Open Access Article
Open Access ArticleDNA dendrimer-based nanocarriers for targeted Co-delivery and controlled release of multiple chemotherapeutic drugs†
Yao Tian‡
ab,
Mengqiu Sun‡ ac,
Rui Songab,
Zhaoqi Yang
ac,
Rui Songab,
Zhaoqi Yang *d and
Hao Zhang
*d and
Hao Zhang *abe
*abe
aSchool of Physical Sciences, Great Bay University, Dongguan 523000, China. E-mail: zhanghao@gbu.edu.cn
bSchool of Chemistry and Chemical Engineering, Northwestern Polytechnical University, Xi'an 710129, China
cDepartment of Materials Science and Engineering, Southern University of Science and Technology, Shenzhen 518055, China
dSchool of Life Sciences and Health Engineering, Jiangnan University, Wuxi 214122, China. E-mail: zhaoqiyang@jiangnan.edu.cn
eResearch & Development Institute of Northwestern Polytechnical University in Shenzhen, Shenzhen 518063, China
First published on 29th January 2025
Abstract
DNA-based nanomaterials have attracted increasing attention over the past decades due to their incomparable programmability and functionality. In particular, dendritic DNA nanostructures are ideal for constructing drug carriers due to their highly branched structure. In this study, an intelligent drug delivery system was constructed based on DNA dendrimers, in which the DNA duplexes were utilized for simultaneously loading both hydrophilic and hydrophobic small molecule drugs. Additionally, cancer microenvironment-responsive and cancer cell-targeting moieties were introduced into the internal framework and surface of the nanostructures, respectively. Our research shows that these DNA-based drug carriers can enter cancer cells through endocytosis and disintegrate under the reduction of cellular glutathione, thereby achieving targeted co-delivery and controlled release of chemotherapeutic agents and antisense oligonucleotides, providing an effective drug delivery strategy for combined treatment of tumors.
DNA-based nanostructures have been widely used in drug delivery and targeted therapy in recent years due to their diverse structures and functions.1–6 Nucleotide sequence design based on the specific structural properties of nucleic acids can accurately construct complex nanostructures.7–11 From a chemical point of view, DNA is composed of a negatively charged phosphate-sugar backbone with complementary hydrophobic base pairs inside the backbone. These base pairs interact with each other through hydrogen bonds to form a double helix structure. Duplex DNA can interact with various small molecules through π–π stacking and electrostatic forces, which makes DNA have potential application value in drug delivery systems.12–14 In addition, gene drugs such as small interfering RNA, antisense oligonucleotides (ASO), deoxyribozymes (DNAzyme) and cytosine guanosine monophosphate (CpG) oligonucleotides can also be integrated into nucleic acid nanostructures through Watson–Crick base pairing.15–17 These properties make DNA an ideal material for building a new generation of smart drug delivery systems.18 Through rational design, the introduction of aptamers and response-controlled release sites on DNA nanostructures can efficiently transport active drug molecules to biological targets and exert their therapeutic effects, while significantly reducing the cytotoxic effect of drugs on normal cells.19,20
Among all types of DNA nanostructures, DNA dendrimers possess unique properties such as hyperbranched porous conformation, high structural stability, and excellent monodispersity.15,21 Because its outer layer has easy to modify functional sites, DNA dendrimers can accommodate imaging agents, drugs, and targeting ligands simultaneously, thus having significant drug loading and directional drug release capabilities.21 In addition, compared with dendrimers made of other organic or inorganic macromolecules, DNA dendrimers have good biocompatibility, significant cellular internalization ability and biodegradability.15,21,22 Furthermore, when used as drug carriers, the binding between DNA dendrimers and small molecule drugs is usually reversible, which helps DNA dendrimers achieve stimuli responsiveness through sequence design, thereby achieving controlled release of drugs. Through continuous design and improvement of sequences, scientists have continuously developed different categories of stimuli-responsive DNA structures.23–28 Since the concentration of L-glutathione (GSH) in tumor cells and tumor microenvironment is much higher than that in normal tissues, this property has been utilized by many scientists to develop GSH-responsive drug delivery systems for tumor treatment.29,30 On this basis, by modifying the sticky ends of DNA dendrimers with aptamers, DNA dendrimers can recognize the corresponding receptors on the cell surface through aptamers, thereby achieving precise targeted delivery of drugs/genes.29,31–33 Although some drug carriers with DNA dendritic structures have been successfully developed in recent years, most of them can only carry a single drug.34 The main reason is that the physical and chemical properties of different types of drugs vary greatly, such as hydrophilicity/hydrophobicity or small molecules/biomacromolecules. Therefore, different types of drugs may repel each other in the same carrier, thus limiting the function of most drug carriers to the delivery of a single type of drug.35
Here, we designed and synthesized a DNA-based dendritic drug carrier system for the co-delivery of multiple different types of chemotherapeutic drugs. At the same time, the surface of this dendritic nanostructure has multiple sticky ends, which can be used to load targeting recognition groups such as DNA aptamers and nucleic acid-based drugs such as antisense oligonucleotides. At the same time, disulfide modification is introduced between some of the phosphodiester bonds that constitute the DNA dendrimer drug carriers. Once these carriers enter cancer cells, disulfide bonds could be reduced under the action of intracellular GSH, thus causing the release of anticancer drugs from the DNA duplexes to play a therapeutic role. In this study, we successfully co-loaded doxorubicin (Dox), Netropsin (Ntp), and an ASO drug into these DNA dendrimers, achieving targeted co-delivery and responsive release of hydrophobic and hydrophilic chemotherapeutic drugs (Fig. 1).
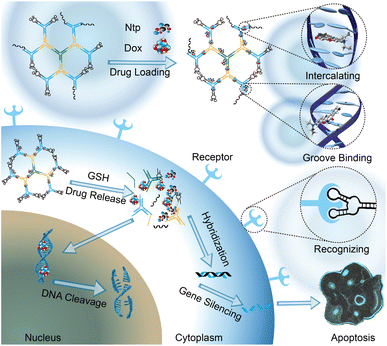 | ||
| Fig. 1 Schematic illustration of the DNA dendrimer-based nanocarriers for targeted co-delivery and controlled release of multiple chemotherapeutic drugs. | ||
Design and synthesis of the drug delivery systems based on DNA dendrimers
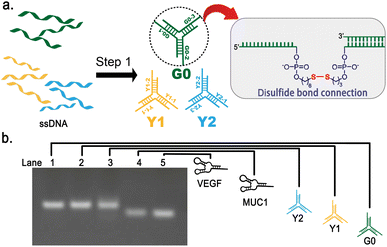
To construct a hierarchical DNA dendrimer structure with controlled drug release capabilities, three Y-shape DNA structures G0, Y1, Y2 were firstly designed and synthesized. The three sticky ends of G0 were identical, while one of the sticky ends of Y1 is designed to be complementary to G0 for the formation of the second layer. The other two sticky ends of Y1 are complementary to Y2 to form the third layer. On this basis, by introducing disulfide bonds into each protruding single strand of the Y-shaped structure (Fig. 2a), the reductive breakage of the DNA single strand under GSH stimulation can be achieved, thereby activating the disintegration of the DNA dendrimer and the controlled release of the drugs. Moreover, to load DNA aptamer onto the dendrimer macromolecules, 13 bases at the 5′ end of aptamer are designed to complement the two identical sticky ends at the outermost layer of DNA Y2.For the construction of Y-shaped DNA structures, three oligonucleotides of equal length were mixed with 50 mM Tris–HCl buffer and 50 mM NaCl in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, incubated at 95 °C for 5 minutes, and cooled to 25 °C over a time period of 40 minutes. After the annealing process, three Y-shaped structure G0, Y1, and Y2 consisting of 13-bp double-stranded central stems and three single-stranded protrusions were formed respectively through base pair hybridization (Fig. 2a).
1, incubated at 95 °C for 5 minutes, and cooled to 25 °C over a time period of 40 minutes. After the annealing process, three Y-shaped structure G0, Y1, and Y2 consisting of 13-bp double-stranded central stems and three single-stranded protrusions were formed respectively through base pair hybridization (Fig. 2a).
To characterize the synthesis of G0, Y1, Y2 and aptamers, the synthesized products were analyzed by agarose gel electrophoresis (Fig. 2b). From the electrophoresis results, the bands shown in Lanes 1–5 indicate that G0, Y1, Y2 and aptamers were successfully synthesized without distinct by-products. The electrophoretic mobility of the bands in Lanes 1–3 are similar and slightly lower than those in Lane 4 and Lane 5. This is consistent with the expectation that G0, Y1 and Y2 have the same number of nucleotide bases, which is greater than that of the aptamers MUC1 and VEGF.
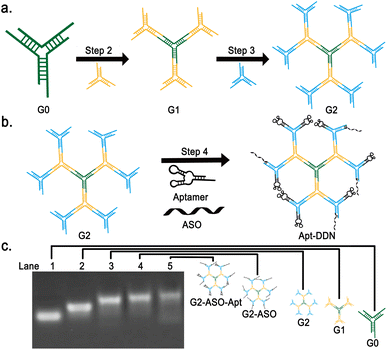
The obtained G0 and Y1 were mixed at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 and incubated at 25 °C for 2 hours to obtain G1. Subsequently, the obtained G1 and Y2 were mixed at a molar ratio of 1
3 and incubated at 25 °C for 2 hours to obtain G1. Subsequently, the obtained G1 and Y2 were mixed at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 under the same condition to synthesize the DNA dendrimer, namely G2 (Fig. 3a). Subsequently, the obtained G2 was functionalized by mixing it with the MUC1 aptamer, VEGF aptamer, and antisense oligonucleotide (ASO) in a 1
6 under the same condition to synthesize the DNA dendrimer, namely G2 (Fig. 3a). Subsequently, the obtained G2 was functionalized by mixing it with the MUC1 aptamer, VEGF aptamer, and antisense oligonucleotide (ASO) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4:4
4:4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratio under identical conditions, resulting in the formation of an aptamer-functionalized DNA dendrimer-based nanocarrier (Apt-DDN) (Fig. 3b).
4 ratio under identical conditions, resulting in the formation of an aptamer-functionalized DNA dendrimer-based nanocarrier (Apt-DDN) (Fig. 3b).
As shown in Lanes 1–3 of Fig. 3c, single band can be clearly observed under 1.5% agarose gel electrophoresis, indicating that G1, G2 and surface functionalized G2 were successfully synthesized (Fig. 3c). These results showed that the overall size of DNA-based dendritic molecules increased with the increase of layers and enrichment of functional elements (aptamers and antisense oligonucleotides), demonstrating the successful synthesis of DNA dendrimers. In addition, it seems that the bands gradually smear with the increase of the number of layers of DNA dendrimers, which suggests that the higher the number of layers, the less yield of the dendrimers can be obtained (Fig. 3c).
Determination of the particle size of DNA dendrimer-based nanocarriers using atomic force microscopy
To characterize the synthesis of DNA dendrimers, sample solutions containing dendritic DNA of various generations were deposited onto a mica surface and subsequently analyzed using tapping mode atomic force microscopy (AFM). AFM observations revealed that DNA dendrimers of each generation were generally dispersed spherical in shape (Fig. 4a). Further analysis gave the average particle diameter of G0, G1, and G2 as 14.398 ± 0.65 nm, 25.153 ± 1.15 nm, 41.671 ± 1.90 nm, respectively, while the average diameter of aptamer-functionalized Apt-DDN was around 53.246 ± 2.43 nm. AFM studies showed that the size of the nanoparticles increased with the number of DNA polymer layers, which was consistent with the design expectations. As a complement to gel electrophoresis, AFM results verified the successful synthesis of dendrimers from another aspect (Fig. 4b).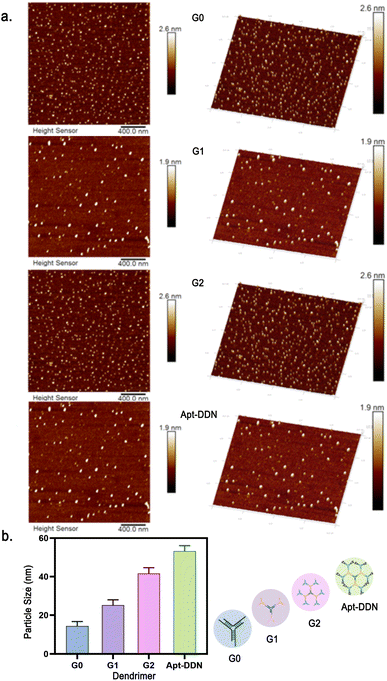 | ||
| Fig. 4 (a) Atomic force microscopic imaging of the DNA-based nanostructures G0, G1, G2, and Apt-DDN by tapping mode. (b) The diameter of DNA-based nanostructures was obtained by NanoScope Analysis. | ||
Verification of GSH-triggered disintegration of the disulfide bond-containing DNA dendrimers
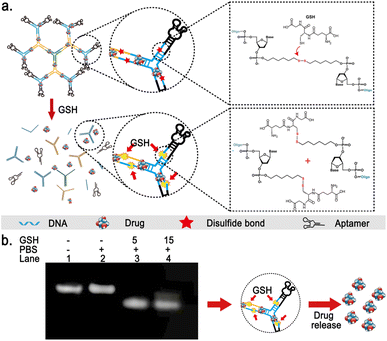
The single strands of each arm of the Y-shaped DNA are modified with disulfide bonds between the phosphodiester bonds near the duplex regions (detailed structures shown in Fig. 5a). These disulfide bonds will theoretically break under the action of high concentrations of reduced glutathione in the tumor microenvironment, triggering the disintegration of the DNA dendritic macromolecular structure, thereby improving the release efficiency of drugs in tumor tissues.In order to evaluate the stability of the synthesized DNA dendrimers under physiological conditions and their responsiveness to GSH, we performed a series of agarose gel electrophoresis experiments on the synthesized Apt-DDN. Results showed that the band intensity of Apt-DDNs after treatment with phosphate-buffered saline is consistent with that of untreated ones, indicating that the DNA dendrimers show good stability under physiological conditions (Fig. 5b). Additionally, when Apt-DDN was incubated in PBS buffer containing 5 mM or 15 mM GSH at 37 °C, the electrophoretic mobility of Apt-DDN was significantly accelerated and the band brightness became smear compared with the untreated DNA dendrimers, suggesting that the modified disulfide bonds in Apt-DDN were reduced by GSH and led to the decomposition of DNA dendrimers to small molecular weight fragments (Fig. 5b).
Determination of the loading capacity and release efficiency of DNA dendrimer-based nanocarriers towards chemotherapeutic drugs
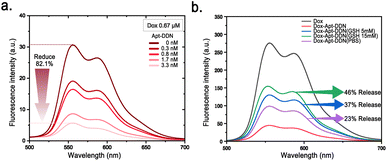
To characterize the drug loading capacity of nanocarriers and their responsiveness to glutathione, fluorescence spectroscopy analysis of the doxorubicin-loaded aptamer-functionalized DNA dendrimer-based nanocarrier (Dox-Apt-DDN) was performed. Doxorubicin (Dox) can intercalate into the double helix structure of cancer cell DNA, disrupting DNA replication and transcription, thereby inhibiting cancer cell proliferation.36 The anthracycline ring within its molecular structure emits a strong red fluorescence under ultraviolet light, making it useful for monitoring drug release behavior.37 When the concentration of doxorubicin was fixed at 0.67 μM, the fluorescence intensity of Dox-Apt-DDN gradually decreased with the increment of DNA dendrimer concentration, indicating that Dox molecules were successfully intercalated in the double helix structure of Apt-DDN and their fluorescence were significantly quenched by their adjacent nucleobases. Fluorescence spectra showed that when 3.3 nM of Apt-DDN was present, the fluorescence intensity decreased by 82.1% compared to Dox alone, demonstrating the great drug loading capacity of the Apt-DDN (Fig. 6a).By analyzing the fluorescence intensity of Apt-DDN samples before and after incubation with different concentrations of GSH (λex = 469 nm), we found that as the GSH concentration increased during the incubation of Apt-DDN, the fluorescence intensity increased as well, which revealed that the drug release was accelerated. Based on the relationship between fluorescence intensity and Dox concentration, we can infer that when the GSH concentration was 5 mM and 15 mM, 37% and 46% of the drug load were released, respectively, which suggested that these DNA-based drug delivery systems are highly sensitive to GSH and can efficiently release drugs under high concentrations of GSH as expected (Fig. 6b).
Studies of the cell internalization efficiency of DNA dendrimer-based nanocarriers
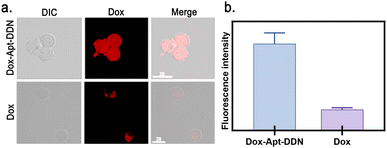
To evaluate whether the constructed Apt-DDN can enhance drug uptake efficiency in living cells, confocal microscopy was used to observe Hela cells after 2 hours of incubation with Dox-Apt-DDN in the presence of doxorubicin (Fig. 7a). The results showed that the fluorescence intensity of Hela cells treated with Dox-Apt-DDN solution was nearly four times than that of the control group, indicating that with the help of the drug carrier Apt-DDN, the cell uptake rate of the anticancer drug doxorubicin was greatly improved compared with itself (Fig. 7b).Examination of the anticancer activity of drug-loaded DNA dendrimer-based nanocarriers
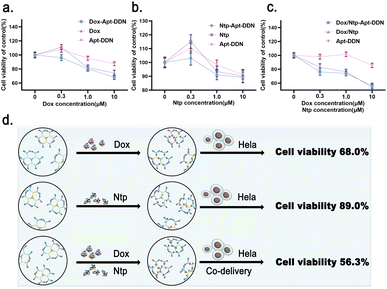
As a commonly used chemotherapeutic agents, netropsin (Ntp) specifically binds to the minor groove of DNA, thereby obstructing the binding of DNA polymerases and transcription factors, which in turn inhibits gene expression.38 On the other hand, Antisense oligonucleotide (ASO) binds complementarily to specific mRNA target sequences, forming RNA-DNA hybrid duplexes that either block the translation process or trigger RNase H-mediated mRNA degradation, thereby reducing the expression of the target gene.39 In this study, the CCK-8 assay was utilized to evaluate the cytotoxicity of Apt-DDN against cancer cell lines. Experimental results showed that the drug-loaded Apt-DDN demonstrated enhanced antitumor activity compared to both the original drug molecule and the ASO-DDN. This indicates that DDN can not only transport therapeutic nucleic acids into tumor cells to exert their effects, but also simultaneously deliver small molecule chemotherapeutic drugs, thereby enhancing the efficacy of tumor cell eradication through combination therapy (Fig. 8a).40When the drug concentration was fixed at 10 μM, the cell viability after treatment with Dox-Apt-DDN, NtP-Apt-DDN, and Dox/NtP-Apt-DDN was 68.0%, 89.0%, and 56.3%, respectively. It can be seen that with the help of DNA-based nanocarriers, the toxicity of two anticancer drugs acting together on target cells is not exactly the sum of the effects of the two drugs acting alone (Fig. 8b). These DNA-based dendrimers is therefore expected to be used for anti-cancer drug screening and drug efficacy evaluation at the cellular level to obtain the optimal combination therapy effects in the future.
Conclusion
In summary, DNA dendrimer-based nanocarriers were successfully prepared for targeted co-delivery and controlled release of multiple chemo-therapeutic drugs in this study. The results showed that Apt-DDNs can achieve targeted drug delivery and controlled drug release through the aptamer on their surface as well as the disulfide bonds incorporated within the DNA backbones. Meanwhile, with efficiently loading of multiple inclusions, these versatile DNA dendrimers achieve targeted co-delivery of various types of anti-cancer drugs (Dox/Ntp/ASO) to overcome single-drug resistance in cancer treatment, which is expected to achieve multimodal treatment on a single platform. The successful synthesis of these versatile DNA dendrimer-based nanocarriers provides a new strategy and tool for tumor treatment, and therefore expected to be further applied in clinical trials of combination therapy.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
Any funds used to support the research of the manuscript should be placed here (per journal style). This research was supported by the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2021A1515220062), the National Natural Science Foundation of China (Grant No. 22405031), Great Bay University (Grant No. YJKY230006), and the Natural Science Foundation of Shaanxi Province (Grant No. 2023-JC-QN-0108).References
- A. Lacroix and H. F. Sleiman, DNA nanostructures: current challenges and opportunities for cellular delivery, ACS Nano, 2021, 15(3), 3631–3645 CrossRef CAS PubMed.
- H. Qi, Y. Xu, P. Hu, C. Yao and D. Yang, Construction and applications of DNA-based nanomaterials in cancer therapy, Chin. Chem. Lett., 2022, 33(3), 1131–1140 CrossRef CAS.
- W. Tang, J. Liu and B. Ding, Nucleic acid nanostructure for delivery of CRISPR/Cas9-based gene editing system, Interdiscip. Med., 2023, 1(1), e20220014 CrossRef.
- T. Wang, Y. Tang, Y. Tao, H. Zhou and D. Ding, Nucleic acid drug and delivery techniques for disease therapy: present situation and future prospect, Interdiscip. Med., 2024, 2(1), e20230041 CrossRef.
- W. Ma, Y. Zhan, Y. Zhang, C. Mao, X. Xie and Y. Lin, The biological applications of DNA nanomaterials: current challenges and future directions, Signal Transduction Targeted Ther., 2021, 6(1), 351 CrossRef CAS.
- Y. Zhu, W. Li, F. Lan, S. Chen, X. Chen and X. Zhang, et al., DNA nanotechnology in tumor liquid biopsy: Enrichment and determination of circulating biomarkers, Interdiscip. Med., 2024, 2(1), e20230043 CrossRef.
- J. Wang, Z. Li and I. Willner, Dynamic reconfigurable DNA nanostructures, networks and materials, Angew. Chem., 2023, 135(18), e202215332 CrossRef.
- R. Li, A. S. Madhvacharyula, Y. Du, H. K. Adepu and J. H. Choi, Mechanics of dynamic and deformable DNA nanostructures, Chem. Sci., 2023, 14(30), 8018–8046 RSC.
- B. Liu, Z. Qi and J. Chao, Framework nucleic acids directed assembly of Au nanostructures for biomedical applications, Interdiscip. Med., 2023, 1(1), e20220009 CrossRef.
- X. Gao, L. Feng, R. Deng, B. Wang, Y. He and L. Zhang, et al., Bottom-up DNA nanostructure-based paper as point-of-care diagnostic: From method to device, Interdiscip. Med., 2024, 2(1), e20230033 CrossRef.
- B. Shen, L. Li, M. Guo, X. Li, Y. Fan and X. Li, et al., Advances in DNA walking nanomachine-based biosensors, Interdiscip. Med., 2024, 2(1), e20230046 CrossRef.
- T. Ramasamy, H. B. Ruttala, S. Munusamy, N. Chakraborty and J. O. Kim, Nano drug delivery systems for antisense oligonucleotides (ASO) therapeutics, J. Controlled Release, 2022, 352, 861–878 CrossRef CAS.
- S. N. Mohammad, Y. S. Choi, J. Y. Chung, E. Cedrone, B. W. Neun and M. A. Dobrovolskaia, et al., Nanocomplexes of doxorubicin and DNA fragments for efficient and safe cancer chemotherapy, J. Controlled Release, 2023, 354, 91–108 CrossRef CAS.
- W.-C. Liao, M. Riutin, W. J. Parak and I. Willner, Programmed pH-responsive microcapsules for the controlled release of CdSe/ZnS quantum dots, ACS Nano, 2016, 10(9), 8683–8689 CrossRef CAS.
- L. A. Kisakova, E. K. Apartsin, L. F. Nizolenko and L. I. Karpenko, Dendrimer-mediated delivery of DNA and RNA vaccines, Pharmaceutics, 2023, 15(4), 1106 CrossRef CAS PubMed.
- Q. Chi, Z. Yang, K. Xu, C. Wang and H. Liang, DNA nanostructure as an efficient drug delivery platform for immunotherapy, Front. Pharmacol., 2020, 10, 1585 CrossRef PubMed.
- Z. Wang, L. Song, Q. Liu, R. Tian, Y. Shang and F. Liu, et al., A tubular DNA nanodevice as a siRNA/chemo-drug co-delivery vehicle for combined cancer therapy, Angew. Chem., 2021, 133(5), 2626–2630 CrossRef.
- Q. Jiang, Y. Shang, Y. Xie and B. Ding, DNA Origami: From Molecular Folding Art to Drug Delivery Technology, Adv. Mater., 2024, 36(22), 2301035 CrossRef CAS PubMed.
- A. Jabbari, E. Sameiyan, E. Yaghoobi, M. Ramezani, M. Alibolandi and K. Abnous, et al., Aptamer-based targeted delivery systems for cancer treatment using DNA origami and DNA nanostructures, Int. J. Pharm., 2023, 123448 CrossRef CAS PubMed.
- C. Ouyang, S. Zhang, C. Xue, X. Yu, H. Xu and Z. Wang, et al., Precision-guided missile-like DNA nanostructure containing warhead and guidance control for aptamer-based targeted drug delivery into cancer cells in vitro and in vivo, J. Am. Chem. Soc., 2020, 142(3), 1265–1277 CrossRef CAS.
- L. Liu, L. Han, Q. Wu, Y. Sun, K. Li and Y. Liu, et al., Multifunctional DNA dendrimer nanostructures for biomedical applications, J. Mater. Chem. B, 2021, 9(25), 4991–5007 RSC.
- J. Le, J. Xu, J. Zheng, B. Li, T. Zheng and Y. Lu, et al., One nanometer self-assembled aptamer-DNA dendrimers carry 350 doxorubicin: Super-stability and intra-nuclear DNA comet tail, Chem. Eng. J., 2020, 388, 124170 CrossRef CAS.
- J. Gao, C. Yang, X. Wu, W. Huang, J. Ye and R. Yuan, et al., A highly sensitive electrochemical biosensor via short-stranded DNA activating LAMP (H+) to regulate i-motif folds for signal transduction, Sens. Actuators, B, 2023, 380, 133354 CrossRef CAS.
- S. Lu, J. Shen, C. Fan, Q. Li and X. Yang, DNA assembly-based stimuli-responsive systems, Advanced Science, 2021, 8(13), 2100328 CrossRef CAS.
- D. Miao, Y. Yu, Y. Chen, Y. Liu and G. Su, Facile construction of i-Motif DNA-conjugated gold nanostars as near-infrared and pH dual-responsive targeted drug delivery systems for combined cancer therapy, Mol. Pharm., 2020, 17(4), 1127–1138 CrossRef CAS.
- M. Nishio, K. Tsukakoshi and K. Ikebukuro, G-quadruplex: Flexible conformational changes by cations, pH, crowding and its applications to biosensing, Biosens. Bioelectron., 2021, 178, 113030 CrossRef CAS.
- I. Serrano-Chacón, B. Mir, Nr Escaja and C. González, Structure of i-motif/duplex junctions at neutral pH, J. Am. Chem. Soc., 2021, 143(33), 12919–12923 CrossRef.
- Q. Wang, S. Yang, F. Li and D. Ling, Nature-inspired K+-sensitive imaging probes for biomedical applications, Interdiscip. Med., 2023, 1(1), e20220004 CrossRef.
- Z. Chen, L. Wan, Y. Yuan, Y. Kuang, X. Xu and T. Liao, et al., pH/GSH-dual-sensitive hollow mesoporous silica nanoparticle-based drug delivery system for targeted cancer therapy, ACS Biomater. Sci. Eng., 2020, 6(6), 3375–3387 CrossRef CAS.
- Y.-F. Chen, M.-W. Hsu, Y.-C. Su, H.-M. Chang, C.-H. Chang and J.-S. Jan, Naturally derived DNA nanogels as pH-and glutathione-triggered anticancer drug carriers, Mater. Sci. Eng. C, 2020, 114, 111025 CrossRef CAS PubMed.
- A. Fraternale, S. Brundu and M. Magnani, Glutathione and glutathione derivatives in immunotherapy, Biol. Chem., 2017, 398(2), 261–275 CrossRef CAS.
- M. P. Gamcsik, M. S. Kasibhatla, S. D. Teeter and O. M. Colvin, Glutathione levels in human tumors, Biomarkers, 2012, 17(8), 671–691 Search PubMed.
- S. Raj Rai, C. Bhattacharyya, A. Sarkar, S. Chakraborty, E. Sircar and S. Dutta, et al., Glutathione: Role in oxidative/nitrosative stress, antioxidant defense, and treatments, ChemistrySelect, 2021, 6(18), 4566–4590 Search PubMed.
- S. Rattanakiat, M. Nishikawa, H. Funabashi, D. Luo and Y. Takakura, The assembly of a short linear natural cytosine-phosphate-guanine DNA into dendritic structures and its effect on immunostimulatory activity, Biomaterials, 2009, 30(29), 5701–5706 CrossRef CAS.
- C. Corsaro, D. Mallamace, G. Neri and E. Fazio, Hydrophilicity and hydrophobicity: Key aspects for biomedical and technological purposes, Phys. A, 2021, 580, 126189 CrossRef CAS.
- M. Kciuk, A. Gielecińska, S. Mujwar, D. Kołat, Ż. Kałuzińska-Kołat and I. Celik, et al., Doxorubicin—an agent with multiple mechanisms of anticancer activity, Cells, 2023, 12(4), 659 CrossRef CAS.
- P. Changenet-Barret, T. Gustavsson, D. Markovitsi, I. Manet and S. Monti, Unravelling molecular mechanisms in the fluorescence spectra of doxorubicin in aqueous solution by femtosecond fluorescence spectroscopy, Phys. Chem. Chem. Phys., 2013, 15(8), 2937–2944 RSC.
- H. M. Berman, S. Neidle, C. Zimmer and H. Thrum, Netropsin, a DNA-binding oligopeptide structural and binding studies, Biochim. Biophys. Acta Nucleic Acids Protein Synth., 1979, 561(1), 124–131 CrossRef CAS.
- E. C. Kuijper, A. J. Bergsma, W. P. Pijnappel and A. Aartsma-Rus, Opportunities and challenges for antisense oligonucleotide therapies, J. Inherited Metab. Dis., 2021, 44(1), 72–87 CrossRef CAS PubMed.
- L. Cai, X. Qin, Z. Xu, Y. Song, H. Jiang and Y. Wu, et al., Comparison of cytotoxicity evaluation of anticancer drugs between real-time cell analysis and CCK-8 method, ACS Omega, 2019, 4(7), 12036–12042 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures including gel electrophoresis, AFM characterization, fluorescence spectroscopy, cytotoxicity assays, confocal images of cells, DNA sequence details. See DOI: https://doi.org/10.1039/d4ra07839j |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |