Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Understanding how the structures of surfactants in hybrid nanoparticles affect the compaction of ct-DNA for cellular uptake: presenting a highly efficient surfactant†
Shalini
Dyagala
a,
Milan
Paul
b,
Vinod K
Aswal
 c,
Swati
Biswas
c,
Swati
Biswas
 *b and
Subit Kumar
Saha
*b and
Subit Kumar
Saha
 *a
*a
aDepartment of Chemistry, Birla Institute of Technology & Science (BITS) Pilani, Hyderabad Campus, Hyderabad, Telangana 500078, India. E-mail: sksaha@hyderabad.bits-pilani.ac.in; sksaha@pilani.bits-pilani.ac.in; subitksaha@gmail.com; Tel: +91-40-66303643
bDepartment of Pharmacy, Birla Institute of Technology & Science (BITS) Pilani, Hyderabad Campus, Hyderabad, Telangana 500078, India. E-mail: swati.biswas@hyderabad.bits-pilani.ac.in; Tel: +91-40-66303630
cSolid State Physics Division, Bhabha Atomic Research Centre (BARC), Trombay, Mumbai, Maharashtra 400085, India
First published on 3rd December 2024
Abstract
Compaction of calf thymus DNA (ct-DNA) by two single-head-double-tailed surfactants with variable tail lengths i.e., Dihexadecyldimethylammonium bromide (DDAB16) and Dioctadecyldimethylammonium bromide (DDAB18), and one triple-head-double-tailed surfactant N1-dodecyl-N2-(2-(dodecyldimethylammonio)ethyl)-N1,N1,N2,N2-tetramethylethane-1,2-diaminium (MQAS12) has been studied. DDAB18 is found to be the most efficient, while MQAS12 is the least efficient for cellular uptake. Hybrid materials of surfactants and silica nanoparticles have better compaction efficiency due to the cooperative binding. Silica nanoparticles (∼100 nm)–DDAB18 hybrid materials can compact ct-DNA at a much lower concentration than a conventional surfactant, addressing the cytotoxicity issues. Hybrid materials formed with smaller silica nanoparticles (∼40 nm) have also been studied. The results obtained have been used to understand whether Coulombic and/or hydrophobic interactions are responsible for DNA compaction. The hydrophobicity per unit surface area (P) of hybrid nanoparticles has a significant role in DNA compaction. The P largely depends on the surfactants’ structures and nanoparticles’ sizes. Single-head-double-tailed surfactants with a comparatively smaller headgroup exhibit a large amount of adsorption on the nanoparticles’ surfaces, producing a large P. DDAB18 appears to be a DNA intercalative binder. Fluorescence anisotropy decay data of 4,6-diamidino-2-phenylindole (DAPI) reveal the dynamics of ct-DNA at different stages of compaction. Cell viability of mouse mammary gland adenocarcinoma cells (4T1) and human embryonic kidney (HEK) 293 cell lines and in vitro cellular uptake of the gene to 4T1 cells have been investigated. This study provides ideas for designing efficient non-viral vectors. Overall, DDAB18-coated silica nanoparticles appear to be safe and effective DNA compaction agents that can carry nucleic acids for biomedical applications.
1. Introduction
Gene therapy tackles genetic diseases by replacing or supplementing defective genes with healthy or novel ones, to produce therapeutic proteins.1 Achieving this involves overcoming barriers like navigating through cell and nuclear membranes during gene transfection, as DNA molecules are large and negatively charged.2,3 For gene transfection, DNA has to be compacted, which is commonly achieved using viral vectors, where the native viral DNA is replaced with the desired DNA.1,4 However, repeated administration of viral vectors may provoke adverse immune responses due to the vector's viral origin.5 To mitigate these limitations, extensive research is underway to devise effective, non-toxic, and non-viral vectors.6 However, the initial non-viral methods developed exhibited lower efficiency and the potential for toxicity at higher concentrations.7,8 Subsequently, extensive research endeavors are underway to address these limitations. Future research will also be exploring the use of DNA-coated colloids and microspheres for the directed self-assembly of diverse crystal and gel structures,9,10 as well as rearranging into crystalline forms through simple DNA conformational changes,11,12 offering promising applications to address many such limitations.One notable approach under investigation is to use cationic surfactants,13–18 which can form various structured assemblies.19–22 Their interactions with nucleic acids due to opposite charges, result in improved gene compaction.21 However, their use in biological systems is hindered by cytotoxicity.23 Efforts to mitigate this challenge include modifying surfactant structures,24,25 such as sizes of headgroups26 or chain lengths.27
Another approach to address the cytotoxicity issues associated with the surfactants is to use nanoparticles with negatively charged surfaces.28 Positively charged surfactants bind with negatively charged DNA through electrostatic interactions, resulting in complex formation that is further stabilized by the hydrophobic interactions between the two components.29 However, in the presence of negatively charged nanoparticles, cationic surfactants first get adsorbed on the nanoparticles’ surfaces, forming a bilayer with positively charged outer surfaces, and then the hybrid nanoparticles bind with the DNA.28,30,31 Thus, a comparatively lower concentration of surfactant is required to compact DNA due to this cooperative binding.28 Silica nanoparticles with negatively charged surfaces have been utilized28,30,31 as they offer ample colloidal stability and biocompatibility and do not lead to any DNA damage as in the case of zinc oxide (ZnO) or titanium dioxide (TiO2) nanoparticles.27
Our curiosity is in understanding the role of chemical structures of cationic surfactants present in the hybrid nanoparticles in the compaction of DNA. While searching the literature, we were inspired by an existing report on a different aspect, i.e., the transfer of negatively charged nanoparticles from the aqueous phase to the organic phase through surfactant adsorption on nanoparticles’ surfaces. Many cationic surfactants with notably higher adsorption efficiency than conventional surfactants (single-head-single-tailed, 1-1 type) have been suggested.32 Wu et al.32 based on their experimental findings and performances of surfactants have presented an empirical rule as given below:
| P = nl/aV | (1) |
Thus, the question arises whether hydrophobicity per unit surface area of silica nanoparticles induced by surfactants forming a hybrid material has a similar role in DNA compaction as well. Understanding the critical roles of cationic surfactants as non-viral vectors adsorbed on the surfaces of negatively charged nanoparticles is vital for designing gene delivery systems. Earlier, we investigated ct-DNA compaction by some 2-2 type cationic (Gemini) surfactants like 12-4-12, 12-6-12, and 12-8-12, and the 1-2 type cationic surfactant, Didodecyldimethylammonium bromide (DDAB12),30,31 and compared their efficiencies with the conventional cationic surfactant, DTAB (1-1 type) in the presence of ∼100 nm silica nanoparticles. Both 2-2 and 1-2 type surfactants were found to be better DNA compacting agents than DTAB in the presence of silica nanoparticles, which must be due to the factor “n,” i.e., the number of alkyl chains per surfactant molecule. It has also been observed that the increasing % of DNA compaction in the presence of silica nanoparticles occurs as follows: 1-1 type < 2-2 type < 1-2 type. Notably, the headgroup size of the 1-2 type is smaller than that of the 2-2 type surfactant. This indicates that the headgroup size, i.e., factor “a” in eqn (1), has an effect on the DNA compaction. To further establish the fact that P decreases with increasing headgroup size (a), and therefore, a lesser extent of DNA compaction occurs, in the present work, we have studied ct-DNA compaction by a 3-2 type surfactant, MQAS12 (Scheme 1). If the empirical rule is in place, then the increasing % of ct-DNA compaction should follow the order of 3-2 type < 1-1 type < 2-2 type < 1-2 type. In fact, our study follows this order.
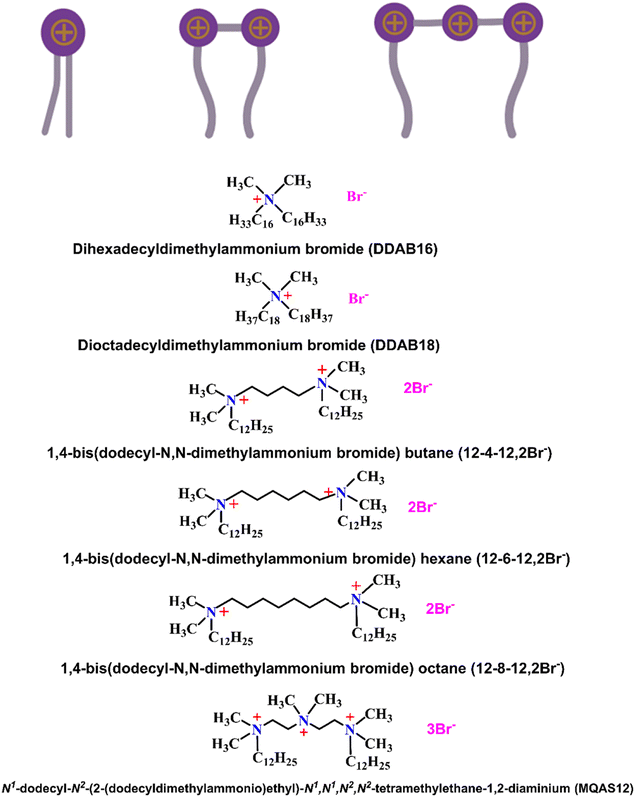 | ||
| Scheme 1 Symbolic representations of 1-2, 2-2, and 3-2 types of surfactants, and the structures of all the surfactants discussed here. | ||
To validate the empirical rule, the third factor that comes into the picture is “l,” i.e., the length of the alkyl chains. An earlier study presented DDAB12 as the most potential DNA compacting agent, being a 1-2 type surfactant. However, cytotoxicity is such a crucial issue that we were looking to further decrease the required concentration of surfactant needed to make DNA compaction happen. Considering the empirical rule, this study was carried out with two more 1-2 type surfactants, DDAB16 and DDAB18 (Scheme 1), with the same headgroup size as DDAB12. As for these surfactants, l is longer than that for DDAB12, therefore, the DNA compaction should be achieved at a further lower concentration of the surfactant due to increased P. This way, we can validate the empirical rule and, at the same time, we can present a DNA compacting agent with great potential. Among all surfactants, DDAB18 should be the most potential surfactant to compact DNA. The bromide form of the surfactant has been used because the bromide ion is known to be a weak fluorescence quencher compared to the chloride ion. ct-DNA is used due to its similarity in structure with that of human DNA.33
The fourth factor that controls P is the volume of the nanoparticles i.e., “V” in eqn (1). If, at all, P has a role in DNA compaction, then one can expect a larger % of compaction with smaller-sized silica nanoparticles. That is why, here, we have used silica nanoparticles of sizes ∼100 and ∼40 nm to validate eqn (1). To establish the mechanism of interactions involved between surfactants and DNA in the presence of silica nanoparticles and demonstrate the kind of interactions responsible for DNA compaction, some data obtained previously30,31 have been cited here.
Multiple techniques have been used to characterize the surfaces of hybrid nanoparticles and to demonstrate binding interactions between ct-DNA and surfactants/hybrid nanoparticles, % compaction of ct-DNA, and morphologies of systems. Biological testing for the cell viability of human embryonic kidney (HEK) 293 and mouse mammary gland adenocarcinoma (4T1) cell lines and in vitro cellular uptake of 4T1 cells through confocal microscopy and flow cytometry measurements for the systems with DDAB18 and MQAS12 has been carried out.
The present study thus provides an idea about the interactions responsible for DNA compaction, the effect of the structures of cationic surfactants and the sizes of negatively charged nanoparticles on it, and a direction for designing surfactants’ structures and developing gene delivery systems in the presence of nanoparticles.
2. Experimental
Comprehensive information regarding the materials and methods employed is provided in Section S1.1. in the ESI.† Details regarding the synthesis of MQAS12,34 and silica nanoparticles,31 as well as the preparation of ct-DNA–surfactant systems, can be found in the ESI,† Sections S1.2., S1.3., and S1.4. respectively.3. Results and discussions
3.1. Energy-dispersive X-ray (EDAX) analysis
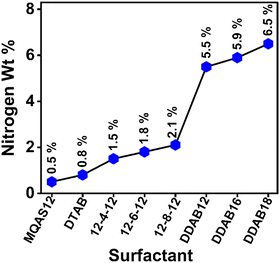
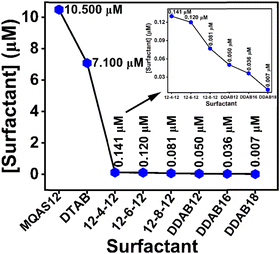
Details of EDAX measurements are given in Section S1.5. in the ESI.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35,36 EDAX analysis is employed to assess the degree of surfactant adsorption onto the surfaces of silica nanoparticles forming hybrid materials. Nitrogen weight percentage (wt%) values are mainly highlighted as nitrogen atom(s) (N) is/are present in the headgroup(s) to describe any difference between the adsorption efficiencies of surfactants. Here, the measurements have been done for the surfactants MQAS12, DDAB16, and DDAB18, and the data attained have been related to those for other surfactants reported previously.30,31 Our primary focus was on samples devoid of ct-DNA, as ct-DNA inherently contains nitrogen, which could potentially lead to erroneous interpretations to explain the adsorption efficiency of surfactants. Fig. S1, and data presented in Table S2 (ESI†), collectively reveal that the DDAB18 + silica nanoparticles (∼100 nm) system exhibits the highest wt% of N (6.5 wt%), followed by the DDAB16 + silica nanoparticles (∼100 nm) system (5.9 wt%). In contrast, the MQAS12 + silica nanoparticles (∼100 nm) system registers a significantly lower nitrogen wt% (0.52 wt%). Fig. 1 substantiates that among the three surfactants tested here, as well as compared to the reported surfactants,30,31 DTAB, 12-4-12, 12-6-12, 12-8-12, and DDAB12, DDAB18 demonstrate the most remarkable adsorption capacities on silica nanoparticle surfaces. The results agree well with the empirical rule,32i.e., P = nl/aV, as discussed in the introduction. Due to its extensive adsorption capability, DDAB18 possesses a higher P when compared to other surfactant categories. The greater the number of surfactant molecules adsorbed onto the surfaces, the more prominent will be the hydrophobicity induced by the hydrocarbon tails of surfactants. This leads to a great extent of DNA compaction, as discussed below. To provide evidence of the binding between ct-DNA and hybrid nanoparticles, with the maximum binding affinity observed for DDAB18, we conducted EDAX analysis even in the presence of ct-DNA. As depicted in Fig. S1 and Table S2 (ESI†), it is obvious that the N wt% increases for all three surfactants in the presence of ct-DNA, with the highest N wt% observed in the DDAB18 system and the lowest in the MQAS12 system when silica nanoparticles are present. Table S2 (ESI†) further substantiates the enhanced binding capacity of surfactants towards ct-DNA in the presence of silica nanoparticles.
35,36 EDAX analysis is employed to assess the degree of surfactant adsorption onto the surfaces of silica nanoparticles forming hybrid materials. Nitrogen weight percentage (wt%) values are mainly highlighted as nitrogen atom(s) (N) is/are present in the headgroup(s) to describe any difference between the adsorption efficiencies of surfactants. Here, the measurements have been done for the surfactants MQAS12, DDAB16, and DDAB18, and the data attained have been related to those for other surfactants reported previously.30,31 Our primary focus was on samples devoid of ct-DNA, as ct-DNA inherently contains nitrogen, which could potentially lead to erroneous interpretations to explain the adsorption efficiency of surfactants. Fig. S1, and data presented in Table S2 (ESI†), collectively reveal that the DDAB18 + silica nanoparticles (∼100 nm) system exhibits the highest wt% of N (6.5 wt%), followed by the DDAB16 + silica nanoparticles (∼100 nm) system (5.9 wt%). In contrast, the MQAS12 + silica nanoparticles (∼100 nm) system registers a significantly lower nitrogen wt% (0.52 wt%). Fig. 1 substantiates that among the three surfactants tested here, as well as compared to the reported surfactants,30,31 DTAB, 12-4-12, 12-6-12, 12-8-12, and DDAB12, DDAB18 demonstrate the most remarkable adsorption capacities on silica nanoparticle surfaces. The results agree well with the empirical rule,32i.e., P = nl/aV, as discussed in the introduction. Due to its extensive adsorption capability, DDAB18 possesses a higher P when compared to other surfactant categories. The greater the number of surfactant molecules adsorbed onto the surfaces, the more prominent will be the hydrophobicity induced by the hydrocarbon tails of surfactants. This leads to a great extent of DNA compaction, as discussed below. To provide evidence of the binding between ct-DNA and hybrid nanoparticles, with the maximum binding affinity observed for DDAB18, we conducted EDAX analysis even in the presence of ct-DNA. As depicted in Fig. S1 and Table S2 (ESI†), it is obvious that the N wt% increases for all three surfactants in the presence of ct-DNA, with the highest N wt% observed in the DDAB18 system and the lowest in the MQAS12 system when silica nanoparticles are present. Table S2 (ESI†) further substantiates the enhanced binding capacity of surfactants towards ct-DNA in the presence of silica nanoparticles.
3.2. Zeta potential analysis
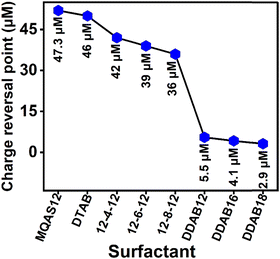
Zeta potential measurement details are available under Section S1.6. in the ESI.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 Zeta potential values can serve as a vital physical parameter for quantifying alterations in surface charges.37 The zeta potential value for the synthesized silica nanoparticles is −31.7 mV, which supports negative charges on their surfaces. Surfactants with positively charged headgroups bind with the negatively charged ct-DNA. The negative zeta potential of ct-DNA progressively changes with increasing surfactant concentration due to the formation of ct-DNA–surfactant aggregates.30,31 Just above a concentration called the charge reversal point (CRP), the zeta potential becomes positive. Fig. S2 and Table S3 (ESI†) illustrate the charge reversal as MQAS12, DDAB16, and DDAB18 concentrations increase progressively in the absence and presence of silica nanoparticles. The data in Table S3 (ESI†) highlight that the DDAB class of surfactants, especially DDAB18, necessitates a substantially lower concentration to reach the CRP compared to the MQAS12 surfactant. The concentration of surfactant required to achieve CRP is diminished in the presence of silica nanoparticles due to cooperative binding.28–31Fig. 2 compares the CRP of different surfactants in the presence of silica nanoparticles. Notably, for comparison, the data for other surfactants that were studied earlier30,31 have been utilized. This concentration is the lowest for DDAB18 and the highest for MQAS12. Also, the zeta potential value is the highest for DDAB18 (∼42.0–58.1 mV) and the lowest for MQAS12 (∼20.0–26.0 mV) in the presence of silica nanoparticles. The order found in Fig. 2 is as per the empirical rule (eqn (1)). If the extent of adsorption of surfactants on silica nanoparticles’ surfaces is high, then the CRP is achieved earlier when the particles with positive charges on the outer layer of the bilayer interact with ct-DNA. The EDAX data discussed above support this order.
37 Zeta potential values can serve as a vital physical parameter for quantifying alterations in surface charges.37 The zeta potential value for the synthesized silica nanoparticles is −31.7 mV, which supports negative charges on their surfaces. Surfactants with positively charged headgroups bind with the negatively charged ct-DNA. The negative zeta potential of ct-DNA progressively changes with increasing surfactant concentration due to the formation of ct-DNA–surfactant aggregates.30,31 Just above a concentration called the charge reversal point (CRP), the zeta potential becomes positive. Fig. S2 and Table S3 (ESI†) illustrate the charge reversal as MQAS12, DDAB16, and DDAB18 concentrations increase progressively in the absence and presence of silica nanoparticles. The data in Table S3 (ESI†) highlight that the DDAB class of surfactants, especially DDAB18, necessitates a substantially lower concentration to reach the CRP compared to the MQAS12 surfactant. The concentration of surfactant required to achieve CRP is diminished in the presence of silica nanoparticles due to cooperative binding.28–31Fig. 2 compares the CRP of different surfactants in the presence of silica nanoparticles. Notably, for comparison, the data for other surfactants that were studied earlier30,31 have been utilized. This concentration is the lowest for DDAB18 and the highest for MQAS12. Also, the zeta potential value is the highest for DDAB18 (∼42.0–58.1 mV) and the lowest for MQAS12 (∼20.0–26.0 mV) in the presence of silica nanoparticles. The order found in Fig. 2 is as per the empirical rule (eqn (1)). If the extent of adsorption of surfactants on silica nanoparticles’ surfaces is high, then the CRP is achieved earlier when the particles with positive charges on the outer layer of the bilayer interact with ct-DNA. The EDAX data discussed above support this order.
3.3. Cyclic voltammetry (CV) studies
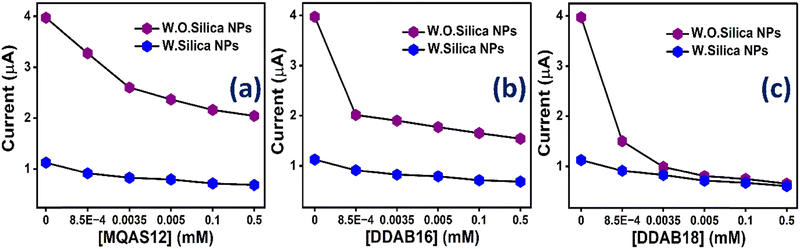
CV measurement details are available in Section S1.7. in the ESI.† Given that surfactants exhibit limited electroactivity, [Fe(CN)6]3−/4− was employed as a redox probe to investigate the interaction between surfactant molecules and ct-DNA.38 The CV measurements illustrated changes in the redox probe with varying concentrations of cationic surfactants at a fixed ct-DNA concentration, as depicted in Fig. S3 (ESI†). As shown in Fig. S3 (ESI†), the cathodic currents (ic) either slightly decreased or remained unaffected, while there was a pronounced deduction in the anodic current (ia) as the surfactant concentration increased. This anodic current ia decreased as a result of the interaction between the surfactants and ct-DNA, leading to the proof of ct-DNA compaction (discussed below). The primary effect of increasing surfactant concentration was neutralizing the net negative charges of ct-DNA, resulting in the observed decrease in ia. Fig. 3 vividly displays notable shifts in anodic current due to various surfactants. This figure directly compares alterations in anodic current caused by different surfactants at various concentrations, both with and without silica nanoparticles. Consider, for instance, a 0.5 mM concentration of surfactants: MQAS12, DDAB16, and DDAB18 recorded approximately 2.04 μA, 1.54 μA and 0.65 μA, respectively, in the absence of silica nanoparticles. In the presence of silica nanoparticles, these values changed to approximately 0.71 μA, 0.64 μA, and 0.60 μA, respectively. This suggests that DDAB18, when combined with silica nanoparticles, notably outperforms other surfactant combinations in effectively neutralizing the negative charges of ct-DNA.3.4. Ethidium bromide (EtBr) exclusion assay
The detailed methods for UV-Vis absorption and fluorescence measurements are given in Section S1.8. in the ESI.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 31 EtBr exclusion assay experiments have been conducted to elucidate the binding interactions between ct-DNA and surfactants.39 In a buffer solution, the fluorescence intensity of EtBr is very low due to solvent-induced quenching.24,40 However, upon intercalative binding of EtBr with ct-DNA, a rapid increase in fluorescence is observed.39 Fig. S4 (ESI†) illustrates fluorescence quenching upon the gradual addition of all three surfactants, both in the absence and presence of silica nanoparticles. The plot of F/Foversus [surfactant] (Fig. S5, ESI†) (where F and Fo are the fluorescence intensities of EtBr with and without surfactant, respectively), demonstrates a minimum point, indicating the maximum displacement of EtBr molecules as a result of surfactant binding to ct-DNA. Beyond this point, the fluorescence intensity remains relatively constant. Table S4 (ESI†) demonstrates the concentrations of surfactants where a minimum point is achieved. The data indicate that this specific point is reached at an exceptionally low concentration for DDAB18 compared to the other surfactants. Therefore, DDAB18 binds strongly with the ct-DNA, so it can potentially compact the DNA at a remarkably low concentration (discussed below). An initial increase in fluorescence intensity (Fig. S5, ESI†) at a low concentration of surfactants indicates that EtBr molecules binding with ct-DNA experience a comparatively less polar/rigid environment. Similar trends are noted in our earlier studies as well.30,31
31 EtBr exclusion assay experiments have been conducted to elucidate the binding interactions between ct-DNA and surfactants.39 In a buffer solution, the fluorescence intensity of EtBr is very low due to solvent-induced quenching.24,40 However, upon intercalative binding of EtBr with ct-DNA, a rapid increase in fluorescence is observed.39 Fig. S4 (ESI†) illustrates fluorescence quenching upon the gradual addition of all three surfactants, both in the absence and presence of silica nanoparticles. The plot of F/Foversus [surfactant] (Fig. S5, ESI†) (where F and Fo are the fluorescence intensities of EtBr with and without surfactant, respectively), demonstrates a minimum point, indicating the maximum displacement of EtBr molecules as a result of surfactant binding to ct-DNA. Beyond this point, the fluorescence intensity remains relatively constant. Table S4 (ESI†) demonstrates the concentrations of surfactants where a minimum point is achieved. The data indicate that this specific point is reached at an exceptionally low concentration for DDAB18 compared to the other surfactants. Therefore, DDAB18 binds strongly with the ct-DNA, so it can potentially compact the DNA at a remarkably low concentration (discussed below). An initial increase in fluorescence intensity (Fig. S5, ESI†) at a low concentration of surfactants indicates that EtBr molecules binding with ct-DNA experience a comparatively less polar/rigid environment. Similar trends are noted in our earlier studies as well.30,31
To support the fact that DDAB18 surfactants bind to the intercalative region of ct-DNA, circular dichroism (CD) spectra (methods are detailed in Section S1.9. of the ESI†) of ct-DNA have been recorded in the presence of different concentrations of DDAB18 in Tris HCl buffer (pH = 7.4, ∼10 mM) solutions in the absence and presence of silica nanoparticles and displayed in Fig. S6(a) and (b) (ESI†), respectively. The CD spectrum of a native DNA e.g. ct-DNA presents a positive band at 273 nm for base stacking and a negative band at 246 nm for right-handed helicity, which is the identity of the B-form of ct-DNA. The ct-DNA's CD spectra do not show any change for minor groove binding; however, both the bands (positive and negative) are affected by intercalative binding.41 The CD spectra in Fig. S6(a) and (b) (ESI†), show that both positive and negative bands are affected upon binding of DDAB18 with ct-DNA, which indicates intercalative binding. The results thus support that the surfactants can replace the EtBr from within the ct-DNA.
3.5. Fluorescence lifetime measurements of DAPI
We conducted fluorescence intensity decay measurements using DAPI in ct-DNA to gain more insights into the binding mechanism between ct-DNA and surfactants (measurement methods are explained in Section S1.8. in the ESI†). One of the two species of DAPI in pure water in a neutral pH medium exhibits solvation of the indole ring in the ground state that undergoes a proton transfer upon excitation, showing the major decay component of a short lifetime (∼0.19 ns).42 The longer lifetime component (∼2.80 ns) corresponds to the DAPI with a particular conformation incapable of experiencing any proton transfer.42 When DAPI binds to AT clusters of DNA, it becomes shielded from the solvent, blocking excited state proton transfer. In contrast, binding to GC clusters exposes it to the solvent, facilitating proton transfer. In this study, we conducted fluorescence intensity decay measurements using DAPI in ct-DNA under several concentrations of each of MQAS12, DDAB16, and DDAB18 surfactants, both with and without silica nanoparticles. The results, as depicted in Fig. S7 and Table S5 (ESI†), consistently revealed bi-exponential fluorescence decay patterns in each system. The average lifetimes (〈τf〉) were calculated using eqn (S1), as shown in the ESI.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 Upon the addition of ct-DNA, the lifetime of the fast component increased from 0.20 to 0.58 ns with a decrease in weighting from 0.71 to 0.67. The average lifetime is increased from 0.98 to 1.30 ns supporting the binding of DAPI with the ct-DNA.
43 Upon the addition of ct-DNA, the lifetime of the fast component increased from 0.20 to 0.58 ns with a decrease in weighting from 0.71 to 0.67. The average lifetime is increased from 0.98 to 1.30 ns supporting the binding of DAPI with the ct-DNA.
Interestingly, the data in Table S5 (ESI†) display that by increasing the concentration of a surfactant, the lifetime of the fast components reduces with an increase in the weighting. There is a concomitant decrease in the weighting of the slow component. The percentage decrease in the lifetime of the fast component and an increase in its weighting (given in parenthesis) at 2.5 μM of surfactant concentration in the absence of silica nanoparticles are ∼21% (∼6%), 88% (46%), and 90% (46%) for MQAS12, DDAB16, and DDAB18, respectively. Whereas, in the presence of silica nanoparticles, these values are 56% (20%), 88% (55%), and 88% (53%) for MQAS12, DDAB16, and DDAB18, respectively. More notably, the average lifetime of DAPI was significantly reduced upon introducing 2.5 μM of each surfactant. This decrease was even more pronounced when silica nanoparticles were added. These results underscore the strong binding of surfactants with ct-DNA, displacing DAPI, which is further increased in the presence of silica nanoparticles. Fig. 4 compares the average lifetimes of DAPI in the absence and presence of silica nanoparticles representing the surfactants’ capabilities to displace DAPI from ct-DNA. Notably, DDAB18 appears to be the most potential surfactant among all studied surfactants as in its presence, the surfaces of nanoparticles gain maximum hydrophobicity and interact strongly with the ct-DNA, showing a large displacement of DAPI. Here, the results for some surfactants reported previously30,31 are also included for comparison.
3.6. Fluorescence anisotropy decay measurements of DAPI. Dynamics of ct-DNA
Fluorescence anisotropy decay measurements (measurement methods are available in Section S1.8. in the ESI†) of DAPI are conducted to assess rotational relaxation times.36,43 These values provide insights into the rigidity or flexibility of the surrounding microenvironment and the dynamics of molecules.30,31,43,44 Time-resolved fluorescence anisotropies (r(t)) of DAPI in various systems for the present three surfactants were calculated using eqn (S2) in the ESI,†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 and the decay curves were obtained by fitting them to eqn (S3) in the ESI.†
43 and the decay curves were obtained by fitting them to eqn (S3) in the ESI.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 Characteristic anisotropy decays for some representative systems are displayed in Fig. S8 (ESI†). The anisotropy decay of DAPI in the buffer is single exponential, irrespective of the presence or absence of silica nanoparticles. Rotational relaxation times of 0.19 and 0.18 ns, respectively, are found under these conditions. However, when DAPI is bound to native ct-DNA and ct-DNA–surfactant complexes, the anisotropy decay exhibits a bi-exponential behavior giving two rotational components in the presence and absence of silica nanoparticles. We used eqn (S4) in the ESI†
43 Characteristic anisotropy decays for some representative systems are displayed in Fig. S8 (ESI†). The anisotropy decay of DAPI in the buffer is single exponential, irrespective of the presence or absence of silica nanoparticles. Rotational relaxation times of 0.19 and 0.18 ns, respectively, are found under these conditions. However, when DAPI is bound to native ct-DNA and ct-DNA–surfactant complexes, the anisotropy decay exhibits a bi-exponential behavior giving two rotational components in the presence and absence of silica nanoparticles. We used eqn (S4) in the ESI†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 to determine the average rotational relaxation times of DAPI in ct-DNA–surfactant systems at various surfactant concentrations with and without silica nanoparticles, as presented in Table S6 (ESI†), including their respective weightings. Notably, the average rotational relaxation time of DAPI, when bound to native ct-DNA, is considerably longer than free DAPI in a buffer medium. This implies that the free-tumbling motions of DAPI in a buffer medium become constrained upon binding with ct-DNA, providing evidence for the binding of DAPI molecules to ct-DNA.
43 to determine the average rotational relaxation times of DAPI in ct-DNA–surfactant systems at various surfactant concentrations with and without silica nanoparticles, as presented in Table S6 (ESI†), including their respective weightings. Notably, the average rotational relaxation time of DAPI, when bound to native ct-DNA, is considerably longer than free DAPI in a buffer medium. This implies that the free-tumbling motions of DAPI in a buffer medium become constrained upon binding with ct-DNA, providing evidence for the binding of DAPI molecules to ct-DNA.
The rotational components are characterized as fast (τ1r) and slow (τ2r). The values of τ1r and τ2r, along with their respective weightings denoted as a1r and a2r, respectively, are provided in Table S6 (ESI†). Notable observations are as follows. In native ct-DNA and complexes, the slow components contribute majorly to depolarization. However, in the presence of a surfactant, the fast component is the major one as some DAPI molecules are displaced from ct-DNA's binding sites. The fast component (relaxation time, τ1r) is ascribed to DAPI's internal motions45 connected with the local structure's relaxation in ct-DNA along with the motions in the bulk. However, the slow component (relaxation time, τ2r) could be due to the ct-DNA double helix's segmental motion42,45,46 or the overall tumbling motions of a big complex like DNA + DAPI or DNA + DAPI + silica nanoparticles or DNA + DAPI + surfactant + silica nanoparticles. The two-step model has been used here to describe the slower (τ2r) motion in a bi-exponential anisotropy decay (details given in Note S1 in the ESI†).47–49 As per the two-step model, the relaxation time for the slow rotational motion i.e. τ2r can be related to the time required for the relaxation through the ct-DNA double helix's segmental motion (τS)42,45,46 and the time for relaxation through the overall tumbling motions of the big complex (τC) according to eqn (S5) (ESI†).47–49 Note S1 in the ESI† also describes the method used to calculate the τC values at 25 °C using eqn (S6) (ESI†) taking the experimentally determined hydrodynamic radii (rh) of the complex. The values of hydrodynamic radii and τC are given in Table S6 (ESI†). The calculated values of τC have been utilized to estimate the τS values using eqn (S5) (ESI†) and all data are tabulated in Table S6 (ESI†). It can be seen from the data in Table S6 (ESI†) that the relaxation time for the slow motion, τ2r, in every system is almost equal to the τS and largely different from the τC value.47 This depicts that the slow motion in the complex is nothing but the ct-DNA double helix's segmental motion,42,45,46 and not the overall motion of the big complex itself.
With a gradual increase in a surfactant's concentration, the fast component's rotational relaxation time primarily decreases with increasing weighting (Table S6, ESI†). The average relaxation time steadily decreases with increasing concentration of surfactant. The extent of this decrease in rotational relaxation time is more in the presence of silica nanoparticles. The results thus depict that the DAPI molecules are replaced by the surfactants, supporting the binding of surfactants and hybrid nanoparticles with ct-DNA. For DDAB16 and DDAB18, both with silica nanoparticles, the weighting of the fast component becomes as large as ∼98%, compared to ∼77% for MQAS12 at their 2.5 μM concentration, indicating the displacement of almost all DAPI molecules from ct-DNA by the former two surfactants. The displacement is substantial in the presence of silica nanoparticles. As per the data in Table S6 (ESI†), for all systems except for the DNA + DAPI + DDAB18 + silica nanoparticles, initially at 1.2 μM, the rotational relaxation time of the helix's segment (longer component) increases,45,50 and then at 2.5 μM it decreases.30,31,45 This could be because initially, for the direct binding of the surfactants or hybrid nanoparticles with ct-DNA, there is an osmotic exclusion of water molecules from the ct-DNA segment's vicinity resulting in an enhancement in its rigidity.50 However, at a higher concentration, the surfactants might bind to a region in the ct-DNA for which there is an osmotic inclusion of water, enhancing the DNA segment's flexibility.50 On the other hand, in the case of DDAB18, in the presence of silica nanoparticles, very strong interactions between surfactants and ct-DNA lead to a rigidification of the ct-DNA helix's segment.
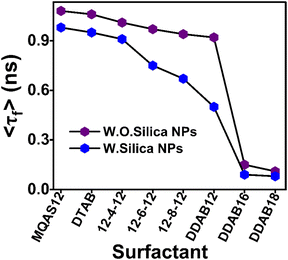
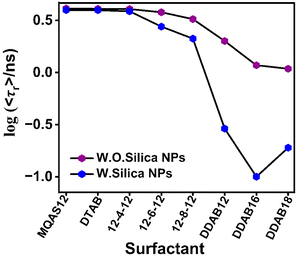
To compare the degree of interactions between ct-DNA and different surfactants, the average rotational relaxation times in the presence of 2.5 μM concentrations of all surfactants in the presence and absence of silica nanoparticles are plotted in Fig. 5. The average rotational relaxation time is found to be shorter for all surfactants in the presence of silica nanoparticles. Comparatively shorter rotational relaxation times for DDAB class surfactants, both in the presence and absence of silica nanoparticles, indicate their robust interactions with the ct-DNA. At 2.5 μM concentration, the 〈τr〉 in the case of DDAB18 is a little longer than that of DDAB16 in the presence of silica nanoparticles, which is due to the comparatively restricted segmental motions of ct-DNA in the presence of the former compared with the latter. This result also supports the strongest interactions between DDAB18 and ct-DNA.
3.7. TEM images of various systems
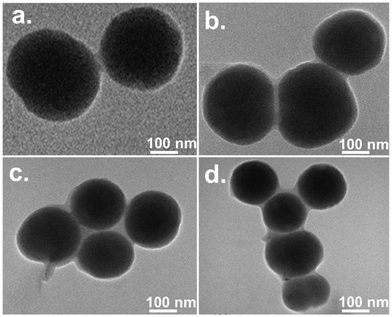
Details regarding TEM measurements can be found in Section S1.5. of the ESI.† As our primary focus revolves around the effect of the structures of surfactants present in the hybrid materials on the DNA compaction, the TEM images of systems with four distinct types of surfactants, one from each of 1-1, 1-2, 2-2, and 3-2 types are presented here. Fig. 6 provides a basis for comparing the sizes of ct-DNA–surfactant–silica nanoparticle complexes, which are roughly ∼300 nm for combinations involving MQAS12 or DTAB, ∼200 nm for 12-6-12, and ∼140 nm for DDAB18. The sizes of particles of surfactant for DTAB/MQAS12-coated silica nanoparticles wrapped by ct-DNA are pretty big compared to those formed by 12-6-12/DDAB18-coated silica nanoparticles and ct-DNA. The smallest size particle is formed in the case of DDAB18 (∼140 mn). The bigger-sized particles compared to the size of silica nanoparticles of ∼100 nm depict the coating of surfactants and ct-DNA on the nanoparticles’ surfaces, as can be seen in the figures with a closer look. The trends are consistent with what is expected from Wu et al.'s empirical rule32 discussed above.3.8. FE-SEM images of various systems
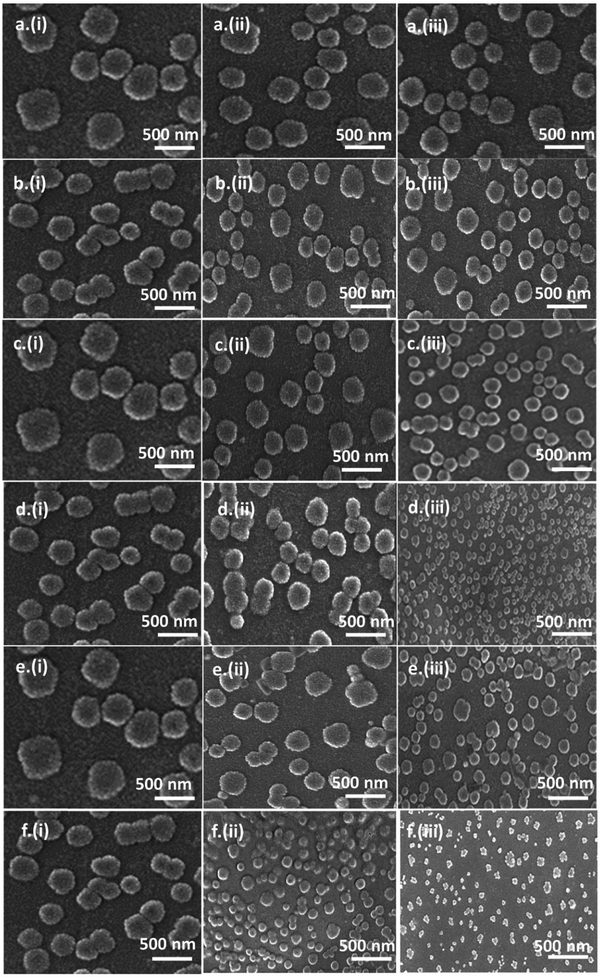
FE-SEM measurement methods are available in Section S1.5. in the ESI.†Fig. 7 and Table S7 (ESI†) provide clear evidence of structural modifications and size variations of ct-DNA at concentrations of 0.0, 1.2, and 2.5 μM of DDAB16, DDAB18, and MQAS12, both in the presence and absence of silica nanoparticles. It can be seen from the data in Table S7 (ESI†) that the reduction in ct-DNA size is evident in all systems, with a notable decrease observed with DDAB18. Significantly, a drop in the size of ct-DNA is also pronounced in the presence of silica nanoparticles but without any surfactant because of the crowding effect.51 For this, the coil depletion of ct-DNA mainly occurs as a result of the excluded volume of silica nanoparticles.51,52 Without silica nanoparticles, the ct-DNA size reduced from approximately ∼650–700 nm (native DNA) to ∼200–245 nm at 2.5 μM of DDAB18, while with silica nanoparticles, it decreased from ∼320–400 nm to ∼120–130 nm at the same concentration of DDAB18. The sizes of particles are almost the same as noted from the TEM images for 2.5 μM of any surfactant in the presence of silica nanoparticles. Notably, for any surfactant at a given concentration, the particles’ sizes are smaller in the presence of silica nanoparticles due to the cooperative binding of surfactants with ct-DNA.28 As per the above data, the size of native ct-DNA is reduced by ∼69% and ∼81% in the absence and presence of silica nanoparticles (∼100 nm), respectively. In the presence of ∼40 nm particles (discussed below), the size is decreased by ∼87%. The literature reports various forms into which DNA molecules can compact/condense, including toroids, flower-like structures, rods/wires, globular arrangements, bead-like, etc.53Fig. 7 depicts FE-SEM images illustrating these morphologies providing clear support for DNA compaction facilitated by surfactants and silica nanoparticles. DDAB16 and DDAB18 are more potential ct-DNA compacting agents than DDAB12 reported earlier, for which bigger particles with a size of ∼135–150 nm were noted under the same condition. This is attributable to the longer hydrophobic tails of the former two surfactants than the latter. Therefore, the surfactant-induced hydrophobicity per unit surface area has a role in DNA compaction.3.9. Dynamic light scattering (DLS) analysis
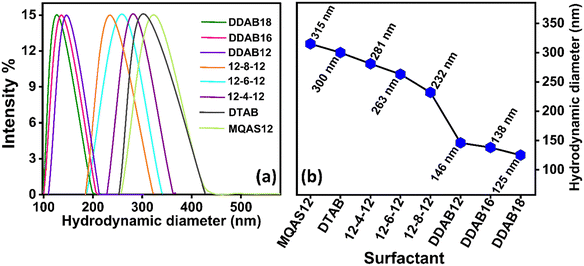
DLS experiments and data analysis (measurement details are given in Section S1.6. in the ESI†) were conducted to measure the hydrodynamic diameter of the ct-DNA–surfactant–silica nanoparticle complexes. The DLS size distribution plots for surfactants MQAS12, DDAB16, and DDAB18, each at different concentrations, are given in Fig. S9 (ESI†), while for other surfactants, they are available elsewhere.30,31 The DLS size distribution plots and hydrodynamic diameters for particles of various systems, each at a 2.5 μM concentration of surfactant in the presence of silica nanoparticles, are presented in Fig. 8a and b, respectively, for comparison. The hydrodynamic diameters and polydispersity index (PDI) values are also presented in Table S8 (ESI†). Notably, these trends are constant with the particle sizes observed in FE-SEM analysis. DDAB18 demonstrates a remarkable reduction in ct-DNA's size compared to other surfactants. Once again, these results support the fact that the number of tails (n), tail length (l), and headgroup size (a), as depicted in the empirical rule by Wu et al.,32 have a noteworthy role in DNA compaction.3.10. Fluorescence microscopic images
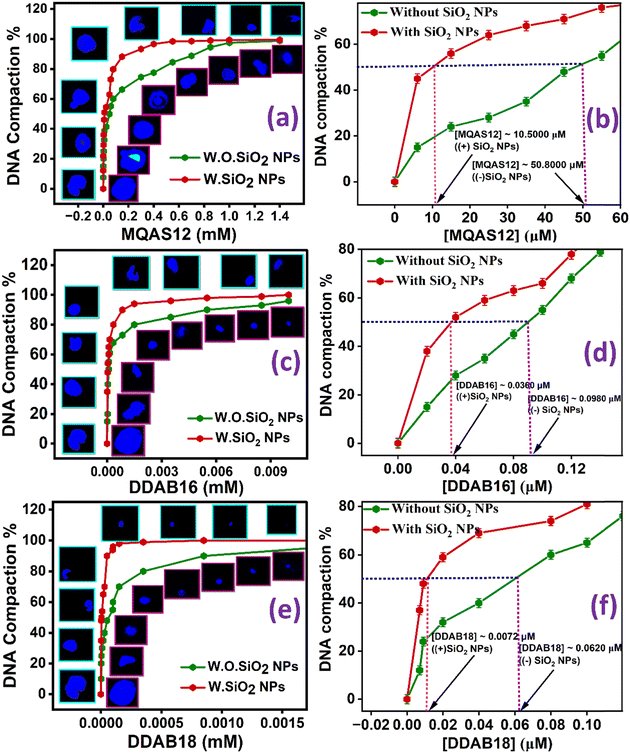
Fluorescence microscopy was employed to quantify the percentage of ct-DNA compaction. The fluorescence microscopic measurement methods are explained in Section S1.10. in the ESI.† These images offer direct visualization of the transition from an elongated form to a compacted one. In this study, the fluorophore DAPI, known for its groove-binding characteristics, was utilized. To determine the compaction percentage, we measured the average long-axis length of a minimum of fifty distinct structures in a given sample. Fig. 9 provides a clear visual representation of the transition of ct-DNA into its compacted form in the presence of all three surfactants. The data presented in Fig. 9(a), (c), and (e) highlight the ct-DNA compaction % and representative images of fluorescence microscopy of a single ct-DNA molecule, showing structural and morphological changes of ct-DNA with increasing [MQAS12], [DDAB16], and [DDAB18], respectively. The data in Fig. 9(b), (d), and (f) specifically display the surfactant concentrations required to achieve 50% compaction of ct-DNA. To compare the potential of the present surfactants and surfactants used before,30,31 the concentrations of surfactants required to achieve 50% compaction of ct-DNA have been plotted and displayed in Fig. 10. A superior performance is shown by DDAB18 apart from DDAB16, MQAS12, as well as previously reported surfactants, establishing it as the most efficient cationic surfactant for ct-DNA compaction at the minimal concentrations essential to address cytotoxicity issues. Notably, while the 50% compaction of ct-DNA is done at ∼0.050 μM of the previously reported DDAB12,30 the same amount of compaction is achieved by as low as ∼0.007 μM of DDAB18.One can argue that at a higher concentration of surfactant, the DNA cannot be seen as DAPI molecules have come out, and the smaller size of DNA is simply due to the smaller number of DAPI remaining attached to DNA. To show that this is not the case, some control experiments have been done by recording fluorescence microscopic images of systems containing 5.0 μM of ct-DNA and 1.0 μM of DDAB18 with increasing concentrations of DAPI (Fig. S10, ESI†). It is expected that with increasing concentration, more and more DAPI will bind with the DNA–surfactant or DNA–surfactant–silica nanoparticle complexes, and if the DNA has not been condensed, then the size of images is expected to be bigger. However, the results in Fig. S10 (ESI†) show no change in the images’ size with increasing dye concentration, supporting that DNA compaction was achieved at a higher surfactant concentration.
3.11. MTT assay
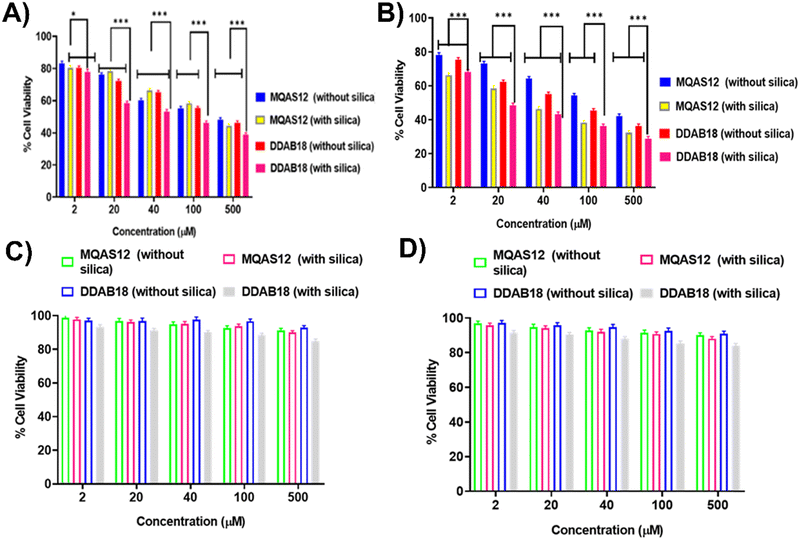
Details of MTT assay procedures are specified in Section S1.11. in the ESI.† Ensuring the safety of biomaterials in vitro is pivotal, particularly regarding their cytotoxicity.The cell viabilities of 4T1 cell lines over 24 and 48 hours of incubation are presented in Fig. 11A and B, respectively, and the same for HEK293 cell lines over 24 and 48 hours of incubation are displayed in Fig. 11C and D, respectively. The cell viability of HEK-293 cells stayed consistently above 88% across sample concentrations from 0 to 500 μM. Additionally, the comparable cell viabilities observed between MQAS12 and DDAB18 suggest strong cytocompatibility for both surfactants, highlighting their similar effects on cell viability. The cell viability study revealed that MQAS12 and DDAB18 with silica nanoparticles demonstrated the highest time and dose-dependent cytotoxicity compared to those without silica nanoparticles in 4T1 cell lines. The reason could be credited to the difference in the cellular uptake of the materials between these two cell lines, which is because of the up-regulations and down-regulations of various markers involved. Moreover, the cell membrane's integrity could be compromised in the case of cancer cells, which caused more internalization of these systems than normal cells. Overall, the cytotoxicity of the MQAS12 and DDAB18 surfactant systems (both in the absence and presence of silica nanoparticles) is comparatively lower.
3.12. Cellular uptake study
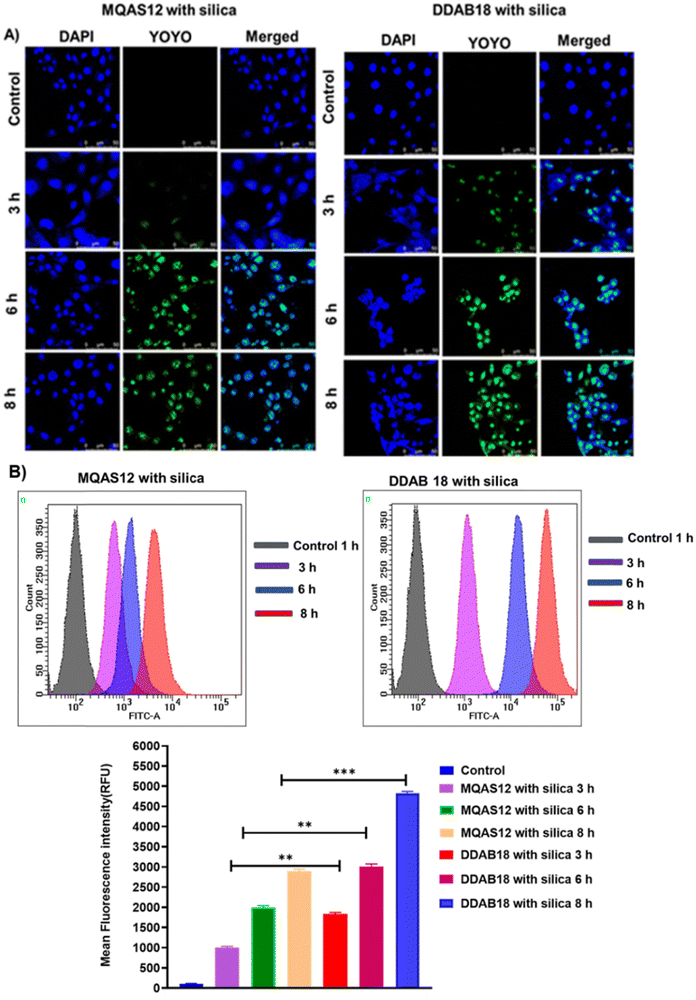
Details of the methods for the cellular uptake studies are given in Sections S1.12. and S1.13. in the ESI.† Confocal fluorescence microscopy imaging and flow cytometry were used to evaluate the cellular internalization of YOYO-1 labeled ct-DNA with each of MQAS12 and DDAB18 both qualitatively and quantitatively in 4T1 cells (Fig. 12). The clear, bright green fluorescence was detected in the cytoplasm and nuclei in YOYO-1 labeled ct-DNA with each of MQAS12 and DDAB18 treated after 6 h incubation with the targeted cells. This suggests quick internalization of YOYO-1 labeled ct-DNA with each of MQAS12 and DDAB18. For 4T1 cells, the red fluorescence intensity of YOYO-1 labeled ct-DNA with each of MQAS12 and DDAB18 increased from 3 to 8 h, indicating time-dependent cellular absorption. Moreover, further flow cytometry studies were performed to validate the increased internalization. The mean fluorescence intensity values for YOYO-1 labeled ct-DNA with each of MQAS12 and DDAB18 were shown to increase from 3 to 8 h in the flow cytometer's histogram. Flow cytometry analysis demonstrated a time-dependent increase in fluorescence intensity in 4T1 cells. The cellular uptake and intracellular release findings showed enhanced cell membrane permeability and release.3.13. Gel retardation assay (agarose gel electrophoresis)
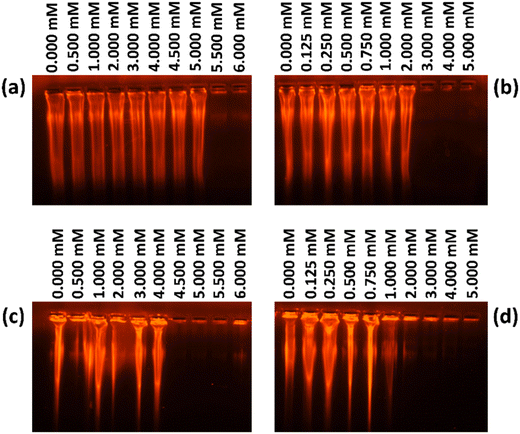
Details of the agarose gel electrophoresis studies are given in Section S1.14. in the ESI.† Electrophoresis experiments were conducted to investigate the interaction between surfactants/hybrid nanoparticles and ct-DNA, as well as the mobility of complexes. Surfactants were added to a fixed concentration of ct-DNA (5 μM) in the 0–6 m range, both with and without silica nanoparticles. Electrophoresis measurements showed that in the presence of silica nanoparticles, surfactants interacted with ct-DNA at lower concentrations compared to systems without nanoparticles. As shown in Fig. 13, with MQAS12, binding occurred at around 5.5 mM (Fig. 13a) without silica nanoparticles, but at 4.5 mM (Fig. 13c) with silica nanoparticles. Similarly, with DDAB18, binding started at 3.0 mM (Fig. 13b) without silica nanoparticles, and at 1.0 mM (Fig. 13d) with silica nanoparticles. This suggests that DDAB18 was more effective in neutralizing negatively charged ct-DNA compared to other surfactant.30,313.14. Discussion on factors responsible for ct-DNA compaction
Surfactant molecules with positively charged headgroups adsorbed on the negatively charged silica nanoparticles’ surfaces will have their tails protruded outward towards the bulk. It is difficult for these particles to be stable in an aqueous medium. Therefore, a bilayer of surfactants is formed as a result of hydrophobic interactions so that the particles with outer positive surface charges will be stable in an aqueous medium.30–32 These positively charged particles then interact with negatively charged DNA, resulting in its compaction through charge neutralization. This way, the negatively charged silica nanoparticles enhance the cationic surfactants’ ability to compact ct-DNA through cooperative binding.28 Notably, here, a pH of 7.4 (biological pH) was set as compared to the pH value of 10.5 chosen by Wu et al.32 in their work on phase transfer. This step has been taken to control the adsorption of a large number of surfactants so that no precipitation takes place and also cytotoxicity can be avoided. No precipitation was observed, and absolute clear solutions for all samples support a bilayer formation even at pH 7.4. A continuous increase in positive zeta potential value with increasing surfactant concentrations indirectly support the bilayer formation. Further experimental supports are discussed below.Based on the results discussed above, the increasing order of ct-DNA compaction efficiency of various surfactants in the presence of silica nanoparticles was found as follows: MQAS12 < DTAB < 12-4-12 < 12-6-12 < 12-8-12 < DDAB12 < DDAB16 < DDAB18. These results are consistent with the empirical rule presented by Wu et al.,32i.e., P = nl/aV discussed in the introduction section. Based on the three factors, n, l, and a, the above-mentioned order can be explained. However, to check the effect of the overall size of the nanoparticles (factor V in the equation) on P and thereby on the compaction of ct-DNA, we have also carried out experiments using silica nanoparticles with a smaller size (∼40 nm) than the ∼100 nm nanoparticles discussed above. It can be seen from the data in Fig. S11 and Table S2 (ESI†) that the N wt% is higher in the presence of ∼40 nm particles as compared to ∼100 nm particles (Table S2, ESI†) for each surfactant, MQAS12, DDAB16, and DDAB18. The N wt% increases in the presence of nucleic acid. The results thus support the binding of ct-DNA with the surfactant-coated nanoparticles. The data in Table S8 (ESI†) show that the compaction of ct-DNA is more in the presence of ∼40 nm particles than ∼100 nm particles. With 2.5 μM DDAB18 and ∼40 nm silica nanoparticles, we could get a particle with a size as low as ∼86 nm. These results give the answer to the question that we raised in the Introduction section, which is whether the hydrophobicity per unit surface area (P) of the silica nanoparticles induced by the surfactants of varying chemical structures has a role in the compaction of ct-DNA.
The greater the number of surfactant molecules adsorbed onto the surfaces, the more prominent the hydrophobicity induced by the hydrocarbon tails of the surfactants will be. The greater the P induced by the first layer, the more significant the number of surfactant molecules taking part in the formation of the second layer due to hydrophobic interactions among the surfactants’ tails with the positive surface charges. Therefore, it is obvious that the P has an indirect effect on the compaction of ct-DNA. Notably, the direct effect of the hydrophobicity of surfactants on increasing compaction of ct-DNA is evidenced by the order: 12-4-12 < 12-6-12 < 12-8-12, where the hydrophobicity of the spacer is increased from left to right.31 Rudiuk et al.'s28 study on DNA's compaction by conventional surfactants gives results aligned in the same direction. To support that P increases with increasing concentration of the surfactant even though the surfaces are charged, the fluorescence spectra and lifetimes of DAPI in the presence of a fixed concentration of silica nanoparticles and varying concentrations of DDAB18 have been recorded (Fig. S12, ESI†). Both fluorescence intensity (Fig. S12a and b, ESI†) and lifetime (Fig. S12c, ESI†) increase with DDAB18 concentration. The increasing number of adsorbed surfactants on silica nanoparticles’ surfaces, is supported by the EDAX data in Table S9 (ESI†) that show increasing N wt% with increasing amounts of DDAB18. Fig. S2 (ESI†) presents an increase in zeta potential with increasing DDAB concentration. As with the increasing hydrophobicity of the layer, the number of positive surface charges also increases; therefore, unless the bilayer is formed, these particles won’t be stable in the aqueous environment. The results also depict that both electrostatic and hydrophobic interactions are responsible for ct-DNA compaction.
4. Conclusion
Our research has revealed that among different types of surfactants, DDAB18, in the presence of silica nanoparticles, serves as an exceptional surfactant for ct-DNA compaction. DDAB18 can undergo fifty percent ct-DNA compaction at a concentration about ninety-nine percent lower than that required for conventional cationic surfactants, DTAB, in the presence of silica nanoparticles (∼100 nm), which addresses the cytotoxicity issues associated with cationic surfactants. This concentration is the minimum compared to other surfactants studied by this group. As per DLS results, in this work, the minimum size of ct-DNA coated hybrid nanoparticles is found to be ∼125 nm in the presence of DDAB18 and silica nanoparticles (∼100 nm). While the size of native ct-DNA is noted to be ∼657 nm. The size of this compacted ct-DNA in the case of DDAB18 is the smallest compared to all other surfactants explored by the same group. Due to the crowding effect, the ct-DNA size is decreased in the presence of silica nanoparticles without any surfactant as well. Fluorescence lifetime data show that DDAB series-surfactants can displace almost all DAPI molecules from ct-DNA because of their strong interactions with nucleic acid. DDAB18 molecules bind in the intercalative region of ct-DNA. Fluorescence anisotropy decay data reveal two components responsible for depolarization in the presence of surfactants with and without silica nanoparticles. In contrast to the systems without surfactants, the fast rotational component is the major one in the systems with surfactants. The slow component is ascribed to the ct-DNA helix's segmental motions, which become slower at a high concentration of DDAB18 in contrast to other surfactants. This infers that at a high concentration of DDAB18, the ct-DNA segments become more rigid. However, in the presence of the same concentrations of DDAB16 and MQAS12, the segments’ rigidity is somewhat reduced. The segment's flexibility is found to be more in the case of the former than in the latter, and also in the systems with silica nanoparticles.In addition to presenting an efficient surfactant for ct-DNA compaction, this work explains the effects of the surfactants’ structure and nanoparticles’ size on ct-DNA compaction in the presence of silica nanoparticles of different sizes (∼100 and ∼40 nm). Electrostatic interactions between positively charged surfactants and negatively charged DNA are necessary. Still, the study shows that the hydrophobicity per unit surface area of the nanoparticles (P) has a great role in the compaction of DNA. DDAB18, among all surfactants, is the most efficient one as it introduces the maximum P because of its small headgroup size and two long hydrophobic tails attached to a single head. The size factors help many DDAB18 molecules to be adsorbed on the nanoparticles’ surfaces, as evidenced by EDAX and zeta potential data producing large P. The cell viability of 4T1 and HEK293 cell lines and in vitro cellular uptake of the ct-DNA to 4T1 cells for the system with DDAB18 surfactant have been carried out, and promising results have been obtained. The present work thus gives a direction for designing and developing potential cationic surfactant-based gene delivery systems in the presence of nanoparticles. DDAB18-coated silica nanoparticles appear to be safe and effective DNA compaction agents that can carry nucleic acid for biomedical applications.
Author contributions
Shalini Dyagala – conceptualization, methodology, investigation, writing original draft. Milan Paul – biological work investigation. Vinod Kumar Aswal – validation. Swati Biswas – biological work supervision. Subit K. Saha – conceptualization, supervision, visualization, project administration, writing review & editing, resources, funding acquisition.Data availability
(1) Data for this article, including the synthetic methods for the MQAS12 surfactant are available at https://doi.org/10.1021/la960669c. (2) Data for this article, including synthesis and characterization of silica nanoparticles and surfactants 1,4-bis(dodecyl-N,N-dimethylammonium bromide)butane (12-4-12,2Br-); 1,4-bis(dodecyl-N,N-dimethylammonium bromide)hexane (12-6-12,2Br-); and 1,4-bis(dodecyl-N,N-dimethylammonium bromide)octane (12-8-12,2Br-) are available at https://doi.org/10.1021/acsabm.3c00256, https://doi.org/10.1021/acsabm.3c00470. (3) The data supporting this article have been included as a part of the ESI.† (4) No software or code has been included as a part of this paper.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was partially carried out using the facilities of UGC-DAE CSR. The authors acknowledge the financial support from UGC-DAE CSR through a Collaborative Research Scheme (CRS) project number CRS/2021-22/03/541. Swati Biswas acknowledges the Indian Council of Medical Research (ICMR) for supporting research through a grant (No. 2021-8945/F1). The authors are very grateful for the Central Analytical Laboratory, Birla Institute of Technology and Science (BITS), Pilani, Hyderabad Campus, India, for providing the instrumentation facilities for different characterizations. S. K. S. thanks the Department of Science and Technology (DST) FIST program, Government of India. S. D. earnestly acknowledges Birla Institute of Technology & Science (BITS), Pilani – Hyderabad campus for the constant financial assistance.References
- M. Ramamoorth and A. Narvekar, J. Clin. Diagn. Res., 2015, 9, 1–6 CrossRef PubMed
.
- P. Carrivain, A. Cournac, C. Lavelle, A. Lesne, J. Mozziconacci, F. Paillusson, L. Signon, J. M. Victor and M. Barbi, Soft Matter, 2012, 8, 9285–9301 RSC
.
- M. E. Dowty, P. Williams, G. Zhang, J. E. Hagstrom and J. A. Wolff, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 4572–4576 CrossRef CAS PubMed
.
- K. Lundstrom, Viruses, 2023, 15, 674–698 CrossRef
.
- A. Fus-Kujawa, P. Prus, K. Bajdak-Rusinek, P. Teper, K. Gawron, A. Kowalczuk and A. L. Sieron, Front. Bioeng. Biotechnol., 2021, 9, 701031 CrossRef
.
- S. U. Khan, M. U. Khan, M. I. Khan, F. Kalsoom and A. Zahra, Curr. Gene Ther., 2023, 23, 135–147 CrossRef CAS PubMed
.
- M. S. Al-Dosari and X. Gao, AAPS J., 2009, 11, 671–681 CrossRef CAS PubMed
.
- A. Rodriguez, A. Del and M. Angeles, Langmuir, 2013, 1, 1–32 Search PubMed
.
- J. T. McGinley, Y. Wang, I. C. Jenkins, T. Sinno and J. C. Crocker, ACS Nano, 2015, 11, 10817–10825 CrossRef
.
- C. L. Porter and J. C. Crocker, Curr. Opin. Colloid Interface Sci., 2017, 30, 34–44 CrossRef CAS
.
- J. Moon, I. S. Jo, E. Ducrot, J. S. Oh, D. J. Pine and G. R. Yi, Macromol. Res., 2018, 12, 1085–1094 CrossRef
.
- F. Cui, S. Marbach, J. A. Zheng, M. Holmes-Cerfon and D. J. Pine, Nat. Commun., 2022, 1, 2304–2319 CrossRef PubMed
.
- M. Cárdenas, T. Nylander, R. K. Thomas and B. Lindman, Langmuir, 2005, 21, 6495–6502 CrossRef PubMed
.
- M. Ud Din Parray, N. Maurya, F. A. Wani, M. S. Borse, N. Arfin, M. A. Malik and R. Patel, J. Mol. Struct., 2019, 1175, 49–55 CrossRef CAS
.
- F. A. Wani, K. Behera, R. A. Padder, M. Husain, M. A. Malik, N. S. Al-Thabaiti, R. Ahmad and R. Patel, Colloid Interface Sci. Commun., 2020, 34, 100221 CrossRef CAS
.
- F. A. Wani, Amaduddin, B. Aneja, G. Sheehan, K. Kavanagh, R. Ahmad, M. Abid and R. Patel, ACS Omega, 2019, 7, 11871–11879 CrossRef PubMed
.
- S. G. Silva, I. S. Oliveira, M. L. C. do Vale and E. F. Marques, Soft Matter, 2014, 10, 9352–9361 RSC
.
- D. Halder and P. Purkayastha, Soft Matter, 2022, 18, 938–942 RSC
.
- R. D. Henderson, C. T. Filice, S. Wettig and Z. Leonenko, Soft Matter, 2021, 17, 826–833 RSC
.
- M. Muñoz-Úbeda, A. Rodríguez-Pulido, A. Nogales, O. Llorca, M. Quesada-Pérez, A. Martín-Molina, E. Aicart and E. Junquera, Soft Matter, 2011, 7, 5991–6004 RSC
.
- R. M. Uda and T. Matsui, Soft Matter, 2015, 11, 8246–8252 RSC
.
- F. M. Menger and C. A. Littau, J. Am. Chem. Soc., 1993, 115, 10083–10090 CrossRef CAS
.
- R. S. Dias, L. M. Magno, A. J. M. Valente, D. Das, P. K. Das, S. Maiti, M. G. Miguel and B. Lindman, J. Phys. Chem. B, 2008, 112, 14446–14452 CrossRef CAS PubMed
.
- F. A. Wani, R. Ahmad and R. Patel, Ind. Eng. Chem. Res., 2020, 37, 16283–16295 CrossRef
.
- T. Sharma, N. Dohare, M. Kumari, U. K. Singh, A. B. Khan, M. S. Borse and R. Patel, RSC Adv., 2017, 7, 16763–16776 RSC
.
- V. Jadhav, S. Maiti, A. Dasgupta, P. K. Das, R. S. Dias, M. G. Miguel and B. Lindman, Biomacromolecules, 2008, 9, 1852–1859 CrossRef CAS PubMed
.
- S. Rudiuk, K. Yoshikawa and D. Baigl, Soft Matter, 2011, 7, 5854–5860 RSC
.
- S. Rudiuk, K. Yoshikawa and D. Baigl, J. Colloid Interface Sci., 2012, 368, 372–377 CrossRef CAS PubMed
.
- L. Karlsson, M. C. P. Van Eijk and O. Soderman, J. Colloid Interface Sci., 2002, 252, 290–296 CrossRef CAS
.
- S. Dyagala, M. Paul, V. K. Aswal, S. Biswas and S. K. Saha, ACS Appl. Bio Mater., 2023, 6, 3848–3862 CrossRef CAS PubMed
.
- S. Halder, M. Paul, S. Dyagala, R. Aggrawal, V. K. Aswal, S. Biswas and S. K. Saha, ACS Appl. Bio Mater., 2023, 6, 2795–2815 CrossRef CAS
.
- L. M. Wu, L. Lai, P. Mei, L. Cheng, Y. Q. Wang and Y. Liu, J. Mol. Liq., 2020, 311, 113323 CrossRef CAS
.
- L. D. Hamilton, R. K. Barclay, M. H. F. Wilkins, G. L. Brown, H. R. Wilson, D. A. Marvin, H. Ephrussi-Taylor and N. S. Simmons, J. Cell Biol., 1959, 5, 397–404 CrossRef CAS
.
- T.-S. Kim, T. Kida, Y. Nakatsuji and I. Ikeda, Langmuir, 1996, 12, 6304–6308 CrossRef CAS
.
- S. Halder, R. Aggrawal, S. Jana and S. K. Saha, J. Photochem. Photobiol., B, 2021, 225, 112351 CrossRef CAS PubMed
.
- R. Aggrawal, S. Halder, S. Dyagala and S. K. Saha, RSC Adv., 2022, 12, 16014–16028 RSC
.
- A. B. Davila-Ibanez, V. Salgueirino, V. Martinez-Zorzano, R. Mariño-Fernández, A. García-Lorenzo, M. Maceira-Campos, M. Muñoz-Ubeda, E. Junquera, E. Aicart, J. Rivas, F. J. Rodriguez-Berrocal and J. L. Legido, ACS Nano, 2012, 6, 747–759 CrossRef CAS
.
- T. Chaudhuri, A. Pan, S. Das and S. P. Moulik, J. Surfactants Deterg., 2018, 21, 127–137 CrossRef CAS
.
- T. Sarwar, S. U. Rehman, M. A. Husain, H. M. Ishqi and M. Tabish, Int. J. Biol. Macromol., 2015, 73, 9–16 CrossRef CAS PubMed
.
- Z.-G. Cui, L.-L. Yang, Y.-Z. Cui and B. P. Binks, Langmuir, 2010, 26, 4717–4724 CrossRef CAS PubMed
.
- A. Manna and S. Chakravorti, J. Phys. Chem. B, 2012, 116, 5226–5233 CrossRef CAS
.
- M. L. Barcellona and E. Gratton, Eur. Biophys. J., 1990, 17, 315–323 CrossRef CAS
.
-
J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, US, 2006 Search PubMed
.
- D. Mathur, Y. C. Kim, S. A. Díaz, P. D. Cunningham, B. S. Rolczynski, M. G. Ancona, I. L. Medintz and J. S. Melinger, J. Phys. Chem. C, 2021, 125, 1509–1522 CrossRef CAS
.
- G. Krishnamoorthy, G. Duportail and Y. Mély, Biochemistry, 2002, 41, 15277–15287 CrossRef CAS PubMed
.
- H. Sehrawat, N. Kumar, S. Panchal, L. Kumar and R. Chandra, Int. J. Biol. Macromol., 2022, 220, 415–425 CrossRef CAS PubMed
.
- R. Aggrawal, S. Halder, B. Gopalan, S. Biswas and S. K. Saha, J. Phys. Chem. C, 2022, 126, 6280–6299 CrossRef CAS
.
- A. Mahata, D. Sarkar, D. Bose, D. Ghosh, A. Girigoswami, P. Das and N. Chattopadhyay, J. Phys. Chem. B, 2009, 113, 7517–7526 CrossRef CAS PubMed
.
- S. Ramadurai, A. Holt, V. Krasnikov, G. Van Den Bogaart, J. A. Killian and B. Poolman, J. Am. Chem. Soc., 2009, 131, 12650–12656 CrossRef CAS PubMed
.
- M. H. Kombrabail and G. Krishnamoorthy, J. Fluoresc., 2005, 15, 741–747 CrossRef CAS PubMed
.
- A. Zinchenko, K. Tsumoto, S. Murata and K. Yoshikawa, J. Phys. Chem. B, 2014, 118, 1256–1262 CrossRef CAS PubMed
.
- D. J. Mc Clements, J. Agric. Food Chem., 2000, 48, 5604–5611 CrossRef CAS PubMed
.
- A. Estévez-Torres and D. Baigl, Soft Matter, 2011, 7, 6746–6756 RSC
.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details including methods, characterization data of the synthesized MQAS12 surfactant, and additional figures and tables related to the EDAX analysis, zeta potential, steady state fluorescence spectra of EtBr, fluorescence intensity decays of DAPI, fluorescence anisotropy decays of DAPI, FE-SEM data, DLS data, and fluorescence microscopic data. See DOI: https://doi.org/10.1039/d4sm01345j |
| This journal is © The Royal Society of Chemistry 2025 |