Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Oral mucosa: anti-inflammatory function, mechanisms, and applications
Yani
Chen†
,
Bicong
Gao†
,
Wenjin
Cai
,
Junhong
Lai
,
Kaichen
Lai
* and
Ying
Wang
 *
*
Stomatology Hospital, School of Stomatology, Zhejiang University School of Medicine Zhejiang Provincial Clinical Research Center for Oral Diseases, Key Laboratory of Oral Biomedical Research of Zhejiang Province, Cancer Center of Zhejiang University, Engineering Research Center of Oral Biomaterials and Devices of Zhejiang Province, Hangzhou 310000, P. R. China. E-mail: laikaichen@zju.edu.cn; 7314032@zju.edu.cn
First published on 10th March 2025
Abstract
Large soft tissue injuries require several weeks to heal and frequently leave fibrotic scars that can negatively impact tissue function. However, the applicability of traditional skin and mucous membrane transplantation for the treatment of lesions in the ocular surface and urethra is limited owing to the unique locations and functions of these tissues. Oral mucosa has been widely used in the repair of such injuries owing to its reduced propensity for inducing an inflammatory response, angiogenesis, and scarring. Enhancing chronic wound healing while avoiding scar formation requires a broader understanding of the cellular and molecular pathways that drive wound repair in the oral mucosa. This review integrates current knowledge on the mechanisms underlying the resistance of the oral mucosa to inflammation and its application as a graft material, highlighting its challenges and potential advancements. The aim of this review is to offer insights into future therapeutic strategies for wound healing and related conditions.
Introduction
Following soft tissue damage, wound contraction and cell growth drive wound closure. While minor wounds heal within a few days, larger tissue trauma typically requires several weeks to heal and can leave fibrous scars that impair tissue function.1 Improper wound management can further complicate healing, leading to chronic wounds that cause significant physical and psychological distress to affected individuals, as well as impose substantial economic and social burdens on their families and healthcare systems. This underscores the need for the development of therapeutic strategies that promote efficient and scar-free healing.Current approaches to repairing soft tissue defects include tissue transplantation, biomaterial implantation, and tissue engineering strategies for tissue reconstruction. Autologous skin grafts, commonly used as a repair material, often exhibit limitations such as mucosal metaplasia, mismatches in color and texture compared to surrounding tissues, and scar hyperplasia. In contrast, the oral mucosa, a non-keratinized squamous epithelium, offers several unique advantages, including abundant availability, ease of harvest, rapid donor site healing, excellent regenerative potential, and the ability to adapt to different epithelial environments2 These properties, combined with the ability to accelerate healing, minimize scarring, and resist inflammation, make oral mucosa an increasingly attractive option as a biomaterial for regenerative medicine.
Oral mucosa and related biomimetic materials have demonstrated significant potential in enhancing wound healing in various tissues. Clinically, they have been successfully applied for diverse purposes, including accelerating skin wound healing with minimal scarring,3 urethral reconstruction and stricture repair,4–6 eyelid7–10 and corneal reconstruction,11–14 and the treatment of tracheal defects. Notably, oral mucosal wounds exhibit faster and scarless healing compared to cutaneous wounds,15–18 a phenomenon closely linked to the rapid and well-regulated resolution of inflammatory responses.19 This unique healing capability positions the oral mucosa as a valuable model for investigating scar-free tissue regeneration.
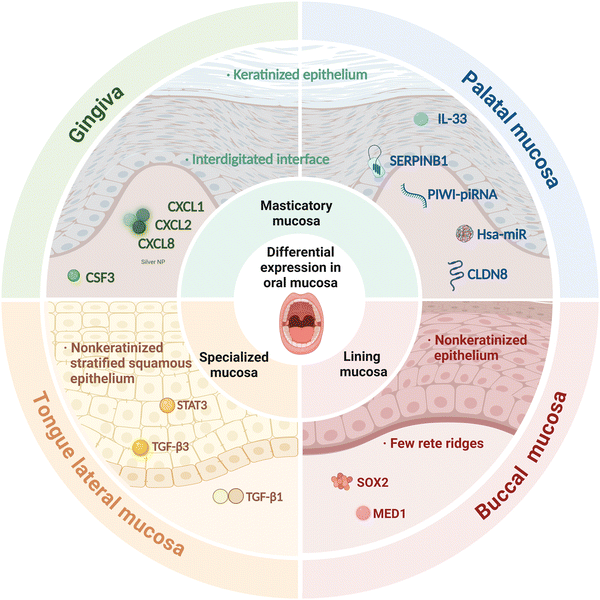
This review explores the mechanisms involved in the ability of the oral mucosa to resist inflammation, contrasting them with those of other mucosal linings during wound healing. We focus on the inflammatory processes in four key oral mucosae—gingival mucosa, hard palatal mucosa, buccal mucosa, and tongue lateral mucosa. Precise immune regulation is critical for achieving low-inflammation, scar-free, and rapid healing in oral mucosal wounds. As summarized in Table 1, recent research has identified key mechanisms and pathways involved in the anti-inflammatory and regenerative properties of the oral mucosa, providing a foundation for further investigation. Leveraging the anti-inflammatory properties of the oral mucosa, cellular therapies and bioengineering approaches are being integrated to develop regenerative treatments that reduce scarring in other tissues. However, the specific mechanisms underlying the inflammation-modulatory and barrier-protective properties of the oral mucosa remain incompletely understood. Recent studies on the cellular and molecular pathways governing oral mucosal inflammation have revealed the roles played by cytokines and alterations in gene expression patterns in this process. Nevertheless, a more comprehensive understanding of these mechanisms is necessary to significantly enhance the safety, efficacy, and applicability of oral mucosa in tissue regeneration, paving the way for innovative therapeutic applications.
| Oral mucosa | Species | Study design | Key findings | Ref. |
|---|---|---|---|---|
| Tongue lateral mucosa | Mice | Animal experiment | Oral wounds demonstrate decreased levels of TGF-β1 by contrast to dermal wounds, accompanied by an increase in the ratio of TGF-β3 to -β1. | 20 |
| Tongue lateral mucosa | Mice | Animal experiment | Dermal and mucosal keratinocytes have different modulation pathways that contribute to variable responses to injury sites. | 21 |
| Gingiva | Human | Animal experiment | Higher levels of inflammation in places with periodontal destruction are related to increased microbial load. | 22 |
| — | Mice, human | Animal experiment | Serum and glucocorticoid-regulated kinase 1 (SGK1) maintains the expression of tumor necrosis factor receptor-associated factor 3 (TRAF3), which inhibits the inflammatory response induced by gingival porphyria. | 23 |
| Gingiva, oral mucosa | Human | Cell experiment | Inflammatory stimuli priming macrophages stimulate the secretion of active TGFβ, leading to autophagy and sustained engagement of myofibroblasts, causing scarring. | 24 |
| Lining mucosa, masticatory mucosa | Mice | Animal experiment | Fine-tuning B7-H1 expression in keratinocytes is critical for mucosal defense against disease. | 25 |
| Gingiva | Mice, human | Cell experiment | Mechanical injury induces IL-6 in epithelial cells, influencing T-cell function and increasing Th17 cells. | 26 |
| Gingiva, buccal mucosa, palatal mucosa, tongue mucosa | Mice | Animal experiment | A high number of Foxp3+ regulatory T cells keep the oral mucosa silent. | 27 |
| Buccal mucosa | Mice, human | Clinical study + Cell experiment | With less differentiation and a more persistent inflammatory response, transcriptional circuits located in oral mucosa help wounds heal more quickly. | 19 |
| Palatal mucosa | Mice | Animal experiment + Bioinformatic analysis | The oral mucosa has a more complex and inherent biological response than the skin, which accelerates repair for being “preactivated”. | 15 |
| Gingiva | Human | Cell experiment | Human saliva promotes wound closure and inflammation in the mouth and skin. | 28 |
| Palatal mucosa | Mice | Animal experiment | Since the mucosa is “preactivated,” a substantial alteration in gene expression is not necessary for the healing process to occur. | 29 |
| Gingiva, palatal mucosa, tongue mucosa | Mice, human | Animal experiment + Cell experiment | In reaction to damage and inflammatory stimulation, human skin keratinocytes and oral keratinocytes express specific TJ genes differentially. | 30 |
| Palatal mucosa | Human | Clinical study + Bioinformatic analysis | The absence of scarless healing in hard palate and gingival wounds is explained by the lack of fibrillar marker expression and autophagy activation. | 16 |
| Gingiva, buccal mucosa | Human | Cell experiment | The mesenchymal compartment may serve a previously unrecognized function in immune reactivity, oral homeostasis, and pathogenesis. | 31 |
| Gingiva | Mice | Animal experiment | SGK1 inhibits the toll-like receptors (TLR)-mediated immune response, which increases inflammation by activating TAK1 and NF-κB in LPS-stimulated immune cells. | 32 |
| Cell line of oral squamous cell carcinoma | Human | Animal experiment | Human oral epithelial cells inhibit T-cell function by secreting prostaglandin E2, which can be evaded when under attack. | 33 |
| Oral mucosa | Mice | Animal experiment | Regulatory T cell treatment is efficient in reducing oral mucosal inflammation in mice. | 34 |
| Buccal mucosa, human oral keratinocytes | Mice | Animal experiment + Cell experiment | Mediator complex subunit 1 (MED1) ablation enhances oral wound healing via an upregulation of keratinocyte proliferation and mobility. | 35 |
| Tongue lateral mucosa | Mice, human | Animal experiment + Cell experiment | Signal transducer and activator of transcription 3 (STAT3)-activated small proline rich protein 1B+ (SPRR1B+) keratinocytes are upregulated in wound healing-related gene clusters, preparing the oral mucosa for rapid healing. | 36 |
In this review, we further highlight the interdisciplinary nature of this research, bridging medicine, materials science, and chemistry, aiming to provide insights into the potential of the oral mucosa as a biomaterial for tissue engineering. We also discuss the challenges and opportunities in scaling up the production of oral mucosa-based grafts and their translation into clinical practice. Ultimately, this review seeks to advance the field by identifying key research directions that will enable the development of innovative therapies for wound healing and tissue regeneration.
Anti-inflammatory mechanisms in different oral mucosae
Gingival mucosa
Following histamine-induced disruption of tight junction protein-encoding genes, oral keratinocytes exhibit superior barrier function compared with skin keratinocytes, as highlighted in Fig. 1, which was hypothesized to result from alterations in tight junction protein levels.30 Unlike other barrier sites, in the gingiva, T-helper 17 (Th17) cells are activated by physiological mechanical injury in the absence of microbial colonization37,38 and accumulate in an interleukin-6 (IL-6)-dependent manner.39 This accumulation leads to enhanced secretion of IL-17, a key mediator of barrier-protective responses, such as those associated with antimicrobial defenses. Additionally, due to mastication26 and commensal microbiota activity,22,40,41 the expression of genes encoding pro-inflammatory and antimicrobial proteins, such as IL-1β, secretory leukocyte peptidase inhibitor (SLPI), and S100 calcium-binding protein A8/A9 complex (S100A8/9), is also elevated in the gingiva following injury, particularly in specific clusters of epithelial cells.The gingival mucosa has unique immune wiring and harbors a stromal cell population in which multiple immune-related pathways and the most upregulated genes, including colony-stimulating factor 3 (CSF3), C-X-C motif chemokine ligand 1 (CXCL1), C-X-C motif chemokine ligand 2 (CXCL2), and C-X-C motif chemokine ligand 8 (CXCL8), are primarily associated with neutrophil recruitment. Serum- and glucocorticoid-regulated kinase 1 (SGK1) inhibits the Porphyromonas gingivalis-induced inflammatory response by maintaining the expression levels of tumor necrosis factor receptor-associated factor 3 (TRAF3), the loss of which significantly increases the production of pro-inflammatory cytokines such as tumor necrosis factor (TNF), IL-6, IL-1, and IL-8 in Porphyromonas gingivalis-stimulated innate immune cells.42,43 Additionally, SGK1 was reported to be a negative regulator of the Toll-like receptor (TLR)-mediated immune response, whose inhibition heightens the inflammatory response by upregulating the activity of transforming growth factor beta (TGF-β)-activated protein kinase 1 (TAK1), which, in turn, heightens that of nuclear factor kappa-B (NF-κB) in lipopolysaccharide (LPS)-stimulated innate immune cells.32
Following damage to oral mucosa, wound healing does not promote autophagy in the attached gingiva. Consequently, no myofibroblast differentiation or collagen deposition is observed. Meanwhile, the inflammatory stimulation of activated macrophages boosts the secretion of activated TGF, which further enhances autophagy and aids in prolonging myofibroblast activation.44
Palatal mucosa
The transcriptional changes occurring in skin were found to be more pronounced than those observed in the palate during the wound-healing process.45 An integrated investigation16 of mRNA/miRNA expression during human oral wound healing indicated that hsa-miR-223 (regulates neutrophil replenishment46 and macrophage function,47), hsa-miR-21, hsa-miR-132 (enhances re-epithelialization, promotes wound closure, and lowers inflammation48), hsa-miR-146b, hsa-miR-124-3p, IL-33 (a cytokine crucial for the resolution of inflammation49), and serpin family b member 1 (SERPINB1; inhibits neutrophil serine proteases and inflammatory caspases) were highly regulated in healing palatal tissue.At the baseline, claudin 8 (CLDN8), a tight junction gene that promotes cancer cell migration and proliferation,50 was highly expressed in palatal mucosa, whereas CLDN1 was markedly upregulated in skin. This suggests that different tight junction genes are involved in barrier property maintenance in palatal and cutaneous epithelia.30
Furthermore, PIWI-interacting RNA (piRNA) and P-element-induced wimpy testis (PIWI) genes (encoding obligate piRNA binding partners) were noted to be highly expressed in both skin and oral mucosal epithelium during wound healing.51 Approximately twice as many protein-coding and miRNA genes were upregulated in skin wounds at rest and during healing compared to that in mucosal wounds,15 suggesting that site-specific healing responses might be impacted by differential PIWI-piRNA complex regulation.52
A human study revealed that scarless healing in hard palatal and gingival wounds could be explained by the lack of fibrillar marker expression and autophagy activation.53 Substantial changes in the mRNA expression levels of key genes were also observed in palatal and gingival tissue following injury. Highly differentially expressed genes included those involved in scar development (ras-related C3 botulinum toxin substrate 1 [RAC1], SERPINE1, and tissue inhibitor of metalloproteinase 1 [TIMP1]), myofibroblast differentiation (CDH1, integrin alpha 4 [ITGA4], and integrin beta 5 [ITGB5]), and inflammation (IL6 and CXCL1).
Buccal mucosa
Wound-activated transcriptional networks are already present in the oral mucosa at a basal state. They enhance antimicrobial defenses and immune responses, allowing the oral mucosa to rapidly control and limit the inflammatory response, resulting in the quick relief of inflammation and the promotion of scarless wound healing.19 In comparison, during the skin healing process, the immune response is overstimulated, culminating in chronic inflammatory responses and scarring.54Iglesias-Bartolome et al.19 demonstrated that the transcription factor SRY-box containing gene 2 (SOX2) plays a key role during healing in oral keratinocytes, regulating a network of genes associated with immune and defense responses.19 Meanwhile, Meng et al. revealed that MED1 ablation promotes oral wound healing by increasing keratinocyte proliferation and mobility.35
Tongue lateral mucosa
Schrementi reported that site-specific variations in TGF-β1 influence oral wound healing. Compared to cutaneous wounds, oral wounds exhibit a substantially higher TGF-β3/-β1 ratio and lower TGF-β1 levels.20 Additionally, TGF-β1 is produced by inflammatory cells drawn to the wound site,55 increasing synthesis and engagement throughout wound healing via autocrine and paracrine feedback loops.56 Xuanyuan et al. identified a STAT3-activated SPRR1B+ keratinocyte subpopulation that was highly enriched for wound healing-associated genes. This subpopulation was shown to prime the oral mucosa for rapid wound healing via STAT3 activation.36Masticatory mucosa and lining mucosa
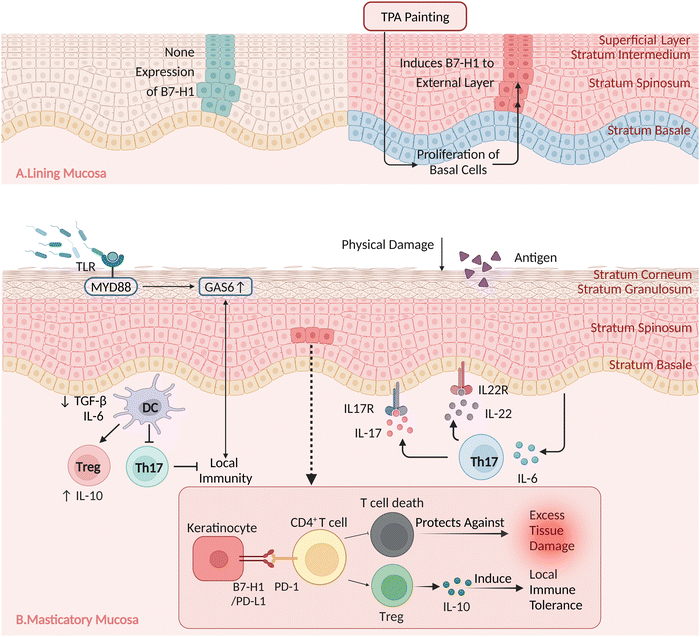
The mechanisms that maintain masticatory mucosal homeostasis comprise two stages. First, at approximately 3–4 weeks postnatally, the oral microbiota stimulates growth arrest-specific 6 (GAS6) expression in the oral epithelium through the activation of TLR2/4 expression, which downregulates pro-inflammatory cytokine secretion and free radical formation, thereby limiting microbiota-mediated epithelial cell activation. In addition, GAS6 stimulates the differentiation of T regulatory (Treg) cells, but not Th17 cells, by regulating IL-6 expression in dendritic cells. In Fig. 2, the key anti-inflammatory mechanisms in the oral mucosa are depicted in chronological order, highlighting the temporal progression of events that contribute to its unique healing properties.When the masticatory mucosa is exposed to extended external stimuli38 such as mastication, epithelial cells are stimulated to produce IL-6,26 which promotes the accumulation of Th17 cells. These release IL-17 and IL-22, thereby mediating a barrier-protective response. IL-17 contributes to immune surveillance and oral mucosa integrity, playing a major role in the regulation of epithelial tight junction protein expression, triggering antimicrobial peptide expression, and inducing the release of neutrophil chemoattractants.57
B7 Homolog 1 (B7-H1; also known as PD-L1) is abundantly expressed in spinous cells but its expression is not detected in the basement membrane. Furthermore, B7-H1 is not consistently expressed in other oral-lining epithelia or other mucosal sites. As depicted in Fig. 3, B7-H1, expressed on keratinocytes, directly interacts with PD-1 expressed on tissue-resident CD4+ T cells that react to exogenous antigens presented through the mucosal surface, thereby protecting against excessive tissue damage and inducing local immune tolerance through the activation of IL-10-secreting T cells.58,59
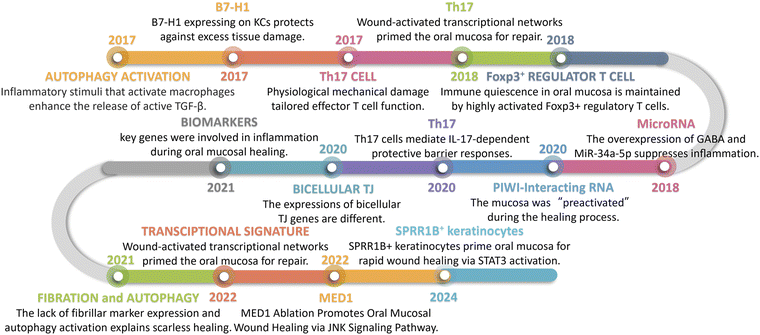 | ||
| Fig. 2 Timeline. The key anti-inflammatory mechanisms in the oral mucosa are depicted in chronological order. | ||
B7-H1 is not expressed in the lining mucosa under physiological conditions. Following 12-O-tetradecanoylphorbol-13-acetate (TPA) painting, basal cell proliferation was observed, along with the induction of B7-H1 in the outer layer; however, B7-H1 enrichment was not detected at the junction with the basement membrane.
Biomedical applications of oral mucosa
Oral mucosa grafts
Soft tissue grafts are the gold standard in clinical practice for filling tissue defects and promoting tissue regeneration.60 Oral mucosa grafting is widely used in periodontal surgery for increasing keratinized tissue.61 Free gingival grafts and connective tissue grafts effectively increase the width of keratinized mucosa and decrease peri-implant mucosal recession.62 A 5-year randomized controlled trial confirmed that free gingival grafts are more effective than collagen matrix in regenerating keratinized mucosa.63 Over the past five years, significant advancements have been made in the biomedical applications of oral mucosa, as summarized in Table 2.| Medical application | Disease | Sources | Form | Outcome | Ref. |
|---|---|---|---|---|---|
| Oral mucosa grafts | Peri-implant disease | Gingival mucosa | Free Gingival graft (FGG) | Increased keratinized mucosa (KM) width and decreased mucosal recession of peri-implant | 62 and 63 |
| Vesicovaginal fistula (VVF) | Buccal mucosal | Buccal mucosal graft (BMG) | Repair the VVF | 64 | |
| Rectovaginal fistula (RVF) | Buccal mucosal | BMG | Repair the RVF | 65 | |
| Ureteral strictures | Lingual mucosa | Robotic ureteroplasty with a lingual mucosal graft (RU-LMG) | Medium-term success in managing ureteral strictures | 66 | |
| Cicatricial entropion | Buccal mucosal | BMG | Reduction of focal cicatricial entropion | 67 | |
| Stem cells suspension and secretions | Diabetic wound | Oral mucosa stem cells (mOMSCs) | Subcutaneous injection of cells | Accelerated wound healing, re-epithelialization, and granulation tissue formation | 3 |
| Periodontitis | Gingival mesenchymal stem cell (GMSC) | Exosome | Reduce M1 macrophage and promote transformation of M1 to M2 | 68 | |
| Skin wound | Oral Mucosa Lamina Propria-Progenitor Cells (OMLP-PCs) | Small extracellular vesicle (sEV) | Minimized scar formation in wound healing | 69 | |
| Bone defect | Periodontal ligament stem cell (PDLSC) | sEV | Accelerated repair of bone defect | 70 | |
| Cell sheets | Periodontitis | PDLSC | Microtissues and monolayers | PDLSC microtissue could be a potential substantial material for guided tissue regeneration (GTR) | 71 |
| Peri-implant disease | Hard palatal-derived mesenchymal stem cells (PMSCs) | Light-controlled scaffold- and serum-free PMSC aggregates | Enhanced bone formation and implant osseointegration | 72 | |
| Congenital esophageal atresia | Buccal mucosa | Cell sheet | Effective prevention of endoscopic balloon dilatation following transplantation | 73 and 74 | |
| Oesophageal cancer | Oral mucosa | Cell sheet | Inhibition of pro-inflammatory cytokines | 75 | |
| Limbal stem cell deficient | Oral mucosa | Cell sheet carried by amniotic membrane | Transplant for corneal recovery | 76 | |
| Periodontitis | PDLSC | Triple-layer cell sheet | Promote osteogenic lineage commitment through improved cell viability and mitochondrial function | 77 | |
| Tissue injury | PDLSC | A Janus bio patch engineered by integrating the bioactive patch with a stem cell sheet of PDLSCs | Combine immunomodulatory effects with osteoblastic capacity for enhanced bone regeneration | 78 | |
| Biomaterial scaffolds | Anterior urethral strictures | Oral mucosa | Tissue-engineered oral mucosa graft MukoCell® | Mukocell® evaluated as a promising alternative for long-segment ureteral reconstruction | 79 and 80 |
| Periodontitis | PDLSC | Electrospinningand collagen hydrogels incorporated PDLSCs and curcumin-loaded ZIF-8 nanoparticles (CURZIF-8) | Regulation of inflammation and promotion of pro-healing factor expression | 81 | |
| Periodontitis | Gingival fibroblasts (GF) | GF-laden β-calcium triphosphate | Reduction in vertical pocket depth and bone regeneration | 82 | |
| Periodontitis | PDLSC | PDLSC-laden bioinks | Mimic native ECM condition to facilitate PDL regeneration | 83 and 84 | |
| Cleft alveolus | GMSC | Predifferentiated GMSCs toward an osteogenic lineage combinated with a self-assembling hydrogel scaffold PuraMatrix™ (PM) and bone morphogenetic protein 2 (BMP2) | Enhanced bone regeneration in alveolar clefts | 85 | |
| Sensorineural hearing loss | GMSC | GMSC-laden hybrid hydrogel scaffold based on peptide modified alginate and Matrigel with growth factors | Enhance the auditory differentiation potential of GMSCs | 86 |
Besides periodontal surgery, oral mucosal grafting techniques are also employed in tissue repair within the respiratory, urinary, and digestive systems, among others.64,65 Buccal and lingual mucosae are widely used for urethroplasty.66,87 The permanent functional restoration of lengthy abnormal tracheal segments was achieved via a reconstructed tracheal replacement strategy that involved cartilage regeneration, microsurgery, and oral mucosa grafting.88 An autologous oral mucosa transplant was utilized to create a sophisticated tracheal substitute with excellent epithelialization and minimal granulation, which helped to reduce airway stenosis and early mortality following tracheoplasty while holding promise for conversion to ciliated epithelium.67 The utilization of lower labial mucosa effectively addresses focal cicatricial entropion by creating a mucosal membrane that prevents scarring and ensures the mechanical separation of the anterior and posterior lamellae.88 A sandwich technique combining ear cartilage and oral mucosal transplants89 with osteochondral flap-plasty is a safe and straightforward surgical treatment for full-thickness eyelid restoration.8
Despite their potential, mucosal grafts have some limitations, such as the difficulty in recovering the donor region under pathological inflammatory conditions and the restricted size and functional capacity of the grafts. Consequently, it is necessary to incorporate biomaterial techniques to enhance treatment efficacy and minimize harm to patients.
Stem cell suspensions and secretions
When directly seeded into wounds, stem cells derived from oral mucosa facilitate the healing process. Kuperman et al. administered subcutaneous injections of mouse oral mucosa stem cells (mOMSCs), suspended in PBS, around and beneath the skin wounds of diabetic mice, providing proof-of-principle for the potential of stem cell injection therapy.3 Stem cells derived from the oral mucosa can undergo gene editing, thereby enhancing their wound-healing capacity. The seeding of oral keratinocytes and fibroblasts transfected with TGF-β1 small interfering RNA (siRNA) onto sterile bladder acellular matrix grafts (BAMGs) reduced the formation of urethral scars.90Factors secreted from cells, including cytokines, exosomes, and miRNAs, are utilized in applied biomedical engineering. After comparing the differences in wound healing between oral mucosa and skin, Kong et al. embedded exogenous epidermal growth factor (EGF) in layered self-assembled microcapsules and basic fibroblast growth factor (bFGF) in hydrogel to mimic the growth factor release pattern of the oral mucosa.91 It has been reported that exosomes obtained from gingiva-derived mesenchymal stem cells (GMSC)-can reduce pro-inflammatory factor production in M1 macrophages, highlighting the anti-inflammatory capacity of exosome therapy.68 Shi et al. loaded exosomes derived from GMSCs into a chitosan/silk hydrogel sponge and observed that this combination enhanced angiogenesis and neuronal ingrowth, thereby facilitating skin repair in diabetic rats.92 Exosomes from oral mucosa epithelial cell (OMEC) sheets effectively accelerated wound healing in both allogeneic and autologous scenarios.93 Knight et al. demonstrated that small extracellular vesicles (sEVs) secreted by oral mucosa lamina propria-progenitor cells (OMLP-PCs) are superior to commonly used MSC-derived sEVs in facilitating rapid and scarless wound healing.69 Skin wound healing can also be accelerated by the overexpression of oral mucosa-specific miRNAs, such as miR-31 and miR-21, that are upregulated during the healing process.94,95
Cell sheets
Oral mucosal cell sheets have exhibited regeneration-promoting effects on skin wound healing and may represent an alternative strategy for tissue regeneration therapy.96,97 Cell sheets maintain the integrity of the extracellular matrix (ECM), thus supporting and guiding cell proliferation and differentiation. Monolayer cultures involve seeding cells onto plastic dishes, collagen membranes, or other biomaterial surfaces, followed by their implantation into tissues to promote the wound-healing effect.71 Jiang et al. fabricated scaffold-free 3D aggregates of hard palate-derived MSCs (PMSCs) using light-controlled cell sheet technology and reported that they promote bone generation.72Tissue-engineered oral mucosal epithelial cell sheets have a stratified, squamous, non-keratinized shape and cytokeratin expression comparable to that of native esophageal epithelium.98 Several studies have established the safety and effectiveness of esophageal regeneration treatment employing autologous oral mucosal epithelial cell sheets, particularly in the field of endoscopic transplantation.73,74 It was reported that cell sheet transplantation can prevent postoperative strictures by decreasing the levels of pro-inflammatory and anti-tumor cytokines in patients with esophageal cancer.75
Oral and corneal epithelia share features such as limited differentiation stages, fast cell renewal, rapid proliferation and expansion, and the absence of a tendency toward keratinization. Nakamura et al. cultivated autologous oral epithelial cell sheets (OMECs) using amniotic membranes as a carrier and found them to be beneficial for the treatment of severe ocular surface disorders following their transplantation.7,99,100 The use of OMECs can decrease the probability of postoperative immunological rejection, block corneal neovascularization, accelerate corneal epithelialization, limit the inflammatory response, promote corneal ulcer repair, and improve vision.17 3T3 or limbal niche feeder cells can reportedly further improve the effect of OMEC transplantation via the secretion of cytokines.76,101
Multi-layered cell sheets have been created for complex tissue regeneration. Fullaondo et al. used plasma rich in growth factors (PRGF) fibrin membranes as the carrier for human periodontal ligament stem cell (hPDLSC) culture to generate a triple-layer cell sheet.77 Iwata et al. built three-layered PDLSC sheets reinforced with woven polyglycolic acid that significantly enhanced periodontal bone regeneration.102 Meanwhile, Zhou et al. isolated adipose-derived stem cells (ADSCs), OMECs, and oral mucosal fibroblasts and combined them to reconstruct full-thickness urethras labeled with ultrasmall super-paramagnetic iron oxide.103
Oral mucosal cell sheets possess substantial developmental potential; however, their utilization requires considerable investment in both time and financial resources.
Biomaterial scaffolds
Advancements in biocompatible materials have expanded our capabilities beyond merely supplying engineered cells, now allowing the construction of a microenvironment that is specifically tailored to promote efficient wound healing and tissue regeneration.104Engineered tissue can be an optimal biomedical scaffold. The use of decellularized extracellular matrix sheets derived from oral mucosa cells represents a promising approach for the development of novel materials, as their microenvironmental properties are beneficial for wound healing in the oral mucosa.105,106 Bhargava et al. seeded keratinocytes and fibroblasts derived from oral mucosa on the surface of de-epidermized dermis (DED) for 7 days, obtaining tissue-engineered buccal mucosa for substitution urethroplasty.107,108
The cellularization of a scaffold containing oral mucosa increases vascularization and the establishment of the urothelial barrier, both of which help lower urine leakage-induced local inflammation and fibrosis.109 Huang et al. constructed a three-dimensional porous bacterial cellulose scaffold seeded with lingual keratinocytes that enhanced urethral tissue regeneration.110 MukoCell®, approved for clinical use in Germany, is a biomedical scaffold mainly consisting of an animal-origin biodegradable scaffold and autologous living OMECs79 The application of MukoCell achieves a similar success rate but with reduced surgery time compared with oral mucosa grafts, indicating that tissue-engineered products can be a feasible alternative for use in tissue reconstruction.80
Biomaterial scaffolds can integrate multiple functions, including proliferation, anti-inflammatory effects, sustained release, and conditional dosing, which enables the dynamic regulation of oral mucosal applications.111,112 Lan et al. combined a polycaprolactone/collagen/cellulose acetate electrospun scaffold with collagen hydrogels containing PDLSCs and curcumin-loaded ZIF-8 nanoparticles, a strategy that enhanced anti-inflammatory and antioxidative capacity.81 Novel pore-forming bioink technology allows for the zonal positioning of stem cells and plasmids encoding genes of interest.113 Collagen-based bioink containing PDLSCs facilitated periodontal ligament regeneration and enhanced interfacial integration.83
The integration of biomaterial scaffolds with oral mucosa-derived stem cells can significantly influence the direction of cellular differentiation. Kandalam et al. combined PuraMatrix (a self-assembling hydrogel scaffold), bone morphogenetic protein 2 (BMP2), and GMSCs pre-differentiated toward an osteogenic lineage and found that this biomaterial scaffold achieved superior bone-regenerating capacity in athymic nude rats compared to the use of transplant material or cells alone.85 It has been reported that oral mucosa-derived stem cells, encapsulated in an optimized three-dimensional scaffold alongside growth factors, display auditory differentiation potential.86 Three-dimensional printing allows for the precise control of biomaterial scaffolds and cells, thereby enabling the arrangement of complex structures within defect regions.114 Using multi-phasic scaffolds, proliferation and differentiation across positions and over time can be controlled, thereby facilitating the regeneration of both soft and osseous tissue in the periodontium.115 For example, Lee et al. manufactured a region-specific, tri-phasic microstructure for generating mineralized tissue, fiber alignment, and bone tissue.116 The multifunctionality of biomaterial scaffolds renders oral mucosa cells an intriguing option for disease treatment.
Advantages of oral mucosa-derived biomaterials
Diverse sources of acquisition
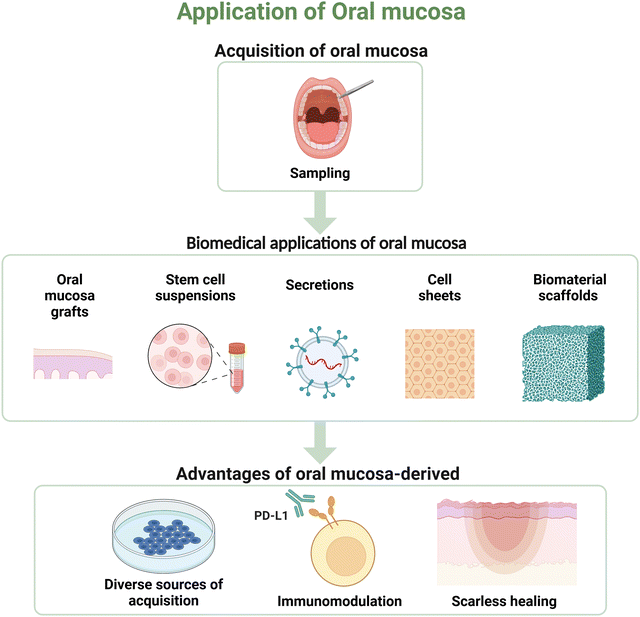
As illustrated in Fig. 4, ease of access is a substantial benefit in the biomaterial application of cells sourced from oral mucosa. All oral mucosa-derived cells can be disposed of enzymatically and cultured in vitro. The oral mucosa, especially the gingival tissue covering the alveolar bone and the third molar area, is frequently incised during dental procedures such as tooth extraction and gingivoplasty. This makes the procurement of oral mucosa relatively straightforward and minimizes ethical concerns.Oral mucosa is an abundant source of both adult and pluripotent stem cells. Compared with stem cells derived from other tissues, those derived from oral mucosa exhibit significantly higher growth rates and regeneration potential.117,118 Commonly used adult stem cells include PDLSCs, oral epithelial progenitor/stem cells (OESCs), and GMSCs.119 MSC-like cells derived from oral mucosa, such as gingival fibroblasts and periodontal ligament fibroblasts, can be effectively reprogrammed into pluripotent stem cells, exhibiting multipotentiality following transduction with a retroviral cocktail. Moreover, the reprogramming efficacy of oral fibroblasts surpasses that of skin fibroblasts.120 The accessibility and the potential for modification of oral mucosa-derived cells are fundamental to their application as biomaterials.
Immunomodulation
The wound healing rate of the oral mucosa is significantly greater than that of skin, which can be attributed to the diminished inflammatory response in the former.121,122 Studies have shown that PDLSCs and gingival fibroblasts possess the immunomodulatory properties of lymphocytes.123,124 Recent studies have demonstrated that PITX1, a transcription factor that maintains the unique balance of immune cells, is highly expressed in oral epithelial tissues, whereas its expression is undetectable in cutaneous keratinocytes.125,126 In addition, oral mucosa contains abundant stem cells and fibroblasts. Fibroblasts are the predominant cell type, which exhibited MSC-like properties and low immunogenicity.127–129 It has also been suggested that MSCs derived from oral mucosa hold potential as tools for use in biomaterial-based immunotherapy.130 Moreover, PDLSC spheroids show increased anti-inflammatory and angiogenic capabilities, possibly due to changes in apoptosis-related signaling.131Scarless healing
The main factors responsible for the scarless healing of oral mucosal wounds include the specific pattern of inflammatory regulation, a high proliferation rate, and efficient ECM remodeling. The expression level and influence of fibromodulin are crucial for scarless repair. During skin wound healing, periostin is highly expressed in fibrotic scars. However, during oral mucosal wound healing, its expression is lower than that in skin, which helps prevent fibrosis.132 In wound healing experiments involving rats, an inverse correlation is observed between fibromodulin expression and scar formation, with adult rats exhibiting reduced fibromodulin levels and enhanced scar formation compared to fetal rats.133The scarless healing property of the oral mucosa renders it an ideal source for biomaterials in tissue regeneration. The buccal mucosa in urethraloplasty in rabbits exhibits marked scarless healing outcomes, characterized by extensive neovascularization, minimal inflammatory cell infiltration, and minimal fibrosis.134
Challenges and future directions
Compared to skin, oral mucosa exhibits superior immune regulation in response to damage owing to its prolonged exposure to mastication stimulation and microbiome symbiosis. Differential modulation of inflammation and immune responses may also assist in site-specific healing. It is unclear if differences between the oral and skin microbiomes influence the healing of oral mucosal wounds because Th17 cells in the gums function independently of symbiotic colonization pathways.The intensity and duration of the inflammatory response play a significant role during wound healing. The substantial antibacterial defenses and anti-inflammatory capabilities of the oral mucosa slow the inflammatory response, encouraging faster healing with minimal scarring. Keratinocytes and stromal cells are involved in this process. Recent research has demonstrated that B7-H1, which is expressed on keratinocytes in the masticatory mucosa, directly interacts with tissue-resident CD4+ T cells that express PD-1, thereby supporting mucosal healing and preventing excessive inflammation. Unlike previous studies that focused on specific regions of the oral mucosa, this study distinguished between the masticatory mucosa and the lining mucosa, offering a better comparison of the mechanisms involved in their control of the inflammatory process.
The oral mucosa is an essential raw material for tissue engineering and regenerative medicine applications. Its main characteristics include ease of access, rapid healing, mild inflammation, strong antibacterial ability, and limited immune rejection. OMECs have been extensively used in a variety of regenerative medicine applications, including eye tissue, tracheal, and esophageal therapies.
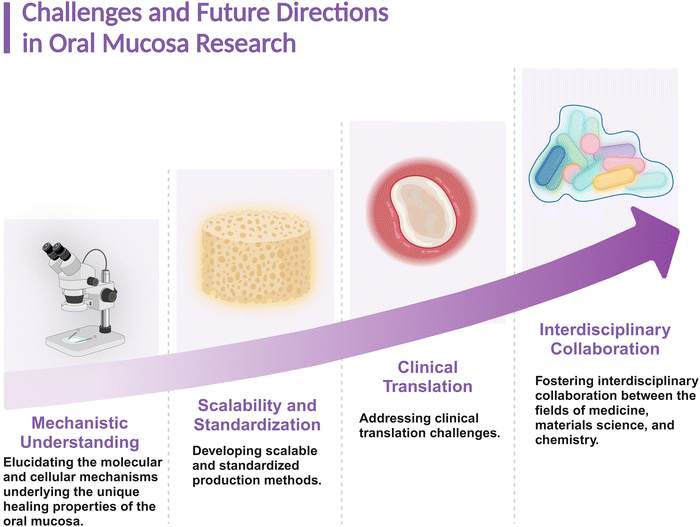
In summary, a rapid immune response, primarily regulated by changes in gene expression, is essential for the superior wound-healing capacity of the oral mucosa. While most recent studies have focused on cytokines and genes, the specific mechanisms underpinning the regulation of inflammation and the barrier-protective properties of the oral mucosa remain underexplored. A deeper understanding of the mechanisms involved in the regulation of inflammation in the oral mucosa, including the influence of the oral microbiota and variations in mucosal structures, will likely enhance the safety, efficacy, and application potential of oral mucosa in tissue regeneration. As outlined in Fig. 5, current challenges and future directions in oral mucosa research include elucidating the molecular and cellular mechanisms underlying its unique healing properties, developing scalable production methods, addressing clinical translation challenges, and fostering interdisciplinary collaboration. Addressing these challenges will be critical for advancing the field of oral mucosa-based therapies.
Conclusions
The oral mucosa exhibits excellent potential for inflammatory and immune regulation during wound healing, thus accelerating recovery and restricting scarring. Additionally, its use as a regenerative material offers several advantages, including an adequate supply, rapid donor site healing, and the absence of mucosal metaplasia after transplantation, presenting excellent regenerative potential. This approach not only furnishes materials for the repair of soft tissue defects but also minimizes the formation of scars, which holds significant clinical value. Future studies should concentrate on key targets and signaling pathways involved in the oral mucosa healing process, alongside preclinical research to support ideal wound healing in the clinic. We anticipate that oral mucosa-based approaches and bionic materials will achieve superior safety and efficacy in the near future, giving patients renewed hope.Author contributions
Yani Chen: writing – original draft. Bicong Gao: writing – review and editing. Wenjin Cai: conceptualization, writing – review and editing. Junhong Lai: writing – review and editing, investigation. Kaichen Lai: writing – review and editing, funding acquisition. Ying Wang: writing – review and editing, project administration.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors declare that there no conflicts of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82370928, 82401157).Notes and references
- O. A. Peña and P. Martin, Nat. Rev. Mol. Cell Biol., 2024, 25, 599–616 Search PubMed.
- A. L. Sánchez, H. A. García-Perdomo and J. A. Robayo, Nat. Rev. Urol., 2023, 20, 259–260 CrossRef PubMed.
- S. Kuperman, R. Efraty, I. Arie, A. Rahmanov, M. Rahmanov Gavrielov, M. Noff, R. Fishel and S. Pitaru, Int. J. Environ. Res. Public Health, 2020, 17, 4854 CrossRef CAS PubMed.
- K. L. Yang, S. B. Fan, J. Wang, L. Yin, Z. H. Li, S. W. Xiong, G. P. Han, C. Meng, P. Zhang, X. S. Li and L. Q. Zhou, Eur. Urol., 2022, 81, 533–540 CrossRef PubMed.
- A. L. Sánchez, H. A. García-Perdomo and J. A. Robayo, Nat. Rev. Urol., 2023, 20, 259–260 CrossRef PubMed.
- C. Q. Liang, J. L. Wang, B. Hai, Y. J. Xu, J. M. Zeng, S. S. Chai, J. W. Chen, H. Zhang, X. C. Gao, G. Cheng, X. Yang, T. Hou, W. C. Li, X. Y. Xiao and B. Li, Eur. Urol., 2022, 82, 193–200 CrossRef PubMed.
- S. Komai, T. Inatomi, T. Nakamura, M. Ueta, G. Horiguchi, S. Teramukai, Y. Kimura, T. Kagimura, M. Fukushima, S. Kinoshita and C. Sotozono, Br. J. Ophthalmol., 2022, 106, 1355–1362 CrossRef PubMed.
- M. J. Jin and Y. Gao, J. Craniofac. Surg., 2021, 32, E660–E661 CrossRef PubMed.
- P. Ngowyutagon, P. Prabhasawat, C. Chirapapaisan, P. Jaru-ampornpan, K. Pornpanich, P. Ekpo, N. Sukon and S. Matamnan, Cornea, 2021, 40, 1482–1486 Search PubMed.
- A. Rahal, D. Meller, A. Manthey, R. Pfoertner, S. Lang, N. Bechrakis and H. Westekemper, Can. J. Ophthalmol., 2023, 58, 543–549 Search PubMed.
- D. H.-K. Ma, Y.-J. Hsueh, K. S.-K. Ma, Y.-J. Tsai, S.-F. Huang, H.-C. Chen, C.-C. Sun, M.-T. Kuo, A.-S. Chao and J.-Y. Lai, Stem Cell Res. Ther., 2021, 12, 524 CrossRef CAS PubMed.
- K. E. Zhurenkov, E. I. Alexander-Sinkler, I. O. Gavrilyik, N. M. Yartseva, S. A. Aleksandrova, T. V. Mashel, J. I. Khorolskaya, M. I. Blinova, A. N. Kulikov, S. V. Churashov, V. F. Chernysh and N. A. Mikhailova, Invest Ophthalmol. Visual Sci., 2022, 63, 16 CrossRef CAS PubMed.
- H. Toshida, T. Kasahara, M. Kiriyama, Y. Iwasaki, J. Sugita, K. Ichikawa, T. Ohta and K. Miyahara, Int. J. Mol. Sci., 2023, 24, 8926 Search PubMed.
- S. Lopez, L. Hoz, E. P. Tenorio, B. Buentello, F. S. Magana, A. Wintergerst, A. Navas, Y. Garfias and H. Arzate, Int. J. Mol. Sci., 2021, 22, 5976 CrossRef CAS PubMed.
- A. Simoes, L. Chen, Z. J. Chen, Y. Zhao, S. Gao, P. T. Marucha, Y. Dai, L. A. DiPietro and X. F. Zhou, Sci. Rep., 2019, 9, 7160 Search PubMed.
- Y. Wang and D. N. Tatakis, J. Periodontol., 2021, 92, 863–874 Search PubMed.
- A. I. Toma, J. M. Fuller, N. J. Willett and S. L. Goudy, Transl. Res., 2021, 236, 17–34 CrossRef CAS PubMed.
- A. M. Overmiller, A. P. Sawaya, E. D. Hope and M. I. Morasso, Cold Spring Harbor Perspect. Biol., 2022, 14, a041244 CrossRef CAS PubMed.
- R. Iglesias-Bartolome, A. Uchiyama, A. A. Molinolo, L. Abusleme, S. R. Brooks, J. L. Callejas-Valera, D. Edwards, C. Doci, M. L. Asselin-Labat, M. W. Onaitis, N. M. Moutsopoulos, J. S. Gutkind and M. I. Morasso, Sci. Transl. Med., 2018, 10, eaap8798 CrossRef PubMed.
- M. E. Schrementi, A. M. Ferreira, C. Zender and L. A. DiPietro, Wound Repair Regen., 2008, 16, 80–86 CrossRef PubMed.
- L. Chen, Z. H. Arbieva, S. Guo, P. T. Marucha, T. A. Mustoe and L. A. DiPietro, BMC Genomics, 2010, 11, 471 Search PubMed.
- L. Abusleme, A. K. Dupuy, N. Dutzan, N. Silva, J. A. Burleson, L. D. Strausbaugh, J. Gamonal and P. I. Diaz, ISME J., 2013, 7, 1016–1025 Search PubMed.
- H. Zhou, S. Gao, X. Duan, S. Liang, D. A. Scott, R. J. Lamont and H. Wang, FASEB J., 2015, 29, 3737–3749 CrossRef CAS PubMed.
- E. Vescarelli, A. Pilloni, F. Dominici, P. Pontecorvi, A. Angeloni, A. Polimeni, S. Ceccarelli and C. Marchese, J. Clin. Periodontol., 2017, 44, 1039–1050 Search PubMed.
- S. Kang, C. Zhang, T. Ohno and M. Azuma, Mucosal Immunol., 2017, 10, 650–660 CrossRef CAS PubMed.
- N. Dutzan, L. Abusleme, H. Bridgeman, T. Greenwell-Wild, T. Zangerle-Murray, M. E. Fife, N. Bouladoux, H. Linley, L. Brenchley, K. Wemyss, G. Calderon, B.-Y. Hong, T. J. Break, D. M. E. Bowdish, M. S. Lionakis, S. A. Jones, G. Trinchieri, P. I. Diaz, Y. Belkaid, J. E. Konkel and N. M. Moutsopoulos, Immunity, 2017, 46, 133–147 Search PubMed.
- J. Y. Park, H. Chung, D. T. DiPalma, X. Tai and J. H. Park, Mucosal Immunol., 2018, 11, 1092–1102 CrossRef CAS PubMed.
- C. R. Neves, J. Buskermolen, S. Roffel, T. Waaijman, M. Thon, E. Veerman and S. Gibbs, J. Tissue Eng. Regener. Med., 2019, 13, 1079–1092 CrossRef PubMed.
- L. Chen, Z. Chen, A. Simões, X. Wu, Y. Dai, L. A. DiPietro and X. Zhou, Int. J. Mol. Sci., 2020, 21, 521 CrossRef CAS PubMed.
- T. R. Leonardo, J. Shi, D. Chen, H. M. Trivedi and L. Chen, Int. J. Mol. Sci., 2020, 21, 2966 Search PubMed.
- D. W. Williams, T. Greenwell-Wild, L. Brenchley, N. Dutzan, A. Overmiller, A. P. Sawaya, S. Webb, D. Martin, G. Hajishengallis, K. Divaris, M. Morasso, M. Haniffa and N. M. Moutsopoulos, Cell, 2021, 184, 4090–4104 CrossRef CAS PubMed.
- X. Han, J. Ren, H. Lohner, L. Yakoumatos, R. Liang and H. Wang, J. Biol. Chem., 2022, 298, 102036 Search PubMed.
- J. L. Sanchez-Trincado, H. F. Pelaez-Prestel, E. M. Lafuente and P. A. Reche, Front. Immunol., 2022, 12, 740613 CrossRef PubMed.
- N. Xue, Y. Wang, H. Cheng, H. Liang, X. Fan, F. Zuo, X. Zeng, N. Ji and Q. Chen, Front. Immunol., 2022, 13, 1009742 Search PubMed.
- Z. Meng, Z. Li, S. Guo, D. Wu, R. Wei, J. Liu, L. Hu and L. Sui, Int. J. Mol. Sci., 2022, 23, 13414 Search PubMed.
- X. Xuanyuan, L. Zhang, Y. Zheng, R. Jiang, Y. Ma, R. Liu, P. Hou, M. Lei, H. Xu and H. Zeng, Commun. Biol., 2024, 7, 1155 CrossRef CAS PubMed.
- F. R. Kirchner and S. LeibundGut-Landmann, Mucosal Immunol., 2021, 14, 455–467 Search PubMed.
- S. L. Gaffen and N. M. Moutsopoulos, Sci. Immunol., 2020, 5, eaau4594 CrossRef CAS PubMed.
- F. Momen-Heravi, R. A. Friedman, S. Albeshri, A. Sawle, M. Kebschull, A. Kuhn and P. N. Papapanou, J. Dent. Res., 2021, 100, 549–556 CrossRef CAS PubMed.
- N. M. Moutsopoulos and J. E. Konkel, Trends Immunol., 2018, 39, 276–287 CrossRef CAS PubMed.
- A. J. Caetano, V. Yianni, A. Volponi, V. Booth, E. M. D'Agostino and P. Sharpe, eLife, 2021, 10, e62810 Search PubMed.
- J. Ren, X. Han, H. Lohner, R. Liang, S. Liang and H. Wang, J. Immunol., 2021, 207, 268–280 CrossRef CAS PubMed.
- K. M. Fawzy El-Sayed, A. Bittner, K. Schlicht, M. Mekhemar, K. Enthammer, M. Hoeppner, M. Es-Souni, J. Schulz, M. Laudes, C. Graetz, C. E. Doerfer and D. M. Schulte, Cells, 2021, 10, 3310 Search PubMed.
- Y. Huang, L. Zhang, L. Tan, C. Zhang, X. Li, P. Wang, L. Gao and C. Zhao, Inflammation, 2023, 46, 1871–1886 CrossRef CAS PubMed.
- T. R. Leonardo, L. Chen, M. E. Schrementi, J. Shi, P. T. Marucha, K. Glass and L. A. DiPietro, Wound Repair Regen, 2023, 31, 156–170 Search PubMed.
- W. Zhou, A. S. Pal, A. Y.-H. Hsu, T. Gurol, X. Zhu, S. E. Wirbisky-Hershberger, J. L. Freeman, A. L. Kasinski and Q. Deng, Cell Rep., 2018, 22, 1810–1823 Search PubMed.
- P. Lopez-Cuevas, C. Xu, C. E. Severn, T. C. L. Oates, S. J. Cross, A. M. Toye, S. Mann and P. Martin, Adv. Sci., 2022, 9, e2202717 CrossRef PubMed.
- A. D. J. Bombin, N. Dunne and H. O. McCarthy, Acta Biomater., 2023, 155, 304–322 CrossRef CAS PubMed.
- M. Faas, N. Ipseiz, J. Ackermann, S. Culemann, A. Grueneboom, F. Schroeder, T. Rothe, C. Scholtysek, M. Eberhardt, M. Boettcher, P. Kirchner, C. Stoll, A. Ekici, M. Fuchs, M. Kunz, B. Weigmann, S. Wirtz, R. Lang, J. Hofmann, J. Vera, D. Voehringer, A. Michelucci, D. Mougiakakos, S. Uderhardt, G. Schett and G. Kroenke, Immunity, 2021, 54, 2531–2546 CrossRef CAS PubMed.
- B. Cheng, A. Rong, Q. Zhou and W. Li, J. Exp. Clin. Cancer Res., 2020, 39, 5 CrossRef CAS PubMed.
- L. Chen, Z. Chen, A. Simoes, X. Wu, Y. Dai, L. A. DiPietro and X. Zhou, Int. J. Mol. Sci., 2020, 21, 521 Search PubMed.
- Y. W. Iwasaki, M. C. Siomi and H. Siomi, in Annual Review of Biochemistry, ed. R. D. Kornberg, 2015, vol. 84, pp. 405–433 Search PubMed.
- M. A. Rojas, S. Ceccarelli, G. Gerini, E. Vescarelli, L. Marini, C. Marchese and A. Pilloni, J. Clin. Periodontol., 2021, 48, 705–720 Search PubMed.
- M. Rodrigues, N. Kosaric, C. A. Bonham and G. C. Gurtner, Physiol. Rev., 2019, 99, 665–706 CrossRef CAS PubMed.
- M. Lodyga and B. Hinz, Semin. Cell Dev. Biol., 2020, 101, 123–139 Search PubMed.
- J. M. Moreau, M. Velegraki, C. Bolyard, M. D. Rosenblum and Z. Li, Sci. Immunol., 2022, 7, eabi4613 Search PubMed.
- L. Abusleme and N. M. Moutsopoulos, Oral Dis., 2017, 23, 854–865 Search PubMed.
- S. Kang, C. Zhang, T. Ohno and M. Azuma, Mucosal Immunol., 2017, 10, 650–660 CrossRef CAS PubMed.
- X. Zhou, Y. Zhou, Q. Ding, Z. Jiao, L. Lu, N. Yang, Y. Ma and K.-Y. Chou, Transplant Immunol., 2009, 21, 192–197 CrossRef CAS PubMed.
- L. Chambrone, J. Botelho, V. Machado, P. Mascarenhas, J. J. Mendes and G. Avila-Ortiz, J. Periodontol., 2022, 93, 1336–1352 CrossRef PubMed.
- E. T. Scheyer, M. Sanz, S. Dibart, H. Greenwell, V. John, D. M. Kim, L. Langer, R. Neiva and G. Rasperini, J. Periodontol., 2015, 86, S73–S76 Search PubMed.
- S. L. Oh, S. Shahami, L. J. Bernal-Cepeda, Y. Fu and M. K. Chung, J. Prosthodont. Res., 2025, 69, 4–11 Search PubMed.
- D. Wei, Q. Wang, H. Sui, Y. Qin, H. Zhang, H. Meng and J. Han, Clin. Implant. Dent. Relat. Res., 2025, 27, e13422 CrossRef PubMed.
- K. M. Taha, M. I. Mohamed, E. Desoky, M. M. Seleem and A. M. Fawzi, Neurourol. Urodyn., 2025, 44, 287–293 CrossRef PubMed.
- C. Cahill, N. Kruger and J. Heine, JMIR Res. Protoc., 2022, 11, e31003 Search PubMed.
- K. Yang, S. Fan, J. Wang, L. Yin, Z. Li, S. Xiong, G. Han, C. Meng, P. Zhang, X. Li and L. Zhou, Eur. Urol., 2022, 81, 533–540 Search PubMed.
- P. S. Saffari, K. A. Roelofs and D. B. Rootman, Indian J. Ophthalmol., 2025, 73, 305–306 Search PubMed.
- R. Wang, Q. Ji, C. Meng, H. Liu, C. Fan, S. Lipkind, Z. Wang and Q. Xu, Int. Immunopharmacol., 2020, 81, 106030 CrossRef CAS PubMed.
- R. Knight, E. Board-Davies, H. Brown, A. Clayton, T. Davis, B. Karatas, J. Burston, Z. Tabi, J. M. Falcon-Perez, S. Paisey and P. Stephens, Stem Cells Transl. Med., 2022, 11, 861–875 CrossRef CAS PubMed.
- B. Zhao, Q. Chen, L. Zhao, J. Mao, W. Huang, X. Han and Y. Liu, Int. J. Nanomed., 2022, 17, 519–536 CrossRef CAS PubMed.
- K. Janjić, H. Agis, A. Moritz, X. Rausch-Fan and O. Andrukhov, J. Periodontol., 2022, 93, 697–708 Search PubMed.
- Z. Jiang, N. Li, Q. Shao, D. Zhu, Y. Feng, Y. Wang, M. Yu, L. Ren, Q. Chen and G. Yang, Bioeng. Transl. Med., 2023, 8, e10334 Search PubMed.
- A. Fujino, Y. Fuchimoto, T. Mori, M. Kano, Y. Yamada, M. Ohno, Y. Baba, N. Isogawa, K. Arai, T. Yoshioka, M. Abe, N. Kanai, R. Takagi, M. Maeda and A. Umezawa, Stem Cell Res. Ther., 2023, 14, 86 CrossRef PubMed.
- A. Fujino, Y. Fuchimoto, Y. Baba, N. Isogawa, T. Iwata, K. Arai, M. Abe, N. Kanai, R. Takagi, M. Maeda and A. Umezawa, Stem Cell Res. Ther., 2022, 13, 35 Search PubMed.
- A. Yoshida, T. Takata, T. Kanda, N. Yamaguchi, H. Minami, K. Nakao, S. Kobayashi, S. Eguchi and H. Isomoto, Sci. Rep., 2021, 11, 15282 CrossRef CAS PubMed.
- J. Oliva, F. Bardag-Gorce and Y. Niihara, Int. J. Mol. Sci., 2020, 21, 411 Search PubMed.
- A. Fullaondo, M. Zalduendo, N. Osinalde, M. H. Alkhraisat, E. Anitua and A. M. Zubiaga, Regener. Med., 2025, 1–14, DOI:10.1080/17460751.2024.2445931.
- S. Liu, W. Wang, Z. Chen, P. Wu, W. Pu, G. Li, J. Song and J. Zhang, Adv. Sci., 2024, 11, e2401882 Search PubMed.
- L. Karapanos, I. Akbarov, V. Zugor, R. Kokx, A. Hagemeier and A. Heidenreich, Int. J. Urol., 2021, 28, 936–942 CrossRef PubMed.
- L. Karapanos, V. Knorr, L. Halbe, M. Schmautz, B. Ergashev and A. Heidenreich, Int. J. Urol., 2023, 30, 1000–1007 CrossRef CAS PubMed.
- X. Lan, Y. Wang and M. Yin, Int. J. Nanomed., 2025, 20, 887–906 CrossRef PubMed.
- M. Abdal-Wahab, K. A. Abdel Ghaffar, O. M. Ezzatt, A. A. A. Hassan, M. M. S. El Ansary and A. Y. Gamal, J. Periodontal Res., 2020, 55, 441–452 Search PubMed.
- I. J. de Souza Araújo, R. S. Perkins, M. M. Ibrahim, G. T. Huang and W. Zhang, ACS Appl. Mater. Interfaces, 2024, 16, 59979–59990 Search PubMed.
- G. T. Yu, W. X. Zhu, Y. Y. Zhao, H. Cui, H. Chen, Y. Chen, T. T. Ning, M. D. Rong, L. Rao and D. D. Ma, Biofabrication, 2024, 16, 025007 Search PubMed.
- U. Kandalam, T. Kawai, G. Ravindran, R. Brockman, J. Romero, M. Munro, J. Ortiz, A. Heidari, R. Thomas, S. Kuriakose, C. Naglieri, S. Ejtemai and S. I. Kaltman, Tissue Eng., Part A, 2021, 27, 424–436 Search PubMed.
- S. Pouraghaei, F. Moztarzadeh, C. Chen, S. Ansari and A. Moshaverinia, ACS Biomater. Sci. Eng., 2020, 6, 2263–2273 CrossRef CAS PubMed.
- G. Tsachouridis, R. Nijman, L. M. O. de Kort and P. de Graaf, Pediatr. Surg. Int., 2025, 41, 79 CrossRef PubMed.
- J. Ren, Y. Xu, Z. Guo, R. Ting, J. Ren, W. Kan, Y. Luo, M. Zhu and T. Qiang, Thorac. Cancer, 2022, 13, 284–295 CrossRef PubMed.
- N. Yamamoto, H. Ogi, S. Yanagibayashi, R. Yoshida, M. Takikawa, A. Nishijima and T. Kiyosawa, Plast. Reconstr. Surg. Glob Open, 2017, 5, e1301 Search PubMed.
- C. Li, Y.-M. Xu, Z.-S. Liu and H.-B. Li, Urology, 2013, 81, 1075–1080 CrossRef PubMed.
- X. Kong, J. Fu, K. Shao, L. Wang, X. Lan and J. Shi, Acta Biomater., 2019, 100, 255–269 CrossRef CAS PubMed.
- Q. Shi, Z. Qian, D. Liu, J. Sun, X. Wang, H. Liu, J. Xu and X. Guo, Front. Physiol., 2017, 8, 904 Search PubMed.
- S. Sjoqvist, T. Ishikawa, D. Shimura, Y. Kasai, A. Imafuku, S. Bou-Ghannam, T. Iwata and N. Kanai, J. Extracell. Vesicles, 2019, 8, 1565264 CrossRef PubMed.
- L. Chen, A. Simoes, Z. Chen, Y. Zhao, X. Wu, Y. Dai, L. A. DiPietro and X. Zhou, Int. J. Mol. Sci., 2019, 20, 3679 Search PubMed.
- J. Shi, X. Ma, Y. Su, Y. Song, Y. Tian, S. Yuan, X. Zhang, D. Yang, H. Zhang, J. Shuai, W. Cui, F. Ren, M. V. Plikus, Y. Chen, J. Luo and Z. Yu, J. Invest. Dermatol., 2018, 138, 2253–2263 Search PubMed.
- J. Lee, D. Shin and J.-L. Roh, Head Neck, 2019, 41, 774–779 CrossRef PubMed.
- J.-L. Roh, J. Lee, E. H. Kim and D. Shin, Oral Oncol., 2017, 75, 81–88 CrossRef PubMed.
- N. Yamaguchi, H. Isomoto, S. Kobayashi, N. Kanai, K. Kanetaka, Y. Sakai, Y. Kasai, R. Takagi, T. Ohki, H. Fukuda, T. Kanda, K. Nagai, I. Asahina, K. Nakao, M. Yamato, T. Okano and S. Eguchi, Sci. Rep., 2017, 7, 17460 CrossRef PubMed.
- T. Nakamura, T. Inatomi, C. Sotozono, T. Amemiya, N. Kanamura and S. Kinoshita, Br. J. Ophthalmol., 2004, 88, 1280–1284 Search PubMed.
- C. Sotozono, T. Inatomi, T. Nakamura, N. Koizumi, N. Yokoi, M. Ueta, K. Matsuyama, H. Kaneda, M. Fukushima and S. Kinoshita, Acta Ophthalmol., 2014, 92, e447–e453 Search PubMed.
- C.-Y. Duan, H.-T. Xie, X.-Y. Zhao and M.-C. Zhang, Regener. Med., 2019, 14, 49–62 Search PubMed.
- T. Iwata, M. Yamato, H. Tsuchioka, R. Takagi, S. Mukobata, K. Washio, T. Okano and I. Ishikawa, Biomaterials, 2009, 30, 2716–2723 Search PubMed.
- S. Zhou, R. Yang, Q. Zou, K. Zhang, T. Yin, W. Zhao, J. G. Shapter, G. Gao and Q. Fu, Theranostics, 2017, 7, 2509–2523 CrossRef CAS PubMed.
- T. Wu, X. Li, J. Xue and Y. Xia, Acc. Mater. Res., 2024, 5, 1507–1519 Search PubMed.
- C. Liu, M. Pei, Q. Li and Y. Zhang, Front. Med., 2022, 16, 56–82 Search PubMed.
- L. Ren, Z. Jiang, H. Zhang, Y. Chen, D. Zhu, J. He, Y. Chen, Y. Wang and G. Yang, Mater. Today Bio, 2023, 22, 100734 Search PubMed.
- S. Bhargava, J. M. Patterson, R. D. Inman, S. MacNeil and C. R. Chapple, Eur. Urol., 2008, 53, 1263–1269 Search PubMed.
- Z. Xuan, V. Zachar and C. P. Pennisi, Int. J. Mol. Sci., 2022, 23, 14074 Search PubMed.
- N. I. Osman, J. M. Patterson, S. MacNeil and C. R. Chapple, Eur. Urol., 2014, 66, 790–791 Search PubMed.
- J. W. Huang, X. G. Lv, Z. Li, L. J. Song, C. Feng, M. K. Xie, C. Li, H. B. Li, J. H. Wang, W. D. Zhu, S. Y. Chen, H. P. Wang and Y. M. Xu, Biomed. Mater., 2015, 10, 055005 Search PubMed.
- S. D. Dutta, J. Hexiu, M. Moniruzzaman, T. V. Patil, R. Acharya, J. S. Kim and K. T. Lim, Biomaterials, 2025, 316, 122991 Search PubMed.
- S. Shahroudi, A. Parvinnasab, E. Salahinejad, S. Abdi, S. Rajabi and L. Tayebi, Carbohydr. Polym., 2025, 349, 123036 CrossRef CAS PubMed.
- T. Gonzalez-Fernandez, S. Rathan, C. Hobbs, P. Pitacco, F. E. Freeman, G. M. Cunniffe, N. J. Dunne, H. O. McCarthy, V. Nicolosi, F. J. O'Brien and D. J. Kelly, J. Controlled Release, 2019, 301, 13–27 CrossRef CAS PubMed.
- I. Roato, B. Masante, G. Putame, D. Massai and F. Mussano, Nanomaterials, 2022, 12, 3878 Search PubMed.
- S. Ivanovski, C. Vaquette, S. Gronthos, D. W. Hutmacher and P. M. Bartold, J. Dent. Res., 2014, 93, 1212–1221 Search PubMed.
- C. H. Lee, J. Hajibandeh, T. Suzuki, A. Fan, P. Shang and J. J. Mao, Tissue Eng., Part A, 2014, 20, 1342–1351 CrossRef CAS PubMed.
- M. E. Cabaña-Muñoz, M. J. Pelaz Fernández, J. M. Parmigiani-Cabaña, J. M. Parmigiani-Izquierdo and J. J. Merino, Pharmaceutics, 2023, 15, 2109 CrossRef PubMed.
- G. B. Tomar, R. K. Srivastava, N. Gupta, A. P. Barhanpurkar, S. T. Pote, H. M. Jhaveri, G. C. Mishra and M. R. Wani, Biochem. Biophys. Res. Commun., 2010, 393, 377–383 Search PubMed.
- H. Egusa, W. Sonoyama, M. Nishimura, I. Atsuta and K. Akiyama, J. Prosthodont. Res., 2012, 56, 151–165 CrossRef PubMed.
- N. Wada, B. Wang, N. H. Lin, A. L. Laslett, S. Gronthos and P. M. Bartold, J. Periodontal Res., 2011, 46, 438–447 CrossRef CAS PubMed.
- A. M. Szpaderska, J. D. Zuckerman and L. A. DiPietro, J. Dent. Res., 2003, 82, 621–626 CrossRef CAS PubMed.
- H. Yuan, G. E. Chlipala, H. I. Bangash, R. Meenakshi, D. Chen, H. M. Trivedi, L. A. DiPietro, P. Gajendrareddy and L. Chen, J. Dent. Res., 2025, 104, 97–105 Search PubMed.
- N. Wada, D. Menicanin, S. Shi, P. M. Bartold and S. Gronthos, J. Cell. Physiol., 2009, 219, 667–676 Search PubMed.
- A. R. Sanz, F. S. Carrión and A. P. Chaparro, Periodontology 2000, 2015, 67, 251–267 CrossRef PubMed.
- A. M. Overmiller, A. Uchiyama, E. D. Hope, S. Nayak, C. G. O’Neill, K. Hasneen, Y. W. Chen, F. Naz, S. Dell'Orso, S. R. Brooks, K. Jiang and M. I. Morasso, JCI Insight, 2024, 9, e182844 CrossRef PubMed.
- R. Iglesias-Bartolome, A. Uchiyama, A. A. Molinolo, L. Abusleme, S. R. Brooks, J. L. Callejas-Valera, D. Edwards, C. Doci, M. L. Asselin-Labat, M. W. Onaitis, N. M. Moutsopoulos, J. S. Gutkind and M. I. Morasso, Sci. Transl. Med., 2018, 10, eaap8798 CrossRef PubMed.
- K. Marynka-Kalmani, S. Treves, M. Yafee, H. Rachima, Y. Gafni, M. A. Cohen and S. Pitaru, Stem Cells, 2010, 28, 984–995 Search PubMed.
- S. Diar-Bakirly and T. El-Bialy, Saudi J. Biol. Sci., 2021, 28, 2518–2526 CrossRef CAS PubMed.
- A. Fadl and A. Leask, Cells, 2023, 12, 2021 Search PubMed.
- O. Andrukhov, A. Blufstein and C. Behm, Front. Oral. Health, 2021, 2, 832976 CrossRef PubMed.
- K. Iwasaki, M. Nagata, K. Akazawa, T. Watabe and I. Morita, J. Periodontal Res., 2019, 54, 364–373 CrossRef CAS PubMed.
- G. Nikoloudaki, K. Creber and D. W. Hamilton, Am. J. Physiol.: Cell Physiol., 2020, 318, C1065–C1077 CrossRef CAS PubMed.
- C. Soo, F. Y. Hu, X. Zhang, Y. Wang, S. R. Beanes, H. P. Lorenz, M. H. Hedrick, R. J. Mackool, A. Plaas, S. J. Kim, M. T. Longaker, E. Freymiller and K. Ting, Am. J. Pathol., 2000, 157, 423–433 CrossRef CAS PubMed.
- G. F. Souza, A. A. Calado, R. Delcelo, V. Ortiz and A. Macedo, Jr., Int. Braz. J. Urol., 2008, 34, 345–351 CrossRef PubMed ; discussion 351–344.
Footnote |
| † These co-first authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |