, R. Yurko, M. Czachowski, Y. L. Wu, N. S. Mason and C. W. Lo, Biomolecules, 2018, 8, 147 CrossRef PubMed.
- K. S. Feldman and M. Zahid, J. Visualized Exp., 2020, 160, e60895 Search PubMed.
- M. Zahid, B. Weber, R. Yurko, K. Islam, V. Agrawal, J. Lopuszynski, H. Yagi and G. Salama, Pharmaceutics, 2023, 15, 2107 CrossRef CAS PubMed.
- R. Yurko, K. Islam, B. Weber, G. Salama and M. Zahid, Front. Chem., 2023, 11, 1220573 CrossRef CAS PubMed.
- G. I. Gallicano, J. Fu, S. Mahapatra, M. V. R. Sharma, C. Dillon, C. Deng and M. Zahid, Pharmaceuticals, 2022, 15, 871 CrossRef CAS PubMed.
- H. Kim, D. Mun, J. Y. Kang, S. H. Lee, N. Yun and B. Joung, Mol. Ther. Nucleic Acids, 2021, 24, 1024–1032 CrossRef CAS PubMed.
- U. M. Avula, H. K. Yoon, C. H. Lee, K. Kaur, R. J. Ramirez, Y. Takemoto, S. R. Ennis, F. Morady, T. Herron, O. Berenfeld, R. Kopelman and J. Kalifa, Sci. Transl. Med., 2015, 7, 311ra172 CrossRef PubMed.
- Y. J. Li, X. Hua, Y. Q. Zhao, H. Mo, S. Liu, X. Chen, Z. Sun, W. Wang, Q. Zhao, Z. Cui, T. An and J. Song, Adv. Healthcare Mater., 2025, 14, e2404068 CrossRef PubMed.
- D. Kehr, J. Ritterhoff, M. Glaser, L. Jarosch, R. E. Salazar, K. Spaich, K. Varadi, J. Birkenstock, M. Egger, E. Gao, W. J. Koch, M. Sauter, M. Freichel, H. A. Katus, N. Frey, A. Jungmann, C. Busch, P. J. Mather, A. Ruhparwar, M. Busch, M. Völkers, R. C. Wade and P. Most, Circulation, 2025, 151(8), 548–565 CrossRef CAS PubMed.
- I. Ruseska and A. Zimmer, Beilstein J. Nanotechnol., 2020, 11, 101–123 CrossRef CAS PubMed.
- M. L. Chiu, J. F. Kronauge and D. Piwnica-Worms, J. Nucl. Med., 1990, 31, 1646–1653 CAS.
- D. M. Chernoff, G. R. Strichartz and D. Piwnica-Worms, Biochim. Biophys. Acta, 1993, 1147, 262–266 CrossRef CAS PubMed.
- J. C. Taubel, N. R. Nelson, A. Bansal, G. L. Curran, L. Wang, Z. Wang, H. M. Berg, C. J. Vernon, H. K. Min, N. B. Larson, T. R. DeGrado, K. K. Kandimalla, V. J. Lowe and M. K. Pandey, Bioconjugate Chem., 2022, 33, 892–906 CrossRef PubMed.
- D. Faria, M. M. Herth, M. Konijnenberg, A. Korde, C. Kuntner-Hannes, R. M. Moresco, P. J. H. Scott, V. Shalgunov and S. Subramanian, Guidance for Preclinical Studies with Radiopharmaceuticals, IAEA Radioisotopes and Radiopharmaceuticals Series No. 8, International Atomic Energy Agency, IAEA, Vienna, 2023 Search PubMed.
- A. Todica, N. L. Beetz, L. Günther, M. J. Zacherl, U. Grabmaier, B. Huber, P. Bartenstein, S. Brunner and S. Lehner, Mol. Imaging Biol., 2018, 20, 268–274 CrossRef PubMed.
- M. Fischer, M. J. Zacherl, L. Weckbach, L. Paintmayer, T. Weinberger, K. Stark, S. Massberg, P. Bartenstein, S. Lehner, C. Schulz and A. Todica, Front. Cardiovasc. Med., 2021, 8, 656742 CrossRef CAS PubMed.
- M. Fischer, T. Weinberger, D. Messerer, M. J. Zacherl, C. Schulz, S. Massberg, P. Bartenstein, S. Lehner, G. Boening and A. Todica, Ann. Nucl. Med., 2022, 36, 533–543 CrossRef CAS PubMed.
- A. S. L. Yu, A. S. L. Yu, G. M. Chertow, V. r. A. Luyckx, P. A. Marsden, K. Skorecki and M. W. Taal, Brenner & Rector's the kidney, Elsevier, Philadelphia, PA, 11th edn, 2020 Search PubMed.
- M. Salerno and G. A. Beller, Circ. Cardiovasc. Imaging, 2009, 2, 412–424 CrossRef PubMed.
- A. Autio, S. Uotila, M. Kiugel, V. Kytö, H. Liljenbäck, N. Kudomi, V. Oikonen, O. Metsälä, S. Helin, J. Knuuti, A. Saraste and A. Roivainen, J. Nucl. Cardiol., 2020, 27, 891–898 CrossRef PubMed.
- E. Berezin and A. A. Berezin, Dis. Markers, 2020, 2020, 1215802 Search PubMed.
- S. A. Leancă, D. Crişu, A. O. Petriş, I. Afrăsânie, A. Genes, A. D. Costache, D. N. Tesloianu and I. I. Costache, II, Life, 2022, 12, 1111 CrossRef PubMed.
- H. Houson, G. Nkepang, H. Gali and V. Awasthi, J. Nucl. Med., 2018, 59, 1534–1534 Search PubMed.
- G. N. Nkepang, H. Gali, H. Houson, A. F. Hedrick, B. Hayes, O. Causey, P. Inman, J. Box, E. Benton, W. Galbraith and V. Awasthi, Appl. Radiat. Isot., 2019, 150, 19–24 CrossRef CAS PubMed.
- H. M. Sherif, A. Saraste, E. Weidl, A. W. Weber, T. Higuchi, S. Reder, T. Poethko, G. Henriksen, D. Casebier, S. Robinson, H.-J. Wester, S. G. Nekolla and M. Schwaiger, Circ. Cardiovasc. Imaging, 2009, 2, 77–84 CrossRef PubMed.
- A. A. Ghotbi, A. Clemmensen, K. Kyhl, B. Follin, P. Hasbak, T. Engstrøm, R. S. Ripa and A. Kjaer, J. Nucl. Cardiol., 2019, 26, 798–809 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5pm00047e |
| This journal is © The Royal Society of Chemistry 2025 |







