 Open Access Article
Open Access ArticlePreferential survival of prebiotic metallopeptides in the presence of ultraviolet light†
Daniele
Rossetto‡
 ab,
Serge
Nader
ab,
Serge
Nader
 b,
Corinna L.
Kufner§
b,
Corinna L.
Kufner§
 c,
Gabriella G.
Lozano
c,
Linda
Cerofolini
c,
Gabriella G.
Lozano
c,
Linda
Cerofolini
 d,
Marco
Fragai
d,
Marco
Fragai
 d,
Vlad
Martin-Diaconescu
d,
Vlad
Martin-Diaconescu
 e,
Barbara
Zambelli
e,
Barbara
Zambelli
 f,
Stefano
Ciurli
f,
Stefano
Ciurli
 f,
Graziano
Guella
f,
Graziano
Guella
 g,
Rafał
Szabla
g,
Rafał
Szabla
 hi,
Dimitar D.
Sasselov
hi,
Dimitar D.
Sasselov
 c and
Sheref S.
Mansy
c and
Sheref S.
Mansy
 *ab
*ab
aDiCIBIO, University of Trento, 38123 Povo, Italy. E-mail: sheref.mansy@unitn.it
bDepartment of Chemistry, University of Alberta, Edmonton, Alberta T6G 2G2, Canada
cDepartment of Astronomy, Harvard University, 60 Garden Street, Cambridge, Massachusetts 02138, USA
dMagnetic Resonance Centre (CERM), Consorzio Interuniversitario Risonanze Magnetiche di Metalloproteine (CIRMMP), Department of Chemistry “Ugo Schiff”, University of Florence, Sesto Fiorentino, Italy
eALBA Synchrotron, Carrer Llum 2-26, Cerdanyola del Valles, Barcelona 08290, Spain
fLaboratory of Bio-Inorganic Chemistry (LBIC), Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy
gDepartment of Physics, University of Trento, 38123 Povo, Italy
hInstitute of Advanced Materials, Faculty of Chemistry, Wrocław University of Science and Technology, Wrocław, Poland
iDepartment of Physics, Faculty of Science, University of Ostrava, 30. dubna 22, 701 03 Ostrava, Czech Republic
First published on 20th May 2025
Abstract
The transition from unregulated, prebiotic chemistry to metabolic-like systems capable of supporting an evolving protocell has remained difficult to explain. One hypothesis is that early catalysts began to prune the chemical landscape in a manner that facilitated the emergence of modern-day enzymes. As enzymes frequently rely on the intrinsic reactivity of metal ions, it follows that these early catalysts may have been metal ions coordinated to prebiotic peptides that have remained as core structures within extant proteins. Here, we demonstrate that UV light directly selects for the types of metal-binding peptide motifs found in biology. This is because bare cysteine is much more susceptible to photolysis than cysteine bound by a metal ion. Therefore, peptides with greater affinity for environmentally available metal ions, such as Fe2+ or Zn2+, are more stable. Our results are supported by mass spectrometry, calorimetry, X-ray absorption, NMR spectroscopy, transient absorption pump probe spectroscopy, and excited-state quantum-chemical calculations. Photostability arises from the ability of the metal ion to engage transiently generated reactive radical centers in a manner that prevents subsequent degradative processes. The data are consistent with the enrichment of a restricted set of high affinity, extant-like metallopeptides in surficial environments on the early Earth.
Introduction
Life as we know it is entirely reliant on metal ions, e.g. nucleic acids and proteins require metals for folding, and central metabolism cannot operate without metal ions. Due to the high abundance of metals on Earth, metals are thought to have shaped prebiotic chemistry in a form that resembles extant metabolism.1 Metal ions have been shown to facilitate the synthesis of a suite of metabolic end-products by incubation with metabolites, often at high temperatures.2,3 However, these reactions largely run unregulated, seemingly at odds with strategies employed by living cells where activation energies are traversed by (metallo)enzymes to impart control. One approach to understanding how such enzymes emerged is to investigate the types of scaffolds that could have coordinated to, and modified the activity of, naturally present metal-based catalysts. Small, catalytically active metallopeptides likely preceded the large protein enzymes found today,4 and these metallopeptides would have emerged as a result of the selective pressures of the early Earth.Although we are far from understanding the precise environmental conditions necessary for the emergence of life,5 the most thoroughly experimentally investigated prebiotic setting thus far is the surface of the early Earth. Such conditions are compatible with the synthesis of all the major building blocks of life.6–12 One unique feature of surface conditions is exposure to ultraviolet (UV) radiation. Sunlight is one of the most abundant energy sources on Earth.11 Just as sunlight drives extant biology, the energy from the young sun could have fueled the prebiotic synthesis of the building blocks of life.13,14 For example, photochemistry has been implicated in the prebiotic synthesis of amino acids, nucleosides, carbohydrates, iron–sulfur clusters, and biopolymers.13,15–17 UV light also provided a strong selective pressure, which may have ensured the emergence of photostable biological nucleotides over their non-photostable, non-biological counterparts.18–22 Prior to the formation of the ozone layer, wavelengths as low as 210 nm could have reached the surface of the early Earth.23 Additional UV shielding could have been provided in water rich in SO2,23 Fe(CN)64−,24 or other small molecules.25 Despite such shielding effects, the flux of photons was sufficient to impact the chemistry of the early Earth.26
Previous work from the Jensen and Thøgersen groups demonstrates that free amino acids decarboxylate upon absorption at 200 nm, while peptides consisting of glycine and alanine are much more resilient to photolysis.27,28 Here, we investigate the potential impact of photochemistry on a peptide containing a prebiotically plausible cysteine (Cys) residue.29,30 Cys possesses a nucleophilic thiol group that can participate in catalysis and serves as a binding site for both structural and catalytic metal ions.31 These features make Cys-containing peptides ideal for forming functional complexes from few ingredients. One potential complication is that the side chain of Cys is more labile than that of other amino acids, degrading at high temperatures32 and upon irradiation with UV light.33 This lability can be useful, as the released hydrosulfide can be incorporated into forming iron–sulfur clusters.33,34 However, prolonged exposure to UV light, for example, would have degraded all the available Cys unless a protection mechanism existed. We find that the binding of metal ions to cysteinyl peptides strongly protects against photolysis. Importantly, mixtures of low affinity and high affinity metal-binding peptides lead to the enrichment of peptides with high affinity for metal ions. The data suggest that the metal-binding motifs found in biology today, such as iron–sulfur clusters and zinc-binding motifs, would have been preferentially stabilized on the prebiotic Earth, perhaps aiding the emergence of (proto)biological function.4,35
Results and discussion
Thiophilic metal ions protect Cys from photolysis
LC-MS was used to quantify the survival of the tripeptide GCG (glycine–cysteine–glycine) upon irradiation with 50 mW cm−2 UV light at 254 nm under anoxic conditions. The intensity of this light source was approximately 4000-fold greater than the solar irradiation on the surface of the early Earth (4.2 μW cm−2).36 The main degradation product of GCG was GAG (glycine–alanine–glycine) ([M + H]+ = 204.0 m/z), consistent with our past work showing photochemical desulfurization (Fig. S1 and S2†).33 Oxidized GCG ([M + H]+ = 469.0 m/z) was also observed, although it was unclear whether oxidized GCG formed during anoxic irradiation or during subsequent LC-MS analysis, which was not performed under anoxic conditions. The half-life (t1/2) of GCG in the absence of metal ions was 3.8 ± 0.5 min in the presence of UV light (Fig. 1). Thiophilic metal ions protected against desulfurization with t1/2 increasing ≥6-fold to 24.1 ± 4.3 min and 36.0 ± 10.6 min in the presence of 5 mM Zn2+ and Cd2+, respectively (Fig. 1A). The increased protection afforded by Cd2+ with respect to Zn2+ was consistent with the increased thiophilicity of Cd2+. To confirm that protection was due to the binding of the cysteinyl side chain, we investigated the impact of the addition of the non-thiophilic metal ion Mg2+ on stability. The t1/2 of GCG in the presence of 5 mM Mg2+ was 3.8 ± 0.4 min, identical to the reaction in the absence of Mg2+ (Fig. 1A). As expected, degradation by UV light proceeded through a different mechanism than thermal degradation, as Mg2+ protected against the latter32 but not the former. Quantification of the effect of Fe2+ was complicated by the fact that Fe2+ can be photooxidized to Fe3+ (ref. 33) and Fe2+/3+ can engage in electron transfer reactions, including the formation of oxidized Cys. Nevertheless, the addition of Fe2+ to GCG provided protection against photolysis with a t1/2 of 6.5 ± 0.6 min (Fig. S3†). The reaction products were more greatly enriched in oxidized GCG in comparison to reactions with non-redox active metals or in the absence of metal ions (Fig. S1 and S4†). As a control, the stability of the non-Cys containing tripeptide GAG was tested. After 8 min of irradiation at 254 nm, 94.7% of the GAG survived (Fig. S5†), much higher than the measured 28.3% for GCG. Peptides that contained Cys residues were more photochemically labile than peptides that lacked Cys. For the remainder of the experiments, the effects of Zn2+ binding were primarily evaluated instead of Fe2+. Zn2+ is a good probe for the binding of Fe2+ as naturally occurring Zn2+-binding sites of proteins are highly similar to rubredoxin-like, mononuclear iron centers in addition to the binding sites of [2Fe–2S] and [4Fe–4S] clusters, and Zn2+ binds to these same sites in vitro.31 Additionally, Zn2+ may have participated in prebiotic chemistry. Zn2+ was likely prebiotically available35,37 and has been invoked in model prebiotic reactions.12When considering a prebiotically plausible flux of photons at 254 nm,26 exposure to UV light for 60 min with our setup translated to approximately 14 months of continuous irradiation. However, survival would have likely been longer on the early Earth, because diurnal cycles limited exposure to irradiation. When factoring in the average exposure at the equator (ca. 4422 h per year), 60 min of irradiation with our setup corresponded to 4.5 years on the prebiotic Earth. Correcting for this lower flux of photons, the t1/2 of GCG was approximately 3 months in the absence of metal ions and greater than 21 months in the presence of Zn2+. Since our measurements were performed using a quartz cuvette with a path length of 10 mm, the data approximate conditions at the immediate surface of exposed water and not more submerged regions better protected from UV light.
Metal-binding motifs outcompete non-metal-binding motifs
Since the binding of Zn2+ protected against photolysis, the extent of protection would be expected to correlate with binding affinity. Short, prebiotically plausible sequences of peptides show differences in affinities for metal ions, including Zn2+ (ref. 38) and two properly placed Cys bind thiophilic metal ions with significantly greater affinity than peptides possessing a single Cys.39 We, therefore, synthesized and evaluated three different peptides containing two Cys residues. One was the hexapeptide GCGGCG, which represented a simple duplication of the GCG peptide sequence used above. Another was the Hook14 peptide (AKGKCPVCGAELTD), which was previously demonstrated to bind Zn2+ with high affinity.40 Finally, a truncated version of Hook14, referred to here as Hook6 (KCPVCG), was designed to possess weaker affinity for Zn2+ by removing conformationally stabilizing interactions (Fig S7†). An experimental assessment of the conformational stability of Hook6 and Hook14 in the presence of Zn2+ was obtained by NMR spectroscopy. Analysis of the spectra confirmed that Hook14 possessed a significantly more rigid fold, giving rise to long-range NOEs, whereas Hook6 assumed multiple conformations consistent with a more dynamic, lower affinity Zn2+–peptide complex (Fig. S8–S12†).The displacement of bound Co2+ by UV-Vis absorption spectroscopy was used to determine the dissociation constant (Kd) of Zn2+, which was found to be 41 ± 15, 18 ± 8, and 5.5 ± 4 μM for GCG, GCGGCG, and Hook6, respectively (Fig. S13†). The Kd for Hook14 was previously reported as 3.5 nM.40 Therefore, these four peptides spanned a four orders of magnitude range of affinities for Zn2+ and thus allowed us to probe whether affinity for Zn2+ impacted protection against photolysis. Increased affinity led to increased protection from photolysis with 84.6 ± 9.3% of Hook14 surviving exposure to UV light for 16 min in the presence of Zn2+ in comparison to 12.7 ± 4.9% for GCG (Fig. 1B).
Since peptides with higher affinity for Zn2+ were protected more efficiently than peptides with lower affinity, we next tested whether mixtures of peptides would lead to selection of high affinity metal-binding motifs. When 5 mM GCG and 2.5 mM Hook6 (both at 5 mM Cys concentration) were co-incubated with 1.25 mM Zn2+ and irradiated with 254 nm light for 16 min, 46.7 ± 6.1% of Hook6 survived and only 15.7 ± 3.0% of GCG survived (Fig. S14†). That is, approximately three-fold more of the peptide with higher affinity for Zn2+ survived in comparison to the peptide with lower affinity for Zn2+ when both were present in the same solution. Selection was even more dramatic when a solution of Hook14 and GCG was irradiated with UV light in the presence of Zn2+. In this case, 84.6 ± 4.2% of Hook14 survived after 16 min of irradiation in comparison to 12.7 ± 2.2% of GCG (Fig. 1C). The survival of GCG was similar to experiments in the absence of Zn2+ (8.52 ± 2.6%), consistent with Hook14 sequestering all the available Zn2+. To corroborate this interpretation, a simulation experiment was performed using AFFINImeter software41 and the experimentally determined dissociation constants. In this simulation, Hook14 was titrated into a solution with (Fig. S17†) and without (Fig. S18†) GCG. The simulation assumed a bidentate coordination for Hook14 and the formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 complex for Zn2+–GCG, as experimentally observed at these concentrations (vide infra). The calculated species distribution at this condition indicated that almost all the Zn2+ bound to Hook14, either in a 1
4 complex for Zn2+–GCG, as experimentally observed at these concentrations (vide infra). The calculated species distribution at this condition indicated that almost all the Zn2+ bound to Hook14, either in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 or a 1
1 or a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry, with essentially no effect of GCG on the equilibrium (Fig. S17 and S18†).
2 stoichiometry, with essentially no effect of GCG on the equilibrium (Fig. S17 and S18†).
To confirm that the presence of Fe2+ would similarly select for a high affinity metal-binding motif, we repeated the competition experiment in the presence of 1.25 mM Fe2+ (Fig. 1D). Also in this case, the survival of the Hook14 peptide was higher than that of GCG, with 75.3 ± 1.2% of Hook14 surviving after 16 min of irradiation compared to 39.6 ± 3.6% of GCG. The data demonstrated that mixtures of small peptides led to the selection of metal-binding motifs with the highest affinity when exposed to UV light.
Peptides are incompletely bound by Zn2+
To gain greater insight into the mechanism of degradation, we used isothermal titration calorimetry (ITC) to further characterize the binding of Zn2+ to GCG. Zn2+ was titrated into a solution containing GCG over three different concentration ranges. The binding reactions were exothermic for all sampled conditions (Fig. S15†) with a small endothermic signal observed as part of the heat of dilution of metal in the control experiments (Fig. S16†). Integration and fitting revealed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GCG
1 GCG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn2+ binding stoichiometry at lower concentrations (75 μM GCG titrated with 0.75 mM ZnSO4 and 150 μM GCG titrated with 1.0 mM ZnSO4) and a 4
Zn2+ binding stoichiometry at lower concentrations (75 μM GCG titrated with 0.75 mM ZnSO4 and 150 μM GCG titrated with 1.0 mM ZnSO4) and a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 GCG
1 GCG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn2+ binding stoichiometry at higher concentrations (300 μM GCG titrated with 2.0 mM ZnSO4). Consistent with an increased number of peptides bound to the metal center, the enthalpic contribution to binding increased from 0.378 kcal mol−1 at the lowest concentration to 2.075 kcal mol−1 at the highest concentration, indicating an increased bonding character of the interaction (Fig. 2, S15 and Table S2†) and a concomitant decrease of the entropy factor with the concentration. As expected from increased steric interference, the value of Kd increased from 60–80 μM at lower concentrations of peptide to 700 μM for 300 μM GCG (Table S2†).
Zn2+ binding stoichiometry at higher concentrations (300 μM GCG titrated with 2.0 mM ZnSO4). Consistent with an increased number of peptides bound to the metal center, the enthalpic contribution to binding increased from 0.378 kcal mol−1 at the lowest concentration to 2.075 kcal mol−1 at the highest concentration, indicating an increased bonding character of the interaction (Fig. 2, S15 and Table S2†) and a concomitant decrease of the entropy factor with the concentration. As expected from increased steric interference, the value of Kd increased from 60–80 μM at lower concentrations of peptide to 700 μM for 300 μM GCG (Table S2†).
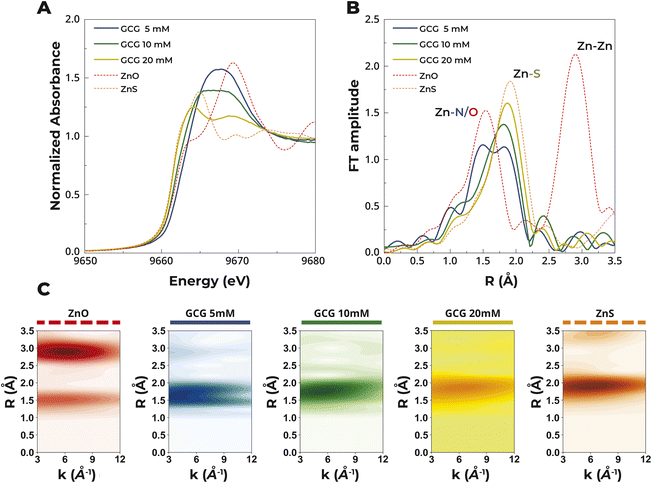
To confirm the difficulty in forming tetrathiolate complexes with Zn2+, X-ray absorption spectroscopy was used to assess the ligand environment. An increase in sulfur coordination in 5 mM Zn2+ solutions was observed when the concentration of GCG was increased from 5 mM to 10 mM and then to 20 mM, as would be expected for complexes with a greater number of coordinated peptides per metal center. The 20 mM GCG sample produced a Zn K-edge XAS (X-ray Absorption Spectroscopy) spectrum consistent with a tetrahedral, tetrathiolate coordinated Zn2+ center (Fig. 3). The rising edge of the XANES region shifted to higher energy with a concomitant increase in the intensity of the white line with higher concentrations of GCG. Such behavior is indicative of a change in the coordination environment, due to replacement of S coordination by N/O atoms (increase in rising edge energy and intensity) as well as an increase in the coordination number (increase in intensity).42,43 Indeed, EXAFS results revealed that the total coordination number increased with decreased thiol concentration consistent with observations from the XANES region. More specifically, at 20 mM GCG, 4S scattering atoms were observed at 2.31 Å. At 10 mM GCG, ∼2.3N/O scattering atoms were observed at 2.05 Å accompanying 2.8S atoms at 2.31 Å. At 5 mM GCG, 3.1N/O atoms and 2.4S atoms were observed at 2.0 Å and 2.31 Å, respectively, completing the first coordination shell of the Zn center (Fig. S1†).
The measured sub-millimolar affinity values indicated that under the experimental conditions of the photolysis experiments, a heterogeneous distribution of Zn2+–tripeptide complexes formed, with a significant fraction of non-metalated species. To determine the distribution of species in solution under the experimental conditions of the photolysis experiments, simulations of ITC binding isotherms were performed using AFFINImeter software,41 as described above, and the thermodynamic parameters obtained from the fits (Table S2†). This procedure allowed us to calculate the concentration of free GCG, as well as that of Zn2+ coordinated by one, two, three, and four GCG (Fig. 4). The model indicated that even at a five-fold molar excess of Zn2+, ca. 40% of GCG remained uncomplexed at equilibrium. The simulation supported the conclusion that low affinity complexes at basic pH were incapable of protection against photolysis, since a significant fraction of the peptide remained free and thus unprotected. These low affinity cysteinyl peptides would have then served as a source of hydrosulfide, potentially supporting the synthesis of iron–sulfur clusters.33
Protonation protects against photolysis
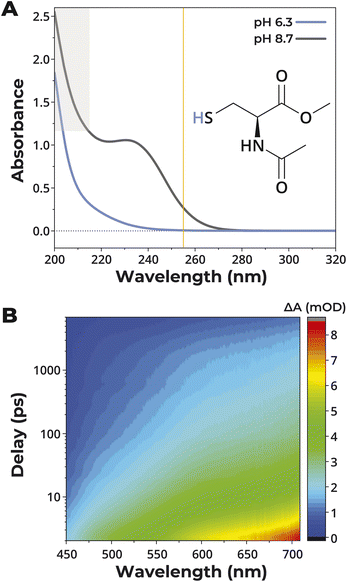
The data were consistent with the coordinate covalent bond between Zn2+ and the thiolate side chain of Cys providing protection against photolysis. As the reactions were at pH 8.7, free Cys side chains were deprotonated. To determine if the formation of a covalent bond to hydrogen, i.e. protonation of the side chain, could similarly protect against photolysis, the stability of GCG at different pH was evaluated. There was a strong correlation between pH and stability (ρ = −0.95), with protonation at lower pH protecting against desulfurization (Fig. 5). Although the presence of Zn2+ facilitated deprotonation by shifting the pKa of the cysteinyl side chain from 8.1 to 6.4 (Fig. S6†), GCG was stable across a broad range of pH in the presence of Zn2+ since both protonation and Zn2+-binding protected the side chain from degradation. That is, at low pH where Zn2+ binding was disfavored, the cysteine residue was protected by protonation. Conversely, at high pH where protonation was disfavored, the thiolate side chain was protected by the binding of Zn2+.Bound Zn2+ diminishes the reactivity of the excited state
UV absorption measurements were made to gain insight into the electronic structure of the ground and excited states. To simplify the system, N-acetyl-L-cysteine methyl ester was used in place of the tripeptide GCG. The primary absorption band (λmax ∼ 232 nm) of N-acetyl-L-cysteine methyl ester at pH 8.7 originated from the thiolate group (Fig. 6A). This absorption band suggested that the photodegradation of the peptide by UV light was initiated by the promotion of an electron from the thiolate to an unoccupied molecular orbital, possibly located on the amide (e.g. a π*-molecular orbital). Such an interpretation was supported by the fact that the band was almost fully suppressed at pH 6.3 (Fig. 6A), a pH where the thiolate groups were almost completely protonated. Similarly, the addition of Zn2+ also led to the suppression of the same band (Fig. S19†), meaning that the excitation of this electron by UV light was less likely when the thiolate was either protonated or coordinated to Zn2+. While the suppression of this band likely contributed to the diminished photodegradation observed under these conditions, the absorption feature at 254 nm remained significant and warranted further investigation.To study the ultrashort-lived intermediates after photoexcitation, we next performed transient absorption (TA) pump probe measurements of N-acetyl-L-cysteine methyl ester at pH 8.7 (Fig. 6B). The transient absorption spectra showed a broad excited-state absorption band in the entire probe range between 450 nm and 700 nm. Control measurements in neat water showed a background signal caused by solvated electrons ≤0.1 mOD (optical density). The transient absorption spectra were consistent with the initial hypothesis of short-range photoinduced electron transfer from the thiolate group to another moiety, thus generating a loosely bound excited-state radical anion. Triexponential global fitting analysis revealed excited state lifetimes of 4 ps, 70 ps, and >7 ns. The >7 ns decay indicated that some portion of the photoexcited molecule could be involved in destructive bimolecular reactions after photoexcitation, such as the formation of disulfide bonds. Consequently, we concluded that the photoprotection of the thiolate-containing molecule was most efficient when the photoinduced electron transfer event from the thiolate group was suppressed by protonation or coordination to Zn2+.
We next performed excited-state quantum-chemical calculations to better understand the photochemistry of Zn2+–peptide complexes in comparison to free, thiolate-containing peptides. For this purpose, we applied the algebraic diagrammatic construction to the second order method [ADC(2)] together with a triple zeta TZVP basis set and the COSMO continuum solvation model of bulk water.44–46 We considered a minimal model of a single GCG peptide that could bind Zn2+ with negatively charged thiolate and carboxylate groups. Additionally, we added two to three water molecules to complete the coordination sphere of the metal ion. Zn2+ was found to adopt either a tetrahedral or a trigonal bipyramidal coordination sphere in the electronic ground state (Fig. 7), as observed with previously reported structures of Zn2+ bound to biological molecules.47 The number of ligands to the metal center was also consistent with the X-ray absorption and ITC data.
The lowest energy excited singlet states found for the peptide both in the presence and absence of Zn2+ possessed a charge transfer character involving electron transfer from the thiolate anion to the π* orbitals of the carbonyl amide groups of the peptide with a major contribution from the N-terminal glycine and a minor contribution from the cysteine, consistent with the UV absorption measurements above (see Tables S3 and S4†). Therefore, the electronic excitation of the peptide resulted in the generation of a radical on the sulfur atom and a radical anion on one of the carbonyl carbon atoms of the peptide. However, as can be seen for the structure corresponding to the minimum of the S1 state, Zn2+ interfered with the reactivity of the sulfur radical by engaging the electron in a strong Zn–S interaction. Additionally, the Zn2+ cation underwent a barrierless reorganization to an octahedral coordination sphere in the excited state by accepting a strong ligand–metal interaction with the negatively charged radical anion of the carbonyl carbon as a consequence of the charge transfer character of the excited state (Fig. 7). This excited-state structure could then undergo proton transfer to the oxygen atom of the negatively charged carbonyl group from the nearest water molecule coordinated to the Zn2+ cation. This process occurred on a flat and nearly barrierless topography of the S1 potential energy surface and eventually led to an S1/S0 conical intersection, which enabled photorelaxation to the electronic ground (S0) state. The Zn2+ cation effectively coordinated the transiently created hydroxyl anion. The observed photostabilizing effect could be complemented by back electron transfer from the transiently hydrogenated carbonyl group to re-create the thiolate anion and back proton transfer to the aforementioned hydroxyl anion.
In the absence of Zn2+, the observed charge transfer excitation present in the free peptide could still result in the transient hydrogenation of the carbonyl group. However, in this case the biradical structure of the excited peptide would not be protected from chemical reactions with other molecules. Such a scenario would likely lead to intermolecular radical recombination between two photoexcited peptides yielding a disulfide bond, as observed in our experiments (Fig. S1†). Alternatively, the C–S bond could undergo dissociation yielding atomic sulfur and a carbon-centered radical. The latter species would subsequently accept the excess hydrogen atom from the hydrogenated carbonyl group, resulting in the formation of alanine, as we observed by LC-MS. Binding of a single Zn2+ engages these reactive radical centers on the sulfur and carbonyl carbon atoms in strong interactions and, consequently, protects the peptide from such deleterious radical recombination events.
Conclusions
Free metal ions are capable of engaging in degradative chemistry that would have interfered with the functioning of a protocell reliant on RNA and lipids. In fact, the catalytic hydrolysis of RNA and the instability of fatty acid vesicles are both mediated by metal ions. Therefore, peptides that sequestered metals from the environment would have been advantageous to early, more labile protocellular systems by removing a ubiquitous hazard. Such peptides could have emerged in regions exposed to UV light, because high affinity peptides are more photostable than their low affinity counterparts, as we show here. Colocalization of lipids and peptides may have been aided by the latter's ability to promote the polymerization of amino acids during wet–dry cycling.48,49 Since extant metal-binding motifs of proteins possess the intrinsic ability to bind metal ions in the absence of more complex scaffolding,50 the early Earth may have contained a pool of peptides enriched in sequences found in modern-day protein enzymes. This also suggests that peptides could have been important before genetically encoded synthesis, because UV light would have selected for decreased diversity and increased affinity for metal ions. That is, few high affinity metal-binding peptides may have been consistently present for protocells to exploit.51 Such an out-of-equilibrium system would require a constant flux of energy, provided by the sun, and the simultaneous synthesis and degradation of peptides.The transition from low to highly active peptides requires folding into tertiary structures. In the absence of strongly coordinated metal ions, peptides typically require a length of 30 residues to reach sufficient stability to be associated with a discrete three-dimensional fold.52 Conversely, the binding of a metal ion can lock in one of several potential folds, providing rigid tertiary structures with much fewer residues. In biology, zinc proteins are among the smallest and most common protein motifs, functioning as individual proteins or as interconnected modules within larger protein folds.53–55 For example, the smallest tertiary protein structure found in nature contains a ββα fold that binds Zn2+.56,57
The ubiquity, size, and remarkable stability of zinc peptides have been used to argue that zinc proteins may be among the most ancient modules found in proteins.31 If proteins emerged from the accretion of smaller available peptides,58 then it seems likely that early proteins were enriched in sequences that survived the harsh, UV exposed conditions of the early Earth, such as metal-binding motifs. These early peptides and resulting proteins may have also been predisposed to catalysis, since it is difficult to form complexes that fully coordinate the metal center with peptidyl ligands. Structural and catalytic metal ions are typically distinguished by complete coordination and partial coordination, respectively, to peptides. The open position, typically filled by displaceable water, then functions as a substrate-binding site. Zinc proteins similar to the motifs we have used here have been observed in oxidoreductases, hydrolases, transferases, and helicases.59 Further, zinc-binding motifs are capable of binding other metal ions in place of Zn2+, including Fe2+ and iron–sulfur clusters,31 thereby increasing the catalytic possibilities and likelihood of metallopeptides on the early Earth. The discovery that high affinity metal-binding peptides would have outcompeted low affinity alternatives suggests that the metal-binding motifs found in extant proteins may have an ancient, prebiotic origin and may have facilitated the emergence of the catalytic, regulatory mechanisms that are the hallmarks of metabolism.60
Data availability
Data associated with this manuscript can be downloaded from Zenodo at [https://doi.org/10.5281/zenodo.15288133].Author contributions
Conceptualization: DR and SSM. Investigation: DR, SN, CLK, GGL, VM-D, BZ, RS, and LC. Supervision: MF, SC, GG, DDS, and SSM. Writing – original draft: DR, SSM. Writing – review & editing: all authors.Conflicts of interest
All authors declare they have no competing interests.Acknowledgements
X-ray absorption experiments were performed at the CLAESS beamline of the ALBA Synchrotron facility with the collaboration of ALBA staff as part of experiment 2019093816. We acknowledge the support of the Protein Technology and the Proteomics and Mass Spectrometry (MS) core facilities of Department CIBIO at the University of Trento. SSM acknowledges support from the Simons Foundation (290358FY19), the Alfred P. Sloan Foundation (G-2022-19518), and the Gordon and Betty Moore Foundation (11479). MF acknowledges support from “Potentiating the Italian Capacity for Structural Biology Services in Instruct Eric (ITACA.SB),” project no. IR0000009. This article has been produced with the financial support of the European Union under the LERCO project number CZ.10.03.01/00/22_003/0000003 via the Operational Programme Just Transition (RS). Computational resources granted by the Wrocław Centre for Networking and Supercomputing (WCSS) under the grant no. 549 are also gratefully acknowledged (RS). SC and BZ acknowledge the support of Consorzio Interuniversitario di Risonanze Magnetiche di Metallo-Proteine (CIRMMP) and the University of Bologna. DDS acknowledges support from the Simons Foundation (290360).References
- S. D. Copley, E. Smith and H. J. Morowitz, Bioorg. Chem., 2007, 35, 430–443 CrossRef CAS PubMed.
- K. B. Muchowska, S. J. Varma and J. Moran, Nature, 2019, 569, 104–107 CrossRef CAS PubMed.
- M. A. Keller, A. V. Turchyn and M. Ralser, Mol. Syst. Biol., 2014, 10, 725 CrossRef PubMed.
- C. Bonfio, E. Godino, M. Corsini, F. Fabrizi de Biani, G. Guella and S. S. Mansy, Nat. Catal., 2018, 1, 616–623 CrossRef CAS.
- C. R. Walton, P. Rimmer and O. Shorttle, Front. Earth Sci., 2022, 10, 1011717 CrossRef.
- B. H. Patel, C. Percivalle, D. J. Ritson, C. D. Duffy and J. D. Sutherland, Nat. Chem., 2015, 7, 301–307 CrossRef CAS PubMed.
- H.-J. Kim and S. A. Benner, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 11315–11320 CrossRef CAS.
- Z. R. Cohen, B. L. Kessenich, A. Hazra, J. Nguyen, R. S. Johnson, M. J. MacCoss, G. Lalic, R. A. Black and S. L. Keller, ChemBioChem, 2022, 23, e202100614 CrossRef CAS PubMed.
- J. D. Toner and D. C. Catling, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 883–888 CrossRef CAS.
- B. M. Tutolo, R. Perrin, R. Lauer, S. Bossaer, N. J. Tosca, A. Hutchings, S. Sevgen, M. Nightingale, D. Ilg, E. B. Mott and T. Wilson, Life, 2024, 14, 1624 CrossRef CAS PubMed.
- D. Deamer and A. L. Weber, Cold Spring Harb. Perspect. Biol., 2010, 2, a004929 Search PubMed.
- S. Becker, J. Feldmann, S. Wiedemann, H. Okamura, C. Schneider, K. Iwan, A. Crisp, M. Rossa, T. Amatov and T. Carell, Science, 2019, 366, 76–82 CrossRef CAS PubMed.
- N. J. Green, J. Xu and J. D. Sutherland, J. Am. Chem. Soc., 2021, 143, 7219–7236 CrossRef CAS PubMed.
- R. Szabla, in Prebiotic Photochemistry: From Urey–Miller-like Experiments to Recent Findings, ed. F. Saija and G. Cassone, The Royal Society of Chemistry, 2021, pp. 79–106 Search PubMed.
- R. J. Rapf and V. Vaida, Phys. Chem. Chem. Phys., 2016, 18, 20067–20084 RSC.
- J. Xu, V. Chmela, N. J. Green, D. A. Russell, M. J. Janicki, R. W. Góra, R. Szabla, A. D. Bond and J. D. Sutherland, Nature, 2020, 582, 60–66 CrossRef CAS PubMed.
- J. Xu, M. Tsanakopoulou, C. J. Magnani, R. Szabla, J. E. Šponer, J. Šponer, R. W. Góra and J. D. Sutherland, Nat. Chem., 2017, 9, 303–309 CrossRef CAS PubMed.
- M. J. Janicki, S. J. Roberts, J. Šponer, M. W. Powner, R. W. Góra and R. Szabla, Chem. Commun., 2018, 54, 13407–13410 RSC.
- Z. R. Todd, R. Szabla, J. W. Szostak and D. D. Sasselov, Chem. Commun., 2019, 55, 10388–10391 RSC.
- L. Bertram, S. J. Roberts, M. W. Powner and R. Szabla, Phys. Chem. Chem. Phys., 2022, 24, 21406–21416 RSC.
- L. Serrano-Andrés and M. Merchán, J. Photochem. Photobiol., C, 2009, 10, 21–32 CrossRef.
- A. A. Beckstead, Y. Zhang, M. S. de Vries and B. Kohler, Phys. Chem. Chem. Phys., 2016, 18, 24228–24238 RSC.
- S. Ranjan and D. D. Sasselov, Astrobiology, 2017, 17, 169–204 CrossRef CAS PubMed.
- S. Ranjan, C. L. Kufner, G. G. Lozano, Z. R. Todd, A. Haseki and D. D. Sasselov, Astrobiology, 2022, 22, 242–262 CAS.
- Z. R. Todd, G. G. Lozano, C. L. Kufner, S. Ranjan, D. C. Catling and D. D. Sasselov, Astrobiology, 2024, 24, 559–569 CrossRef CAS.
- P. B. Rimmer, S. J. Thompson, J. Xu, D. A. Russell, N. J. Green, D. J. Ritson, J. D. Sutherland and D. P. Queloz, Astrobiology, 2021, 21, 1099–1120 CrossRef CAS.
- M. M. Madsen, F. Jensen and J. Thøgersen, Phys. Chem. Chem. Phys., 2020, 22, 2307–2318 RSC.
- J. Thøgersen, A. S. Chatterley, T. Weidner and F. Jensen, J. Am. Chem. Soc., 2023, 145, 9777–9785 CrossRef PubMed.
- C. S. Foden, S. Islam, C. Fernández-García, L. Maugeri, T. D. Sheppard and M. W. Powner, Science, 2020, 370, 865–869 CrossRef CAS PubMed.
- S. Wehbi, A. Wheeler, B. Morel, N. Manepalli, B. Q. Minh, D. S. Lauretta and J. Masel, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2410311121 CrossRef CAS PubMed.
- L. Belmonte and S. S. Mansy, J. Chem. Inf. Model., 2017, 57, 3162–3171 CrossRef CAS PubMed.
- D. Rossetto, L. Valer, N. Yeh Martín, G. Guella, Y. Hongo and S. S. Mansy, ACS Earth Space Chem., 2022, 6(5), 1221–1226 CrossRef CAS PubMed.
- C. Bonfio, L. Valer, S. Scintilla, S. Shah, D. J. Evans, L. Jin, J. W. Szostak, D. D. Sasselov, J. D. Sutherland and S. S. Mansy, Nat. Chem., 2017, 9, 1229–1234 CrossRef CAS PubMed.
- S. Scintilla, D. Rossetto, M. Clémancey, J. Rendon, A. Ranieri, G. Guella, M. Assfalg, M. Borsari, S. Gambarelli, G. Blondin and S. S. Mansy, Chem. Sci., 2025, 16, 4614–4624 RSC.
- A. Aithal, S. Dagar and S. Rajamani, ACS Omega, 2023, 8, 5197–5208 CrossRef CAS.
- S. Ranjan, R. Wordsworth and D. D. Sasselov, Astrophys. J., 2017, 843, 110 CrossRef.
- R. M. Hazen, Am. J. Sci., 2013, 313, 807–843 CrossRef CAS.
- L. Belmonte, D. Rossetto, M. Forlin, S. Scintilla, C. Bonfio and S. S. Mansy, Phys. Chem. Chem. Phys., 2016, 18, 20104–20108 RSC.
- L. Valer, D. Rossetto, T. Parkkila, L. Sebastianelli, G. Guella, A. L. Hendricks, J. A. Cowan, L. Sang and S. S. Mansy, ChemBioChem, 2022, 23, e202200202 CrossRef CAS PubMed.
- T. Kochańczyk, P. Jakimowicz and A. Krężel, Chem. Comm., 2013, 49, 1312–1314 RSC.
- Software for Science Developments (S4SD), 2023, http://www.affinimeter.com.
- K. Clark-Baldwin, D. L. Tierney, N. Govindaswamy, E. S. Gruff, C. Kim, J. Berg, S. A. Koch and J. E. Penner-Hahn, J. Am. Chem. Soc., 1998, 120, 8401–8409 CrossRef CAS.
- R. A. Kelly, J. C. Andrews and J. G. DeWitt, Microchem. J., 2002, 71, 231–245 CrossRef CAS.
- A. Dreuw and M. Wormit, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2015, 5, 82–95 CAS.
- S. Karbalaei Khani, A. Marefat Khah and C. Hättig, Phys. Chem. Chem. Phys., 2018, 20, 16354–16363 RSC.
- C. Hättig, in Adv. Quantum Chem., ed. H. J. Å. Jensen, Academic Press, 2005, vol. 50, pp. 37–60 Search PubMed.
- M. Laitaoja, J. Valjakka and J. Jänis, Inorg. Chem., 2013, 52, 10983–10991 CrossRef CAS PubMed.
- S. Sarkar, S. Dagar and S. Rajamani, ChemSystemsChem, 2021, 3, e2100014 CrossRef CAS.
- S. Rajamani, A. Vlassov, S. Benner, A. Coombs, F. Olasagasti and D. Deamer, Origins Life Evol. B., 2008, 38, 57–74 CrossRef CAS PubMed.
- D. Rossetto, L. Sebastianelli, S. Oberegger, S. Todorovic, H. Haas and S. S. Mansy, Adv. Biol., 2024, 8, e2300545 CrossRef PubMed.
- D. Rossetto, N. Cvjetan, P. Walde and S. S. Mansy, Acc. Chem. Res., 2024, 57, 2293–2302 CrossRef CAS PubMed.
- H. I. Merritt, N. Sawyer and P. S. Arora, Pept. Sci., 2020, 112, e24145 CrossRef CAS PubMed.
- M. Fu and P. J. Blackshear, Genome Res., 2017, 17, 130–143 CAS.
- W. J. Friesen and M. K. Darby, J. Biol. Chem., 2001, 276, 1968–1973 CrossRef CAS.
- A. L. Zerkle, C. H. House and S. L. Brantley, Am. J. Sci., 2005, 305, 467–502 CrossRef CAS.
- M. Isalan, in Encyclopedia of Biological Chemistry, ed. W. J. Lennarz and M. D. Lane, Academic Press, Waltham, 2nd edn, 2013, pp. 575–579 Search PubMed.
- J. Rodríguez, J. Mosquera, O. Vázquez, M. E. Vázquez and J. L. Mascareñas, Chem. Commun., 2014, 50, 2258–2260 RSC.
- V. Alva and A. N. Lupas, Curr. Opin. Struct. Biol., 2018, 48, 103–109 CrossRef CAS.
- N. J. Pace and E. Weerapana, Biomolecules, 2014, 4, 419–434 CrossRef.
- N. Lauber, C. Flamm and K. Ruiz-Mirazo, BioEssays, 2021, 43, 1–12 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5sc02170g |
| ‡ Current address: Synthetic Biology Group, University of Saarland, 66123 Saarbrucken, Germany. |
| § Current address: Leibniz Institute of Photonic Technology, 07745 Jena, Germany. |
| This journal is © The Royal Society of Chemistry 2025 |






![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H distance associated with the proton transfer process from one of the water molecules coordinating the Zn2+ to the negatively charged carbonyl group. The excited-state and ground-state energies were calculated with the ADC(2) and MP2 methods, respectively, including the TZVP basis set.
O⋯H distance associated with the proton transfer process from one of the water molecules coordinating the Zn2+ to the negatively charged carbonyl group. The excited-state and ground-state energies were calculated with the ADC(2) and MP2 methods, respectively, including the TZVP basis set.