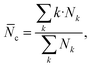Open Access Article
Open Access ArticleA dual antibacterial action of soft quaternary ammonium compounds: bacteriostatic effects, membrane integrity, and reduced in vitro and in vivo toxicity†
Doris Crnčevićab,
Lucija Krcec,
Zlatko Brkljačad,
Mislav Cvitković c,
Sanja Babić Brčićef,
Rozelindra Čož-Rakovacef,
Renata Odžaka and
Matilda Šprung
c,
Sanja Babić Brčićef,
Rozelindra Čož-Rakovacef,
Renata Odžaka and
Matilda Šprung *a
*a
aUniversity of Split, Faculty of Science, Department of Chemistry, R. Bošković 33, Split, Croatia. E-mail: dcrncevic@pmfst.hr; rodzak@pmfst.hr; msprung@pmfst.hr
bUniversity of Split, Faculty of Science, Doctoral Study in Biophysics, R. Bošković 33, Split, Croatia
cUniversity of Split, Faculty of Science, Department of Physics, R. Bošković 33, Split, Croatia. E-mail: lucija.krce@pmfst.hr; mcvitkovi@pmfst.hr
dSelvita Ltd., Prilaz Baruna Filipovića 29, Zagreb, Croatia. E-mail: zlatko.brkljaca@selvita.com
eRuđer Bošković Institute, Laboratory for Biotechnology in Aquaculture, Division of Materials Chemistry, Bijenička 54, Zagreb, Croatia. E-mail: sanja.babic@irb.hr; rrakovac@irb.hr
fCenter of Excellence for Marine Bioprospecting (BioProCro), Ruđer Bošković Institute, Bijenička 54, Zagreb, Croatia
First published on 16th January 2025
Abstract
Quaternary ammonium compounds (QACs) have served as essential antimicrobial agents for nearly a century due to their rapid membrane-disrupting action. However, the emergence of bacterial resistance and environmental concerns have driven interest in alternative designs, such as “soft QACs”, which are designed for enhanced biodegradability and reduced resistance potential. In this study, we explored the antibacterial properties and mechanisms of action of our newly synthesized soft QACs containing a labile amide bond within a quinuclidine scaffold. Our findings revealed that these compounds primarily exhibit a bacteriostatic mode of action, effectively suppressing bacterial growth even at concentrations exceeding their minimum inhibitory concentrations (MICs). Unlike traditional QACs, fluorescence spectroscopy and microscopy demonstrated membrane preservation during treatment, with reduced membrane integration compared to cetylpyridinium chloride (CPC), as corroborated by parallel artificial membrane permeability assays. Additionally, molecular dynamics simulations revealed “hook-like” conformations that limit lipid bilayer penetration and promote the formation of larger aggregates, reducing their effective concentration and minimizing cytotoxic effects. Interestingly, secondary antibacterial mechanisms, including inhibition of protein synthesis, were observed, further enhancing their activity. Zebrafish embryotoxicity and in vitro cytotoxicity studies confirmed significantly lower toxicity compared to CPC. By addressing limitations associated with conventional QACs, including toxicity, resistance, and environmental persistence, these soft QACs provide a promising foundation for next-generation antimicrobials. This work advances the understanding of QAC mechanisms while paving the way for safer, eco-friendly applications in healthcare, agriculture, and industrial settings.
1 Introduction
Quaternary ammonium compounds (QACs) are a class of cationic surfactants that have been employed for nearly a century across a wide range of industries, including healthcare, agriculture, and the production of household cleaning supplies.1–3 Their most prominent characteristic is antibacterial efficacy, first identified in the early 1930s with benzalkonium chloride (BAC), which quickly became a key active ingredient in disinfectants and surface sanitizers commonly used in hospital settings.4–6 The antibacterial activity of QACs is primarily attributed to their distinct structural elements, which facilitate interactions with bacterial cell membrane components. Although the precise mechanism of action is not fully understood, it is proposed that QACs act as membranolytic agents in a few steps. The electrostatic interaction of QAC's positively charged nitrogen atom and negatively charged residues of teichoic and lipoteichoic acids within the peptidoglycan matrix and lipid bilayer causes initial membrane destabilization.7 This is further followed by the insertion of hydrophobic substituents into the membrane matrix causing the leakage of cytoplasmic contents and ultimately leading to cell lysis.8–10 In addition to their role as “biological detergents” that act on biological membranes, some QACs have also demonstrated antibacterial activity by targeting intracellular components, such as DNA or pyridoxal-dependent enzymes targting.11–13Due to their strong antibacterial activity, the demand for these compounds has surged, particularly during the SARS-CoV-2 pandemic which was followed by an urgent need for effective topical antiseptic formulations.14 At the same time, the inherent chemical stability of commercially available QACs has become a subject of considerable concern within the scientific community, as these compounds have the potential to simultaneously activate bacterial resistance mechanisms and pose significant risks to human health.15–17 These findings underscore the need for further new-generation QACs development, exploration of their antibacterial mechanism of action, as well as a comprehensive investigation of their structure–activity relationship.
In pursuit of environmentally friendly yet biologically effective QAC variants, contemporary synthetic strategies involve the functionalization of the compound backbone with hydrolysable groups, such as ester or amide, enabling the controlled degradation of so-called “soft QACs”. Among these, QACs with amide groups are considered promising new derivatives, as ester-containing QACs tend to self-degrade, diminishing the reliability of their antibacterial effectiveness.18–20 We have previously reported the new soft amido-QACs prone to protease degradation derived from heterocyclic backbone of naturally occurring quinuclidine (Scheme 1).21 The assessment of their biological activity, ranging from non-active to moderately active structures (Scheme 1) led us to hypothesize that the polar amide functional group at the C-3 atom of the quinuclidine scaffold significantly impacts the antibacterial efficacy of the new QAC derivatives potentially hindering the electrostatic interactions with the bacterial membrane.
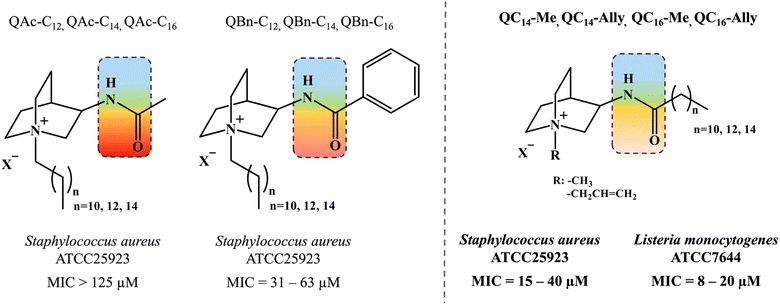 | ||
| Scheme 1 Previous investigations on soft quaternary ammonium compounds (QACs) derived from heterocyclic backbone of quinuclidine taken from ref. 21. Amide functional group is located at third carbon atom of quinuclidine scaffold depicted in the colored squares reflecting the polar character. Different substituents in extension of amide functionality (QAc – acetamide, QBn – benzamide and QCn – aklylamide) were found to exhibit differential antimicrobial activity. | ||
In this study, we provide a comprehensive investigation of the antibacterial mechanisms and structure–activity relationships of four newly synthesized soft QACs, incorporating a polar amide group within a quinuclidine scaffold. Unlike conventional QACs, these compounds exhibit a dual mode of action, combining membrane interaction with protein synthesis inhibition, which may help address bacterial resistance. Time-kill assays revealed a bacteriostatic effect, while microscopy and fluorescence spectroscopy confirmed preserved membrane integrity at their respective minimum inhibitory concentrations (MICs).
Molecular dynamics (MD) simulations revealed that these compounds adopt unique “hook-like” conformations that limit membrane penetration compared to the commercial standard, CPC, a finding supported by parallel artificial membrane permeability assays (PAMPA). This reduced interaction correlates with significantly lower cytotoxicity and embryotoxicity, as demonstrated through in vitro and in vivo testing.
By linking structure to activity, this study advances the understanding of QAC mechanisms and highlights the potential of these soft QACs as safer, environmentally friendly antimicrobials for applications in healthcare and beyond.
2 Results and discussion
2.1. Bacteriostatic activity
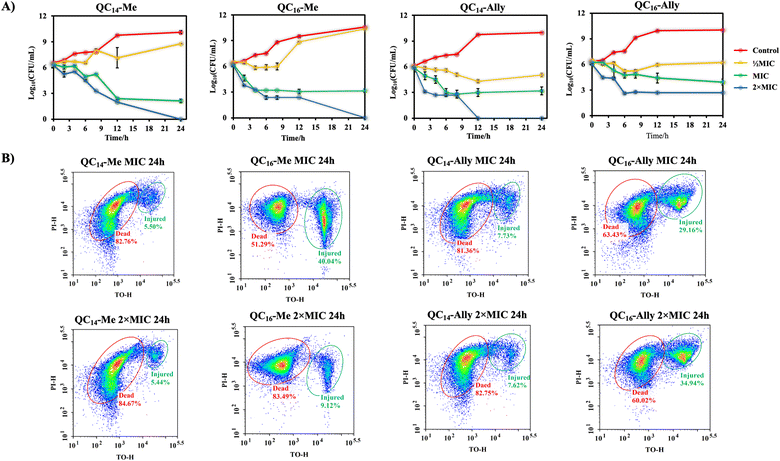
Examination of bacterial viability during the treatment over a certain incubation time provides initial insight into the mode of action of the tested agent. Namely, antibacterial agents can generally exhibit bactericidal or bacteriostatic effect.22,23 While bactericidal agents kill 99.99% of cells during a 24 hours treatment, the bacteriostatic mode of action implies keeping the population in the stationary growth phase without directly causing cell death. Due to their membranolytic potency, quaternary ammonium compounds (QACs) are most known as bactericidal agents causing almost immediate membrane damage and cell death. Although some QACs are also prone to cellular uptake followed by DNA or proteins targeting, such antibacterial activity requires further exploration.12,13,24,25The cell viability of Staphylococcus aureus ATCC25923 during the 24 hours treatment at ½MIC, MIC and 2 × MIC is shown in Fig. 1. Time-resolved curves of log10 of colony forming units per mililiter (CFU mL−1) point out subtle differences between methyl and allyl analogues of QAC candidates at ½MIC treatment. The results show that methylated QACs (QC14-Me and QC16-Me) keep cells in stationary phase for six hours, in contrast to more effective allyl analogues (QC14-Ally and QC16-Ally) that manage to stop cells from dividing during the entire incubation time. Treatment with concentrations of candidate compounds corresponding to MIC, despite resulting in diminished log10 (CFU mL−1) over time, surprisingly revealed the same effect of each compound on bacterial population. Namely, after six hours of MIC treatment with candidate compounds, S. aureus ATCC25923 population was kept in the stationary phase with no indicative cell division. Furthermore, the drop in the number of bacterial cells of ≤3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 (CFU mL−1) after 24 hours of exposure further supported that these compounds are indeed bacteriostatic agents that rather inhibit bacterial growth without damaging bacteria to the extent of cell death. On the contrary, treatment with 2 × MIC resulted in almost complete bacterial eradication upon 12 or 24 hours of treatment with only QC16-Ally displaying reduced potential to annihilate bacteria as further confirmed by flow cytometry (Fig. 1B).
log10 (CFU mL−1) after 24 hours of exposure further supported that these compounds are indeed bacteriostatic agents that rather inhibit bacterial growth without damaging bacteria to the extent of cell death. On the contrary, treatment with 2 × MIC resulted in almost complete bacterial eradication upon 12 or 24 hours of treatment with only QC16-Ally displaying reduced potential to annihilate bacteria as further confirmed by flow cytometry (Fig. 1B).
These results clearly show that even at prolonged MICs exposure (24 h) at least half of the population is dead with a similar fraction of injured cells that can potentially recover in favorable conditions. It seems those candidates bearing C14 alkyl chains are better than candidates with longer chains which can be due to several reasons. Firstly, QACs with longer chains tend to be less soluble, or secondly, they tend to form larger aggregates that diminish their antibacterial potential. This trend is visible at 2 × MIC, which also did not result in complete bacterial eradication even at 24 h of exposure to the antibacterial agent. However, as opposed to MIC treatment, treatment with 2 × MIC clearly shows a larger fraction of dead cells, leaving just about 5–10% of injured cells that can potentially recover. Together, these results confirm our initial observation about a bacteriostatic mode of action which provides a new insight into the different mechanism that has not been documented for these compounds up until now.
2.2. Membranolytic activity
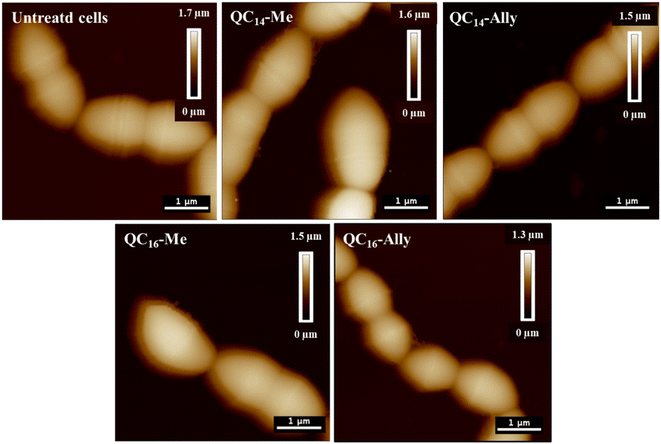
The untreated L. monocytogenes cells, shown in Fig. 2, exhibit the characteristic densely packed rod-shaped morphology, even post-fixation.
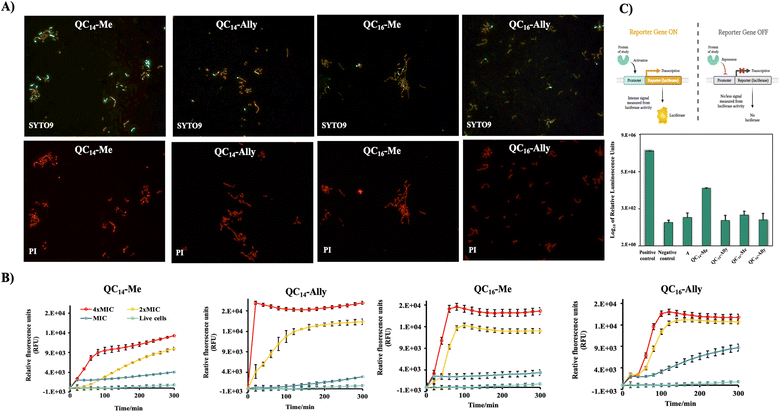
Treatment with 2 × MIC concentrations of QC14-Me, QC14-Ally, QC16-Me, and QC16-Ally resulted in no detectable loss of distinct cell morphology, although a consistent increase in cell height was observed across all treated cells. Furthermore, a 3 hours long exposure to 2 × MIC of these compounds did not induce significant membrane damage, which aligns with prior findings in Staphylococcus aureus, another Gram-positive representative. Instead, the cell membranes in treated L. monocytogenes cells appeared preserved, with only minor irregularities along the cell edges. To further investigate membrane integrity, both bacterial strains were subjected to optical fluorescence microscopy and fluorescence spectroscopy analysis.
Furthermore, the uptake of PI by Staphylococcus aureus ATCC25923 cells in treatment was measured spectrofluorimetically at different time intervals over six hours of treatment with 4 × MIC, 2 × MIC and MIC concentrations of the selected compounds (Fig. 3B). The treatment with 4 × MIC and 2 × MIC concentrations showed the same effect for all tested QACs. An almost immediate steep increase of the relative fluorescent units (RFUs) of PI suggested pronounced membrane disruption. On the other hand, at concentrations corresponding to MIC, RFUs were slightly higher for all compounds when compared to the untreated control cells indicating an absence of pronounced membrane damage over six-hours long period of treatment.
Obtained results underlined the previously observed implication of the bacteriostatic activity of QC14-Me, QC14-Ally, QC16-Me and QC16-Ally suggesting the need for further investigation of their mode of action through alternative pathways, primarily including the inactivation of the protein synthesis pathways.
2.3. Inhibition of protein synthesis
Antibacterial agents are generally classified in two large groups regarding their mechanism of combating pathogens. Bactericidal agents, such as carbapenems and polymyxins, are known for their membrane and cell wall targeting effect.28 Besides membrane disorganization, bactericidal agents can also act as blockers of DNA replication or inhibitors of cell wall synthesis.29,30 In both cases the result is extermination of the entire population during 24 hours long incubation time. On the other hand, bacteriostatic activity implies prevention of bacterial division resulting in the stationary growth phase of the population in treatment which is attributed to the potency of protein synthesis inhibition.23,31 In most cases, this effect is caused by irreversible binding to the 30S ribosomal subunit which is a common pattern for aminoglycosides, such as kanamycin and spectinomycin 32.Quaternary ammonium compounds (QACs) are categorized as potent membranolytic agents, especially when it comes to Gram-positive pathogens given the simplicity of membrane composition in contrast to Gram-negative bacterial isolates.8,10 The unique structure of QAC's scaffold comprised of permanently positively charged nitrogen plays a pivotal role in their proposed mechanism of antibacterial action, exploiting electrostatic interaction with components of bacterial cell membrane. Further membrane disorganization is achieved by incorporating hydrophobic substituents into the membrane matrix resulting in membrane perforation and ultimately cell lysis. Although QACs are known for their membrane-targeting antibacterial approach, some QACs employ different antibacterial potency acting on intracellular components such as DNA and/or proteins.12,33,34
Previously observed bacteriostatic effect prompted us to examine the potential of QC14-Me, QC14-Ally, QC16-Me and QC16-Ally to inhibit the protein synthesis pathways. For this purpose, we employed an assay based on in vitro synthesis of luciferase protein – expression product of a reporter sequence under the control of T7 promoter. If the expression is successful, luciferase protein degrades its natural substrate luciferin to the product coelenteramide followed by the emission of an intense luminescent signal expressed in relative luminescence units (RLUs). In contrast, if the addition of the tested compound prevents the transcription of protein of interest, a luminescent signal is absent.
The reaction mixture that served as a positive control for luciferase protein expression, apart from T7 extract and S30 ribosomal mixture, contained nuclease-free water and a DNA template that contained luciferase reporter gene downstream of the T7 promoter site. On the other hand, the composition of the negative control differed in absence of the DNA template, preventing the expression of luciferase and therefore emission of luminescent signal. The effect of tested compounds QC14-Me, QC14-Ally, QC16-Me and QC16-Ally on in vitro luciferase protein expression was compared with the effect of kanamycin which is known to act as the protein synthesis inhibitor employing a bacteriostatic mode of action.35 Fig. 3C shows RLUs detected in each reaction mixture upon one hour of incubation. As expected, the highest RLUs were observed in positive control, indicating a successful expression of luciferase protein. When compared to positive control, all the tested candidate compounds showed 100–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold decrease in RLUs, pointing out the absence of expressed luciferase. This is further confirmed in comparison with the RLUs detected in negative control and kanamycin respectively. Furthermore, QC14-Ally and QC16-Ally exhibited greater inhibitory effects in regard to methylated QACs, which is consistent with previously observed trend in biological activity of selected candidate compounds. The evaluation of their ability to inhibit protein synthesis pathways indicates a potential new antibacterial mechanism for selected QACs, simultaneously highlighting a promising direction for future research into the mode of action of this class of compounds.
000-fold decrease in RLUs, pointing out the absence of expressed luciferase. This is further confirmed in comparison with the RLUs detected in negative control and kanamycin respectively. Furthermore, QC14-Ally and QC16-Ally exhibited greater inhibitory effects in regard to methylated QACs, which is consistent with previously observed trend in biological activity of selected candidate compounds. The evaluation of their ability to inhibit protein synthesis pathways indicates a potential new antibacterial mechanism for selected QACs, simultaneously highlighting a promising direction for future research into the mode of action of this class of compounds.
2.4. Membrane interaction analysis
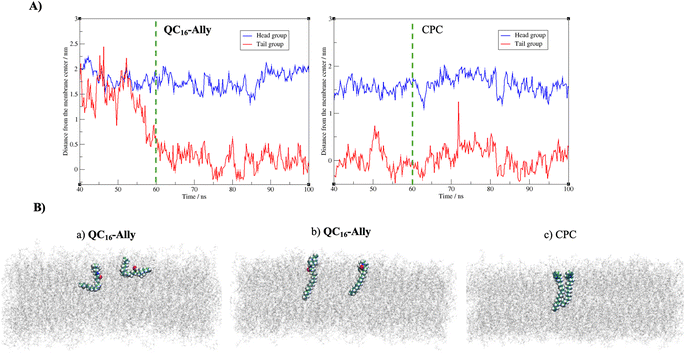
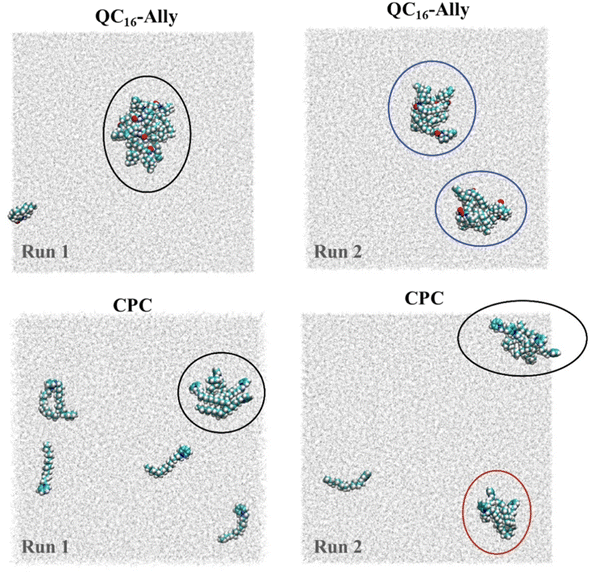
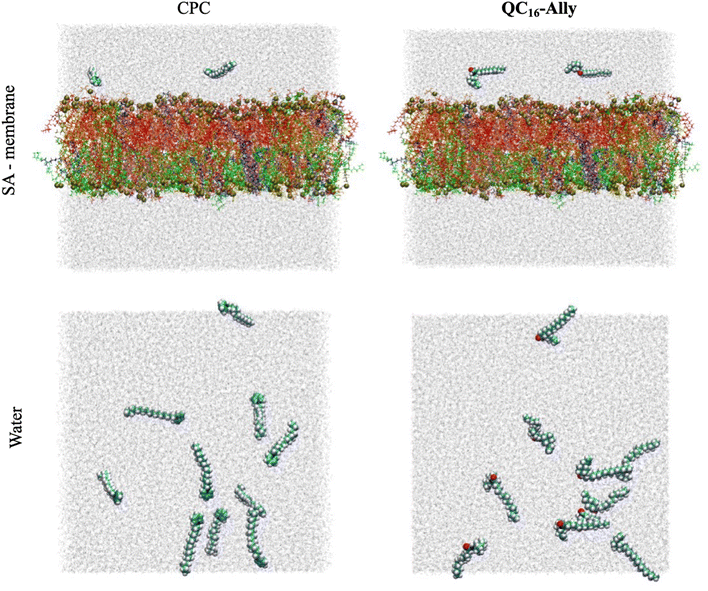
Fig. 4A reveals that both compounds incorporate into the membrane rather swiftly, likely due to their long hydrophobic tails. CPC integrates into the membrane fully by approximately 40 ns (and partially as early as 20 ns, data not shown), whereas QC16-Ally requires about 60 ns to achieve full integration, suggesting that CPC may penetrate the membrane more readily than QC16-Ally. Additionally, we compared the average positions of the head and tail groups of each compound, focusing on the behavior after 60 ns – when both compounds are fully incorporated (highlighted in Fig. 4A) to the right of the green dashed line. While the distance of the tail groups of CPC and QC16-Ally from the membrane center is rather similar, a notable difference can be observed in the positioning of their head groups. Specifically, the head group of CPC is located 16.0 Å from the membrane center, whereas this distance for QC16-Ally is 17.8 Å. This indicates that the head group of CPC penetrates deeply into the membrane compared to QC16-Ally which seems to protrude more prominently toward the membrane: water interface. This might be explained by the favorable interactions of the polar amide group with the water molecules at the membrane surface.
Next, we examined the conformational behavior of CPC and QC16-Ally within the model membrane. QC16-Ally occupies a significantly broader conformational phase space than CPC, adopting both “hook” or “L-shaped” and elongated conformations (Fig. 4B). In contrast, CPC shows a much more restricted phase space, predominantly existing in the elongated form (Fig. 4B). To quantify these observations, we monitored the distance (dAC) between the terminal carbon atom and the C-2 atom of the aliphatic chain over the last 40 ns (see Fig. S1†). In QC16-Ally, 40% of the conformations had a dAC of less than 14.5 Å whereas dAC < 14.5 Å is regarded as approximate cut-off distance indicating the “L-shaped” form. On the contrary, only about 13% of CPC conformations met this criterion with majority of conformers displaying dAC ≥ 14.5 Å indicating elongated conformations. These results clearly show that both CPC and QC16-Ally exist in two conformations namely elongated and “L-shaped”, with significantly higher percentage of “L-shaped” conformers in QC16-Ally (40%) compared to CPC (13%).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 water molecules and chloride counterions, with the molecules randomly placed using Packmol36 (Fig. 10, bottom panel). To monitor the potential aggregation, several aggregation-related metrics were calculated, namely the number of clusters and the average cluster size as the functions of time, and a histogram of cluster sizes. These indicators allow us to observe both how the systems approach equilibrium and the typical size of aggregates formed.37 Molecules were considered part of the same cluster if the minimum distance between them was less than 3.5 Å. The average cluster size,
000 water molecules and chloride counterions, with the molecules randomly placed using Packmol36 (Fig. 10, bottom panel). To monitor the potential aggregation, several aggregation-related metrics were calculated, namely the number of clusters and the average cluster size as the functions of time, and a histogram of cluster sizes. These indicators allow us to observe both how the systems approach equilibrium and the typical size of aggregates formed.37 Molecules were considered part of the same cluster if the minimum distance between them was less than 3.5 Å. The average cluster size, ![[N with combining macron]](https://www.rsc.org/images/entities/i_char_004e_0304.gif) c, was calculated as
c, was calculated aswhere Nk is the number of clusters containing k molecules. The average cluster size is calculated from k = 2, i.e., the monomers are purposely not accounted for.37 On the other hand, the histogram of cluster sizes was obtained by averaging the number of clusters of k molecules over the last 50 ns of respective simulation.
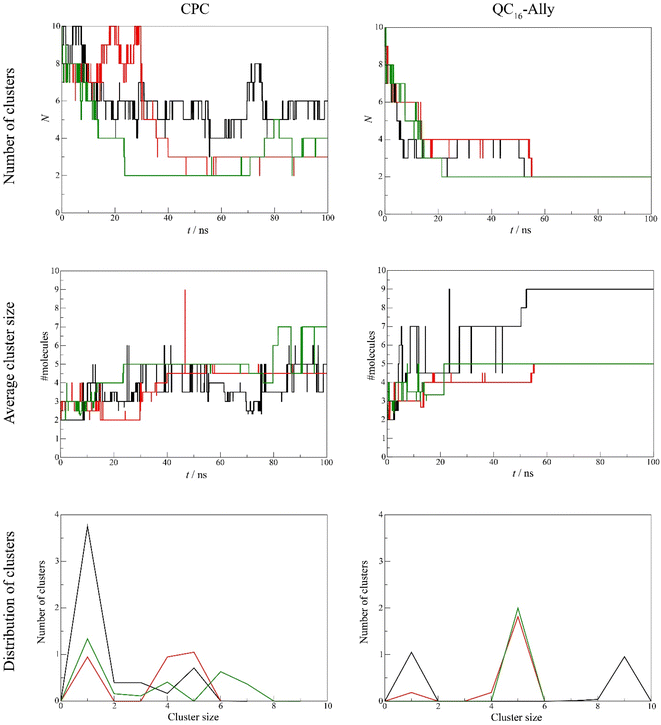
Our analysis revealed clear differences in aggregation tendencies between commercial standard CPC, and QC16-Ally, with our candidate compound exhibiting greater tendency to form clusters. The number of QC16-Ally clusters dropped to ≤4 within 20 ns and stabilized at 2 clusters by 50 ns across all three simulations. In contrast, CPC does not reach such convergence, with cluster numbers ranging from 3 to 6 throughout the simulations. Similarly, QC16-Ally forms larger and more stable clusters reaching up to 9 molecules, in contrast to CPC with approximately 7 molecules in aggregate (Fig. 5).
Regarding cluster morphology, QC16-Ally exhibited more spherical and thus more stable clusters, whereas CPC predominantly formed elongated, less defined clusters (Fig. 6).
2.5. In vitro and in vivo toxicity
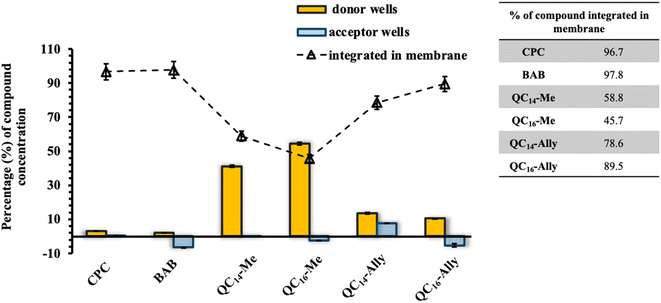
Given the potential applications of our synthesized soft QAC derivatives, we aimed to further evaluate their toxicity on two healthy human cell lines, retinal pigment epithelial (RPE1) and keratinocytes (HaCaT). The in vitro half maximal inhibitory concentration, IC50, values obtained were compared to the toxicity profiles of commercially available QACs, cetylpyridinium chloride (CPC) and dimethyldodecylbenzylammonium bromide (BAB) respectively. As depicted in Fig. 8A, the commercial QACs, CPC and BAB, exhibit cytotoxic activity towards both RPE1 and HaCaT cell lines at low, single digit micromolar concentrations, corresponding to their MIC values. In contrast, the synthesized derivatives, QC14-Me, QC14-Ally, QC16-Me and QC16-Ally, demonstrated reduced toxicity compared to CPC and BAB, with QC14-Me exhibiting the lowest cytotoxicity against both cell lines. Coupled with their low minimal inhibitory concentration (MIC) and the potential for degradation into non-toxic products, these findings suggest that our soft QACs could be considered as promising candidates for the development of new QACs that are safe for both human health and the environment.
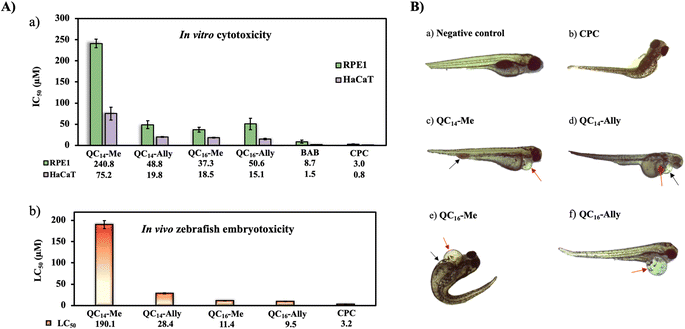 | ||
| Fig. 8 (A) (a) Cytotoxicity of candidate compounds QC14-Me, QC14-Ally, QC16-Me and QC16-Ally towards healthy human cell lines retinal pigment epithelial (RPE1) and keratinocytes (HaCaT). Obtained values of half maximal inhibitory concentration, IC50, (μM) were compared to the commercially available standards dimethyldodecylbenzylammonium bromide (BAB) and cetylpyridinium chloride (CPC) respectively. (b) Graphical representation of lethal concentration of 50% (LC50) Danio rerio embryos with table containing corresponding LC50 values. Determined LC50 values are within 95% confidence interval (Fig. S2†). (B) Representation of morphological abnormalities upon treatment with concentration higher than LC50 for each tested compound: (a) negative control – a normally developed individual without morphological abnormalities, (b) CPC – scoliosis, (c) QC14-Me – pericardial edema (red arrow), blood accumulation in the tail area (black arrow); (d) QC14-Ally – pericardial edema (black arrow), yolk sac edema (red arrow); (e) QC16-Me – pericardial edema (red arrow), yolk sac edema (black arrow), scoliosis; (f) QC16-Ally – pericardial edema (red arrow), scoliosis. | ||
3 Conclusions
This study provides new insights into the bacteriostatic properties and dual mechanisms of action of soft quaternary ammonium compounds (QACs) designed with a labile amide bond in the quinuclidine scaffold. In contrast to traditional QACs, which rapidly kill bacteria by disrupting membranes, our findings indicate that these soft QACs primarily inhibit bacterial growth through a combination of membrane interaction and protein synthesis interference. This dual mode of action positions them as promising alternatives for addressing challenges associated with bacterial resistance.Key experiments, including time-kill assays and membrane integrity analyses, demonstrated that our soft QACs primarily maintain bacterial populations in a stationary phase without immediate cell death. Molecular dynamics (MD) simulations further revealed their unique “hook-like” conformations that restrict deep membrane penetration and facilitate aggregate formation, reducing their effective concentration at the membrane surface. This reduced permeability aligns with their lower cytotoxicity, as validated by zebrafish embryotoxicity assays and human cell line studies, highlighting their safety advantages over conventional QACs. Furthermore, evidence of protein synthesis inhibition underscores their ability to target intracellular bacterial processes, providing a secondary antibacterial mechanism.
These findings emphasize the potential of soft QACs as safer, environmentally friendly alternatives for antimicrobial applications. While the current study establishes their antibacterial properties and safety profile, further investigations could focus on optimizing their chemical scaffolds to enhance antibacterial potency while retaining biodegradability and low toxicity. Expanding efficacy testing against a broader range of bacterial strains, including antibiotic-resistant pathogens, would strengthen their relevance as next-generation antimicrobials. Additionally, exploring their potential for synergistic combinations with existing antimicrobials could help improve overall treatment efficacy while mitigating resistance development.
Beyond their antimicrobial properties, future research should address the environmental degradation pathways of soft QACs to confirm their eco-friendly nature under real-world conditions. Moreover, efforts to incorporate these compounds into practical formulations, such as surface disinfectants, medical device coatings, and personal care products, will be essential to bridge the gap between laboratory findings and industrial applications.
The reduced toxicity, biodegradability, and dual antibacterial activity demonstrated by these soft QACs underscore their promise as innovative, sustainable solutions to the challenges posed by bacterial resistance and environmental concerns. These insights provide a strong foundation for further development and highlight the potential for soft QACs to play a pivotal role in advancing safer, more effective antimicrobials across healthcare, agriculture, and industry.
4 Materials and methods
4.1. Viability of bacterial cells in treatment over time (time-kill assay)
The time kill assay was performed with a representative Gram-positive bacterium, Staphylococcus aureus ATCC25923. The overnight culture was diluted in fresh temperature-controlled Mueller–Hinton broth (MHB) and propagated in a shaking incubator at the temperature of 37 °C with the constant rotation at 220 rpm. The pre-exponentially grown culture was diluted to a final concentration of 5 × 105 CFU mL−1 and an aliquot of 50 μL of the prepared cell culture suspension was added to the 96-well microtiter plate containing 50 μL of the compounds to be tested at final concentrations corresponding to the minimum inhibitory concentration (MIC) and 2 × MIC. The untreated S. aureus ATCC25923 cells served as growth control. At the start of the experiment, after the addition of the cell culture suspension, a 10 μL aliquot was pipetted from each well, diluted in 990 μL MHB and further serially diluted up to 106-fold. A 20 μL aliquot of each cell suspension dilution was plated on MHB agar using a sterile loop. The 96-well plate containing the treated cells was incubated at 37 °C and the previously described dilution and plating of the treated culture was repeated at the desired time intervals (2, 4, 6, 8, 12 and 24 hours). The Petri dishes were incubated for 24 hours. The bacterial colonies formed were then counted and the number of viable cells after treatment was calculated at each time interval. The results were plotted graphically as log10 of colony forming units per mililiter (CFU mL−1) versus time.4.2. Flow cytometry measurement of the cells in treatment
The cell viability of the representative Gram-positive bacterium Staphylococcus aureus ATCC25923 during treatment with candidate compounds was further tested by flow cytometry (NovoCyte Advanteon). Two overnight cultures were diluted 10 times and incubated in a shaking incubator at 220 rpm and the temperature of 37 °C. Pre-exponentially grown cells were centrifuged at 4500 g for 10 minutes at room temperature. After discarding the supernatant of the culture medium, one cell pellet was resuspended in an equal volume of the staining buffer (phosphate buffer, pH = 7.4, 1 mM EDTA, 0.1% Tween-20) and the other in an equal volume of the absolute ethanol. The cell pellet to which absolute ethanol has been added was incubated for 30 minutes and then centrifuged again under the same conditions. After centrifugation, the supernatant of absolute ethanol was discarded, and the cell pellet was resuspended in the staining buffer. Both prepared cultures were diluted in the desired volume of staining buffer to the final cell concentration of 1 × 106 colony forming units per mililiter (CFU mL−1) and served as fluorescently labeled compensation specimens along with the same concentration of unstained cells. BD Biosciences (BD™ Cell Viability) kit containing two fluorescent dyes (thiazole orange, TO and propidium iodide, PI) was used for cell labelling and detection according to the manufacturer's instructions. 50 μL cell suspension in staining buffer was added to the wells of 96-well plate containing tested compounds in staining buffer in MIC and 2 × MIC final concentrations and incubated at 37 °C. At the desired time intervals, the treated cells were stained with a mixture of TO and PI dyes followed by flow cytometric detection of cell viability.4.3. Atomic force microscopy (AFM)
29.5 μL of the Cell-Tak solution in 0.1 M NaHCO3, pH = 8.00 was pipetted into the center of a sterile Petri dish and incubated in the laminar flow hood for 30 minutes. Petri dishes were further washed with at least ten portions of mQ water. After thorough rinsing, the Petri dishes were air dried in a sterile laminar flow hood for about 40 minutes, or longer if necessary. The overnight culture of Listeria monocytogenes ATCC7644 was diluted 10-fold and propagated in culture medium in a shaking incubator at 35 °C and 180 rpm. After thirty minutes, an aliquot of the culture was added to the previously coated surface of the Petri dish and incubated at 35 °C for 30 minutes. After incubation, the Petri dish was thoroughly rinsed with fresh culture medium and then with the sterile phosphate buffer (PBS). The cells were fixed with a 2.5% solution of glutaraldehyde (GT) in PBS for a total of four hours at 4 °C. After fixation and rinsing in sterile PBS, the untreated cells were measured using the atomic force microscope (AFM). To image bacterial cells treated with candidate compounds using the AFM, a solution of the corresponding compound was added to the cells in the culture medium before the fixation, so that the final concentration of the compound in the treatment is 2 × MIC. After the treated cells have been exposed to the treatment for three hours, they were fixed in GT in the same manner as described above. The AFM measurements of treated and untreated L. monocytogenes ATCC7644 cells were performed using a Nano-wizard IV system (JPK/Bruker, Berlin, Germany) in quantitative imaging mode with a ScanAsyst Fluid probe (Bruker, Billerica, MA, USA). During the measurements, the set point was maintained at 0.7 nN, the length along the Z-axis was 1200 nm, and the images were acquired at a resolution of 256 × 256 pixels. The collected data was analyzed using the JPK software for data processing.4.4. Optical fluorescence microscopy
A Petri dish (FluoroDish) with adhesive (Cell-Tak in 0.1 M NaHCO3, pH = 8.00) was prepared in the same way as for the AFM measurements. The overnight culture of Listeria monocytogenes ATCC7644 was diluted 10-fold in tempered nutrient culture medium and propagated in an incubator shaker at 35 °C and 170 rpm. After incubation, 30 μL of the bacterial cell suspension was pipetted onto the surface of the coated Petri dish and incubated for 10 minutes at appropriate temperature to ensure the highest possible number of immobilized bacterial cells. The contents of the Petri dish was washed with tempered culture medium and the immobilized cells were further incubated at 35 °C on the stand of an optical microscope. At the beginning and after incubation, optical images of the immobilized cells were taken to confirm their initial viability. At the end of the incubation, the contents of the Petri dish were washed again, and the culture medium was replaced with a solution of the selected compounds in culture medium at a final concentration of 2 × MIC. The cells were exposed to the treatment for a total of three hours. At the end of the treatment, the Petri dish was washed with sterile physiological solution. The immobilized treated cells were stained with a mixture of fluorescent SYTO9 and propidium iodide (PI) dyes (1.5 μL dye per mL), which are components of the LIVE/DEAD BacLight Bacterial Viability Kit L7012. The images of the fluorescently labelled treated bacterial cells were taken after 30 minutes of incubation in the dark.4.5. Propidium iodide (PI) uptake assay
An overnight culture of Staphylococcus aureus ATCC25923 was diluted 20-fold in tempered Mueller-Hinton broth (MHB) and propagated for one hour in a shaking incubator at 37 °C and 220 rpm. The compounds were diluted in sterile filtered phosphate buffer (PBS), pH = 7.4, to a final concentration of 8 × MIC. The prepared solutions were serially diluted to concentrations of 4 × MIC, 2 × MIC and MIC. The optical density of the bacterial suspension was measured at 600 nm and the cell pellet was precipitated by centrifugation at 4500g for ten minutes. The supernatant was discarded, and the cell pellet was resuspended in sterile filtered PBS buffer. The resuspended bacterial cells were further diluted in PBS buffer to a final concentration of 1 × 106 colony forming units per mililiter (CFU mL−1). An aliquot of 500 μL of the diluted suspension of bacterial cells was added to microtubes containing previously prepared solutions of the tested compounds in PBS buffer. 1 μL of propidium iodide solution (3 mg mL−1) was added to each microtube so that the final concentration of fluorescent dye was 5 μM. Fluorescence intensity (ex: 536 nm; em: 617 nm) was measured over six hours of treatment at 37 °C using Tecan Infinite 200 Pro plate reader.4.6. In vitro inhibition of protein synthesis
The potential of tested candidate compounds to inhibit protein synthesis was evaluated using a commercially available S30 T7 High Yield Protein Expression System kit (Promega) based on Escherichia coli cell lysate containing T7 polymerase for transcription of the gene of interest. In addition to the T7 extract, the expression was additionally ensured by the S30 mixture containing amino acids, ribonucleoside triphosphates (rNTPs), transfer RNA (tRNA), ATP-regenerating enzymes and isopropyl-β-D-1-thiogalactopyranoside (IPTG). The expression vector was pFN6A plasmid DNA which contains the hRluc reporter gene downstream of the T7 promoter site. After preparation of mastermix, reaction tubes were incubated in a thermomixer for one hour at a temperature of 37 °C with rotation of 1200 rpm. The commercially available Renilla Luciferase Assay System kit (Promega) was used to evaluate the results after incubation. An aliquot of each mastermix reaction was diluted 40 times in Renilla Luciferase Lysis Assay buffer. A total of 50 μL of the samples prepared in this way was added to the wells of the white microtiter plate. Renilla Luciferase Assay substrate was diluted 5-fold in Renilla Luciferase Assay Buffer and 50 μL was added to each well of 96-well plate. After the addition of the substrate solution, the relative luminescence intensity units, RLUs, were measured using Tecan Infinite 200 Pro plate reader with a signal integration time of ten seconds.4.7. Molecular dynamics (MD) simulation studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 water molecules (TIP3P water model), 360 sodium ions and 52 chloride ions. CHARMM-GUI membrane builder minimization and equilibration procedure was used to obtain equilibrated simulation box of the S. aureus model membrane at T = 30 °C. Upon initial equilibration, the membrane was sinuated for 100 ns using unbiased all-atom molecular dynamics (MD). The snapshot corresponding to t = 100 ns was extracted and then employed in preparation of the cetylpyridinium chloride (CPC) + S. aureus (CPC/SA-membrane) and QC16-Ally + S. aureus (QC16-Ally/SA-membrane) membrane systems. Both CPC and QC16-Ally were parameterized consistently with the lipid forcefield, namely using CHARMM36m force field. To prepare the initial simulation boxes containing both the lipid bilayer and the chosen molecules, exactly two molecules of each compound were placed in the previously equilibrated simulation box of the model membrane. Tested compounds were placed inside the water layer, above the outer leaflet of the Staphylococcus aureus model membrane. Upon the initial placement, the simulation boxes were firstly minimized and equilibrated for 5 ns in the NPT ensemble, using Nosé–Hoover thermostat55 with Berendsen barostat56 (p = 1 bar, semiisotropic pressure coupling). The duration of each production run was 100 ns.
000 water molecules (TIP3P water model), 360 sodium ions and 52 chloride ions. CHARMM-GUI membrane builder minimization and equilibration procedure was used to obtain equilibrated simulation box of the S. aureus model membrane at T = 30 °C. Upon initial equilibration, the membrane was sinuated for 100 ns using unbiased all-atom molecular dynamics (MD). The snapshot corresponding to t = 100 ns was extracted and then employed in preparation of the cetylpyridinium chloride (CPC) + S. aureus (CPC/SA-membrane) and QC16-Ally + S. aureus (QC16-Ally/SA-membrane) membrane systems. Both CPC and QC16-Ally were parameterized consistently with the lipid forcefield, namely using CHARMM36m force field. To prepare the initial simulation boxes containing both the lipid bilayer and the chosen molecules, exactly two molecules of each compound were placed in the previously equilibrated simulation box of the model membrane. Tested compounds were placed inside the water layer, above the outer leaflet of the Staphylococcus aureus model membrane. Upon the initial placement, the simulation boxes were firstly minimized and equilibrated for 5 ns in the NPT ensemble, using Nosé–Hoover thermostat55 with Berendsen barostat56 (p = 1 bar, semiisotropic pressure coupling). The duration of each production run was 100 ns.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 water molecules and cetylpyridinium chloride, CPC and QC16-Ally in final concentration of approximately 15 mM (10 molecules per simulation box). All the MD simulations were performed using GROMACS 2021.5 simulation package,53 with a time step of 2 fs, short-range Coulomb and van der Waals cut-offs set at 10 Å, three-dimensional periodic boundary conditions, and incorporating the PME procedure.57 All MD simulations were propagated using Nosé-Hoover thermostat55 with the pressure being monitored via Parrinello–Rahman barostat58 (p = 1 bar, isotropic pressure coupling) in the duration of t = 100 ns. The starting snapshots of the propagated systems containing CPC and QC16-Ally, respectively, are shown in Fig. 10.
000 water molecules and cetylpyridinium chloride, CPC and QC16-Ally in final concentration of approximately 15 mM (10 molecules per simulation box). All the MD simulations were performed using GROMACS 2021.5 simulation package,53 with a time step of 2 fs, short-range Coulomb and van der Waals cut-offs set at 10 Å, three-dimensional periodic boundary conditions, and incorporating the PME procedure.57 All MD simulations were propagated using Nosé-Hoover thermostat55 with the pressure being monitored via Parrinello–Rahman barostat58 (p = 1 bar, isotropic pressure coupling) in the duration of t = 100 ns. The starting snapshots of the propagated systems containing CPC and QC16-Ally, respectively, are shown in Fig. 10.
 | ||
| Fig. 10 Starting configurations of cetylpyridinium chloride (CPC)/SA-membrane (left) and QC16-Ally/SA-membrane (right) systems are shown in the top panel, with the initial configurations of the CPC/water (left) and QC16-Ally/water (right) systems shown in the bottom panel. Water molecules are shown in light gray, CPC and QC16-Ally using van der Waals sphere representation, with the different lipids constituting the model Staphylococcus aureus membrane shown using licorice representation, and in different colors. More precisely, FFPG and IFPG are shown in blue, PFPG. JFPG, ZFPG, JIPG and ZIPG are shown in red, SFPG, VFPG, TFPG and TIPG are colored orange, XFPG is colored yellow, OIPG and OFPG are shown in tan, JFGK, ZFGK, SFGK and TFGK are colored green, and ZFCL is colored pink (consult Tables S1 and S2† for further clarification of the lipid codenames). Phosphorous atoms belonging to the lipid headgroups are shown in van der Waals sphere representation and colored yellow. | ||
4.8. Parallel artificial membrane permeability assay (PAMPA assay)
The simulated membrane was prepared by dissolving lecithin in dodecane while heating in an ultrasonic bath to a final mass fraction of 1%. An aliquot of 5 μL of the prepared solution was pipetted onto the surface of the donor wells (Multiscreen-IP Filter Plate, 0.45 μm, Clear, Hydrophobic PVDF membrane, Millipore). Before the lecithin solution was dried out, 150 μL of the tested compound solution was added to the donor wells. A “sandwich” composed of donor/acceptor wells was assembled so that the simulated membrane is constantly immersed in the solvent in the acceptor wells during incubation. The plate was incubated at 37 °C for 24 hours. After incubation, the absorption spectrum of donor and acceptor wells was recorded in the wavelength range from 190 to 400 nm. The concentration of the tested compound in each well, both donor and acceptor, was calculated from the maximum absorbance. The integrity of the simulated membrane was tested using two different dyes, brilliant cresyl blue and lucifer yellow.4.9. In vitro cytotoxicity
The cytotoxicity of selected compounds was tested on two healthy human cell lines – human retinal pigment epithelial (RPE-1) and human keratinocytes (HaCaT) cells. The results were compared with the results of the commercially available quaternary ammonium salts, cetylpyridinium chloride (CPC) and benzidodecyldimethylammonium bromide (BAB). Human cells were grown in Dulbecco's Modified Eagle medium (DMEM) at 37 °C in a humidified atmosphere with 5% CO2. The tested compounds were dissolved in DMEM medium and serially diluted in a microtiter plate in the concentration range from 250 to 0.25 μM. 5000 human cells were pipetted into each well of the plate and the prepared plate was incubated for 48 hours. At the end of the incubation, 20 μL of the CellTiter 96® AQueous MTS reagent (Promega) was added to the wells of the microtiter plate according to the manufacturer's instructions. After three hours of incubation with the reagent, the absorbance was measured at a wavelength of 490 nm. The half maximal inhibitory concentration, IC50,value for each compound was calculated by plotting the percentage of viable cells versus concentration of compound using the GraFit 6.0 software.4.10. In vivo zebrafish embryotoxicity test
Zebrafish Danio rerio (WIK type) were obtained from the European Zebrafish Resource Center of the Karlsruhe Institute of Technology. Zebrafish maintenance and embryo production are described in detail in our previous study.59 A zebrafish embryotoxicity test (ZET) was performed in accordance with OECD 236.60 In dose range-finding experiments, embryos (N = 10) were exposed to a wide range of concentrations in serial dilutions: QC14-Me (500–31 μM), QC14-Ally (500–15.6 μM), QC16-Me (500–3.9 μM), QC16-Ally (500–1.56 μM), and cetylpyridinium chloride, CPC (0.5–0.0078 μM). Once a relevant range of concentrations was identified, a refined concentration range was tested using 48 embryos per concentration. E3 medium was used as a negative control. Plates were kept at 27.5 ± 0.5 °C (Innova 42 incubator, New Brunswick). Mortalities and abnormalities were recorded 120 hours post fertilization (hpf) using an inverted microscope (Olympus CKX41), equipped with a Leica EC3 digital camera and LAS EZ 3.2.0 digitizing software statistical program.4.11. The zebrafish locomotor assessment
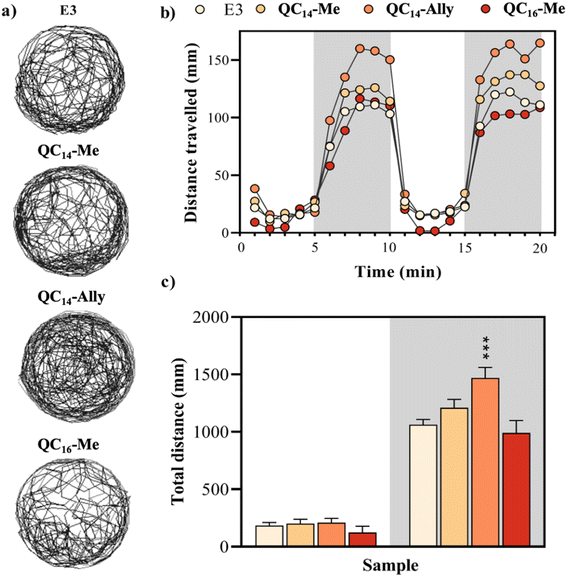
The swimming behavior of zebrafish larvae (N = 48) was assessed following 120 h of exposure to tested samples in a 96-well plate using the DanioVision system (Noldus Information Technology, Netherlands). Concentrations chosen for locomotor assessment were as follows: QC14-Me (20 μM), QC14-Ally (8 μM), and QC16-Me (8 μM). Zebrafish activity was recorded using the EthoVision XT software (Noldus Information Technology, Netherlands) over a 20 min period, including three periods of light and darkness (5 min in the darkness and 10 min in the light). To minimize the background noise, a smoothing profile with a minimum distance moved threshold of 0.2 cm was applied. The temperature during the measurement was maintained at 27.5 °C. After tracking, larvae were examined under inverted microscope to identify malformed or dead specimens, which were excluded from statistical analysis.4.12. Statistical analysis
Statistical analysis and graphical representations were conducted using GraphPad Prism 6.01 software. Results are expressed as means ± SD, with a significance treshold set at p ≤ 0.05 for all data. Prior to determination of the median lethal concentration (LC50), data were subjected to logarithmic transformation. One-way analysis of variance (ANOVA) and Tukey's post hoc test were employed to assess the significance of differences between treatments. In cases where the assumption of normality was violated, the Kruskal–Wallis one-way analysis of variance on ranks was utilized.Ethical statement
Animal housing and spawning were performed in aquaria units approved by the Croatian Ministry of Agriculture and in compliance with Directive 2010/63/EU. All experiments in this study were conducted on the non-protected embryonal stages (up to 96 hpf), which do not require approval from the animal welfare commissions.61Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
D. C. performed experiments providing results about bacteriostatic activity and cytotoxicity; L. K. and D. C. collected and analyzed AFM and fluorescence microscopy data; Z. B. and M. C. collected and analysed MD simulation data, S. B. B. and R. Č.-R. conducted in vivo studies, R. O. designed compounds, M. Š. directed the study, D. C. and M. Š. analyzed all data and wrote the manuscript. All authors contributed to and approved the final version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was supported by the the Croatian Science Foundation grant number UIP-2020-02-2356 (M. Š.) and institutional projects 641-01/23-02/0008 (M. Š.), 641-01/23-02/0010 (R. O.) funded by the Faculty of Science, University of Split.References
- W. A. Arnold, et al., Quaternary Ammonium Compounds: A Chemical Class of Emerging Concern, Environ. Sci. Technol., 2023, 7645–7665, DOI:10.1021/acs.est.2c08244.
- F. Bureš, Quaternary Ammonium Compounds: Simple in Structure, Complex in Application, Top. Curr. Chem., 2019, 14, DOI:10.1007/s41061-019-0239-2.
- C. P. Gerba, Quaternary ammonium biocides: Efficacy in application, Appl. Environ. Microbiol., 2015, 464–469, DOI:10.1128/AEM.02633-14.
- G. Domagk, Eine neue Klasse von Desinfektionsmitteln, Dtsch. Med. Wochenschr., 1935, 61, 829–832 CrossRef CAS.
- P. B. Price, BENZALKONIUM CHLORIDE (ZEPHIRAN CHLORIDE®) AS A SKIN DISINFECTANT, Arch. Surg., 1950, 61(1), 23, DOI:10.1001/archsurg.1950.01250020026004 . [Online]. Available: http://archsurg.jamanetwork.com/.
- B. M. P. Pereira and I. Tagkopoulos, Benzalkonium chlorides: Uses, regulatory status, and microbial resistance, Appl. Environ. Microbiol., 2019, e00377, DOI:10.1128/AEM.00377-19.
- S. Brown, J. P. Santa Maria and S. Walker, Wall teichoic acids of gram-positive bacteria, Annu. Rev. Microbiol., 2013, 67, 313–336, DOI:10.1146/annurev-micro-092412-155620.
- M. R. J. Salton, Lytic Agents, Cell Permeability, and Monolayer Penetrability, J. Gen. Physiol., 1968, 227–252 CrossRef CAS.
- P. Gilbert and L. E. Moore, Cationic antiseptics: Diversity of action under a common epithet, J. Appl. Microbiol., 2005, 703–715, DOI:10.1111/j.1365-2672.2005.02664.x.
- S. Alkhalifa, et al., Analysis of the Destabilization of Bacterial Membranes by Quaternary Ammonium Compounds: A Combined Experimental and Computational Study, ChemBioChem, 2020, 21(10), 1510–1516, DOI:10.1002/cbic.201900698.
- M. R. Garipov, et al., Targeting pathogenic fungi, bacteria and fungal-bacterial biofilms by newly synthesized quaternary ammonium derivative of pyridoxine and terbinafine with dual action profile, Bioorg. Chem., 2020, 104, 104306, DOI:10.1016/j.bioorg.2020.104306.
- W. B. Hugo and M. Frier, Mode of action of the antibacterial compound dequalinium acetate, Appl. Microbiol., 1969, 17(1), 118–127, DOI:10.1128/am.17.1.118-127.1969.
- M. Tischer, G. Pradel, K. Ohlsen and U. Holzgrabe, Quaternary ammonium salts and their antimicrobial potential: Targets or nonspecific interactions?, ChemMedChem, 2012, 22–31, DOI:10.1002/cmdc.201100404.
- A. L. Frantz, Chronic quaternary ammonium compound exposure during the COVID-19 pandemic and the impact on human health, J. Toxicol. Environ. Health Sci., 2023, 199–206, DOI:10.1007/s13530-023-00173-w.
- A. R. Mahoney, M. M. Safaee, W. M. Wuest and A. L. Furst, iScience The silent pandemic: Emergent antibiotic resistances following the global response to SARS-CoV-2, iScience, 2021, 24, 102304, DOI:10.1016/j.isci.
- J. M. Boyce, Quaternary ammonium disinfectants and antiseptics: tolerance, resistance and potential impact on antibiotic resistance, Antimicrob. Resist. Infect. Control, 2023, 32, DOI:10.1186/s13756-023-01241-z.
- T. G. Osimitz and W. Droege, Quaternary ammonium compounds: perspectives on benefits, hazards, and risk, Toxicol. Res. Appl., 2021, 5, 239784732110490, DOI:10.1177/23978473211049085.
- S. V. Sapozhnikov, et al., Design, synthesis, antibacterial activity and toxicity of novel quaternary ammonium compounds based on pyridoxine and fatty acids, Eur. J. Med. Chem., 2021, 211, 113100, DOI:10.1016/j.ejmech.2020.113100.
- R. A. Allen, M. C. Jennings, M. A. Mitchell, S. E. Al-Khalifa, W. M. Wuest and K. P. C. Minbiole, Ester-and Amide-containing MultiQACs: Exploring Multicationic Soft Antimicrobial Agents, 2017, [Online]. Available: http://www.elsevier.com/open-access/userlicense/1.0/.
- D. S. S. M. Uppu, et al., Side Chain Degradable Cationic-Amphiphilic Polymers with Tunable Hydrophobicity Show in Vivo Activity, Biomacromolecules, 2016, 17(9), 3094–3102, DOI:10.1021/acs.biomac.6b01057.
- R. Odžak, D. Crnčević, A. Sabljić, I. Primožič and M. Šprung, Synthesis and Biological Evaluation of 3-Amidoquinuclidine Quaternary Ammonium Compounds as New Soft Antibacterial Agents, Pharmaceuticals, 2023, 16(2), 187, DOI:10.3390/ph16020187.
- P. F. McDermott, R. D. Walker and D. G. White, Antimicrobials: modes of action and mechanisms of resistance, Int. J. Toxicol., 2003, 22(2), 135–143, DOI:10.1080/10915810305089.
- G. A. Pankey and L. D. Sabath, Clinical Relevance of Bacteriostatic versus Bactericidal Mechanisms of Action in the Treatment of Gram-Positive Bacterial Infections, [Online]. Available: https://academic.oup.com/cid/article/38/6/864/320723.
- S. V. Sapozhnikov, et al., Design, synthesis, antibacterial activity and toxicity of novel quaternary ammonium compounds based on pyridoxine and fatty acids, Eur. J. Med. Chem., 2021, 211, 113100, DOI:10.1016/j.ejmech.2020.113100.
- J. Fedorowicz, et al., Synthesis and biological evaluation of hybrid quinolone-based quaternary ammonium antibacterial agents, Eur. J. Med. Chem., 2019, 179, 576–590, DOI:10.1016/j.ejmech.2019.06.071.
- H. Bierne and P. Cossart, Listeria monocytogenes Surface Proteins: from Genome Predictions to Function, Microbiol. Mol. Biol. Rev., 2007, 71(2), 377–397, DOI:10.1128/mmbr.00039-06.
- L. Boulos, M. Prevost, B. Barbeau, J. Coallier, R. Desjardinś and D. Desjardinś, LIVE/DEAD BacLightE: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water, J. Microbiol. Methods, 1999, 77–86 CrossRef CAS PubMed.
- C. A. Moubareck, Polymyxins and bacterial membranes: A review of antibacterial activity and mechanisms of resistance, Membranes, 2020, 181, DOI:10.3390/membranes10080181.
- P. Sarkar, V. Yarlagadda, C. Ghosh and J. Haldar, A review on cell wall synthesis inhibitors with an emphasis on glycopeptide antibiotics, Medchemcomm, 2017, 516–533, 10.1039/c6md00585c.
- D. C. Hooper, Mechanisms of Action and Resistance of Older and Newer Fluoroquinolones, [Online]. Available: http://cid.oxfordjournals.org/.
- R. I. Aminov, A brief history of the antibiotic era: Lessons learned and challenges for the future, Front. Microbiol., 2010, 1, 134, DOI:10.3389/fmicb.2010.00134.
- M. E. O'Sullivan, et al., Aminoglycoside ribosome interactions reveal novel conformational states at ambient temperature, Nucleic Acids Res., 2018, 46(18), 9793–9804, DOI:10.1093/nar/gky693.
- J. Fedorowicz, et al., Synthesis and biological evaluation of hybrid quinolone-based quaternary ammonium antibacterial agents, Eur. J. Med. Chem., 2019, 179, 576–590, DOI:10.1016/j.ejmech.2019.06.071.
- S. V. Sapozhnikov, et al., Design, synthesis, antibacterial activity and toxicity of novel quaternary ammonium compounds based on pyridoxine and fatty acids, Eur. J. Med. Chem., 2021, 211, 113100, DOI:10.1016/j.ejmech.2020.113100.
- R. Chulluncuy, C. Espiche, J. A. Nakamoto, A. Fabbretti and P. Milón, Conformational response of 30S-bound IF3 to A-site binders streptomycin and kanamycin, Antibiotics, 2016, 5(4), 38, DOI:10.3390/antibiotics5040038.
- L. Martinez, R. Andrade, E. G. Birgin and J. M. Martínez, PACKMOL: A package for building initial configurations for molecular dynamics simulations, J. Comput. Chem., 2009, 30(13), 2157–2164, DOI:10.1002/jcc.21224.
- T. F. Headen, E. S. Boek, G. Jackson, T. S. Totton and E. A. Müller, Simulation of Asphaltene Aggregation through Molecular Dynamics: Insights and Limitations, Energy Fuel., 2017, 31(2), 1108–1125, DOI:10.1021/acs.energyfuels.6b02161.
- S. Otasevic and T. Vojinovic, Lecithin and anionic lipids as an imitation of the lipid membrane in Parallel Artificial Membrane Permeation Assay (PAMPA) blood -brain barrier Models, Prog. Nutr., 2020, 22(3), e2020035, DOI:10.23751/pn.v22i3.9720.
- M. Kansy, F. Senner and K. Gubernator, Physicochemical high throughput screening: parallel artificial membrane permeation assay in the description of passive absorption processes, J. Med. Chem., 1998, 41(7), 1007–1010, DOI:10.1021/jm970530e.
- W. A. Arnold, et al., Quaternary Ammonium Compounds: A Chemical Class of Emerging Concern, Environ. Sci. Technol., 2023, 7645–7665, DOI:10.1021/acs.est.2c08244.
- G. Zheng, E. Schreder, S. Sathyanarayana and A. Salamova, The first detection of quaternary ammonium compounds in breast milk: Implications for early-life exposure, J. Expo. Sci. Environ. Epidemiol., 2022, 32(5), 682–688, DOI:10.1038/s41370-022-00439-4.
- A. Salamova, G. Zheng and T. F. Webster, Quaternary ammonium compounds: Bioaccumulation potentials in humans and levels in blood before and during the covid-19 pandemic, Environ. Sci. Technol., 2021, 55(21), 14689–14698, DOI:10.1021/acs.est.1c01654.
- R. M. Basnet, D. Zizioli, M. Guarienti, D. Finazzi and M. Memo, Methylxanthines induce structural and functional alterations of the cardiac system in zebrafish embryos, BMC Pharmacol. Toxicol., 2017, 18(1), 72, DOI:10.1186/s40360-017-0179-9.
- A. V. Kalueff, D. J. Echevarria and A. M. Stewart, Gaining translational momentum: More zebrafish models for neuroscience research, Prog. Neuro-Psychopharmacol. Biol. Psychiatry, 2014, 1–6, DOI:10.1016/j.pnpbp.2014.01.022.
- K. Howe, et al., The zebrafish reference genome sequence and its relationship to the human genome, Nature, 2013, 496(7446), 498–503, DOI:10.1038/nature12111.
- R. M. Basnet, D. Zizioli, S. Taweedet, D. Finazzi and M. Memo, Zebrafish larvae as a behavioral model in neuropharmacology, Biomedicines, 2019, 23, DOI:10.3390/BIOMEDICINES7010023.
- W. Norton, Towards developmental models of psychiatric disorders in zebrafish, Front. Neural Circuits, 2013, 79, DOI:10.3389/fncir.2013.00079.
- J. A. Morris, Zebrafish: A model system to examine the neurodevelopmental basis of schizophrenia, Prog. Brain Res., 2009, 179(C), 97–106, DOI:10.1016/S0079-6123(09)17911-6.
- S. Jo, T. Kim and W. Im, Automated builder and database of protein/membrane complexes for molecular dynamics simulations, PLoS One, 2007, 2(9), e880, DOI:10.1371/journal.pone.0000880.
- E. L. Wu, et al., CHARMM-GUI membrane builder toward realistic biological membrane simulations, J. Comput. Chem., 2014, 1997–2004, DOI:10.1002/jcc.23702.
- J. Lee, et al., CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field, J. Chem. Theory Comput., 2016, 12(1), 405–413, DOI:10.1021/acs.jctc.5b00935.
- F. Joodaki, L. M. Martin and M. L. Greenfield, Supporting Information: Generation and computational characterization of a complex Staphylococcus aureus lipid bilayer, Langmuir, 2022, 38(31), 9481–9499 CrossRef CAS PubMed.
- M. J. Abraham, et al., Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1–2, 19–25, DOI:10.1016/j.softx.2015.06.001.
- J. Huang, et al., CHARMM36m: An improved force field for folded and intrinsically disordered proteins, Nat. Methods, 2016, 14(1), 71–73, DOI:10.1038/nmeth.4067.
- S. Nosé, A molecular dynamics method for simulations in the canonical ensemble, Mol. Phys., 1984, 52(2), 255–268, DOI:10.1080/00268978400101201.
- H. J. C. Berendsen, J. P. M. Postma, W. F. Van Gunsteren, A. Dinola and J. R. Haak, Molecular dynamics with coupling to an external bath, J. Chem. Phys., 1984, 81(8), 3684–3690, DOI:10.1063/1.448118.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, A smooth particle mesh Ewald method, J. Chem. Phys., 1995, 103(19), 8577–8593, DOI:10.1063/1.470117.
- M. Parrinello and A. Rahman, Polymorphic transitions in single crystals: A new molecular dynamics method, J. Appl. Phys., 1981, 52(12), 7182–7190, DOI:10.1063/1.328693.
- S. Babić, et al., Utilization of the zebrafish model to unravel the harmful effects of biomass burning during Amazonian wildfires, Sci. Rep., 2021, 11(1), 2527, DOI:10.1038/s41598-021-81789-1.
- S. 2, 1-22. OECD 236 (2013). Fish embryo acute toxicity (FET) test. OECD Guidelines for the Testing of Chemicals, “OECD/OCDE 2”..
- “DIRECTIVE 2010/63/EU OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL of 22 September 2010 on the protection of animals used for scientific purposes (Text with EEA relevance)”..
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra07975b |
| This journal is © The Royal Society of Chemistry 2025 |