Open Access Article
Open Access ArticleEngineering a hypoxia-tolerant Saccharomyces cerevisiae for rapid ethanol production via co-utilization of glucose and acetic acid and redox-enhanced flocculation†
Sadat Mohamed Rezk
Khattab‡
 *abc,
Mohammed Oksh Mohammed
Mousa‡
*abc,
Mohammed Oksh Mohammed
Mousa‡
 ad,
Takashi
Nagata
ad,
Takashi
Nagata
 ade,
Takashi
Watanabe
ade,
Takashi
Watanabe
 bf and
Masato
Katahira
bf and
Masato
Katahira
 *adef
*adef
aInstitute of Advanced Energy, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan. E-mail: khattab.sadatmohamedrezk.7c@kyoto-u.ac.jp; katahira.masato.6u@kyoto-u.ac.jp
bResearch Institute for Sustainable Humanosphere, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan
cFaculty of Science, Al-Azhar University, Assiut 71524, Egypt
dGraduate School of Energy Science, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan
eIntegrated Research Center for Carbon Negative Science, Institute of Advanced Energy, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan
fBiomass Product Tree Industry-Academia Collaborative Research Laboratory, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan
First published on 28th May 2025
Abstract
Enhancing the robustness of microbial cell factories is essential for improving both first- and second-generation bioethanol production. During fermentation, Saccharomyces cerevisiae produces acetic acid as a by-product under certain conditions, which inhibits cellular functions and reduces fermentation efficiency. Additionally, pretreatment of lignocellulosic biomass releases acetic acid, further exacerbating fermentation stress toward the yeast. Hypoxic fermentation, combined with metabolic engineering, offers an alternative strategy to mitigate these challenges. To address this, we used CRISPR-Cas9 gene editing to sequentially delete NADH-dependent glycerol-3-phosphate dehydrogenase 1 (GPD1), cytosolic aldehyde dehydrogenase (ALD6), and mitochondrial external NADH dehydrogenase isoforms (NDE1 and NDE2), while integrating an empty plasmid into the LEU2 locus to generate control strains C1 to C5. Notably, strain C5 (GPD1Δ ALD6Δ NDE1Δ NDE2Δ), exhibited a 150% increase in the fermentation rate compared to strain C1 when fermenting a minimal medium containing 10% glucose and 0.4% acetic acid under hypoxic conditions. To further enhance acetic acid utilization and ethanol production, we integrated a plasmid containing acetylating acetaldehyde dehydrogenase from Salmonella enterica (SeEutE) into the LEU2 locus, generating EutE strains E1 to E5. Strain E5 (GPD1Δ ALD6Δ NDE1Δ NDE2Δ [SeEutE]) exhibited a 200% increase in fermentation rate compared to strain C5, with 75% ethanol-induced flocculation. Strain E5 consumed approximately 25% of the supplemented acetic acid and achieved near-theoretical ethanol yields from the total consumed glucose and acetic acid. Furthermore, strain E5 exhibited a 9% improvement in the fermentation rate under hypoxic conditions compared to hyperoxic conditions. These enhancements together represent an overall improvement of more than 343% compared to the parent strain. Thus, by integrating quadruple deletion (GPD1Δ ALD6Δ NDE1Δ NDE2Δ) with the heterologous expression of SeEutE integration, we introduce a novel strategy to construct a hypoxia and acetate tolerant S. cerevisiae strain. This engineered strain achieves rapid, redox-balanced fermentation and ethanol-induced flocculation, offering a significant advance by overcoming limitations in glucose fermentation rate, redox imbalance, and weak acetate tolerance.
1. Introduction
Addressing climate change, which has a significant impact on the biosphere and ecosystems, is a global priority. The United Nations Sustainable Development Goals (SDGs) emphasize the need to balance environmental, economic, and social requirements, making the sustainable production of biofuels both essential and highly valued.1Saccharomyces cerevisiae, a model microbial cell factory, plays a key role in first-generation bioethanol production, contributing to an annual yield exceeding 100 billion liters.2 However, acetic acid is produced as a by-product during bioethanol fermentation, inhibiting cell growth and reducing both fermentation rate and ethanol yield, particularly under conditions that lead to elevated acetic acid accumulation. While excessive acetic acid can be inhibitory, its toxicity varies depending on strain tolerance and environmental conditions.3,4 High concentrations of acetic acid induce apoptosis in yeast cells through mechanisms involving mitochondrial dysfunction, reactive oxygen species (ROS) accumulation, and cytochrome c release. The target of rapamycin (TOR) pathway plays a regulatory role in stress responses and metabolic adaptation to acetic acid toxicity, influencing processes such as amino acid and nucleotide biosynthesis, protein turnover, carbohydrate metabolism, cell wall and membrane integrity, signaling, and cell cycle regulation.5In second-generation bioethanol production, S. cerevisiae strains face additional challenges, including reduced robustness and efficiency in handling inhibitors released from lignocellulosic biomass, such as furfural, 5-hydroxy methyl furfural (5-HMF), and acetic acid. Acetic acid, commonly released during the deacylation of hemicellulose and lignin in lignocellulosic hydrolysates, exacerbates these challenges and poses a significant obstacle to bioethanol production from lignocellulosic biomass.6 The co-utilization of acetic acid as an electron acceptor by S. cerevisiae was first demonstrated using the heterologous expression of E. coli acetylating acetaldehyde dehydrogenase (EcMhpF) along with double deletion of both NADH-dependent glycerol-3-phosphate dehydrogenases (GPD1 and GPD2) under anaerobic conditions. This strategy successfully prevented glycerol biosynthesis, enabled acetic acid co-utilization, and enhanced ethanol production. However, the engineered strain exhibited a 2.6-fold reduction in fermentation speed and showed reduced tolerance to higher sugar concentrations.7 In a subsequent report, recovery of tolerance was achieved through evolutionary engineering.8 The conversion of acetic acid to ethanol requires the transport of acetic acid into the cell, its conversion to acetyl CoA by ATP-dependent acetyl-CoA synthase (ACS1, ACS2), and subsequent reductions to acetaldehyde and ethanol by EcMhpF and alcohol dehydrogenase isoforms (ADHs), respectively.7 A key limitation for acetic acid co-utilization in this pathway is the availability of NADH during glucose fermentation. Heterologous expression of NADPH-dependent alcohol dehydrogenase from Entamoeba histolytica (EhADH) in industrial diploid S. cerevisiae strains increased NADH levels, supporting the activity of two copies of acetylating acetaldehyde dehydrogenase from Bifidobacterium adolescentis (BaAdhE). This modification replaced GPD1 and GPD2 with BaAdhE, thereby preventing glycerol biosynthesis and increasing the co-utilization of acetate.9 Replacing the NADP+-dependent 6-phosphogluconate dehydrogenase (GND) with a NAD+-dependent variant, combined with substituting glycerol biosynthesis with E. coli EutE, improves acetic acid co-utilization with glucose and enhances ethanol production, though it results in a lower fermentation rate.10 Similarly, heterologous replacement of the native NADH-dependent ScGPD1 with an archaeal NADPH-preferring gene (GpsA) and heterologous replacement of the native ScGPD2 with EcEutE, along with the deletion of cytosolic aldehyde dehydrogenase (ALD6), successfully blocked glycerol biosynthesis and increased ethanol production and acetic acid co-utilization under hyperosmotic conditions.11 However, this also led to a significant reduction in fermentation rates.
Xylose, the second most abundant sugar in lignocellulosic biomass after glucose, provides additional NADH through the cofactor recycling between xylose reductase (XR) and xylitol dehydrogenase (XDH). The redox imbalance between XR and XDH has been utilized to improve acetic acid co-utilization with xylose, confirming that NADH deficiency constrains acetic acid co-utilization. Co-fermentation of xylose and acetic acid also helps reduce xylitol accumulation.12 Optimization of the acetic acid reduction pathway, including expression of three copies of codon-optimized EcAdhE and Salmonella enterica acetyl-CoA synthetase (SeACS), has significantly improved anaerobic xylose fermentation and acetate co-utilization.13 Under aerobic conditions without glucose, when the target product does not involve oxidoreductase reactions, acetic acid was co-consumed with xylose at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratio, underscoring the importance of NADH availability and the inhibitory effect of glucose on acetic acid co-consumption.14
4 ratio, underscoring the importance of NADH availability and the inhibitory effect of glucose on acetic acid co-consumption.14
Despite these advances, most existing redox engineering strategies present significant trade-offs—such as reduced fermentation speed, impaired osmoadaptation, or diminished strain robustness—especially under hypoxic or industrially relevant conditions. Designs based on full deletion of GPD1/GPD2, overexpression of NADH-consuming enzymes, or anaerobic xylose–acetate co-fermentation often result in slow ethanol production, excessive glycerol accumulation, and poor tolerance to process-related stresses. Furthermore, ethanol-induced flocculation—an important trait for stress protection and biomass recovery—has not been previously integrated into acetate-utilizing yeast strains.
In this study, we addressed these limitations by developing a modular, stepwise metabolic engineering strategy. This involved deleting mitochondrial external NADH dehydrogenases (NDE1 and NDE2)—enzymes typically active under normoxic or hyperoxic conditions—and introducing redox balancing via heterologous expression of Salmonella enterica acetylating acetaldehyde dehydrogenase (SeEutE), as illustrated in Fig. 1. To our knowledge, this is the first report to link ethanol-induced flocculation with enhanced acetate co-utilization.
Specifically, we constructed a hypoxia- and acetate-tolerant S. cerevisiae strain through sequential deletion of GPD1, ALD6, NDE1, and NDE2 (Fig. 1), which significantly accelerated glucose fermentation under hypoxia. Subsequent integration of SeEutE improved NADH recycling, promoted acetate assimilation, enhanced ethanol production, and triggered flocculation in response to ethanol accumulation. This strategy led to more than a threefold increase in fermentation rate, near-theoretical ethanol yield, and superior performance under hypoxic versus hyperoxic conditions. Collectively, this work establishes a robust platform for efficient glucose–acetate co-fermentation and scalable bioethanol production under oxygen-limited industrial settings.
2. Materials and methods
2.1 Media components
YPD medium (10 g yeast extract, 20 g peptone, and 20 g glucose per liter) was used for routine cultivation and preservation of yeast strains. Agar was added at 1.8% to solidify the medium for agar plates. For positive selection in the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) system, YPD medium was supplemented with geneticin (G418), resulting in YPDG418 medium. Positive colonies with chromosomal integration at the LEU2 locus were selected on Yeast Nitrogen Base (YNB) medium without amino acids (Invitrogen™), supplemented with 6.7 g L−1 YNB, 20 g L−1 glucose and 20 mg L−1 histidine, designated as YNBD20.15 The composition of YNB is listed in Table S1.†For fermentations with glucose as the sole carbon source, YNB was supplemented with 100 g L−1 glucose, 4 g L−1 acetic acid, and 0.2 g L−1 histidine, with the pH adjusted to 5 using NaOH (referred to as YNBD100-AC4). To prevent interference from degradation products during autoclaving, all liquid media used in this study were filter-sterilized by TPP vacuum filtration (rapid-Filtermax, 0.2 μm PES, Switzerland). Escherichia coli NEB® 10-beta (New England Biolabs) was cultivated in Luria–Bertani (LB) medium, containing 1% tryptone, 1% sodium chloride (NaCl), and 0.5% yeast extract. Ampicillin (150 μg mL−1) was added to the LB medium to select for E. coli carrying ampicillin-resistant plasmids, while kanamycin (75 μg mL−1) was used for selecting E. coli harboring the pCas9 multiplex plasmid, as previously described.16,17
2.2 Primers, cassettes, and plasmids constructions
All primers were designed based on the S. cerevisiae S288C sequence from the Saccharomyces Genome Database (Table S2†). Primers were synthesized by FASMAC, Japan.2.3 Engineering the recombinant strains
The engineering strategy is summarized in Fig. S1.† The haploid D452-2 strain (MATα leu2 his3 ura3 can1) served as the parental strain (D).20 The URA3 locus in the D strain was restored in a previous report,16 resulting in the base strain B1 (Table 1). Strain B2, with GPD1 deleted,4 was used for sequential deletions of ALD6, NDE1, and NDE2, following a method reported by Ryan et al.18 with a few modifications.16,17| Strains | Relevant genotype | Reference |
|---|---|---|
| D452-2 (D) | MATα leu2 his3 ura3 can1 | 20 |
| DURA3 (B1) | D, URA3::TDH3 promoter and DIT1d22 terminator | 16 |
| B2 | B1, ΔGPD1:: TDH3p-DIT1d22t | 4 |
| B3 | B2, ΔALD6:: TDH3p-DIT1d22t | This study |
| B4 | B3, ΔNDE1:: TDH3p-DIT1d22t | This study |
| B5 | B4, ΔNDE2:: TDH3p-DIT1d22t | This study |
| C1 | B1, LEU2:: TDH3p-DIT1d22t | This study |
| C2 | B2, LEU2:: TDH3p-DIT1d22t | This study |
| C3 | B3, LEU2:: TDH3p-DIT1d22t | This study |
| C4 | B4, LEU2:: TDH3p-DIT1d22t | This study |
| C5 | B5, LEU2:: TDH3p-DIT1d22t | This study |
| E1 | B1, LEU2:: TDH3p-SeEutE-DIT1d22t | This study |
| E2 | B2, LEU2:: TDH3p-SeEutE-DIT1d22t | This study |
| E3 | B3, LEU2:: TDH3p-SeEutE-DIT1d22t | This study |
| E4 | B4, LEU2:: TDH3p-SeEutE-DIT1d22t | This study |
| E5 | B5, LEU2:: TDH3p-SeEutE-DIT1d22t | This study |
A repair cassette composed of TDH3p-DIT1d22t was generated by PCR from the YEpM4-TD plasmid, incorporating homologous regions (50–60 bp) flanking the ALD6 promoter and terminator. The repair cassette and a CRISPR-Cas9 multiplex plasmid were introduced into competent cells using a lithium acetate transformation protocol detailed previously.17 Transformants were selected on YPDG418 medium, and colonies were screened for successful ALD6 deletions via PCR verification. At least four positive colonies were re-cultured on YPDG418 plates to eliminate false positives, and their reproducibility was confirmed through fermentation experiments. Strain B3 was propagated in YPD medium with shaking to remove the CRISPR-Cas9 plasmid and was subsequently used to generate strains B4, C3, and E3 (Table 1).
Similar procedures were applied to delete NDE1 and NDE2, resulting in strains B4 and B5. The plasmid YEpM4-TD was linearized with ClaI and introduced into B1–B5 strains to generate control strains (C1–C5) (Fig. S1†). Concurrently, the linearized YEpM4-SeEutE plasmid was integrated into the same locus in B1–B5 strains, generating EutE-expressing strains (E1–E5) (Fig. S1†).
Transformants were selected on YNBD20 agar plates, and the first 12 colonies were transferred to and re-cultivated on fresh YNBD20 plates to prevent parental strain overgrowth. Four colonies were randomly selected to verify fermentation reproducibility, and all strains and their genotypes are listed in Table 1.
2.4 Pre-culture and fermentation conditions
Under sterile conditions, strains were pre-cultured in 10 mL of YPD medium in 50 mL TPP® TubeSpin bioreactor tubes (Switzerland) with orbital shaking at 180 rpm (EYELA, LTI-601SD, Japan) for 18 hours at an approximately 45° sitting angle during the orbital shaking. The cells were harvested by centrifugation at 5800 g, washed twice with sterile Milli-Q water, and resuspended to an OD600 of 2 in 50 mL of YNBD100-4AC (pH 5) in a 100 mL Erlenmeyer flask under hypoxic conditions [volume of yeast culture (Vc)/volume of flask (Vf) at a ratio of 50/100] to initiate the fermentation process. Fermentation was conducted at 150 rpm with 1 mL samples taken at intervals to measure growth and other fermentation characteristics.2.5 Procedures of fermentation analysis
Fermentation analyses were conducted following the method described by Khattab and Watanabe (2021). Briefly, 100 μL samples were diluted with 900 μL of Milli-Q water, mixed, and centrifuged at 15 300 g for 5 minutes using a Tomy centrifuge (Japan). The supernatants were filtered through a hydrophilic FILTSTAR PTFE 0.45 μm syringe filter (Hawach Scientific, China) and transferred to 2 mL HPLC glass vials using a 1 mL syringe. An autosampler injected the samples into an HPLC system equipped with an Aminex HPX-87H column and a refractive index detector. The mobile phase was 5 mM H2SO4 in aqueous solution and analyses were run at a flow rate of 0.6 mL min−1. Concentrations of glucose, acetic acid, glycerol, ethanol, and other intermediate metabolites such as acetoin, 2,3-BDO, succinic acid, pyruvate, and acetaldehyde were quantified using high-grade standards.2.6 Calculations of fermentation efficiency
Fermentation efficiency was calculated based on the theoretical yield (TY) using the following equations:TY = molar mass of product/molar mass of substrate (glucose and/or acetic acid)
TY of bioethanol from glucose:
| C6H12O6 → C2H5OH |
| 1 g → 0.51 g |
TY of bioethanol from acetic acid:
| CH3COOH → C2H5OH |
| 1 g → 0.77 g |
| TY of bioethanol from glucose (g L−1) = [initial concentration (IC) of glucose (g L−1) − residual concentration (RC) of glucose (g L−1)] × 0.51 |
| TY of bioethanol from acetic acid (g L−1) = [IC of acetic acid (g L−1) − RC of acetic acid (g L−1)] × 0.77 |
2.7 Enzyme activity
Cell pellets were harvested for enzyme activity measurements at 15 h of fermentation by centrifugation at 3 000 g for 5 min at 4 °C. The cell lysate was prepared as previously described:21 cell pellets were resuspended in 100 mM potassium phosphate (pH 7.5) containing 1 mM MgCl2 and 10 mM 2-mercaptoethanol, followed by vortexing with approximately 400 mg of glass beads. The lysing process involved six cycles of vigorous shaking, each followed by 30 seconds of chilling on ice. The cell-free extract (CFE) was separated by centrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 g for 5 min at 4 °C and further clarified by additional centrifugation. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) using bovine serum albumin standards and measurements were made at an absorbance of 562 nm on an Infinite M200 PRO plate reader (Tecan, Switzerland).
400 g for 5 min at 4 °C and further clarified by additional centrifugation. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) using bovine serum albumin standards and measurements were made at an absorbance of 562 nm on an Infinite M200 PRO plate reader (Tecan, Switzerland).
The specific activity of SeEutE was assayed as described by Efxtance et al.,22 in a solution of 50 mM citrate buffer (pH 6.0), 100 μM zinc acetate, 240 μM NADH, and 140 μM acetyl-CoA. A 10 μL aliquot of the crude extract was used to monitor the decrease in NADH concentration over time at 340 nm. Background rates were subtracted in the calculations. Enzyme activity was expressed as μmol of NADH oxidized per min per mg of CFE.
2.8 Flocculation assay
Flocculation was measured as previously described,23 with modifications. YNBD100-AC4 cell pellets were harvested after 21 h of fermentation by centrifugation at 2 300 g for 5 min at 25 °C and washed twice with 0.1 M EDTA to remove any residue that could interfere with flocculation. The washed cells were resuspended in a de-flocculating buffer (50 mM sodium acetate and 0.1 M EDTA), and the optical density was adjusted to an OD600 of approximately 2.0 (initial optical density, A). 10 mL of this suspension was centrifuged, and the pellet was washed with deionized water and flocculating buffer (50 mM sodium acetate and 0.1% CaCl2). The cells were then resuspended in 10 mL of flocculating buffer, vortexed for 5 min, and allowed to rest for 5 min. The OD600 of the supernatant (final optical density, B) was then measured. Flocculation percentages were calculated as follows:| Flocculation percentage = (1 − B/A) × 100% |
2.9 Varying the fermentation conditions from hyperoxic to hypoxic in strains E4 and E5
Oxygen availability was varied by adjusting the Vc relative to the Vf in 100 mL Erlenmeyer flasks. Fermentation of YNBD100-AC4 was performed with Vc/Vf ratios of 10/100 (hyperoxic conditions), 30/100, and 70/100 (hypoxic conditions).2.10 Ethanol-induced engineered yeast flocculation
To evaluate ethanol-induced flocculation, the standard fermentation procedures were used with 50 mL of YNBD100-AC4 medium and ethanol was added to the medium to a final concentration of 3%, and 4% after three hours fermentation. Flocculation was observed four hours post-addition and compared with a control experiment without ethanol supplementation.2.11 Analysis of cellular NADH/NAD+ ratio
Intracellular NADH and NAD+ levels were determined photometrically using the NAD+/NADH Assay Kit-WST (N509, Dojindo, Japan), following the manufacturer's instructions with modifications to the initial sample preparation step. At 9 hours of fermentation in YNBD100-AC4 medium, cells corresponding to 10 mL at OD600 = 6 were harvested and lysed as described in Section 2.7. The supernatant was subsequently processed using Amicon Ultra centrifugal filters (10 kDa molecular weight cutoff) to isolate low-molecular-weight metabolites, including NADH and NAD+. A 10% aliquot of the filtrate was used for the quantitative determination of total NADH and NAD+ concentrations.2.12 Statistical analysis
A completely randomized design (CRD) was used with three replications for all fermentation characteristics, except for the enzyme activity of SeEutE, which had four replications. Analysis of variance (ANOVA) was performed using the Proc Mixed procedure in the SAS software package version 9.2.24 Means were compared using Duncan's multiple range test at a 5% level of probability.253. Results and discussions
3.1 Selection basis for target gene deletion in this study
Studies on anaerobic fermentation using engineered S. cerevisiae with double deletions of GPD1 and GPD2 have successfully demonstrated the oxidation of excess NADH through the introduction of acetylating acetaldehyde dehydrogenase, facilitating the co-utilization of acetic acid with glucose.7–9 While this strategy increases NADH availability for enzymes like SeEutE, it also eliminates G-3P, which is crucial for glycerophospholipid formation. This disruption can affect various cellular processes, including cell wall integrity, signaling, and regulation.26,27 G-3P is normally dephosphorylated to glycerol, serving as an osmoadaptive regulator,28 and its absence exacerbates the inhibitory effects of acetic acid.4 The double substitution of ScGPD1 with GpsA and ScGPD2 with EcEutE, coupled with the deletion of ScALD6, effectively halted glycerol biosynthesis and enabled the co-utilization of acetic acid and glucose under osmotic fermentation conditions. However, this modification resulted in a significant decline in the fermentation rate.10,11GPD1 is well known for being expressed under aerobic, and GPD2 under anaerobic conditions.28 Furthermore, gene expression profiles under anaerobic conditions reveal that 140 genes have transcript levels more than threefold higher than under aerobic conditions, while 219 genes are more expressed under aerobic conditions.29 These metabolic shifts may limit the fermentation rate, indicating a need for alternative metabolic engineering strategies tailored to hypoxic conditions. Indeed, under aerobic conditions, the exclusive co-conversion of acetic acid and xylose into triacetic acid lactone or vitamin A demonstrated that a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratio between acetic acid and xylose was optimal. This serves as a strong example of proper design where ATP requirements, reducing equivalents, substrate co-utilization, and feeding patterns were optimized.14
4 ratio between acetic acid and xylose was optimal. This serves as a strong example of proper design where ATP requirements, reducing equivalents, substrate co-utilization, and feeding patterns were optimized.14
In the presence of oxygen which serves as the terminal electron acceptor in the electron transport chain, NDE1 and NDE2 play a crucial role in oxidizing cytosolic NADH, facilitating ATP generation (Fig. 1). It has been observed that NDE1 compensates for the loss of NDE2 (NDE2Δ). Additionally, the deletion of both NDE1 and NDE2 (NDE1Δ NDE2Δ) results in more than a sevenfold increase in glycerol biosynthesis.30 Removing these genes also enhances the enzyme activity of mitochondrial glycerol-3-phosphate dehydrogenase (GUT2) and decreases its apparent Km for G-3P. G-3P is the substrate of GUT2, and also a product of GPD isoforms.31 Deleting GPD1 in the base strain D452-2 (B2) reduced glycerol biosynthesis by 79% under hypoxic conditions.4 Based on these findings, we selected GPD1 for deletion while leaving GPD2 to balance the need for osmoadaptation with the provision of NADH for SeEutE. Deleting GPD1, NDE1, and NDE2 genes decreases oxidative phosphorylation and respiration, generating hypoxia-tolerant cells. Hypoxia is known for spatial reorganization and accelerating fermentation rate under certain conditions.32,33
Additionally, during co-utilization with glucose, acetic acid biosynthesis can form a futile cycle, despite its essential role in the pyruvate dehydrogenase (PDH) bypass pathway for supplying acetyl-CoA (Fig. 1). To disrupt this futile cycle, ALD6 was selected for deletion in this study. Previous studies have shown that ALD6Δ can be effectively combined with the xylose-fermenting yeast strain SR8 to enhance acetic acid co-utilization.12,13 Also, deletion of ScALD6 improved anaerobic co-fermentation of acetic acid with 1 M glucose in strain IMX901, which had its native NADH-dependent ScGPD1 replaced with the archaeal NADPH-preferring GpsA, and ScGPD2 substituted by EcEutE.11 Therefore, combining NDE1Δ, NDE2Δ with GPD1Δ could potentially provide additional NADH for the heterologous expression of SeEutE. Based on this rationale, GPD1, ALD6, NDE1, and NDE2 were selected for sequential deletion in this study (Fig. S1†). The sequential deletion of GPD1, ALD6, NDE1, and NDE2 was implemented not only to redirect NADH toward acetate reduction but also to establish a controlled redox and respiratory rewiring under hypoxic-like conditions. Specifically, the deletion of NDE1 and NDE2, which encode the primary mitochondrial external NADH dehydrogenases functional under normoxic or hyperoxic environments, restricts cytosolic NADH oxidation via the electron transport chain, thereby simulating an O2-independent, hypoxia-mimicking state. This genetic intervention is expected to suppress oxidative phosphorylation and ATP-coupled respiration, thereby promoting fermentative metabolism to compensate for the energetic deficit, particularly in the presence of acetic acid, while simultaneously generating a strong redox driving force for acetate reduction. Notably, hypoxia-induced metabolic adaptation in S. cerevisiae has been shown to involve the spatial reorganization of glycolytic enzymes, such as enolase, into cytoplasmic foci—a phenomenon that enhances glucose conversion to pyruvate and oxaloacetate, ultimately boosting fermentative carbon flux under conditions of restricted respiration.32 Furthermore, the deletion of GPD1 reduces glycerol biosynthesis and impacts osmoadaptation by limiting glycerol-3-phosphate availability, potentially increasing membrane stress under acetate exposure. Deletion of ALD6, a key enzyme in the acetate-producing PDH bypass, prevents futile cycling of acetaldehyde to acetate, thereby conserving reducing equivalents and reinforcing the redox shift. Together, these deletions not only redirect metabolic flux but also alleviate oxidative and membrane-associated stress, enhancing robustness under hypoxic fermentation.
3.2 Expression of SeEutE in the parent strain and its effects on fermentation
The codon-optimized SeEutE was successfully expressed in S. cerevisiae for the first time, achieving significant specific activity of 0.063 ± 0.01 U in the CFE for strain E1 (P < 0.05), while the control strain C1 showed no detectable activity (Table 2). Previously, SeEutE expressed in E. coli demonstrated reversable activities, with acetaldehyde dehydrogenase activity at 68.1 ± 2 U and acetyl-CoA reductase activity at 49.2 ± 3 U for the purified protein.34| Strain name | Specific activityb |
|---|---|
| a CFE were obtained from cells at 15 h of fermentation in YNBD100-Ac4 medium, pH 5. Error values represent standard deviation from the mean (n = 4). b Indicate μmole/min mg CFE. c Not detected. Statistical analysis indicated that means sharing the same letter (A, B, C, D) are not significantly different from each other (P < 0.05). | |
| C1 (native) | NDcE |
| E1 (EutE) | 0.063 ± 0.01 D |
| C2 (GPD1Δ) | NDcE |
| E2 (GPD1Δ + EutE) | 0.146 ± 0.02 B |
| C3 (GPD1Δ + ALD6Δ) | NDcE |
| E3 (GPD1Δ + ALD6Δ + EutE) | 0.074 ± 0.01 C |
| C4 (GPD1Δ + ALD6Δ + NDE1Δ) | NDcE |
| E4 (GPD1Δ + ALD6Δ + NDE1Δ + EutE) | 0.072 ± 0.01 C&D |
| C5 (GPD1Δ + ALD6Δ + NDE1Δ + NDE2Δ) | NDcE |
| E5 (GPD1Δ + ALD6Δ + NDE1Δ + NDE2Δ + EutE) | 0.158 ± 0.02 A |
Incorporation of the SeEutE gene into the parent strain (E1) led to an 11% increase in the glucose consumption rate (1.99 ± 0.05 g (L−1 h−1) vs. 1.8 ± 0.04 g (L−1 h−1) in Fig. 2A) and a 4% increase in ethanol yield ratio (0.47 ± 0.01 g g−1vs. 0.45 ± 0.01 ge/gs in Fig. 2B) compared to the control strain C1. Meanwhile, acetic acid and glycerol production decreased by 6% (AcAP of 0.67 ± 0.02 g L−1vs. 0.71 ± 0.01 g L−1 in Table 3) and 19% (GP of 0.8 ± 0.02 g L−1vs. 0.99 ± 0.02 g L−1 in Table 3), respectively. Cell growth increased slightly by 7% (CG of 9 ± 0.72 OD vs. 8.4 ± 0.40 OD) (Table 3, and Fig. 2 and 3). Duncan's multiple range test at the P < 0.05 level revealed a significant difference between E1 and C1 only in glycerol production, with no significant change observed in other parameters (Table S5B†).
| Strains | Fermentation parameters | |||||||
|---|---|---|---|---|---|---|---|---|
| GCa (g L−1) | AcACb (g L−1) | AcAPc (g L−1) | EPd (g L−1) | EP/Te (%) | GPf (g L−1) | TPRog (gp/gs) | CGh (OD) | |
| a Glucose consumption. b Acetic acid consumption. c Acetic acid production. d Ethanol production. e Ethanol product/theoretical. f Glycerol production. g Total products ratio (gproducts/gsubstrates). h Cell growth. Values represent the averages ± SD, n = 3. i CG was measured after de-flocculation using a buffer containing 50 mM sodium acetate and 0.1 M EDTA. | ||||||||
| C1 (native) | 43.5 ± 1.99 | 0.0 ± 0.00 | 0.71 ± 0.01 | 19.6 ± 0.18 | 88.0 ± 0.1 | 0.99 ± 0.01 | 0.49 ± 0.01 | 8.4 ± 0.40 |
| E1 (EutE) | 48.3 ± 0.86 | 0.0 ± 0.00 | 0.67 ± 0.10 | 22.7 ± 0.3 | 92.2 ± 0.2 | 0.80 ± 0.02 | 0.50 ± 0.01 | 9.0 ± 0.72 |
| C2 (GPD1Δ) | 35.99 ± 0.77 | 0.0 ± 0.00 | 0.67 ± 0.02 | 15.6 ± 0.22 | 85.0 ± 0.2 | 0.19 ± 0.01 | 0.46 ± 0.10 | 8.3 ± 0.23 |
| E2 (GPD1Δ + EutE) | 46.7 ± 0.19 | 0.0 ± 0.00 | 0.42 ± 0.12 | 21.9 ± 0.7 | 91.9 ± 0.1 | 0.19 ± 0.01 | 0.48 ± 0.01 | 8.7 ± 0.94 |
| C3 (GPD1Δ + ALD6Δ) | 29.8 ± 1.10 | 0.0 ± 0.00 | 0.02 ± 0.04 | 12.7 ± 0.6 | 83.0 ± 0.2 | 0.13 ± 0.01 | 0.43 ± 0.19 | 7.9 ± 0.23 |
| E3 (GPD1Δ + ALD6Δ + EutE) | 27.5 ± 0.13 | 0.26 ± 0.02 | 0.00 ± 0.00 | 13.1 ± 0.02 | 92.9 ± 0.2 | 0.16 ± 0.01 | 0.48 ± 0.01 | 9.0 ± 0.72 |
| C4 (GPD1Δ + ALD6Δ + NDE1Δ) | 31.0 ± 0.34 | 0.00 ± 0.00 | 0.33 ± 0.03 | 13.6 ± 0.47 | 86.0 ± 0.2 | 0.26 ± 0.01 | 0.46 ± 0.01 | 8.1 ± 0.23 |
| E4 (GPD1Δ + ALD6Δ + NDE1Δ + EutE) | 98.0 ± 0.25 | 1.04 ± 0.10 | 0.00 ± 0.0 | 50.0 ± 0.5 | 98.5 ± 0.3 | 0.50 ± 0.01 | 0.50 ± 0.01 | 10.2i ± 0.35 |
| C5 (GPD1Δ + ALD6Δ + NDE1Δ + NDE2Δ) | 45.6 ± 0.70 | 0.0 ± 0.00 | 0.03 ± 0.01 | 21.2 ± 0.07 | 91.4 ± 0.18 | 0.46 ± 0.02 | 0.48 ± 0.01 | 8.6i ± 0.20 |
| E5 (GPD1Δ + ALD6Δ + NDE1Δ + NDE2Δ + EutE) | 98.7 ± 0.96 | 0.83 ± 0.07 | 0.00 ± 0.0 | 49.8 ± 0.34 | 97.7 ± 0.3 | 0.54 ± 0.04 | 0.51 ± 0.0 | 10.7i ± 0.23 |
Although the integration of SeEutE resulted in the improvement of ethanol production, competition with native NADH-dependent pathways with enzymes such as GPD and NDE limited the extent of the enhancements, as expected. In our recent study, the heterologous expression of other NADH oxidases, such as water-forming LlNoxE from Lactococcus lactis, in the same base strain B1 under hypoxic conditions led to an increase in acetic acid biosynthesis.4 This outcome underscores the advantage of using SeEutE over LlNoxE for reducing acetic acid production.
3.3 Integration of SeEutE with GPD1Δ in strain E2
To assess the effect of GPD1 deletion on the activity of SeEutE, we engineered strain E2. This strain exhibited a significantly larger specific activity than E1 in the CFE (P < 0.05); activity of E2 (0.146 ± 0.02 U) is 2.3 times higher than that of strain E1 (0.063 ± 0.01 U) (Table 2). The differences in specific activities of SeEutE between strains remain unclear. This enhancement in activity led to a further decrease in acetic acid concentration in E2 (AcAP of 0.42 ± 0.12 g L−1) by 37% compared to E1 (AcAP of 0.67 g L−1 ± 0.02 g L−1) (Table 3).Although the decrease in AcAP was not significant at P < 0.05 level as well as the growth (Table S5B†), the ethanol production rate in E2 is significantly higher than C2 by 40% (0.91 ± 0.03 g (L−1 h−1vs. 0.65 ± 0.01 g (L−1 h−1, Fig. 2A). Ethanol yield ratio in E2 was enhanced by 9% compared to C2 (0.47 ± 0.01 vs. 0.43 ± 0.01 ge/gs, Fig. 2B), although no further improvement was seen over E1 (Fig. 2).
3.4 Abolishing the acetic acid futile cycle in strain E3 and beyond
Acetic acid biosynthesis serves as an intermediate step in the formation of acetyl-CoA via the PDH bypass pathway, which involves pyruvate decarboxylase PDC isoforms 1, 5, and 6, ALD6, ACS1, and ACS2.35–37 The S. cerevisiae genome contains five alternative isoforms for ALD associated with acetic acid production. Acetic acid production in the C3 strain (GPD1Δ ALD6Δ) was very low (AcAP of 0.02 ± 0.01 g L−1, Table 3), confirming the predominance of ALD6 in acetic acid production.Additionally, strain E3 (GPD1Δ ALD6Δ [SeEutE]) further consumed acetic acid by 0.26 g L−1 (AcAC, Table 3). These significant changes in the AcAP (P < 0.05, Table S5B†) clearly demonstrated that the deletion of ALD6Δ, in combination with incorporation of SeEutE can eliminate the futile cycle of acetic acid production during the co-utilization of acetic acid and glucose (Table 3 and Fig. S2 and S3†).
In strain E3, bioethanol production significantly increased to 92.9% of the theoretical yield, compared to 88% in strain C1 (EP/T, Table 3). However, glucose consumption in E3 was significantly reduced at P < 0.05 to 27.5 ± 0.13 g L−1 compared to C1 (43.5 ± 1.99) at 24 h (GC, Table 3, Fig. S2 and S3, S5B†). Thus, while the elimination of the acetic acid futile cycle is promising, it is not sufficient on its own for effective co-utilization.
3.5 Triple and quadruple deletion of GPD1, ALD6, and NDE1 (and NDE2) with SeEutE expression: achieving ethanol-induced flocculation and fermentation robustness
Strain E4 consumed 98.0 ± 0.25 g L−1 of glucose within 24 h, compared to 31.0 ± 0.34 g L−1 consumed by strain C4 and 43.5 ± 1.99 g L−1 by strain C1(GC, Table 3), indicating the increased vigour of strain E4. The glucose consumption rates for C1, C4 and E4 were 1.8 ± 0.04 g (L−1 h−1), 1.3 ± 0.01 g (L−1 h−1), and 4.1 ± 0.01 g (L−1 h−1) (Fig. 2A), respectively, with corresponding bioethanol production rates of 0.82 ± 0.01 g (L−1 h−1), 0.57 ± 0.01 g (L−1 h−1), and 2.08 ± 0.02 g (L−1 h−1) (Fig. 2A). Furthermore, the ethanol product/theoretical of E4 reached 98.5 ± 0.3%, compared to 86.0 ± 0.2% for C4 and 88.0 ± 0.1% for C1 (EP/T, Table 3). Glycerol production was 0.99 ± 0.01 g L−1 in C1, 0.26 ± 0.01 g L−1 in C4, and 0.50 ± 0.01 g L−1 in E4 (GP, Table 3). Acetic acid consumption by strain E4 increased to 1.04 ± 0.10 g L−1 compared to 0.26 ± 0.02 g L−1 by strain E3 (AcAC, Table 3 and Fig. S2 and S3†).
Ethanol production by C5 at 48 h reached 46.3 ± 0.2 g L−1, compared to 36.3 ± 0.2 g L−1 produced by strain C1 (Fig. S2D†). Strain E5 produced ethanol even more efficiently, reaching 49.8 ± 0.34 g L−1 in just 24 h (EP, Table 3). The ethanol production rate in E5 rose to 2.07 ± 0.01 g (L−1 h−1), compared to 0.88 ± 0.0 g (L−1 h−1) in C5 and 0.82 ± 0.01 g (L−1 h−1) in C1 (Fig. 2B). C5 did not consume any acetic acid, whereas E5 consumed 0.83 ± 0.07 g L−1 (AcAC, Table 3). Glycerol production in C5 and E5 increased to 0.46 ± 0.02 g L−1 and 0.54 ± 0.04 g L−1, respectively, compared to 0.26 ± 0.01 g L−1 in C4 and 0.50 ± 0.01 g L−1 in E4 (GP, Table 3). These enhancements in the fermentation characteristics of E5 compared to C1 and C5 were further validated through statistical analysis (P < 0.05; Table S5†). The increased acetic acid consumption and glycerol production in E5 compared to C5 suggest a balance between ScGPD2 and SeEutE for NADH oxidation. This balance enabled the generation of G-3P and glycerol, along with the co-utilization of glucose and acetic acid. This balance between ScGPD2 and SeEutE was demonstrated for the first time. In earlier studies, both ScGPD1 and ScGPD2 were deleted to fully direct NADH toward acetylating acetaldehyde dehydrogenase.7,9 However, this approach inhibited G-3P and glycerol biosynthesis, leading to deficiencies in cell wall viability, integrity, signaling, and regulation.26,27 These factors may explain the sluggish fermentation rate observed in previous studies that deleted both GPD1 and GPD2 under anaerobic conditions. Our alternative approach with hypoxic conditions, which retains GPD2, has produced both superior fermentation rates and efficiencies compared to previous reports.
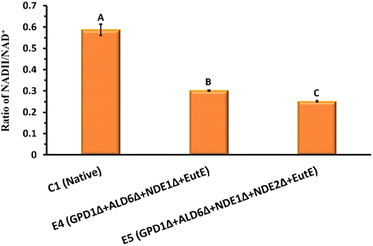
The intracellular NADH/NAD+ ratio is a critical indicator of redox balance during fermentation. As shown in Fig. 3, the control strain C1 exhibited a high NADH/NAD+ ratio 0.59 ± 0.03, indicating significant cytosolic NADH accumulation under hypoxic conditions. In contrast, both engineered strains E4 and E5 showed markedly reduced NADH/NAD+ ratios 0.3 ± 0.002 and 0.25 ± 0.003, respectively (Fig. 3). This significant decline (P < 0.05) demonstrates the effectiveness of the introduced modifications in rebalancing the redox state.
Specifically, the deletions of GPD1 and ALD6 reduce competing NADH-consuming pathways (glycerol and acetate formation), while deletion of NDE1 and NDE2 restricts mitochondrial NADH oxidation, creating a more reduced cytosolic environment. Introduction of SeEutE, provides an alternative NADH oxidation route directly linked to ethanol production. Together, these modifications redirect excess NADH toward ethanol synthesis, improving fermentation efficiency and reducing the need for byproduct formation. The enhanced redox homeostasis in E5, as reflected by the lowest NADH/NAD+ ratio, correlates well with its superior ethanol productivity (Fig. 2). Taken together, the engineered acetate co-utilization pathway in strain E5 combines (i) enhanced NADH availability via GPD1, NDE1, and NDE2 deletions, (ii) disruption of the acetic acid futile cycle through ALD6 deletion, and (iii) redox-balanced reduction of acetyl-CoA to ethanol via SeEutE. This configuration enables robust acetate assimilation under hypoxic conditions, overcoming the limitations of previous systems.
Specifically, the deletions of GPD1 and ALD6 reduce competing NADH-consuming pathways (glycerol and acetate formation), while deletion of NDE1 and NDE2 restricts mitochondrial NADH oxidation, creating a more reduced cytosolic environment. Introduction of SeEutE, provides an alternative NADH oxidation route directly linked to ethanol production. Together, these modifications redirect excess NADH toward ethanol synthesis, improving fermentation efficiency and reducing the need for byproduct formation. The enhanced redox homeostasis in E5, as reflected by the lowest NADH/NAD+ ratio, correlates well with its superior ethanol productivity (Fig. 2). Taken together, the engineered acetate co-utilization pathway in strain E5 combines (i) enhanced NADH availability via GPD1, NDE1, and NDE2 deletions, (ii) disruption of the acetic acid futile cycle through ALD6 deletion, and (iii) redox-balanced reduction of acetyl-CoA to ethanol via SeEutE. This configuration enables robust acetate assimilation under hypoxic conditions, overcoming the limitations of previous systems.
In S. cerevisiae, flocculin genes (FLO1, FLO5, FLO8, FLO9, FLO10, and FLO11) play key roles in enhancing cell wall hydrophobicity, flocculation, and cell adhesion. These traits facilitate yeast cell separation, which is advantageous in industrial processes.38–43 Moreover, flocculation provides protection under stress conditions by shielding cells from inhibitors.6,44 A transcriptional repressor and activator MOT3 has regulates cell wall hydrophobicity by modulating the expression of the hydrophobic cell wall protein YPG1. This regulation enhances resistance to inhibitory chemical compounds (ICCs), even in the absence of acetic acid co-utilization.6
With the quadruple deletions of GPD1, ALD6, NDE1, and NDE2 in C5, flocculation reached 65.0 ± 4.1% after 21 h of fermentation (Fig. 4). Glucose consumption in C5 was enhanced by 150% compared to C1 (Fig. 2A). In E5, the expression of SeEutE alongside these quadruple deletions further increased the flocculation to 75.0 ± 2.0% (Fig. 4), with larger flocs observed compared to those formed by E4 (Fig. S4†). E5 exhibited over 200% higher glucose consumption and ethanol production rates than C5, as well as slightly faster fermentation compared to E4 (Fig. S2 and S3†). We hypothesize that the more pronounced flocculation in E5 is associated with enhanced protection against multiple stresses, including ethanol, as previously reported.40,43,44
In E5, flocculation began 15 h into the fermentation, after ethanol production reached 30.16 ± 0.47 g L−1. It was reported that the exposure to 3% ethanol induces 70% flocculation in strains overexpressing FLO1 and 28% in strains overexpressing FLO5.40 Ethanol can trigger flocculation through controlled FLO1 expression regulated by the trehalose-6-phosphate synthase 1 (TPS1) promoter, which responds to ethanol accumulation during fermentation. Ethanol-induced flocculation has been shown to enhance biomass recovery.43 These observations led us to hypothesize that ethanol promoted flocculation. To test this, we supplemented the medium with 3% and 4% ethanol after 3 h of initial fermentation using strain E5. As a result, the flocculation increased by 80% and 210%, respectively, compared to the control without ethanol supplementation (Fig. 5). These results support our hypothesis and are consistent with previous reports demonstrating that 3% ethanol induces flocculation under certain conditions as reported elsewhere.40,43
Moreover, the integration and overexpression of biofilm-forming genes (FLO5, FLO8, and FLO10) in S. cerevisiae have been shown to improve biofilm formation. Engineered strains 1308-FLO5 and 1308-FLO10 demonstrated significant increases in adhesion and ethanol production compared to the wild type. These strains also reduced broth turbidity and enhanced membrane separation efficiency, increasing ethanol flux through the membrane by 36.3% during biofilm-immobilized fermentation.42
Interestingly, our engineering approach did not involve the overexpression of any FLO genes by design, suggesting that ethanol-induced flocculation in E5 may be driven by an indirect mechanism, warranting further investigation. In future studies, we will conduct molecular characterization of key regulatory genes associated with accelerated fermentation rates, as well as flocculation-related genes such as FLO1, FLO5, FLO8, FLO10, and YGP1. We will also examine their transcriptional responses to ethanol exposure to elucidate the molecular basis of this phenotype.
Acetic acid consumption was lower under the 70/100 Vc/Vf ratio conditions, but glycerol production increased to 1.3 ± 0.01 g L−1, approximately 2.4 times higher than under hyperoxic conditions (Fig. 6). This suggests further activation of GPD2 activity under near-anaerobic conditions (Vc/Vf 70/100), consistent with previous studies highlighting the role of GPD2 under anaerobic conditions.45 On the other hand, glycerol production slightly increased in E4 at a 70/100 Vc/Vf ratio, suggesting that NDE2 may play a role in sharing NADH oxidation with GPD2 (Fig. S3B and Table S7†). The increased glycerol production in E5 under stronger hypoxia may reflect a redox-balancing response in which GPD2 compensates for impaired mitochondrial respiration due to the deletion of NDE1 and NDE2. As SeEutE and GPD2 both consume NADH, their co-function under hypoxic stress likely contributes to redox homeostasis. This cooperative NADH oxidation enables E5 to maintain higher fermentation efficiency and cell viability, whereas complete GPD deletions in previous studies often led to fermentation defects. This indicates a distinct metabolic response between strains E4 and E5 under varying oxygen levels, with E5 showing a more robust adaptation to hypoxic conditions. These findings underscore the importance of preserving GPD2-mediated redox flexibility in engineered strains, as its activity appears to complement SeEutE under hypoxic stress to sustain efficient fermentation.
3.6 Cross-study comparison and application context
To further contextualize the potential of acetate co-utilization and address strain performance relative to the literature, we compared the sugar consumption rates, ethanol production rates, and yield efficiencies of our current strain (E5) with previously engineered S. cerevisiae strains (Table 4). A defining feature of our strategy is the induction of O2-independent hypoxia via triple deletion of GPD1, NDE1, and NDE2, along with ALD6 deletion. The sequential deletion of GPD1, ALD6, NDE1, and NDE2, combined with SeEutE expression, significantly accelerated glucose fermentation under hypoxic conditions in this study, resulting in a 3.42-fold increase in fermentation rate compared to the control strain C1 (Fig. S2 and S3†). Strain E5 reached 97.7% of the theoretical ethanol yield within 24 h when fermenting 100 g L−1 glucose supplemented with 4 g L−1 acetate, representing a significant improvement over the 88% yield observed in C1. Ethanol-induced flocculation was also triggered in E5 when ethanol concentration reached approximately 3%.| Carbons consumption rate g (L−1 h−1) | Ethanol production rate g (L−1 h−1) | Acetic acid co-utilized g L−1 | Fermentation time (h) | Efficiency of ethanol yield (%) a | Key genetic modifications | Media and conditions | Reference |
|---|---|---|---|---|---|---|---|
| a Efficiency was calculated based on the total consumption of sugars and acetate. The theoretical maximum yield is 0.51 g g−1 for glucose and xylose, and 0.77 g g−1 for acetate. b Experimental data were estimated approximately from figure visualizations. | |||||||
| 0.53b | 0.48b | 0.50b | 38b | 89.2b | GPD1Δ, GPD2Δ + EcMhpF | 20 g glucose + 2 g acetic acid; anaerobic | 7 |
| 0.99 | 0.46 | 5.30 | 120 | 87.8 | GPD1Δ, GPD2Δ, EhADH1, ACS2, ZWF1, 2× BaAdhE | 117 g glucose + 8 g acetic acid; anaerobic | 9 |
| 0.95b | 0.45b | 0.80b | 23b | 90.9b | GPD1Δ, GPD2Δ, ALD6Δ, GND2Δ, GND1Δ + MfGNDa, EcEutE | 20 g glucose + 3 g acetic acid; anaerobic | 10 |
| 3.03 | 1.45 | 4.86b | 61b | 92.3b | GPD1Δ, GPD2Δ, ALD6Δ + AfGpsA, EcEutE | 180 g glucose + 3 g acetic acid (maintained >1.5 g L−1); anaerobic | 11 |
| 1.22b | 0.5* | 1.70b | 88 | 82.3b | ALD6Δ, XR, XDH, XK (2 copies), adapted strain + EcEutE | 80 g xylose + 20 g glucose + 2 g acetic acid; anaerobic | 12 |
| 1.42 | 0.60 | 7.1 | 72 | 80.4 | ALD6Δ, XR, XDH, XK (2 copies), adapted strain + 3× EcAdhE + SeACS | 80 g xylose + 20 g glucose + 8 g acetic acid; anaerobic | 13 |
| 4.15 | 2.07 | 0.83 | 24 | 97.7 | ΔGPD1, ΔALD6, ΔNDE1, ΔNDE2 + SeEutE | 100 g glucose + 4 g acetic acid; hypoxic | E5 (this study) |
We respectfully highlight the study by Papapetridis et al.,11 in which a GPD1Δ GPD2Δ ALD6Δ strain co-expressing AfGpsA and EcEutE co-utilized 4.86 g L−1 acetate with 180 g L−1 glucose under anaerobic conditions, achieving a 92.3% ethanol yield over 61 h. In contrast, our hypoxia-tolerant strain E5—retaining GPD2 to preserve osmoadaptation—achieved a higher yield in less than half the time (Table 4). These findings suggest that future integration of AfGpsA into E5 may further enhance acetate assimilation efficiency. Acetate co-utilization has also been validated in glucose–xylose co-fermentation systems. For example, integration of three copies of EcAdhE and SeACS enabled consumption of 7.1 g L−1 acetate alongside 80 g L−1 xylose, significantly improving ethanol yield.13 When NADH limitation was alleviated by expressing EhADH1, up to 5.3 g L−1 acetate was co-utilized with 114 g L−1 glucose.9 In a system targeting triacetic acid lactone (TAL)—a product whose biosynthesis does not require NADH—acetate-to-xylose co-utilization reached a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratio.14 Notably, these acetate concentrations (1–15 g L−1) are consistent with levels typically found in lignocellulosic hydrolysates.12
4 ratio.14 Notably, these acetate concentrations (1–15 g L−1) are consistent with levels typically found in lignocellulosic hydrolysates.12
Collectively, these findings confirm that acetate co-utilization enhances ethanol production across diverse substrate types (glucose, xylose, and mixtures), and the performance of E5 aligns well with industrially relevant acetate concentrations (Table 4). Its hypoxia and acetate tolerant phenotype make E5 a promising and robust chassis for scalable bioethanol production from lignocellulosic biomass.
3.7 Future direction and metabolic engineering
The metabolic engineering strategy applied in E5 (GPD1Δ ALD6Δ NDE1Δ NDE2Δ [SeEutE]) represents a novel approach for developing a S. cerevisiae strain that prefers hypoxic conditions associated with ethanol-induced flocculation and achieves unprecedented rates (342% faster than the native strain) of co-fermenting acetic acid with glucose under the tested conditions. These results were achieved with the integration of a single copy of SeEutE. An additional strategy to enhance performance involves the “tugging” approach, successfully applied in previous studies. Henningsen et al. employed two copies of BaAdhE to co-utilize acetic acid with glucose, followed by the introduction of NADPH-dependent alcohol dehydrogenase to supply additional NADH to EhADH.9 Therefore, further improvements could possibly be realized by introducing multiple copies of SeEutE or combining it with more efficient acetyl-CoA synthase enzymes.Acetic acid metabolism is regulated by carbon catabolite repression (CCR) through the ACS1 and ACS2 genes in response to glucose.14,46 Zhang et al. optimized acetate reduction in the presence of glucose and xylose by expressing three copies of both mutated SeACS and codon-optimized EcAdhE, which enhanced xylose fermentation and reduced by-products such as xylitol and glycerol while enhancing acetate co-utilization. A similar approach could be applied to E5 to further enhance co-fermentation.13
4. Conclusions
A metabolically engineered S. cerevisiae strain, E5, was developed for glucose fermentation and acetic acid co-utilization under hypoxic conditions. The quadruple deletion of GPD1, ALD6, NDE1, and NDE2, combined with SeEutE expression, enhanced flocculation triggered by ethanol production at ∼3%. E5 fermented 10% glucose and 0.4% acetic acid 342% faster than C1, achieving 98% of the theoretical ethanol yield—11% higher than C1. Additionally, it utilized 25% of the supplemented acetic acid and exhibited a 9% higher fermentation rate under hypoxic conditions. This engineering strategy demonstrates significant potential for bioethanol and bio-based chemical production from glucose and acetic acid.Data availability
All necessary data required to assess our findings are available in this manuscript or its ESI.† Further details related to this article may be requested from the authors.Author contributions
Sadat M. R. Khattab: conceptualization, methodology, validation, formal analysis, investigation, resources, data curation, writing – original draft, writing – review & editing, supervision, project administration, funding acquisition. Mohamed O. Mohamed: methodology, validation, formal analysis, investigation, data curation, writing – review & editing. Takashi Nagata: validation, writing – review & editing, supervision. Takashi Watanabe: conceptualization, writing – review & editing, project administration, funding acquisition. Masato Katahira: conceptualization, data curation, writing – review & editing, supervision, project administration, funding acquisition.Conflicts of interest
The authors declare that they have no financial interests or personal relationships that could have influenced the work presented in this paper.Acknowledgements
This work was supported by a Mission 5–2 Research Grant from the Research Institute for Sustainable Humanosphere, Kyoto University [Project No. 5-2-3], the Joint Usage/Research Program on Zero-Emission Energy Research, Kyoto University [ZE2024A-01], e-ASIA Joint Research Program [e-ASIA JRP, JPMJSC18E1], the Research Unit for Realization of Sustainable Society (RURSS), Research Coordination Alliance, Kyoto University, and collaborative research with DAICEL Co. Ltd. We would like to thank Prof. Alhosein Hamada, Faculty of Agriculture and Food Science, King Faisal University, KSA, for his assistance in verifying the statistical analysis.References
- R. Cavicchioli, W. J. Ripple and K. N. Timmis, et al., Scientists' warning to humanity: microorganisms and climate change, Nat. Rev. Microbiol., 2019, 17, 569–586, DOI:10.1038/s41579-019-0222-5.
- S. M. R. Khattab and T. Watanabe, Bioethanol from sugarcane bagasse: status and perspectives, in Bioethanol Production from Food Crops: Sustainable Sources, Interventions, and Challenges, ed. C. R. Ramesh and S. Ramachandran, Elsevier, London, 2019, pp. 187–212: DOI:10.1016/B978-0-12-813766-6.00010-2.
- N. Saha, S. Swagatika and R. S. Tomar, Investigation of the acetic acid stress response in Saccharomyces cerevisiae with mutated H3 residues, Microb. Cell, 2023, 10(10), 217–232, DOI:10.15698/mic2023.10.806.
- S. M. R. Khattab and T. Watanabe, Replacing glycerol-3-phosphate dehydrogenase with NADH Oxidase: Effects on glucose fermentation and product formation in Saccharomyces cerevisiae, Arch. Microbiol., 2025, 207, 3 CrossRef CAS PubMed , https://doi-org.kyoto-u.idm.oclc.org/10.1007/s00203-024-04187-x.
- S. R. Chaves, A. Rego, V. M. Martins and M. J. Sousa, Regulation of Cell Death Induced by Acetic Acid in Yeasts, Front. Cell Dev. Biol., 2021, 9, 642375, DOI:10.3389/fcell.2021.642375.
- P. Kahar, A. Itomi, H. Tsuboi, M. Ishizaki, M. Yasuda, C. Kihira, H. Otsuka, N. B. Azmi, H. Matsumoto, C. Ogino and A. Kondo, The flocculant Saccharomyces cerevisiae strain gains robustness via alteration of the cell wall hydrophobicity, Metab. Eng., 2022, 72, 82–96, DOI:10.1016/j.ymben.2022.03.001.
- V. Guadalupe-Medina, M. J. Almering, A. J. van Maris and J. T. Pronk, Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor, Appl. Environ. Microbiol., 2010, 76(1), 190–195, DOI:10.1128/AEM.01772-09.
- V. Guadalupe-Medina, B. Metz, B. Oud, C. M. van Der Graaf, R. Mans, J. T. Pronk and A. J. van Maris, Evolutionary engineering of a glycerol-3-phosphate dehydrogenase-negative, acetate-reducing Saccharomyces cerevisiae strain enables anaerobic growth at high glucose concentrations, Microb. Biotechnol., 2014, 7(1), 44–53, DOI:10.1111/1751-7915.12080.
- B. M. Henningsen, S. Hon, S. F. Covalla, C. Sonu, D. A. Argyros, T. F. Barrett, E. Wiswall, A. C. Froehlich and R. M. Zelle, Increasing anaerobic acetate consumption and ethanol yields in Saccharomyces cerevisiae with NADPH-specific alcohol dehydrogenase, Appl. Environ. Microbiol., 2015, 81(23), 8108–8117, DOI:10.1128/AEM.01689-15.
- I. Papapetridis, M. van Dijk, A. P. Dobbe, B. Metz, J. T. Pronk and A. J. A. van Maris, Improving ethanol yield in acetate-reducing Saccharomyces cerevisiae by cofactor engineering of 6-phosphogluconate dehydrogenase and deletion of ALD6, Microb. Cell Fact., 2016, 15, 67, DOI:10.1186/s12934-016-0465-z.
- I. Papapetridis, M. van Dijk, A. J. A. van Maris and J. T. Pronk, Metabolic engineering strategies for optimizing acetate reduction, ethanol yield, and osmotolerance in Saccharomyces cerevisiae, Biotechnol. Biofuels, 2017, 10, 107, DOI:10.1186/s13068-017-0791-3.
- N. Wei, J. Quarterman and S. Kim, et al., Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast, Nat. Commun., 2013, 4, 2580, DOI:10.1038/ncomms3580.
- G. C. Zhang, I. I. Kong, N. Wei, D. Peng, T. L. Turner, B. H. Sung, J. H. Sohn and Y. S. Jin, Optimization of an acetate reduction pathway for producing cellulosic ethanol by engineered yeast, Biotechnol. Bioeng., 2016, 113(12), 2587–2596, DOI:10.1002/bit.26021.
- L. Sun, J. W. Lee, S. Yook, S. Lane, Z. Sun, S. R. Kim and Y. S. Jin, Complete and efficient conversion of plant cell wall hemicellulose into high-value bioproducts by engineered yeast, Nat. Commun., 2021, 12(1), 4975, DOI:10.1038/s41467-021-25241-y.
- S. M. R. Khattab and T. Kodaki, Efficient bioethanol production by overexpression of endogenous Saccharomyces cerevisiae xylulokinase and NADPH-dependent aldose reductase with mutated strictly NADP+-dependent Pichia stipitis xylitol dehydrogenase, Process Biochem., 2014, 49, 1838–1842, DOI:10.1016/j.procbio.2014.07.017.
- S. M. R. Khattab and T. Watanabe, Efficient conversion of glycerol to ethanol by an engineered Saccharomyces cerevisiae strain, Appl. Environ. Microbiol., 2021, 87(23), e0026821, DOI:10.1128/AEM.00268-21.
- S. M. R. Khattab, M. A. Abdel-Rahman, M. Katahira and T. Watanabe, Engineering Saccharomyces cerevisiae and controlling conditions for 2,3-butanediol production from glycerol, Sustain. Energy Fuels, 2024, 8, 4297–4310, 10.1039/D4SE00912F.
- W. Ryan, S. Poddar and J. H. Cate, CRISPR-Cas9 genome engineering in Saccharomyces cerevisiae cells, Cold Spring Harb. Protoc., 2016 DOI:10.1101/pdb.prot086827.
- Y. Ito, T. Kitagawa and M. Yamanishi, et al., Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae, Sci. Rep., 2016, 6, 36997, DOI:10.1038/srep36997.
- K. Hosaka, J. Nikawa, T. Kodaki and S. Yamashita, A dominant mutation that alters the regulation of INO1 expression in Saccharomyces cerevisiae, J. Biochem., 1992, 111(3), 352–358, DOI:10.1093/oxfordjournals.jbchem.a123761.
- S. M. R. Khattab, M. Saimura and T. Kodaki, Boost in bioethanol production using recombinant Saccharomyces cerevisiae with mutated strictly NADPH-dependent xylose reductase and NADP+-dependent xylitol dehydrogenase, J. Biotechnol., 2013, 165(3), 153–156, DOI:10.1016/j.jbiotec.2013.03.009.
- J. Extance, M. J. Danson and S. J. Crennell, Structure of an acetylating aldehyde dehydrogenase from the thermophilic ethanologen Geobacillus thermoglucosidasius, Protein Sci., 2016, 25(11), 2045–2053, DOI:10.1002/pro.3027.
- O. Kobayashi, N. Hayashi, R. Kuroki and H. Sone, Region of Flo1 Proteins responsible for sugar recognition, J. Bacteriol., 1998, 180(24), 6503–6510 CrossRef CAS PubMed.
- SAS Institute. The SAS System for Windows, Release 9.2. SAS Institute, Cary, NC, USA, 2008 Search PubMed.
- R. G. Steel and J. Torrie, Principles and Procedures of Statistics: A Biological Approach. 2nd edn, McGraw-Hill, New York, USA, 1980 Search PubMed.
- J. Lee and N. D. Ridgway, Substrate channeling in the glycerol-3-phosphate pathway regulates the synthesis, storage, and secretion of glycerolipids, Biochim. Biophys. Acta Mol. Cell Biol. Lipids, 2020, 1865(1), 158438, DOI:10.1016/j.bbalip.2019.03.010.
- E. de Nadal and F. Posas, The HOG pathway and the regulation of osmoadaptive responses in yeast, FEMS Yeast Res., 2022, 22(1), foac013, DOI:10.1093/femsyr/foac013.
- R. Ansell, K. Granath, S. Hohmann, J. M. Thevelein and L. Adler, The two isoenzymes for yeast NAD+-dependent glycerol-3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation, EMBO J., 1997, 16, 2179–2187, DOI:10.1093/emboj/16.9.2179.
- J. J. ter Linde, H. Liang, R. W. Davis, H. Y. Steensma, J. P. van Dijken and J. T. Pronk, Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae, J. Bacteriol., 1999, 181(24), 7409–7413, DOI:10.1128/JB.181.24.7409-7413.1999.
- M. A. Luttik, K. M. Overkamp, P. Kötter, S. de Vries, J. P. van Dijken and J. T. Pronk, The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH, J. Biol. Chem., 1998, 273(38), 24529–24534, DOI:10.1074/jbc.273.38.24529.
- I. L. Pahlman, C. Larsson, N. Averet, O. Bunoust, S. Boubekeur, L. Gustafsson and M. Rigoulet, Kinetic regulation of the mitochondrial glycerol-3-phosphate dehydrogenase by the external NADH dehydrogenase in Saccharomyces cerevisiae, J. Biol. Chem., 2002, 277(31), 27991–27995, DOI:10.1074/jbc.M204079200.
- N. Miura, M. Shinohara, Y. Tatsukami, Y. Sato, H. Morisaka, K. Kuroda and M. Ueda, Spatial reorganization of Saccharomyces cerevisiae enolase to alter carbon metabolism under hypoxia, Eukaryot. Cell, 2013, 12(8), 1106–1119, DOI:10.1128/EC.00093-13.
- Z. Fu, T. D. Verderame, J. M. Leighton, B. P. Sampey, E. R. Appelbaum, P. S. Patel and J. C. Aon, Exometabolome analysis reveals hypoxia at the up-scaling of a Saccharomyces cerevisiae high-cell density fed-batch biopharmaceutical process, Microb. Cell Fact., 2014, 13(1), 32, DOI:10.1186/1475-2859-13-32.
- H. Zhu, R. Gonzalez and T. A. Bobik, Coproduction of acetaldehyde and hydrogen during glucose fermentation by Escherichia coli, Appl. Environ. Microbiol., 2011, 77(18), 6441–6450, DOI:10.1128/AEM.05358-11.
- S. Boubekeur, O. Bunoust, N. Camougrand, M. Castroviejo, M. Rigoulet and B. Guérin, A mitochondrial pyruvate dehydrogenase bypass in the yeast Saccharomyces cerevisiae, J. Biol. Chem., 1999, 274(30), 21044–21048, DOI:10.1074/jbc.274.30.21044.
- F. Saint-Prix, L. Bönquist and S. Dequin, Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: the NADP+-dependent Ald6p and Ald5p isoforms play a major role in acetate formation, Microbiology, 2004, 150(Pt 7), 2209–2220, DOI:10.1099/mic.0.26999-0.
- C. Lei, X. Guo, M. Zhang, X. Zhou, N. Ding, J. Ren, M. Liu, C. Jia, Y. Wang, J. Zhao, Z. Dong and D. Lu, Regulating the metabolic flux of the pyruvate dehydrogenase bypass to enhance lipid production in Saccharomyces cerevisiae, Commun. Biol., 2024, 7(1), 1399, DOI:10.1038/s42003-024-07103-7.
- K. Shida-Fujii, S. Goto, H. Sugiyama, Y. Takagi, T. Saiki and M. Takagi, Breeding of flocculent industrial alcohol yeast strains by self-cloning of the flocculation gene FLO1 and repeated-batch fermentation by transformants, J. Gen. Appl. Microbiol., 1998, 44(5), 347–353, DOI:10.2323/jgam.44.347.
- A. F. Cunha, S. K. Missawa, L. H. Gomes, S. F. Reis and G. A. Pereira, Control by sugar of Saccharomyces cerevisiae flocculation for industrial ethanol production, FEMS Yeast Res., 2006, 6(2), 280–287, DOI:10.1111/j.1567-1364.2006.00038.x.
- P. Govender, J. L. Domingo, M. C. Bester, I. S. Pretorius and F. F. Bauer, Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae, Appl. Environ. Microbiol., 2008, 74(19), 6041–6052, DOI:10.1128/AEM.00394-08.
- S. E. Van Mulders, E. Christianen, S. M. Saerens, L. Daenen, P. J. Verbelen, R. Willaert, K. J. Verstrepen and F. R. Delvaux, Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae, FEMS Yeast Res., 2009, 9(2), 178–190, DOI:10.1111/j.1567-1364.2008.00462.x.
- Z. Wang, W. Xu and Y. Gao, et al., Engineering Saccharomyces cerevisiae for improved biofilm formation and ethanol production in continuous fermentation, Biotechnol. Biofuels, 2023, 16, 119, DOI:10.1186/s13068-023-02356-6.
- Q. Li, X. Q. Zhao, A. K. Chang, Q. M. Zhang and F. W. Bai, Ethanol-induced yeast flocculation directed by the promoter of TPS1 encoding trehalose-6-phosphate synthase 1 for efficient ethanol production, Metab. Eng., 2012, 14(1), 1–8, DOI:10.1016/j.ymben.2011.12.003.
- S. Smukalla, M. Caldara, N. Pochet, A. Beauvais, S. Guadagnini, C. Yan, M. D. Vinces, A. Jansen, M. C. Prevost, J. P. Latgé, G. R. Fink, K. R. Foster and K. J. Verstrepen, FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast, Cell, 2008, 135(4), 726–737, DOI:10.1016/j.cell.2008.09.037.
- T. L. Nissen, C. W. Hamann, M. C. Kielland-Brandt and J. N. pand J. Villadsen, Anaerobic and aerobic batch cultivations of Saccharomyces cerevisiae mutants impaired in glycerol synthesis, Yeast, 2000, 16(5), 463–474, DOI:10.1002/(sici)1097-0061(20000330)16:5<463::aid-yea535>3.3.co;2-v.
- S. Kratzer and H. J. Schuller, Carbon source-dependent regulation of the acetyl-coenzyme A synthetase-encoding gene ACS1 from Saccharomyces cerevisiae, Gene, 1995, 161(1), 75–79, DOI:10.1016/0378-1119(95)00289-i.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5se00258c |
| ‡ Joint first authors. |
| This journal is © The Royal Society of Chemistry 2025 |