Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ultra-high resolution magnetic resonance microscopy of in situ gadolinium gold nanoparticle-labeled cells in the rat brain†
Alena
Kisel
 a,
Minrui
Luo
a,
Minrui
Luo
 d,
Matthew D.
Bailey
d,
Harman
Ghuman
d,
Matthew D.
Bailey
d,
Harman
Ghuman
 b,
Matthew
Rotz
d,
Vinícius P.
Campos
b,
Matthew
Rotz
d,
Vinícius P.
Campos
 g,
Marcelo A. C.
Vieira
g,
Marcelo A. C.
Vieira
 g,
T. Kevin
Hitchens
c,
Thomas J.
Meade
g,
T. Kevin
Hitchens
c,
Thomas J.
Meade
 *def and
Michel
Modo
*def and
Michel
Modo
 *ab
*ab
aDepartment of Radiology, McGowan Institute for Regenerative Medicine, University of Pittsburgh, 3025 East Carson St, Pittsburgh, Pennsylvania, USA. E-mail: mmm154@pitt.edu; Tel: +1 (412) 383 7200
bDepartment of Bioengineering, University of Pittsburgh, Pittsburgh, Pennsylvania, USA
cDepartment of Neurobiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA
dDepartment of Chemistry, Evanston, Illinois 60208, USA. E-mail: tmeade@northwestern.edu; Tel: +1 (847) 491 2481
eDepartment of Neurobiology, Evanston, Illinois, USA
fDepartment of Molecular Biosciences, Northwestern University, Evanston, Illinois, USA
gDepartment of Electrical and Computer Engineering, São Carlos School of Engineering University of São Paulo, São Carlos, SP, Brazil
First published on 3rd June 2025
Abstract
Mapping the distribution of cells within a tissue using MR imaging has remained a significant challenge for the field. Cellular MRI can trace cells within tissue, but typically does not achieve the resolution necessary to define a cell's precise anatomical location. To detect cells with ultra-high resolution MRI, a high r1 relaxivity intracellular contrast agent is required. Localizing this contrast within its biological context also necessitates an isotropic spatial resolution corresponding to the size of a cell's cytoplasm (∼20 μm) to place it within its biological context. We here demonstrate that gadolinium gold nanoparticles (GdAuNP) induce a high T1-weighted cellular MRI contrast at ultra-high magnetic fields (9.4 T, and 11.7 T) that affords in situ labelled cell detection at very high resolutions (150, 100, 50, and 20 μm). A 20 μm 3D gradient-echo image (400 minutes scan) combined with MR image denoising robustly visualized the distribution of in situ labeled cells in the rat brain. Signal averaging (NA = 5) also consistently afforded the detection of labeled cells. Positive T1-weighted contrast was confirmed to be caused by GdAuNP using histology. Immunohistochemistry confirmed the presence of GdAuNP almost entirely inside cells, primarily those of the neuronal lineage. Histology verified that the MR images accurately visualized individual cells' distribution within their anatomical context. Cellular resolution MRI of GdAuNP-labeled cells hence affords new avenues to investigate how individual cells contribute to the development, repair, and regeneration of tissues.
Introduction
1H Magnetic Resonance Imaging (MRI) has excellent soft tissue contrast due to its high water density.1 Akin to histology, to visualize the distribution or migration of cells, however, an exogenous contrast material, such as iron oxide or gadolinium-based contrast agents, is required to distinguish the cells of interest from their surrounding tissue.2 Cellular MRI has been used to track the migration of stem cells, as well as distinguish different cellular fractions within the liver.3 The MR detection of single micron-sized particles of iron oxide (MPIOs) is feasible4 and affords the detection of single cells.5,6 The superparamagnetic effects of iron oxide cause local magnetic field gradients that are observed as areas of signal hypointensities on -weighted images. This does not only affect the immediate location of the contrast particle, but the so-called blooming effect can extend over 50× the particle size.7,8 The presence of multiple cells can, hence, lead to a total signal loss in tissue and prevent the anatomical localization of the cell(s) of interest. To localize a cell to an individual voxel, negative image contrast and blooming effects should, therefore be minimized.
-weighted images. This does not only affect the immediate location of the contrast particle, but the so-called blooming effect can extend over 50× the particle size.7,8 The presence of multiple cells can, hence, lead to a total signal loss in tissue and prevent the anatomical localization of the cell(s) of interest. To localize a cell to an individual voxel, negative image contrast and blooming effects should, therefore be minimized.
Unequivocal positive contrast confined to a single voxel would be desirable to localize cells in their anatomical context. For instance, T1-weighted (T1w) MRI can accurately detect in vivo neural stem cells (NSCs) implanted in the rat brain using hyperintense contrast due to their in vitro labeling with gadolinium-gold nanoparticles (GdAuNP).9 Conjugation of gadolinium chelates to gold nanoparticles is an efficient means to dramatically increase and maintain their T1 relaxivity upon intracellular uptake, as well as to improve their thermokinetic stability in a biological milieu.10–12 Attachment of DNA on GdAuNP improves cellular uptake and traps nanoparticles in a peri-nuclear location, which prevents exocytosis and transfer to other cells.9,13 A highly selective T1w visualization of GdAuNP-labeled cells after intracerebral implantation can hence be achieved in vivo whilst affording their anatomical localization.9
Although the blooming effect of iron oxide particles has afforded the detection of single cells/particles by MRI at a 100 μm resolution.5,6 This cellular MRI to date has not achieved a cellular resolution that would afford a precise localization of a cell's positioning, especially amongst a population of contrast agent-labeled cells. To achieve a cellular resolution for a population of contrast agent-labeled cells, MRI will require a spatial resolution that can potentially detect a cell within one isotropic voxel. We previously measured in vitro the average diameter of NSCs to be 19.29 ± 0.75 μm.14 We here therefore propose that an isotropic voxel size ≤20 μm is required to achieve a cellular-resolution MRI, a resolution 125× smaller than previously used to detect individual cells labeled with iron oxide-based contrast agents.6,15,16 Isotropic resolutions <100 μm are considered ultra-high resolution and consequently are often referred to as MR microscopy (MRM).17
Achieving a cellular resolution MRM will be important to monitor the distribution and migration of a population of transplanted cells through a tissue,18 and also provide deeper insights into the spatio-temporal dynamics of neurogenesis and its response to tissue damage.19–21 Considering the widespread migration of NSCs, as well as the varied topology of brain damage, as occurs after a stroke or traumatic brain injury, non-invasive whole brain imaging is required to monitor the distribution of cells. We here demonstrate that GdAuNP labeling of cells in situ combined with a 20 μm isotropic resolution affords a cellular resolution MRM to investigate the distribution of individual cells in the neurogenic subventricular zone (SVZ), as well as within striatal tissue.
Results
Optimization of T1-weighted MRI to distinguish GdAuNP from brain tissue
GdAuNP are designed to achieve a high relaxivity that is efficiently delivered into cells for cellular MRI. The highly negative charge of ssDNA renders AuNPs exceptionally stable. The ssDNA facilitates endocytosis and the localization of GdAuNP in the peri-nuclear space within cells. The random DNA sequence cannot integrate into the nuclear DNA and hence traps the GdAuNP in the peri-nuclear space. By Gd-labeling ssDNA on AuNPs and labeling the remaining nanoparticle surface with additional Gd, we are able to achieve a GdAuNP r1 of over 3300 mM−1 s−1. These ssDNA-GdAuNP are suitable for long-term cellular imaging studies, as their subcellular localization and anchoring prevents their clearance from labeled cells and provides a signal specific to these cells. However, to efficiently exploit their excellent relaxivity and labeling, optimal MRI acquisition parameters are required.Measurement of T1 relaxivity in situ of GdAuNP-labeled cells afforded a specific intracellular environment to optimize contrast between labeled cells and brain tissue (Fig. 1A). GdAuNP decreased T1 by 24.1% at room temperature (22 °C) and by 25.3% at physiological temperature (37 °C) (Fig. 1B). Based on these measurements, optimal flip angles (FA) were calculated for different repetition times (TR) using the Ernst equation at 9.4 T and 11.7 T (Fig. 1C). A contour plot arraying both FA and TR revealed an optimal TR = 1000 ms with a FA of 90° achieving a T1 contrast >22% (Fig. 1D). However, a shorter TR = 500 ms with an FA of 70° already achieved a contrast >17% within half the acquisition time, providing a faster acquisition and the opportunity for more signal averages within the same unit of time. A greater reduction in T1 due to GdAuNP relaxivity further translates into a greater T1 contrast between GdAuNP and brain tissue with these optimized acquisition parameters (Fig. 1E).
An experimental verification of these parameters was achieved by injection of GdAuNP into two distinct microenvironments, notably the lateral ventricle of a rat brain (liquid environment with abundant access to water), as well as rat striatal tissue (limited water access and increased cellular uptake). Strong contrast was evident at both TR = 500 ms with FA > 70°, as well as TR = 1000 ms with an FA of 90° (Fig. 2A). This was further confirmed by quantification of contrast revealing up to 30% T1w contrast with a TR = 500 ms and 20% with a TR = 1000 ms (Fig. 2B). The T1w contrast for TR = 500 ms was consistently ∼10% higher than for TR = 1000 ms (Fig. 2C). The T1 signal of both GdAuNP and brain tissue was increased with rising temperature, achieving a higher signal difference at physiological temperature (37 °C) (Fig. 2D). Field strength further amplified the effects of temperature on T1 signal measurements (Fig. 2E). However, T1w contrast between GdAuNP and brain tissue was consistent at 22 °C and 37 °C, as well as at 9.4 T and 11.7 T (Fig. 2F).
Achieving a cellular resolution MR microscopy
The key challenge in cellular resolution MRM is to achieve a sufficiently high spatial resolution to be able to resolve cells within their biological context. Increasing spatial resolution provided a means to ascertain the presence of GdAuNP in both the lateral ventricle, as well as striatal tissue (Fig. 3A). T1w contrast was evident at a 150, 100, and 50 μm isotropic voxel resolution with a single signal average (i.e. NA = 1). However, the signal-to-noise ratio (SNR) decreased considerably and at a 20 μm resolution, no robust detection of GdAuNP was evident, nor was tissue contrast. To compensate for the decreasing signal, MR images were denoised to lower the background noise level. This dramatically improved the visualization of the residual tissue and GdAuNP signal. A robust T1w contrast between GdAuNP and brain tissue was achieved (Fig. 3B). Even at 20 μm isotropic voxel resolution, a distinct lining of the lateral ventricle was evident in the denoised image compared to both the original 50 and 20 μm resolution scans (Fig. 3C). Within striatal tissue, a clear signal contrast was evident and more robustly distinguishable from surrounding tissue, even when compared to the 50 μm resolution.To further optimize the detection of GdAuNP by MRM at a ultra-high resolution (i.e. 20 μm isotropic voxels), signal averaging can increase signal intensities and improve detection, as well as contrast (Fig. 4A). A major improvement in signal intensity is evident with 5 signal averages. Combined with different degrees of denoising, signal averaging can achieve a high T1w contrast of GdAuNP with a highly specific localization within the tissue (Fig. 4B). A quantitative analysis of signal changes revealed marked reductions in the noise intensity by increasing signal averaging, but this tapered off beyond 5 signal averages (Fig. 4C). Denoising achieved a major decrease in noise measurements in line with the improvements observed with signal averaging. Strikingly, denoising also achieved a considerable reduction in variance. The same pattern of results was evident for the T1w signal from GdAuNP. SNR of denoised images was >20 times higher than the original images and improved linearly with more signal averages (Fig. 4D). A stronger denoising had a more marked gain in SNR. CNR was also dramatically improved with denoising, but followed a logarithmic trend that dramatically declined beyond 5 signal averages. A significant improvement in GdAuNP detection using MRM can hence be achieved using signal averaging combined with image denoising.
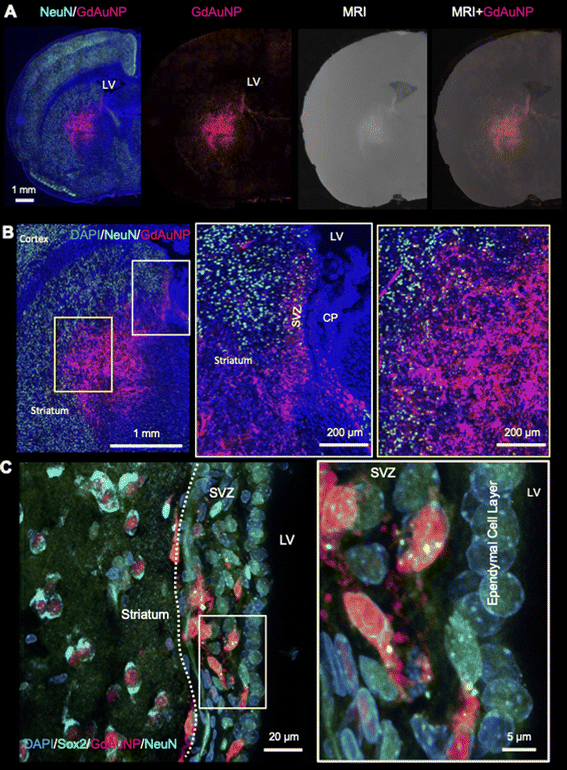
A robust cellular detection using this approach was achieved in 3 rats (Fig. 5A), affording the precise 3D localization of GdAuNP-labeled cells in the coronal, sagittal and axial plane (Fig. 5B). T1w thresholding afforded the separation of the brain background signal from the GdAuNP T1w contrast to create a 3D reconstruction of its distribution (Fig. 5C). The injection tracts are readily visible, as well as the distribution of GdAuNP labeled cells throughout the striatum. The lateral ventricle is also visible and less granular, due to GdAuNP being homogeneously distributed through the cerebrospinal fluid (CSF) after its intracerebroventricular injection. However, the region also encompasses cells in the SVZ, which are outlining the ventricular space. This approach hence provides a unique means to visualize the 3D distribution of GdAuNP-labelled cells in tissue. The 20 μm isotropic voxel resolution also visualizes the in situ distribution of labeled cells using T1w thresholding to reveal, for instance, the unique organization of the patch-matrix in the striatum (Fig. 5D). The unique pattern of T1w contrast distribution in this area indicates that GdAuNP are contained within the cellular compartments of the patch-matrix, rather than being homogenously distributed throughout the extracellular space.
Verification of cellular MRM by histology
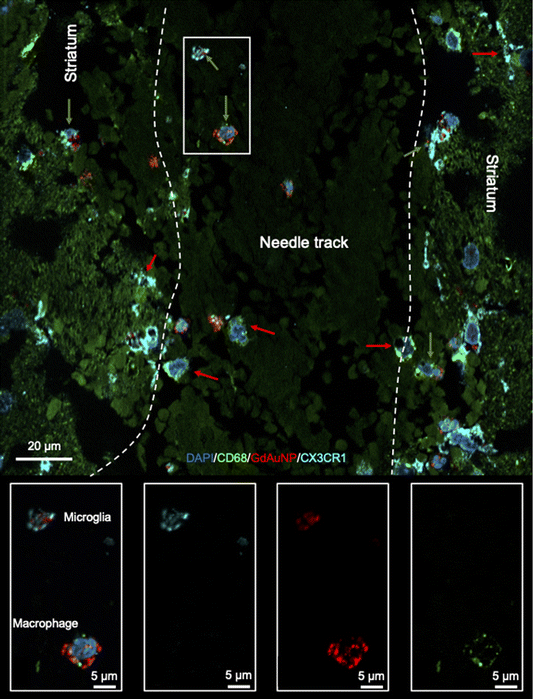
To verify that the T1w signal is indeed due to the presence of GdAuNP in cells, the T1w MRI signal was verified using immunohistochemistry. The red fluorophore conjugated on the DNA attached to the GdAuNP affords their detection using fluorescence microscopy. A macroscopic comparison between the T1w thresholded image and the red fluorescent whole histological slices indicates that the T1 signal change is reflecting the presence of GdAuNP on histological slices (Fig. 6A). An overlay further corroborated the correspondence of both imaging modalities (Fig. 6B). A closer inspection of the patch-matrix also verifies that T1 changes are a reflection of the differential distribution of GdAuNP in tissue (Fig. 6C). It is important to note that histological slices (50 μm) are thicker than MRI slices (20 μm).Immunohistochemistry revealed individual cells, such as neurons (i.e. NeuN+ cells) in conjunction with GdAuNP to compare with their T1w localization (Fig. 7A). A cellular localization is evident in both the striatum and the SVZ (Fig. 7B). Within the SVZ, GdAuNP are observed within SOX2+ NSCs, but not ependymal cells which constitute the first layer of cells surrounding the lateral ventricle (Fig. 7C). Even within the SVZ, SOX2+ cells endocytosed GdAuNP, whereas other neighboring cells did not or only contained negligible amounts. Although macroscopically, the GdAuNP covers a large area of the striatum and SVZ, very little GdAuNP remains in the extracellular space (Fig. 8A). GdAuNP are mostly localized to the cytoplasm of cells, especially neurons, in a perinuclear location (Fig. 8B). This indicates that the T1w signal on MRI is due to the presence of GdAuNP inside individual cells. Although there is preferential uptake of GdAuNP into SOX2+ and NeuN+, astrocytes (Fig. 9A) and endothelial cells (Fig. 9B) also occasionally contain smaller quantities of GdAuNP. GdAuNP were also found within peripheral macrophages (CD68+) that infiltrated in response to the injection tract damage, as well as brain-resident microglia (CX3CR1+) (Fig. 10). However, within the same location there were also macrophages and microglia that did not contain GdAuNP. In all cases, almost all GdAuNP were located intracellularly, with very few GdAuNP evident in the extracellular space. These results indicate that T1w contrast on MR images is due to intracellular GdAuNP, predominantly in cells of the neuronal lineage.
Discussion
Visualizing cells within tissue has remained a major challenge and requires contrast material that distinguishes cells of interest from the surrounding tissue. We here demonstrated that to detect cells at a cellular resolution using MRM, an isotropic spatial resolution of a maximum of 20 μm is required, as this is the diameter of a cells' cytoplasm, but additionally a highly efficient contrast material that is endocytosed by cells is necessary to achieve a distinction from its surrounding tissue. The design of cell-penetrating high relaxivity contrast agents, notably DNA-conjugated GdAuNP,9,13,22 here demonstrated an extensive in situ endocytosis, while affording a high level of unequivocal positive T1-weighted MR contrast from the surrounding tissue. Post-acquisition denoising of the MR images proved to be a valuable process to dramatically increase SNR in cellular resolution images. These multidisciplinary advances afford a paradigm shift to image individual cells in their anatomical context using MRM.Cellular MRI versus cellular resolution MRM
Although cellular MRI has been well established to visualize the distribution and migration of cells within tissue, this is typically for a population of cells at a lower spatial resolution.3 The detection of individual cells can be achieved using micron-sized iron oxide nanoparticles (MIONs) to create a blooming effect 50× the size of the particle and hence affords the visualization of a single labeled cell,4,6,15,16 but it does not afford the precise localization of that cell and the presence of a large number of cells will efface the anatomical context.A high relaxivity contrast material is required to provide a sufficient MRI signal change that distinguishes the cell from surrounding tissue. To reduce the spatial resolution down to the size of an individual cell (i.e. 20 μm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14), a blooming effect is disadvantageous and typically T2-based contrast agents compete with other contrast sources (e.g. small bleeds, air bubbles) that can easily be mistaken for labeled cells. The design of a high relaxivity cell-penetrating T1 contrast agent is hence favorable to cellular resolution MRM.13,22,23 However, an extensive optimization of imaging parameters was required to ensure that a maximum contrast is achieved.
14), a blooming effect is disadvantageous and typically T2-based contrast agents compete with other contrast sources (e.g. small bleeds, air bubbles) that can easily be mistaken for labeled cells. The design of a high relaxivity cell-penetrating T1 contrast agent is hence favorable to cellular resolution MRM.13,22,23 However, an extensive optimization of imaging parameters was required to ensure that a maximum contrast is achieved.
A physiological temperature was favorable, whereas a reduced T1 effect was evident close to the Curie point, as previously reported for other gadolinium based materials.24–26 However, T1-weighted contrast between GdAuNP and brain tissue was only minimally affected by temperatures above 22 °C. Although T1 for both brain and GdAuNP was slightly higher at 11.7 T compared to 9.4 T, the contrast between GdAuNP and brain tissue was equivalent, highlighting the potential for these agents at high field strengths. A single average of image acquisition was sufficient to achieve a good T1-weighted contrast at a 50 μm isotropic resolution, but at a 20 μm resolution, the GdAuNP was difficult to discern against the brain background signal.
Nevertheless, denoising of these cellular resolution images during post-processing afforded a very robust recouperation of T1 signal from brain tissue, as well as GdAuNP-labled cells. Increasing the number of averages for image acquisition also improved the detection of the T1-weighted contrast of GdAuNP, with a diminishing return beyond 5 signal averages. Denoising of these averaged images further improved detection, but as the signal in images improved, denoising exerted less dramatic effects and came at the cost of sharpness. Signal averaging is therefore advantageous over denoising for cellular resolution MRI, but denoising is extremely powerful to accommodate a limited image acquisition time.
Verification of GdAuNP-labled cell detection using MRM
To ensure that MR images indeed reflect the presence and distribution of GdAuNP, brains were sliced to afford a visualization of GdAuNP in tissue using whole slice microscopy of the red fluorescent moiety conjugated on the GdAuNP. Fluorescence microscopy verified that the hyperintense MR signal followed the same distribution as the histological distribution of GdAuNP. To ensure that these GdAuNP were indeed inside cells, rather than in extracellular space, immunohistochemistry further visualized individual cells in brain slices and afforded a co-localization with GdAuNP. Almost all GdAuNP found within these brain slices were contained intracellularly, confirming that our MR images indeed reflected individual cells labeled in situ with GdAuNP.GdAuNP efficiently endocytosed in situ, mostly within cells of the neuronal lineage, including neural stem/progenitor cells in the SVZ. Smaller nanoparticles are preferable for neuronal uptake (<50 nm), whereas larger particles are taken-up by microglia.27,28 However, the AuNP corona also plays a major role in endocytosis. Conjugation of random DNA strands to GdAuNP improves endocytosis and contributes to its perinuclear localization. The perinuclear localization, rather than lysosomal sequestration, and attachment of Gd to these DNA strands maintains GdAuNP's intracellular relaxivity,9 whereas endocytosis of other Gd contrast materials results in significant T1 quenching.29–31 DNA-GdAuNP previously afforded the in vivo imaging of transplanted NSCs using T1-weighted MRI9 and demonstrated the utility of these agents for cellular MRI. Achieving a cellular resolution using MR microscopy (MRM) can potentially usher in a new era for in vivo cellular and molecular imaging.
Potential and limits of cellular resolution MR microscopy
Since the initial development of MR imaging, increasing signal and resolution has been one of the main technological challenges.32 However, as spatial resolution is increased the MR signal is decreased following a cubic root function. The main focus of MRI has therefore been on tissue rather than cell imaging. However, as demonstrated here, the low signal at cellular resolution can be overcome by post-processing to denoise images and improve the signal recovery.33 Additional improvements in acquisition can further enhance the detection of individual cells. Application of compressed sensing to the sparse signal of GdAuNP can potentially reduce image acquisition for a single average of 400 minutes to within an hour, which is feasible for in vivo MR microscopy in rats.34 To separate individual neighboring cells, an even higher spatial resolution will be required to satisfy the Abbe or Rayleigh limit of separation.35 An isotropic voxel at a 5 μm resolution (i.e. 64× higher resolution than 20 μm) would be 1/8 of a cell and sufficient to separate neighboring cells. Post-processing using super-resolution techniques, such as convex optimization,36 could improve cell separation and the spatial localization of individual cells within a population of labeled cells. Using extensive averaging and specialized micro-coils at high field, a 3 μm isotropic resolution is commonly considered the limit of MR microscopy due to signal detection sensitivity, translational diffusion and thermal noise.37,38 However, recent progress in MR imaging using dynamic nuclear polarization (DNP) at 5K indicates that at least for ex vivo imaging of brain samples, isotropic spatial resolutions of <2 μm might become feasible.39 There is an ongoing effort to bridge the gap between current microscopic MRI techniques and nanoscale tools, such as MR force microscopy, to achieve nanoscale MRI (nanoMRI) that will further improve spatial resolution and signal measurements for contrast material.40Achieving a cellular resolution MRM will be especially impactful for cellular and molecular MRI, which aims to visualize how individual cells and their gene expression affect their biological context in situ rather than in thin histological slices. Applications that can be envisaged range from cellular positioning of cells during brain development,41 tracing the response of neural progenitors to brain damage,42 monitoring the migration of implanted NSCs,9 the infiltration of cells into brain tumors,43 to gene expression by individual cells in response to a disease stimulus.44 As demonstrated here, the combination of a contrast agent that has a high intracellular relaxivity and exhibits efficient in vivo endocytosis with a cellular resolution will offer new opportunities to investigate how individual cells contribute to tissue development and repair. MRI is a unique versatile tool that can herewith report on cellular dynamics in the intact brain that currently no other imaging technique can provide.
Methods
DNA–gadolinium–gold nanoparticle (DNA–Gd@Au) synthesis
Citrate-stabilized gold nanoparticles (AuNPs) with a core diameter of 13 ± 1.4 nm were synthesized by reduction of HAuCl4 in refluxing water using trisodium citrate using a previously published procedure.13,22 Gd(III)-labeled, single-stranded DNA was synthesized using standard controlled pore glass (CPG) solid-phase support protocols with a MerMade 12 oligonucleotide synthesizer (Bio Automation) under dry Ar. All synthesizing reagents and protected 3′-Thiol modifier CPGs were purchased from Glen Research. The synthesized sequence was 3′ S-S-TTT-TTT-TTT-T*TT-T*TT-T*TT-T*TT-T*TT–Cy3 5′ where T* indicates C6 amino modifier dT modified bases and a cyanine dye (Cy3) was added as the final base using a Cy3 phosphonamidite coupled under standard conditions. Oligonucleotides were deprotected from the solid phase using AMA conditions (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methylamine/saturated ammonium hydroxide) for 1.5 hours, followed by azide modification with azidobutyrate NHS-ester in DMSO for 12 hours at room temperature. The inorganic Gd(III) complexes Gd-609 and Gd-828 were synthesized using previously reported procedures and HPLC purified using X-bridge C18 column9,22 (Scheme 1). Conjugation of the Gd-609 to the azide modified DNA was performed using Cu(I)-catalyzed 1,3 dipolar cycloaddition (click) chemistry. Each step mentioned before is purified by desalting with Sephadex G25 column (NAP 5, GE Life Sciences) and characterized by MALDI with a matrix of 2,5-dihydroxyacetophenone (DHAP) and ammonium citrate in methanol. The final DNA was HPLC purified using DNAPac™ PA200 Analytical column (4 × 250 nm) employing a gradient elution comprising tris buffer and linear gradient of salt over 17 minutes. Quantity of DNA was determined by UV-vis Cary 60 at OD 260. DNA conjugation to nanoparticles occurred after 3′ thiol deprotection with dithiothreitol (DTT) and desalting (Scheme 2). 100 equivalents of DNA were conjugated to citrate AuNPs with salt aging to 600 mM salt using five aliquots of 5 M NaCl over six hours with end-over-end rotation following a previously reported procedure,9,22 at which time the particles were left to rotate overnight. With 1× purification at 20
1 methylamine/saturated ammonium hydroxide) for 1.5 hours, followed by azide modification with azidobutyrate NHS-ester in DMSO for 12 hours at room temperature. The inorganic Gd(III) complexes Gd-609 and Gd-828 were synthesized using previously reported procedures and HPLC purified using X-bridge C18 column9,22 (Scheme 1). Conjugation of the Gd-609 to the azide modified DNA was performed using Cu(I)-catalyzed 1,3 dipolar cycloaddition (click) chemistry. Each step mentioned before is purified by desalting with Sephadex G25 column (NAP 5, GE Life Sciences) and characterized by MALDI with a matrix of 2,5-dihydroxyacetophenone (DHAP) and ammonium citrate in methanol. The final DNA was HPLC purified using DNAPac™ PA200 Analytical column (4 × 250 nm) employing a gradient elution comprising tris buffer and linear gradient of salt over 17 minutes. Quantity of DNA was determined by UV-vis Cary 60 at OD 260. DNA conjugation to nanoparticles occurred after 3′ thiol deprotection with dithiothreitol (DTT) and desalting (Scheme 2). 100 equivalents of DNA were conjugated to citrate AuNPs with salt aging to 600 mM salt using five aliquots of 5 M NaCl over six hours with end-over-end rotation following a previously reported procedure,9,22 at which time the particles were left to rotate overnight. With 1× purification at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 min, the AuNPs surface was subsequently backfilled with excess Gd-828 for 24 h. Functionalized particles were purified by spin filtration (30 kDa MWCO, Amicon® Ultra 15) in three cycles of 1500 × g, 10 min each. Functionalized particles were then characterized by inductively coupled plasma-mass spectroscopy (ICP-MS) for [Au] and [Gd]. The molar relaxivity (r1) of GdAuNPs was 3398 mM−1 s−1.
000 × g for 30 min, the AuNPs surface was subsequently backfilled with excess Gd-828 for 24 h. Functionalized particles were purified by spin filtration (30 kDa MWCO, Amicon® Ultra 15) in three cycles of 1500 × g, 10 min each. Functionalized particles were then characterized by inductively coupled plasma-mass spectroscopy (ICP-MS) for [Au] and [Gd]. The molar relaxivity (r1) of GdAuNPs was 3398 mM−1 s−1.
Animals and DNA–Gd@Au injections
All animal procedures complied with the US Animals Welfare Act (2010) and were approved by the University of Pittsburgh Institutional Animal Care and Use Committee (IACUC). Sprague-Dawley rats (male, n = 3, 280 ± 15 g, Charles River Laboratories, USA) were maintained on a 12 hour light/dark schedule, with food and water available ad libitum. 30 μL of GdAuNP solution were injected into right lateral ventricle and 4 μL of GdAuNP solution were injected into the right striatum. Stereotaxic coordinates were determined in accordance with the rat brain atlas:45 anterior–posterior −0.5 mm from bregma, medial–lateral −1.5 mm from the middle line, dorsal–ventral −4.0 mm from the brain surface for the lateral ventricle; and anterior–posterior −0.5 mm from bregma, medial–lateral −3.5 mm from the middle line, dorsal–ventral −4.5 mm from the brain surface for the striatum. Nanoparticle solution was injected under isoflurane anesthesia (4% induction, 1% maintenance in 30% O2) using a frame-mounted injection pump (World Precision Instruments, USA) through a 50 μL Hamilton syringe with a 32 G beveled stainless-steel needle (Hamilton, USA). The injection rate was 10 μL per min for the intraventricular injection and 1 μL per min for the striatal injection; after injection the needle was staying in the brain for 10 min and then was slowly removed to minimize a chance of a liquid backflow. Lidocaine topical cream (4%, Generic) was applied as an analgesic, as well as buprenorphine (0.05 mg kg−1 i.p., every 12 hours) to provide sustained pain relief. After 24 hours post-implantations rats were perfusion-fixed under terminal anesthesia (1 mL kg−1 Fatal Plus). Blood was first flushed transcardially using ×1 PBS, then tissue was fixed by transcardial perfusion with 4% paraformaldehyde.MR imaging
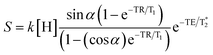 | (1) |
For simplicity, we assume a constant spin density k(H), and that the  effects
effects  are negligible for short TE experiments. Signal intensities were simulated for flip angles (α) up to 90° and TR values of 50, 100, 500 and 1000 ms. Contour maps were computed to illustrate how both TR and FA affected contrast. Verification of calculated optimal TR and FA parameters was achieved by scanning ex vivo brains injected with GdAuNP into the lateral ventricle and striatum at 9.4 T. A FLASH sequence (TR = 500 or 1000 ms, TE = 2.65 ms, NA = 6, Flip Angles of 10°, 30°, 50°, 70°, 90°, FOV 30 × 30 mm, 256 × 256 matrix, 0.117 mm in plane resolution, 35 slices at 0.5 mm thickness, 9 min 36 s or 19 min 20 s) was used to acquire T1w MR images. Mean intensities for ROIs in brain and GdAuNP areas, as well as T1w contrast were plotted for FA for TR = 500 ms and TR = 1000 ms.
are negligible for short TE experiments. Signal intensities were simulated for flip angles (α) up to 90° and TR values of 50, 100, 500 and 1000 ms. Contour maps were computed to illustrate how both TR and FA affected contrast. Verification of calculated optimal TR and FA parameters was achieved by scanning ex vivo brains injected with GdAuNP into the lateral ventricle and striatum at 9.4 T. A FLASH sequence (TR = 500 or 1000 ms, TE = 2.65 ms, NA = 6, Flip Angles of 10°, 30°, 50°, 70°, 90°, FOV 30 × 30 mm, 256 × 256 matrix, 0.117 mm in plane resolution, 35 slices at 0.5 mm thickness, 9 min 36 s or 19 min 20 s) was used to acquire T1w MR images. Mean intensities for ROIs in brain and GdAuNP areas, as well as T1w contrast were plotted for FA for TR = 500 ms and TR = 1000 ms.
| Resolution (μm) | Volume (mm3) | Matrix | Voxels | Time (min) | SNR (original/denoised) | CNR (original/denoised) |
|---|---|---|---|---|---|---|
| 150 | 0.003375 | 80 × 119 × 108 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 028 028![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 160 160 |
7 | 47.83/161.66 | 14.52/147.11 |
| 100 | 0.001000 | 120 × 178 × 160 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 417 417![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
16 | 29.25/126.06 | 6.26/60.99 |
| 50 | 0.000125 | 240 × 356 × 320 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340 340![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
64 | 10.16/147.61 | 1.53/53.30 |
| 20 | 0.000008 | 600 × 889 × 800 | 426![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 720 720![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
400 | 3.15/85.01 | 0.36/12.81 |
| Parameter | Description | NA = 1 (Strong) | NA = 5 (Medium) | NA = 10 (Weak) |
|---|---|---|---|---|
| N1 | Cube size (N1 × N1 × N1) | 8 | 6 | 5 |
| N2 | Maximum number of similar blocks | 64 | 64 | 32 |
| Ns | Search neighborhood size (Ns × Ns × Ns) | 11 | 11 | 11 |
| lambda_thr4D | Threshold value | 4.5 | 3.9 | 3.2 |
| Thresholding | Hard or soft thresholding | ‘Soft’ | ‘Soft’ | ‘Soft’ |
| do_wiener | Flag to perform (1) or not (0) the wiener filtering step | 0 | 0 | 0 |
Histological analyses
After removal from the skull, brains were post-fixed in 4% paraformaldehyde for 24 hours prior to being cryopreserved in 30% sucrose with sodium azide (Sigma) at 4 °C. Histological sections (50 μm thickness) were cut on a cryostat (Leica) directly onto microscopic slides to preserve the tissue morphology. Brain sections were washed 3 times with 0.01 M PBS, followed by 1 hour permeabilization and blocking in PBS + 0.1% Triton X-100 + 5% BSA (Sigma) at room temperature (21 °C). Sections were incubated overnight at 21 °C with primary antibodies (Table 3) diluted in PBS + 0.1% Triton X-100 + 1% BSA. After 18 hours, primary antibodies were washed off (3× PBS) and appropriate secondary AlexaFluor 488 or 647 antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Table 4) were applied for 1 hour at room temperature. Antibodies were removed by 3× washes with PBS before counterstaining with the nuclear marker Hoechst 33342 (1 μg mL−1, Thermo Scientific). After a further 3× washes, sections were coverslipped with Vectashield mounting medium for fluorescence and stored at 4 °C prior to imaging. Visualization of antibodies was performed with a fluorescence microscope (Axioimager M2, Zeiss) interfaced with a monochrome camera (Axiocam 820, Zeiss) driven by Zen Blue software (Zeiss) using a motorized stage.
1000; Table 4) were applied for 1 hour at room temperature. Antibodies were removed by 3× washes with PBS before counterstaining with the nuclear marker Hoechst 33342 (1 μg mL−1, Thermo Scientific). After a further 3× washes, sections were coverslipped with Vectashield mounting medium for fluorescence and stored at 4 °C prior to imaging. Visualization of antibodies was performed with a fluorescence microscope (Axioimager M2, Zeiss) interfaced with a monochrome camera (Axiocam 820, Zeiss) driven by Zen Blue software (Zeiss) using a motorized stage.
| Antibody (host) | Clone | Dilution | Company | Cat. Ref. |
|---|---|---|---|---|
| Hoechst 33342 | A-T regions of DNA | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Thermo Scientific | 62249 |
| SOX2 (rabbit) | SP76 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Abcam | Ab93689 |
| NeuN (mouse) | 1B7 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Abcam | Ab104224 |
| GFAP (chicken) | Polyclonal | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Abcam | Ab4674 |
| RECA-1 (mouse) | RECA-1 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Abcam | Ab9774 |
| CD68 (mouse) | ED1 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Abcam | Ab31630 |
| CX3CR1 (rabbit) | EPR24267-2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Abcam | Ab303613 |
| Antibody (host) | Clone | Fluorochrome | Dilution | Company | Cat. Ref. |
|---|---|---|---|---|---|
| Anti-rabbit (donkey) | Polyclonal | 488 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Abcam | Ab150061 |
| Anti-mouse (donkey) | Polyclonal | 647 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Invitrogen | A31571 |
| Anti-goat (donkey) | Polyclonal | 488 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Abcam | Ab150129 |
| Anti-chicken (goat) | Polyclonal | 647 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Invitrogen | A21449 |
| Anti-goat (donkey) | Polyclonal | 647 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
Invitrogen | A21447 |
Data availability
MRI scans are available from the authors upon reasonable request.Conflicts of interest
The authors have no personal financial or institutional interest in any of the drugs, materials, or devices described in this article.Acknowledgements
The study was mainly funded by NINDS (R01NS08226) and in part by the Department of Radiology, University of Pittsburgh. This work used Advanced Imaging Center (RRID:SCR_025139), a core research facility partially supported by the office of the Senior Vice Chancellor for Health Sciences at the University of Pittsburgh. VPC and MACV acknowledge the support by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001 for the denoising.References
- K. Kose, Physical and technical aspects of human magnetic resonance imaging: present status and 50 years historical review, Adv. Phys., 2021, 6, 1885410 Search PubMed.
- M. Modo, J. Kolosnjaj-Tabi, F. Nicholls, W. Ling, C. Wilhelm, O. Debarge, F. Gazeau and O. Clement, Considerations for the clinical use of contrast agents for cellular MRI in regenerative medicine, Contrast Media Mol. Imaging, 2013, 8, 439–455 CrossRef CAS PubMed.
- M. Modo, M. Hoehn and J. W. Bulte, Cellular MR imaging, Mol. Imaging, 2005, 4, 143–164 CrossRef PubMed.
- E. M. Shapiro, S. Skrtic, K. Sharer, J. M. Hill, C. E. Dunbar and A. P. Koretsky, MRI detection of single particles for cellular imaging, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10901–10906 CrossRef CAS PubMed.
- E. M. Shapiro, L. N. Medford-Davis, T. M. Fahmy, C. E. Dunbar and A. P. Koretsky, Antibody-mediated cell labeling of peripheral T cells with micron-sized iron oxide particles (MPIOs) allows single cell detection by MRI, Contrast Media Mol. Imaging, 2007, 2, 147–153 CrossRef CAS PubMed.
- E. M. Shapiro, K. Sharer, S. Skrtic and A. P. Koretsky, In vivo detection of single cells by MRI, Magn. Reson. Med., 2006, 55, 242–249 CrossRef PubMed.
- J. B. Williams, Q. Ye, T. K. Hitchens, C. L. Kaufman and C. Ho, MRI detection of macrophages labeled using micrometer-sized iron oxide particles, J. Magn. Reson. Imag., 2007, 25, 1210–1218 CrossRef PubMed.
- M. H. Mendonca Dias and P. C. Lauterbur, Ferromagnetic particles as contrast agents for magnetic resonance imaging of liver and spleen, Magn. Reson. Med., 1986, 3, 328–330 CrossRef CAS PubMed.
- F. J. Nicholls, M. W. Rotz, H. Ghuman, K. W. MacRenaris, T. J. Meade and M. Modo, DNA-gadolinium-gold nanoparticles for in vivo T1 MR imaging of transplanted human neural stem cells, Biomaterials, 2016, 77, 291–306 CrossRef CAS PubMed.
- M. F. Ferreira, B. Mousavi, P. M. Ferreira, C. I. Martins, L. Helm, J. A. Martins and C. F. Geraldes, Gold nanoparticles functionalised with stable, fast water exchanging Gd3+ chelates as high relaxivity contrast agents for MRI, Dalton Trans., 2012, 41, 5472–5475 RSC.
- M. F. Warsi and V. Chechik, Strategies for increasing relaxivity of gold nanoparticle based MRI contrast agents, Phys. Chem. Chem. Phys., 2011, 13, 9812–9817 RSC.
- V. S. Marangoni, O. Neumann, L. Henderson, C. C. Kaffes, H. Zhang, R. Zhang, S. Bishnoi, C. Ayala-Orozco, V. Zucolotto, J. A. Bankson, P. Nordlander and N. J. Halas, Enhancing T(1) magnetic resonance imaging contrast with internalized gadolinium(III) in a multilayer nanoparticle, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 6960–6965 CrossRef CAS PubMed.
- Y. Song, X. Xu, K. W. MacRenaris, X. Q. Zhang, C. A. Mirkin and T. J. Meade, Multimodal gadolinium-enriched DNA-gold nanoparticle conjugates for cellular imaging, Angew Chem. Int. Ed. Engl., 2009, 48, 9143–9147 CrossRef CAS PubMed.
- T. Rossetti, F. Nicholls and M. Modo, Intracerebral Cell Implantation: Preparation and Characterization of Cell Suspensions, Cell Transplant., 2016, 25, 645–664 Search PubMed.
- P. Smirnov, F. Gazeau, J. C. Beloeil, B. T. Doan, C. Wilhelm and B. Gillet, Single-cell detection by gradient echo 9.4 T MRI: a parametric study, Contrast Media Mol. Imaging, 2006, 1, 165–174 CrossRef CAS PubMed.
- P. Smirnov, M. Poirier-Quinot, C. Wilhelm, E. Lavergne, J. C. Ginefri, B. Combadiere, O. Clement, L. Darrasse and F. Gazeau, In vivo single cell detection of tumor-infiltrating lymphocytes with a clinical 1.5 Tesla MRI system, Magn. Reson. Med., 2008, 60, 1292–1297 CrossRef PubMed.
- H. Benveniste and S. J. Blackband, Translational neuroscience and magnetic-resonance microscopy, Lancet Neurol., 2006, 5, 536–544 CrossRef PubMed.
- M. Modo, K. Mellodew, D. Cash, S. E. Fraser, T. J. Meade, J. Price and S. C. Williams, Mapping transplanted stem cell migration after a stroke: a serial, in vivo magnetic resonance imaging study, Neuroimage, 2004, 21, 311–317 CrossRef PubMed.
- D. D. Shuboni-Mulligan, S. Chakravarty, C. L. Mallett, A. M. Wolf, P. M. Dmitriev, S. M. Forton and E. M. Shapiro, In vivo serial MRI of age-dependent neural progenitor cell migration in the rat brain, Neuroimage, 2019, 199, 153–159 CrossRef PubMed.
- E. M. Shapiro, O. Gonzalez-Perez, J. Manuel Garcia-Verdugo, A. Alvarez-Buylla and A. P. Koretsky, Magnetic resonance imaging of the migration of neuronal precursors generated in the adult rodent brain, Neuroimage, 2006, 32, 1150–1157 CrossRef PubMed.
- B. Iordanova and E. T. Ahrens, In vivo magnetic resonance imaging of ferritin-based reporter visualizes native neuroblast migration, Neuroimage, 2012, 59, 1004–1012 CrossRef CAS PubMed.
- M. W. Rotz, R. J. Holbrook, K. W. MacRenaris and T. J. Meade, A Markedly Improved Synthetic Approach for the Preparation of Multifunctional Au-DNA Nanoparticle Conjugates Modified with Optical and MR Imaging Probes, Bioconjug. Chem., 2018, 29, 3544–3549 CrossRef CAS PubMed.
- N. Rammohan, R. J. Holbrook, M. W. Rotz, K. W. MacRenaris, A. T. Preslar, C. E. Carney, V. Reichova and T. J. Meade, Gd(III)-Gold Nanoconjugates Provide Remarkable Cell Labeling for High Field Magnetic Resonance Imaging, Bioconjug. Chem., 2017, 28, 153–160 CrossRef CAS PubMed.
- F. Settecase, M. S. Sussman and T. P. L. Roberts, A new temperature-sensitive contrast mechanism for MRI: Curie temperature transition-based imaging, Contrast Media Mol. Imaging, 2007, 2, 50–54 CrossRef CAS PubMed.
- C. D. Graham, Magnetic Behavior of Gadolinium near Curie Point, J. Appl. Phys., 1965, 36, 1135–1136 CrossRef CAS.
- E. A. Tereshina, S. Khmelevskyi, G. Politova, T. Kaminskaya, H. Drulis and I. S. Tereshina, Magnetic ordering temperature of nanocrystalline Gd: enhancement of magnetic interactions via hydrogenation-induced "negative" pressure, Sci. Rep., 2016, 6, 22553 CrossRef CAS PubMed.
- S. H. Wang, C. W. Lee, A. Chiou and P. K. Wei, Size-dependent endocytosis of gold nanoparticles studied by three-dimensional mapping of plasmonic scattering images, J. Nanobiotechnol., 2010, 8, 33 CrossRef CAS PubMed.
- A. Stojiljkovic, K. Kuehni-Boghenbor, V. Gaschen, G. Schupbach, M. Mevissen, C. Kinnear, A. M. Moller and M. H. Stoffel, High-content analysis of factors affecting gold nanoparticle uptake by neuronal and microglial cells in culture, Nanoscale, 2016, 8, 16650–16661 RSC.
- E. Terreno, S. Geninatti Crich, S. Belfiore, L. Biancone, C. Cabella, G. Esposito, A. D. Manazza and S. Aime, Effect of the intracellular localization of a Gd-based imaging probe on the relaxation enhancement of water protons, Magn. Reson. Med., 2006, 55, 491–497 CrossRef CAS PubMed.
- M. Modo, J. S. Beech, T. J. Meade, S. C. Williams and J. Price, A chronic 1 year assessment of MRI contrast agent-labelled neural stem cell transplants in stroke, Neuroimage, 2009, 47(Suppl. 2), T133–T142 CrossRef PubMed.
- C. Brekke, S. C. Morgan, A. S. Lowe, T. J. Meade, J. Price, S. C. Williams and M. Modo, The in vitro effects of a bimodal contrast agent on cellular functions and relaxometry, NMR Biomed., 2007, 20, 77–89 CrossRef CAS PubMed.
- A. Viard, F. Eustache and S. Segobin, History of Magnetic Resonance Imaging: A Trip Down Memory Lane, Neuroscience, 2021, 474, 3–13 CrossRef CAS PubMed.
- P. K. Mishro, S. Agrawal, R. Panda and A. Abraham, A Survey on State-of-the-Art Denoising Techniques for Brain Magnetic Resonance Images, Ieee Rev. Biomed. Eng., 2022, 15, 184–199 Search PubMed.
- M. Lustig, D. Donoho and J. M. Pauly, Sparse MRI: the application of compressed sensing for rapid MR imaging, Magn. Reson. Med., 2007, 58, 1182–1195 CrossRef PubMed.
- H. Ghuman, T. K. Hitchens and M. Modo, A systematic optimization of (19)F MR image acquisition to detect macrophage invasion into an ECM hydrogel implanted in the stroke-damaged brain, Neuroimage, 2019, 202, 116090 CrossRef CAS PubMed.
- N. Kawamura, T. Yokota and H. Hontani, Super-Resolution of Magnetic Resonance Images via Convex Optimization with Local and Global Prior Regularization and Spectrum Fitting, Int. J. Biomed. Imaging, 2018, 2018, 9262847 Search PubMed.
- M. Weiger, D. Schmidig, S. Denoth, C. Massin, F. Vincent, M. Schenkel and M. Fey, NMR Microscopy with isotropic resolution of 3.0 μm using dedicated hardware and optimized methods, Concepts Magn. Reson. B Magn. Reson. Eng., 2008, 33, 84–93 CrossRef.
- L. Ciobanu, D. A. Seeber and C. H. Pennington, 3D MR microscopy with resolution 3.7 microm by 3.3 microm by 3.3 microm, J. Magn. Reson., 2002, 158, 178–182 CrossRef CAS PubMed.
- H. Y. Chen, C. B. Wilson and R. Tycko, Enhanced spatial resolution in magnetic resonance imaging by dynamic nuclear polarization at 5 K, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2201644119 CrossRef CAS PubMed.
- L. Rosa, J. Blackledge and A. Boretti, Nano-Magnetic Resonance Imaging (Nano-MRI) Gives Personalized Medicine a New Perspective, Biomedicines, 2017, 5(1), 7 CrossRef PubMed.
- R. E. Jacobs and S. E. Fraser, Imaging neuronal development with magnetic resonance imaging (NMR) microscopy, J. Neurosci. Methods, 1994, 54, 189–196 CrossRef CAS PubMed.
- J. P. Sumner, E. M. Shapiro, D. Maric, R. Conroy and A. P. Koretsky, In vivo labeling of adult neural progenitors for MRI with micron sized particles of iron oxide: quantification of labeled cell phenotype, Neuroimage, 2009, 44, 671–678 CrossRef PubMed.
- M. Gutova, J. A. Frank, M. D'Apuzzo, V. Khankaldyyan, M. M. Gilchrist, A. J. Annala, M. Z. Metz, Y. Abramyants, K. A. Herrmann, L. Y. Ghoda, J. Najbauer, C. E. Brown, M. S. Blanchard, M. S. Lesniak, S. U. Kim, M. E. Barish, K. S. Aboody and R. A. Moats, Magnetic resonance imaging tracking of ferumoxytol-labeled human neural stem cells: studies leading to clinical use, Stem Cells Transl. Med., 2013, 2, 766–775 CrossRef CAS PubMed.
- A. Y. Louie, M. M. Huber, E. T. Ahrens, U. Rothbacher, R. Moats, R. E. Jacobs, S. E. Fraser and T. J. Meade, In vivo visualization of gene expression using magnetic resonance imaging, Nat. Biotechnol., 2000, 18, 321–325 CrossRef CAS PubMed.
- G. Paxinos and C. Watson, The rat brain in stereotaxic coordinates, Elsevier, Amsterdam, NL, 2007 Search PubMed.
- A. Foi, Noise estimation and removal in MR imaging: the variance-stabilization approach, IEEE International symposium on biomedical imaging: from nano to macro, IEEE, 2011, pp. 1809–1814 Search PubMed.
- G. Vegas-Sánchez-Ferrero and S. Aja-Fernández, Statistical analysis of noise in MRI: modeling, filtering and estimation, Springer, 2016 Search PubMed.
- M. Maggioni, V. Katkovnik, K. Egiazarian and A. Foi, Nonlocal transform-domain filter for volumetric data denoising and reconstruction, IEEE Trans. Image Process., 2012, 22, 119–133 Search PubMed.
- D. Szczupak, D. J. Schaeffer, X. Tian, S. H. Choi, C. Fang, P. M. Iack, V. P. Campos, J. P. Mayo, J. Patsch, C. Mitter, A. Haboosheh, M. A. C. Vieira, G. Kasprian, F. Tovar-Moll, R. Lent and A. C. Silva, Direct interhemispheric cortical communication via thalamic commissures: a new white-matter pathway in the primate brain, Cereb. Cortex, 2023, 34(1), bhad394 CrossRef PubMed.
- T. Santini, M. Koo, N. Farhat, V. P. Campos, S. Alkhateeb, M. A. C. Vieira, M. A. Butters, C. Rosano, H. J. Aizenstein, J. Mettenburg, E. M. Novelli and T. S. Ibrahim, Analysis of hippocampal subfields in sickle cell disease using ultrahigh field MRI, Neuroimage Clin., 2021, 30, 102655 CrossRef PubMed.
- K. Panetta, Y. Zhou, S. Agaian and H. Jia, Nonlinear unsharp masking for mammogram enhancement, IEEE Trans. Inf. Technol. Biomed, 2011, 15, 918–928 Search PubMed.
- S. Nercessian, S. S. Agaian and K. A. Panetta, Multi-scale image enhancement using a second derivative-like measure of contrast, Image Processing: Algorithms and Systems X; and Parallel Processing for Imaging Applications II, SPIE, 2012, pp. 213–221 Search PubMed.
- S. DelMarco and S. Agaian, The design of wavelets for image enhancement and target detection, Proc. SPIE, 2009, 11–22 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5sc01588j |
| This journal is © The Royal Society of Chemistry 2025 |