Open Access Article
Open Access ArticleGold nanoparticles synthesized from soil-derived Streptomyces sp. ASM19: characterization, antimicrobial, anticancer potency targeted G2/M phase cell-cycle arrest, and in silico studies
Sultan Aati†
a,
Hanan Y. Aati† *b,
Ahmed A. Hamedc,
Sherine El-Shamyd,
Shahad H. Aatie,
Usama Ramadan Abdelmohsen
*b,
Ahmed A. Hamedc,
Sherine El-Shamyd,
Shahad H. Aatie,
Usama Ramadan Abdelmohsen fg,
Gerhard Bringmannh,
Mostafa N. Tahai,
Hossam M. Hassan
fg,
Gerhard Bringmannh,
Mostafa N. Tahai,
Hossam M. Hassan *jk and
Hebatallah S. Bahr
*jk and
Hebatallah S. Bahr l
l
aDental Health Department, College of Applied Medical Sciences, King Saud University, Riyadh, Saudi Arabia. E-mail: sati@ksu.edu.sa
bDepartment of Pharmacognosy, College of Pharmacy, King Saud University, Riyadh 11495, Saudi Arabia. E-mail: hati@ksu.edu.sa
cNational Research Centre, Microbial Chemistry Department, 33 El-Buhouth Street, Dokki, Giza 12622, Egypt. E-mail: ahmedshalbio@gmail.com
dPharmacognosy Department, Faculty of Pharmacy, Modern University for Technology and Information, Cairo, Egypt. E-mail: s_elshamy@hotmail.com
eCollege of Dentistry, King Saud University, Riyadh, Saudi Arabia. E-mail: Shahadaati0@gmail.com
fDeraya Center for Scientific Research, Deraya University, New Minia City, 61111, Egypt. E-mail: Usama.Ramadan@mu.edu.eg
gDepartment of Pharmacognosy, Faculty of Pharmacy, Minia University, Minia, Egypt
hInstitute of Organic Chemistry, University of Würzburg, Am Hubland, Würzburg, 97074, Germany. E-mail: gerhard.bringmann@uni-wuerzburg.de
iDepartment of Microbiology and Immunology, Faculty of Pharmacy, Nahda University, Beni-Suef 62513, Egypt. E-mail: moustafa.nasr@nub.edu.eg
jDepartment of Pharmacy, Kut University College, Wasit, 52001, Iraq. E-mail: abuh20050@yahoo.com
kDepartment of Pharmacognosy, Faculty of Pharmacy, Beni-Suef University, Beni-Suef 62514, Egypt. E-mail: abuh20050@yahoo.com
lDepartment of Pharmacognosy, Faculty of Pharmacy, Nahda University, Beni-Suef 62513, Egypt. E-mail: hebatallah.samir@nub.edu.eg
First published on 6th February 2025
Abstract
Gold nanoparticles (Au) have attracted considerable attention in the field of biomedicine in recent years. The present work was designed to investigate gold nanoparticles obtained using a soil-associated actinomycetes, Streptomyces sp. ASM19. This microorganism was isolated and identified using DNA sequencing. The chemical profile of the Streptomyces sp. ASM19 extract was analyzed using LC-HRES-MS. Streptomyces sp. ASM19 extract was utilized to synthesize actinomycetes-based gold nanoparticles (Ac-AuNPs), which were analyzed using ultraviolet-visible (UV-vis) spectrophotometry, Fourier transform infrared (FTIR) spectroscopy, and atomic force microscopy (AFM). In addition, the antibacterial, and anti-biofilm, as well as, the anti-proliferative properties of Ac-AuNPs against seven cancer lines were investigated. LC-HRES-MS analysis traced a total of 111 peaks, 14 of them are key peaks belonging to the chemical classes, alkaloids, steroids, and polyketides. Analysis of the synthesized Ac-AuNPs revealed that they exhibited a wine-red color and a plasmon band appeared at 540 nm, confirming the formation of the Ac-AuNPs. Further, FTIR confirmed various functional groups present in Ac-AuNPs. The crude extract of Streptomyces sp. ASM19 demonstrated consistent antibacterial activity in contrast to Ac-AuNPs. The anti-proliferative properties of Ac-AuNPs demonstrated encouraging anticancer properties against SCC9 and SCC25 cell lines with IC50 values of 3.77 and 1.56 μg mL−1. Furthermore, Ac-AuNPs had total apoptotic percentages of 26.37% (SCC9) and 32.08% (SCC25), which are around 25 times higher than the control (0.95%). Additionally, it caused a notable G2/M phase cell-cycle arrest. On the other hand, molecular docking study carried out for the annotated compounds; tomaymycin (8) and nocapyrone P (5), showed considerable binding affinities compared to the co-crystallized inhibitor (fisetin) against the cyclin-dependent kinase 6 active site. Overall, the present study could be useful for nano drug delivery and may be applied for clinical studies in the future.
1. Introduction
Cancer is a complicated disease characterized by aberrant cell proliferation, high degree of invasiveness, and resistance to apoptosis. Reports revealed that cancer is the second-leading cause of death globally and it is regrettably anticipated that the incidence of cancer will rise by about 50% in the upcoming years.1 On the other hand, cancer patients are suffering from the harmful effects of the majority of anticancer medications on the market on normal cells; they exhibit serious side effects and are not as effective against certain cancer types. Additionally, medication resistance was evident in some cancer types.2 Therefore, discovering possible cytotoxic medications that enhance patient outcomes has long piqued the curiosity of scientists and researchers. Recently, researchers have investigated chemotherapeutics that target the signaling pathways that regulate the tumor cell cycle and its microenvironment, causing cell death, reducing tumor cell proliferation, or preventing the growth of tumor masses. Cyclin-dependent kinases (CDKs) are well-investigated biological targets that control signaling pathways within cells. The family of serine/threonine protein kinases, known as CDKs, is involved in many physiological processes, including the regulation of the cell cycle. Certain CDKs (1–7) are only active when they are linked to cyclins A–H, which are their regulator proteins. Since abnormal cell division is a hallmark of cancer, controlling CDK activity has been shown to be a promising treatment approach for the pathophysiology of the illness.2Nanoparticles are the fundamental building blocks of specific materials in nanoscale structures.3 This distinguishing property of nanomaterials has fueled intense research into the synthesis, characterization, and application of functional nanomaterials for antibacterial, catalytic, pharmacological, electrochemical, and imaging applications.4
Metal nanoparticles based on silver, gold, iron, platinum, copper, or zinc have captivated scientists for millennia due to their numerous applications in nanotechnology.5 Gold nanoparticles have garnered a lot of interest and numerous investigations because of their unique optical, catalytic, and biological capabilities, as well as their low toxicity and biocompatibility.6,7
Physical and chemical methods have been widely investigated for the synthesis of nanoparticles; nevertheless, their use in therapeutic applications is limited by the employment of toxic chemicals and solvents, strict synthetic conditions, an environmentally unfriendly protocol, and greater energy consumption. As a result, there is an increased demand for the development of non-toxic, dependable, and environmentally friendly nanoparticle synthesis for biomedical applications.8,9 In recent years, a wide range of microorganisms, including bacteria, viruses, algae, actinomycetes, and fungi, have been used to synthesize nanoparticles. Nonetheless, actinomycetes have garnered significant attention because they are least investigated and stand as an efficient candidate for the manufacture of metal nanoparticles.10,11 However, only a small number of prokaryotic species (bacteria and actinomycetes) have been assessed for their capacity to generate NPs; hence, the synthesis must be expanded. Actinomycetes are widely used to produce antibiotics and other valuable metabolites, but they have received less attention when it comes to nanoparticle research. The free amino groups and cysteine residues of the actinomycete proteins attach to silver or gold nanoparticles and stabilize the produced particles. Enzymes secreted from cells may contain proteins that aid in the reduction of silver or chloroauric ions and cap the metal nanoparticles. On the surface of metal nanoparticles, several proteins may exist as a capping and stabilizing substance.12 It is unclear exactly which enzymes are involved in the creation of gold. It has recently been discovered that phenol oxidase is involved in this process.13 This research aims to synthesize Streptomyces mediated nano-gold, its characterization using different analytical tools, and the evaluation of its biological activities.
2. Experimental
2.1. Sample collection
The soil samples were obtained in May 2023 from Mansoura, Egypt. They were photographed with proper photographic equipment and securely stored at the Microbial Chemistry Department, National Research Center.2.2. Actinomycetes isolation from soil
To isolate actinomycetes from soil samples, 1.0 g of the sample was suspended in water and diluted by serial dilution method and 100 μL placed on starch nitrate agar media (PDA) medium supplemented with 100 mg L−1 ampicillin and 50 mg L−1 streptomycin. Actinomycetes colonies were allowed to grow on the plates by incubating them at 25 °C.2.3. Genetic identification of actinomycetes strains
The actinomycetes strain was sub-cultured in ISP2 for three days at 25 °C. A 0.5 mL sterile saline solution was used to spread pure, separate colonies. After that, the suspension was centrifuged for 10 minutes at ambient temperature at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. Following the manufacturer's instructions, a DNeasy Blood & Tissue Kit was used to extract the DNA. Two primers were used: 1492R (5′GGTTACCTTGTTACGACTT′3) and 27 F (5AGAGTTTGATCCTGGCTCAG-3). Using primers from SolGent and Macrogen, two commercial sequencing services in South Korea, the PCR products were run through 35 cycles of 30 s at 94 °C, 30 s at 55 °C, 90 s at 72 °C, and a final step of 5 min at 72 °C. The sequencing results were resolved using the automated DNA-sequencing device from Applied Biosystems, model 3730-XL.14,15
000 rpm. Following the manufacturer's instructions, a DNeasy Blood & Tissue Kit was used to extract the DNA. Two primers were used: 1492R (5′GGTTACCTTGTTACGACTT′3) and 27 F (5AGAGTTTGATCCTGGCTCAG-3). Using primers from SolGent and Macrogen, two commercial sequencing services in South Korea, the PCR products were run through 35 cycles of 30 s at 94 °C, 30 s at 55 °C, 90 s at 72 °C, and a final step of 5 min at 72 °C. The sequencing results were resolved using the automated DNA-sequencing device from Applied Biosystems, model 3730-XL.14,15
2.4. Actinomycetes filtrate preparation
The ISP2 broth medium was used to cultivate the actinomycetes. Following autoclaving, colonies from the starch nitrate plate were moved into the ISP2 broth using a sterile loop to inoculate the actinomycetes. After that, the flasks were continuously shaken for eight days at 31 °C. The filtrate was centrifuged for 10 minutes at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm following 8 days of culture. Additional analysis was conducted using the resultant supernatant.
000 rpm following 8 days of culture. Additional analysis was conducted using the resultant supernatant.
2.5. Biosynthesis of AuNPs
The biosynthesis of AuNPs experiment was carried out by combining 25 mL of actinomycetes supernatant with 25 mL of 1 mM Au solution (prepared in deionized water). The mixture was then placed in a shaker incubator at 200 rpm in a dark environment at 37 °C for 24 h and pH 8.0. Two control groups were simultaneously established under the same conditions: a positive control where the actinomycetes supernatant was incubated with deionized water, and a negative control containing only the Au solution.162.6. Characterization of AuNPs
2.7. Biological assessment
SigmaPlot 12.0 software was used to analyze the viability percentages of the cell line at different concentrations of Au-nanoparticles to calculate the IC50 value, which reflects the dosage that decreases cell survival by 50%. This study contributed to our knowledge about the potential of the biosynthesized Au-nanoparticles as anticancer agents by offering insightful information on their possible lethal effects on human tumor cells.21
2.8. Virtual target identification
Retinol's potential target characterisation was accomplished by pharmacophore-based virtual screening with PharmMapper.23,24 According to this platform, any molecule in the PDB that most closely matches a pharmacophore model that has been extracted and saved as a library ligand dataset in mol2 format is given a score. An approximately 77 score matrix is created by this process.25 Following that, each pharmacophore's fit score is determined when a new molecule is supplied, and each fit score is compared to the fit score matrix to ascertain the molecule's position on the scale of all pharmacophore scores. The pure fit score that emerges from this process has significantly more weight and confidence than random pharmacophore matching. The platform received the query structure in pdb format, and the results were output as an Excel sheet that arranged the protein targets based on fit scores.2.9. Molecular docking study
Utilizing an AutoDock Vina docking machine 25, the docking analysis was performed utilizing the crystal structures of the active site of human cyclin-dependent kinase 6 (PDB ID: 1XO2). The docking grid box and binding site were identified using the co-crystallized ligand fisetin, a naturally occurring flavonol. The grid box's coordinates were set to be: x = 1.303, y = 37.229, z = 139.064.The root mean square (RMSD) criterion for ligand to binding site shape matching was chosen at 2.0 Å. CHARRM force field (v.1.02) with distance-dependent dielectric and non-bonded cutoff distance of 10.0 Å was used to calculate the interaction energies. An energy grid that extended from the binding point was then established at 5.0 Å. Retinol, the studied molecule, had the lowest energy within the chosen binding region. PyMOL software was used to edit and visualize the generated binding postures.26
2.10. Molecular dynamics simulation (MDS)
The program utilized to do molecular dynamics simulation (MDS) was NAMD 3.0.0.26 The Charmm36 force field is applied by this software. The VMD software's QwikMD toolkit was used to construct protein systems, and any missing hydrogens in the protein structure were examined.27 The co-crystallized water molecules were eliminated, and the protonation states of the amino acid residues were adjusted to a pH of 7.4. The entire structure, along with 0.15 M Na+ and Cl− ions in 20 Å solvent buffer, was then immersed in an orthorhombic box of TIP3P water. After then, the systems were equilibrated for 5 ns and their energy was decreased. The VMD plugin Force Field Toolkit (ffTK) was used to generate the ligands' parameters and topologies. The produced parameters and topology files were then put into VMD so that the simulation steps could be carried out and the protein–ligand complexes could be read easily and accurately.2.11. Binding free energy calculations
The MMPBSA incorporates the Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA). The binding free energy of the docked complex was determined using the AMBER18 py module.28 In total, 100 frames were processed from the trajectories, and the net energy of the system was estimated using the following equation:| ΔGbinding = ΔGcomplex − ΔGreceptor − ΔGinhibitor |
Multiple energy components, such as van der Waals energy, electrostatic energy, internal energy from molecular mechanics, and polar contribution to solvation energy, must be calculated for each of the terms described above.
3. Results
3.1. Molecular identification of Streptomyces sp. (ASM19)
Sequencing the soil isolate Streptomyces sp. ASM19, which demonstrated a strong potential to biosynthesize gold nanoparticles,14 exhibited a high similarity (99.89%) to Streptomyces sp. (Fig. 1). ASM19, a novel Streptomyces-specific sequence, has been deposited in Gene Bank under the name Streptomyces sp. ASM19 (accession number PP946394). The phylogenetic tree of Streptomyces sp. ASM19, using the neighbor joining technique is shown in Fig. 1. The evolutionary history was inferred by using the Maximum Likelihood method and Tamura-Nei model.29 The tree with the highest log probability (−48601.53) is displayed in that figure. By applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances computed using the Tamura-Nei model, the initial tree or trees for the heuristic search were automatically generated. The topology with the highest log likelihood value was then chosen. Branch lengths are expressed in terms of the number of substitutions per site, and the tree is depicted to scale. Thirty-one nucleotide sequences were analyzed. Included were the first, second, third, and noncoding codon locations. The resulting dataset contained 1504 locations in total. In MEGA11, evolutionary analyses were performed.303.2. LC-HRESIMS-assisted chemical profiling
LC-HRESIMS analysis resulted in tracing a total of 111 peaks using the positive ionization mode. Out of these, 14 key peaks were putatively identified (Table 1 and Fig. 2, 3). During the dereplication procedure, a taxonomic filter was used to choose only hits that were connected to the strains of actinomycetes under study. The identified metabolites belonged to different chemical classes e.g., alkaloids, steroids, and polyketides (Fig. 3).| No. | RT | Mass | Molecular formula | Dereplication | References |
|---|---|---|---|---|---|
| 1 | 5.45 | 210.1364 | C11H18N2O2 | Cyclo(pro-leu) | 31 |
| 2 | 7.54 | 191.0943 | C11H13NO2 | Streptopyrrolidine | 32 |
| 3 | 9.83 | 162.1007 | C6H14N2O3 | L-2-Amino-4-(2-aminoethoxy)-butanoic acid | 33 |
| 4 | 10.81 | 157.0368 | C6H7NO4 | Clavam-2-carboxylic acid | 34 |
| 5 | 13.12 | 194.0949 | C11H14O3 | Nocapyrone P | 35 |
| 6 | 13.78 | 174.0887 | C8H14O4 | Vertimycin | 36 |
| 7 | 14.79 | 390.2776 | C24H38O4 | 12α-Hydroxy-3-ketocholanic acid | 37 |
| 8 | 17.14 | 304.1428 | C16H20N2O4 | Tomaymycin | 38 |
| 9 | 22.25 | 314.1626 | C18H22N2O3 | Streptomycindole | 39 |
| 10 | 24.86 | 412.2609 | C26H36O4 | Janthinopolyenemycin B | 40 |
| 11 | 28.47 | 604.3971 | C35H56O8 | Bafilomycin D | 41 |
| 12 | 28.73 | 622.4075 | C35H58O9 | Bafilomycin A1 | 42 |
| 13 | 29.36 | 287.1527 | C17H21NO3 | Trichostatic acid | 43 |
| 14 | 31.26 | 316.2619 | C18H36O4 | Aggreceride A | 44 |
3.3. Biogenic synthesis of gold nanoparticles
When the Streptomyces sp. ASM19 supernatant was added to the HAuCl4 aqueous solution, the color change made it evident that AuNPs had formed. After actinomycetes species supernatants were added, the production of gold nanoparticles (AuNPs) was mediated at 30 °C in a dark environment. When Streptomyces sp. ASM19 supernatant was added, the supernatant became yellow to purple, confirming the production of AuNPs (Fig. 4). | ||
| Fig. 4 (A) UV spectrum; (B) transmission electron micrograph; (C) scanning electron micrograph of biogenic Ac-AuNP. | ||
3.4. Biosynthesized AuNPs characterization
In the presence of Streptomyces sp. ASM19 supernatants, AuNPs were produced. The 400–550 nm range of the surface plasmon resonance (SPR) absorption spectra suggests that AuNPs were produced at various times. The generated AuNPs' surface plasmon resonance (SPR) absorption spectrum range was between 450 and 650 nm, and their SPR band was located at 550 nm (Fig. 4a). The size and shape of the green AuNPs and AuNP particles were examined using field emission scanning electron microscopy (FESEM) and transmission electron microscopy (TEM) (Fig. 4c). According to the TEM micrograph (Fig. 4b), the AuNPs had a sphere-like form with an average particle size of 6.28 ± 0.78 to 100.2 ± 0.25 nm.3.5. FTIR and XRD
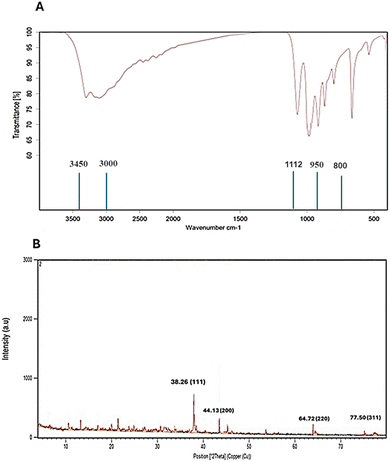
Additionally, Fig. 5a illustrates the FTIR spectra of AuNPs synthesized by Streptomyces sp. ASM19. The peaks that appear at 3000–3450 cm−1 are the characteristic bands for NH2 and OH groups, the peaks at 1112 cm−1 are the characteristic bands for C–O, and the peaks appearing at 800 to 950 cm−1 are the characteristic bands for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C. Additionally, the XRD examination of Streptomyces sp. ASM19 gold nanoparticles revealed a crystalline structure of the biogenic nanoparticles, as evidenced by four peaks matching typical Bragg reflections 38(111), 44(200), 64(220), and 77(311) of the face centers cubic lattice.
C. Additionally, the XRD examination of Streptomyces sp. ASM19 gold nanoparticles revealed a crystalline structure of the biogenic nanoparticles, as evidenced by four peaks matching typical Bragg reflections 38(111), 44(200), 64(220), and 77(311) of the face centers cubic lattice.
3.6. Colloidal and surface properties of as-prepared AuNPs
The produced AuNPs showed colloidal and surface stability. The AuNPs sample, which included Streptomyces sp. ASM19, had zeta potentials of −28.35. A study45 claims that metal nanoparticles are more stable when their zeta potential value is closer to −30 mV; these findings suggest that there was an electrostatic interaction between the AuNPs and the particles in the solution (Table 2).| Sample | Colloidal properties dynamic light scattering (DLS) | Zeta potential, mV |
|---|---|---|
| Streptomyces sp. ASM19 AuNPs | 68.45 ± 12.55 | −28.35 ± 7.35 |
3.7. Biological evaluation
| HepG2 | Caco2 | MCF7 | A549 | SNB-75 | SCC9 | SCC25 | |
|---|---|---|---|---|---|---|---|
| a The data in the table represent mean ± SD. Samples are significantly different from the positive control (staurosporine) at p < 0.05. Hepatocellular carcinoma (HepG2), colorectal adenocarcinoma (Caco2), human breast cancer (MCF7), non-small cell lung cancer (A549), human brain tumor (SNB-75), and squamous cell carcinoma (SCC-9 and SCC-25). | |||||||
| Ac-AuNPs | 38.809 ± 2.3 | 11.658 ± 0.69 | 14.312 ± 0.85 | 70.094 ± 4.15 | 14.009 ± 0.83 | 3.777 ± 0.13 | 1.566 ± 0.05 |
| Staurosporine | 6.330 ± 0.37 | 5.469 ± 0.32 | 4.721 ± 0.28 | 10.027 ± 0.59 | 5.698 ± 0.34 | 7.933 ± 0.27 | 11.85 ± 0.4 |
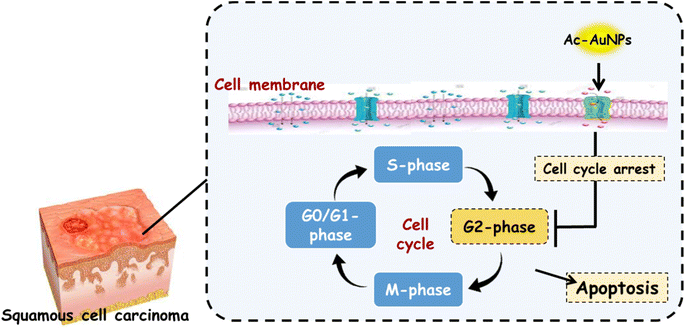
3.7.4.1. Apoptosis analysis. Propidium iodide (PI) and annexin-V-FITC staining was utilized in flow cytometry assay to assess the induction of apoptosis in SCC9 and SCC25 cell lines. Following treatment of SCC9 and SCC25 cell lines with Ac-AuNPs extract, Ac-AuNPs/SCC9 showed a significant increase in total apoptosis, from 1.95% (control SCC9) to 26.37% (approximately 13.5 time higher than the control). Additionally, an increase in late cellular apoptosis was noted, rising from 0.15% (control propidium iodide SCC9) to 7.22% (i.e. nearly 50-fold). Furthermore, the percentage of early apoptosis increased from 0.39% (control SCC9) to 15.13% (i.e. by a factor of nearly 39). On the other hand, the Ac-AuNPs/SCC25 sample analysis revealed that late cellular apoptosis increased from 0.13% (control SCC25) to 8.85%, and total apoptosis increased significantly from 2.13% (control SCC25) to 32.08%. Additionally, early apoptosis increased from 0.65% (control SCC25) to 19.51% (Fig. 9 and 10). These findings suggest that the examined extract (Ac-AuNPs) greatly accelerate the death of cancer cells and induce apoptosis.
 | ||
| Fig. 9 Graphical presentation of apoptosis induced on SCC9 cells by (A) control, (B) Ac-AuNPs extract; and cell cycle profile induced by (C) control, and (D) Ac-AuNPs extract. | ||
 | ||
| Fig. 10 Graphical presentation of apoptosis induced on SCC25 cells by (A) control, (B) Ac-AuNPs extract; and cell cycle profile induced by (C) control, and (D) Ac-AuNPs extract. | ||
3.7.4.2. Cell cycle analysis. As shown in Fig. 9–11, cell cycle analysis was carried out for the cell lines, SCC9 and SCC25 using Ac-AuNPs as a treatment. It was disclosed that Ac-AuNPs caused cell cycle arrest in the G2/M phase for both cancer cell lines at a value of 24.16% and 38.11%, respectively. On the other hand, the untreated control SCC9 and SCC25 cell lines showed cell accumulation in the G2/M phase at values of 10.24% and 8.41, respectively. Therefore, in comparison to the control cells, one can conclude that Ac-AuNPs can cause a considerable cell-cycle arrest in the G2/M phase.
3.8. Virtual screening-based target identification
To investigate the binding interaction and stability of both, tomaymycin (8) and nocapyrone P (5) inside the active site of cyclin-dependent kinase 6, the modeled structures of both compounds were re-docked into the predetermined active site of the crystal structure of cyclin-dependent kinase 6 (PDB ID: 1XO2).As shown in Fig. 12, both compounds aligned with the co-crystallized inhibitor fisetin, which was able to form multiple H-bonds with ILE-19, LYS-43, GLU-61, GLU-99, VAL-101, ASP-104, GLN-149, and ASP-163, in addition to hydrophobic interactions with VA-27, PHE-98, and LEU-152. Hence, fisetin showed significant stability inside the active site of human cyclin-dependent kinase 6, where it deviated from the initial crystallized binding pose with small values over the course of 100 ns long MD simulation (average RMSD = 0.6 Å) (Fig. 12C). Consequently, its MM-PBSA-calculated absolute binding free energy (ΔGbind) was −11.56 kcal mol−1.
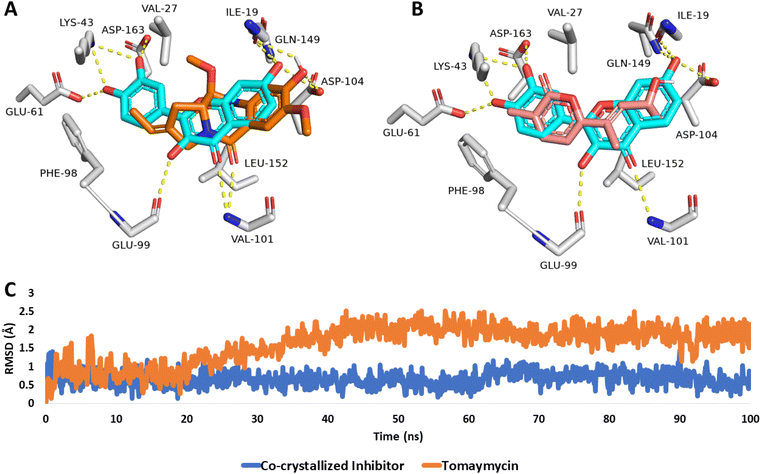 | ||
| Fig. 12 Binding modes of (A) tomaymycin (8) and (B) nocapyrone P (5) in alignment with the co-crystallized inhibitor fisetin inside the active site of the human cyclin-dependent kinase 6 (PDB ID: 1XO2). (C) RMSDs of both, tomaymycin and the co-crystallized inhibitor fisetin over the course of the 100 ns long MD simulation. | ||
Similarly, tomaymycin (8) was able to establish H-bonds with ILE-19, GLU-99, ASP-104, and GLN-149 together with two hydrophobic interactions with VA-27 and LEU-152 (Fig. 12A), while nocapyrone P (5), and due to its lower molecular size in comparison to both, fisetin and tomaymycin (8), was not able to establish any H-bond or even hydrophobic interactions except for the π-stacking with PHE-98 (Fig. 12B). Accordingly, nocapyrone P (5) could not achieve stable binding inside the active site of the enzyme and dissociated from it shortly after starting the MD simulation (at 25 ns). Tomaymycin (8), on the other hand, was much more stable over the course of 100 ns long MD simulation showing low a RMSD value (∼1.8 Å) (Fig. 12C) and ΔGbind of −9.33 kcal mol−1.
4. Discussion
The use and development of nanotechnology has increased dramatically since its inception to meet the growing need for healthcare solutions.46 Recently, the movement towards green industries and sustainable development has become a global necessity to protect environmental resources. Thus, it is crucial for scientists to investigate the toxicity of these nanomaterials. Consequently, the ecologically friendly and green synthesis of nanoparticles is a significant way to give lower toxicity levels and enable unrestricted use in products meant for human consumption. In this sense, creating biocompatible and affordable compounds with potential uses in medicine delivery and healthcare through the production of metal oxide nanoparticles utilizing microorganisms such as, bacteria, fungi, algae, and plant extracts has significant promise.47 Actinomycetes can be used in the green biogenic synthesis of nanoparticles. They were widely used for producing non-toxic, ecologically friendly nanoparticles that are quick, cheap, and simple. Reports revealed that actinomycetes-mediated biosynthesized nanoparticles can be small and stable, with antimicrobial, antioxidant, and cytotoxic properties that help them fight a variety of illnesses and infections.45,48,49 Thermomonospora, Rhodococcus, and Streptomyces species are among the few actinomycetes that have been linked to the creation of silver and gold nanoparticles.45 Streptomyces is a well-known fungus that produces a large range of secondary metabolites, including immunosuppressants, antibiotics, and numerous other physiologically active substances that are widely used in the enzyme and pharmaceutical industries.45 The synthesis of Au nanoparticles (NPs) has been reported for Streptomyces cyaneus,50 Streptomyces hygroscopicus,51 and Streptomycetes viridogens (HM10),52 Streptomycetes monashensis.53In the present study, it was found that, Streptomyces sp. strain ASM19 isolated from soil have the ability to synthesize gold nanoparticles that were characterized using UV-vis spectrophotometry, FT-IR spectroscopy, atomic force microscopy (AFM), as well as, zeta potential analysis that revealed details regarding the stability of synthesized nanostructures. Furthermore, the biological efficacy including, the antibacterial, anti-biofilm and cytotoxic activities of the synthesized gold nanoparticles were evaluated.
4.1. Antibacterial activity
Our study results revealed that the biogenic AuNPs showed minimal antibacterial and biofilm inhibitory action against the tested bacteria, S. aureus, E. coli, and B. subtilis, unlike the crude extract of Streptomyces sp. ASM19, which demonstrated higher antibacterial activity (Fig. 6 and 7). After reviewing the literature, it was found that many studies support our findings viz. Ramamurthy et al. compared between the antibacterial activities of the silver and gold nanoparticles produced using the aqueous extract of Solanum torvum fruits and noticed that silver nanoparticles show strong zone of inhibition against Pseudomonas aeruginosa, Escherichia coli, and Bacillus subtilis whereas, gold nanoparticles exhibit less zone of inhibition.54 Składanowski et al., 2017 evalutaed the antibacterial activities of silver (Ag) and gold (Au) nanoparticles using Streptomyces sp. strain NH21 against a number of Gram +ve and −ve bacteria, and they found that while AgNPs exhibited antibacterial activity, AuNPs did not.45 (Hamed & Abdelftah, 2019) antibacterial study showed no antibacterial activity of AuNPs against certain strains such as, Staphylococcus aureus 25923 and Bacillus cereus.49 (Nishanthi & Palani, 2016) and (Rizvi et al., 2023) declared that AuNPs is better to be loaded with antibiotic for better antibacterial activity.55,56 Soliman et al., 2023 AgNPs were more effective than AuNPs against Gram-positive and Gram-negative pathogenic bacteria.57 However, gold nanoparticles mediated by Streptomycetes viridogens shown strong antibacterial action against S. aureus and E. coli, with a 20 mm zone of inhibition for each, according to a study by Balagurunathan et al.52 Bindhu et al. have also reported that gold nanoparticles have antibacterial action against both Gram-positive and Gram-negative bacteria,58 and recently by Kerdtoob et al., 2024;53 those studies presented antibacterial nature of gold nanoparticles. According to Składanowski et al. (2017), gold nanoparticles are more well-known for their possible use in medicine as an antiviral or drug delivery system than for their antibacterial qualities.45 (Nishanthi & Palani, 2016) and (Rizvi et al., 2023) revealed that the antibacterial action of gold nanoparticles is better in the presence of antibiotics.55,56 Therefore, we can concluded that the antibacterial action of some AuNPs is due to the presence of antibiotic or antibacterial metabolites within. The chemical profile of gold nanoparticles generated from Streptomyces and their antibacterial properties need to be further investigated.4.2. Cytotoxic activity
The results of the cytotoxic investigation demonstrated that Streptomyces ASM19 mediated gold nanoparticles showed greater cytotoxicity against squamous cell carcinoma, SCC-9 and SCC-25 cell lines, better than staurosporine, the positive control medication, with IC50 values of 3.77 and 1.56 μg mL−1, respectively. These results encouraged us to conduct further analysis regarding apoptosis and cell cycle analysis because the capacity of cancerous cells to avoid apoptosis is a defining characteristic of the disease. Thus, inducing cancer cell death by activating apoptotic signaling pathways is the main mechanism of many anticancer medicines.59 Results revealed that the examined extract (ASM19) greatly accelerate the death of cancer cells and induce apoptosis. As well as, can cause a considerable cell-cycle arrest in the G2/M phase. In order to determine the key metabolites in ASM19 extract that are responsible for this observed anticancer activity, the identified compounds from LC-HRESMS-assisted chemical profiling were literally reviewed. According to the previously reported literature, bafilomycin A1 (12) and trichostatic acid (13) acid have been found to exert anti-cancer and cytotoxic activities via different mechanisms e.g., DNA binding, induction of autophagy, and epigenetic modulation, respectively.60–62Since the identified metabolites from LC-HRESMS-assisted chemical profiling might contribute to the observed anticancer properties of ASM19 extract. Therefore, they were submitted to a comprehensive in silico analysis as illustrated in the following section.
4.3. Virtual screening-based target identification
Identification of the biological target of a molecule is difficult. Meanwhile, the success rate of finding suitable molecular targets has been greatly enhanced by the ongoing development of in silico tools like molecular modeling and virtual screening.Structure-based and ligand-based virtual screening protocols are used by many of the currently available online target identification platforms. PharmMapper is one such platform, it uses the pharmacophore model of a query molecule to screen for and recommends the most likely protein targets.23,24 The central tenet of pharmacophore-based screening is that the spatial arrangements of structural features (i.e., the pharmacophore maps) are largely responsible for the binding of specific molecules to their protein targets. Therefore, the likelihood of chemicals binding to the same protein target is highest when their forms correspond to these pharmacophore maps. In light of this, PharmMapper was utilized to recommend a potential protein target or targets for the metabolites found in the ASM19 extract. Fit ratings were used to order the results that were retrieved. Only targets linked to cancer were eliminated. As a result, human cyclin-dependent kinase 6 (PDB ID: 1XO2)63 was found to be among the top-scoring (Fit scores = 8.51 and 7.45) cancer-relevant hits for tomaymycin (8) and nocapyrone P (5), respectively, and hence, it was suggested to be a probable target.
Interestingly, cyclin-dependent kinase 6 has been reported as a key oncoprotein in squamous cell cancers, and this matches well with the in vitro testing illustrated above in Table 3,64–66 indicating that both tomaymycin (8) and nocapyrone P (5) play a major role in the ASM19 anticancer activity besides that of the previously reported ones (bafilomycin A1 and trichostatic acid).
5. Conclusion
The current study outlined a simple, cost-effective, and environmentally friendly method for the biofabrication of AuNPs using the extract of the actinomycetes Streptomyces species ASM19. The successful AuNP synthesis was confirmed by the UV-vis spectra, which showed a characteristic peak at 540 nm. The significant zeta potential value indicated the stability of the Ac-AuNPs. The synthesized Ac-AuNPs demonstrated a notable cytotoxic capability against the squamous cell cancer cell lines, SCC9 and SCC25, outperforming the usual medication, staurosporine. Furthermore, the flow cytometry technique was employed to evaluate the rates of neoplastic cell apoptosis and necrosis. The acquired results showed a considerable cell population undergoing early apoptosis (15.13% and 19.51%) and late apoptosis (7.22% and 8.85%) towards the SCC9 and SCC25 cell lines, respectively. Additionally, they caused a notable G2/M phase cell-cycle arrest. The observable functions demonstrated by Ac-AuNPs are crucial for the development of new therapeutic drugs for the management of cancer. Because actinomycetes-mediated synthesis reduces metal ions more quickly and steadily than other approaches, it is considered to be more beneficial. In biomedical applications, including drug/gene delivery and multifunctional therapy, gold nanoparticles are thus promising new tools. In light of recent studies, such as the review by Yao et al.,67 which examined over 20 clinical trials involving gold nanoparticles (GNPs) and found no major safety concerns, our findings contribute to the growing body of evidence supporting the safe application of Streptomyces-derived nanoparticles. However, as Yao et al. emphasized the necessity for multi-dose, multi-center masked trials to deepen understanding of GNPs in clinical settings, we similarly advocate for comprehensive toxicological evaluations and large-scale studies to fully ascertain the safety and efficacy of Streptomyces-derived nanoparticles across various applications.67Data availability
All data generated or analysed during this study are included in this article.Author contributions
Conceptualization, H. A. and H. M. H.; methodology, A. A. H. and S. S.; soft-ware, M. N. T. and H. S. B.; validation, H. M. H., S. A. and A. T. A.; formal analysis, S. A.; investigation, G. B. and U. R. A.; resources, H. A. and S. H. A.; data curation, H. M. H.; writing—original draft preparation, G. B., U. R. A., and H. S. B.; writing—review and editing, H. M. H.; visualization, M. N. T.; super-vision, H. M. H.; project administration, H. M. H.; funding acquisition, H. A. All authors have read and agreed to the published version of the manuscript. “All authors have read and agreed to the published version of the manuscript”.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Researchers Supporting Project number (RSP2025R504), King Saud University, Riyadh, Saudi Arabia, for supporting this work.References
- M. M. Hammoud, A. S. Nageeb, M. A. Morsi, E. A. Gomaa, A. A. Elmaaty and A. A. Al-Karmalawy, New J. Chem., 2022, 46, 11422–11436 RSC.
- S. S. El-Hawary, A. S. Moawad, H. S. Bahr, E. Z. Attia, M. M. H. El-Katatny, M. Mustafa, A. A. Al-Karmalawy, M. E. Rateb, J. Y. Zhang, U. R. Abdelmohsen and R. Mohammed, J. Appl. Microbiol., 2023, 134, 1–12 CrossRef PubMed.
- F. J. Heiligtag and M. Niederberger, Mater. Today, 2013, 16, 262–271 CrossRef CAS.
- A. Schröfel, G. Kratošová, I. Šafařík, M. Šafaříková, I. Raška and L. M. Shor, Acta Biomater., 2014, 10, 4023–4042 CrossRef PubMed.
- A. A. Yaqoob, H. Ahmad, T. Parveen, A. Ahmad, M. Oves, I. M. I. Ismail, H. A. Qari, K. Umar and M. N. Mohamad Ibrahim, Front. Chem., 2020, 8, 341 CrossRef CAS PubMed.
- C. Singh, R. K. Baboota, P. K. Naik and H. Singh, Adv. Mater. Lett., 2012, 3, 279–285 CrossRef CAS.
- S. Ahmed and S. Ikram, J. Photochem. Photobiol., B, 2016, 161, 141–153 CrossRef CAS PubMed.
- P. M. Tiwari, K. Vig, V. A. Dennis and S. R. Singh, Nanomaterials, 2011, 1, 31–63 CrossRef CAS PubMed.
- B. S. Srinath and V. Ravishankar Rai, 3 Biotech, 2015, 5, 671–676 CrossRef CAS PubMed.
- P. Golinska, M. Wypij, A. P. Ingle, I. Gupta, H. Dahm and M. Rai, Appl. Microbiol. Biotechnol., 2014, 98, 8083–8097 CrossRef CAS PubMed.
- V. R. Ranjitha and V. R. Rai, 3 Biotech, 2017, 7, 1–7 CrossRef.
- L. Xu, Y.-Y. Wang, J. Huang, C.-Y. Chen, Z.-X. Wang and H. Xie, Theranostics, 2020, 10, 8996 CrossRef CAS PubMed.
- L. Blaschek and E. Pesquet, Front. Plant Sci., 2021, 12, 754601 CrossRef PubMed.
- A. A. Hamed, M. S. Abdel-Aziz, M. Fadel and M. F. Ghali, J. Innovations Pharm. Biol. Sci., 2016,(2), 155–165 CAS.
- A. A. Hamed, M. S. Abdel-Aziz, M. Fadel and M. F. Ghali, Bull. Natl. Res. Cent., 2018, 42, 1–10 CrossRef.
- E. M. Mostafa, M. A. Abdelgawad, A. Musa, N. H. Alotaibi, M. H. Elkomy, M. M. Ghoneim, M. S. E. M. Badawy, M. N. Taha, H. M. Hassan and A. A. Hamed, Antibiotics, 2022, 11, 668 CrossRef CAS.
- D. M. Eskander, S. M. M. Atalla, A. A. Hamed and E.-D. A. El-Khrisy, Pharmacogn. J., 2020, 12, 636–644 CrossRef CAS.
- A. A. Hamed, H. Kabary, M. Khedr and A. N. Emam, RSC Adv., 2020, 10, 10361–10367 RSC.
- M. S. Hifnawy, H. M. Hassan, R. Mohammed, M. M. Fouda, A. M. Sayed, A. A. Hamed, S. F. AbouZid, M. E. Rateb, H. A. Alhadrami and U. R. Abdelmohsen, Mar. Drugs, 2020, 18, 243 CrossRef CAS PubMed.
- S. S. Mohamed, M. E. El Awady, S. A. Abdelhamid, A. A. Hamed, A. A. A. Salama and M. S. Selim, J. Genet. Eng. Biotechnol., 2023, 21, 12 CrossRef PubMed.
- A. A. Ghfar, M. M. El-metwally, M. Shaaban, S. A. Gabr, N. S. Gabr, M. S. M. Diab, A. Aqel, M. A. Habila, W. H. Al-qahtani, M. Y. Alfaifi, S. E. I. Elbehairi and B. A. Aljumah, Molecules, 2021, 26, 1–8 CrossRef PubMed.
- A. I. Hassaballah, A. M. AboulMagd, M. M. Hemdan, M. H. Hekal, A. A. El-Sayed and P. S. Farag, RSC Adv., 2024, 14, 1995–2015 RSC.
- X. Liu, S. Ouyang, B. Yu, Y. Liu, K. Huang, J. Gong, S. Zheng, Z. Li, H. Li and H. Jiang, Nucleic Acids Res., 2010, 38, W609–W614 CrossRef CAS PubMed.
- Y. Dong-Ping, L. Jun, L. U. Yin, L. I. N. Jie and T. Li, Chin. J. Nat. Med., 2014, 12, 443–448 Search PubMed.
- R. Ghosh, A. Chakraborty, A. Biswas and S. Chowdhuri, J. Biomol. Struct. Dyn., 2020, 39, 1–13 Search PubMed.
- A. M. Sayed, H. A. Alhadrami, A. O. El-Gendy, Y. I. Shamikh, L. Belbahri, H. M. Hassan, U. R. Abdelmohsen and M. E. Rateb, Microorganisms, 2020, 8, 1–17 Search PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- B. S. Cavada, V. J. S. Osterne, V. R. Pinto-Junior, L. A. G. Souza, C. F. Lossio, M. T. L. Silva, C. Correia-Neto, M. V. Oliveira, J. L. A. Correia and A. H. B. Neco, J. Mol. Model., 2020, 26, 1–9 CrossRef PubMed.
- K. Tamura and M. Nei, Mol. Biol. Evol., 1993, 10, 512–526 CAS.
- K. Tamura, G. Stecher and S. Kumar, Mol. Biol. Evol., 2021, 38, 3022–3027 CrossRef CAS PubMed.
- F. Fdhila, V. Vázquez, J. L. Sánchez and R. Riguera, J. Nat. Prod., 2003, 66, 1299–1301 CrossRef CAS.
- H. J. Shin, T. S. Kim, H.-S. Lee, J. Y. Park, I.-K. Choi and H. J. Kwon, Phytochemistry, 2008, 69, 2363–2366 CrossRef CAS.
- J. P. Scannell, D. L. Pruess, H. A. Ax, A. Jacoby, M. Kellett and A. Stempel, J. Antibiot., 1976, 29, 38–43 CrossRef CAS PubMed.
- V. A. López-Agudelo, D. Gómez-Ríos and H. Ramirez-Malule, Antibiotics, 2021, 10, 84 CrossRef.
- X.-M. Zhang, M.-W. Sun, H. Shi and C.-H. Lu, Nat. Prod. Res., 2017, 31, 2245–2249 CrossRef CAS.
- T. Haneishi, A. Terahara, K. Hamano and M. Arai, J. Antibiot., 1974, 27, 400–407 CrossRef CAS.
- H. Li, P. B. Shinde, H. J. Lee, E. S. Yoo, C.-O. Lee, J. Hong, S. H. Choi and J. H. Jung, Arch. Pharmacal Res., 2009, 32, 857–862 CrossRef CAS PubMed.
- S. K. Arora, J. Antibiot., 1981, 462–464 CrossRef CAS.
- N. Netz and T. Opatz, Mar. Drugs, 2015, 13, 4814–4914 CrossRef CAS PubMed.
- K. Anjum, I. Sadiq, L. Chen, S. Kaleem, X. C. Li, Z. Zhang and X. Y. Lian, Tetrahedron Lett., 2018, 59, 3490–3494 CrossRef CAS.
- Z. Yu, L. X. Zhao, C. L. Jiang, Y. Duan, L. Wong, K. C. Carver, L. A. Schuler and B. Shen, J. Antibiot., 2011, 64, 159–162 CrossRef CAS.
- K. Kobayashi, T. Fukuda, T. Usui, Y. Kurihara, A. Kanamoto and H. Tomoda, J. Antibiot., 2015, 68, 126–132 CrossRef CAS PubMed.
- K. Georgousaki, N. Tsafantakis, S. Gumeni, I. Gonzalez, T. A. Mackenzie, F. Reyes, C. Lambert, I. P. Trougakos, O. Genilloud and N. Fokialakis, Bioorg. Med. Chem. Lett., 2020, 30, 126952 CrossRef CAS PubMed.
- S. Ōmura, A. Nakagawa, N. Fukamachi, K. Otoguro and B. Kobayashi, J. Antibiot., 1986, 39, 1180–1181 CrossRef.
- M. Składanowski, M. Wypij, D. Laskowski, P. Golińska, H. Dahm and M. Rai, J. Cluster Sci., 2017, 28, 59–79 CrossRef.
- A. Salman, C. Soukkarieh and S. Alzeer, Bull. Pharm. Sci., 2021, 44, 81–88 Search PubMed.
- A. Rasool, S. Sri, M. Zulfajri and F. S. H. Krismastuti, Inorg. Chem. Commun., 2024, 112954 CrossRef CAS.
- J. Vinay Gopal, M. Thenmozhi, K. Kannabiran, G. Rajakumar, K. Velayutham and A. A. Rahuman, Mater. Lett., 2013, 93, 360–362 CrossRef CAS.
- M. M. Hamed and L. S. Abdelftah, Egypt. J. Aquat. Biol. Fish., 2019, 23, 173–184 CrossRef.
- A. I. El-Batal and M. S. Al Tamie, J. Chem. Pharm. Res, 2015, 7, 1020–1036 CAS.
- S. Sadhasivam, P. Shanmugam, M. Veerapandian, R. Subbiah and K. Yun, BioMetals, 2012, 25, 351–360 CrossRef CAS.
- R. Balagurunathan, M. Radhakrishnan, R. B. Rajendran and D. Velmurugan, Indian J. Biochem. Biophys., 2011, 48, 331–335 CAS.
- S. Kerdtoob, P. Chanthasena, W. Limphirat, W. Penkhrue, P. Ganta, W. Srisakvarangkool, M. Yasawong and N. Nantapong, RSC Adv., 2024, 14, 4778–4787 RSC.
- C. H. Ramamurthy, M. Padma, R. Mareeswaran, A. Suyavaran, M. S. Kumar, K. Premkumar and C. Thirunavukkarasu, Colloids Surf., B, 2013, 102, 808–815 CrossRef CAS PubMed.
- R. Nishanthi and P. Palani, in 2016 IEEE 16th International Conference on Nanotechnology (IEEE-NANO), 2016, pp. 431–434 Search PubMed.
- S. M. D. Rizvi, A. S. A. Lila, A. Moin, T. Hussain, M. A. Kamal, H. Sonbol and E.-S. Khafagy, Pharmaceutics, 2023, 15, 430 CrossRef CAS.
- M. K. Y. Soliman, S. S. Salem, M. Abu-Elghait and M. S. Azab, Appl. Biochem. Biotechnol., 2023, 195, 1158–1183 CrossRef CAS PubMed.
- M. R. Bindhu and M. Umadevi, Mater. Lett., 2014, 120, 122–125 CrossRef CAS.
- K. N. Shashiraj, A. Hugar, R. S. Kumar, M. Rudrappa and S. Nayaka, Bioengineering, 2023, 10, 2–19 CrossRef PubMed.
- C. Lin, Z. Wang, L. Li, Y. He, J. Fan, Z. Liu, S. Zhao and D. Ju, Appl. Microbiol. Biotechnol., 2015, 99, 8487–8494 CrossRef CAS PubMed.
- Y. Yan, K. Jiang, P. Liu, X. Zhang, X. Dong, J. Gao, Q. Liu, M. P. Barr, Q. Zhang and X. Hou, Sci. Rep., 2016, 6, 37052 CrossRef CAS PubMed.
- K. Georgousaki, N. Tsafantakis, S. Gumeni, I. Gonzalez, T. A. Mackenzie, F. Reyes, C. Lambert, I. P. Trougakos, O. Genilloud and N. Fokialakis, Bioorg. Med. Chem. Lett., 2020, 30, 126952 CrossRef CAS PubMed.
- H. Lu, D. J. Chang, B. Baratte, L. Meijer and U. Schulze-Gahmen, J. Med. Chem., 2005, 48, 737–743 CrossRef CAS PubMed.
- S. Timmermann, P. W. Hinds and K. Münger, Cell Growth Differ., 1997, 8, 361–370 CAS.
- G. van Caloen and J.-P. Machiels, Curr. Opin. Oncol., 2019, 31, 122–130 CrossRef CAS PubMed.
- O. Kujan, G. Huang, A. Ravindran, M. Vijayan and C. S. Farah, J. Oral Pathol. Med., 2019, 48, 560–565 CrossRef PubMed.
- L. Yao, D. Bojic and M. Liu, J. Pharm. Anal., 2023, 13, 960–967, DOI:10.1016/j.jpha.2023.06.001.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |