Open Access Article
Open Access ArticleGenetically engineered gas vesicle proteins with proliferative potential for synergistic targeted tumor therapy†
Li Linab,
Yan Dua,
Yaotai Wanga,
Yong Luoac,
Fujie Jianga,
Haiyan Yangad,
Li Rena and
Jianzhong Zou *a
*a
aState Key Laboratory of Ultrasound in Medicine and Engineering, College of Biomedical Engineering, Chongqing Medical University, Chongqing, 400016, China. E-mail: zoujzh@cqmu.edu.cn; Tel: +86-13708302390
bDepartment of Ultrasound Medicine, Chongqing Shapingba Hospital, School of Medicine, Chongqing University, Chongqing, 400033, China
cDepartment of Ultrasound, The People's Hospital of Chongqing Liang Jiang New Area, Chongqing 400010, China
dDepartment of Ultrasound, Chongqing General Hospital, Chongqing University, 401147, China
First published on 2nd January 2025
Abstract
Nanomedicine enables precision-targeted therapies through a non-invasive approach, and nanoparticles may be biologically affected during their colonization in vivo. Ensuring the efficient expression of their ex vivo performance in vivo, while ensuring biosafety, is of great significance. Previous studies have employed genetically engineered E. coli following in vivo entry as a genetically engineered targeting synergist, to enhance the effect of focused ultrasound ablation by exploiting its targeted colonization of tumor tissue. However, the proliferation process of the actual potentiating nanomaterials, i.e., the aerosol proteins produced by genetically engineered E. coli, in vivo has not been precisely observed. The authors of this paper demonstrate this spatiotemporal change in the expression of gas vesicle proteins while genetically engineered E. coli reproduces following tumor colonization. Based on their targeting and proliferative properties, the authors chose to intervene in the treatment at the maximal gas vesicle protein count to enhance the monitoring and utilization of the potentiator. By examining the therapeutic potential of the novel combination of genetic engineering and focused ultrasound, we present a robust strategy that improves the efficiency of non-invasive treatments.
1. Introduction
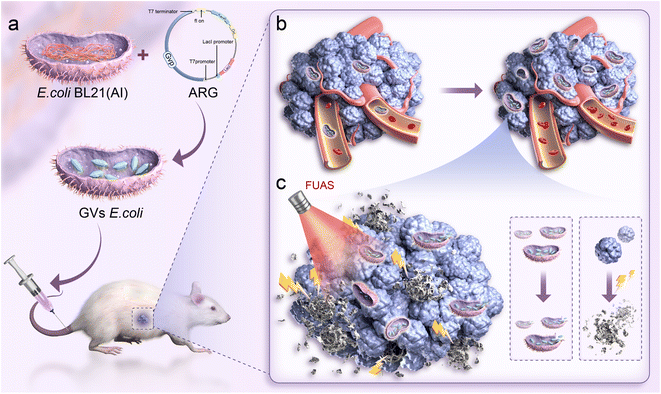
Focused ultrasound ablation (FUAS), a non-invasive therapy widely used in clinical settings, allows real-time visualization and monitoring of the targeted area, primarily employing thermal and mechanical mechanisms1–3 and tailoring treatment plans to match tumor morphology. However, the penetration limitations of ultrasound waves in deep or complexly positioned tissues can affect the efficiency and precision of the treatment.4 Research on synergism has been conducted to unblock the external spatial control mechanisms and to enhance therapeutic effects.5,6Phase-transition nanoparticles have emerged as a popular research subject for synergistic agents due to their ability to enhance ablation under ultrasound and serve as a visual monitoring medium,7 although they lack precise targeting capabilities.8,9 Researchers have developed the use of probiotics or engineered bacteria as carriers to piggyback drugs and release them via nano microballoon phase change.10,11 For instance, utilizing electrostatic adsorption to attach Bifidobacterium to phase-transition materials targets tumors and enhances nanosphere-enhanced ablation.12 This combination of therapeutic techniques provides not only a highly efficient piggyback rate and tumor-targeting specificity but also a clinically controllable safety profile.
Similarly, E. coli, when appropriately engineered, reduces bacterial toxicity to the host, mitigates severe immune responses, and enhances biocompatibility, thus serving as a reliable gene engineering carrier.13,14 E. coli can colonize tumor tissues, avoiding damage to healthy tissues and maintaining stability for soluble expression, making it a widely used genetic engineering vector.15–17
Mikhail G. Shapiro and colleagues reported genetically engineered acoustic reporter genes (ARGs) that specifically produce a unique type of acoustic nanoscale gas vesicle (GV), which was transferred into E. coli to generate Genetically Engineered Bacteria (GEB) harboring the target plasmid as a novel biomolecular material.18 GV are biconical nanostructures with an outer protein layer and an inner gas-filled cavity, growing by excluding water internally and absorbing gas from the extracellular cytoplasm, eventually forming a filled vesicle that maintains its structure.19,20 The discovery of these unique protein shells, which exhibit high external hydrophilicity and molecular interactions such as hydrogen bonding, ensures the structural stability of the nanoparticles in cellular fluids.21,22 E. coli can efficiently express plasmids, with GEBs visibly filled with expressed GV. Compared to biological carriers that transport exogenous phase-transition nanoparticles, this biomolecular material carrying a significant amount of GV provides the cavitation nuclei needed for FUAS, making it a novel biological targeting synergistic agent for cancer therapy using FUAS.23–25
Previous studies have shown that genetically engineered targeting synergist (GVs-E) injected into mice with tumor models after in vitro purification reveal bacterial metabolism in vital organs and targeted enrichment and proliferation at tumor sites. When the synergistic agents are in the tumor, FUAS ablation is performed, ultimately confirming the synergistic ability of the genetically engineered targeting synergist.26,27 However, beyond plasmid gene mutations preventing plasmid replication,28,29 literature reports suggest that excessive expression of gas vesicles may affect plasmid replication and distribution, leading to overload, both of which can cause rapid loss from the bacterial population, thus impacting the synergistic effect.30–32 As shown in Fig. 1, this experiment aims to find more direct evidence proving the enrichment and proliferation process of genetically engineered targeting synergists at tumor sites, replicating and carrying the target gene ARG, as the main target cavitation nucleus GV is fully expressed in the body, thus synergistically combining FUAS and genetically engineered targeting agents for precise tumor treatment, enhancing treatment efficiency, stability, and controllability.
2. Materials and methods
2.1. Materials and equipment
The plasmid pET28a_T7-ARG1 was requested access from the State Key Laboratory of Ultrasound in Medicine and Engineering; E. coli BL21 (AI) was purchased from Vidi Biotechnology Co. Ltd (Shanghai, China); PBS (0.01 M, pH 7.2–7.4) was purchased from Biochannel Co. Ltd (Nanjing, China); Luria–Bertani (LB), and L-arabinose were purchased from Solario Co. Ltd (Beijing, China). The equipment included the following: focused ultrasound tumor therapeutic system (model-JC200, Chongqing Haifu Medical Technology Co., Ltd, Chongqing, China, hereinafter referred to as “Haifu system”), optical microscope (Bx51tf, Olympus Corporation, Tokyo, Japan), transmission electron microscope (TEM, H-7600, Hitachi, Tokyo, Japan), microplate reader (Infinite M200 pro, Tecan Austria GmbH, Grodig, Austria), UV-vis spectrophotometer (NanoDrop-2000, Thermo Scientific, Wilmington, DE, USA).2.2. Cell culture and animal model
The mouse mammary cancer 4T1 cells were applied to the Cell Bank of the Chinese Academy of Sciences to obtain. Cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum and 1% streptomycin/penicillin at 37 °C in a 5% CO2 atmosphere. Tumor cells (1 × 106) were suspended in 0.1 mL PBS and subsequently injected subcutaneously into the left and right flanks of the mice. All animal experiments were conducted according to the guidelines for the Care and Use of Laboratory Animals of Chongqing Medical University and under the supervision of the Animal Ethics Committee of Chongqing Medical University.2.3. Preparation of GVs-E
The plasmid pET28a_T7-ARG1 was transformed into chemically competent Escherichia coli BL21 (AI) cells and incubated at 37 °C for 24 hours. Single colonies were then grown in LB medium at 37 °C with shaking, transferred to antibiotic-containing LB medium, and grown until OD600 reached 0.5. Induction was carried out with L-arabinose and IPTG, followed by shaking at 30 °C for 22 hours. Antibiotics were removed by centrifugation and diluted for further experiments.2.4. Characterization of GVs-E
We observed the morphological characteristics of successfully transformed GVs-E colonies on the agar plates. The induced bacterial cultures were stained with Gram stain and examined under a light microscope. After centrifugation of the bacterial fluid, the bacteria that successfully expressed ARG were observed in the upper liquid layer of the bacterial fluid. Next, the characteristic morphology and structure of GVs-E itself and GVs-E-expressed aerosol proteins were observed by electron transmission electron microscopy. Plasmids from GVs-E were extracted using a small plasmid extraction kit, and their concentrations were measured with a spectrophotometer.2.5. In vivo biodistribution of the genetically engineered targeting synergist
25 4T1 tumor-bearing mice were divided into five groups (n = 5) and were injected intravenously with 0.1 mL of GVs-E suspension (1 × 108 CFU mL−1) once daily for three consecutive days. At various time points after the completion of the injections (days 0, 1, 3, 7, and 14), five mice from each group were sacrificed to harvest major organs (heart, liver, spleen, lungs, and kidneys) and tumors. Each tissue was homogenized in PBS and serially diluted, then from each dilution, 0.1 mL was inoculated onto LB agar plates, incubated at 37 °C for 8–24 h, and the number of colonies was counted.2.6. Proliferation characteristics of the genetically engineered targeting synergist
The homogenized tumor tissues were plated on LB medium and incubated at 37 °C for 22 hours to induce expression. Finally, the expressed GV-containing Escherichia coli were harvested by centrifugation at 350g for 4 hours at 4 °C. Plasmids from the homogenized plates containing GVs-E were extracted, and their concentrations were measured with a spectrophotometer for quantitative analysis, all plasmids were verified by sequencing (Beijing Luhe Huada Gene Technology Co., Ltd, Wuhan, China). Twenty tumor-bearing mice with injected GVs-E (1 × 108 CFU mL−1) every day for three consecutive days were divided into four groups (n = 5) based on time points (days 1, 3, 7, and 14), euthanized, and their tumors were collected. The tumors were fixed overnight at 4 °C with 2% paraformaldehyde, washed with PBS, fixed for 2 hours at 4 °C with 1% osmium tetroxide, stained with uranyl acetate at 4 °C, dehydrated through a graded series of acetone (70% → 80% → 90% → 100%), embedded, and ultra-thin sections were observed under a transmission electron microscope to study the structure, morphology, and distribution of gas vesicle proteins produced by GVs-E in the tumors at different time points.2.7. Genetically engineered targeting for synergistic FUAS
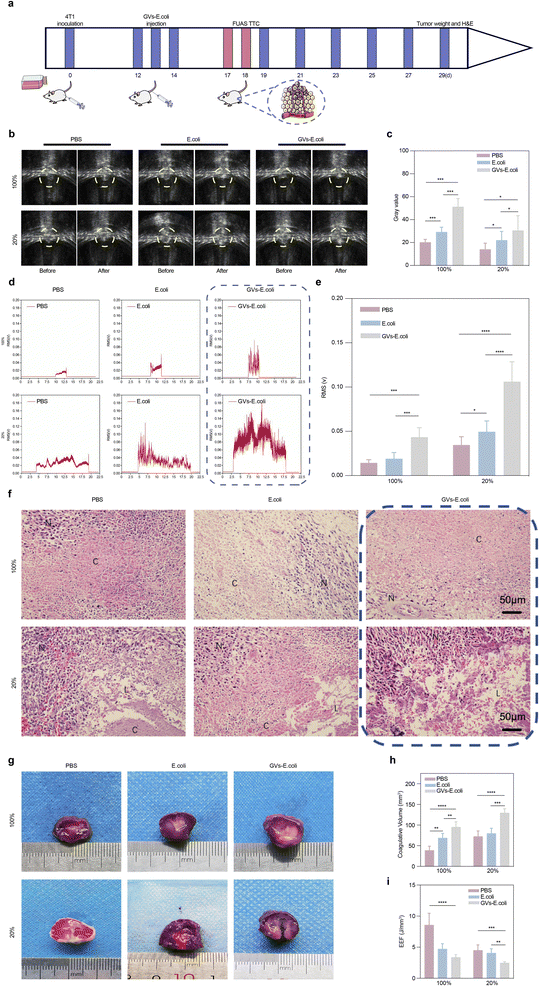
Ninety tumor-bearing mice were randomly divided into three groups (n = 30) and injected intravenously with PBS, E. coli, or GVs-E for three consecutive days. After 72 hours, each group was further divided into two subgroups (n = 15) to receive different duty cycles (20%, 100%) of focused ultrasound tumor treatment using the Haifu system. The system was set with a pulse ultrasound frequency of 1.0 MHz, PRF 200 Hz, ultrasound power of 150 W, and an effective exposure time of 3 seconds, focused to a depth of approximately 6 mm within the mouse tumor tissue. The inertial cavitation noise signals during exposure were detected using a Passive Cavitation Detector (PCD). Before and after ablation, the ablation area's grayscale was automatically measured by the focused ultrasound tumor treatment system.Twenty-four hours later, ten mice from each of the six groups (a total of 60 mice) were euthanized to collect tumor tissues. Five tumor tissues from each group were immersed in a 2% 2,3,5-triphenyl tetrazolium chloride (TTC) solution at 37 °C for 30 minutes to obtain the coagulative necrosis volume (V, m3) and the Energy Efficiency Factor (EEF, J mm−3), which indicates the ultrasound energy required per unit volume of tumor ablation. These were calculated using the formula: V = (π/6) × length × width × depth, EEF = ηPt/V; where η is the transducer focus factor of 0.7; P is the ultrasound irradiation power (W); t is the effective ultrasound exposure time (s). Lower EEF values indicate lower energy required per unit volume for tumor ablation, hence higher ablation efficiency.
The remaining 30 tumor tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned for Hematoxylin and Eosin (H&E) staining to evaluate tumor ablation caused by FUAS for each group.
We divided 40 mice with 4T1 tumors into 4 groups (n = 10) and treated them with PBS, GVS-E, and FUAS at two duty cycles (20%, 100%). Following single exposure, we euthanized 5 mice from each group, homogenized their tumor tissues, and incubated them for 24 hours before counting the viable CFUs. The remaining mice received a second exposure immediately after the first, with treatment foci as far apart as possible within the tumor. The mice were euthanized 24 hours post-treatment, and the tumor tissues were stained with TTC.
2.8. Biosafety testing
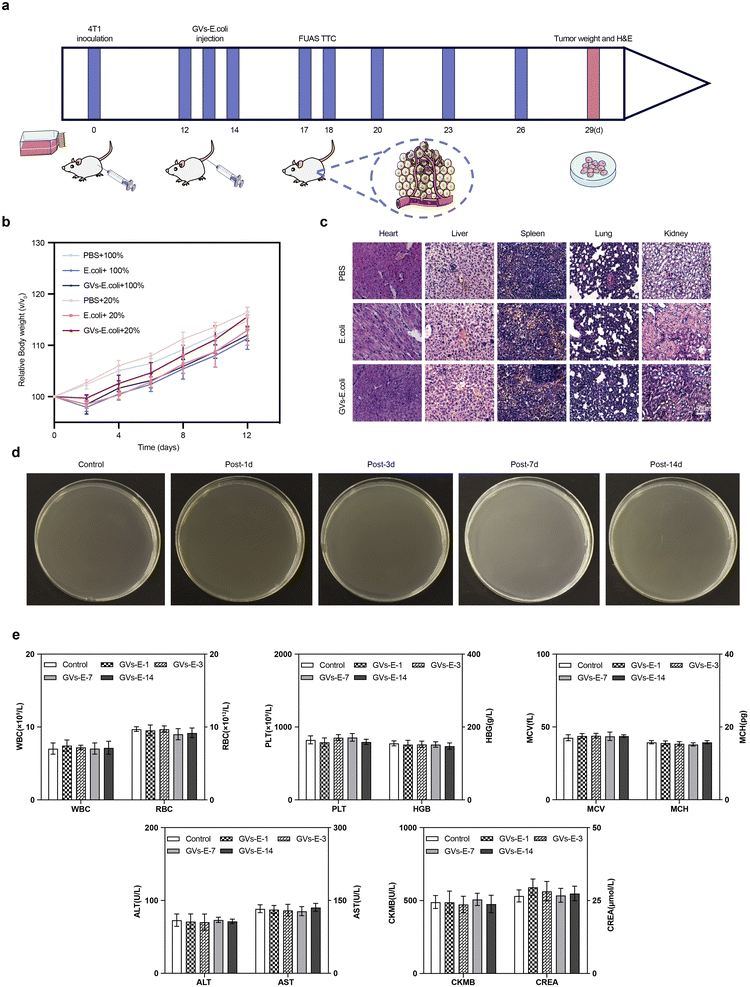
To further verify the safety of the treatment regimen, the remaining 30 tumor-bearing mice (n = 5) had their body weight changes monitored every other day from day 0 to day 12 post-irradiation to assess metabolic status. On day 14, the mice were euthanized, and major organs were collected and stained with H&E to demonstrate the safety of tumor ablation mediated by genetically engineered targeting agents in conjunction with FUAS.25 mice were randomly divided into five groups, with four groups receiving an intravenous injection of 0.1 mL of GVs-E and one group receiving 0.1 mL of PBS as a control. Blood samples were taken from the GVs-E-injected mice on days 1, 3, 7, and 14 post-injection, and from the PBS-injected group on day 1. Collection was done via orbital puncture under anesthesia, using a 0.05 mL pipette, and samples were promptly diluted with 0.1 mL of PBS. The diluted blood (0.15 mL total) was spread onto LB agar plates and incubated at 37 °C for 24 hours to monitor microbial growth.
To assess the in vivo biocompatibility of GVs-E, four groups of mice (n = 5 per group) were injected with 0.1 mL of a GVs-E suspension at a concentration of 1 × 108 CFU mL−1. In contrast, a control group (n = 5) received 0.1 mL of PBS. Orbital blood samples were collected at 1, 3, 7, and 14 days post-injection from both GVs-E and PBS groups to evaluate complete blood count (WBC, RBC, PLT, HGB, MCH, and MCV) and serum biochemical (ALT, AST, CREA, CK-MB).
2.9. Antitumor efficacy evaluation
Besides monitoring the body weight, tumor volumes were recorded every three days from day 0 to day 12 post-ablation in the 30 tumor-bearing mice used in the biosafety testing, with five mice per group. After euthanasia, tumors were excised, weighed, and recorded.2.10. Statistical analysis
Analysis was performed using GraphPad Prism version 9.2.0. Quantitative data are expressed as mean ± standard deviation. The Student's t-test, one-way ANOVA, and two-way ANOVA were used for data analysis. P-Values < 0.05 were considered statistically significant, with *P < 0.05, **P < 0.01, ***P < 0.001.3. Results
3.1. Characterization of GVs-E
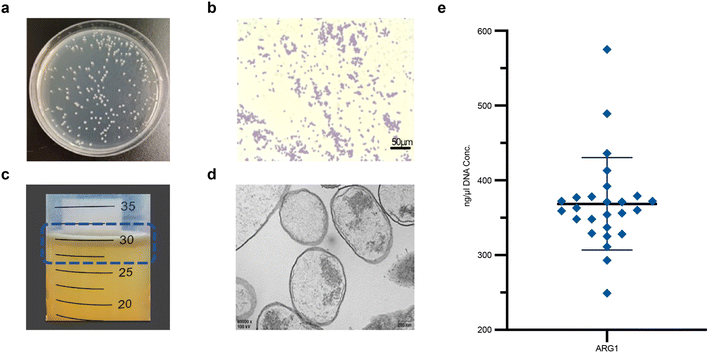
After 24 hours of incubation, colonies formed were 2–3 mm in diameter, smooth, moist, raised with neat edges, and exhibited a milky white turbidity (Fig. 2a). Gram staining and light microscopy of GVs-E showed pale pink, rod-shaped bacteria, confirming the Gram-negative characteristics of Escherichia coli (Fig. 2b). Under induced conditions, ARG1 was high-level expressed, producing a large quantity of gas-containing GV, which appeared as hollow structures with biconical ends, measuring approximately 27 nm to 44 nm in diameter and 100 nm to 200 nm in length. Due to buoyancy, the GVs-E bacterial fluid floated on the top layer after centrifugation, visible to the naked eye as milky white (Fig. 2c). TEM revealed successful construction of GV within GVs-E, with a hollow spindle-like structure inside the GV (Fig. 2d), providing a basis for synergistic targeted tumor therapy. Plasmid concentrations ranged from 240–580 ng μl−1, with the majority falling between 290–400 ng μl−1 (Fig. 2e).3.2. In vivo biodistribution of the genetically engineered targeting synergist
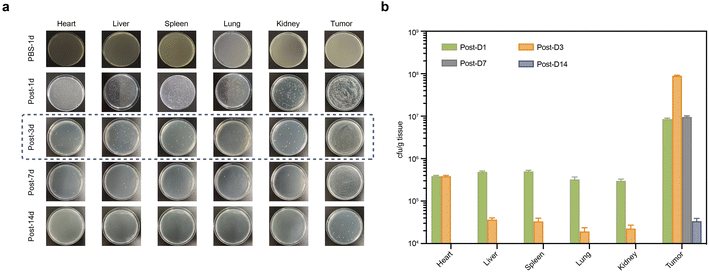
According to the colony counts from homogenized samples, GVs-E E. coli was present in the heart, liver, spleen, kidneys, lungs, and tumors as early as the first day after injection, but predominantly colonized tumor areas (Fig. 3a). While bacteria in major organs were gradually cleared, the density of GVs-E in tumor areas increased, peaking on the third day and then gradually decreasing. By day 14, only a minimal number of bacterial colonies remained in tumor homogenates (Fig. 3b). The proliferation of GVs-E in vivo demonstrated that as a genetically engineered targeting synergist, it differs from conventional synergists by achieving a stable quantity through proliferation processes, specifically retaining in tumor targets. The peak bacterial levels in tumors corresponded with significant clearance levels in other major organs, suggesting immune activity, which confirms the specificity and unique biosafety profile of GVs-E as a targeting synergist.3.3. Proliferation characteristics of the genetically engineered targeting synergist
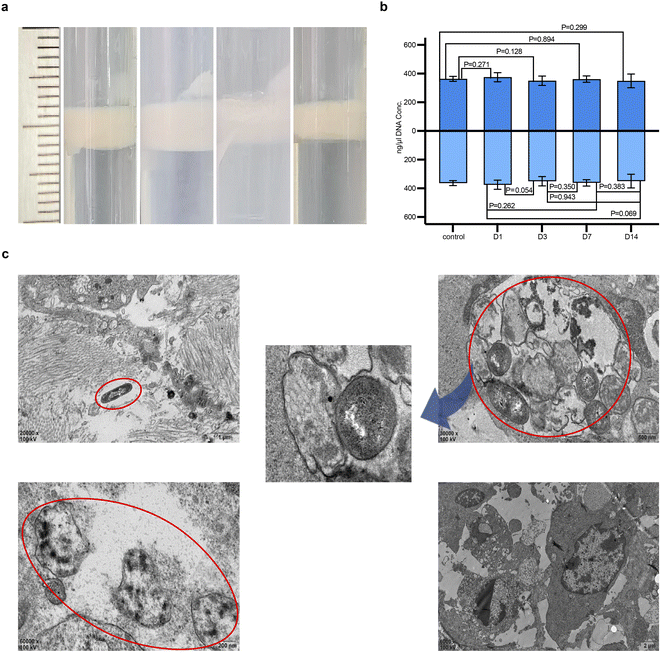
At various time points, induced bacterial fluids from tumor homogenates were observed floating on the top layer, indicating sufficient bacterial quantity, as shown in Fig. 4a after PBS washing and purification. This confirmed that during in vivo tissue proliferation, ARG was also replicated within the transformed bacterial cytoplasm, enabling the progeny bacteria to stably express GV. Plasmid concentrations at all four time points were above 100 ng μl−1, indicating effective expression of the gene product and GV in the tumor tissues by the constructed plasmid. Sequencing confirmed no mutation sites in the ARG1 sequence among the samples. Comparison of plasmid concentrations across groups showed no significant differences, indicating synchronous inheritance with bacterial proliferation and no instability in progeny plasmid transfer affecting concentration during the colonization period, although a gradual deviation in concentration was observed on day 14 as shown in Fig. 4b, possibly due to plasmid loss during replication. Confirmation that ARG could be stably inherited in sufficient quantities in vivo was further supported by TEM observations of its expression within tumor tissues (Fig. 4c). On day 1 post-injection, a few bacteria were distributed interstitially within tumor tissues, with the visible formation of several translucent conical GV within the cytoplasm, while some bacteria appeared smaller with partial GV expression showing the specific hollow characteristic. By day 3, more bacteria were observed under the microscope with densely packed hollow biconical spindle-shaped GV arranged within the bacterial cytoplasm, with no significant differences compared to ex vivo-induced forms. Some GVs-E were also observed being phagocytosed. By day 7, only a few bacteria were visible, with many showing incomplete or absent cell walls, and their GV could no longer be observed. By day 14, the distribution of bacteria was no longer accessible in the samples.After single exposure (Fig. S1†), some GVS-E remained viable in the tumor tissue. The percentage of viable GVS-E was between 1 × 106 and 1 × 107 CFU mL−1. After double exposure (Fig. S2†), the ablation volume in the PBS group was smaller than in the GVS-E group under the same duty cycle (Fig. S2†). This observation preliminarily assesses the potential for synergistic ablation with multiple exposures of GVS-E, confirming its significant role as a synergistic agent in conjunction with FUAS.
However, factors such as the limited ablation space within animal models, the original parameters' unsuitability for multiple exposures, and overlapping ablation volumes in double exposure, hinder the accurate assessment of ablation efficiency and synergistic effects.
3.4. Genetically engineered targeting for FUAS treatment
Under ultrasound monitoring, the GVs-E. coli group showed significant grayscale changes compared to the first two groups. With a duty cycle of 20% relative to 100%, the grayscale changes before and after ablation were not as significant as those with the continuous wave but still statistically significant (Fig. 5b and c). The inertial cavitation during the ablation process was indicated by broadband noise in the PCD signals, with significant differences exhibited by the GVs-E. coli group (Fig. 5d and e). It indicates that regardless of the mechanical effect-dominated low space-occupancy group or the thermal effect-dominated continuous high-intensity focused ultrasound, comparing the PBS group and the E. coli group, the GVs-E. coli group promoted the cavitation activity in the target area by propagating GVs-E. coli in vivo to replenish the cavitation nuclei, which markedly enhanced cavitation capacity and led to a larger ablation area.In Fig. 5f, H&E-stained paraffin sections showed cellular shrinkage, nuclear fragmentation, or pyknosis in the 100% duty cycle group and distinctive tissue fissures or cavities in the 20% duty cycle group; Fig. 5g and h show the gray-white area of coagulative necrosis in the 100% duty cycle group and areas of coagulative and some liquefactive necrosis appearing as cavities in the 20% duty cycle group, all visible under a microscope and to the naked eye, corresponding to the amplitude of inertial cavitation signals collected by PCD. With the same radiation total dose and exposure time, the larger the volume of necrosis, the lower the unit volume of energy required for ablation (Fig. 5i), proving that a large number of GV as cavitation nuclei participating in the ablation process significantly optimizes the ablation effect.
3.5. Biosafety testing
To further assess the biosafety of the genetically engineered targeting synergist treatment, weight changes were monitored every two days from day 0 to day 12 post-intravenous injection (Fig. 6a). Apart from the PBS group, the body weight of the tumor-bearing mice in the experimental groups fluctuated slightly within the first two days post-injection but returned to a growth trend similar to the PBS group after two days (Fig. 6b), indicating that the genetically engineered targeting synergist has a limited and reversible impact on basal metabolism. Fig. 6c shows post-ablation tissue sections observed under a microscope, where the genetically engineered targeting synergist and the other two groups showed plump cellular morphology with homogeneous interstitial spaces, without evident signs of cellular damage, proving its stable safety in the metabolism and major organs of tumor-bearing mice. Blood culture experiments (Fig. 6d) showed that bacteria were cleared from the blood circulation. No colonies were observed on the agar plates one day after injection, indicating that GVs-E was nearly completely cleared from the blood circulation within 24 hours. No bacterial growth was detected in the blood over the 14 day observation period. The analysis of blood count and serum biochemical parameters (Fig. 6e) revealed no significant differences in whole blood cell counts, liver function, kidney function, and myocardial enzymes between the GVs-E and control groups.3.6. Anti-tumor efficacy evaluation
To evaluate the anti-tumor efficacy of the genetically engineered targeting synergist treatment, tumor tissue growth inhibition was used as an evaluation metric. The tumor-bearing mice in each group were injected intravenously with the genetically engineered targeting synergist, PBS, and a carrier fluid comparator. In the group injected with carrier fluid compared to the PBS group, significant inhibition was only observed in the first three days post-injection, yet the rate of tumor growth in the later period was not significant in comparison with the PBS group. However, the genetically engineered targeting synergist, under the same ablation conditions, showed more stable tumor growth, demonstrating significant anti-tumor efficacy (Fig. 7a). On day 12 post-treatment, tumors were harvested from each group, and tumor weights were measured; under the same treatment conditions, the GVs-E group had the lowest mass, consistent with the volume outcomes (Fig. 7b).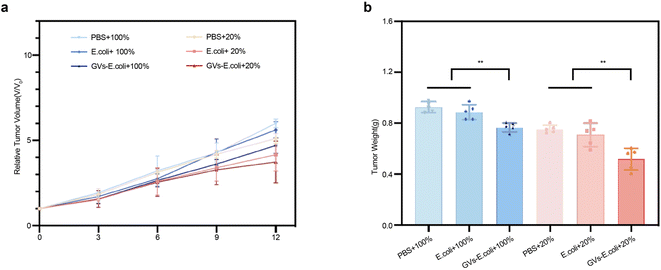 | ||
| Fig. 7 Anti-tumor efficacy evaluation (a) tracking tumor volume changes during the treatment cycle. (b) Termination of treatment, ex vivo measurement of tumor weight per group. | ||
4. Conclusion
The genetically engineered targeting synergist is a multifunctional agent that integrates bacterial targeting, rapid proliferation, genetic synthesis, bubble imaging, and mediated focused ablation. This newly genetically engineered product, as a safe targeting synergist, is selectively taken up and accumulated by tumor tissues. This work demonstrates that, in contrast to other nanoparticle synergists, it is capable of targeting and colonising tumour sites, and of proliferating extensively when introduced into mammals. During proliferation, it retains a complete gene-encoding core and, following autonomous replication of GVs-E, stably expresses an adequate volume of fixed-size GV. These GVs can be effectively used to mediate focused ultrasound, significantly enhancing the cytotoxic effect and achieving optimized spatially restricted treatment. The genetically engineered targeting synergist offers broad translational prospects for achieving millimeter-level precision in clinical diagnosis and treatment.The current study was constrained from conducting in-depth microscopic observations and documentation of the ablation process due to the limitations imposed by the experimental conditions. This restricted the scope for elucidating the underlying mechanical mechanisms and for refining the therapeutic parameters. Consequently, the genetically engineered targeting synergist did not achieve its maximum therapeutic potential. Further research might focus on the cellular level, such as the mechanical and thermal ablation mechanisms produced by ultrasound, which could still be further optimized or equipped as necessary. Given the widespread application of ultrasound therapy, the future clinical application of the genetically engineered targeting synergist also becomes feasible.
Ethical statement
All animal experiments were conducted according to the guidelines for the Care and Use of Laboratory Animals of Chongqing Medical University and under the supervision of the Animal Ethics Committee of Chongqing Medical University (IACUC-CQMU-2023-0070).Data availability
The main data supporting the findings of this study are available within the paper. Plasmid sequences will be available from Addgene. Further information (like comparative sequence analysis) and requests for resources and reagents should be directed to the corresponding author.Author contributions
Li Lin, Yan Du and Fujie Jiang carried out the studies, participated in collecting data, and drafted the manuscript. Jianzhong Zou, Yaotai Wang and Haiyan Yang performed the statistical analysis and participated in its design. Yong Luo and Li Ren participated in acquisition, analysis, or interpretation of data and draft the manuscript. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Our study was sponsored by two grants. The first is from Chongqing Natural Science Foundation (Postdoctoral Science Foundation), for a project entitled Study on the Effect and Mechanism of Multi-functional Targeted Nanoparticles Combined with Genetically Engineered Bacteria in Collaborative High-intensity Focused Ultrasound for Tumor Ablation (Grant No. CSTB2023NSCQ-BHXO132). The second grant was provided by Chongqing Women and Children General Project Fund (Grant No. 2020FY108).References
- Y. Meng, K. Hynynen and N. Lipsman, Nat. Rev. Neurol., 2021, 17, 7–22 CrossRef.
- P. M. Dillon, B. J. Horton, T. Bullock, C. Brenin and D. R. Brenin, J. Clin. Oncol., 2018, 36, TPS19 Search PubMed.
- J. R. Sukovich, C. A. Cain, A. S. Pandey, N. Chaudhary, S. Camelo-Piragua, S. P. Allen, T. L. Hall, J. Snell, Z. Xu, J. M. Cannata, D. Teofilovic, J. A. Bertolina, N. Kassell and Z. Xu, J. Neurosurg., 2018, 131, 1331–1338 Search PubMed.
- H. Y. Ge, L. Y. Miao, L. L. Xiong, F. Yan, C. S. Zheng, J. R. Wang, J. W. Jia, L. G. Cui and W. Chen, Ultrasound Med. Biol., 2014, 40, 947–955 CrossRef PubMed.
- R. Tang, H. He, X. Lin, N. Wu, L. Wan, Q. Chen, Y. Hu, C. Cheng, Y. Cao, X. Guo, Y. Zhou, X. Xiong, M. Zheng, Q. Wang, F. Li, Y. Zhou and P. Li, Journal for ImmunoTherapy of Cancer, 2023, 11, 006226 Search PubMed.
- S. Suehiro, T. Ohnishi, D. Yamashita, S. Kohno, A. Inoue, M. Nishikawa, S. Ohue, J. Tanaka and T. Kunieda, J. Neurosurg., 2018, 129, 1416–1428 CAS.
- R. Wang, Y. Yao, Y. Gao, M. Liu, Q. Yu, X. Song, X. Han, D. Niu and L. Jiang, Int. J. Nanomed., 2023, 18, 2539–2552 CrossRef CAS PubMed.
- J. Yan, W. Fei, Q. Song, Y. Zhu, N. Bu, L. Wang, M. Zhao and X. Zheng, Drug Delivery, 2022, 29, 2296–2319 CrossRef CAS PubMed.
- V. O. Shipunova, E. N. Komedchikova, P. A. Kotelnikova, I. V. Zelepukin, A. A. Schulga, G. M. Proshkina, E. I. Shramova, H. L. Kutscher, G. B. Telegin, A. V. Kabashin, P. N. Prasad and S. M. Deyev, ACS Nano, 2020, 14, 12781–12795 CrossRef CAS.
- Q. Hu, M. Wu, C. Fang, C. Cheng, M. Zhao, W. Fang, P. K. Chu, Y. Ping and G. Tang, Nano Lett., 2015, 15, 2732–2739 CrossRef CAS.
- S. Chowdhury, S. Castro, C. Coker, T. E. Hinchliffe, N. Arpaia and T. Danino, Nat. Med., 2019, 25, 1057–1063 CrossRef CAS PubMed.
- X. Gao, W. Zou, B. Jiang, D. Xu, Y. Luo, J. Xiong, S. Yan, Y. Wang, Y. Tang, C. Chen, H. Li, H. Qiao, Q. Wang and J. Zou, Sci. Rep., 2019, 9, 6423 CrossRef PubMed.
- T. Danino, A. Prindle, G. A. Kwong, M. Skalak, H. Li, K. Allen, J. Hasty and S. N. Bhatia, Sci. Transl. Med., 2015, 7, 289ra284 Search PubMed.
- D. T. Riglar and P. A. Silver, Nat. Rev. Microbiol., 2018, 16, 214–225 CrossRef CAS PubMed.
- C. K. Baban, M. Cronin, D. O'Hanlon, G. C. O'Sullivan and M. Tangney, Bioengineered Bugs, 2010, 1, 385–394 CrossRef.
- G. L. Rosano and E. A. Ceccarelli, Front. Microbiol., 2014, 5, 172 Search PubMed.
- Y. Lu, N. Mei, Y. Ying, D. Wang, X. Li, Y. Zhao, Y. Zhu, S. Shen and B. Yin, Int. J. Nanomed., 2024, 19, 759–785 CrossRef CAS.
- R. W. Bourdeau, A. Lee-Gosselin, A. Lakshmanan, A. Farhadi, S. R. Kumar, S. P. Nety and M. G. Shapiro, Nature, 2018, 553, 86–90 CrossRef CAS PubMed.
- G. J. Lu, A. Farhadi, J. O. Szablowski, A. Lee-Gosselin, S. R. Barnes, A. Lakshmanan, R. W. Bourdeau and M. G. Shapiro, Nat. Mater., 2018, 17, 456–463 CrossRef CAS.
- S. T. Huber, D. Terwiel, W. H. Evers, D. Maresca and A. J. Jakobi, Cell, 2023, 186, 975–986.e913 CrossRef CAS PubMed.
- P. Dutka, D. Malounda, L. A. Metskas, S. Chen, R. C. Hurt, G. J. Lu, G. J. Jensen and M. G. Shapiro, Protein Sci., 2021, 30, 1081–1086 CrossRef CAS PubMed.
- F. Pfeifer, Life, 2022, 12, 1455 CrossRef CAS.
- Y. Tashiro, R. E. Monson, J. P. Ramsay and G. P. Salmond, Environ. Microbiol., 2016, 18, 1264–1276 CrossRef CAS.
- D. P. Sawyer, A. Bar-Zion, A. Farhadi, S. Shivaei, B. Ling, A. Lee-Gosselin and M. G. Shapiro, Nat. Methods, 2021, 18, 945–952 CrossRef CAS.
- Y. Yang, Y. Yang, D. Liu, Y. Wang, M. Lu, Q. Zhang, J. Huang, Y. Li, T. Ma, F. Yan and H. Zheng, Nat. Commun., 2023, 14, 3297 CrossRef CAS PubMed.
- J. O. Szablowski, A. Bar-Zion and M. G. Shapiro, Acc. Chem. Res., 2019, 52, 2427–2434 CrossRef CAS.
- A. Bar-Zion, A. Nourmahnad, D. R. Mittelstein, S. Shivaei, S. Yoo, M. T. Buss, R. C. Hurt, D. Malounda, M. H. Abedi, A. Lee-Gosselin, M. B. Swift, D. Maresca and M. G. Shapiro, Nat. Nanotechnol., 2021, 16, 1403–1412 CrossRef CAS PubMed.
- J. P. J. Hall, R. C. T. Wright, E. Harrison, K. J. Muddiman, A. J. Wood, S. Paterson and M. A. Brockhurst, PLoS Biol., 2021, 19, e3001225 CrossRef CAS.
- A. B. Reams and J. R. Roth, Cold Spring Harbor Perspect. Biol., 2015, 7, a016592 CrossRef.
- J. Nielsen and J. D. Keasling, Cell, 2016, 164, 1185–1197 CrossRef CAS PubMed.
- S. Snoeck, C. Guidi and M. De Mey, Microb. Cell Fact., 2024, 23, 96 CrossRef PubMed.
- O. Borkowski, F. Ceroni, G. B. Stan and T. Ellis, Curr. Opin. Microbiol., 2016, 33, 123–130 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra07532c |
| This journal is © The Royal Society of Chemistry 2025 |