Open Access Article
Open Access ArticleAn eco-friendly and cost-effective approach for the synthesis of a novel GA@CuO–ZnO nanocomposite: characterization, antimicrobial and anticancer activities
Gharieb S. El-Sayyad *ab,
El-Sayed R. El-Sayedc,
Samar H. Rizkd,
Mostafa A. Abdel-Maksoude,
Adel M. Zakrif,
Abdul Malikg,
Mohamed N. Malashh and
Amr H. Hashem
*ab,
El-Sayed R. El-Sayedc,
Samar H. Rizkd,
Mostafa A. Abdel-Maksoude,
Adel M. Zakrif,
Abdul Malikg,
Mohamed N. Malashh and
Amr H. Hashem *i
*i
aMedical Laboratory Technology Department, Faculty of Applied Health Sciences Technology, Badr University in Cairo (BUC), Badr city, Cairo 11829, Egypt. E-mail: Gharieb.El-Saied@buc.edu.eg
bDrug Microbiology Lab, Drug Radiation Research Department, National Center for Radiation Research and Technology (NCRRT), Egyptian Atomic Energy Authority (EAEA), Cairo, Egypt
cDepartment of Food Chemistry and Biocatalysis, Wrocław University of Environmental and Life Sciences, Norwida 25, 50-375 Wrocław, Poland
dDepartment of Biochemistry, Faculty of Pharmacy, Ahram Canadian University (ACU), Giza, Egypt
eDepartment of Botany and Microbiology, College of Science, King Saud University, P.O. Box 2455, Riyadh 11451, Saudi Arabia
fPlant Production Department, College of Food and Agricultural Sciences, King Saud University, Saudi Arabia
gDepartment of Pharmaceutics, College of Pharmacy, King Saud University, Saudi Arabia
hMicrobiology and Immunology Department, Faculty of Pharmacy, Ahram Canadian University (ACU), 6th October City, Giza, Egypt
iBotany and Microbiology Department, Faculty of Science, Al-Azhar University, Nasr City, Cairo 11884, Egypt. E-mail: amr.hosny86@azhar.edu.eg
First published on 6th January 2025
Abstract
In this study, a nanocomposite based on copper oxide–zinc oxide nanoparticles and Gum Arabic (GA@CuO–ZnO nanocomposite) was successfully synthesized using green method. Characterization results revealed that the prepared nanocomposite appeared at the nanoscale level, showed excellent dispersion, and formed stable colloidal nano-solutions. The bimetallic GA@CuO–ZnO nanocomposite was evaluated for its anticancer, antibacterial, and antifungal properties. Cytotoxicity testing of the synthesized GA@CuO–ZnO nanocomposite against Wi38 and Vero normal cell lines showed IC50 values of 143.3 and 220.9 μg ml−1, respectively, indicating that the nanocomposite is safe for use. Moreover, the synthesized GA@CuO–ZnO nanocomposite showed anticancer activity against HepG2 and MCF7 cell lines, with IC50 values of 54.7 and 79.2 μg ml−1, respectively. The nanocomposite also showed promising antibacterial activity against Escherichia coli, Salmonella typhimurium, Staphylococcus aureus, S. epidermis, and Bacillus subtilis, with MIC values ranging from 31.25 to 62.5 μg ml−1. Furthermore, the prepared nanocomposite exhibited antifungal activity against Candida albicans, Cryptococcus neoformans, Aspergillus fumigatus and A. brasiliensis, with an MIC of 31.25 μg ml−1 against all tested fungal strains, except A. fumigatus (MIC 125 μg ml−1). In conclusion, the synthesized GA@CuO–ZnO nanocomposite was successfully synthesized, and it exhibited promising antibacterial, antifungal and anticancer activities at safe doses.
Introduction
Antimicrobial resistance (AMR) refers to the ability of microorganisms to develop resistance to medications that were once effective in treating infections. It represents a significant global health threat as it hampers the efficacy of antimicrobial medications and complicates the treatment of infections. Multiple factors contribute to the emergence of antibiotic resistance.1 A key factor is the overuse and misuse of antibiotics in human and animal healthcare, as well as in agriculture. Improper or unnecessary use of antibiotics can drive the adaptation and development of resistance mechanisms in bacteria, enabling their survival. The transfer of resistance can occur among several bacterial species via mobile genetic components, exacerbating the problem.2 Antimicrobial resistance presents substantial obstacles in healthcare, leading to extended and more intricate treatment regimens, elevated healthcare expenses, and higher mortality rates.3 Infections caused by antibiotic-resistant bacteria are often associated with extended periods of hospitalization and increased rates of treatment failure. Moreover, the development of novel antimicrobial medications is progressing at a slower rate than the rapid emergence of resistance, underscoring the urgent need for responsible and judicious use of antimicrobial agents.4Cancer is a multifaceted collection of illnesses distinguished by the unregulated proliferation and dissemination of aberrant cells. These cells possess the capacity to infiltrate and destroy healthy tissues, and they can also disseminate to different regions of the body via a phenomenon known as metastasis. Cancer can manifest in any anatomical region and has the potential to affect various organs and tissues. Cancer formation typically follows a complex series of genetic alterations that disrupt the normal control of cell growth and division.5 These mutations can be acquired through exposure to specific risk factors, such as tobacco smoke, radiation, certain chemicals, infectious agents, and genetic predisposition, during an individual's lifespan. Nevertheless, it is crucial to acknowledge that not all cancers can be avoided, and certain types may arise without any identifiable risk factors.6
Traditional anticancer agents, also known as conventional chemotherapy, encompass a broad range of drugs that have been used for many years in cancer treatment. These agents are designed to kill or inhibit the growth of cancer cells by interfering with their ability to divide and multiply.7 Cancer is a significant contributor to global mortality, with rising incidences observed on an annual basis.
The application of nanobiotechnology in pathogen resistance and fighting the cancer pandemic has demonstrated significant and robust outcomes in the fight against microbial infections and tumor treatment.8,9 This is because the technology allows better attraction to the bacterial, fungal, and cancer cells and ROS production stimulates their inhibition.10,11 Entire biological components have been included in the scope of biological methods for nanoparticles and nanocrystal production.12–15 Due to its safety and environmental friendliness, the biological production of metal nanoparticles utilizing macromolecules, plants, bacteria, and fungi has drawn a lot of attention recently.8,16–22
Natural polymeric cap agents, such as chitosan and gum Arabic, are used to regulate the inorganic polymer NPs' particle size and improve their stability.23,24 Gum Arabic is different from other macromolecules in biology in that it has a variety of attractive features that make it a preferred option for the production of nanomaterials. These crucial attributes are lower toxicity, biological compatibility, affordability, and availability.25
Bimetallic nanomaterials are receiving a lot of attention because of the distinctive chemical, physical, and physiological characteristics that result from the combination of several metal elements.24,26–29 Copper oxide–zinc oxide (CuO–ZnO) NPs are the most well-known bimetallic nanoparticles (NPs) because of their chemically controllable plasmonic properties and prospective uses in the detection, catalysis, and combating microbial plant pathogens.27,30 Throughout the review literature,24,26–28,31–34 the produced bimetallic nanoparticles have shown encouraging antibacterial and antibiofilm action versus a particular category of extremely pathogenic and multidrug-resistant pathogens.
The bimetallic nanoparticles' electrostatic interaction, which attaches proteins to interfere with homeostasis (disturbing the chain of electron transport), inhibits signal transduction, harms the membranes of cells, interferes with enzymes and proteins, creates reactive oxygen species, also known as ROS, and oxidative stress, and causes genotoxicity, may be involved in the mechanism of action of antimicrobials.35,36
In addition to their biological properties as antibacterial, antifungal and anticancer agents, the characterization of the synthesized bimetallic GA@CuO–ZnO NPs and one of the environmentally friendly methods for their production are the main topics of this study. GA@CuO–ZnO NPs may be prepared using a range of chemical, biological, and physical methods because of the remarkable progress in nanotechnology. Therefore, the purpose of this study was to synthesize and characterize a novel nanocomposite (GA@CuO–ZnO) and to assess its anticancer, antibacterial and antifungal activities.
Materials and methods
Chemicals and reagents
Analytical-grade chemicals, including gum Arabic (Meron Company, India), and zinc acetate and copper acetate (Sigma Aldrich, UK) were used for the production of bimetallic nanoparticles in the study.Synthesis of GA@CuO–ZnO nanocomposite
The precise amounts of the salts were employed in the biosynthesis of the nanocomposite. Specifically, 10 ml of (2.0 mM) Zn (CH3COO)2(H2O)2 and 10 ml of (2.0 mM) Cu(CH3COO)2 (2.0 mM) were combined at room temperature for half an hour. After that, they received around 80 milliliters of the 20% gum Arabic that serves as a reducing and stabilizing agent. After combining the two solutions, the combination's pH was 7.8. To achieve the most effective synthesis of GA@CuO–ZnO nanocomposite, the reaction conditions were chosen to include a temperature-controlled incubation at 30 °C and a reaction duration of 24 h under agitation (500 rpm) in a shaking incubator, as shown in Scheme 1.37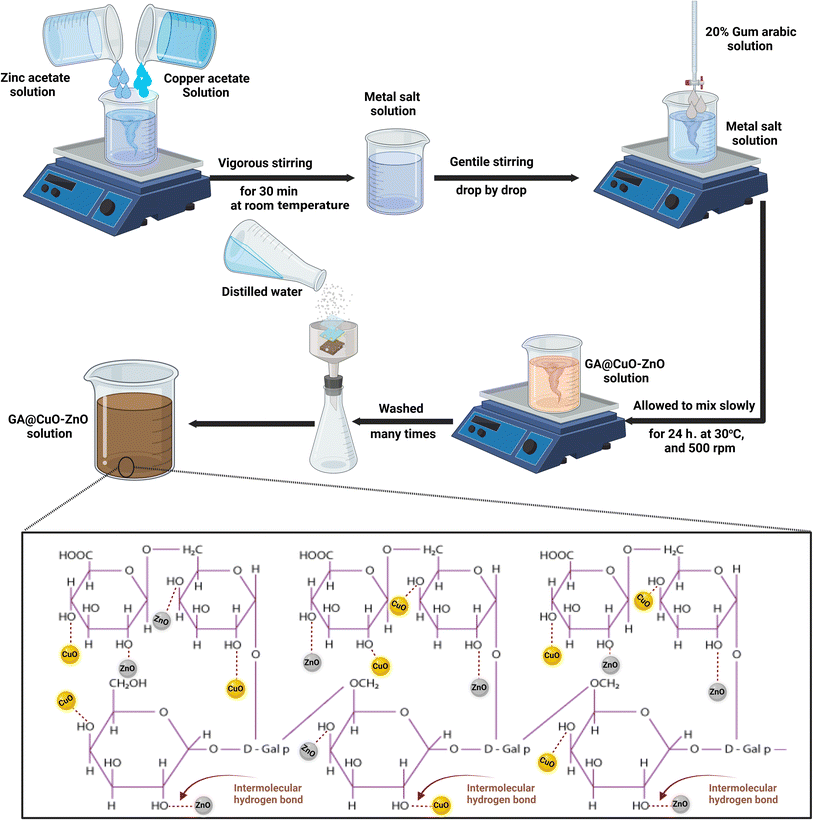 | ||
| Scheme 1 Schematic for the synthesis of GA@CuO–ZnO nanocomposite and the possible binding sites in GA. | ||
According to the literature,24 FTIR results revealed peak absorption values at 3291 cm−1 (attributed to –OH), 2985 cm−1 (symmetric and asymmetric –CH vibration), 1642 cm−1 (attributed to –COOH), 1462 cm−1 (uronic acid symmetrically stretched by carboxylic groups), 1066 cm−1 (perhaps due to arabinogalactan), and 890.0 cm−1 (attributed to galactose 1–4 linkage and mannose 1–6 linkage). The same peaks in the metal NPs spectra indicate that GA was successful in capping. Moreover, the galactose/mannose peak became less prominent and the arabinogalactan peak lost its doublet. Additionally, a clear peak at 715.25 cm−1 was seen, which may be the outcome of metal NPs interacting and combining with hydroxyl groups to create metal–O.38
According to the results of El-Batal et al.,24 all of the peaks were in identical wavenumbers, suggesting that the synthesized NPs were similarly incorporated and/or conjugated through the primary functional groups of the stabilizer GA. The lack of noise and additional unidentified peaks indicated the high purity of the synthesized sample.39
UV-Vis spectroscopy was used to compute the optical density (OD) of GA@CuO–ZnO nanocomposite at a given wavelength in order to ascertain the likelihood of the preparation.
Characterization of the synthesized GA@CuO–ZnO nanocomposite
A UV-Vis spectrophotometer (JASCO V-560) was used to examine the absorbance and spectral properties of the produced GA@CuO–ZnO nanocomposite at particular wavelengths. An extra experiment devoid of any metal ions was offered for Auto-zero reasons. First, each specimen's optical characteristics were examined to ascertain the overall number of wavelengths required to estimate absorbance.The mean size distribution of the prepared GA@CuO–ZnO nanocomposite was determined using the dynamic light scattering measurements on a DLS-PSS-NICOMP 380-ZLS particle-sized system (St. Barbara, California, USA). A tiny cuvette was used to contain 100 μL of the tested GA@CuO–ZnO nanocomposite. Following 2 min of equilibrium at 25.0 ± 2 °C ambient temperature, five procedures were carried out.
The generated GA@CuO–ZnO nanocomposite was further evaluated through a high-resolution transmission electron microscope (HR-TEM, JEM2100, JEOL, Japan) to determine their size, shape, and overall appearance. Furthermore, it was possible to examine the crystallization, crystallite diameters, and/or morphology of the generated GA@CuO–ZnO nanocomposite by using the XRD-6000 instrument (Shimadzu equipment, and SSI, Japan). The produced GA@CuO–ZnO nanocomposite surrounding GA was analyzed utilizing a scanning electron microscope (SEM, ZEISS, EVO-MA10, Germany) to assess their surface form and arrangement. Finally, the surface charges of the produced GA@CuO–ZnO nanocomposite were informally evaluated at the pH of preparation using a zeta potential analyzer from Malvern Device, UK.40
Antibacterial and antifungal activity
Antibacterial activity of the synthesized GA@CuO–ZnO nanocomposite was evaluated toward Escherichia coli ATCC 25922, Salmonella typhimurium ATCC 14028, Staphylococcus aureus ATCC 25923, S. epidermis ATCC 14990 and Bacillus subtilis ATCC 6051. Furthermore, antifungal activity was assessed against Candida albicans ATCC 90028, Cryptococcus neoformans ATCC14116, Aspergillus fumigatus ATCC 204305 and A. brasiliensis ATCC 16404. All steps of the agar well diffusion method were performed in strict adherence to CLSI document M51-A2.41100 μL of the synthesized GA@CuO–ZnO nanocomposite, zinc acetate, copper acetate, standard antibiotic (ampicillin/sulbactam; SAM 20), and antifungal drug (amphotericin B) were placed in agar wells (7 mm) seeded with bacterial and fungal strains separately, and then incubating at 37 °C for 24 h for bacterial and unicellular fungal strains, and at 30 °C for 72 h for filamentous fungal strains. After the incubation process, the inhibition zone for each treatment was measured,42–44 and MIC was determined using the microdilution method.45–47
Cytotoxicity and anticancer activity
Cytotoxicity of the synthesized GA@CuO–ZnO nanocomposite was evaluated according to the MTT protocol.48,49 To check the biosafety of the synthesized GA@CuO–ZnO nanocomposite, a Wi38 normal cell line was used. Moreover, both MCF7 and HepG2 cancerous cell lines were used to assess the anticancer activity of the synthesized GA@CuO–ZnO nanocomposite. The optical density was measured at 560 nm. To determine cell viability, the following formula was used (1), and to determine cell inhibition, eqn (2) was applied:
 | (1) |
| Inhibition% = 100 − viability% | (2) |
Results and discussion
Synthesis and characterization of the synthesized GA@CuO–ZnO nanocomposite
Gum Arabic's ability to create GA@CuO–ZnO nanocomposite as a capping and reducing agent was assessed. When the bimetallic GA@CuO–ZnO nanocomposite was produced, the resulting solution's initially pale-yellow color was turned into a brown hue. The resulting brown color was attributed to the activation of the surface plasmon resonance that occurs within the GA@CuO–ZnO nanocomposite, and the change in particle size which provides a reliable spectroscopic indicator of their presence.50The detected absorbance of 2.23 created the experimental peak in the spectra (Fig. 1), and the UV-Vis studies showed that the generated GA@CuO–ZnO nanocomposite was small and had a visible absorption peak at 420 nm. Conversely, Fig. 1 shows the UV-Vis spectra of the initial precursors, zinc acetate and copper acetate, which had distinct wavelengths; zinc acetate at 290 nm (ref. 51) and copper acetate at 210 nm.52 It must be emphasized that a noted peak at 245 nm in the synthesized GA@CuO–ZnO nanocomposite spectrum corresponds to the GA spectrum.53
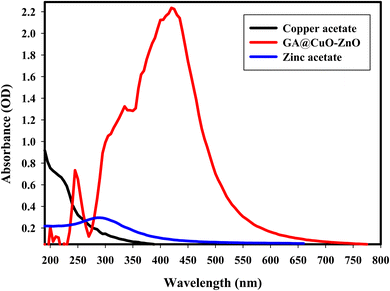 | ||
| Fig. 1 UV-Vis spectra of copper acetate, zinc acetate, and the synthesized GA@CuO–ZnO nanocomposite. | ||
The intensity of the brown hue correlated with the ability for the biosynthesis of CuO–ZnO NPs that were generated.54 Surface Plasmon resonance (SPR) is often influenced by the intensity, size, morphology, structure, and dielectric characteristics of any produced nanoparticles.55,56
The peak spectra generated for the synthesized GA@CuO–ZnO nanocomposite are broad, ranging from 270 to 500 nm, this is due to the formation of poly-dispersed GA@CuO–ZnO nanocomposite and other factors such as particle size distribution, and polydispersity index (PDI) (according to the result of DLS), surface defects (SEM result), particle size variation (HRTEM result), or the nature of the bimetallic interaction (Scheme 1).
The synthesized GA@CuO–ZnO nanocomposite was characterized by its hydrodynamic diameter, particle size dispersion, and polydispersity index (PDI) using Dynamic Light Scattering (DLS) analysis. By comparing the collected data with HR-TEM investigations, we were able to establish the average size of the created nanoparticles.57
The HR-TEM image in Fig. 2a demonstrated that the produced GA@CuO–ZnO nanocomposite was bound to stabilizing gum Arabic and had a semi-spherical shape. Fig. 2b shows that the particles' sizes ranged from 18.34 nm to 90.45 nm, with a median diameter of 50.28 ± 1.5 nm. The produced gum Arabic filtrate was rich in active functional groups and the supplied polydispersed NPs acted as capping agents, stabilizing, and reducing the concentration of these groups.58 Zn and Cu may concurrently diminish due to the radical multilocation of gum Arabic, as elucidated and contrasted in our work. The HR-TEM image (Fig. 2b) demonstrated the uniform line spacing, which resulted in a single-grade system and copper was equally distributed throughout the zinc matrix, resulting in a unique alloy.24
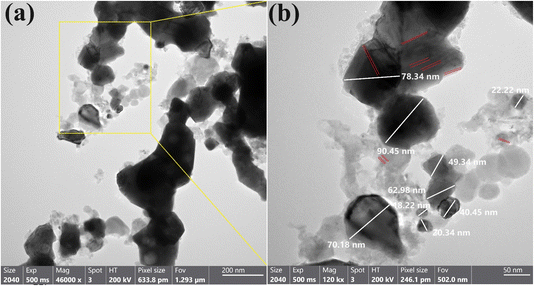 | ||
| Fig. 2 HR-TEM images of the synthesized GA@CuO–ZnO nanocomposite (a), and their corresponding magnification (b). | ||
Upon comparison with literature on average particle size and shape, our GA@CuO–ZnO nanocomposite was discovered to be polydispersed, with varying sizes and a preponderance of spherical particles. The NPs that were created had a spherical or orbicular shape, although the asymmetrical form could be due to the fact that they were created from green extract, which could have resulted in a variety of morphologies. Polydisplayed NPs are a long-term solution since we used gum Arabic, the most practicable reducing and covering agent, in our investigation.59
As shown in Fig. 3a, the usual size of the particle's dispersion for GA@CuO–ZnO nanocomposite, which was produced using gum Arabic, was determined using the DLS technique to be 80.34 nm. Samples are considered monodisperse according to the International Standards Organizations (ISOs) when the polydispersity index (PDI) readings are less than 0.05. Conversely, when PDI findings are more than 0.7, particles with a variable distribution are expected to be generated.60 The PDI values of the GA@CuO–ZnO nanocomposite were 0.91, according to our findings. The prepared nanocomposite exhibited an acceptable polymorphism range according to the current values. Results demonstrated that HR-TEM imaging showed smaller particle estimations than the average and predominant sizes obtained by DLS analysis. According to the following citation,61 the major factors contributing to the nanocomposite's enormous size are its internal hydrodynamic radius and the water layers immediately surrounding it. Fig. 3b shows the results of analyzing the GA@CuO–ZnO nanocomposite's zeta potential at a pH of 7.8 in the preparation. This study shows that the zeta potential of the fabricated GA@CuO–ZnO nanocomposite remains negative at the investigated material's pH. Because gum Arabic has a negative electrical charge, the preparation's zeta potential in the neutral medium (pH 7.8) was −30.5 mV, as shown in Fig. 3b.
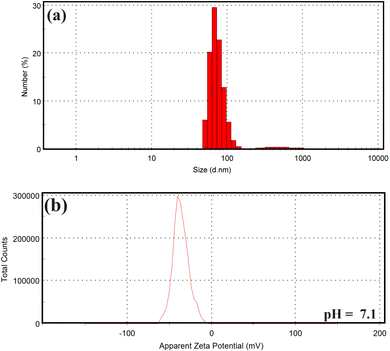 | ||
| Fig. 3 Particle size distribution, and surface charge of the synthesized GA@CuO–ZnO nanocomposite: (a) DLS analysis, and (b) zeta potential. | ||
Using scanning electron microscopy (SEM) the surface form and characteristics of the synthesized nanoparticles can be analyzed. Fig. 4a shows that the synthesized GA@CuO–ZnO nanocomposite surfaces seem clear when viewed in conjunction with the gum Arabic SEM data, and the generated gum Arabic also had the corresponding bright spherical particles. Gum Arabic, which resembles fused and capped illuminated NPs, successfully partitioned the GA@CuO–ZnO nanocomposite into its spheroidal particle component (Fig. 4b).
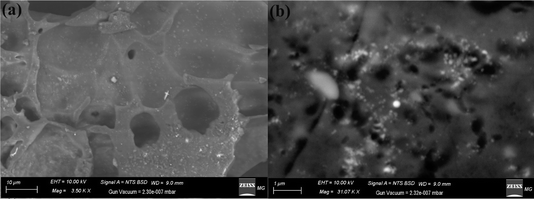 | ||
| Fig. 4 SEM images of the synthesized GA@CuO–ZnO nanocomposite at different magnifications (a and b). | ||
In contrast to previous studies on the topic of morphological form, the synthesized GA@CuO–ZnO nanocomposite developed in this work exhibited uniform distribution, controlled size, and exact spherical shape. Mohsin et al.62 synthesized bimetallic silver and gold core–shell NPs by varying the pH and temperature settings of the citrate reduction process. The synthetic process is heavily dependent on pH and temperature because the approved morphological shape and border size specifications stipulate that the particles should appear spheroidal and maintain a size range of 50 to 65 nm.
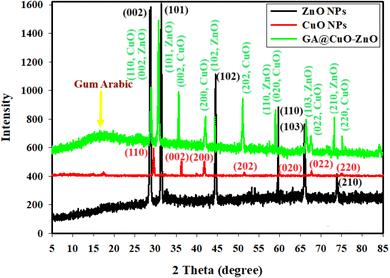
Fig. 5 shows the results of XRD analyses conducted on a synthetic GA@CuO–ZnO nanocomposite. Both amorphous and crystalline structures are present in the final nanocomposite. In order to quickly identify the distinctive properties of the manufactured GA@CuO–ZnO nanocomposite, we analyze the XRD data of our newly produced ZnO NPs and CuO NPs as separate patterns (Fig. 5). It must be noted that 2θ represent the gum Arabic was in the exact range, which ranged from 5° to 21°.24 Fig. 5 displays the XRD results of the produced bimetallic CuO–ZnO NPs and draws attention to the XRD diffraction peaks of the ZnO NPs. Using the standard card JCPDS number 36-1451, these peaks at 2θ = 27.40°, 31.22°, 45.54°, 56.56°, 67.17°, and 75.56° correspond to Bragg's reflections at angles (002), (101), (102), (110), (103), and (201), respectively.63
Along with the typical card JCPDS number 89-2531, they additionally include the CuO NP diffraction peaks at 2θ = 30.22°, 36.11°, 40.75°, 52.72°, 58.27°, 67.83°, and 71.45°; these peaks match Bragg's reflections at degrees (110), (002), (200), (202), (020), (022), and (220),64 respectively. There was crystallization in the synthesized GA@CuO–ZnO nanocomposite, and the XRD data that was available showed that it had a face-centered cubic (fcc) crystal structure (Fig. 5). The production of highly crystalline bimetallic nanoparticles and their linking with amorphous gum Arabic, as confirmed from the XRD data, improved their dispersion in the solution and their potential use.65
Ultimately, the midway crystallite size of bimetallic CuO–ZnO NPs was determined using the Williamson–Hall (W–H) equation,66,67 and it was found to be 29.46 nm.
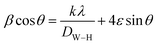 | (3) |
Antibacterial activity
In the current study, GA@CuO–ZnO nanocomposite and their starting materials were evaluated for antibacterial activity toward some pathogenic strains as shown in Table 1 and Fig. 6. Results revealed that GA@CuO–ZnO nanocomposite exhibited outstanding antibacterial activity against all tested bacterial strains. Table 1 illustrates the antibacterial activity of GA@CuO–ZnO nanocomposite where results showed that GA@CuO–ZnO nanocomposite exhibited antibacterial activity against E. coli and S. typhimurium as Gram-negative bacteria where inhibition zones were 21 ± 1.0 and 25.3 ± 1.52 mm, respectively, at a concentration of 1000 μg ml−1. Also, GA@CuO–ZnO nanocomposite showed antibacterial activity toward B. subtilis, S. aureus and S. epidermis as Gram-positive bacteria where inhibition zones were 21.4 ± 1.53, 23.5 ± 1.80 and 24.4 ± 0.57 mm, respectively, at a concentration 1000 μg ml−1. Standard antibiotic SAM 20 exhibited weak antibacterial activity toward and S. typhimurium and S. aureus only, but did not show any activity on other bacterial strains. Also, copper acetate and zinc acetate did not exhibit antibacterial activity toward all tested bacterial strains as the tested strains are multidrug-resistant (Fig. 6).| Bacterial strain | Cu acetate IZ (mm) | Zn acetate IZ (mm) | GA@CuO–ZnO nanocomposite | SAM 20 IZ (mm) | |
|---|---|---|---|---|---|
| IZ (mm) | MIC (μg ml−1) | ||||
| Data within the groups are analyzed using a one-way analysis of variance (ANOVA) followed by a,b Duncan’s multiple range test (DMRT). | |||||
| E. coli | ND | ND | 21 ± 1.0b | 62.5 | ND |
| S. typhi | ND | ND | 25.3 ± 1.52a | 31.25 | 16.0 ± 060a |
| B. subtilis | ND | ND | 21.4 ± 1.53b | 62.5 | ND |
| S. aureus | ND | ND | 23.5 ± 1.80 ab | 62.5 | 14.13 ± 0.32b |
| S. epidermis | ND | ND | 24.4 ± 0.57 ab | 62.5 | ND |
Furthermore, MICs of the GA@CuO–ZnO nanocomposite against all tested bacterial strains were evaluated, where the results revealed that the MIC of bimetallic CuO–ZnO NPs toward all strains was 62.5 μg ml−1 except for S. typhimurium it was 31.25 μg ml−1. Turabik et al.68 reported that bimetallic Cu/Zn NPs showed antibacterial activity where MIC was in the range of 16 and 256 μg ml−1. Additionally, Cissus quadrangularis stem was used for the synthesis of bimetallic Cu–Zn NPs, where results illustrate that it exhibited antibacterial activity against E. coli (MTCC-5704), P. aeruginosa (MTCC-2295), B. subtilis (MTCC-121), S. aureus (MTCC-3160) and Streptococcus mutans (MTCC-497).69 Furthermore, in other study, bimetallic Zn–CuO NPs based nanocomposite was prepared, and exhibited high antibacterial activity toward E. coli, S. aureus and B. subtilis with MICs measured as 7.81 and 62.5 μg ml−1, respectively.30
When GA@CuO–ZnO nanocomposite interacts with cells, they can engage in electrostatic interactions with the cell membrane. This can lead to damage to the cell membrane, compromising its integrity and functionality. Disruption of the cell membrane can result in the leakage of the cellular components and ions, which can have detrimental effects on cellular processes. Moreover, the GA@CuO–ZnO nanocomposite can disrupt proteins and enzymes within the cells. Proteins play critical roles in various cellular processes, and their disruption can lead to malfunctioning of these processes. Enzymes, in particular, are essential for catalyzing biochemical reactions, and their disruption can interfere with metabolic pathways.
The formation of reactive oxygen species (ROS) is yet another effect that occurs as a result of the interaction between the GA@CuO–ZnO nanocomposite and different types of cells. ROS are chemicals that are extremely reactive and have the potential to contribute to oxidative stress within cells. There is a possibility that oxidative stress will cause damage to cellular components such as DNA, proteins, and lipids, which may ultimately result in cellular malfunction or even cell death. Furthermore, GA@CuO–ZnO nanocomposite has the ability to bind to proteins. This binding can disturb cellular homeostasis by interfering with normal protein functions. One example is the disruption of the electron transport chain, which is a crucial process for generating energy in cells. Interference with the electron transport chain can impair cellular respiration and overall energy production.35,36
Antifungal activity
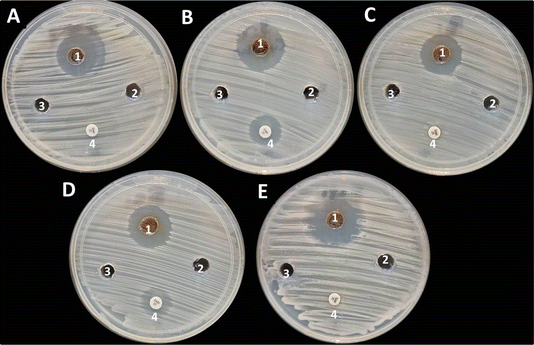
The antifungal activity of GA@CuO–ZnO nanocomposite against both unicellular and multicellular fungi was assessed, as shown in Fig. 7. Results revealed that GA@CuO–ZnO nanocomposite showed antifungal activity toward all tested fungal strains. Moreover, the GA@CuO–ZnO nanocomposite showed antifungal activity toward C. albicans, C. neoformans and A. niger with inhibition zones of 23.86 ± 1.60, 21.96 ± 1.05 and 21.16 ± 1.25 mm, respectively, but showed weak antifungal activity toward A. fumigatus where inhibition zone was 15.3 ± 1.27 mm (Table 2).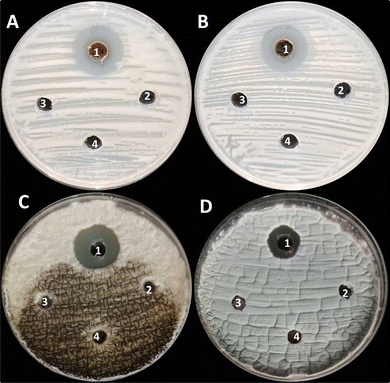 | ||
| Fig. 7 Antifungal activity of GA@CuO–ZnO nanocomposite (1), copper acetate (2), zinc acetate (3) and amphotericin B (4) toward C. albicans (A), C. neoformans (B), A. niger (C) and A. fumigatus (D). | ||
| Fungal strain | Cu acetate IZ (mm) | Zn acetate IZ (mm) | GA@CuO–ZnO nanocomposite | Amphotericin B IZ (mm) | |
|---|---|---|---|---|---|
| IZ (mm) | MIC (μg ml−1) | ||||
| Data within the groups are analyzed using a one-way analysis of variance (ANOVA) followed by a,b Duncan’s multiple range test (DMRT). | |||||
| C. albicans | ND | ND | 23.86 ± 1.60a | 31.25 | ND |
| C. neoformans | ND | ND | 21.96 ± 1.05a | 31.25 | ND |
| A. niger | ND | ND | 21.16 ± 1.25a | 31.25 | ND |
| A. fumigatus | ND | ND | 15.3 ± 1.27b | 125 | ND |
Furthermore, MICs of GA@CuO–ZnO nanocomposite against all tested fungal strains were tested. Results in Table 2 illustrated that the MIC of GA@CuO–ZnO nanocomposite toward all tested fungal strains was 31.25 μg ml−1 except for A. fumigatus it was 125 μg ml−1. In a previous study, bimetallic copper–zinc nanoparticles exhibited significant antifungal activity toward C. parapsilosis and C. tropicalis with MICs of 32 and 64 μg ml−1, respectively.68 Also, the nanocomposite based on bimetallic zinc–copper oxide nanoparticles showed weak antifungal activity with MIC 250 μg ml−1 against C. neoformans and C. albicans.30
Gaber et al.22 synthesized bimetallic CuO–ZnO NPs using Aspergillus fumigatus where it exhibited antifungal activity toward Fusarium oxysporum with MIC of 125 μg ml−1. Gaber et al.22 examined the impact of bimetallic ZnO–CuO on F. oxysporum using TEM ultrastructure analysis. They observed a complete destruction of all cellular components, disintegration of the cell wall, and damage to the plasma membrane. Furthermore, the nucleus exhibited a diminutive size and a distorted shape, while the chromatin components were dispersed throughout the cytoplasm, accompanied by many intensely pigmented bodies.
Cytotoxicity and anticancer activity
In order to assess the safety of the produced compounds, it is essential to evaluate their cytotoxicity towards normal cell lines. The present work evaluated the cytotoxic effect of GA@CuO–ZnO nanocomposite on the normal cell lines Wi38 and Vero, as depicted in Fig. 8A. Results revealed that the IC50 of GA@CuO–ZnO nanocomposite toward Wi38 was 143.3 μg ml−1. Cell viability of Wi38 at concentrations 31.25 and 62.5 μg ml−1 were 99.28 and 88.49% respectively. Moreover, the IC50 of the GA@CuO–ZnO nanocomposite toward the Vero normal cell line was 220.9 μg ml−1. Therefore, the prepared GA@CuO–ZnO nanocomposite in this study can be considered safe for use, as it exhibits a non-cytotoxic classification when the IC50 value exceeds 90 μg ml−1.70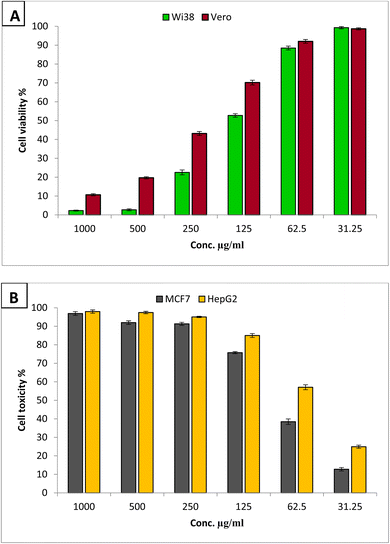 | ||
| Fig. 8 Cytotoxicity of GA@CuO–ZnO nanocomposite against Wi38 and Vero normal cell lines (A), and anticancer activity against MCF7 and HepG2 cancerous cell lines (B). | ||
Fig. 8B shows that the GA@CuO–ZnO nanocomposite exhibited promising anticancer activity toward MCF7 and HepG2 and the activity against HepG2 was greater than that against the MCF7 cell line. The GA@CuO–ZnO nanocomposite showed IC50s of 54.7 and 79.2 μg ml−1 toward HepG2 and MCF7, respectively.
When leaf extract of Artemisia abyssinica was used for biosynthesis of bimetallic Zn–Cu NPs, and evaluated for anticancer activity, it was found that the Zn–Cu NPs had anticancer activity 89% at 500 μg ml−1 with an IC50 value of 33.12 μg ml−1 toward MCF7 cell line.71 Another study72 demonstrated the successful synthesis of Ag–ZnO nanoparticles using an extract derived from Chonemorpha grandiflora leaves, the efficacy of these nanoparticles was evaluated on MCF7, HCT116, and A549 cell lines. Previously, laser ablation was utilized to synthesize ZnO–Ag nanoparticles73 and their anticancer capabilities were evaluated against HCT116 and HeLa cancer cell lines. In their study, Pandiyan et al.74 (2019) found that the nanocomposite (Ag–Au/ZnO) exhibited promising anticancer properties against HeLa cells, which are human cervical cancer cells.
Bimetallic nanoparticles can exhibit synergistic effects, where the combination of two metals enhances their individual anticancer activities. For example, the presence of multiple metals can lead to increased reactive oxygen species (ROS) generation, which can induce oxidative stress and subsequent cancer cell death. Also, bimetallic nanoparticles often possess improved stability and biocompatibility compared to individual metal nanoparticles. This allows for their effective delivery and interaction with cancer cells, leading to enhanced therapeutic efficacy.75
Conclusions
In the current study, GA@CuO–ZnO nanocomposite was successfully synthesized through an eco-friendly green method. Characterization results revealed that the synthesized GA@CuO–ZnO nanocomposite showed a UV absorption peak at 420 nm, and was semi-spherical in shape with an average diameter of 50.28 ± 1.5 nm. Antibacterial and antifungal activities were evaluated toward different microbial strains. The GA@CuO–ZnO nanocomposite showed significant antibacterial efficacy against both Gram-positive and Gram-negative bacteria. In addition, the GA@CuO–ZnO nanocomposite demonstrated antifungal properties against both single-celled and multi-celled fungi. Furthermore, the safety of the GA@CuO–ZnO nanocomposite was established on the Wi38 normal cell line through cytotoxicity data. The IC50 value, which indicates the concentration at which 50% of the cells are affected, was found to be 143.3 μg ml−1. In addition, the GA@CuO–ZnO nanocomposite demonstrated significant potential in inhibiting the growth of MCF7 and HepG2 cancer cell lines, with IC50 values of 54.7 and 79.2 μg ml−1, respectively. Finally, the synthesized GA@CuO–ZnO nanocomposite using gum Arabic showed promising different biological activities, thus it can be used for combating pathogenic microbes and cancer in the medical field after extensive in vivo studies. One of the limitations of our work is that optimizing the synthetic process for NP synthesis was necessary to create extremely concentrated, small-sized bimetallic NPs. Our next work to validate the production of bimetallic CuO–ZnO NPs must involve numerous confirmations, including XPS and EDX analysis. This is necessary to establish the composition and assure repeatability of the synthesis procedure.Data availability
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.Author contributions
Gharieb S. El-Sayyad: conceptualization, methodology, writing – original draft preparation, writing – review and editing; El-Sayed R. El-Sayed: methodology, writing – original draft preparation; Samar H. Rizk: methodology, writing – original draft preparation; Mostafa A. Abdel-Maksoud: methodology, writing – original draft preparation; Adel M. Zakri: methodology, writing – review and editing, critically revising; Abdul Malik: methodology, writing – review and editing, critically revising; Mohamed N. Malash: methodology, writing – review and editing; Amr H. Hashem: conceptualization, methodology, writing – original draft preparation, writing – review and editing.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
The authors extend their appreciation to King Saud University, Riyadh, Saudi Arabia, for Supporting Project number (RSP2025R376).References
- F. Prestinaci, P. Pezzotti and A. Pantosti, Pathog. Global Health, 2015, 109, 309–318 CrossRef.
- L. M. Bebell and A. N. Muiru, Glob. Heart, 2014, 9, 347–358 CrossRef PubMed.
- M. A. Salam, M. Y. Al-Amin, M. T. Salam, J. S. Pawar and N. Akhter, Healthcare, 2023, 11(13), 1946 CrossRef PubMed.
- G. Muteeb and M. T. Rehman, Pharmaceuticals, 2023, 16(11), 1615 CrossRef CAS PubMed.
- A. U. Khan, H. S. Dagur, M. Khan, N. Malik, M. Alam and M. Mushtaque, Eur. J. Med. Chem. Rep., 2021, 3, 100010 CAS.
- M. Pandi, R. SenthilKumaran, P. Rajapriya, S. Yogeswari and J. Muthumary, Biores. Bull., 2013, 2, 1–9 Search PubMed.
- S. Dasari and P. B. Tchounwou, Eur. J. Pharmacol., 2014, 740, 364–378 CrossRef CAS PubMed.
- E. Saied, A. H. Hashem, O. M. Ali, S. Selim, M. S. Almuhayawi and M. A. Elbahnasawy, Life, 2022, 12, 1331 CrossRef CAS.
- M. A. Albalawi, A. M. Abdelaziz, M. S. Attia, E. Saied, H. H. Elganzory and A. H. Hashem, Antioxidants, 2022, 11, 2323 CrossRef CAS.
- J. M. Rajwade, R. Chikte and K. Paknikar, Appl. Microbiol. Biotechnol., 2020, 104, 1437–1461 CrossRef CAS PubMed.
- L. Fu, Z. Wang, O. P. Dhankher and B. Xing, J. Exp. Bot., 2020, 71, 507–519 CrossRef CAS PubMed.
- P. K. Tyagi, Int. J. Curr. Microbiol. Appl. Sci., 2016, 5, 548–558 CrossRef CAS.
- M. Abd Elkodous, H. M. El-Husseiny, G. S. El-Sayyad, A. H. Hashem, A. S. Doghish, D. Elfadil, Y. Radwan, H. M. El-Zeiny, H. Bedair and O. A. Ikhdair, Nanotechnol. Rev., 2021, 10, 1662–1739 CrossRef.
- A. H. Hashem, E. Saied, O. M. Ali, S. Selim, S. K. Al Jaouni, F. M. Elkady and G. S. El-Sayyad, Appl. Biochem. Biotechnol., 2023, 195, 5753–5776 CrossRef CAS.
- M. Saied, M. Hasanin, T. M. Abdelghany, B. H. Amin and A. H. Hashem, Int. J. Biol. Macromol., 2023, 242, 124709 CrossRef CAS.
- N. H. Kamaruzaman, N. N. M. Noor, R. M. S. R. Mohamed, A. Al-Gheethi, S. K. Ponnusamy, A. Sharma and D.-V. N. Vo, Environ. Res., 2022, 209, 112831 CrossRef CAS PubMed.
- A. H. Hashem, T. A. Selim, M. H. Alruhaili, S. Selim, D. H. M. Alkhalifah, S. K. Al Jaouni and S. S. Salem, J. Funct. Biomater., 2022, 13, 112 CrossRef CAS PubMed.
- O. M. Ali, M. S. Hasanin, W. B. Suleiman, E. E.-H. Helal and A. H. Hashem, Biomass Convers. Biorefin., 2024, 14, 6987–6998 CrossRef CAS.
- E. Saied, S. S. Salem, A. A. Al-Askar, F. M. Elkady, A. A. Arishi and A. H. Hashem, Bioengineering, 2022, 9, 397 CrossRef CAS PubMed.
- A. H. Hashem, E. Saied, B. H. Amin, F. O. Alotibi, A. A. Al-Askar, A. A. Arishi, F. M. Elkady and M. A. Elbahnasawy, J. Funct. Biomater., 2022, 13, 242 CrossRef CAS PubMed.
- A. H. Hashem, M. E. El-Naggar, A. M. Abdelaziz, S. Abdelbary, Y. R. Hassan and M. S. Hasanin, Int. J. Biol. Macromol., 2023, 249, 126011 CrossRef CAS PubMed.
- S. E. Gaber, A. H. Hashem, G. S. El-Sayyad and M. S. Attia, Biomass Convers. Biorefin., 2024, 14, 25395–25409 CrossRef CAS.
- M. G. Naseri, E. B. Saion, H. A. Ahangar and A. H. Shaari, Mater. Res. Bull., 2013, 48, 1439–1446 CrossRef CAS.
- A. I. El-Batal, M. Abd Elkodous, G. S. El-Sayyad, N. E. Al-Hazmi, M. Gobara and A. Baraka, Int. J. Biol. Macromol., 2020, 165, 169–186 CrossRef CAS PubMed.
- P. Chawla, N. Kumar, A. Bains, S. B. Dhull, M. Kumar, R. Kaushik and S. Punia, Int. J. Biol. Macromol., 2020, 146, 232–242 CrossRef CAS PubMed.
- R. R. El-Behery, E.-S. R. El-Sayed and G. S. El-Sayyad, BMC Microbiol., 2023, 23, 224 CrossRef CAS.
- M. S. Attia, G. S. El-Sayyad, A. M. Abdelaziz, S. E. Gaber, A. M. A. Khalil, A. M. Saleh, O. M. Al zoubi and A. H. Hashem, Agronomy, 2023, 13, 2725 CrossRef CAS.
- A. H. Hashem, G. S. El-Sayyad, A. A. Al-Askar, S. A. Marey, H. AbdElgawad, K. A. Abd-Elsalam and E. Saied, Plants, 2023, 12, 3288 CrossRef CAS.
- A. I. El-Batal, M. I. Eisa, M. A. M. Saad, H. M. Fakhry, W. M. El-Neshwy, S. S. Abdel-Fatah, F. M. Mosallam and G. S. El-Sayyad, Int. J. Biol. Macromol., 2024, 262, 130010 CrossRef CAS.
- M. S. Hasanin, A. H. Hashem, A. A. Al-Askar, J. Haponiuk and E. Saied, Electron. J. Biotechnol., 2023, 65, 45–55 CrossRef CAS.
- A. H. Hashem and G. S. El-Sayyad, Biomass Convers. Biorefin., 2023, 1–13 Search PubMed.
- A. I. El-Batal, B. M. Al-shammari, G. S. El-Sayyad, S. H. Rizk, A. M. Abdelaziz, M. M. Nofel and M. S. Attia, Biomass Convers. Biorefin., 2023, 1–18 Search PubMed.
- B. H. Elkhodary, M. S. Attia, G. S. El-Sayyad and M. S. Salem, Biomass Convers. Biorefin., 2023, 1–24 Search PubMed.
- H. Y. Mostafa, G. S. El-Sayyad, H. G. Nada, R. A. Ellethy and E. Zaki, Arch. Biochem. Biophys., 2023, 736, 109539 CrossRef CAS PubMed.
- P. V. Baptista, M. P. McCusker, A. Carvalho, D. A. Ferreira, N. M. Mohan, M. Martins and A. R. Fernandes, Front. Microbiol., 2018, 9, 1441 CrossRef PubMed.
- N. Arora, K. Thangavelu and G. N. Karanikolos, Front. Chem., 2020, 8, 412 CrossRef CAS.
- A. M. Saad, M. T. El-Saadony, A. M. El-Tahan, S. Sayed, M. A. Moustafa, A. E. Taha, T. F. Taha and M. M. Ramadan, Saudi J. Biol. Sci., 2021, 28, 5674–5683 CrossRef CAS.
- A. Ashour, A. I. El-Batal, M. A. Maksoud, G. S. El-Sayyad, S. Labib, E. Abdeltwab and M. El-Okr, Particuology, 2018, 40, 141–151 CrossRef CAS.
- A. I. El-Batal, N. M. Balabel, M. S. Attia and G. S. El-Sayyad, J. Cluster Sci., 2020, 31, 1021–1040 CrossRef CAS.
- A. Said, M. Abu-Elghait, H. M. Atta and S. S. Salem, Appl. Biochem. Biotechnol., 2023, 1–14 Search PubMed.
- W. Pa, Reference method for broth dilution antifungal susceptibility testing of yeasts, Approved standard, CLSI document M27-A2, Clinical and Laboratory Standards Institute, 2nd edn, 2002, https://cir.nii.ac.jp/crid/1570854176048718848 Search PubMed.
- A. M. Shehabeldine, A. H. Hashem, A. R. Wassel and M. Hasanin, Appl. Biochem. Biotechnol., 2022, 194, 783–800 CrossRef CAS.
- A. H. Hashem, M. Hasanin, S. Kamel and S. Dacrory, Colloids Surf., B, 2022, 209, 112172 CrossRef CAS.
- M. Hasanin, A. H. Hashem, A. A. El-Rashedy and S. Kamel, Cellulose, 2021, 28, 8355–8374 CrossRef CAS.
- C. Valgas, S. M. D. Souza, E. Smânia and A. Smânia, Braz. J. Microbiol., 2007, 38, 369–380 CrossRef.
- A. H. Hashem, A. M. A. Khalil, A. M. Reyad and S. S. Salem, Biol. Trace Elem. Res., 2021, 1–11 Search PubMed.
- A. H. Hashem, A. M. Shehabeldine, A. M. Abdelaziz, B. H. Amin and M. H. Sharaf, Appl. Biochem. Biotechnol., 2022, 194, 3468–3482 CrossRef CAS.
- A. Van de Loosdrecht, R. Beelen, G. Ossenkoppele, M. Broekhoven and M. Langenhuijsen, J. Immunol. Methods, 1994, 174, 311–320 CrossRef CAS PubMed.
- A. Khalil, A. Abdelaziz, M. Khaleil and A. Hashem, Lett. Appl. Microbiol., 2021, 72(3), 263–274 CrossRef CAS.
- M. Sui, S. Kunwar, P. Pandey and J. Lee, Sci. Rep., 2019, 9, 1–14 CrossRef CAS.
- A. A. Barzinjy and H. H. Azeez, SN Appl. Sci., 2020, 2, 991 CrossRef CAS.
- H. Rani, S. P. Singh, T. P. Yadav, M. S. Khan, M. I. Ansari and A. K. Singh, Mater. Chem. Phys., 2020, 239, 122052 CrossRef CAS.
- M. S. Attia, N. M. Balabel, I. M. Ababutain, M. S. Osman, M. M. Nofel, M. Abd Elkodous, W. F. Elkhatib, G. S. El-Sayyad and A. I. El-Batal, J. Cluster Sci., 2022, 33, 1373–1386 CrossRef CAS.
- R. Munir, K. Ali, S. A. Z. Naqvi, M. A. Maqsood, M. Z. Bashir and S. Noreen, Sep. Purif. Technol., 2023, 306, 122527 CrossRef CAS.
- K. L. Kelly, E. Coronado, L. L. Zhao and G. C. Schatz, J. Phys. Chem. B, 2023, 107(3), 668–677 CrossRef.
- K. S. Prasad and K. Selvaraj, Biol. Trace Elem. Res., 2014, 157, 275–283 CrossRef CAS.
- A. Lawrie, A. Albanyan, R. Cardigan, I. Mackie and P. Harrison, Vox Sang., 2009, 96, 206–212 CrossRef CAS.
- P. Monika, M. Chandraprabha, R. Hari Krishna, M. Vittal, C. Likhitha, N. Pooja and V. Chaudhary, Biotechnol. Genet. Eng. Rev., 2022, 1–29 Search PubMed.
- E. Castro-Longoria, A. R. Vilchis-Nestor and M. Avalos-Borja, Colloids Surf., B, 2011, 83, 42–48 CrossRef CAS PubMed.
- M. Nissen, R. Förster, T. Wieduwilt, A. Lorenz, S. Jiang, W. Hauswald and M. A. Schmidt, Small, 2022, 18, 2202024 CrossRef CAS.
- T. G. Souza, V. S. Ciminelli and N. D. S. Mohallem, J. Phys.:Conf. Ser., 2016, 733, 012039 CrossRef.
- M. Mohsin, M. Jawad, M. A. Yameen, A. Waseem, S. H. Shah and A. J. Shaikh, Plasmonics, 2020, 15, 1599–1612 CrossRef CAS.
- F. Bigdeli and A. Morsali, Mater. Lett., 2010, 64, 4–5 CrossRef CAS.
- M. A. Siddiquee, M. ud din Parray, M. R. Kamli, M. A. Malik, S. H. Mehdi, K. Imtiyaz, M. M. A. Rizvi, H. K. Rajor and R. Patel, J. Mater. Res. Technol., 2021, 13, 2066–2077 CrossRef CAS.
- S. Poyraz, I. Cerkez, T. S. Huang, Z. Liu, L. Kang, J. Luo and X. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 20025–20034 CrossRef CAS.
- P. Belavi, G. Chavan, L. Naik, R. Somashekar and R. Kotnala, Mater. Chem. Phys., 2012, 132, 138–144 CrossRef CAS.
- K. Pal, M. A. Elkodous and M. M. Mohan, J. Mater. Sci.: Mater. Electron., 2018, 29, 10301–10310 CrossRef CAS.
- M. Turabik, S. Özdemir, G. Akinbingol, S. Gonca and C. Gecgel, Inorg. Chem. Commun., 2023, 155, 111072 CrossRef CAS.
- S. Khatak, N. Wadhwa and P. Jain, Biosci., Biotechnol. Res. Asia, 2021, 17, 763–774 Search PubMed.
- J.-R. Ioset, R. Brun, T. Wenzler, M. Kaiser and V. Yardley, A Training Manual for Screening in Neglected Diseases, 2009 Search PubMed.
- T. A. Orshiso, E. A. Zereffa, H. C. A. Murthy, T. B. Demissie, O. Pardeshi, L. S. Avhad and S. Ghotekar, ACS Omega, 2023, 8, 41039–41053 CrossRef CAS PubMed.
- N. Ambujakshi, H. Raveesha, S. Manohara, N. Dhananjaya, S. Pratibha and C. Shivakumara, Mater. Res. Express, 2019, 6, 095068 CrossRef CAS.
- K. A. Elsayed, M. Alomari, Q. A. Drmosh, M. Alheshibri, A. Al Baroot, T. S. Kayed, A. A. Manda and A. L. Al-Alotaibi, Alexandria Eng. J., 2022, 61, 1449–1457 CrossRef.
- N. Pandiyan, B. Murugesan, M. Arumugam, J. Sonamuthu, S. Samayanan and S. Mahalingam, J. Photochem. Photobiol., B, 2019, 198, 111559 CrossRef CAS PubMed.
- H. Makada, S. Habib and M. Singh, Sci. Afr., 2023, 20, e01700 CAS.
| This journal is © The Royal Society of Chemistry 2025 |