Open Access Article
Open Access ArticleApoptotic cell death of stomach cancer lines (AGS) induced by Co-NTB complex through cellular organelles and DNA damage†
Sri Renukadevi Balusamya,
Mani Balamurugan b,
Sumitha Purushothamanc,
Sivaraman Somasundaramd,
Mohamed Farouk Elsadeke,
Daewon Sohnf,
Saeedah Musaed Almutairig,
Ivan Mijakovichi,
Shadi Rahimi
b,
Sumitha Purushothamanc,
Sivaraman Somasundaramd,
Mohamed Farouk Elsadeke,
Daewon Sohnf,
Saeedah Musaed Almutairig,
Ivan Mijakovichi,
Shadi Rahimi *h and
Haribalan Perumalsamy
*h and
Haribalan Perumalsamy *jk
*jk
aDepartment of Food Science and Biotechnology, Sejong University, Gwangjin-gu, Seoul, Republic of Korea
bDepartment of Materials Science and Engineering, Seoul National University (SNU), 1 Gwanak ro, Seoul 08826, Republic of Korea
cDepartment of Microbiology, Bioprocess Engineering Division, Smykon Biotech, Kanniyakumari, India
dDepartment of Chemistry, Saveetha School of Engineering, SIMATS, Chennai, Tamil Nadu 600124, India
eDepartment of Biochemistry, College of Science, King Saud University, P.O. 2455, Riyadh 11451, Saudi Arabia
fDepartment of Chemistry, College of Natural Sciences, Hanyang University, Seoul 04763, Republic of Korea
gDepartment of Botany and Microbiology, College of Science, King Saud University, P.O. 2455, Riyadh 11451, Saudi Arabia
hSystems and Synthetic Biology Division, Department of Life Sciences, Chalmers University of Technology, Gothenburg SE-412 96, Sweden. E-mail: shadir@chalmers.se
iThe Novo Nordisk Foundation, Center for Biosustainability, Technical University of Denmark, Kongens Lyngby DK-2800, Denmark
jResearch Institute for Convergence of Basic Science, Hanyang University, Seoul 04763, Republic of Korea. E-mail: harijai2004@gmail.com
kCenter for Creative Convergence Education, Hanyang University, Seoul 04763, Republic of Korea
First published on 9th January 2025
Abstract
Given that stomach cancer is the fourth leading cause of cancer-related death, there is a need to develop new drugs. Among various methods, metal-based coordination compounds are considered as an efficient strategy against this type of cancer. Similarly, the benzimidazole moiety plays a crucial role in biology; thus, various benzimidazole-based compounds have been found to be active as potential anticancer drugs and are currently used in clinical trials. In this study, we explored the benzimidazole-based cobalt(II) complex as an anticancer agent against AGS stomach cancer cell lines. Interestingly, the MTT assay of the Co-NTB complex shows a lower IC50 value of 4.25 μg mL−1 compared to cisplatin, which has an IC50 of 7.5 μg mL−1 against AGS cell lines. Light microscopy and Hoechst/propidium iodide dye staining clearly indicate that the complex damages DNA, leading to cell death through an apoptotic pathway. The apoptotic cell death pathway was further complemented by Lysotracker and Mitotracker staining, as well as transmission electron microscopy (TEM) imaging. Overall, the Co-NTB complex acts as an effective anticancer agent against AGS stomach cancer cell lines, with apoptotic cell death induced by targeting cellular organelles and DNA.
1. Introduction
Stomach cancer is the fourth leading cause of death by cancer, accounting for over 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths, and with approximately 1.1 million new cases in 2020. It was also considered as the fifth most common malignant tumour in the world.1,2 Metal compounds are applicable in the medical field dating back to the XVI century with reports on the therapeutic effect of metals and their compounds in the treatment of diseases such as cancer.3 However, substantial side effects, lack of specificity, and undesirable accumulation in tumor sites are associated to these chemicals, and they do not demonstrate enough selectivity and have shown toxicity to normal cells including skin, lung, and epithelial cells.4 Particularly, metals such as cobalt(II), zinc(II) and tin(IV) in complexes demonstrated considerable cytotoxicity to human normal skin fibroblast (HFF) cells.5,6 On the other hand, a nickel(II) complex exhibited moderate cytotoxicity in mouse beta pancreatic cell line β-TC3.7 Further, conventionally modified metallodrugs have become an intriguing research area in medicinal chemistry as potential cancer treatments.8,9 In particular, platinum-based metallodrugs, specifically cisplatin and its analogues, are the most widely and successfully used therapeutic treatments for a variety of cancers.10–12 Despite their successful use, the platinum-based drug cisplatin causes undesired therapeutic cytotoxicity, such as neurotoxicity, nephrotoxicity, hepatotoxicity, and drug resistance.13,14
000 deaths, and with approximately 1.1 million new cases in 2020. It was also considered as the fifth most common malignant tumour in the world.1,2 Metal compounds are applicable in the medical field dating back to the XVI century with reports on the therapeutic effect of metals and their compounds in the treatment of diseases such as cancer.3 However, substantial side effects, lack of specificity, and undesirable accumulation in tumor sites are associated to these chemicals, and they do not demonstrate enough selectivity and have shown toxicity to normal cells including skin, lung, and epithelial cells.4 Particularly, metals such as cobalt(II), zinc(II) and tin(IV) in complexes demonstrated considerable cytotoxicity to human normal skin fibroblast (HFF) cells.5,6 On the other hand, a nickel(II) complex exhibited moderate cytotoxicity in mouse beta pancreatic cell line β-TC3.7 Further, conventionally modified metallodrugs have become an intriguing research area in medicinal chemistry as potential cancer treatments.8,9 In particular, platinum-based metallodrugs, specifically cisplatin and its analogues, are the most widely and successfully used therapeutic treatments for a variety of cancers.10–12 Despite their successful use, the platinum-based drug cisplatin causes undesired therapeutic cytotoxicity, such as neurotoxicity, nephrotoxicity, hepatotoxicity, and drug resistance.13,14
In attempt to replace, there is a need to search for novel metal-based compounds with antitumour activity that is more specific and less cytotoxicity to normal cells. In recent years, novel metal derivatives, coordination compounds with different metals and their potential anticancer and antitumor activities have been investigated.15 There are new therapeutic designs that are metal-based coordination compounds with ligands, such as benzimidazoles that exhibit potential biological activity. The privilege of benzimidazole is its structural analogy to the purines which are part of DNA. Furthermore, there are several benzimidazole-based compounds with pharmacological importance and several of its chemical derivatives are being used as prospective anticancer medicines in clinical studies.15
Cobalt is an essential element for the human body. It is found at the metal centre of vitamin B12 that is an essential nutrient for humans because of that various cobalt derivatives are employed in cancer thermotherapy.16–19 Previously, a few benzimidazole-based cobalt complexes have been reported that exhibit potential anticancer activity.15,20–39 For instance, Yilmaz et al.20 reported that the (dichlorobis(1-benzyl)-2-(4-chlorophenyl)-1H-benzimidazole-KN3)cobalt(II)) (LogIC50 – 0.38 μM) was more effective than docetaxel (LogIC50 – 1.13 μM) against DU-145 human prostate cancer cells after 24 hours.28 Another study found that the cobalt(II) combination with 2,6-bis(1-phenylbenzimidazol-2-yl)pyridine inhibited cell proliferation in Eca9706 and Eca109 esophageal cancer cell lines but not in HepG2 and Huh7 liver cancer cells.40 Indeed, the same complex may not exhibit antiproliferative activity against all malignant cell lines; hence, it is critical to investigate the anticancer activity of known Co-complexes with various cell lines.
The tris(2-benzimidazolylmethyl) amine (NTB) acts as biomimic agents for the active sites of metelloproteins such as histidine37,41 and histidine plays an important role in protein synthesis and reactive species scavenging. The NTB ligand is known for its complexation with various metal ions for a long time and plays a crucial role in various biological activity. Therefore, in this study, we employed the ligand NTB in conjunction with cobalt(II), which is crucial to enhance the cytotoxic properties towards cancer cell lines.
Notably most of the reported Co-complexes against gastric cancer shows high IC50 values, for instance benzimidazole-based complex [Co(1-((2-(pyridine-4-yl)-1H-benzoimidazol-1-yl)methyl)-1H-pyridine)Cl2] exhibited poor cytotoxic activity (IC50 93.80 ± 8.04 μM) against human gastric carcinoma cells compared to cisplatin (IC50, 4.151 ± 0.05 μM).34 In addition, various benzimidazole-based cobalt complexes were tested in gastric carcinoma cells, however poor activity was reported against human gastric carcinoma cells.24,42 Notably, Co-NTB complex have been used as anticancer agent against Hela and HCT-15 cell lines and found inhibitory effect (IC50 values of 54 mg mL−1 for both cell line). However, Co-NTB complex have not been tested as anticancer agent against AGS cell lines previously. Henceforth, the objectives of this study to explore the anticancer efficacy of a conventionally modified novel Co-NTB complex on stomach cancer cell lines were studied. Furthermore, efficacy of Co-NTB complex was compared to most widely used platinum-based drug cisplatin. Briefly, in our study we explored Co-NTB complex as anticancer agent against AGS cell lines using MTT viability assay, and morphological assessment. Then, Co-NTB complex induced apoptotic mediated cell death was confirmed by performing FACS analysis and different fluorescence staining analysis including Hoechst, propidium iodide (PI), LysoTracker, MitoTracker. Furthermore, localization and specificity of Co-NTB complex in AGS cell organelles was observed by TEM analysis. Overall, our investigation revealed intriguing fact that, Co-NTB acts as an efficient metallodrug against gastric cancer cell lines (AGS) with higher antiproliferative activity compared to cisplatin.
2. Results and discussion
2.1. Synthesis and characterization
The [Co(NTB)Cl]Cl complex was synthesised as reported earlier43,44 (Fig. 1a). The UV-spectrum of the complex in water showed a strong absorption at 530–550 nm and another weak and broad d–d transition between 700–1000 nm denoting the formation of cobalt complex with NTB (Fig. S1†). The spectral pattern resembles with octahedral high-spin Co(II) complexes, possibly due to the dissolution of the complex in water leading to the coordination of the water molecule by replacing the chloride ions.45 The IR-spectrum of Co-NTB show peak in the region of 3400 cm−1, which confirms the presence of benzimidazolyl N–H group and peaks in the range of 2700–2900 cm−1 confirms the alkane C–H stretching corresponding to the –CH2 groups in NTB. (Fig. S2†). The 1H NMR spectrum of Co-NTB shows shift of all the ligand peaks while complexation with cobalt. The N–H peak shift from 12.2 ppm to 7.2 ppm and the aromatic peaks shifted from 7.50 and 7.1 ppm to 3.6 and 1.7 ppm, likewise the aliphatic –CH2 peak shift from 4.15 to 2.1 ppm upon complexation with Co(II) (Fig. S3†). All the changes clearly confirm the complexation of NTB with cobalt(II) center.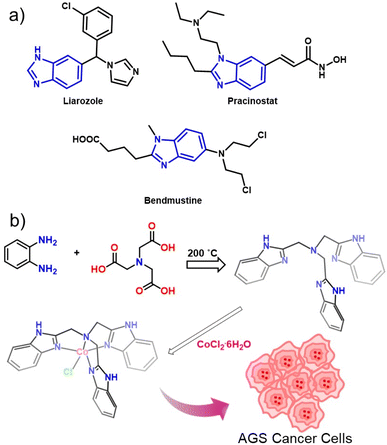 | ||
| Fig. 1 (a) A few examples of benzimidazole containing anti-cancer drugs. (b) Schematic representation of Co-NTB synthesis. | ||
2.2. Cytotoxicity analysis of Co-NTB towards AGS cell lines
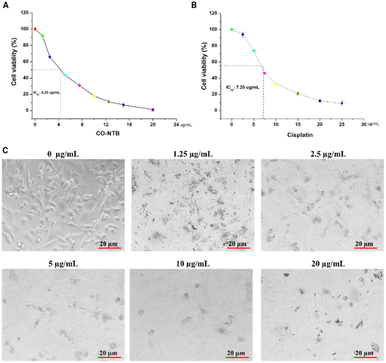
The cytotoxicity analysis was performed to demonstrate the antiproliferative activity of Co-NTB complex against AGS stomach cancer cell lines. The benzimidazole derivative that targets DNA minor groove binders showed cytotoxicity in a variety of cell lines, including ovarian (SKOV-3; 1.30 mg mL−1), cervical (HeLa; 15 mg mL−1), and gastric (BGC-823; 5.09 mg mL−1).46 Similarly, another derivative, methyl 2-(5-fluoro-2-hydroxyphenyl)-1H-benzo[d]imidazole-5-carboxylate (MBIC), demonstrated selective toxicity in HeLa cells, with an IC50 of 0.54 μg mL−1.47 Another benzimidazole analog, BZD9L1, when treated with colorectal cancer cell lines, produced apoptosis by reducing cell proliferation, migration, and invasion, with IC50 values of 5.4 mg L−1 for HCT116 and 6.46 mg L−1 for HT-29.48 Likewise, another benzimidazole analogue inhibitor, AC1-004, showed cytotoxicity (3.2 μg mL−1) in colorectal cancer HCT-116 cells.49 In our study, we performed MTT assay and exhibited with IC50 value was 4.25 μg mL−1. The MTT analysis proved that the Co-NTB complex showed anti-proliferative effects on the stomach cancer cell line in a dose-dependent manner (Fig. 2A). Whereas the IC50 value of AGS cells treated with cisplatin for 12 hours was 7.20 μg mL−1 (Fig. 2B). Fig. 2C depicts light microscopical observation on dose-dependent efficacy of Co-NTB complex on morphology of AGS cells when the cells treated with various concentrations of Co-NTB complex (1.25–20 μg mL−1) for 24 h. It could demonstrate that destruction of morphological features leads to cell death by addition of Co-NTB complex at all tested concentrations (1.25–20 μg mL−1), compared to the control without any treatment (0 μg mL−1). However, the significant damage could be appeared at higher dosage of complex (2.5–20 μg mL−1).Although significant anticancer activity was observed in Co-NTB complex, cytotoxicity observation on non-cancerous cell lines must be assessed to know the specificity of the therapeutic efficacy. Therefore, in our study, we also confirmed toxicity of non-cancerous cell lines, including the human keratinocyte cell line (HaCaT) and murine mouse macrophage (RAW 246.7) cells. The dose-dependent concentration of Co-NTB on both non-cancerous cells showed 50% of toxicity at >100 μg mL−1. Our results demonstrated that both cells showed comparatively less toxicity than AGS demonstrated that Co-NTB complex may have more specificity over their toxicity. The data is provided in ESI (Fig. S4).†
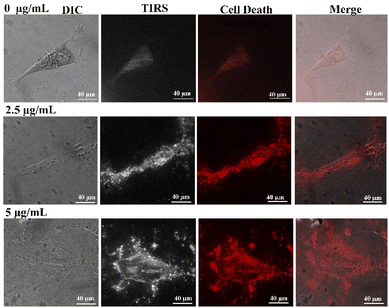
We also investigated the cell death using PI staining and phase contrast microscopic analysis of cells exposed to the complex at two different concentrations of 2.5 and 5 μg mL−1 for 24 h. As it is shown in Fig. 3, the cell death could be found in the samples treated with both tested concentrations. Furthermore, we observed a dose-dependent effect of the Co-NTB complex on AGS cell lines by phase-contrast microscopy. At 2.5 μg mL−1, the Co-NTB complex triggered the cytotoxic effects, and the cell-death increased with increasing concentration. Overall, the CoNTB complex showed strong cytotoxicity against AGS cells, which was supported by reduced number of viable cells, destruction of cell morphological features, and evidence on cell death.
2.3. Investigation of AGS cell death pathway
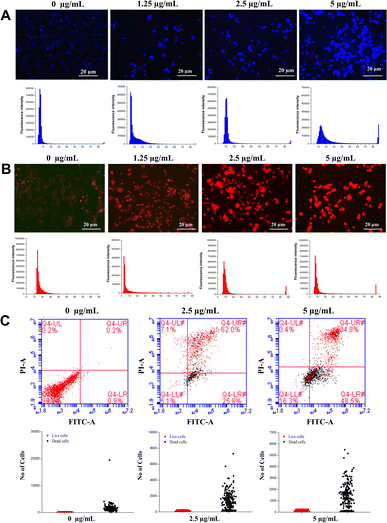
Given reduced AGS cells viability and cell death detection upon treatment with the Co-NTB complex that was observed in the previous section (Fig. 2), we aimed to use Hoechst/PI dyes to observe the rise in the cell death rate induced by Co-NTB complex, that are presented by blue and red colours, respectively (Fig. 3, 4A and B). A cell that undergoes apoptosis, demonstrates nuclear condensation and DNA fragmentation that can be detected by Hoechst stain and fluorescence microscopy.50PI is also a fluorescent DNA-binding dye, that freely penetrates cell membranes of dead or dying cells but is excluded from viable cells. As it is indicated, compare to the control without any treatment (0 μg mL−1), there was an increased number of apoptotic cells caused by 2.5 and 5 μg mL−1 of Co-NTB complex treated to the cells for 24 h (Fig. 4A and B). However, the higher concentration of 5 μg per mL Co-NTB complex induced a greater number of cells undergoing apoptosis as evidenced by more blue color fluorescence in this sample (Fig. 4A). Similarly, in case of PI (Fig. 4B), high number of dead cells were found in the samples treated with 2.5 and 5 μg per mL complex.
Besides Hoechst/PI dyes, we also used FACS analysis to show the effect of different concentrations of Co-NTB complex (2.5 and 5 μg mL−1) on classifyingn the live and dead cells, shown by red and black dots, respectively (Fig. 4C). High number of live cells are present in the control sample without treatment with the Co-NTB complex (0 μg mL−1). However, 2.5 and 5 μg mL−1 concentrations of Co-NTB complex induced cell death and we could find a high number of dead cells present in the treated samples with both tested concentrations.
2.4. TUNEL assay of Co-NTB complex
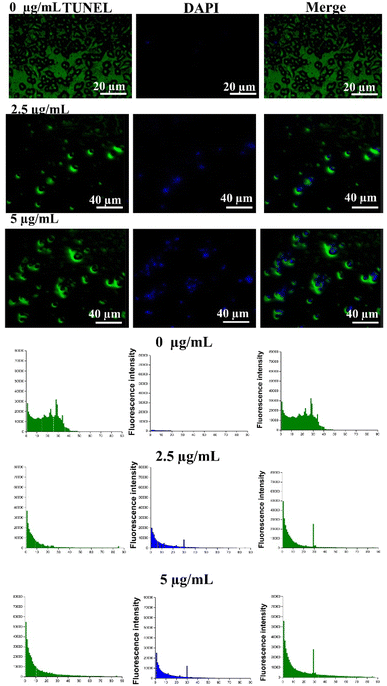
DNA is considered as the primary target for metal-based drugs. In fact, the metal-based anticancer drugs can form specific lesions on DNA that induces apoptosis.51 Given observed apoptosis caused by Co-NTB complex in the previous section, we investigated if the apoptosis was caused by DNA damage. To demonstrate the DNA damage in the AGS cells, we treated the cells with different concentrations of Co-NTB complex (2.5 and 5 μg mL−1) for 24 h and we further performed the TUNEL assay (Fig. 5).The TUNEL assay detects the DNA breakage that normally arises during early and late stages of apoptosis. In TUNEL assay, terminal deoxynucleotidyl transferase (TdT) reacts with fluorescein (FITC)-labeled dUTP to attach uridine to 3′-hydroxyl (3′OH) terminus in DNA strand breaks. Double-stranded DNA is also counterstained by DAPI that intercalates between strands of double-stranded DNA. DAPI is a cell impermeant dye, and it cannot pass through the membrane of live cells as no DAPI signal was found in control sample without any treatment (0 μg mL−1). Positive TUNEL signal colocalized with DAPI signal were found in the samples treated with 2.5 and 5 μg mL−1 concentrations of Co-NTB complex compared to the control, verifying the DNA damage by tested concentrations of Co-NTB complex in dead cells.
It was reported that the Co(II) complex of bidentate Schiff base ligand (L) 2-((1H-benzo [d]imidazole-4ylimmino) methyl phenol interact with DNA externally that may be through the formation of hydrogen bond between the phenolic hydroxyl groups of the Schiff base and the nucleotides.52,53 The association constant for Co(II) complex in their study is reported as 2.22 × 103 M.54 It was also shown that the mononuclear specie (Co-24) containing the pyrazole-based ligand could be bound with DNA through grooves. In case of binuclear compounds (Co-25 and Co-26) with same ligand, they could be bound with DNA through both grooves and intercalation before inducing oxidative cleavage of DNA.
2.5. Investigation of cell death pathway
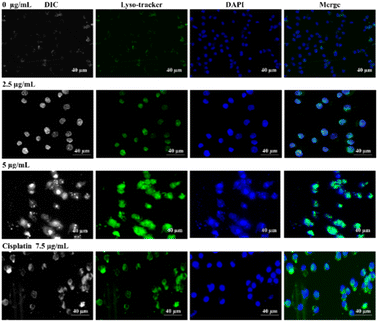
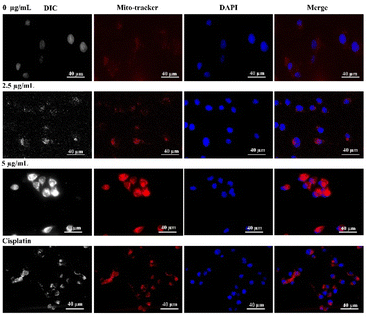
The molecular target pathway of platinum based metallodrugs may involve apoptotic mediated cell death when enters immediately to the cells. In particular, most extensively used cisplatin directly interacts with DNA and interrupt DNA function, resulting cellular death.55,56 Likewise, in this study, our synthesized Co-NTB complex plays a major role in apoptotic cell death, similar to cisplatin with lower concertation. In addition, we provided, TEM images that clearly demonstrated, the localization and organelle-specific target of Co-NTB, which may target lysosomes or mitochondria, potentially leading to further ROS-mediated cell death.To demonstrate the cell death induced by Co-NTB complex, we treated the cells with different concentrations of Co-NTB complex (2.5 and 5 μg mL−1) for 24 h and we further stained the cells with LysoTracker and MitoTracker (Fig. 6 and 7). Cells treated with cisplatin (7.2 μg mL−1) was used as the positive control. Lysotracker dye, a highly soluble small molecule, is retained in acidic subcellular compartments such as the lysosome. LysoTracker staining is suggested as a great complement to other methods for detection of cell death.57
This staining is particularly useful to look for changes in the overall pattern of apoptosis. Treatment of cells with Co-NTB complex (2.5 and 5 μg mL−1) induced LysoTracker signal compared to the control samples without any treatment (0 μg mL−1). Double-stranded DNA is also counterstained by DAPI (in blue colour). In merge images, LysoTracker-positive puncta appeared where DAPI signals were found (Fig. 6). In fact, LysoTracker marked the regions of apoptosis that colocalized with double-stranded DNA. Similar to the samples treated with the complex, the treated cells with cisplatin also induced colocalized LysoTracker and DAPI signals.
Mitotracker dye is a cation that distributes to the mitochondrial matrix as a function of the Nernst equation. Cells undergoing apoptosis show a decrease in mitochondrial membrane potential. MitoTracker signal is dependent on the electro-chemical properties of the mitochondria, which are different for live and dead cells.58 There was no MitoTracker signal in the control samples with live cells without any treatment (0 μg mL−1). However, MitoTracker signal was detected in the samples treated with 2.5 and 5 μg per mL Co-NTB complex as well as 7.2 μg per mL cisplatin. Double-stranded DNA is also counterstained by DAPI (in blue colour) and the MitoTracker and DAPI signals were colocalized as indicated in Fig. 7.
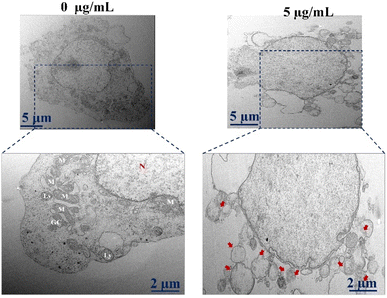
Besides staining with Lyso-tracker and Mito-tracker, we also investigate the cell death using TEM (Fig. 8). TEM images also proved the evidence of cell death as shown by red arrows in the sample treated with 5 μg per mL Co-NTB complex compared to the control sample without any treatment (0 μg mL−1).
3. Conclusions
In this study, we explored the anticancer activity of the conventionally modified novel Co-NTB complex against AGS stomach cancer cell lines. The complex shows a lower IC50 value of 4.25 μg mL−1 compared to standard drug cisplatin. The Co-NTB complex efficiently kills the AGS cancer cells by accumulating in the nucleus, mitochondria and causing structural changes, DNA damage and cleavage, leading to apoptotic mediated cell death. Our findings demonstrate the potential of NTB-based cobalt complexes as a promising anticancer treatment for stomach cancer with an apoptotic pathway of cell death generated by targeting cellular organelles. This study provides insights for the possibility of utilisation of other benzimidazole based molecular complexes as efficient anticancer agents against stomach cancer. Despite the strong anticancer activity of Co-NTB, toxicity on different normal cells lines should be investigated to improve more specificity of target cells. Furthermore, additional in vivo studies on Co-NTB are required to improve their consistency for development as a potential anticancer drug. Further toxicity profiling on non-target organisms could be considered for aquatic organisms with environmental safety concerns.4. Materials and methods
4.1. Reagents and chemicals
o-Phenylenediamine, nitrilotriacetic acid, cobalt(II) chloride hexahydrate were purchased from Sigma-Aldrich. All other chemicals and solvents were obtained from commercially available sources and used as received unless noted. Pre-coated Silica gel TLC plates from Merck Co. were used to monitor the reaction progress and UV light at 254 nm and 365 nm is used to visualize the TLC. 1H NMR (Nuclear Magnetic Resonance) spectra was acquired on a Bruker DRX-300 (300 MHz) instrument using TMS as an internal standard.4.2. Synthesis of NTB (tris(2-benimidazolylmethyl)amine)
The ligand, NTB was synthesized as reported earlier43,59 with slight modification in equivalence and the structural data was confirmed by comparing with the literature. Briefly, o-phenylenediamine (13.5 g, 125 mmol) and nitrilotriacetic acid (7.65 g, 40 mmol) were mixed well and powdered together and heated to 190–200 °C using oil bath for 1 h. After cooling the reaction mixture was powdered and refluxed with decolourising charcoal in methanol and hot filtered. While cooling and evaporation pink coloured crystal appeared, which was separated by filtration and washed with cold methanol to get pure NTB. Yield 60%. Melting range 270–275 °C. δH/ppm (300 MHz, DMSO-d6) 12.2 (s, 3H), 7.57 (m, 6H), 7.17 (m, 6H), 4.15 (s, 6H); m/z (ES-ToF); 407.1 ([M + H]+).4.3. Synthesis of [Co(NTB)Cl)]Cl (Co-NTB)
The Co-NTB was synthesized as reported earlier43,44 with slightly modified procedure. Cobalt(II) chloride hexahydrate (0.237 g 1 mmol) dissolved in 10 mL of ethanol and stirred well for 5 min. The mixture solution turned into purple denotes ready for further process. Later, the NTB ligand (0.407 g 1 mmol) added in 20 mL of ethanol and thoroughly mixed for 5 min. Then cobalt(II) chloride hexahydrate solution were added slowly into NTB ligand solution and stirred well for 1 h. Then, the solution mixture was slowly evaporated for overnight at room temperature, yielding purple precipitate as the end product. After that, the purple precipitate was thoroughly dried in a vacuum desiccator (yield: 76%) to eliminate any remaining solvent. The final Co-NTB complex was subjected to further physiochemical and biological analysis.4.4. Cell line culture
The human stomach cancer cell line (AGS) used in this study was purchased from Korean Cell Line Bank (Seoul, Republic of Korea). The cell lines were grown in RPMI 1640, which has 1% antibiotic–antimycotic solution and 10% FBS. The cells were cultured in a CO2 incubator maintained at 37 °C with a 5% CO2 supply. The most promising cells were chosen for further treatment having passaged over several times as fully grown cells.4.5. Anti-proliferation assay
The cells (5 × 103) were seeded onto 96-well plates and grown for more than 24 hours to reach 80% confluence prior to treatment. The dose dependent concentration ranging from 0 μg mL−1 to 100 μg per mL of Co-NTB was prepared to analyze the minimal inhibition concentration against stomach cancer cell line AGS. Similarly, the positive control drug cisplatin also prepared different concentration (0 μg mL−1 to 100 μg mL−1) to compare the efficacy of Co-NTB for further molecular studies. After 24 hours, both treated and untreated cells were washed twice with 1× PBS (pH 7.4) and added 100 μl of 0.05% of MTT solution (5 mg mL−1) prepared in cultured medium. After 4 hours, the MTT solution from all wells removed and added 200 μl of dimethyl sulfoxide (DMSO). Furthermore, 96 well plates were placed on the rotary sacker to dissolve formazan crystal salt, which had developed in a purple color. The negative control received no treatment, while cisplatin was employed as the positive control. All the sample groups were triplicated. The VersaMax microplate reader (Molecular Devices, USA) was utilized to observe the responses at 560 nm and 670 nm, respectively.4.6. Morphological observation
The AGS cells (3 × 104) seeded into 6 well plates for the treatment of Co-NTB, positive control cisplatin to compare the morphological difference between them. Control well received no treatment and observed well grown morphology to interpret with treated wells. To observe clear morphology differences, we used inverted light microscope (Leica DMIL LED) to reveal the effect of Co-NTB and compared cis-platin.4.7. Fluorescence staining analysis
To detect apoptotic mediated cell death, different types of fluorescent staining techniques were used including Hoechst and propidium iodide (PI) staining was used as described previously.60,61 Briefly, for Hoechst, PI and staining analysis, 2 × 104 of AGS cells were seeded into 6 well plates and waited for more than 12 h. Then, based on IC50 value, we prepared two different concentrations of Co-NTB (2.5 and 5 μg mL−1) to treat AGS cell line in the 6-well plates along with untreated negative control. After 24 h of incubation, both treated and untreated cells were washed twice with 1× PBS buffer to remove dead cells along with debris. Then cells were fixed with 4% paraformaldehyde and performed both Hoechst and PI staining separately as described previously by Perumalsamy et al.,60,61 Similarly, for the terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay, we prepared 2 × 104 of AGS cells and treated for 24 h with two different concentrations of Co-NTB (2.5 and 5 μg mL−1) and performed TUNEL staining as previously described60 for both treated and untreated cells to detect Co-NTB induced cell death. All the sample groups were performed with three replicates.4.8. Flow cytometry analysis of cell sorting (FACS)
The detection of apoptotic mediated cell death, we performed FACS by Annexin V/PI staining as previously described.60,61 5 × 104 of AGS cells were seeded in 6 well plates and waited for the cells to reach 80% confluency before treatment. The two different doses of Co-NTB (2.5 and 5 μg mL−1) were treated cells for 24 h. Then the untreated and treated cells were washed twice with 1× PBS buffer to remove dead cells before preparing the staining for the FACS analysis. Then, good, conditioned cells were removed from the 6-well plates without damaging and centrifuged for 3 min at 1000 rpm. Further, the cell pellets dissolved in 500 μL 1× PBS and prepared sample group with or without Annexin V/PI staining based on protocol as described for the FACS analysis.4.9. Lyso/Mito tracker
AGS cells 2 × 105 cells per well were seeded in a 6-well plate and incubated for 24 h for further treatment. After 24 h, different concentration of Co-NTB (2.5 and 5 μg mL−1) was treated to AGS cells and the cisplatin (7.5 μg mL−1) was served as positive control to compare the efficiency of Co-NTB. Further, cultured plates with or without treatment of Co-NTB or cisplatin were washed twice with 1× PBS buffer to remove dead or damaged cells. Then cells were stained with commercially available kits such as, Mitotracker®Red (Cell signaling, #9082) and Lysotarcker@Green (Cell signaling, 8783#) and visualized the cells using phase-contrast microscope.4.10. Transmission electron microscopic analysis
2 × 104 cells were seeded in 6-well plate for overnight and then cells were treated with or without Co-NTB. To determine the localization and specificity of Co-NTB in AGS cell organelle, we performed transmission scanning electron microscope (TEM) to visualize organelle damage by previously described protocol.60 In brief, after the treatment of Co-NTB, the cells were washed twice with 1× PBS buffer and well-grown cells were detached from culture dish and centrifuged for 3 min at 1000 rpm. Then cells pellets were placed in 10% glycerin and maintained for further TEM sample preparation. Based on protocol described previously,60 we prepared the samples for TEM analysis.4.11. Statistical analysis
All experiments were performed in triplicates, and the IC50 value calculation, we used GraphPad Prism 5 (GraphPad Software, La Jolla, CA) software. Furthermore, the SAS 9.13 software (SAS Institute, Cary, NC) was used to perform statistical analyses.Data availability
Data are available on request from the authors.Author contributions
H. P. and S. B. has done conceptualization, design, carried out biological experiments, and drafted the manuscript. S. R. and M. B. wrote the manuscript. M. B., S. P. and S. S. carried out experiments related to chemistry part. D. S. and I. M. project administration and funding. M. E. and H. P. has done editing and formatting the manuscript. All authors have written, reviewed and revised the manuscript. All the authors have seen and agreed to submit it.Conflicts of interest
There is no conflict of interest to disclose.Acknowledgements
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) (grant numbers 2020R1A6A1A06046728 and 2022R1A6C101A779). The authors would like to extend their sincere appreciation for funding this work through the Researchers Supporting Project number (RSP2025R349), King Saud University, Riyadh, Saudi Arabia.References
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, CA A Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- J. Ferlay, M. Colombet, I. Soerjomataram, D. M. Parkin, M. Piñeros, A. Znaor and F. Bray, Int. J. Cancer, 2021, 149, 778–789 CrossRef CAS PubMed.
- G. V Suárez-Moreno, D. Hernández-Romero, Ó. García-Barradas, Ó. Vázquez-Vera, S. Rosete-Luna, C. A. Cruz-Cruz, A. López-Monteon, J. Carrillo-Ahumada, D. Morales-Morales and R. Colorado-Peralta, Coord. Chem. Rev., 2022, 472, 214790 CrossRef.
- N. J. Wheate, S. Walker, G. E. Craig and R. Oun, Dalton Trans., 2010, 39, 8113–8127 RSC.
- S. Abdolmaleki, M. Ghadermazi and A. Aliabadi, Inorg. Chim. Acta, 2021, 527, 120549 CrossRef CAS.
- R. Heydari, E. Motieiyan, S. Abdolmaleki, A. Aliabadi, M. Ghadermazi, F. Bagheri and H. Amiri Rudbari, J. Coord. Chem., 2020, 73, 2347–2362 CrossRef CAS.
- S. Abdolmaleki, M. Ghadermazi, A. Fattahi, S. Shokraii, M. Alimoradi, B. Shahbazi and A. R. Judy Azar, J. Coord. Chem., 2017, 70, 1406–1423 CrossRef CAS.
- M. A. Girasolo, A. Attanzio, P. Sabatino, L. Tesoriere, S. Rubino and G. Stocco, Inorg. Chim. Acta, 2014, 423, 168–176 CrossRef CAS.
- V.-T. Nguyen, T.-K.-C. Huynh, G.-T.-T. Ho, T.-H.-A. Nguyen, T. Le Anh Nguyen, D. Q. Dao, T. V. T. Mai, L. K. Huynh and T.-K.-D. Hoang, R. Soc. Open Sci., 2022, 9, 220659 CrossRef CAS PubMed.
- G. Giaccone, T. A. Splinter, C. Debruyne, G. S. Kho, P. Lianes, N. van Zandwijk, M. C. Pennucci, G. Scagliotti, J. van Meerbeeck, Q. van Hoesel, D. Curran, T. Sahmoud and P. E. Postmus, J. Clin. Oncol., 1998, 16, 2133–2141 CrossRef CAS PubMed.
- Ö. Tarı, F. Gümüş, L. Açık and B. Aydın, Bioorg. Chem., 2017, 74, 272–283 CrossRef PubMed.
- A. du Bois, H.-J. Lück, W. Meier, H.-P. Adams, V. Möbus, S. Costa, T. Bauknecht, B. Richter, M. Warm, W. Schröder, S. Olbricht, U. Nitz, C. Jackisch, G. Emons, U. Wagner, W. Kuhn and J. Pfisterer, J. Natl. Cancer Inst., 2003, 95, 1320–1329 CrossRef CAS PubMed.
- X. Yao, K. Panichpisal, N. Kurtzman and K. Nugent, Am. J. Med. Sci., 2007, 334, 115–124 CrossRef PubMed.
- R. Oun, Y. E. Moussa and N. J. Wheate, Dalton Trans., 2018, 47, 6645–6653 RSC.
- D. Hernández-Romero, S. Rosete-Luna, A. López-Monteon, A. Chávez-Piña, N. Pérez-Hernández, J. Marroquín-Flores, A. Cruz-Navarro, G. Pesado-Gómez, D. Morales-Morales and R. Colorado-Peralta, Coord. Chem. Rev., 2021, 439, 213930 CrossRef.
- A. K. Renfrew, E. S. O'Neill, T. W. Hambley and E. J. New, Coord. Chem. Rev., 2018, 375, 221–233 CrossRef CAS.
- A. H. Habib, C. L. Ondeck, P. Chaudhary, M. R. Bockstaller and M. E. McHenry, J. Appl. Phys., 2008, 103, 07A307 CrossRef.
- E. Fantechi, C. Innocenti, M. Zanardelli, M. Fittipaldi, E. Falvo, M. Carbo, V. Shullani, L. Di Cesare Mannelli, C. Ghelardini, A. M. Ferretti, A. Ponti, C. Sangregorio and P. Ceci, ACS Nano, 2014, 8, 4705–4719 CrossRef CAS PubMed.
- Z. Abdullaeva, E. Omurzak, C. Iwamoto, H. S. Ganapathy, S. Sulaimankulova, C. Liliang and T. Mashimo, Carbon, 2012, 50, 1776–1785 CrossRef CAS.
- E. Apohan, U. Yilmaz, O. Yilmaz, A. Serindag, H. Küçükbay, O. Yesilada and Y. Baran, J. Organomet. Chem., 2017, 828, 52–58 CrossRef CAS.
- Z. Zhao, J. Zhang, S. Zhi, W. Song and J. Zhao, J. Inorg. Biochem., 2019, 197, 110696 CrossRef CAS PubMed.
- S. A. Galal, K. H. Hegab, A. M. Hashem and N. S. Youssef, Eur. J. Med. Chem., 2010, 45, 5685–5691 CrossRef CAS PubMed.
- C. Icsel, V. T. Yilmaz, Ş. Aydinlik and M. Aygun, Eur. J. Med. Chem., 2020, 202, 112535 CrossRef CAS PubMed.
- J.-A. Zhao, H.-B. Yu, S.-C. Zhi, R.-N. Mao, J.-Y. Hu and X.-X. Wang, Chin. Chem. Lett., 2017, 28, 1539–1546 CrossRef CAS.
- A. Abdullah Al Awadh, Saudi Pharm. J., 2023, 31, 101698 CrossRef CAS PubMed.
- N. El-wakiel, M. El-keiy and M. Gaber, Spectrochim. Acta Mol. Biomol. Spectrosc., 2015, 147, 117–123 CrossRef CAS PubMed.
- I. D. Vlaicu, R. Olar, C. Maxim, M. C. Chifiriuc, C. Bleotu, N. Stănică, G. Vasile Scăeţeanu, C. Dulea, S. Avram and M. Badea, Appl. Organomet. Chem., 2019, 33, e4976 CrossRef.
- Ü. Yılmaz, E. Apohan, H. Kucukbay, Ö. Yılmaz, E. Tatlici and O. Yesilada, J. Heterocycl. Chem., 2022, 59(7), 1241–1246 CrossRef.
- G. Prakasha, H. D. Revanasiddappa, B. Jayalakshmi, B. T. Prabhakar, C. Shivamallu, P. M. Viswanath, R. R. Achar, E. Silina, V. Stupin, N. Manturova, A. A. Shati, M. Y. Alfaifi, S. E. I. Elbehairi, S. J. Armaković, S. Armaković and S. P. Kollur, Pharmaceuticals, 2023, 16, 125 CrossRef PubMed.
- M. Hou, H. C. Li, N. An, S. Y. Pang, W. G. Li and J. Tong, J. Mol. Struct., 2023, 1294, 136500 CrossRef CAS.
- K. R. Tay, C. R. Flavell, L. Cassini, M. Wimber and J. L. C. Lee, Neurosci. J., 2019, 39, 1109–1118 CrossRef CAS PubMed.
- R.-K. Pan, J.-L. Song, G.-B. Li, S.-Q. Lin, S.-G. Liu and G.-Z. Yang, Transit. Met. Chem., 2017, 42(3), 253–262 CrossRef CAS.
- Q.-W. Huang, S.-X. Wang, S.-G. Liu, W.-Y. Su, G.-B. Li and Y.-M. He, J. Struct. Chem., 2016, 57, 188–193 CrossRef CAS.
- J. Zhao, S. Zhi, H. Yu, J. Zhang, J. Zhang and J. Hu, J. Coord. Chem., 2017, 70, 3110–3131 CrossRef CAS.
- V. Kamat, A. Kotian, A. Nevrekar, K. Naik, D. Kokare and V. K. Revankar, Inorg. Chim. Acta, 2017, 466, 625–631 CrossRef CAS.
- I. I. Dyukova, L. G. Lavrenova, T. A. Kuz’menko, V. Y. Komarov, T. S. Sukhikh and E. V. Vorontsova, Inorg. Chim. Acta, 2019, 486, 406–411 CrossRef CAS.
- H.-L. Wu, J.-G. Liu, P. Liu, W.-B. Lv, B. Qi and X.-K. Ma, J. Coord. Chem., 2008, 61, 1027–1035 CrossRef CAS.
- H. López-Sandoval, M. E. Londoño-Lemos, R. Garza-Velasco, I. Poblano-Meléndez, P. Granada-Macías, I. Gracia-Mora and N. Barba-Behrens, J. Inorg. Biochem., 2008, 102, 1267–1276 CrossRef PubMed.
- O. Sánchez-Guadarrama, H. López-Sandoval, F. Sánchez-Bartéz, I. Gracia-Mora, H. Höpfl and N. Barba-Behrens, J. Inorg. Biochem., 2009, 103, 1204–1213 CrossRef PubMed.
- G. B. Li, S. X. Wang, W. Y. Su, R. K. Pan, K. D. Liu and S. G. Liu, Russ. J. Coord. Chem., 2014, 40, 764–767 CrossRef CAS.
- Y. Nakao, M. Onoda, T. Sakurai, A. Nakahara, I. Kinoshita and S. Ooi, Inorg. Chim. Acta, 1988, 151, 55–59 CrossRef CAS.
- S. Zhi, Y. Li, J. Qiang, J. Hu, W. Song and J. Zhao, J. Inorg. Biochem., 2019, 201, 110816 CrossRef CAS PubMed.
- L. K. Thompson, B. S. Ramaswamy and E. A. Seymour, Can. J. Chem., 1977, 55, 878–888 CrossRef CAS.
- R. M. Soo and M. Mu-shin, Bull. Korean Chem. Soc., 1997, 18, 406–409 Search PubMed.
- J. G. Martins, P. Gameiro, M. T. Barros and H. M. V. M. Soares, J. Chem. Eng. Data, 2010, 55, 3410–3417 CrossRef CAS.
- Y. T. Lee, Y. J. Tan and C. E. Oon, Acta Pharm. Sin. B, 2023, 13, 478–497 CrossRef CAS PubMed.
- M. Hasanpourghadi, C. Karthikeyan, A. K. Pandurangan, C. Y. Looi, P. Trivedi, K. Kobayashi, K. Tanaka, W. F. Wong and M. R. Mustafa, J. Exp. Clin. Cancer Res., 2016, 35, 58 CrossRef PubMed.
- Y. J. Tan, Y. T. Lee, K. Y. Yeong, S. H. Petersen, K. Kono, S. C. Tan and C. E. Oon, Future Med. Chem., 2018, 10, 2039–2057 CrossRef CAS PubMed.
- M.-S. Won, N. Im, S. Park, S. K. Boovanahalli, Y. Jin, X. Jin, K.-S. Chung, M. Kang, K. Lee, S.-K. Park, H. M. Kim, B. M. Kwon, J. J. Lee and K. Lee, Biochem. Biophys. Res. Commun., 2009, 385, 16–21 CrossRef CAS PubMed.
- J. G. Smith, X. Liu, R. M. Kaufhold and M. J. Caulfield, Clin. Diagnostic Lab. Immunol., 2001, 8(5), 871–879 CrossRef CAS PubMed.
- A. M. Pizarro and P. J. Sadler, Biochimie, 2009, 91, 1198–1211 CrossRef CAS PubMed.
- A. Favier, M. Blackledge, J.-P. Simorre, S. Crouzy, V. Dabouis, A. Gueiffier, D. Marion and J.-C. Debouzy, Biochemistry, 2001, 40, 8717–8726 CrossRef CAS PubMed.
- V. C. da Silveira, H. Benezra, J. S. Luz, R. C. Georg, C. C. Oliveira and A. M. da C. Ferreira, J. Inorg. Biochem., 2011, 105, 1692–1703 CrossRef PubMed.
- R. Kalarani, M. Sankarganesh, G. G. V. Kumar and M. Kalanithi, J. Mol. Struct., 2020, 1206, 127725 CrossRef CAS.
- S. Dasari and P. B. Tchounwou, Eur. J. Pharmacol., 2014, 740, 364–378 CrossRef CAS PubMed.
- S. Kumar and P. B. Tchounwou, Oncotarget, 2015, 6, 40734–40746 CrossRef PubMed.
- J. L. Fogel, T. Z. T. Thein and F. V Mariani, J. Visualized Exp., 2012, e4254 Search PubMed.
- L. C. Santos, R. Vogel, J. E. Chipuk, M. R. Birtwistle, G. Stolovitzky and P. Meyer, Nat. Commun., 2019, 10, 1313 CrossRef PubMed.
- M. Balamurugan, R. Mayilmurugan, E. Suresh and M. Palaniandavar, Dalton Trans., 2011, 40, 9413–9424 RSC.
- H. Perumalsamy, K. Sankarapandian, K. Veerappan, S. Natarajan, N. Kandaswamy, L. Thangavelu and S. R. Balusamy, Phytomedicine, 2018, 46, 119–130 CrossRef CAS PubMed.
- S. R. Balusamy, H. Perumalsamy, K. Veerappan, M. A. Huq, S. Rajeshkumar, T. Lakshmi and Y. J. Kim, BioMed Res. Int., 2020, 2020, 6040727 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06377e |
| This journal is © The Royal Society of Chemistry 2025 |