Open Access Article
Open Access ArticleA chemically modified DNAzyme-based electrochemical sensor for binary and highly sensitive detection of reactive oxygen species
Baoting
Dou
 a,
Hui
Shen
a,
Zhimin
Li
a,
Huanyu
Cheng
a,
Hui
Shen
a,
Zhimin
Li
a,
Huanyu
Cheng
 b and
Po
Wang
b and
Po
Wang
 *a
*a
aSchool of Chemistry and Materials Science, Jiangsu Normal University, Xuzhou 221116, China. E-mail: wangpo@jsnu.edu.cn
bDepartment of Engineering Science and Mechanics, The Pennsylvania State University, University Park, State College, PA 16802, USA
First published on 8th January 2025
Abstract
Reactive oxygen species (ROS) play a critical role in regulating various physiological processes. To gain a comprehensive understanding of their distinct functions in different physiological events, it is imperative to detect binary ROS simultaneously. However, the development of the sensing method capable of binary ROS detection remains a significant challenge. In this study, we address this challenge by integrating chemically modified DNAzyme probes with a functionalized metal–organic framework (MOF) to create an efficient electrochemical sensing platform for the binary detection of ROS. ROS targets would activate the DNAzyme cleavage activity by removing the phenylboronate (BO) and phosphorothioate (PS) modifications, leading to the controlled release of doxorubicin (DOX) and methylene blue (MB) enclosed within MOF nanocomposites. This process generates two distinct voltammetric current peaks, with their potentials and intensities reflecting the identity and concentration of the ROS targets. The sensor demonstrates simultaneous detection of multiple ROS (H2O2 and HClO) produced by cancer cells with high sensitivity across a broad linear range of 1 to 200 nM and a low detection limit in the sub-nanomolar range. The design strategies behind the developed ROS sensing system could also be exploited to create other biosensors with highly sensitive and binary detection to promote clinical research and revolutionize disease diagnostics.
Introduction
As a class of reactive, neutral, and anionic small molecules, representative reactive oxygen species (ROS) include superoxide (O2˙−), hydrogen peroxide (H2O2), singlet oxygen (1O2), and hypochlorous acid/hypochlorite (HClO/ClO−).1 These molecules are often involved in cellular immunity, migration, differentiation, and signal transduction.2,3 For instance, HClO generated by a myeloperoxidase-catalyzed reaction is involved in a variety of physiological and pathological events.4,5 As a powerful nucleophilic non-radical oxidant, HClO is essential for the immune system to fight off invading microorganisms. However, excessive use of HClO as a disinfectant in domestic water and food production can lead to the oxidative damage of various biological substances and induce oxidative stress and cell damage.6–8 Additionally, an excessive amount of hydrogen peroxide (H2O2) has been linked to numerous pathological disorders, such as Alzheimer's disease, angiocardiopathy, neurodegeneration, diabetes, and cancer, and can cause harm to health in industrial, biological, and environmental systems.9–11 Therefore, it is of great biomedical significance to develop an effective strategy for ROS detection.Different analytical techniques have been developed for the detection of ROS, including electrochemical,12–14 Raman spectroscopy,15 and fluorescence16 methods. Of these, electrochemical detection exhibits high sensitivity, low cost, quick response, environmental friendliness, and ease of use.17 Electrochemical ROS sensing systems can be divided into enzyme-immobilized and enzyme-free sensors.18 The former utilize protein enzyme catalysts like hemoglobin,19 catalase,20 and horseradish peroxidase21 to monitor the electronic transfer of ROS redox reactions; however, their catalytic activity can be affected by temperature, pH, and reaction medium, presenting challenges in practical applications with complicated matrices.22,23 On the other hand, enzyme-free sensors employ catalytic nanomaterials to modify the sensing interface and achieve electrocatalytic redox reactions, benefitting from the unique electrical, catalytic, and mechanical properties of nanomaterials.24–26 For instance, we prepared a trimetallic hybrid nanoflower/MoS2 nanocomposite for in situ determination of H2O2, which obtained a detection limit of 0.3 nM, and the mean number of H2O2 secreted per cancer cell was determined to be 1011.27 Although these methods are highly advanced, they primarily focus on nanomaterial catalysts for ROS detection, with fewer applications using biometric probes. Importantly, the high reactivity of ROS has magnified the difficulty of binary detection in biological and environmental systems, making it a significant challenge to develop a sensing system for binary ROS detection.
In this regard, this study develops ROS stimuli-responsive DNAzyme probes and an electrochemical signal-encapsulated metal–organic framework (MOF) to achieve binary ROS detection. As a class of deoxynucleotide oligomers, DNAzymes can be identified as deoxyribozymes or catalytic DNA through in vitro selection to exhibit enzyme-like activities.28,29 Stimuli-responsive modifications on DNAzymes contribute to their specific applications in sensing metal ions, proteins, and DNA, as well as intracellular gene regulation.30,31 Despite these promising developments, there is still a scarcity in the study and application of ROS-activatable DNAzyme probes. Based on the combination of the good biocompatibility, high loading ability, and versatile designability of MOFs,32–35 this work incorporates phenylboronate (BO) and phosphorothioate (PS) modifications to design H2O2-activable and HClO-activable DNAzyme probes, which utilize ROS targets to control the release of electrochemical signals encapsulated in MOF nanocomposites for binary ROS detection. The study would exhibit two major advantages. First, the binary detection of ROS targets was accomplished through selective recognition and the corresponding signal output. Second, the sensitivity was enhanced through an efficient amplification method, allowing for a minimal detection limit and revealing its immense practical applicability for cancer detection and diagnostics.
Experimental
Chemicals and materials
Oligonucleotides supplied by Sangon Biotechnology Co., Ltd. (Shanghai, China) have been purified by HPLC, with the sequences provided as follows. Substrate probe (rA-SP): 5′-SH-(CH2)6-ACTCAC TAT/rA/GG AAGAGA TG-3′; PS modified DNAzyme probe 1 (PS-DP1): 5′-CATCTC TTCTCC GAGCCG GTCGAA ATAGTG AGTAPSAA ACTCAC TAT-3′; BO modified DNAzyme probe 2 (BO-DP2): 5′-CATCTC TTCTCC GBOABOGBOCCG GTCGAA ATAGTG AGT-3′, where rA refers to a ribonucleotide, APS refers to the phosphorothioate version of A, and ABO and GBO refer to BO-modified nucleotides. All the chemicals required for the experiment, including zirconium tetrachloride (ZrCl4), phorbol 12-myristate-13-acetate (PMA), benzoic acid (BzOH), N,N-dimethylformamide (DMF), ascorbic acid (AA), doxorubicin hydrochloride (DOX), uric acid (UA), sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (Sulfo-SMCC), sodium nitrite (NaNO2), lipopolysaccharide (LPS), methylene blue (MB), glutathione (GSH), sodium nitrate (NaNO3), 2-aminoterephthalic acid (NH2-BDC), dopamine (DA), acetylcholine (ACh), noradrenaline (NE), glycine (Gly), serotonin (ST), sodium peroxynitrite (NaONOO) and 3,4-dihydroxyphenylacetic acid (DOPAC), were supplied by Aladdin Chemical Co., Ltd. (Shanghai, China). The standard H2O2 assay kit (colorimetric method) was purchased from Sangon Biotechnology Co., Ltd. (Shanghai, China). The Amplite fluorimetric HClO assay kit was supplied by AmyJet Scientific Co., Ltd. (Wuhan, China). The HeLa (human cervical carcinoma) cell line was purchased from the cell bank of the type culture collection of the Chinese Academy of Sciences (Shanghai, China), and Dulbecco's Modified Eagle's Medium (DMEM) was obtained from Thermo Fisher Scientific Co., Ltd (Shanghai, China).Preparation of the UiO MOF
The UiO MOF was synthesized with a hydrothermal method based on a reported method with minor modifications.36 In brief, ZrCl4 (250 mg), benzoic acid (4 g), and NH2-BDC (230 mg) were first added and dissolved in DMF (20 mL) under ultrasonication for 3 min. The above solution was subsequently transferred into a stainless steel-lined Teflon autoclave and heated at 120 °C for 24 h. After cooling to 25 °C, the obtained product was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The precipitate was washed using DMF and anhydrous ethanol alternately to remove residual chemicals. Finally, the resulting product UiO MOF was dried under reduced pressure at 100 °C for further use.
000 rpm for 10 min. The precipitate was washed using DMF and anhydrous ethanol alternately to remove residual chemicals. Finally, the resulting product UiO MOF was dried under reduced pressure at 100 °C for further use.
Electrochemical detection of binary ROS
The prepared UiO MOF (1 mg) was first dispersed in 450 μL of 20 mM phosphate-buffered saline (PBS, 5 mM MgCl2, 100 mM NaCl, pH 7.4) under ultrasonic treatment for 10 min and conjugated with Sulfo-SMCC (50 μL, 10 mM) for 120 min. The excess Sulfo-SMCC was removed through centrifugation at 8000 rpm for 5 min. Next, the resulting products were redispersed in 500 μL PBS buffer consisting of 100 μM rA-SP and incubated for 5 h, followed by centrifugation and rinsing to remove unbound rA-SP, resulting in the formation of rA-SP/UiO. Further incubation of DOX (10 mM) with rA-SP/UiO (1 mg L−1) for 12 h under continuous agitation loaded the DOX into UiO. After adding PS-DP1 (150 μM) and incubating for 4 h for pore sealing, the resulting complex was rinsed using PBS buffer to remove residual DOX and DNA, defining it as (DNA–DOX)/UiO. (DNA–MB)/UiO was prepared through the same procedures but rA-SP/UiO (1 mg L−1) was incubated with MB (10 mM) and BO-DP2 (150 μM) in sequence. Furthermore, different concentrations of HClO and H2O2 were added into the PBS solution consisting of (DNA–MB)/UiO (1 mg L−1) and (DNA–DOX)/UiO (1 mg L−1) in the presence of 100 μM Zn2+, followed by reacting at 25 °C with gentle agitation for 50 min for electrochemical testing.Pretreatment of the working electrode
The gold electrode (AuE, 2 mm diameter), utilized as the working electrode, was pretreated by immersing it in a freshly prepared piranha solution (consisting of concentrated 98% H2SO4/30% H2O2 in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio) for 30 min, followed by rinsing with ultrapure water. Subsequently, the electrode underwent polishing using 0.3 μm and 0.05 μm alumina slurries to achieve a mirror-like surface. After sonication in ethanol and ultrapure water, the AuE was subjected to electrochemical cleaning through potential scanning in 0.5 M H2SO4 solution ranging from −0.3 to 1.55 V until stable voltammetric peaks were observed. Following this procedure, the electrode was rinsed with ultrapure water and dried under a nitrogen stream for subsequent detection.
1 volume ratio) for 30 min, followed by rinsing with ultrapure water. Subsequently, the electrode underwent polishing using 0.3 μm and 0.05 μm alumina slurries to achieve a mirror-like surface. After sonication in ethanol and ultrapure water, the AuE was subjected to electrochemical cleaning through potential scanning in 0.5 M H2SO4 solution ranging from −0.3 to 1.55 V until stable voltammetric peaks were observed. Following this procedure, the electrode was rinsed with ultrapure water and dried under a nitrogen stream for subsequent detection.
Detection of ROS released from cancer cells
HeLa cells were grown in a DMEM solution that contained 10% fetal bovine serum and 1% penicillin/streptomycin, in a humidified atmosphere of 5% CO2 at 37 °C. After reaching 90% confluence, the cells were collected and counted using a hemacytometer and then packed in PBS buffer. The final packed cells (5 × 105) were redispersed in PBS buffer and subsequently mixed with PMA (20 mg L−1) and LPS (20 mg L−1) to conduct electrochemical measurements after the addition of (DNA–MB)/UiO (1 mg L−1) and (DNA–DOX)/UiO (1 mg L−1).Apparatus and measurement
Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) images were acquired using an FEI Tecnai G2 Spirit Twin microscope operated at 120 kV and a Hitachi SU8010 scanning electron microscope (Tokyo, Japan) at an acceleration voltage of 5 kV, respectively. Differential pulse voltammetry (DPV) was carried out using a CHI 660E electrochemical workstation (Shanghai Chenhua Instrument, China) equipped with a platinum wire counter electrode, a gold working electrode, and an Ag/AgCl reference electrode. X-ray diffraction (XRD) was analyzed using a D8 Advance X-ray diffractometer. Brunauer–Emmett–Teller (BET) measurements were taken at −196 °C using nitrogen on an automatic volumetric adsorption apparatus (Micrometrics ASAP, 2020). X-ray photoelectron spectroscopy (XPS) spectra were acquired using an XPS ESCALAB 250Xi (Thermo Fisher Scientific, USA). The native polyacrylamide gel electrophoresis (PAGE) experiments were performed on a DYCZ-24DN electrophoresis instrument (Beijing LIUYI Biotechnology Co., Ltd). The prepared DNA samples were mixed with loading buffer (6×) at a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio and loaded onto 16% PAGE. The electrophoresis process was conducted in 1× TBE buffer (pH 8.0) at 110 V for 60 min, followed by staining with ethidium bromide for approximately 30 min to enable visualization using the Bio-Rad imaging system in Hercules, CA, U.S.A.
1 volume ratio and loaded onto 16% PAGE. The electrophoresis process was conducted in 1× TBE buffer (pH 8.0) at 110 V for 60 min, followed by staining with ethidium bromide for approximately 30 min to enable visualization using the Bio-Rad imaging system in Hercules, CA, U.S.A.
Ethical statement
The blood samples were acquired from consenting healthy volunteers in the affiliated hospital of Xuzhou Medical University (Jiangsu, China). All experiments associated with blood samples were performed in accordance with the World Health Organization (WHO) guidelines (WHO Publication ISBN-13: 978-92-4-159922-1, 2010) and approved by the ethics committee at the affiliated hospital of Xuzhou Medical University. Informed consent was obtained from the human participants of this study.Results and discussion
Working principle of HClO and H2O2 detection
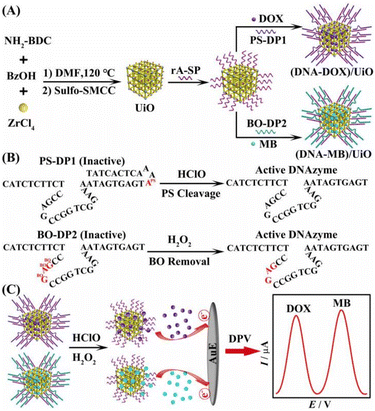
The design of the sensor involves two electrochemical reporters (DOX and MB), in which DOX is a dual-functioning compound that acts as both an anticancer medication and an electroactive marker.37–39 Its intrinsic electroactive properties, stemming from its quinone and hydroquinone groups, enable the direct and rapid electrochemical analysis of DOX in biological samples without the need for complex pretreatment. MB is an active electron transmitter that is widely utilized in sensing applications because of its remarkable stability and exceptional electrochemical properties.40,41 The distinct current signals of MB and DOX with well-resolved potentials at −0.22 V and −0.62 V enable the precise identification of the binary ROS targets. The preparation of the UiO MOF was based on the self-assembly of Zr4+ and NH2-BDC in DMF solution, followed by modification with Sulfo-SMCC via a reaction between the primary amine groups of the MOF and the NHS esters of Sulfo-SMCC (Scheme 1A). After conjugating the sulfhydryl-modified rA-SP to the UiO MOF surface, the electron donors (DOX and MB) would be incorporated into the UiO framework. In the hybridization of DNAzyme probes with rA-SP to seal the hole, the DNA duplexes (PS-DP1/rA-SP and BO-DP2/rA-SP) act as gatekeepers to encapsulate the electron donors within the UiO framework. Self-blocked by a blocking sequence with an inserted PS linkage, PS-DP1 is initially an inactive DNAzyme strand (Scheme 1B). The release of the blocking sequence from the cleavage of the PS linkage by HClO activates the cleavage activity of the DNAzyme. Similarly, the addition of H2O2 removes the disruptive BO modified in catalytic core sequences in BO-DP2, thus recovering the DNAzyme activity. The addition of HClO and H2O2 activates PS-DP1 and BO-DP2 cleavage activity, leading to the release of DOX and MB, where the cycle cleavage of rA-SP results in an amplified electrochemical signal for binary detection of HClO and H2O2 with ultrahigh sensitivity and a low limit of detection (Scheme 1C).Characterization of different nanocomposites
The individual UiO MOF particles display the natural regular octahedron nanostructure, with sizes ranging from 90 to 160 nm (Fig. 1A). The structure observed in the SEM image is also verified by the TEM image with enhanced magnification (Fig. 1B). The distribution curve of the UiO pore diameter in Fig. 1C demonstrates relatively concentrated pore sizes with a mean aperture range from 5 to 15 Å. The type-II N2 adsorption and desorption isotherms of UiO in the inset of Fig. 1C further suggest the porous structure of the UiO MOF framework, with an estimated surface area of approximately 1328 m2 g−1 according to the BET. As can be seen from the representative XRD patterns in Fig. 1D, the sharp diffraction peaks at 2θ = 7.36, 8.48, 12.04, 17.08, 22.25, and 25.68 are assigned to the (111), (002), (022), (004), (115), and (224) crystal planes of UiO, respectively. There is also no obvious difference in the XRD patterns of (DNA–MB)/UiO and (DNA–DOX)/UiO when compared to that of pristine UiO, indicating the minimal influence of these modifications.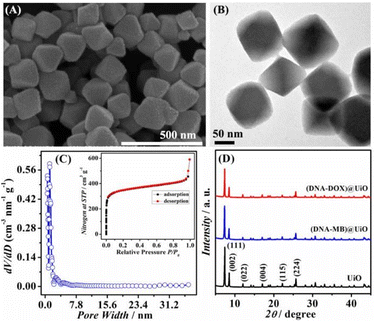 | ||
| Fig. 1 (A) SEM and (B) TEM images and (C) BET characterization of the UiO MOF framework. (D) XRD patterns of (DNA–MB)/UiO, (DNA–DOX)/UiO, and pristine UiO. | ||
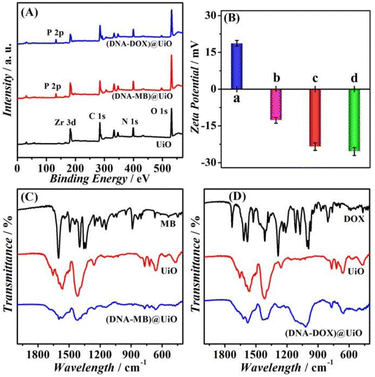
The XPS survey spectra of UiO in Fig. 2A exhibit characteristic peaks at 185, 284, 399, and 531 eV, revealing the presence of Zr 3d, C 1s, N 1s, and O 1s, respectively. In addition, the P 2p at 144 eV originating from the phosphate backbones of DNA manifests the successful preparation of (DNA–MB)/UiO and (DNA–DOX)/UiO. Assessment of the surface functionalization of these prepared samples with zeta potential shows an increased negative charge from +18.73 mV for the pristine UiO to −12.66 mV for the UiO modified with the rA-SP linkage, which further increases to −25.43 mV after the hybridization with the DNAzyme probes (Fig. 2B). The coexisting peaks of MB and UiO in the FT-IR spectra of (DNA–MB)/UiO indicate the effective encapsulation of MB in UiO (Fig. 2C). Similarly, the effective encapsulation of DOX is also evident in (DNA–DOX)/UiO (Fig. 2D).
The activation mechanism of the DNAzyme probe
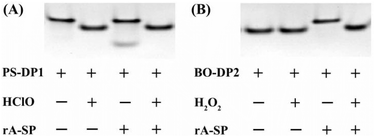
ROS-induced activation of the DNAzyme activity is verified by PAGE characterization. Compared to the distinct band with low mobility in PS-DP1, the active DNAzyme strand (33 nt) produced after the addition of HClO to cleave PS-DP1 exhibits much higher mobility (Fig. 3A). It should be noted that the simultaneously released blocking sequence (12 nt) is too short to be stained and imaged. Two separated bands obtained from the direct mixture of PS-DP1 and rA-SP indicate no hybridization between the two due to the presence of a blocking sequence. In contrast, further addition of HClO into the mixture results in a new band at the position of the active DNAzyme strand and the disappearance of the rA-SP band, demonstrating activated DNAzyme activity by HClO. Similarly, the lower mobility band from the mixture of BO-DP2 and rA-SP when compared to BO-DP2 indicates no cleavage, whereas the vanished BO-DP2/rA-SP band in the presence of H2O2 indicates the activation of BO-DP2 DNAzyme activity by H2O2 (Fig. 3B). (Note: The cleavage products of rA-SP were only 10 bases, making it difficult to stain and image in the two PAGE experiments.)Feasibility testing of the sensor
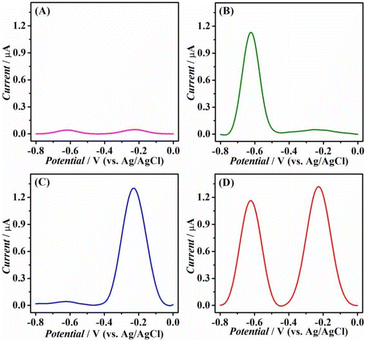
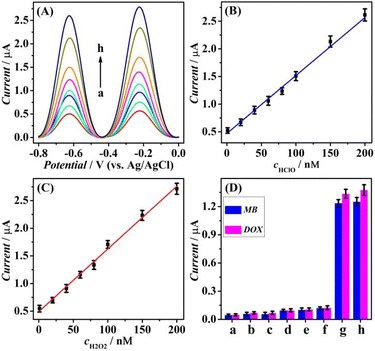
The feasibility of the sensor for simultaneous detection of HClO and H2O2 was further investigated with DPV. The mixture of (DNA–MB)/UiO (1 mg L−1) and (DNA–DOX)/UiO (1 mg L−1) exhibited minor current responses from −0.8 to 0 V (Fig. 4A, SDDOX = 2.1%, SDMB = 1.8%, and n = 3). The small background current was attributed to the nonspecific adsorption of signal substances on the UiO MOF surface, which is a common phenomenon in similar studies.42–44 Compared with the small background response of DOX, the addition of HClO (80 nM) led to a significantly increased current at −0.62 V (Fig. 4B, SDDOX = 2.9%, SDMB = 2.2%, and n = 3). The increased current from 0.0764 μA to 1.162 μA was ascribed to the release of DOX based on the activation of DNAzyme cleavage activity (PS-DP1). Similarly, the DNAzyme cleavage activity of BO-DP2 was activated and MB was released when BO was removed from BO-DP2 upon the addition of H2O2 (80 nM), resulting in an enhanced peak current from 0.0861 μA to 1.311 μA at −0.22 V (Fig. 4C, SDDOX = 2.5%, SDMB = 3.3%, and n = 3). More importantly, the presence of HClO and H2O2 can be simultaneously detected from the two distinct and well-resolved peak potentials at −0.62 V and −0.22 V, respectively (Fig. 4D, SDDOX = 3.9%, SDMB = 3.6%, and n = 3).The optimization of experimental conditions
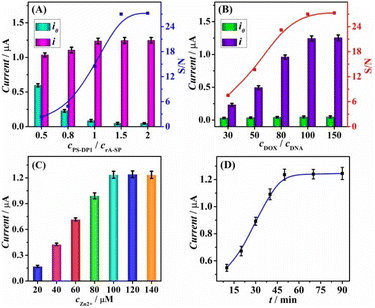
The sensing performance of the sensor was influenced by various experimental conditions, such as the concentration ratio of DNAzyme probe to rA-SP, the concentration ratio of signal reporter to DNA probe, the concentration of Zn2+, and reaction time. To explore the effect of the concentration ratio of DNAzyme probe to rA-SP on the sensing performance of the sensor, the detection of HClO was investigated as a model. As displayed in Fig. 5A, the background current (i0) decreased obviously with the increase in cPS-DP1/crA-SP from 0.5 to 1.5, while the signal current (i) increased gradually with the increase in cPS-DP1/crA-SP from 0.5 to 1.0, and levelled off thereafter. As a result, the concentration ratio of 1.5 for PS-DP1 to rA-SP was chosen for further studies due to the highest signal-to-noise ratio (S/N = i/i0). Besides, it was observed that the signal-to-noise ratio increased gradually with the increase in the concentration ratio of DOX to DNA probe from 30 to 100 and reached a plateau thereafter (Fig. 5B). Therefore, the concentration ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for DOX to DNA probe was selected as the optimal ratio for HClO detection. The influence of Zn2+ concentration on the signal response of the sensor is displayed in Fig. 5C. It was demonstrated that the current response of the sensor increased gradually with the increase in Zn2+ concentration from 20 to 100 μM and levelled off between 100 and 140 μM, indicating that the DNAzyme cleavage reaction reached saturation at 100 μM Zn2+, which was the optimized concentration in this work. Moreover, the current response increased with the increase in reaction time from 10 to 50 min before saturation (Fig. 5D), and the reaction time of 50 min was used in the subsequent studies unless specified otherwise. The similar optimization results were obtained for H2O2 analysis.
1 for DOX to DNA probe was selected as the optimal ratio for HClO detection. The influence of Zn2+ concentration on the signal response of the sensor is displayed in Fig. 5C. It was demonstrated that the current response of the sensor increased gradually with the increase in Zn2+ concentration from 20 to 100 μM and levelled off between 100 and 140 μM, indicating that the DNAzyme cleavage reaction reached saturation at 100 μM Zn2+, which was the optimized concentration in this work. Moreover, the current response increased with the increase in reaction time from 10 to 50 min before saturation (Fig. 5D), and the reaction time of 50 min was used in the subsequent studies unless specified otherwise. The similar optimization results were obtained for H2O2 analysis.
Performance characterization of the sensor
The DPV current responses of DOX and MB increase with the increasing concentrations of HClO and H2O2 (Fig. 6A), with an excellent linear fit between DOX current and HClO concentration: IDOX = 0.01048cHClO + 0.4697 (R2 = 0.9931) (Fig. 6B) and between MB current and H2O2 concentration: IMB = 0.01135cH2O2 + 0.4915 (R2 = 0.9949) (Fig. 6C) in the range from 1 to 200 nM. Using the slope m from the calibration plot and the standard deviation Sb of background noise, the limits of detection (LOD = 3Sb/m) for HClO and H2O2 are determined to be 0.27 and 0.35 nM, respectively.45–47 The reported biosensor to simultaneously detect binary ROS with high sensitivity significantly outperforms other previously reported studies based on different detection methods (Table 1). Compared to the minimal increase (23–95%) in the current responses of DOX and MB to interferents (i.e., NaNO2, UA, AA, NaNO3, and GSH) at 800 nM over that of the blank, HClO and H2O2 at a low concentration (80 nM) lead to significantly enhanced current responses of 1445% for DOX and 1517% for MB (Fig. 6D). The response currents only undergo a negligibly small change of 3.1% after mixing HClO and H2O2 with the above interferents. Moreover, the impacts of neurotransmitters and ROS by-products on the detection of HClO (80 nM) and H2O2 (80 nM) were also investigated. It was demonstrated that common neurotransmitters in physiological fluids displayed almost no interference with the ROS detection at a 200-fold concentration, such as DA, ACh, NE, Gly, and ST. The influences of the by-products of ROS on the determination were tested, including NaONOO and DOPAC. After the introduction of 50-fold concentrations of NaONOO and DOPAC into HClO (80 nM) and H2O2 (80 nM), the current changes of the sensor were less than 3.8% of the original signal response. These findings revealed the high specificity and good applicability of the proposed sensor for the assay of ROS in real samples.| Technique | Detection strategy | Linear range | Detection limit | Simultaneous detection | Real sample application | Ref. |
|---|---|---|---|---|---|---|
| a SERS: surface-enhanced Raman scattering; LSPR: localized surface plasmon resonance; PEDOT/PSS: poly(3,4-ethylenedioxythiophene)/poly(styrene sulfonate). | ||||||
| Fluorescence | Ratiometric detection using an all-in-one fluorescent semiconducting polymer based far-red to near-infrared (NIR) nanoprobe | 1 μM to 50 μM | 500 nM | NO | Mouse mononuclear macrophage RAW264.7 cells | 48 |
| LSPR | Oxidative etching of AgNPs/hematoporphyrin monomethyl ether (HMME)/Au nanoflowers | 0.2 μM to 10 μM | 66.7 nM | NO | Human liver hepatocellular carcinoma HepG2 cells | 49 |
| Fluorescence | ROS responsive 2-mercaptohydroquinone modified MOF probe | 50 nM to 5 μM | 13 nM | NO | Human cervical carcinoma HeLa cells | 50 |
| SERS | Seleno-phenylboronic acid pinacol ester/AuNP nanoprobe targeting | 1 μM to 100 μM | 200 nM | NO | Human liver hepatocellular carcinoma HepG2 cells | 51 |
| Electrochemistry | Trimetallic hybrid nanoflower-decorated MoS2 nanosheet catalyst | 1 nM to 100 nM | 0.3 nM | NO | Human breast cancer MCF-7 cells | 27 |
| Electrochemistry | Iron single atomic site catalyst anchored on 2-D N-doped graphene | 10 μM to 920 μM | 200 nM | NO | Human bronchial epithelial 16HBEC cells | 52 |
| Electrochemistry | PEDOT/PSS film functionalized with a cobalt phthalocyanine (CoPc) catalyst | 200 nM to 50 μM | 95 nM | NO | Human lung adenocarcinoma A549 cells | 53 |
| Electrochemistry | Chemically modified DNAzyme probe sensing | 1 nM to 200 nM | 0.27 nM | YES | Human cervical carcinoma HeLa cells and blood samples | This work |
The sensor's long-term stability is examined following storage at 4 °C. Experimental findings show that the current intensity decreases by 3.2% and 5.1% of its initial response after 15 and 30 days, respectively, indicating the feasibility for continuous operation in practical applications. Furthermore, the eminent reproducibility of this sensor is validated by the small relative standard deviations (RSDs) of 3.7% and 4.5% obtained from six repetitive measurements of HClO (80 nM) and H2O2 (80 nM), respectively.
The detection application in real samples
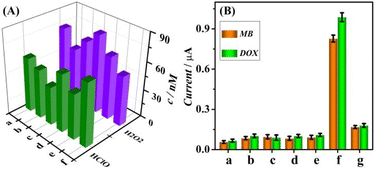
It was revealed that ROS are not stable in physiological fluids.54–56 The specific procedures of serum separation from blood could diminish the levels of ROS due to their instability,57–59 which impeded the quantification of HClO and H2O2 in human serum samples. To circumvent this challenge, freshly collected whole blood samples were employed to evaluate the practicality of the sensor for the assay of ROS in real samples. After collection in ethylenediaminetetraacetic acid tubes, the whole blood samples were diluted 20-fold with PBS buffer for electrochemical measurements following incubation with (DNA–MB)/UiO (1 mg L−1) and (DNA–DOX)/UiO (1 mg L−1). The blood samples from six volunteers were detected three times in parallel, and the assay results are displayed in Fig. 7A. The concentration ranges of HClO and H2O2 in six fresh blood samples were obtained to be 40.5–67.2 nM and 53.4–82.9 nM, respectively, which were in good agreement with some previous reports.18,60,61 To verify the accuracy and reliability of the sensor for ROS detection, the same blood samples were determined using a commercially available standard H2O2 assay kit and an Amplite fluorimetric HClO assay kit. The detection results of the proposed sensor were compared with those of commercial standard kits using Student's t-test, which exhibited a statistical variance of 0.46% at the 95% confidence level, indicating the accuracy and effectiveness of the sensing system for the assay of real biological samples.The proof-of-the-concept demonstration of the designed sensor to analyze ROS released from cancer cells highlighted its potential use in practical applications. In contrast to negligible DOX and MB peak currents in the mixture of MOF probes (column a, Fig. 7B), there was no clear change in current responses upon the addition of LPS (23% for column b) and PMA (18% for column c) without cells. The influence of solvent on the detection signal of the sensor was also tested. Compared with the current response of blank buffer (column a), no significant signal alteration was observed for DMSO (column d), indicating its inability to stimulate the generation of HClO and H2O2 from the HeLa cells. The results confirmed that the DMSO solvent did not influence the detection signal. Meanwhile, the incubation with HeLa cells in the absence of PMA and LPS also did not lead to an obvious current response (22% for column e). In contrast, PMA and LPS injected into the cell solution induced the secretion of H2O2 and HClO,27,62 resulting in significantly increased DOX and MB current responses of 1012% (column f). The numbers of H2O2 and HClO molecules released from per cell were determined using the formula: N0 = [(ΔR/k) × NA]/Ncell, where ΔR represents the current response, k is the sensor sensitivity, NA denotes the Avogadro constant (6.02 × 1023 mol−1), and Ncell is the number of cells.21,27,63,64 With DPV currents of 987.4 nA for HClO and 827.6 nA for H2O2, sensor sensitivities of 10.48 nA nM−1 for HClO and 11.35 nA nM−1 for H2O2, and a cell count of 5 × 105, the N0 values of HClO and H2O2 were calculated to be 1.1 × 1011 and 0.9 × 1011, respectively. These results closely matched a previously reported N0 value of approximately 1011 for HClO and H2O2.27,64,65 The further addition of catalase (5000 U mL−1) results in decreased current responses by 81% (column g), ascribed to the selective scavenging of ROS.27
Conclusions
In conclusion, the manuscript presents an innovative electrochemical biosensor that combines the chemical modifications of DNAzyme probes with a MOF-based electrochemical homogenous detection strategy. The presence of representative ROS such as HClO and H2O2 activates the DNAzyme activity with cyclic cleavage to result in significantly enhanced electrochemical signals. The current signals of DOX and MB exhibit well-resolved potential peaks, achieving the selective discrimination of multiple different ROS. As a result, the reported sensor provides binary and highly sensitive detection of ROS over a wide linear range with an ultralow limit of detection. The proof-of-the-concept demonstration of the designed sensor on the binary detection of ROS released from living cells highlights promising application potential in various cancer diagnoses.Data availability
The data that support the findings of this study are available in the main article. Also, other relevant data for this study are available from the corresponding authors, upon reasonable request.Author contributions
Baoting Dou: conceptualization, project administration, validation, and writing – original draft. Hui Shen: data curation and methodology. Zhimin Li: formal analysis and validation. Huanyu Cheng: writing – review & editing. Po Wang: supervision, project administration, conceptualization, funding acquisition, and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21675067 and 22404067), the Natural Science Foundation of Jiangsu Province (BK20210917 and BE2019645), the Natural Science Foundation of Xuzhou City (KC23238), and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions. H. C. acknowledges the support provided by the NSF (Grant No. 2309323) and Penn State University.Notes and references
- C. Lennicke and H. M. Cochemé, Mol. Cell, 2021, 81, 3691–3707 CrossRef CAS PubMed.
- X. Teng, L. Qi, T. Liu, L. Li and C. Lu, TrAC, Trends Anal. Chem., 2023, 162, 117020 CrossRef CAS.
- A. Li, X. Luo, L. Li, D. Chen, X. Liu, Z. Yang, L. Yang, J. Gao and H. Lin, Anal. Chem., 2021, 93, 16552–16561 CrossRef CAS PubMed.
- P. Huang, W. Zhang, J. Wang, F. Huo and C. Yin, Chin. Chem. Lett., 2025, 36, 109778 CrossRef CAS.
- W. Qu, R. Tian, B. Yang, T. Guo, Z. Wu, Y. Li, Z. Geng and Z. Wang, Anal. Chem., 2024, 96, 5428–5436 Search PubMed.
- Z. Xu, Y. Wu, X. Wang, J. Chen, H. Luo and N. Li, Food Chem., 2022, 397, 133754 CrossRef CAS PubMed.
- Y. Dai, Z. Zhan, L. Chai, L. Zhang, Q. Guo, K. Zhang and Y. Lv, Anal. Chem., 2021, 93, 4628–4634 CrossRef CAS PubMed.
- B. Cao, H. Zhang, M. Sun, C. Xu, H. Kuang and L. Xu, Adv. Mater., 2024, 36, 2208037 CrossRef CAS PubMed.
- H. Sies, R. J. Mailloux and U. Jakob, Nat. Rev. Mol. Cell Biol., 2024, 25, 701–719 CrossRef CAS.
- S. Srinivas and A. Senthil Kumar, Langmuir, 2024, 40, 10634–10647 CrossRef CAS.
- Q. Jia and D. Sieburth, Nat. Commun., 2021, 12, 2304 CrossRef CAS.
- V. Rahemi, S. Trashin, Z. Hafideddine, S. Van Doorslaer, V. Meynen, L. Gorton and K. De Wael, Anal. Chem., 2020, 92, 3643–3649 CrossRef CAS PubMed.
- K. Dong, Y. Lei, H. Zhao, J. Liang, P. Ding, Q. Liu, Z. Xu, S. Lu, Q. Li and X. Sun, J. Mater. Chem. A, 2020, 8, 23123–23141 RSC.
- R. Pan, K. Hu, R. Jia, S. A. Rotenberg, D. Jiang and M. V. Mirkin, J. Am. Chem. Soc., 2020, 142, 5778–5784 CrossRef CAS PubMed.
- S. Kumar, A. Kumar, G. H. Kim, W. K. Rhim, K. L. Hartman and J. M. Nam, Small, 2017, 13, 1701584 CrossRef.
- D. Srikun, A. E. Albers, C. I. Nam, A. T. Iavaron and C. J. Chang, J. Am. Chem. Soc., 2010, 132, 4455–4465 CrossRef CAS PubMed.
- L. Yang, X. Yin, B. An and F. Li, Anal. Chem., 2021, 93, 1709–1716 CrossRef CAS.
- L. Xing, W. Zhang, L. Fu, J. M. Lorenzo and Y. Hao, Food Chem., 2022, 385, 132555 CrossRef CAS PubMed.
- J. G. Kleingardner and K. L. Bren, Acc. Chem. Res., 2015, 48, 1845–1852 CrossRef CAS PubMed.
- S. G. Rhee, H. A. Woo and D. Kang, Antioxid. Redox Signaling, 2018, 28, 537–557 CrossRef CAS.
- M. Lian, X. Chen, Y. Lu and W. Yang, ACS Appl. Mater. Interfaces, 2016, 8, 25036–25042 CrossRef CAS PubMed.
- S. Rattanopas, A. Schulte and S. Teanphonkrang, Anal. Chem., 2022, 94, 4919–4923 CrossRef CAS.
- P. Coatsworth, Y. Cotur, A. Naik, T. Asfour, A. S. P. Collins, S. Olenik, Z. Zhou, L. Gonzalez-Macia, D. Chao, T. Bozkurt and F. Güder, Sci. Adv., 2024, 10, eadj6315 CrossRef CAS.
- Z. Yang, J. Guo, L. Wang, J. Zhang, L. Ding, H. Liu and X. Yu, Small, 2024, 20, 2307815 CrossRef CAS.
- Y. Sun, L. Han and P. Strasser, Chem. Soc. Rev., 2020, 49, 6605–6631 RSC.
- S. M. Kondengadan and B. Wang, Angew. Chem., Int. Ed., 2024, 63, e202403880 CrossRef CAS PubMed.
- B. Dou, J. Yang, R. Yuan and Y. Xiang, Anal. Chem., 2018, 90, 5945–5950 CrossRef CAS PubMed.
- C. Shi, D. Yang, X. Ma, L. Pan, Y. Shao, G. Arya, Y. Ke, C. Zhang, F. Wang, X. Zuo, M. Li and P. Wang, Angew. Chem., Int. Ed., 2024, 63, e202320179 CrossRef CAS PubMed.
- J. Yan, R. Bhadane, M. Ran, X. Ma, Y. Li, D. Zheng, O. M. H. Salo-Ahen and H. Zhang, Nat. Commun., 2024, 15, 3684 CrossRef CAS PubMed.
- B. Dou, H. Zhou, Y. Hong, L. Zhao and P. Wang, Chem. Commun., 2021, 57, 7116–7119 RSC.
- H. Zhao, D. Yi, L. Li, Y. Zhao and M. Li, Angew. Chem., Int. Ed., 2024, 63, e202404064 CrossRef CAS.
- B. P. Carpenter, A. R. Talosig, B. Rose, G. Di Palma and J. P. Patterson, Chem. Soc. Rev., 2023, 52, 6918–6937 RSC.
- B. Shen, X. Chen, K. Shen, H. Xiong and F. Wei, Nat. Commun., 2020, 11, 2692 CrossRef CAS PubMed.
- C. Mao, S. Wang, J. Li, Z. Feng, T. Zhang, R. Wang, C. Fan and X. Jiang, ACS Nano, 2023, 17, 2840–2850 CrossRef CAS.
- X. Chen, Q. Tang, J. Wang, Y. Zhou, F. Li, Y. Xie, X. Wang, L. Du, J. Li, J. Pu, Q. Hu, Z. Gu and P. Liu, Adv. Mater., 2023, 35, 2210440 CrossRef CAS PubMed.
- C. Li, X. Feng, S. Yang, H. Xu, X. Yin and Y. Yu, ACS Appl. Mater. Interfaces, 2021, 13, 52406–52416 CrossRef CAS.
- D. Nieciecka, A. Joniec, G. J. Blanchard and P. Krysinski, Langmuir, 2013, 29, 14560–14569 CrossRef CAS.
- B. P. Ting, J. Zhang, Z. Gao and J. Y. Ying, Biosens. Bioelectron., 2009, 25, 282–287 Search PubMed.
- X. Zeng, C. Wu, Y. Xiong, Z. Zhan, C. Shen, F. Lin, J. Zhang and P. Chen, Biosens. Bioelectron., 2024, 256, 116273 CrossRef CAS PubMed.
- M. Zhu, S. Li, H. Li and F. Xia, Anal. Chem., 2020, 92, 12437–12441 CrossRef CAS.
- Q. Zhang, S. Liu, C. Du, Y. Fu, K. Xiao, X. Zhang and J. Chen, Anal. Chem., 2021, 42, 14272–14279 CrossRef PubMed.
- B. Dou, L. Xu, B. Jiang, R. Yuan and Y. Xiang, Anal. Chem., 2019, 91, 10792–10799 CrossRef CAS.
- Y. Li, H. Meng, J. Huang and C. Zhan, ACS Appl. Mater. Interfaces, 2020, 12, 2087–2094 CrossRef PubMed.
- J. Chang, X. Wang, J. Wang, H. Li and F. Li, Anal. Chem., 2019, 91, 3604–3610 CrossRef CAS PubMed.
- A. E. Radi, J. L. Acero Sánchez, E. Baldrich and C. K. O'Sullivan, J. Am. Chem. Soc., 2006, 128, 117–124 CrossRef CAS.
- Q. Feng, M. Wang, X. Han, Q. Chen, B. Dou and P. Wang, ACS Appl. Bio Mater., 2020, 3, 1276–1282 CrossRef CAS PubMed.
- B. Dou, K. Wang, Y. Chen and P. Wang, Anal. Chem., 2024, 96, 10594–10600 CrossRef CAS.
- L. Wu, I. C. Wu, C. C. DuFort, M. A. Carlson, X. Wu, L. Chen, C. T. Kuo, Y. Qin, J. Yu, S. R. Hingorani and D. T. Chiu, J. Am. Chem. Soc., 2017, 139, 6911–6918 CrossRef CAS PubMed.
- K. Wang, F. Zhang, Y. Wei, W. Wei, L. Jiang, Z. Liu and S. Liu, Anal. Chem., 2021, 93, 7870–7878 CrossRef CAS PubMed.
- L. Jiang, H. Chen, C. He, H. Xu, Z. Zhou, M. Wu, E. K. Fodjo, Y. He, M. E. Hafez, R. Qian and D. Li, Anal. Chem., 2023, 95, 3507–3515 CrossRef CAS PubMed.
- X. Li, X. Duan, P. Yang, L. Li and B. Tang, Anal. Chem., 2021, 93, 4059–4065 CrossRef CAS PubMed.
- J. Li, C. Wu, C. Yuan, Z. Shi, K. Zhang, Z. Zou, L. Xiong, J. Chen, Y. Jiang, W. Sun, K. Tang, H. Yang and C. Li, Anal. Chem., 2022, 94, 14109–14117 CrossRef CAS PubMed.
- J. Yan, Y. Qin, W. Fan, W. Wu, S. Lv, L. Yan, Y. Liu and W. Huang, Chem. Sci., 2021, 12, 14432–14440 RSC.
- S. G. Rhee, Science, 2006, 312, 1882–1883 CrossRef PubMed.
- C. Tapeinos and A. Pandit, Adv. Mater., 2016, 28, 5553–5585 CrossRef CAS PubMed.
- S. Lin, H. Ze, X. G. Zhang, Y. J. Zhang, J. Song, H. Zhang, H. L. Zhong, Z. L. Yang, C. Yang, J. F. Li and Z. Zhu, Angew. Chem., Int. Ed., 2022, 61, e202203511 CrossRef CAS PubMed.
- F. Lacy, D. A. Gough and G. W. Schmid-Schönbein, Free Radicals Biol. Med., 1998, 25, 720–727 CrossRef CAS.
- E. Deskur, L. Przywarska, P. Dylewicz, L. Szczęśniak, T. Rychlewski, M. Wilk and H. Wysocki, Int. J. Cardiol., 1998, 67, 219–224 CrossRef CAS.
- G. Wang, J. Li, L. Wang, Y. Yang, J. Wu, W. Tang, H. Lei and L. Cheng, ACS Nano, 2024, 18, 10885–10901 CrossRef CAS PubMed.
- Y. D. Lee, C. K. Lim, A. Singh, J. Koh, J. Kim, I. C. Kwon and S. Kim, ACS Nano, 2012, 6, 6759–6766 CrossRef CAS PubMed.
- B. Wang, F. Zhang, S. Wang, R. Yang, C. Chen and W. Zhao, Chem. Commun., 2020, 56, 2598–2601 RSC.
- H. Zhu, J. Fan, J. Wang, H. Mu and X. Peng, J. Am. Chem. Soc., 2014, 136, 12820–12823 CrossRef CAS.
- C. X. Guo, X. T. Zheng, Z. S. Lu, X. W. Lou and C. M. Li, Adv. Mater., 2010, 22, 5164–5167 CrossRef CAS.
- Y. Zhang, J. Xiao, Q. Lv, L. Wang, X. Dong, M. Asif, J. Ren, W. He, Y. Sun, F. Xiao and S. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 38201–38210 CrossRef CAS.
- X. Jiang, H. Wang, R. Yuan and Y. Chai, Anal. Chem., 2018, 90, 8462–8469 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |