Open Access Article
Open Access ArticleSpatially-resolved characterization of the metabolic and N-glycan alterations in colorectal cancer using matrix-assisted laser desorption/ionization mass spectrometry imaging†
Yaqi Zhang‡
ab,
Qiangjun Chen‡d,
Lei Wangab,
Haoyuan Gengab,
Zihan Zhua,
Cancan Lvb,
Yisheng Zhaoab,
Xiao Wang ab,
Chenglong Sun
ab,
Chenglong Sun ab,
Panpan Chen
ab,
Panpan Chen *ab and
Chao Zhang*c
*ab and
Chao Zhang*c
aShandong Engineering Research Center for Innovation and Application of General Technology for Separation of Natural Products, Shandong Analysis and Test Center, Qilu University of Technology (Shandong Academy of Sciences), Jinan, 250014, China. E-mail: chenpanpan1203@163.com
bKey Laboratory for Natural Active Pharmaceutical Constituents Research in Universities of Shandong Province, School of Pharmaceutical Sciences, Qilu University of Technology (Shandong Academy of Sciences), Jinan, 250014, China
cDepartment of Pediatrics, Qilu Hospital of Shandong University, Jinan 250012, China
dDepartment of Breast and Thyroid Surgery, Yi Du Central Hospital of Weifang, Shangdong Province, No. 5168 Jiangjunshan Road, Weifang 262500, China
First published on 21st January 2025
Abstract
Colorectal cancer is the second leading cause of cancer-related deaths worldwide, and its development typically involves complex metabolic reprogramming. By mapping the spatial distributions of metabolites and N-glycans in heterogeneous colorectal cancer tissues, we can elucidate cancer-associated metabolic and N-glycan changes. Herein, we combine mass spectrometry imaging-based metabolomics and N-glycomics to characterize the spatially resolved reprogramming of metabolites and N-glycans in colorectal cancer tissues. The metabolic characteristics of different regions of colorectal cancer were evaluated through the utilization of orthogonal partial least squares discriminant analysis. In combination with metabolic pathway enrichment analysis, significant alterations were identified in the fatty acid metabolism, arginine and proline metabolism of colorectal cancer. Cancer cell regions exhibited a marked upregulation of saturated fatty acids, monounsaturated fatty acids, polyamines, and histidine. Additionally, we discovered that the high-mannose N-glycans were predominantly distributed in tumor tissue regions, whereas complex N-glycans were more commonly found in the normal tissue regions adjacent to the tumor. Such findings provide new insights into the spatial signatures of metabolites and N-glycans in colorectal cancer, thereby offering a crucial basis for the diagnosis of colorectal cancer and potential vulnerabilities that might be targeted for cancer therapy.
Introduction
Colorectal cancer is the second leading cause of cancer deaths worldwide, claiming approximately 930![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 lives annually.1,2 The initiation and progression of colorectal cancer are closely associated with its complex biological characteristics, often involving intricate metabolic alterations. Endogenous metabolites, including fatty acids, amino acids, polyamines and nucleotides, are essential nutrients required for tumor cell growth, proliferation and invasion. They also regulate biological processes such as cell signaling, energy metabolism, and immune evasion.3–6 Previous studies have shown that tumor cells undergo significant metabolic reprogramming, altering their metabolism to meet the demands of rapid growth and invasion.7–10 Protein glycosylation is a prevalent post-translational modification in which proteins covalently attach to sugar molecules, forming glycoproteins, encompassing N-glycosylation and O-glycosylation.11–15 Growing evidence suggests that the reprogramming of N-glycans in tumor cells can be used to discriminate normal and tumor tissues, potentially providing stage specificity for different kinds of tumors.16–19 However, it should be noted that metabolites and N-glycans are closely related in metabolic pathways. For example, uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc) is a key nucleotide sugar precursor in the process of N-glycosylation, while asparagine is a small molecule metabolite involved in the synthesis of UDP-GlcNAc. Thus, metabolic changes in metabolites may indirectly affect the efficiency of N-glycosylation.20,21 Characterizing the alterations of metabolites and N-glycans during tumor initiation and progression will contribute to the understanding of tumor molecular mechanisms, offering potential diagnostic and therapeutic targets for tumor treatments.22,23
000 lives annually.1,2 The initiation and progression of colorectal cancer are closely associated with its complex biological characteristics, often involving intricate metabolic alterations. Endogenous metabolites, including fatty acids, amino acids, polyamines and nucleotides, are essential nutrients required for tumor cell growth, proliferation and invasion. They also regulate biological processes such as cell signaling, energy metabolism, and immune evasion.3–6 Previous studies have shown that tumor cells undergo significant metabolic reprogramming, altering their metabolism to meet the demands of rapid growth and invasion.7–10 Protein glycosylation is a prevalent post-translational modification in which proteins covalently attach to sugar molecules, forming glycoproteins, encompassing N-glycosylation and O-glycosylation.11–15 Growing evidence suggests that the reprogramming of N-glycans in tumor cells can be used to discriminate normal and tumor tissues, potentially providing stage specificity for different kinds of tumors.16–19 However, it should be noted that metabolites and N-glycans are closely related in metabolic pathways. For example, uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc) is a key nucleotide sugar precursor in the process of N-glycosylation, while asparagine is a small molecule metabolite involved in the synthesis of UDP-GlcNAc. Thus, metabolic changes in metabolites may indirectly affect the efficiency of N-glycosylation.20,21 Characterizing the alterations of metabolites and N-glycans during tumor initiation and progression will contribute to the understanding of tumor molecular mechanisms, offering potential diagnostic and therapeutic targets for tumor treatments.22,23
Metabolomics analysis can investigate the differential expression of metabolites between tumor and normal tissues, identify tumor-associated metabolic alterations, and highlight the complex metabolic reprogramming in tumor microenvironment.24–28 Researchers have successfully carried out metabolomics analysis on colorectal cancer tissues using liquid chromatography-mass spectrometry (LC-MS). For example, Shen et al. employed LC-MS to conduct metabolomics analysis on human colon tissues, and they found that glutamate, glutathione, creatine, proline and tryptophan were significantly upregulated in colorectal cancer tissue compared to normal tissue.29 Professor Owen J. Sansom's team utilized LC-MS to analyze the molecular mechanisms of colorectal cancer from a metabolomics perspective, revealing the potential of adenosyl homocysteinase as a novel therapeutic target for colorectal cancer.30 The Sansom team utilized mouse models and human colorectal cancer patient samples, employing LC-MS technology to reveal the key role of the amino acid transporter SLC7A5 in KRAS-mutant colorectal cancer and propose SLC7A5 as a potential therapeutic target.31 However, this method requires homogenization of the tissue samples, which severely disrupts the spatial information of metabolites in highly heterogeneous cancer tissues, and cannot achieve in situ analysis of metabolites. Matrix-assisted laser desorption/ionisation mass spectrometry imaging (MALDI–MSI) is a state-of-the-art molecular imaging technique used for tissue profiling, which allows for in situ mapping the spatial and temporal dynamics of metabolites in biological tissues.32–35
In this study, we proposed a multi-omics strategy combining MSI-based metabolomics and N-glycomics to visualize the spatial distributions of metabolites and N-glycans in different regions of heterogeneous colorectal cancer tissues. Data-driven unsupervised probabilistic latent semantic analysis (PLSA) suggests significant molecular heterogeneity in colorectal cancer tissues at both metabolite and N-glycan levels. Furthermore, we screened different expressed metabolites in distinct tissue microregions including tumor tissue, normal mucosa and muscularis tissues. Combined with metabolic pathway enrichment analysis, it was found that fatty acid metabolism, arginine and proline metabolism, and histidine metabolism were significantly reprogrammed in tumor cells of colorectal cancer. Then, we explored the different expressions of N-glycans in distinct microregions of colorectal cancer tissues and found that high-mannose N-glycans were predominantly distributed in tumor regions, whereas complex N-glycans were more abundant in tumor-adjacent normal tissues. These results broaden our understanding of metabolic reprogramming in colorectal cancer tumors and provide potential tumor diagnostic molecules at the metabolic and N-glycan levels.
Materials and methods
Chemicals and reagents
1,5-naphthalenediamine (1,5-DAN) was purchased from Shanghai Aladdin Bio-chemical Technology Co., Ltd (Shanghai, China). HPLC-grade acetonitrile (ACN) and ethanol were provided by Merck (Muskegon, USA). Purified water was afforded by Hangzhou Wahaha Group Co., Ltd (Hangzhou, China). PNGase F was obtained from New England Biolabs (MA, USA). The citrate buffer was provided by Servicebio (Wuhan, China). Tissue-Tek optimal cutting temperature (OCT) compound was purchased from Sakura Finetek Japan Co., Ltd (Tokyo, Japan). The indium tin oxide-coated slides were provided by Bruker (Daltonics, Billerica, MA, USA).Sample preparation for MALDI-MSI analysis
The research was conducted according to the guidelines of the Declaration of Helsinki, and the protocol was approved by the Medical Ethics Committee of Yi Du Central Hospital of Weifang (no. 2023-100). A total of seven postoperative colorectal cancer tissues were collected and embedded in OCT and then sectioned into 14 μm consecutive slices at −20 °C using a Thermo CryoStar NX50 NOVPD cryostat (Bremen, Germany). The tissue sections were mounted on the conductive side of ITO-coated slides for MALDI-MSI analysis.Sample preparation for N-glycan MALDI-MSI
The colorectal cancer tissue sections were subjected to gradient elution using 100% ethanol (2 × 3 min), 95% ethanol (1 × 3 min), 70% ethanol (1 × 3 min), and water (1 × 6 min) to remove interfering molecules and enhance the sensitivity of N-glycan detection. After hydration, the tissue slides were heated for 25 min in citrate buffer at 100 °C for antigen retrieval. Afterward, the slides were dried in a vacuum pump for 10 min. PNGase F (500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1, NEB) was sprayed over the slides using a Matrix Builder automatic matrix sprayer to release N-glycans from glycoproteins. 20 μL of PNGase F was first dissolved in 1 mL of water, and then the dissolved PNGase F was sprayed on the tissue sections. The flow rate of the PNGase F solution was 20 μL min−1, the spray pressure was set at 10 psi, the nozzle speed was 1000 mm min−1, the nozzle temperature was maintained at 37 °C, the distance between the nozzle and the sample was 4 cm, the nozzle movement spacing was 3 mm, and 40 spray cycles were applied. Following this, tissue sections were incubated at 37 °C for overnight in a humidified chamber.
000 U mL−1, NEB) was sprayed over the slides using a Matrix Builder automatic matrix sprayer to release N-glycans from glycoproteins. 20 μL of PNGase F was first dissolved in 1 mL of water, and then the dissolved PNGase F was sprayed on the tissue sections. The flow rate of the PNGase F solution was 20 μL min−1, the spray pressure was set at 10 psi, the nozzle speed was 1000 mm min−1, the nozzle temperature was maintained at 37 °C, the distance between the nozzle and the sample was 4 cm, the nozzle movement spacing was 3 mm, and 40 spray cycles were applied. Following this, tissue sections were incubated at 37 °C for overnight in a humidified chamber.
Matrix coating
1,5-DAN in ACN/H2O (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, v
20, v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) solution at a concentration of 3.5 mg mL−1 was sprayed onto the tissue section at a flow rate of 100 μL min−1. The nitrogen jet pressure and temperature were set at 0.4 MPa and 50 °C, the temperature and speed settings for matrix coating were 30 °C and 0.1 mL min−1, respectively. A total of ten spray cycles were carried out over each of the tissue sections.
v) solution at a concentration of 3.5 mg mL−1 was sprayed onto the tissue section at a flow rate of 100 μL min−1. The nitrogen jet pressure and temperature were set at 0.4 MPa and 50 °C, the temperature and speed settings for matrix coating were 30 °C and 0.1 mL min−1, respectively. A total of ten spray cycles were carried out over each of the tissue sections.
MALDI-MSI analysis
MALDI-MSI was conducted with a Rapiflex MALDI tissuetyper TOF/TOF MS (Bruker Daltonics, Billerica, MA) in both positive- and negative-ion modes. The spatial resolution was set to 100 μm. The ion source voltage was 16 kV, the laser frequency was 5000 Hz with a laser shot frequency of 200 Hz. The laser energy was set at 75%, the laser spot size was adjusted to 100 μm, and scan settings were configured in reflectron mode. The spatial distributions of metabolites were visualized using SCiLS Lab 2018b software (GmbH, Bremen, Germany).Data analysis
Imaging data files were imported into the SCiLS software to construct MS images and perform segmentation analysis. In the tissue segmentation map, we applied the Bisecting k-Means method, using Correlation Distance as the metric and setting the Minimal Interval Width to ±11.795 mDa. For the PLSA, we first conducted peak detection on the tumor tissue data without applying any scaling. Subsequently, we employed PLSA with random initialization to identify and analyze three major components. By matching the MS images with H&E staining images, regions of interest were selected and the MS data for these regions were obtained. After pre-processing the MS data, the matrix data were further imported into SIMCA 14.0 software, where Pareto scaling and mean-centering were applied to the data. Supervised data analysis was then conducted using orthogonal partial least squares discriminant analysis (OPLS-DA) to visualize the distribution and relationships between samples. Additionally, R 4.2.3 and MetaboAnalyst 5.0 software were used for statistical analysis and data processing.Analyte identification
Different metabolite ions were extracted to carried out high resolution MS/MS analysis on tissue sections using Q-Exactive Orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany). Then, the metabolite ions were identified based on accurate mass value (<5 ppm for Orbitrap-MS, <20 ppm for MALDI TOF-MS) in Table S1.† The representative MS/MS spectra were demonstrated in Fig. S5–S9.† The structures of N-Glycans were obtained by comparison with the free database (Glycoshape, https://glycoshape.io/).Results and discussion
Spatially resolved multi-omics reveals tumor heterogeneity of colorectal cancer
To explore the histological heterogeneity of colorectal cancer, we first stained the cancer section from different patients. As shown in Fig. 1a, in addition to tumor cells, we also discovered normal mucosa and muscularis tissues on the H&E images. Furthermore, we investigated the metabolic heterogeneity of colorectal cancer tissues using MALDI-MSI (Fig. S4†). Fig. 1b demonstrates the metabolites-driven tissue segmentation maps according to the metabolite fingerprints of each scanning spot in cancer sections. Different tissue regions are categorized according to their metabolite characteristics and given a specific color. Remarkably metabolic heterogeneity can be inferred from the metabolites-driven segmentation maps. Then, we conducted unsupervised probabilistic latent semantic analysis (PLSA) based on the spatial distribution of tissue metabolites. Fig. 1c shows that the detected metabolites were segmented into three fundamental components. Component 1 represents a class of metabolites abundantly expressed in tumor tissue. Component 2 indicates metabolites that exhibit a notable increase in normal intestinal mucosal tissue. Furthermore, metabolites distributed separately in muscularis are segmented into component 3. These results illustrate the molecular heterogeneity of colorectal cancer at the metabolite level.Furthermore, aberrant expression of glycosylation is closely associated with the onset and progression of numerous diseases, especially in certain tumors where abnormal N-glycosylation patterns have been observed.36,37 Accordingly, further investigation of N-glycans will aid in elucidating the relationship between N-glycan components in glycoproteins and specific diseases. In this study, we employed peptide N-glycosidase F to cleave the glycoside bond between N-glycan and asparagine to release N-glycan from glycoproteins. Fig. 1d presents the N-glycan-driven tissue segmentation maps according to the N-glycan fingerprints of each pixel point in cancer sections. In a similar manner, we employed unsupervised probabilistic latent semantic analysis to classify N-glycans into three fundamental components. Fig. 1e illustrates the spatial distribution of each N-glycan group within tumor tissue, normal mucosa, and muscularis tissue. We discovered that N-glycan components have different spatial distributions, suggesting heterogeneity in N-glycan levels.
In situ screening of altered metabolites in colorectal cancer tissue
To pinpoint regionally differentiated metabolites in distinct colorectal cancer tissue regions, we constructed an orthogonal partial least squares discriminant analysis (OPLS-DA) model for MALDI-MSI data of colorectal cancer. Fig. 2a–d show the OPLS-DA model established based on MALDI-MS profiles of tumor, intestinal mucosa, and muscularis in positive and negative ion modes. Obvious clustering and segregation were discovered in OPLS-DA model, representing a striking metabolic difference between tumor and intestinal mucosa (Fig. 2b and d), tumor and muscularis (Fig. 2a and c), muscularis and intestinal mucosa (Fig. S1†). Differently expressed metabolites between tumor and adjacent normal tissues can be screened by the S-Plot of OPLS-DA model, where the top right spots in S-plot indicate up-regulated metabolites in tumor tissue and the bottom left spots in S-plot represents down-regulated metabolites in the tumor tissue (Fig. 2e–h). In positive MALDI-MSI ion mode, we found that polyamines and phosphatidylcholines (PCs) were the most significantly altered metabolites in tumor vs. muscularis groups and in tumor vs. intestinal mucosa groups (Fig. 2e and g). Similarly, fatty acids (FAs), glutamine, and aspartate were screened as the most significantly altered metabolites in tumor vs. muscularis groups and in tumor vs. intestinal mucosa groups in negative MALDI-MSI ion mode (Fig. 2h and f). Furthermore, we carried out clustering heat map analysis to characterize the global clustering trend of different metabolite ions in distinct regions of colorectal cancer tissues. As shown in Fig. 2i, we discovered that saturated fatty acids and monounsaturated fatty acids cluster significantly in tumor tissues under the negative ion mode. Fig. 2j reveals that most PCs, especially monounsaturated PCs, are highly enriched in tumor tissue in positive ion mode. The discriminating metabolites were then imported into MetaboAnalyst 6.0 (https://www.metaboanalyst.ca/) to perform metabolic pathway analysis, and we found that fatty acid metabolism, histidine metabolism, arginine and proline metabolism, alanine, aspartate, and glutamate metabolism were remarkably disrupted during tumorigenesis (Fig. 2k). These results re-emphasize significant metabolic reprogramming in colorectal cancer tissue.Imaging the altered polyamine synthesis and metabolism in colorectal cancer
Polyamines including spermine and spermidine were screened as tumor-associated metabolites in this study. In terms of cellular pathways, polyamines are endogenously produced polycationic small molecules derived from the decarboxylation of the non-proteinogenic amino acid ornithine by ornithine decarboxylase (Fig. 3a). Extensive studies indicated that the disorder of polyamine metabolism is closely related to the occurrence of different cancers. Spermidine is a putrescine metabolite catalyzed by spermidine synthase (SRM), spermine is then generated from spermidine via spermine synthase (SMS).36–38 The imaging and results of colorectal cancer tissue revealed that spermidine is much higher than that in muscularis and intestinal mucosa tissues, with spermine also showing a noticeable increase in tumor tissue compared to both muscularis and intestinal mucosa tissues (Fig. 3b and c). This result further reveals that the synthesis and metabolism of polyamines are significantly disrupted in colorectal cancer.Fatty acids metabolism disorders in colorectal cancer
Tumor cells rely on the large amounts of energy and small molecule metabolites released through fatty acid β-oxidation to meet the demands of their rapid proliferation.39,40 Fatty acid synthase catalyzes the de novo synthesis of fatty acids from acetyl-CoA. Through the actions of elongates (ELOVL1-7) and desaturases (Δ4,5,6,9D), fatty acids of different carbon chain lengths and degrees of saturation were produced (Fig. 4a). Specifically, we observed elevated levels of saturated fatty acids (FA-C14:0, FA-C18:0, FA-C20:0) in tumor tissue, whereas their expression was downregulated in muscularis and intestinal mucosa tissues (Fig. 4b–d). Similarly, monounsaturated fatty acids (FA-C16:1, FA-C18:1) exhibited higher expression in tumor tissue compared to muscularis and intestinal mucosa tissues (Fig. 4e and f). Polyunsaturated fatty acids (PUFAs), which included ω-3 and ω-6 fatty acids, were synthesized through a series of enzyme-catalyzed reactions. Oleic acid (FA-C18:1) was converted into polyunsaturated fatty acids such as FA-C20:4, FA-C22:5, and FA-C22:6 through the catalytic actions of elongate (ELOVL), fatty acid desaturase (FAD), and stearoyl-CoA desaturase 1 (SCD1). A striking observation was that FA-C20:4 (ω-6 polyunsaturated fatty acid) and FA-C20:5 (ω-3 polyunsaturated fatty acid) were expressed in tumor tissues, as well as in the muscularis and intestinal mucosa tissues (Fig. 4h and i). Leucine, as a precursor for the synthesis of acetyl-CoA, is expressed higher in the muscularis than in the tumor tissue (Fig. S2†). These studies emphasized the expression patterns of polyunsaturated fatty acids in different regions of colorectal cancer, which was crucial for exploring their functional differences between cancerous and normal tissues. However, it should be noticed that due to the presence of in-source fragmentation, the visualized fatty acids here include both free fatty acids and in source fragmentation fatty acids from larger lipids.Phospholipids are essential components of cell membranes, and tumor cells typically require various metabolic pathways to acquire the energy needed to construct new cell membranes.41 Mass spectrometry imaging results reveal the spatial distribution of different phospholipid (PC) molecules in colorectal cancer tissues. Mono-unsaturated phospholipids PC-32:1, PC-34:1, PC-36:1 were highly expressed in tumor regions, while PC-38:4 showed increased expression at the junction of tumor and muscularis areas (Fig. S3†).
Imaging the altered histidine metabolism in colorectal cancer
Histidine is an essential amino acid in organisms, which can be decarboxylated by histidine decarboxylase to form histamine.42 Increasing evidence suggests that cancer often undergoes prolonged and repeated inflammatory responses before its onset.43,44 In this study, we found that the level of histidine in cancerous tissues was higher than that in the muscularis and intestinal mucosa tissues (Fig. 5a). Correspondingly, the decarboxylation product of histidine, histamine, was lower in tumor tissues compared to muscularis and intestinal mucosa tissues, with no difference between muscularis and intestinal mucosa tissues (Fig. 5b). These results may suggest that cancerous tissues, after undergoing prolonged inflammation, deplete large amounts of inflammatory mediators, including histamine, resulting in a decrease in histamine levels within the cancerous tissues.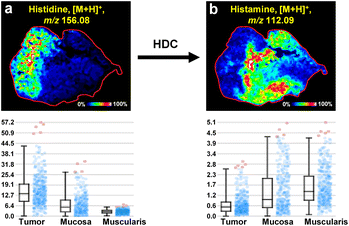 | ||
| Fig. 5 Imaging the altered histidine metabolism in colorectal cancer. (a) MALDI-MSI image and expression levels of histidine. (b) MALDI-MSI image and expression levels of histamine. | ||
MALDI-MSI of N-glycans in colorectal cancer
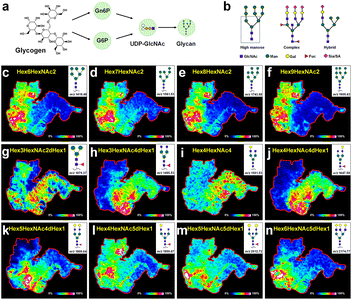
Biomacromolecule N-glycans are mainly synthesized by cells and participate in signal transduction on the cell surface and interactions between cells.45,46 Glycogen is degraded into glucose-6-phosphate (G6P) in the cytoplasm, which is then converted into N-acetylglucosamine-6-phosphate (Gn6P). Gn6P is further converted to UDP-GlcNAc, which is used to synthesize the core oligosaccharide. This core oligosaccharide is transferred to asparagine residues on nascent polypeptide chains by oligosaccharyltransferase and undergoes further modifications in the endoplasmic reticulum and Golgi apparatus, ultimately forming complex N-glycan structures (Fig. 6a). Asparagine linked oligosaccharides (N-glycans) have a core pentasaccharide composed of mannose and glucosamine, which can be classified into three types: high mannose, hybrid sugars, and complex sugars (Fig. 6b). Currently, an increasing number of studies indicated that cancer transformation was often accompanied by abnormal cellular glycation characteristics during tumor development. As shown in Fig. 6c–f, the high mannose N-glycans, such as Hex6HexNAc2 (m/z, 1419.48), Hex7HexNAc2 (m/z, 1581.53), Hex8HexNAc2 (m/z, 1743.58), and Hex9HexNAc2 (m/z, 1905.63) were present in the tumor tissues. N-glycans Hex4HexNAc4 (m/z, 1501.53, Fig. 6i) with a dual N-acetylglucosamine structure, as well as other complex N-glycans containing fucose and galactose structures such as Hex3HexNAc4dHex1 (m/z, 1485.53, Fig. 6h) and Hex4HexNAc4dHex1 (m/z, 1647.59, Fig. 6j), had higher content in the muscularis and intestinal mucosa tissues. Particular interest was that the dual antenna composite N-glycan Hex5HexNAc4dHex1 (m/z, 1809.64, Fig. 6k) and the triple antenna composite N-glycan Hex6HexNAc4dHex1 (m/z, 2174.77, Fig. 6n) mainly existed at the junction of tumors and normal tissues. These findings further supported the correlation between high-mannose type N-glycosylation and tumor progression. The abnormal growth rate of cancer cells may be due to the inability of N-glycans to form complex or hybrid structures.Conclusions
To sum up, we proposed a multi-omics approach that integrates metabolomic and N-glycomics analyses, coupled with MALDI-MSI imaging, to visualize the spatial distributions of metabolites and N-glycans across distinct regions of colorectal cancer. Region-specific metabolites and N-glycans in cancer and normal cells were screened. Further combined with metabolic pathway enrichment analysis, we revealed that fatty acid metabolism, arginine and proline metabolism, and histidine metabolism underwent significant reprogramming. The expression of polyamines, saturated and monounsaturated fatty acids, and histidine were markedly elevated in the tumor area. Conversely, polyunsaturated fatty acids and histamine metabolites were more abundant in normal tissue than in the tumor. Additionally, in situ spatial characterization showed that high mannose N-glycans were mainly distributed in tumor areas, whereas complex N-glycans were more abundant in normal tissue areas. The research provides a molecular basis for the diagnosis and pathological mechanism of colorectal cancer at the metabolic level and provides potential targets for its diagnosis and treatment intervention.Data availability
The data presented in this study are available in article and ESI.†Author contributions
Formal analysis, investigation, writing – original draft, visualization: Yaqi Zhang. Conceptualization, methodology, resources, project administration, foundation acquisition: Qiangjun Chen. formal analysis, investigation: Lei Wang, Zihan Zhu and Yisheng Zhao. Investigation: Haoyuan Geng and Cancan Lv. Conceptualization, methodology, supervision, project administration, foundation acquisition: Xiao Wang. Conceptualization, methodology, resources, writing – reviewing and editing, supervision, project administration, foundation acquisition: Chenglong Sun. Conceptualization, methodology, resources, writing – reviewing and editing, supervision, project administration, foundation acquisition: Panpan Chen. Conceptualization, methodology, resources, supervision: Chao Zhang. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was funded by the Natural Science Foundation of Shandong Province (no. ZR2022QH257, ZR2021QC056 and ZR2021QB174), the Youth Excellent Talents Program of Qilu University of Technology (Shandong Academy of Sciences) (no. 2024QZJH04), and Talent Research Project of Qilu University of Technology (no. 2023RCKY103).References
- E. Dekker, P. J. Tanis, J. L. A. Vleugels, P. M. Kasi and M. B. Wallace, Lancet, 2019, 394, 1467–1480 CrossRef.
- R. L. Siegel, K. D. Miller, N. S. Wagle and A. Jemal, Ca-Cancer J. Clin., 2023, 73, 17–48 CrossRef PubMed.
- T. Tsukidate, Q. Li and H. C. Hang, Curr. Opin. Chem. Biol., 2020, 54, 19–27 CrossRef CAS PubMed.
- D. S. Wishart, Physiol. Rev., 2019, 99, 1819–1875 CrossRef CAS PubMed.
- W. Cui, M. Guo, D. Liu, P. Xiao, C. Yang, H. Huang, C. Liang, Y. Yang, X. Fu, Y. Zhang, J. Liu, S. Shi, J. Cong, Z. Han, Y. Xu, L. Du, C. Yin, Y. Zhang, J. Sun, W. Gu, R. Chai, S. Zhu and B. Chu, Nat. Cell Biol., 2024, 26, 124–137 CrossRef CAS PubMed.
- M. Vaghari-Tabari, G. A. Ferns, D. Qujeq, A. N. Andevari, Z. Sabahi and S. Moein, J. Cell. Physiol., 2021, 236, 5512–5532 CrossRef CAS PubMed.
- X. Xu, Q. Peng, X. Jiang, S. Tan, Y. Yang, W. Yang, Y. Han, Y. Chen, L. Oyang, J. Lin, L. Xia, M. Peng, N. Wu, Y. Tang, J. Li, Q. Liao and Y. Zhou, Exp. Mol. Med., 2023, 55, 1357–1370 CrossRef CAS PubMed.
- Z. E. Stine, Z. T. Schug, J. M. Salvino and C. V. Dang, Nat. Rev. Drug Discovery, 2021, 21, 141–162 CrossRef PubMed.
- L. Li, Y. Qin and Y. Chen, Biochim. Biophys. Acta, Mol. Cell Res., 2024, 1871, 119697 CrossRef CAS PubMed.
- R. Fujiwara-Tani, T. Sasaki, T. Takagi, S. Mori, S. Kishi, Y. Nishiguchi, H. Ohmori, K. Fujii and H. Kuniyasu, Int. J. Mol. Sci., 2022, 23, 7824 CrossRef CAS.
- E. Makrydaki, R. Donini, A. Krueger, K. Royle, I. Moya Ramirez, D. A. Kuntz, D. R. Rose, S. M. Haslam, K. M. Polizzi and C. Kontoravdi, Nat. Chem. Biol., 2024, 20, 732–741 CrossRef CAS PubMed.
- M. M. Hasan, M. A. Mimi, M. A. Mamun, A. Islam, A. S. M. Waliullah, M. M. Nabi, Z. Tamannaa, T. Kahyo and M. Setou, Front. Neuroanat., 2021, 15, 1662–5129 Search PubMed.
- L. Zhang, C. Wang, Y. Wu, Q. Sha, B.-F. Liu, Y. Lin and X. Liu, J. Chromatogr. A, 2020, 1619, 460934 CrossRef CAS PubMed.
- Y. Guan, M. Zhang, J. Wang and H. Schlüter, J. Proteome Res., 2021, 20, 2914–2922 CrossRef CAS PubMed.
- E. Rodriguez, D. V. Lindijer, S. J. van Vliet, J. J. Garcia Vallejo and Y. van Kooyk, iScience, 2024, 27, 109037 CrossRef CAS.
- R. Sun, A. M. J. Kim and S.-O. Lim, Cells, 2021, 10, 1100 CrossRef CAS PubMed.
- Ö. F. Koçak, H. M. Kayili, M. Albayrak, M. E. Yaman, Y. Kadıoğlu and B. Salih, Anal. Biochem., 2019, 584, 113389 CrossRef PubMed.
- M. C. Ryczko, J. Pawling, R. Chen, A. M. Abdel Rahman, K. Yau, J. K. Copeland, C. Zhang, A. Surendra, D. S. Guttman, D. Figeys and J. W. Dennis, Sci. Rep., 2016, 6, 2045–2322 Search PubMed.
- M. M. Vicente, I. Alves, Â. Fernandes, A. M. Dias, B. Santos-Pereira, E. Pérez-Anton, S. Santos, T. Yang, A. Correia, A. Münster-Kühnel, A. R. M. Almeida, S. Ravens, G. A. Rabinovich, M. Vilanova, A. E. Sousa and S. S. Pinho, Cell, Mol. Immunol., 2023, 20, 955–968 CrossRef CAS.
- W. Igl, O. Polašek, O. Gornik, A. Knežević, M. Pučić, M. Novokmet, J. Huffman, C. Gnewuch, G. Liebisch, P. M. Rudd, H. Campbell, J. F. Wilson, I. Rudan, U. Gyllensten, G. Schmitz and G. Lauc, Mol. BioSyst., 2011, 7, 1852 RSC.
- B. Ebert, C. Rautengarten, H. E. McFarlane, T. Rupasinghe, W. Zeng, K. Ford, H. V. Scheller, A. Bacic, U. Roessner, S. Persson and J. L. Heazlewood, Nat. Plants, 2018, 4, 792–801 CrossRef CAS PubMed.
- A. M. Vibhute, H.-n. Tanaka, S. K. Mishra, R. F. Osuka, M. Nagae, C. Yonekawa, H. Korekane, R. J. Doerksen, H. Ando and Y. Kizuka, Biochim. Biophys. Acta, Gen. Subj., 2022, 1866, 130118 CrossRef CAS.
- A. Grigorian, S.-U. Lee, W. Tian, I. J. Chen, G. Gao, R. Mendelsohn, J. W. Dennis and M. Demetriou, J. Biol. Chem., 2007, 282, 20027–20035 CrossRef CAS.
- M. Jacob, A. L. Lopata, M. Dasouki and A. M. Abdel Rahman, Mass Spectrom. Rev., 2017, 38, 221–238 CrossRef.
- Q. Zang, C. Sun, X. Chu, L. Li, W. Gan, Z. Zhao, Y. Song, J. He, R. Zhang and Z. Abliz, Anal. Chim. Acta, 2021, 1155, 338342 CrossRef CAS PubMed.
- L. Bendall, S. Tiziani, Y. Kang, R. Harjanto, J. Axelrod, C. Piermarocchi, W. Roberts and G. Paternostro, PLoS One, 2013, 8, 1932–6203 Search PubMed.
- S. Zou, B. Qin, Z. Yang, W. Wang, J. Zhang, Y. Zhang, M. Meng, J. Feng, Y. Xie, L. Fang, L. Xiao, P. Zhang, X. Meng, H. H. Choi, W. Wen, Q. Pan, B. Ghesquière, P. Lan, M.-H. Lee and L. Fang, Cancer Res., 2023, 83, 414–427 CrossRef CAS PubMed.
- I. Vitale, G. Manic, L. M. Coussens, G. Kroemer and L. Galluzzi, Cell Metab., 2019, 30, 36–50 CrossRef CAS PubMed.
- Y. Shen, M. Sun, J. Zhu, M. Wei, H. Li, P. Zhao, J. Wang, R. Li, L. Tian, Y. Tao, P. Shen and J. Zhang, Mol. Omics, 2021, 17, 464–471 RSC.
- A. K. Najumudeen, F. Ceteci, S. K. Fey, G. Hamm, R. T. Steven, H. Hall, C. J. Nikula, A. Dexter, T. Murta, A. M. Race, D. Sumpton, N. Vlahov, D. M. Gay, J. R. P. Knight, R. Jackstadt, J. D. G. Leach, R. A. Ridgway, E. R. Johnson, C. Nixon, A. Hedley, K. Gilroy, W. Clark, S. B. Malla, P. D. Dunne, G. Rodriguez-Blanco, S. E. Critchlow, A. Mrowinska, G. Malviya, D. Solovyev, G. Brown, D. Y. Lewis, G. M. Mackay, D. Strathdee, S. Tardito, E. Gottlieb, C. R. G. C. Consortium, Z. Takats, S. T. Barry, R. J. A. Goodwin, J. Bunch, M. Bushell, A. D. Campbell and O. J. Sansom, Nat. Genet., 2021, 53, 16–26 CrossRef CAS PubMed.
- V. Voorde, R. T. Steven, A. K. Najumudeen, C. A. Ford, A. Dexter, A. Gonzalez-Fernandez, C. J. Nikula, Y. Xiang, L. Ford, S. Maneta Stavrakaki, K. Gilroy, L. B. Zeiger, K. Pennel, P. Hatthakarnkul, E. A. Elia, A. Nasif, T. Murta, E. Manoli, S. Mason, M. Gillespie, T. R. M. Lannagan, N. Vlahov, R. A. Ridgway, C. Nixon, A. Raven, M. Mills, D. Athineos, G. Kanellos, C. Nourse, D. M. Gay, M. Hughes, A. Burton, B. Yan, K. Sellers, V. Wu, K. De Ridder, E. Shokry, A. Huerta Uribe, W. Clark, G. Clark, K. Kirschner, B. Thienpont, V. S. W. Li, O. D. K. Maddocks, S. T. Barry, R. J. A. Goodwin, J. Kinross, J. Edwards, M. O. Yuneva, D. Sumpton, Z. Takats, A. D. Campbell, J. Bunch and O. J. Sansom, Nat. Metab., 2023, 5, 1303–1318 CrossRef.
- Y. Dong, B. Li and A. Aharoni, Trends Plant Sci., 2016, 21, 686–698 CrossRef CAS PubMed.
- R. M. Caprioli, J. Am. Soc. Mass Spectrom., 2015, 26, 850–852 CrossRef CAS PubMed.
- N. E. Mascini, M. Cheng, L. Jiang, A. Rizwan, H. Podmore, D. R. Bhandari, A. Römpp, K. Glunde and R. M. A. Heeren, Anal. Chem., 2016, 88, 3107–3114 CrossRef CAS PubMed.
- C. Sun, F. Wang, Y. Zhang, J. Yu and X. Wang, Theranostics, 2020, 10, 7070–7082 CrossRef CAS PubMed.
- X. Han, D. Wang, L. Yang, N. Wang, J. Shen, J. Wang, L. Zhang, L. Chen, S. Gao, W.-X. Zong and Y. Wang, Proc. Natl. Acad. Sci. U.S.A., 2024, 121, 0027–8424 Search PubMed.
- O. Phanstiel, Int. J. Cancer, 2017, 142, 1968–1976 CrossRef.
- D. Murthy, K. S. Attri, V. Suresh, G. H. Rajacharya, C. A. Valenzuela, R. Thakur, J. Zhao, S. K. Shukla, N. V. Chaika, D. LaBreck, C. V. Rao, M. A. Hollingsworth, K. Mehla and P. K. Singh, Proc. Natl. Acad. Sci. U.S.A., 2014, 121, 0027–8424 Search PubMed.
- J. F. J. Bogie, M. Haidar, G. Kooij and J. J. A. Hendriks, Adv. Drug Delivery Rev., 2020, 159, 198–213 CrossRef CAS.
- Y. Cheng, G. Xie, T. Chen, Y. Qiu, X. Zou, M. Zheng, B. Tan, B. Feng, T. Dong, P. He, L. Zhao, A. Zhao, L. X. Xu, Y. Zhang and W. Jia, J. Proteome Res., 2011, 11, 1354–1363 CrossRef.
- A. Pakiet, J. Kobiela, P. Stepnowski, T. Sledzinski and A. Mika, Lipids Health Dis., 2019, 18, 1476–511X CrossRef.
- J. Moro, D. Tomé, P. Schmidely, T.-C. Demersay and D. Azzout-Marniche, Nutrients, 2020, 12, 1414 CrossRef CAS.
- A. Martner, H. G. Wiktorin, B. Lenox, F. Ewald Sander, E. Aydin, J. Aurelius, F. B. Thorén, A. Ståhlberg, S. Hermodsson and K. Hellstrand, J. Immunol., 2015, 194, 5014–5021 CrossRef CAS PubMed.
- G. S. Hotamisligil, Nature, 2017, 542, 177–185 CrossRef CAS PubMed.
- M. Takahashi, Y. Hasegawa, K. Maeda, M. Kitano and N. Taniguchi, Glycoconjugate J., 2022, 39, 167–176 CrossRef CAS PubMed.
- L. R. Conroy, H. A. Clarke, D. B. Allison, S. S. Valenca, Q. Sun, T. R. Hawkinson, L. E. A. Young, J. E. Ferreira, A. V. Hammonds, J. B. Dunne, R. J. McDonald, K. J. Absher, B. E. Dong, R. C. Bruntz, K. H. Markussen, J. A. Juras, W. J. Alilain, J. Liu, M. S. Gentry, P. M. Angel, C. M. Waters and R. C. Sun, Nat. Commun., 2023, 14, 2041–1723 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra08100e |
| ‡ These authors contributed equally to this study. |
| This journal is © The Royal Society of Chemistry 2025 |