Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Web-based graphical user interface for visualizing and analyzing chemical reaction networks
Akihiro Hondaa,
Mikael Kuwahara a,
Yoshiki Hasukawa
a,
Yoshiki Hasukawa a,
Keisuke Takahashi
a,
Keisuke Takahashi *ab and
Lauren Takahashi
*ab and
Lauren Takahashi *a
*a
aDepartment of Chemistry, Hokkaido University, North 10, West 8, Sapporo 060-0810, Japan. E-mail: keisuke.takahashi@sci.hokudai.ac.jp; lauren.takahashi@sci.hokudai.ac.jp
bList Sustainable Digital Transformation Catalyst Collaboration Research Platform, Institute for Chemical Reaction Design and Discovery, Hokkaido University, Sapporo 001-0021, Japan
First published on 29th May 2025
Abstract
A graphical user interface for network visualization and analysis is proposed, integrated within the Catalyst Acquisition by Data Science (CADS) platform to simplify the study of chemical reaction networks. The interface enables users to upload CSV data and perform centrality calculations, clustering, shortest path searches, and more, without requiring programming skills. It offers intuitive features such as node highlighting, navigation, and visualization in force-directed or circular layouts. Additionally, networks can be represented as Petri nets or directed graphs, providing deeper insights into reaction dynamics. This platform aims to make network theory more accessible and accelerate its application in chemical research.
1 Introduction
Network theory has garnered significant attention in the field of chemistry due to its ability to represent chemical reactions in the form of networks.1–5 In this framework, chemical reactions are described as transformations involving reactants, which are substances that undergo chemical changes, and products, which are the substances formed as a result. Within a chemical reaction network, nodes represent reactants, intermediates, and products, while edges denote the reactions that connect these species. The intricate web of intermediates that often mediate the transformation from reactants to products is of particular interest, as understanding these connections can unveil the underlying mechanisms governing the reaction.6,7 Graph theory-based analyses, such as centrality analysis, are powerful tools for identifying key intermediates that play pivotal roles in controlling reaction path ways.7–9 Similarly, shortest path analysis facilitates the identification of the most efficient routes between specific reactants and products, providing insights into reaction efficiency and selectivity.10 By utilizing network visualization and analysis techniques, researchers can intuitively explore the complex interplay of species within a reaction, enabling the design of optimized reaction pathways and novel catalysts. Consequently, the integration of network theory into chemical research has become an indispensable approach for elucidating reaction mechanisms and advancing the understanding of chemical processes.Network visualization and analysis have garnered significant interest in chemistry, offering insights into complex datasets and relationships. However, the adoption of these techniques is often hindered by the need for programming expertise and foundational knowledge of graph theory, presenting a barrier for researchers in general. Efforts have been made to improve the accessibility of network analysis and have resulted in the development of several tools to help navigate the complexity of reaction networks.11–14 However, despite the ability to navigate the reactions found in reaction platforms such as SCAN and their relevant chemical data, the data used for network analysis are fixed or otherwise tied to the platform, restricting use to those who which to analysis reaction networks consisting of computation data.10 While software tools such as NetworkX and Gephi provide robust capabilities for network visualization and analysis, they typically require a certain level of programming and graph theory proficiency, limiting accessibility for beginners and non-specialists.15–17 Other resources like Cytoscape require additional installation and are designed for specific areas of research.18 To address these challenges, a user-friendly, web-based graphical user interface for network visualization and analysis is proposed, integrated within the Catalyst Acquisition by Data Science (CADS) platform (https://cads.eng.hokudai.ac.jp/).19 Network graphical user interface in this work aims to accelerate research by providing an intuitive, accessible environment that enables researchers—regardless of their programming experience—to perform initial assessments and generate impactful visualizations with minimal effort.
2 Architecture
Graphical user interface of network visualization and graph theory analysis is constructed. This tool is developed with the goal of improving accessibility to network analysis tools to users who are unfamiliar with network analysis, graph theory, and programming but wish to explore network analysis or otherwise apply these tools towards their research. Network visualization is created based on D3.js, a Javascript library used for network visualization. Networks are constructed where node positions are determined by how a node relates to all other nodes within the network. The basic graph is created by applying a repulsive force between nodes to all nodes, plus an attractive force between nodes connected by links. In addition, a force towards the center of the canvas is set on all nodes, aligning the graph to the center. The software is mainly based on the D3 force simulations algorithm for analysis. The D3 force simulations require a list of edges consisting of two columns, the source node and the target node, for the creation of the graph. When the user enters and submits this list in a form-style page, the data is first sent to the back-end, which is written in Python. Here, Python libraries NetworkX and Pandas are used to calculate various metrics for the network diagram according to the user's desired configuration. The indicators are then classified and linked to each node, and sent back to the front-end side. D3.js is used to define the D3 force simulations settings, mouse click handler functions, for instance. Based on the calculated data, nodes and edges are drawn from the list of edges, omitting duplicates. For cases where an edge has a weight, the strength of the attraction of the edge is determined by how great the weight is. This places nodes of greater importance more centrally in the network.In network analysis, centrality is often calculated as it can give insight into which nodes control certain aspects of the network or otherwise play a central role in how paths are created in a network. Centrality is a measure that quantifies the importance or influence of a node within a network based on its position and connectivity. In a chemical reaction network, centrality can help identify key intermediates or species that play crucial roles in connecting multiple reaction pathways or controlling the overall reaction flow. Here, if centrality is calculated, the software automatically sets the size of the node larger as the centrality value increases. Centrality is calculated according to the number of connections and the value of the weights of the edges to be connected. These visualisation performance-enhancing features, together with SVG drawing, enable the generation of intuitive and highly mobile network diagrams.
3 Components
Users can upload network data in CSV format, where the file contains source and target nodes. The source node represents the starting point, and the target node indicates the endpoint. The system automatically connects these nodes to generate the corresponding network components. The input interface for network visualization and analysis is initially constructed, as shown in Fig. 1, with the assumption that users have already uploaded their data. As illustrated in Fig. 1, users can select source and target nodes via a pull-down menu. Additionally, edge weights can be assigned through a pull-down menu, provided the uploaded data includes the corresponding values. The interface also supports centrality analysis, with six centrality metrics available: degree, eigenvector, Katz, PageRank, betweenness, and closeness. An attraction parameter is included to adjust the force dynamics for constructing the network. Users can choose between visualizing the network as a Petri net or a directed graph, with the default setting being a non-Petri network with a directed graph. A node highlighting feature allows specific nodes to be marked, while a clustering option supports four algorithms: Greedy, Louvain, Girvan–Newman, and label propagation, with customizable clustering force parameters. Two types of network graph layouts are offered: force-directed and circular layouts. Options are also provided to remove networks that are disconnected from the largest network or nodes without connections to others. Overall, the network visualization and analysis tools are designed to enable comprehensive functionality through an intuitive, click-based interface.Upon submitting the input data, the corresponding chemical reaction network is generated and displayed. The methane oxidation chemical reaction network from a previous study7 is used as the dataset. One of the key features of CADS is its ability to simultaneously display different components, as illustrated in Fig. 2. This figure demonstrates the network data presented in tabular form alongside multiple network visualizations, each analyzed using distinct network analysis techniques. The use of tabular data facilitates precise comprehension of numerical details. In addition, parallel visualization of results obtained from different analytical methods allows for a clearer and more intuitive identification of their differences. This functionality significantly benefits researchers by enabling direct comparison and evaluation of the network from diverse perspectives.
The network visualization and analysis component incorporates several essential functions, as illustrated in Fig. 3. Notably, this component offers interactive network visualization, providing users with a range of tools to explore the network effectively. The first feature is a zoom-in and zoom-out functionality, enabling users to adjust the view using the mouse wheel. This allows for both a comprehensive overview of the entire network structure and a detailed examination of local features. Additionally, a pop-up function is included, which displays detailed information about a selected node when the mouse pointer hovers over it. As shown in Fig. 3, this information includes the node name, the number of links it possesses, and the nodes connected to it. The visualized network can also be freely navigated by holding the left mouse button and dragging to reposition it. Lastly, when a user clicks on a node, the selected node and its connected nodes are highlighted and emphasized. This feature enables users to instantly identify all connections associated with the selected node, enhancing the analysis and interpretation of the network.
A shortest path search function is implemented and can be accessed using the steps illustrated in Fig. 4. This algorithm is an adaptation of Dijkstra's shortest path algorithm, enhanced with constraints to limit traversal to specific nodes and directed edges. The graph is treated as a directed, weighted structure, where movement between nodes is only allowed in the specified direction of each edge. The shortest path is determined by iteratively selecting the node with the lowest tentative distance from the source and updating the distances of its valid neighbors. At each step, only neighbors reachable through permitted directed edges and satisfying user-provided input conditions are considered. Edge weights are interpreted such that connections with greater weight are favored during pathfinding. The algorithm maintains a record of the previous node for each visited node, allowing the path to be constructed once the destination is reached. The process continues until the destination is reached or no further progress can be made. If a valid path to the destination exists under the imposed constraints, the algorithm returns the sequence of nodes comprising the shortest path. If no such path exists, an empty result is returned.
In this function, user can select the starting node by holding control key while holding key, user can click end node. When users select two node, the shortest path to connect two nodes are instantly highlighted. Required characters can be set in the search, and if set, the route is searched only at nodes that contain those characters. In the case of directed graphs, routes are searched for only in the direction of the arrows, but as with undirected graphs, it is possible to search ignoring the arrows by configuration. This function is powerful to unveil the reaction path.
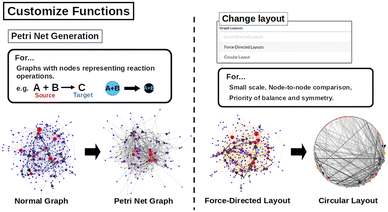
Various network layout techniques are implemented in the network component, as shown in Fig. 5. One of the notable feature is to convert networks into Petri graphs. Petri graphs are network representations with two types of nodes and are suitable for representing chemical reactions. For example, when considering the reaction A + B → C, where a certain molecule A is transformed into molecule C by reagent B, easy expressions such as A → C or B → C can lead to the misunderstanding that molecule C is produced from reagent B. To solve this problem, the reactant is once connected to it as a node of a chemical species pair, creating an A + B → C link. This chemical species pair node can be said to indicate a reaction operation. By using these two types of node, the chemical species pair and the chemical species, the chemical reaction can be translated into a network in a more accurate way. Two types of network layouts are offered: force-directed layout and circular layout. The force-directed layout arranges nodes in a way that mimics physical forces, such as attraction and repulsion. This layout is particularly useful for visualizing the structure of complex networks, as it naturally separates clusters and highlights relationships among nodes. On the other hand, the circular layout positions nodes along the circumference of a circle. This layout is advantageous for representing hierarchical relationships or when comparing multiple networks, as it provides a uniform and symmetrical arrangement. These layout techniques enable researchers to analyze and interpret network structures effectively from different perspectives.
4 Conclusion
The proposed graphical user interface for network visualization and analysis simplifies the exploration of chemical reaction networks, making advanced tools accessible without requiring programming expertise. With features like centrality analysis, clustering, shortest path searches, and multiple layout options, the platform enables intuitive and interactive exploration by transforming complex network analysis tools into a more user-friendly format. The ability to represent networks as Petri nets or directed graphs further enhances its utility in understanding reaction dynamics. Given its ability to model reaction network data into a dynamic network, it is expected that further development will include support for modeling reaction data generated from other open-source reaction network frameworks as well as incorporate microkinetics-specific functions. Thus, this user-friendly interface lowers technical barriers and promotes the broader adoption of network theory in chemical research.5 Code
The complete source code for the network visualization and analysis component can be found together with the public source code for the whole CADS system on GitHub at the following web address: https://github.com/Material-MADS/mads-app under MIT license.Data availability
All data and source codes are opened at github https://github.com/Material-MADS/mads-app.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is funded by the Japan Science and Technology Agency (JST), ERATO grant number (JPMJER1903), PRESTO grant number (JPMJPR24T5), JST Mirai Program grant number (JP-MJMI25G1), JSPS KAKENHI Grant-in-Aid for Scientific Research (B) Grant Number (JP23H01762) and (24K01241).Notes and references
- R. Garcia-Domenech, J. Galvez, J. V. de Julian-Ortiz and L. Pogliani, Some new trends in chemical graph theory, Chem. Rev., 2008, 108, 1127–1169 CrossRef CAS PubMed.
- S. Maeda, K. Ohno and K. Morokuma, Systematic exploration of the mechanism of chemical reactions: the global reaction route mapping (GRRM) strategy using the ADDF and AFIR methods, Phys. Chem. Chem. Phys., 2013, 15, 3683–3701 RSC.
- C. A. Grambow, A. Jamal, Y.-P. Li, W. H. Green, J. Zador and Y. V. Suleimanov, Unimolecular reaction pathways of a γ-ketohydroperoxide from combined application of automated reaction discovery methods, J. Am. Chem. Soc., 2018, 140, 1035–1048 CrossRef CAS PubMed.
- R. Van de Vijver and J. Zador, KinBot: Automated stationary point search on potential energy surfaces, Comput. Phys. Commun., 2020, 248, 106947 CrossRef CAS.
- J. M. Shambhawi, J. M. Weber and A. A. Lapkin, Micro-kinetics analysis based on partial reaction networks to compare catalysts performances for methane dry reforming reaction, Chem. Eng. J., 2023, 466, 143212 CrossRef CAS.
- C. W. Gao, J. W. Allen, W. H. Green and R. H. West, Reaction Mechanism Generator: Automatic construction of chemical kinetic mechanisms, Comput. Phys. Commun., 2016, 203, 212–225 CrossRef CAS.
- L. Takahashi, J. Ohyama, S. Nishimura and K. Takahashi, Representing the Methane Oxidation Reaction via Linking First Principles Calculations and Experiment with Graph Theory, J. Phys. Chem. Lett., 2020, 12, 558–568 CrossRef PubMed.
- M. J. McDermott, S. S. Dwaraknath and K. A. Persson, A graphbased network for predicting chemical reaction pathways in solid-state materials synthesis, Nat. Commun., 2021, 12, 3097 CrossRef CAS PubMed.
- L. Takahashi, S. Yoshida, J. Fujima, H. Oikawa and K. Takahashi, Unveiling the reaction pathways of hydrocarbons via experiments, computations and data science, Phys. Chem. Chem. Phys., 2022, 24, 29841–29849 RSC.
- M. Kuwahara, Y. Harabuchi, S. Maeda, J. Fujima and K. Takahashi, Searching chemical action and network (SCAN): an interactive chemical reaction path network platform, Digital Discovery, 2023, 2, 1104–1111 RSC.
- U. Gupta and D. G. Vlachos, Reaction Network Viewer (ReNView): An open-source framework for reaction path visualization of chemical reaction systems, SoftwareX, 2020, 11, 100442 CrossRef.
- C. H. Muller, M. Steiner, J. P. Unsleber, T. Weymuth, M. Bensberg, K.-S. Csizi, M. Morchen, P. L. Turtscher and M. Reiher, Heron: Visualizing and Controlling Chemical Reaction Explorations and Networks, J. Phys. Chem. A, 2024, 128, 9028–9044 CrossRef CAS PubMed.
- E. Martinez-Nunez, G. L. Barnes, D. R. Glowacki, S. Kopec, D. Pelaez, A. Rodriguez, R. Rodriguez-Fernandez, R. J. Shannon, J. J. Stewart and P. G. Tahoces, et al., AutoMeKin2021: An open-source program for automated reaction discovery, J. Comput. Chem., 2021, 42, 2036–2048 CrossRef CAS PubMed.
- G. N. Simm and M. Reiher, Context-driven exploration of complex chemical reaction networks, J. Chem. Theory Comput., 2017, 13, 6108–6119 CrossRef CAS PubMed.
- A. Hagberg, P. J. Swart and D. A. Schult, Exploring network structure, dynamics, and function using NetworkX, 2008 Search PubMed.
- M. Bastian, S. Heymann and M. Jacomy, Gephi: an open source software for exploring and manipulating networks, Proceedings of the International AAAI Conference on Web and Social Media, 2009, pp. 361–362 Search PubMed.
- A. Rodriguez, R. Rodriguez-Fernandez, S. A. Vazquez, G. L. Barnes, J. J. P. Stewart and E. Martinez-Nunez, tsscds2018: A code for automated discovery of chemical reaction mechanisms and solving the kinetics, J. Comput. Chem., 2018, 39, 1922–1930 CrossRef CAS PubMed.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res., 2003, 13, 2498–2504 CrossRef CAS PubMed.
- J. Fujima, Y. Tanaka, I. Miyazato, L. Takahashi and K. Takahashi, Catalyst Acquisition by Data Science (CADS): a webbased catalyst informatics platform for discovering catalysts, React. Chem. Eng., 2020, 5, 903–911 RSC.
| This journal is © The Royal Society of Chemistry 2025 |