Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploration of lignin-binding synthetic polymers with pendant hydrophobic amino acids†
Tomonari
Tanaka
 *a,
Rika
Hinohara
a,
Oscar Abraham
Carias Duron
a,
Yuji
Aso
*a,
Rika
Hinohara
a,
Oscar Abraham
Carias Duron
a,
Yuji
Aso
 a,
Naoko
Kobayashi
a,
Naoko
Kobayashi
 b,
Kaori
Saito
b and
Takashi
Watanabe
b,
Kaori
Saito
b and
Takashi
Watanabe
 b
b
aDepartment of Biobased Materials Science, Graduate School of Science and Technology, Kyoto Institute of Technology, Matsugasaki, Sakyo-ku, Kyoto 606-8585, Japan. E-mail: t-tanaka@kit.ac.jp
bBiomass Product Tree Industry-Academia Collaborative Research Unit, Research Institute for Sustainable Humanosphere, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan
First published on 1st November 2024
Abstract
Polymers bearing amino acid methyl esters with a terminus fluorescein group were synthesized through controlled radical polymerization employing a fluorescein-labeled chain transfer agent and subsequent post-polymerization modification with amino acid methyl esters. The binding properties of polymers bearing hydrophobic amino acids with milled wood lignin in water were investigated through spectroscopic analysis as screening tests and surface plasmon resonance (SPR) analysis as quantitative measurements. The polymers bearing tryptophan and phenylalanine moieties were observed to exhibit a high binding affinity. The binding parameters of polymers bearing tryptophan and phenylalanine moieties with wood lignin were determined using SPR.
Sustainability spotlightLignin is a primary component of plant cell walls along with polysaccharides such as cellulose and hemicelluloses, and it is an aromatic heteropolymer that constitutes woody biomass. Lignin degradation is vital in lignocellulosic biorefineries. However, it is challenging because of the stability of its chemically and physically aromatic-rich structure. The lignin-binding synthetic polymers developed in this study can be conjugated with lignin degradation enzymes and catalysts to increase their affinity for lignin and further improve their catalytic activity, thus contributing to the advancement of lignin degradation technology. This work contributes to the following UN sustainable development goals: industry, innovation and infrastructure (SDG 9) and responsible consumption and production (SDG 12). |
Introduction
Lignin is an aromatic heteropolymer that constitutes woody biomass, and it is a primary component of plant cell walls along with polysaccharides such as cellulose and hemicelluloses.1 The monomers that constitute lignin are collectively known as monolignols; they include coniferyl alcohol, 4-hydroxycinnamyl alcohol, and sinapyl alcohol and are connected by ether and carbon–carbon bonds.2 The removal of lignin is essential in refining cellulose and pulp from wood. Further, lignin degradation is vital in lignocellulosic biorefineries; however, this is challenging because of the stability of its chemically and physically aromatic-rich structure. The thermal degradation of lignin can yield valuable chemicals but necessitates high temperatures, often resulting in carbonization and the release of volatile byproducts.3–6 Laccases and peroxidases are lignin degradation enzymes.7–11 Recently, alternative enzyme-mimicking catalysts, such as self-assembled peptides and artificial metalloenzymes, have been reported; however, they cannot degrade under mild conditions.12–14 Effective lignin degradation relies on the binding affinity of enzymes and catalysts to the lignin. Thus, the pursuit of molecules capable of binding to wood lignin is pivotal. A lignin-binding peptide (amino acid sequence: HFPSPIFQRHSH) has been found using a phage display technique, and a 10−4 molar-range of the equilibrium dissociation constant (KD) value with wood lignin has been reported.15 Moreover, its dimer exhibits strong binging to wood lignin with a 10−5 molar-range of the KD value.16 The molecular modeling of the lignin-binding peptide suggests the formation of a hydrophobic surface comprising side chains of hydrophobic and aromatic amino acids. These reports have led us to hypothesize that a synthetic polymer bearing hydrophobic amino acids effectively binds to wood lignin through a clustering of amino acids on a polymer backbone. In this study, polymers bearing hydrophobic amino acid methyl esters are synthesized, and their binding affinities with wood lignin have been investigated. Two binding tests are performed to assess the binding properties with wood lignin: the spectroscopic analysis of sample solutions and molecular interaction measurements using surface plasmon resonance (SPR).Results and discussion
Synthesis of amino acid-bearing polymers
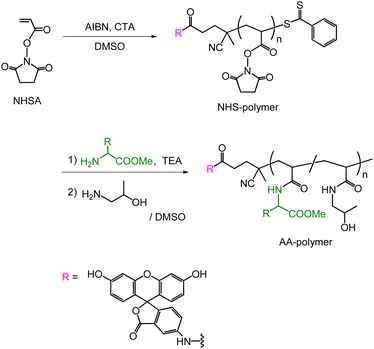
The synthetic procedure for polymers bearing amino acid methyl esters (AA-polymers) is presented in Scheme 1. The reversible addition–fragmentation chain-transfer (RAFT) polymerization17,18 of the activated ester-bearing monomer, N-succinimidyl acrylate (NHSA), was performed using fluorescein-labeled 4-cyano-4-(phenylcarbonothioylthio)pentanoic acid as the chain-transfer agent (CTA) in dimethyl sulfoxide (DMSO). A polymer bearing N-hydroxysuccinimide (NHS) groups (NHS-polymer) with a degree of polymerization of 82 and a terminus fluorescein group was obtained. Subsequently, the desired AA-polymers were synthesized through the post-polymerization modification19–22 of the NHS-polymer reacting with amino acid methyl ester, followed by a reaction with 1-amino-2-propanol (AP) to substitute the remaining NHS groups on the polymer backbone. Table 1 summarizes the synthesis results of the AA-polymers. By changing the type of amino acid methyl esters, eight different types of AA-polymers with 22–29% degree of amino acid substitution were obtained. Six kinds of methyl esters of hydrophobic amino acids (valine (Val), leucine (Leu), isoleucine (Ile), phenylalanine (Phe), tryptophan (Trp), and tyrosine (Tyr)) were used (entries 4–9). For comparison, polymers without amino acid and bearing methyl esters of glycine (Gly) and alanine (Ala) were synthesized (entries 1–3).| Entry | AA-polymer | Amino acida | Yield/% | DSb/% |
|---|---|---|---|---|
| a 0.3 equivalence of amino acid methyl ester was fed against NHS esters at the polymer side chains. b Degree of amino acid substitution determined by 1H NMR. c No amino acids are present at the polymer side chains. | ||||
| 1 | No AA | — | 58 | 0 |
| 2 | Gly29 | Gly | 41 | 29 |
| 3 | Ala28 | Ala | 44 | 28 |
| 4 | Val26 | Val | 66 | 26 |
| 5 | Leu29 | Leu | 88 | 29 |
| 6 | Ile25 | Ile | 60 | 25 |
| 7 | Phe27 | Phe | 75 | 27 |
| 8 | Trp29 | Trp | 96 | 29 |
| 9 | Tyr22 | Tyr | 50 | 22 |
Screening tests for binding affinity with wood lignin
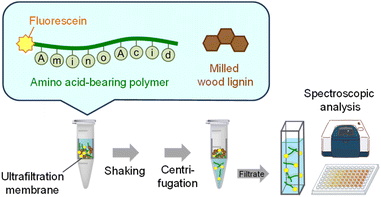
The binding properties of the AA-polymers to milled wood lignin derived from Japanese cedar (CMWL) were investigated in water. First, a simple spectroscopic analysis was developed to qualitatively assess the lignin–polymer binding affinity. After mixing a polymer with CMWL in water for 24 h at room temperature, the mixture was subjected to ultrafiltration to remove the polymer–lignin conjugates. The differences in the visible light absorbance (ΔA) and fluorescence intensity (ΔF) of the filtrate, which contained a free fluorescein-labeled AA-polymer, were measured before and after mixing with CMWL (Fig. 1). When Phe27 was assayed with CMWL, both ΔA and ΔF values were approximately 80% (Fig. 2). However, no changes in ΔA and ΔF were observed without CMWL. Additionally, no changes were observed when the polymer without amino acid (No AA) was assayed, indicating that 80% of Phe27 bound to the lignin, which is consistent with the results from both visible light and fluorescence spectroscopic analyses.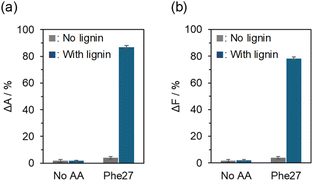 | ||
| Fig. 2 Binding tests with CMWL by analyzing the changes in the (a) visible light absorbance and (b) fluorescence intensity. | ||
Second, the binding properties of eight AA-polymers were investigated by analyzing the changes in visible light absorbance (Fig. 3a). When binding tests using six polymers bearing hydrophobic amino acids (Val26, Leu29, Ile25, Phe27, Trp29, and Tyr22) were performed, ΔA values of over 50% were observed. Particularly, Phe27 and Trp29 obtained relatively high ΔA values. In contrast, no change in absorbance was observed for No AA, and low ΔA values were observed for Gly29 and Ala28. These results were plotted against the hydropathy index of amino acids23 (Fig. 3b). Phe27 and Trp29 bearing hydrophobic aromatic amino acids revealed high binding affinities. In nonaromatic amino acids, the binding affinity of Ile25 was higher than that of Val26 and Leu29, suggesting that the high hydropathy index of Ile was critical. Additionally, Tyr22 revealed a relatively high binding affinity, suggesting that the presence of the aromatic group was efficient. In summary, high hydrophobicity or aromatic groups clustered at polymer side chains are essential for a high binding affinity with wood lignin.
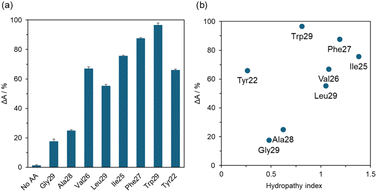 | ||
| Fig. 3 (a) Binding tests with CMWL by analyzing the changes in visible light absorbance and (b) their plots against the hydropathy index of amino acids. | ||
Lignin-binding analysis using SPR
The binding affinities of the AA-polymers with CMWL were quantitatively investigated via SPR measurements24,25 in water. A high response change was observed when Trp29 was analyzed using the CMWL-immobilized SPR sensor chip (Fig. 4a). Medium response changes were observed when Phe27 and IIe25 were analyzed, and there were little to no changes when Gly29, Ala28, and No AA were used as analytes. These results are consistent with those obtained from the spectroscopic binding tests. Further, tryptophan methyl ester (TrpOMe) and phenylalanine methyl ester (PheOMe), which were not attached to polymers, were used as the analytes in the same concentration as the amino acids at the polymer side chains. However, there were fewer changes in the responses than those of the polymers (Fig. 4b). This indicates that the clustering of the hydrophobic aromatic amino acid results in a high binding affinity of the AA-polymers to CMWL because of the multivalent π–π stacking. For Ile and Leu, there were little differences in the response between the polymer and the amino acid methyl ester.Finally, the KD values of Trp29 and Phe27 with CWML in water were determined by equilibrium analysis, with varied concentrations of the AA-polymers (Fig. 5). The equilibrium response increased with high concentrations of AA-polymers. Based on these SPR results, the maximum resonance (Rmax) and KD values of the AA-polymers with CMWL were calculated and are summarized in Table 2. The Rmax value of Trp29 was higher than that of Phe27. Thus, the KD value of Trp29 was lower than that of Phe27 and was in the micromolar range, suggesting that the binding affinity of Trp29 with CMWL was higher than that of Phe27. This indicates that clustering aromatic and hydrophobic amino acids results in a high binding affinity to wood lignin in water.
| AA-polymer | R max/RU | K D/μM |
|---|---|---|
| Trp29 | 255 ± 18 | 0.616 ± 0.23 |
| Phe27 | 110 ± 6.1 | 1.26 ± 0.44 |
Experimental
Materials
N-Succinimidyl acrylate (NHSA), 5-amino fluorescein, glycine methyl ester hydrochloride, L-alanine methyl ester hydrochloride, L-valine methyl ester hydrochloride, L-leucine methyl ester hydrochloride, L-tryptophan methyl ester hydrochloride, and L-tyrosine methyl ester hydrochloride were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). α,α′-Azobisisobutyronitrile was purchased from Nacalai Tesque Inc. (Kyoto, Japan). L-Isoleucine methyl ester hydrochloride, L-phenylalanine methyl ester hydrochloride, triethylamine (TEA), and 1-amino-2-propanol (AP) were purchased from Fujifilm Wako Pure Chemical Industries (Osaka, Japan). 4-Cyano-4-(phenylcarbonothioylthiol)pentanoic acid N-succinimidyl ester and poly(ethylene glycol)methyl ether thiol (PEG-SH, average molecular weight: 6000) were purchased from Sigma-Aldrich Co., LLC. (St Louis, USA). Milled wood lignin was prepared by aqueous dioxane extraction from finely ball-milled softwood Japanese cedar (CMWL) as previously described.26 All organic solvents were commercially available and used without further purification.Measurements
Nuclear magnetic resonance (NMR) spectra were recorded using Bruker (MA, USA) BioSpin AV-300 and AV-600 spectrometers. Electrospray ionization mass spectrometry was performed using a Bruker Daltonics micrOTOF Q-III spectrometer. Size exclusion chromatography (SEC) analysis utilized a system comprising a JASCO (Tokyo, Japan) PU-4180 pump, a CO-4060 column oven, and an RI-4030 refractive index detector. A Shodex (RESONAC (Tokyo, Japan)) KD-804 column (ϕ 8.0 mm, 300 mm) was employed, with 10 mM LiBr/N,N-dimethyl formamide (DMF) as the eluent at a flow rate of 0.5 mL min−1 at 50 °C. Shodex poly(methyl methacrylate) standard samples were used to determine molecular weights. Visible light absorbance was measured using a Thermo Fisher Scientific K.K. (MA, USA) microplate spectrophotometer, Multiscan SkyHigh, on a 96-well plate. Fluorescent intensity was recorded using a JASCO FP-6500 spectrometer. SPR analysis was conducted using a Biacore (Uppsala, Sweden) T200 instrument.Synthesis of fluorescein-bearing CTA
A solution of 4-cyano-4-(phenylcarbonothioylthiol)pentanoic acid N-succinimidyl ester (100 mg, 0.265 mmol) and 5-amino fluorescein (184 mg, 0.530 mmol) in DMF (1.0 mL) was stirred for 18 h at room temperature. After concentrating the reaction mixture under reduced pressure, the residue was purified by silica gel column chromatography (hexane/ethyl acetate = 1/2), concentrated, and dried under reduced pressure to yield fluorescein-bearing CTA (18.4 mg, 0.0303 mmol, 11% yield).
1H NMR (300 MHz, DMSO-d6, δ (ppm)): 10.1 (s, 1H, amide), 7.9 (d, J = 7.5 Hz, 2H, Ph), 7.7 (t, J = 7.3 Hz, 1H, Ph), 7.5 (d, J = 7.8 Hz, 2H, Ph), 7.2 (d, J = 2.1 Hz, 1H, Ph of fluorescein), 6.8–7.0 (m, 6H, Ph of fluorescein), 6.6–6.7 (m, 2H, Ph of fluorescein), 5.8 (s, 2H, OH), 3.0 (t, J = 6.6 Hz, 2H, CH2), 2.6–2.7 (m, 2H, CH2), 2.0 (s, 3H, CH3); 13C NMR (75 MHz, DMSO-d6, δ (ppm)): 169.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of amide), 169.2 (C
O of amide), 169.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of ester), 159.4 and 151.6 (
O of ester), 159.4 and 151.6 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH in Ph), 151.4 and 151.2 (
–OH in Ph), 151.4 and 151.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –O–
–O–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) in Ph), 150.7 (
in Ph), 150.7 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –C(PhOH)2O in Ph), 144.0 (
–C(PhOH)2O in Ph), 144.0 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –C
–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S in Ph), 139.4 (C–NH in Ph), 133.6, 129.1, 126.5, and 124.2 (Ph), 121.8 (C
S in Ph), 139.4 (C–NH in Ph), 133.6, 129.1, 126.5, and 124.2 (Ph), 121.8 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 118.6, 117.9, 117.8, 112.9, 110.2, 110.1, 106.2, and 102.1 (Ph), 81.6 (O–
N), 118.6, 117.9, 117.8, 112.9, 110.2, 110.1, 106.2, and 102.1 (Ph), 81.6 (O–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) (Ph)3), 45.9 (
(Ph)3), 45.9 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) (CN)CH3–S), 32.0 (–
(CN)CH3–S), 32.0 (–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2–C(CN)CH3–S), 29.3 (–CH2–
H2–C(CN)CH3–S), 29.3 (–CH2–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2–C
H2–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 23.1 (CH3); ESI-MS m/z: [M–H]–, calcd for C33H24N2O6S2, 607.10; found, 607.07.
O), 23.1 (CH3); ESI-MS m/z: [M–H]–, calcd for C33H24N2O6S2, 607.10; found, 607.07.
Synthesis of the NHS-polymer
A solution of NHSA (50 mg, 0.6 mmol), AIBN (0.48 mg, 6 μmol), and fluorescein-bearing CTA (3.6 mg, 12 μmol) in DMSO (0.6 mL) was prepared in a glass tube, which was degassed over three freeze–thaw cycles. The solution in the glass tube was then sealed under vacuum and stirred for 6 h at 65 °C. The resulting polymer product was purified by reprecipitation in acetone and dried under reduced pressure to yield PNHSA (30 mg, 60%).
1H NMR (300 MHz, DMSO-d6, δ (ppm)): 8.0–6.6 (Ar), 3.3–3.1 ((–CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –)n), 3.0–2.7 (CH2 of NHS), 2.4–1.7 ((–C
–)n), 3.0–2.7 (CH2 of NHS), 2.4–1.7 ((–C –CH–)n).
–CH–)n).
Synthesis of amino acid-bearing polymers
A solution of NHS-polymer (10 mg) in DMSO (0.1 mL), along with 0.3 equivalence of amino acid methyl ester hydrochloride against NHS esters at the polymer side chains and TEA (same molar ratio as amino acid methyl ester hydrochloride), was stirred for 24 h at room temperature. Subsequently, AP (twice the molar amount of NHS esters) was introduced to the mixture and stirred for another 24 h at room temperature. The resulting AA-polymers were then purified by dialysis (using Spectra/Por7 with a molecular weight cut-off of 3500) against two liters of deionized water, repeated three times, followed by freeze-drying to obtain AA-polymers.No AA
1H NMR (600 MHz, D2O, δ (ppm)): 4.0–3.7 (CH of AP), 3.3–2.8 (CH2 of AP), 2.3–1.8 ((–CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –)n), 1.8–1.3 ((–C
–)n), 1.8–1.3 ((–C –CH–)n), 1.2–1.0 (CH3 of AP).
–CH–)n), 1.2–1.0 (CH3 of AP).
Gly29
1H NMR (600 MHz, D2O, δ (ppm)): 4.1–3.7 (CH2 of Gly, CH3 of COOMe, and CH of AP), 3.3–2.9 (CH2 of AP), 2.3–1.8 ((–CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –)n), 1.8–1.3 ((–C
–)n), 1.8–1.3 ((–C –CH–)n), 1.2–1.0 (CH3 of AP).
–CH–)n), 1.2–1.0 (CH3 of AP).
Ala28
1H NMR (600 MHz, D2O, δ (ppm)): 4.4–4.2 (CH of Ala), 4.1–3.7 (CH3 of COOMe and CH of AP), 3.3–2.9 (CH2 of AP), 2.3–1.8 ((–CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –)n), 1.8–1.4 ((–C
–)n), 1.8–1.4 ((–C –CH–)n), 1.4–1.3 (CH3 of Ala), 1.2–1.0 (CH3 of AP).
–CH–)n), 1.4–1.3 (CH3 of Ala), 1.2–1.0 (CH3 of AP).
Val26
1H NMR (600 MHz, D2O, δ (ppm)): 4.3–4.0 (N–CH of Val), 4.1–3.7 (CH3 of COOMe and CH of AP), 3.3–2.9 (CH2 of AP), 2.4–1.9 (C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) (CH3)2 of Val and (–CH2–C
(CH3)2 of Val and (–CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –)n), 1.8–1.4 ((–C
–)n), 1.8–1.4 ((–C –CH–)n), 1.2–1.0 (CH3 of AP), 1.0–0.8 (CH3 of Val).
–CH–)n), 1.2–1.0 (CH3 of AP), 1.0–0.8 (CH3 of Val).
Leu29
1H NMR (600 MHz, D2O, δ (ppm)): 4.5–4.2 (N–CH of Leu), 4.0–3.7 (CH3 of COOMe and CH of AP), 3.3–2.9 (CH2 of AP), 2.3–1.8 (CH2 of Leu and (–CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –)n), 1.8–1.4 (C
–)n), 1.8–1.4 (C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) (CH3)2 of Leu and (–C
(CH3)2 of Leu and (–C –CH–)n), 1.3–1.0 (CH3 of AP), 1.0–0.8 (CH3 of Leu).
–CH–)n), 1.3–1.0 (CH3 of AP), 1.0–0.8 (CH3 of Leu).
Ile25
1H NMR (600 MHz, D2O, δ (ppm)): 4.0–3.7 (CH3 of COOMe and CH of AP), 3.3–2.8 (CH2 of AP and Ch of Ile), 2.3–1.8 ((–CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –)n), 1.8–1.4 (CH2 of Ile and (–C
–)n), 1.8–1.4 (CH2 of Ile and (–C –CH–)n), 1.3–1.0 (CH3 of AP), 1.0–0.8 (CH3 of Ile).
–CH–)n), 1.3–1.0 (CH3 of AP), 1.0–0.8 (CH3 of Ile).
Phe27
1H NMR (600 MHz, D2O, δ (ppm)): 7.4–7.0 (Ph), 4.1–3.7 (CH3 of COOMe and CH of AP), 3.3–2.9 (CH2 of Phe and AP), 2.3–1.9 ((–CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –)n), 1.8–1.3 ((–C
–)n), 1.8–1.3 ((–C –CH–)n), 1.2–1.0 (CH3 of AP).
–CH–)n), 1.2–1.0 (CH3 of AP).
Trp29
1H NMR (600 MHz, D2O, δ (ppm)): 7.7–6.7 (Ar), 4.0–3.7 (CH3 of COOMe and CH of AP), 3.3–2.9 (CH2 of Trp and AP), 2.3–1.9 ((–CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –)n), 1.8–1.3 ((–C
–)n), 1.8–1.3 ((–C –CH–)n), 1.2–1.0 (CH3 of AP).
–CH–)n), 1.2–1.0 (CH3 of AP).
Tyr22
1H NMR (600 MHz, D2O, δ (ppm)): 7.2–6.6 (Ph), 4.1–3.7 (CH3 of OMe and CH of AP), 3.3–2.9 (CH2 of Tyr and AP), 2.3–1.9 ((–CH2–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) –)n), 1.8–1.3 ((–C
–)n), 1.8–1.3 ((–C –CH–)n), 1.2–1.0 (CH3 of AP).
–CH–)n), 1.2–1.0 (CH3 of AP).
Lignin binding test using spectrometers
A solution containing AA-polymer (2 mg) in water (0.5 mL), along with a DMSO solution (0.1 mL) of CMWL (1 mg), was stirred for 24 h at room temperature. Subsequently, the solution was filtered through an ultrafiltration membrane (Amicon Ultra-0.5 100 kDa). The resulting filtrate was analyzed using a microplate spectrophotometer at 490 nm, as well as a fluorescence spectrometer with excitation at 495 nm and emission at 520 nm.Lignin binding test using SPR
After thoroughly washing a SPR sensor chip (SIA Kit Au) three times with piranha solution (a mixture of H2SO4 and 30% H2O2 in a 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio), an aqueous solution of PEG-SH (1 mM) was applied onto the chip surface and allowed to incubate for 2 h. Following this, the chip was rinsed with Milli-Q water, and a solution of CMWL in DMSO (10 mg mL−1) was applied onto the chip surface and allowed to incubate for 0.5 h. After removing the DMSO solution of CMWL from the sensor chip, a new solution of CMWL in DMSO was applied onto the sensor chip. This procedure for immobilizing lignin was repeated three times. During the association phase, a solution of AA-polymer (0.2 mg mL−1) was injected, and during the dissociation phase, Milli-Q water was flowed through. To regenerate the sensor chip, a 10 mM aqueous solution of glycine-HCl (pH 2.0) was injected for 5 min. Throughout the measurement, the flow rate was maintained at 30 μL min−1. Amino acid methyl esters were injected as analytes at the same concentration as the amino acid methyl esters of the polymer side chains. The KD values were determined using equilibrium response data obtained by varying the concentration of analytes in a Microsoft Excel file containing macro programs for nonlinear regression of the following equation, which automatically calculated the parameters by minimizing standard error.27 The KD is defined as the concentration of analyte at which the resonance is half of Rmax.
3 ratio), an aqueous solution of PEG-SH (1 mM) was applied onto the chip surface and allowed to incubate for 2 h. Following this, the chip was rinsed with Milli-Q water, and a solution of CMWL in DMSO (10 mg mL−1) was applied onto the chip surface and allowed to incubate for 0.5 h. After removing the DMSO solution of CMWL from the sensor chip, a new solution of CMWL in DMSO was applied onto the sensor chip. This procedure for immobilizing lignin was repeated three times. During the association phase, a solution of AA-polymer (0.2 mg mL−1) was injected, and during the dissociation phase, Milli-Q water was flowed through. To regenerate the sensor chip, a 10 mM aqueous solution of glycine-HCl (pH 2.0) was injected for 5 min. Throughout the measurement, the flow rate was maintained at 30 μL min−1. Amino acid methyl esters were injected as analytes at the same concentration as the amino acid methyl esters of the polymer side chains. The KD values were determined using equilibrium response data obtained by varying the concentration of analytes in a Microsoft Excel file containing macro programs for nonlinear regression of the following equation, which automatically calculated the parameters by minimizing standard error.27 The KD is defined as the concentration of analyte at which the resonance is half of Rmax.| Req = (Rmax [analyte])/(KD + [analyte]) |
Conclusions
We reported that synthetic polymers bearing hydrophobic and aromatic amino acid methyl esters strongly bound to wood lignin. Their lignin-binding properties were investigated through spectroscopic analysis and SPR measurements. The binding affinities of the AA-polymers were evaluated by monitoring the changes in visible light absorbance in the filtrate obtained from the polymer and milled wood lignin mixture. The binding parameters of polymers bearing tryptophan methyl esters and phenylalanine methyl esters with milled wood lignin were determined using SPR measurements, revealing KD values in the micromolar range. The clustered hydrophobic aromatic amino acid on the polymer results in a high binding affinity to wood lignin because of the multivalent π–π stacking. The density and orientation of aromatic rings are also suggestable.28 Conjugating lignin-binding synthetic polymers bearing amino acids with lignin degradation catalysts, such as enzymes and artificial catalysts, could enhance their affinity for wood lignin, consequently boosting their catalytic activity. Such polymers hold significant promise for advancing lignin degradation technology.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Tomonari Tanaka conceived the project, designed the experiments, directed the research, and wrote the paper. Rika Hinohara and Oscar Abraham Carias Duron synthesized and analyzed the polymers and other samples. Rika Hinohara, Naoko Kobayashi, and Kaori Saito prepared the lignin sample. Yuji Aso and Takashi Watanabe discussed the results and reviewed the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by Iketani Science and Technology Foundation and by the Mission 5-2 Research Grant of the Research Institute for Sustainable Humanosphere, Kyoto University. We thank Daicel corporation for their support.Notes and references
- C. Chio, M. Sain and W. Qin, Renewable Sustainable Energy Rev., 2019, 107, 232 CrossRef CAS.
- J. Ralph, C. Lapierre and W. Boerjan, Curr. Opin. Biotechnol., 2019, 56, 240 CrossRef CAS.
- M. Brebu and C. Vasile, Cellul. Chem. Technol., 2010, 44, 353 CAS.
- J. Zhao, W. Xiuwen, J. Hu, Q. Liu, D. Shen and R. Xiao, Polym. Degrad. Stab., 2014, 108, 133 CrossRef CAS.
- C. L. Waters, R. R. Janupala, R. G. Mallinson and L. L. Lobban, J. Anal. Appl. Pyrolysis, 2017, 126, 380 CrossRef CAS.
- H. Kawamoto, J. Wood Sci., 2017, 63, 117 CrossRef CAS.
- J. Zakzeski, P. C. A. Bruijnincx, A. L. Jongerius and B. M. Weckhuysen, Chem. Rev., 2010, 110, 3552 CrossRef CAS PubMed.
- L. Pollegioni, F. Tonin and E. Rosini, FEBS J., 2015, 282, 1190 CrossRef CAS PubMed.
- L. F. Longe, J. Couvreur, M. Leriche Grandchamp, G. Garnier, F. Allais and K. Saito, ACS Sustain. Chem. Eng., 2018, 6, 10097 CrossRef CAS.
- N. Kamimura, S. Sakamoto, N. Mitsuda, E. Masai and S. Kajita, Curr. Opin. Biotechnol., 2019, 56, 179 CrossRef CAS PubMed.
- M. Zhou, O. A. Fakayode, M. Ren, H. Li, J. Liang, A. E. A. Yagoub, Z. Fan and C. Zhou, Bioresour. Bioprocess., 2023, 10, 21 CrossRef PubMed.
- Q. Liu, A. Kuzuya and Z.-G. Wang, iScience, 2023, 26, 105831 CrossRef CAS PubMed.
- M. V. Doble, A. G. Jarvis, A. C. C. Ward, J. D. Colburn, J. P. Götze, M. Bühl and P. C. J. Kamer, ACS Sustain. Chem. Eng., 2018, 6, 15100 CrossRef CAS.
- T. Jian, Y. Zhou, P. Wang, W. Yang, P. Mu, X. Zhang, X. Zhang and C.-L. Chen, Nat. Commun., 2022, 13, 3025 CrossRef CAS PubMed.
- A. Yamaguchi, K. Isozaki, M. Nakamura, H. Takaya and T. Watanabe, Sci. Rep., 2016, 6, 21833 CrossRef CAS PubMed.
- S. Oshiro, A. Yamaguchi and T. Watanabe, RSC Adv., 2017, 7, 31338 RSC.
- X. Tian, J. Ding, Y. Chen, B. Zhang, F. Qiu, X. Zhuang and Y. Chen, Polymers, 2018, 10, 318 CrossRef PubMed.
- M. D. Nothling, Q. Fu, A. Reyhani, S. A. -Logan, K. Jung, J. Zhu, M. Kamigaito, C. Boyer and G. G. Qiao, Adv. Sci., 2020, 7, 2001656 CrossRef CAS.
- E. Blasco, M. B. Sims, A. S. Goldmann, B. S. Sumerlin and C. B. Kowollik, Macromolecules, 2017, 50, 5215 CrossRef CAS.
- T. Kubo, C. P. Easterling, R. A. Olson and B. S. Sumerlin, Polym. Chem., 2018, 9, 4605 RSC.
- X. Chen and T. Michinobu, Macromol. Chem. Phys., 2021, 223, 2100370 CrossRef.
- T. Tanaka, Polymers, 2024, 16, 1100 CrossRef CAS PubMed.
- D. Eisenberg, E. Schwarz, M. Komaromy and R. Wall, J. Mol. Biol., 1984, 179, 125 CrossRef CAS.
- Q. Wang, Z.-H. Ren, W.-M. Zhao, L. Wang, X. Yan, A.-s. Zhu, F.-m. Qiu and K.-K. Zhang, Nanoscale, 2022, 14, 564 RSC.
- A. Philip and A. R. Kumar, Coord. Chem. Rev., 2022, 458, 214424 CrossRef CAS.
- Y. Tokunaga, T. Nagata, T. Suetomi, S. Oshiro, K. Kondo, M. Katahira and T. Watanabe, Sci. Rep., 2019, 9, 1977 CrossRef.
- M. Kitaoka, J. Appl. Glycosci., 2023, 70, 33 CrossRef PubMed.
- Y. Tokunaga, T. Nagata, K. Kondo, M. Katahira and T. Watanabe, Biotechnol. Biofuels, 2020, 13, 164 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: SEC chromatogram and NMR spectra. See DOI: https://doi.org/10.1039/d4su00433g |
| This journal is © The Royal Society of Chemistry 2025 |