Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Detection of C-reactive protein using single cluster analysis of gold nanoparticle aggregates using a dark-field microscope equipped with a smartphone†
Nanami
Fukuzumi‡
a,
Takako
Nakagawa‡
b,
Gen
Hirao
a,
Atsushi
Ogawa
 c,
Mizuo
Maeda
d,
Tsuyoshi
Asahi
a and
Tamotsu
Zako
c,
Mizuo
Maeda
d,
Tsuyoshi
Asahi
a and
Tamotsu
Zako
 *ab
*ab
aDepartment of Chemistry and Biology, Graduate School of Science and Engineering, Ehime University, 2-5 Bunkyo, Matsuyama, Ehime 790-8577, Japan. E-mail: zako.tamotsu.us@ehime-u.ac.jp
bDepartment of Chemistry, Faculty of Science, Ehime University, 2-5 Bunkyo, Matsuyama, Ehime 790-8577, Japan
cProteo-Science Center, Ehime University, 2-5 Bunkyo, Matsuyama, Ehime 790-8577, Japan
dRIKEN Cluster for Pioneering Research, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan
First published on 23rd December 2024
Abstract
Gold nanoparticles (AuNPs), which have been used as colorimetric biosensors, show strong light scattering, allowing individual AuNPs to be identified using a dark-field microscope (DFM). In this study, we developed a method of observing the target molecule-derived aggregation of AuNPs modified with DNA aptamers at the single-cluster level using the DFM. C-Reactive protein (CRP) is an important clinical biomarker of inflammatory and cardiovascular diseases, for which a simple, inexpensive, and sensitive detection method is needed. In this study, the CRP-mediated aggregate formation of CRP aptamer-modified AuNPs was evaluated with single-cluster analysis using the DFM, and the detection limit was 17 nM, which was sufficient as a diagnostic indicator for CRP. We also developed a portable DFM equipped with a smartphone and a stage adjustment system, which enables single-cluster observation of AuNPs, and showed that 50 nM of CRP could be detected, indicating that this approach is suitable for point-of-care diagnosis. With the selection of appropriate aptamers, this method can be applied for the detection of various molecules.
Introduction
Gold nanoparticles (AuNPs) have unique chemical and physical properties, which vary with size and shape.1–4 In particular, the AuNP solution undergoes a colour change from red to blue or purple because of the formation of aggregates. This is caused by a change in the surface plasmon coupling between the particles. The presence of the target molecule can be visualized by designing the AuNP surface such that the solution colour change due to this aggregation triggered by the target molecule. Thus, AuNP-based colorimetric sensors have been developed to detect various molecules, DNA, proteins, metal ions, and organic/inorganic compounds.2,5–11C-Reactive protein (CRP) is a major acute-phase reactant protein produced in the liver that rises rapidly in the plasma when the human body is infected or damaged.12,13 The serum CRP levels are an early clinical indicator of inflammation and infection, ranging from 10–40 μg mL−1 during viral infection, 40–200 μg mL−1 during bacterial infection, and exceeding 200 μg mL−1 during sepsis. Individuals with blood CRP levels below 10 μg mL−1 are considered free of inflammation, but even chronic CRP levels around 5 μg mL−1 are not considered safe because of the high risk of developing coronary artery disease, acute myocardial infarction, and even ischaemic stroke.14–16 Current technologies for the clinical detection of CRP are mainly antibody-based techniques such as enzyme-linked immunosorbent assay (ELISA), which require complex operations, time, expensive reagents and equipment, and highly trained personnel.17,18 Therefore, there is a need for a sensitive method that can detect 5 μg mL−1 of CRP easily and inexpensively. Sensors based on aptamer-CRP interactions have been developed to avoid the use of expensive reagents.19–21 In addition, CRP detection methods by combining a CRP-recognising aptamer with a AuNP-based sensor have been reported.22,23 Zhao et al. also fabricated a magnetic bead-aptamer-AuNP sandwich structure and developed a CRP detection method based on single-molecule counting of AuNPs that dissociate upon the addition of CRP.24
AuNPs scatter light very strongly owing to the plasmon resonance effect, so that each AuNP can be identified individually using a dark-field microscope (DFM).25,26 We have previously detected thrombin and estradiol by combining an AuNP-based sensor with DFM observations of AuNP aggregates.7,27 Furthermore, we succeeded in detecting DNA, amyloids, and antibodies with higher sensitivity with the observation of AuNP aggregates at the single-cluster level using the DFM.28–30 CRP detection by observing the colour change of single AuNPs with the DFM upon formation of antibody-modified AuNP sandwich structure has also been reported.31
In this study, a DFM-based single-cluster detection method using a target-derived aggregation observation technique was developed to achieve facile CRP detection with a sufficient sensitivity for recognising inflammation. Fig. 1 shows a schematic diagram of the preparation of AuNPs modified with the CRP aptamer (CRP aptamer-AuNPs) and the CRP detection mechanism. First, the CRP aptamer, to which a sequence complementary to the modified thiolated ssDNA was added, was immobilised onto ssDNA-modified AuNPs (ssDNA-AuNPs) via complementary strand formation to create CRP aptamer-AuNPs. CRP was then added to the CRP aptamer-AuNP solution and the samples were imaged using the DFM. The number and intensity of spots in the DFM images were determined using ImageJ software, and the aggregation ratio of the AuNPs was calculated.28 In the DFM images, a decrease in the number of spots and an increase in their intensity were observed as the concentration of added CRP increased. The detection limit using the benchtop DFM was 17 nM, which was sufficient as a diagnostic indicator for CRP and 10-fold more sensitive than that using the same aptamer and AuNP-based colorimetric method in this study.
 | ||
| Fig. 1 Schematic illustration of CRP detection by observation of AuNPs modified with CRP aptamers with the DFM. | ||
In this study, we also developed a portable DFM equipped with a smartphone and a stage adjustment system (smartphone DFM) which enables single cluster observation of AuNPs. The results showed that 50 nM (∼5 μg mL−1) of CRP was successfully detected, similar to the result of the benchtop DFM image. This is the first demonstration of a molecular detection based on single-molecule analysis of AuNPs using a smartphone DFM. Finally, we demonstrated that this strategy is applicable to samples containing complex biomolecules such as fetal bovine serum (FBS), as 50 nM CRP was detectable using the smartphone DFM. The development of this method is expected to further expand target detection using the DFM and contribute to point-of-care (POC) diagnosis because of its portability. In addition, this method may be applied to various molecular detection methods by selecting appropriate aptamers.
Experimental
Materials
AuNPs (40 nm) were obtained from BBI Solutions (Cardiff, UK). Thiolated ssDNA (thiol-ssDNA, 5′-SH-AAGGGATTCCTGGGAAAACTGGAC-3′) was purchased from Integrated DNA Technologies (Coralville, IA, USA). Complementary sequence added CRP aptamer (CRP aptamer, 5′GTGGTTTACCTCGTGCGTTAGTGGGGAGCTTAGGGGAAGCTTTTTTGTCCAGTTTTCCCAGGAATCCCTT-3′) and non-thiolated ssDNA (5′-AAGGGATTCCTGGGAAAACTGGAC-3′) were purchased from Eurofins Genomics (Tokyo, Japan).20 Dithiothreitol (DTT), and ethylene glycol (EG) were purchased from Wako (Osaka, Japan). CRP was purchased from ORIENTAL YEAST Co. Ltd. (Tokyo, Japan). NAP-5 columns (Sephadex G-25 DNA grade) were purchased from Cytiva (Little Chalfont, UK). Silane-coated glass slides were purchased from Muto Pure Chemicals Co., Ltd. (Tokyo, Japan). FBS was purchased from Thermo Fisher Scientific (Waltham, MA, USA). All aqueous solutions used in this study were prepared with Milli-Q water (MQ water) (Millipore, Burlington, VT, USA).Preparation of ssDNA-AuNPs
Thiol-ssDNA was immobilised on the AuNP surface via a thiol–Au bond using the freezing method, as previously described.10 In brief, thiol-ssDNA was treated with DTT and purified using NAP-5 columns. The purified thiol-ssDNA was mixed with AuNPs (concentration ratio of 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (DNA): 1 (AuNP)), and frozen at a temperature of −80 °C for 40 min in the presence of 10 mM EG.10,32 After thawing, the unreacted probe ssDNA was removed by centrifugation, and the ssDNA-AuNPs were re-dispersed in MQ water at a concentration of 750 pM as a stock solution. The amount of ssDNA on the AuNPs was estimated by the DTT displacement method as described10 and determined to be 264 ± 18 molecules per particle.
000 (DNA): 1 (AuNP)), and frozen at a temperature of −80 °C for 40 min in the presence of 10 mM EG.10,32 After thawing, the unreacted probe ssDNA was removed by centrifugation, and the ssDNA-AuNPs were re-dispersed in MQ water at a concentration of 750 pM as a stock solution. The amount of ssDNA on the AuNPs was estimated by the DTT displacement method as described10 and determined to be 264 ± 18 molecules per particle.
Detection of CRP using ssDNA-AuNPs
ssDNA-AuNPs were mixed with the CRP aptamer (concentration ratio of 400 (aptamer): 1 (ssDNA-AuNP)). After incubation in TN buffer (10 mM Tris HCl buffer, pH 7.5 and 50 mM NaCl) at a temperature of 90 °C for 1 min, the sample was cooled at room temperature. Modification of AuNPs with thiol-ssDNA and the CRP aptamer was confirmed by size measurement of the AuNP samples using a Zetasizer-Nano ZS (Malvern Worcestershire, UK) (ESI,† Fig. S1(a)). Attachment of the CRP aptamer to ssDNA-AuNPs was also confirmed by the higher stability of CRP aptamer-AuNPs against salt than that of ssDNA-AuNPs possibly due to higher electrostatic repulsion by modification of the CRP aptamer (ESI,† Fig. S1(b)). Measurement of zeta potential values with the Zetasizer-Nano ZS showed that CRP aptamer-AuNPs exhibited a higher negative charge than ssDNA-AuNPs, supporting the attachment of the CRP aptamer (ESI,† Fig. S1(c)). The number of the attached CRP aptamer was evaluated by measuring the amount of the released aptamer after incubation of CRP aptamer-AuNP at 90 °C for 1 min and subsequent centrifugation, and was estimated to be approximately 100 ± 6 molecules per particle. It is noted that the scattering intensities of AuNPs, ssDNA-AuNPs and CRP aptamer-AuNPs observed using the DFM at the single molecule level were similar, supporting that aggregates were not formed during the modification process (ESI,† Fig. S1(d)).For the detection of CRP, various concentrations (0–2000 nM) of CRP (5 μL) were added to 300 pM CRP aptamer-AuNP solution (10 μL) and incubated for 60 min at room temperature. A total of 5 μL of 4× binding buffer (40 mM Tris-HCl buffer, pH 7.5, 200 mM NaCl, 40 mM MgCl2, and 20 mM KCl) was added to the sample before incubation.
For CRP detection in the presence of serum, FBS diluted four times by MQ water was filtered using a 0.22 μm pore size syringe filter (Millipore). Then the diluted FBS samples (5 μL) mixed with various concentrations of CRP (5 μL) were added to 300 pM CRP aptamer-AuNP solution (10 μL) and incubated for 60 min at room temperature as described above.
For the colorimetric analysis, images were captured with a digital camera, and digital colour analysis of the solution colour images was performed using ImageJ software. Briefly, the images were split into red, green, and blue components. The redness of the solution was quantitatively evaluated by calculating the 1/redness value (1/(red − (green + blue)/2)) from the intensity of each colour component, as described previously.10
DFM analysis
After dilution of the samples to 1/30 with MQ water, 6 μL samples were deposited onto glass slides and covered with cover glass. DFM images were obtained using a BX53 microscope (Olympus, Tokyo, Japan) equipped with a dark-field condenser (U-DCW), a 60× objective lens (UPlanFLN), and a CCD camera (DP73). Images were taken using CellSens standard software version 1.6 (Olympus). The intensity of each spot in the DFM images was obtained using ImageJ software.Smartphone DFM
Smartphone DFM images were captured using the home-build smartphone DFM equipped with a dark-field condenser (U-DCW, Olympus), a 50× objective lens (SLMPlan, Olympus), a white-LED light source (MCWHLP2, Thorlabs, Inc., NJ, USA), and a smartphone (HUAWEI P30, Huawei Device Co., Ltd, Shenzhen, China). The glass slides and smartphone were placed on commercial manual positioning devices and built with the optics into the main body fabricated by a 3D printer (Adventurer 4, FLASHFORGE) (ESI,† Fig. S2). Dark-field images were taken using the smartphone. The intensity of each spot in the smartphone DFM images was obtained using ImageJ software.Results and discussion
CRP-mediated CRP aptamer-AuNP aggregate formation
To verify whether CRP-mediated CRP aptamer-AuNP aggregates could be formed, we performed CRP detection using a colorimetric method. As shown in Fig. 2(a), the colour of CRP aptamer-AuNP solution was red in the absence of CRP. This indicated that the CRP aptamer-AuNPs remained dispersed in the binding buffer which contained a high salt concentration, possibly owing to the electrostatic repulsive force derived from the negative charge retained by the modified CRP aptamer and the immobilised ssDNA. In contrast, the colour of the solution changed to light purple when 500 nM CRP was added (Fig. 2(a)). The colour change was quantitatively evaluated using the redness of the solution in the images shown in Fig. 2a and b. An increase in 1/redness values above the 3σ line was observed in the presence of 500 nM CRP. This result supported the formation of a CRP-mediated cross-linked aggregation of AuNPs and indicates that CRP aptamer-AuNPs can be used for CRP detection.CRP detection by estimating AuNP aggregation ratio using the DFM
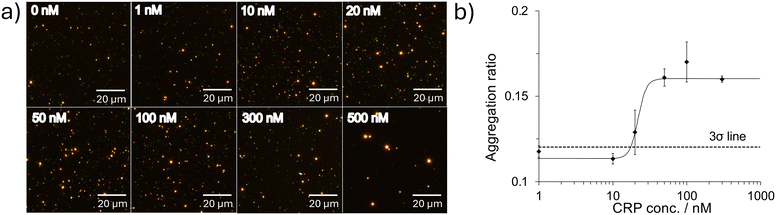
To investigate the sensitivity of CRP detection using the DFM, we performed detection of CRP at various concentrations from 0 to 500 nM (0, 1, 10, 20, 50, 100, 300, 500 nM). Fig. 3(a) shows the DFM images of CRP aptamer-AuNPs incubated with CRP at each concentration (0–500 nM). The dispersed CRP aptamer-AuNPs were observed as dark green spots, while aggregated CRP aptamer-AuNPs were observed as bright orange spots, owing to the change in the surface plasmon due to aggregation. Importantly, at higher CRP concentrations, the number of bright orange spots increased, and that of dark green spots decreased.The size of the AuNP aggregates was estimated at the single-cluster level based on the intensity of each spot in the DFM images, and histograms were obtained at each CRP concentration (ESI,† Fig. S3). Based on the histograms of the scattered light intensity, the ratio of aggregates formed in the sample with each concentration of CRP was estimated as previously described;28 spots with intensities higher than 87 a.u. were calculated as aggregates because 90% of the dispersed control sample in the absence of CRP showed intensities lower than 87 a.u. This threshold was determined as a value by which a better detection limit can be obtained than other thresholds. As shown in Fig. 3(b), the aggregation ratio increased with increasing CRP concentration. The detection limit was evaluated using the 3σ method as previously reported and determined to be 17 nM.33 These results also indicated that CRP detection is possible by observing the aggregates of CRP aptamer-AuNPs in a single-cluster analysis of the intensity of each spot using the DFM.
CRP detection using the smartphone DFM
In this study, the use of the smartphone DFM for on-site CRP detection was investigated. Previously, our group made a portable DFM using a smartphone as a detector and reported concentration-dependent changes in the RGB component ratios of the whole DFM images upon the addition of target molecules.7 Sun et al. also reported a portable DFM for gold nanorods measurement.34 However, single-cluster analysis using these previous portable DFMs was not possible because of the lack of a stage-adjustment function. In this study, to enable single-cluster analysis, we developed a new smartphone DFM with a stage-adjustment mechanism and a light source which resembles that of a benchtop DFM (Fig. 4(a)).Fig. 4(b) shows the DFM images of the CRP aptamer-AuNP samples incubated with CRP at various concentrations (0–500 nM) obtained using the smartphone DFM. The number of spots decreased with increasing CRP concentration, whereas the number of brighter spots increased. It should be noted that the spots captured using the smartphone DFM showed a variety of colours, which were not similar to those captured using the benchtop DFM (Fig. 3(a)), possibly due to chromatic aberration. The aggregation ratio was then calculated using single-cluster analysis of the intensity of each spot, as described above (ESI,† Fig. S4). As shown in Fig. 4(c), the aggregation ratio increased with increasing CRP concentration. There was a significant difference (*, p < 0.05) between the control sample in the absence of CRP and the sample containing 50 nM CRP, indicating that the smartphone DFM can be used to detect CRP at concentrations as low as 50 nM and is expected to make CRP detection feasible outside laboratories.
Detection of CRP in the presence of serum
To further evaluate the practical feasibility of this method, detection of CRP in the presence of FBS, which contains various ingredients, was performed as a model of complex biological fluids. First, CRP was detected in the presence of 10% FBS using the benchtop DFM. As shown in Fig. 5(a), a CRP concentration-dependent increase was observed in the bright orange spots derived from the CRP aptamer-AuNPs in an aggregated state. As shown in Fig. 5(b), the aggregation ratio increased with the CRP concentration (ESI,† Fig. S5(a)). There was a significant difference (*, p < 0.05) between the control sample in the absence of CRP and the sample containing 50 nM CRP. This result indicated that 50 nM CRP in serum can be detected using this approach. Importantly, similar results were obtained using the smartphone DFM (Fig. 5(c) and (d), and ESI,† Fig. S5(b)): a CRP concentration-dependent decrease in the number of spots and an increase in brighter spots were observed, and 50 nM CRP was detected in the presence of FBS. 50 nM CRP could also be detected in the presence of 1% bovine serum albumin (ESI,† Fig. S6). These results supported the use of our approach for detecting CRP in samples containing complex biomolecules. For further application of CRP detection in real samples, pre-treatments of human serum samples such as removal of endogenous CRP for preparation of background samples to make a standard curve for the quantification of CRP and removal of co-contaminants that would affect aggregation of AuNPs using antibody-modified magnetic nanoparticles would be necessary.Conclusions
In this study, we developed a method to detect CRP by evaluating the CRP-mediated aggregates of CRP aptamer-AuNPs using a DFM. In the presence of CRP, the colour of the solution changed from red to light purple, suggesting the capture of CRP by the CRP aptamer and the formation of CRP-mediated cross-linked aggregation of AuNPs. Single-cluster analysis of the bright spots observed using the DFM showed that the detection sensitivity of this method was 10-fold higher than that of the colorimetric method shown in this study. Single-cluster analysis using the smartphone DFM detected 50 nM of CRP in the presence of serum. This presented sufficiently high sensitivity as a diagnostic indicator for CRP.To date, many efforts have been made to develop biosensors for sensitive CRP detection using optical, electrochemical or mass-based methods.17,18 For example, Kim et al. reported surface-enhanced Raman spectroscopy-based sensitive CRP detection using the DNA aptamer modified with biotin and methylene blue, AuNPs and Au-Te nanostructure.35 An electrochemical aptasensor using carbon nanofiber-chitosan nanocomposites and RNA aptamers has also been developed.36 Although sensitivity of these methods are high, the current method using the smartphone DFM provides a simple and inexpensive CRP detection. It is also noted that no significant difference was observed under light–dark conditions (ESI,† Fig. S7). These characteristics of this method are expected to contribute to POC diagnosis. In addition, this method may be applied to various molecular detection techniques such as biomedical diagnosis and environmental assessment, by selecting appropriate aptamers.
Data availability
The data used in the current study are available from the corresponding author on reasonable request. The data supporting this article have been included as part of the ESI.†Author contributions
T. Z. conceived the project. T. Z., N. F., T. N., G. H., A. O., T. A. and M. M. designed the experiments. N. F. and T. N. performed the experiments. T. A. manufactured the smartphone DFM. N. F. and T. Z. wrote the initial manuscript, and all the authors discussed the results and contributed to the manuscript preparation. N. F. and T. N. contributed this work equally.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to the financial support by JSPS KAKENHI (19H02527, 22K19114 and 23K26471 to TZ) and Ehime University (Research Unit for Advanced Nano-Bioanalysis).Notes and references
- S. K. Ghosh and T. Pal, Chem. Rev., 2007, 107, 4797–4862 CrossRef CAS PubMed.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- K. M. Mayer and J. H. Hafner, Chem. Rev., 2011, 111, 3828–3857 CrossRef CAS PubMed.
- M.-C. Daniel and D. Astruc, Chem. Rev., 2004, 104, 293–346 CrossRef CAS PubMed.
- H. Aldewachi, T. Chalati, M. N. Woodroofe, N. Bricklebank, B. Sharrack and P. Gardiner, Nanoscale, 2018, 10, 18–33 RSC.
- W. Zhao, M. A. Brook and Y. Li, ChemBioChem, 2008, 9, 2363–2371 CrossRef CAS PubMed.
- Y. Yano-Ozawa, N. Lobsiger, Y. Muto, T. Mori, K. Yoshimura, Y. Yano, W. J. Stark, M. Maeda, T. Asahi, A. Ogawa, A. Ogawa and T. Zako, RSC Adv., 2021, 11, 11984–11991 RSC.
- N. Kanayama, T. Takarada and M. Maeda, Chem. Commun., 2011, 47, 2077–2079 RSC.
- Z. He, G. Wang, X. Liang, T. Takarada and M. Maeda, Anal. Sci., 2021, 37, 415–423 CrossRef CAS PubMed.
- G. Hirao, N. Fukuzumi, A. Ogawa, T. Asahi, M. Mizuo and T. Zako, RSC Adv., 2023, 13, 30690–30695 RSC.
- Z. Yuan, F. Lu, M. Peng, C.-W. Wang, Y.-T. Tseng, Y. Du, N. Cai, C.-W. Lien, H.-T. Chang, Y. He, Y. He and E. S. Yeung, Anal. Chem., 2015, 87, 7267–7273 CrossRef CAS PubMed.
- N. R. Sproston and J. J. Ashworth, Front. Immunol., 2018, 9, 754 CrossRef PubMed.
- M. B. Pepys and G. M. Hirschfield, J. Clin. Invest., 2003, 111, 1805–1812 CrossRef CAS PubMed.
- B. Clyne and J. S. Olshaker, J. Emerg. Med., 1999, 17, 1019–1025 CrossRef CAS PubMed.
- T. A. Pearson, G. A. Mensah, R. W. Alexander, J. L. Anderson, R. O. Cannon III, M. Criqui, Y. Y. Fadl, S. P. Fortmann, Y. Hong, G. L. Myers, R. P. Tracy and F. Vinicor, Circulation, 2003, 107, 499–511 CrossRef PubMed.
- E. B. Windgassen, L. Funtowicz, T. N. Lunsford, L. A. Harris and S. L. Mulvagh, Postgrad. Med., 2011, 123, 114–119 CrossRef PubMed.
- H. H. Khanmiri, F. Yazdanfar, A. Mobed, F. Rezamohammadi, M. Rahmani and T. Haghgouei, Biomed. Microdevices, 2023, 25, 27 CrossRef CAS PubMed.
- S. K. Vashist, A. G. Venkatesh, E. Marion Schneider, C. Beaudoin, P. B. Luppa and J. H. T. Luong, Biotechnol. Adv., 2016, 34, 272–290 CrossRef CAS PubMed.
- C.-J. Huang, H.-I. Lin, S.-C. Shiesh and G.-B. Lee, Biosens. Bioelectron., 2010, 25, 1761–1766 CrossRef CAS PubMed.
- B. Wu, R. Jiang, Q. Wang, J. Huang, X. Yang, K. Wang, W. Li, N. Chen and Q. Li, Chem. Commun., 2016, 52, 3568–3571 RSC.
- N. L. Phung, J. G. Walter, R. Jonczyk, L. K. Seiler, T. Scheper and C. Blume, ACS Comb. Sci., 2020, 22, 617–629 CrossRef CAS PubMed.
- M. António, R. Ferreira, R. Vitorino and A. L. Daniel-da-Silva, Talanta, 2020, 214, 120868 CrossRef PubMed.
- A.-M. Hada, S. Suarasan, M. Muntean, M. Potara and S. Astilean, Anal. Chim. Acta, 2024, 1307, 342626 CrossRef CAS PubMed.
- Y. Zhao, J. Zhao, T. Jin, S. Sun, W. Liu and Y. Tan, RSC Adv., 2019, 9, 34293–34298 RSC.
- A. Tcherniak, J. W. Ha, S. Dominguez-Medina, L. S. Slaughter and S. Link, Nano Lett., 2010, 10, 1398–1404 CrossRef CAS PubMed.
- C. L. Nehl, N. K. Grady, G. P. Goodrich, F. Tam, N. J. Halas and J. H. Hafner, Nano Lett., 2004, 4, 2355–2359 CrossRef CAS.
- Y. Yano, M. Nisougi, Y. Yano-Ozawa, T. Ohguni, A. Ogawa, M. Maeda, T. Asahi and T. Zako, Anal. Sci., 2019, 35, 685–690 CrossRef CAS PubMed.
- T. Bu, T. Zako, M. Fujita and M. Maeda, Chem. Commun., 2013, 49, 7531–7533 RSC.
- T. Bu, T. Zako and M. Maeda, Anal. Sci., 2016, 32, 307–311 CrossRef CAS PubMed.
- K. Yoshimura, P. Patmawati, M. Maeda, N. Kamiya and T. Zako, Anal. Sci., 2021, 37, 507–511 CrossRef CAS PubMed.
- M. Ouyang and D. Di Carlo, Biosens. Bioelectron., 2019, 132, 162–170 CrossRef CAS PubMed.
- B. Liu and J. Liu, J. Am. Chem. Soc., 2017, 139, 9471–9474 CrossRef CAS PubMed.
- K. Sato, K. Hosokawa and M. Maeda, Nucleic Acids Res., 2005, 33, e4 CrossRef PubMed.
- D. Sun and T. Y. Hu, Biosens. Bioelectron., 2018, 99, 513–518 CrossRef CAS PubMed.
- S. M. Kim, J. Kim, G. Yim, H. J. Ahn, M. Lee, T.-H. Kim, C. Park, J. Min, H. Jang and T. Lee, Anal. Bioanal. Chem., 2022, 414, 3197–3204 CrossRef CAS PubMed.
- M. A. Tabrizi and P. Acedo, Nanomaterials, 2022, 12, 415 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00329b |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2025 |