Open Access Article
Open Access ArticlePolymeric nanocarriers for cancer treatment: the promise of sensitive poly(2-(diisopropylamino)ethyl methacrylate)
Joana S. Ferreira†
 a,
João P. Vareda†
a,
João P. Vareda† a,
A. S. Oliveira
a,
A. S. Oliveira a,
Jéssica S. Barbosab,
Francisca Bastosb,
Patrícia V. Mendonça
a,
Jéssica S. Barbosab,
Francisca Bastosb,
Patrícia V. Mendonça *a and
A. C. Fonsecaa
*a and
A. C. Fonsecaa
aUniversity of Coimbra, Centre for Mechanical Engineering, Materials and Processes, Department of Chemical Engineering, Rua Sílvio Lima-Polo II, 3030-790 Coimbra, Portugal. E-mail: patmend@eq.uc.pt; anafs@eq.uc.pt
bBluepharma, Indústria Farmacêutica, SA, São Martinho do Bispo, 3045-016 Coimbra, Portugal
First published on 30th May 2025
Abstract
Polymeric nanoparticles are extremely valuable carriers for drug/gene delivery to treat cancer, as they can protect different therapeutic agents during blood circulation while being able to deliver them at desired locations. Owing to the versatility of polymers, it is possible to fine-tune the performance of nanocarriers by changing different properties, such as chemical structure, architecture, composition and molecular weight or even by functionalising the polymers with targeting molecules. The use of pH-sensitive polymers is a very popular strategy to prepare smart carriers, taking advantage of the acidic intratumoural environment to induce hydrophobic/hydrophilic transitions that allow fast and efficient release of small drugs or genetic material. This review summarizes the contributions of the use of promising pH-sensitive poly(2-(diisopropylamino)ethyl methacrylate) (PDPA), with pKa around 6.2, in the preparation of nanocarriers for the treatment of different types of cancer through gene therapy, drug delivery or photodynamic therapy. Interest in PDPA-based copolymers for biomedical applications is increasing, as different studies have reported successful encapsulation and delivery of different therapeutic molecules with PDPA-based smart nanocarriers. In vivo studies have shown that tumour growth can be suppressed, revealing the potential of new cancer therapies.
1. Introduction
The World Health Organization reports that cancer is a leading cause of death worldwide. The therapeutic outcome depends on the type of cancer, the stage/extent of the disease and its progression rate, the condition of the patient and the patient's response to the therapy (e.g., development of resistance to chemotherapeutic agents).1–3 Therefore, it becomes clear that there is a pressing need for novel, less invasive, less harmful and more selective treatments.In this context and driven by the COVID-19 pandemic, nanoparticle-based drug delivery systems have materialised as a valuable therapeutic approach to achieve more targeted and effective drug and gene delivery.3,4 These delivery approaches play a crucial role not only by enhancing the solubility and stability of therapeutic compounds, thereby improving their bioavailability, but also by facilitating targeted delivery, minimising off-target effects and reducing dose-limiting side effects.5 Particularly, gene therapy represents next-generation therapeutic approaches, as it targets the root causes of various inherited or acquired diseases, aiming to mitigate their progression by replacing or silencing faulty genes. Nanoparticle-based delivery systems can be categorised as viral or non-viral vectors. Viral vectors, such as retroviruses, lentiviruses, adenoviruses and adeno-associated viruses, are highly efficient at introducing specific genes into target cells through transfection.6 However, despite their strong replication capacity, these vectors are often associated with cytotoxicity, immunogenicity and complex and expensive production processes.7–9 On the other hand, non-viral vectors comprising polymer-, lipid- or inorganic-based nanoparticles have emerged as promising alternatives, due to their improved safety profile, lower immune response and high loading capacity, along with the advantage of a more cost-effective large-scale production.
Among non-viral vectors, polymeric nanoparticles (PNPs) have been extensively investigated as therapeutic delivery systems, particularly for cancer treatment. This is owed to their unique features, namely biocompatibility and versatility,5 as it is possible to tune the physicochemical properties of PNPs, such as size, shape, degradation profile and surface chemistry, to optimize their therapeutic efficiency.10–12 PNPs can be prepared from natural, synthetic or semi-synthetic polymers, which can be further engineered to provide specific drug or gene encapsulation, to enhance blood circulation time and to control the release of different therapeutic compounds.10,11
Amphiphilic block copolymers, which possess both hydrophilic and hydrophobic segments, are amongst the most investigated carriers for controlled drug delivery because of their ability to self-assemble in aqueous media allowing encapsulation of hydrophilic and/or hydrophobic drugs (Fig. 1).12,13 By changing the chemical nature and chain length of each polymeric segment, it is possible to fine-tune the morphology of the resulting highly organised nanostructures (e.g., micelles, rods, etc.), making block copolymers very versatile carriers.14 For instance, polymersomes (polymeric vesicles) can be loaded simultaneously with hydrophilic and hydrophobic drugs in different regions of the block copolymer.15 Importantly, several self-assembled polymers approved by the Food and Drug Administration, such as poly(ε-caprolactone)-b-poly(ethylene glycol)-b-poly(ε-caprolactone) (PCL-b-PEG-b-PCL) or poly(γ-L-glutamic acid)-b-poly(L-phenylalanine ethyl ester) (PGA-b-PAE) block copolymers, have been explored for oncology treatment to date.16 Regarding gene delivery, cationic polymers have been used as polymeric carriers, because they can encapsulate anionic nucleic acids through electrostatic interactions, forming stable polyplexes (Fig. 1). Ionizable cationic polymers (e.g., polymers containing tertiary amines, β-amino esters, etc.) are preferred over permanently charged ones, to facilitate the release of the cargo in target locations and according to the characteristics of the surrounding environment.17,18
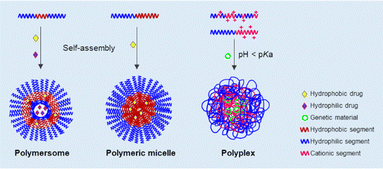 | ||
| Fig. 1 Schematic representation of the formation of different types of PNPs-based drug/gene delivery systems. | ||
Moreover, PNPs can be functionalised with targeting moieties that promote specific binding (ligand–receptor interactions) to cells or tissues of interest. This strategy limits the non-specific accumulation of therapeutic compounds in healthy tissues while promoting its targeted accumulation, uptake and release in cancer cells or tissues.19
Considering the tumour microenvironment, the therapeutic efficiency of PNPs can be further potentiated by using polymers that are responsive to stimuli, such as pH or oxidative stress, especially when working with hydrophobic or poorly soluble drugs.20 Among these, pH-responsive polymers are particularly valuable in oncology due to their ability to facilitate PNPs internalisation into tumour cells via endosomal vesicles, which possess an acidic pH. This enables the release of the therapeutic compound into the cytosol through endosomal escape. Additionally, the tumour microenvironment typically has a lower pH (5.0–6.5)21 compared to that of surrounding healthy tissues (7.35–7.45), providing a trigger for the efficient release of therapeutic agents, due to the sharp hydrophobic/hydrophilic transition of pH-sensitive polymers.22 Such responsive character is a powerful tool to achieve more targeted and controlled delivery of therapeutic agents, including the specific delivery of nucleic acids to the cytosol, which has historically been a major challenge. Notably, a wide range of pH-responsive polymers and/or block copolymers have been used for drug/gene delivery systems, including chitosan, poly(ethyleneimine) (PEI), poly(amino acid)s, poly(β-amino ester)s (PBAEs), poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA) and poly(2(diisopropylamino)ethyl methacrylate) (PDPA)-based copolymers, among others.17,23,24
Polymers that respond to more than one stimulus are rare but even more interesting, as their structure conformation can be adjusted in a subtle and well-controlled manner.25 Within these, PDMAEMA has been the most investigated temperature- and pH-responsive polymer for gene delivery. However, concerns about its toxicity have limited the translation of PDMAEMA-based materials from laboratory into clinical practice.26 PDPA, whose chemical structure resembles the one of PDMAEMA, has been regarded in the past two decades as a promising sensitive polymer for drug/gene delivery. Despite the main attention being focused on the polymer's pH-sensitivity, PDPA can also exhibit temperature-responsive behaviour, thus opening exciting future perspectives. This review summarizes the state-of-the-art development of PNPs for cancer treatment using PDPA as a sensitive polymer. The application of this polymer in several therapeutic strategies, namely gene therapy, small-drug molecule delivery and photodynamic therapy, is discussed.
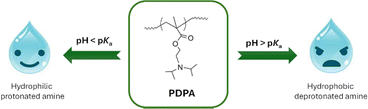
2. pH-sensitive poly(2-(diisopropylamino)ethyl methacrylate)
PDPA is a pH-responsive polymer with a pKa of about 6.2, given by the protonation/deprotonation of the tertiary amino groups.27,28 The rapid transition from a hydrophobic to a hydrophilic state with decreasing pH values below the pKa (Fig. 2), makes this cationic polymer very attractive for biomedical applications, namely for targeted drug delivery.24,29–31 Moreover, the responsive behaviour of PDPA can be adjusted by manipulating the composition, molecular weight, architecture or functionality of the (co)polymer to tailor pKa values according to the desired application.32 The precise control over these parameters is typically achieved using reversible deactivation radical polymerisation (RDRP) methods,33 being the most employed techniques atom transfer radical polymerisation (ATRP) and reversible addition–fragmentation chain transfer (RAFT) polymerisation.29,31,34–36 This powerful toolbox enables the straightforward synthesis of (multi) block copolymers and other complex sequence-controlled copolymers with targeted molecular weight and low dispersity (Đ = Mw/Mn < 1.5). RDRP methods can be explored to prepare PDPA-based copolymers with different macromolecular architectures, which has a direct impact on the size and shape of the resulting nanocarriers. For example, star-shaped polymers were obtained by direct polymerisation of DPA using multifunctional initiators37 or by the combination of RDRP and “click” chemistry methods.38 PDPA brushes were obtained by the combination of this responsive polymer with hydrophilic poly(oligo(ethylene oxide) methyl ether methacrylate) (POEOMA), which has long pendant side-chains or by the surface-initiated RDRP of DPA from solid substrates.39,40 Recently, hyperbranched PDPA-co-POEOMA structures haven been synthesised using RAFT polymerisation in the presence of a divinyl monomer as the branching agent, to prepare drug nanocarriers.41,42 RDRP techniques are also compatible with different types of polymer functionalisation methods, which is essential for preparing materials with targeting ability or even for combining RDRP-derived segments with biodegradable and natural polymers43 to improve the performance of the resulting nanocarriers. Our research group has contributed to the development of ATRP methods to improve control over PDPA's molecular weight and chain-end functionality, aiming the preparation of well-defined PDPA-based block copolymers under eco-friendly conditions.44,45The pH-responsiveness of PDPA is undoubtedly useful for cancer treatment, as the pH gradient of the tumour environment can act as a trigger for the effective release of therapeutic molecules inside cancer cells. The first work investigating PDPA-containing nanocarriers for cancer therapy was reported in 2008,46 and the interest in this polymer has been increasing over the years owing to the promising results demonstrated. To prepare PDPA-based carriers, typically PDPA is linked to a permanent hydrophilic block (e.g., PEG, poly(N-(2-hydroxypropyl) methacrylamide) (PHPMA), poly(2-methacryloyloxyethyl phosphorylcholine) (PMPC)) to afford amphiphilic block copolymers, which can self-assemble into nanostructures of different shapes. These are very versatile, as the pH-sensitivity of PDPA allows the encapsulation of several types of therapeutic molecules, such as small molecules (e.g., doxorubicin (DOX),47 paclitaxel (PTX)48) and genetic material,24,29,49 simply by changing the type of solvent used for the preparation of the nanosystem.
Although most works reported in the literature focus on PDPA as a mono-pH-responsive polymer, some studies have demonstrated that PDPA can also present temperature-sensitivity.25,50,51 Specifically, PDPA presents a lower critical solution temperature (phase separation upon heating) in an aqueous medium containing salts and the value is dependent on the type and concentration of salt.25 To the best of our knowledge, the dual-responsive nature of PDPA has not yet been explored in cancer treatment.
2.1. PDPA-based drug carriers for cancer treatment
Several works have demonstrated that the use of PDPA-based amphiphilic block copolymers containing hydrophilic segments, such as PEG,30,47,52 PHPMA,48 PMPC53 or zwitterionic poly(carboxybetaine methacrylate) (PCBMA),54 can further improve the efficacy of tumour therapy by reducing the non-specific absorption of proteins onto the hydrophilic outer shells and increasing colloidal stability of the assembled nanosystems.The functionalisation of PDPA-based copolymers with targeting molecules has also been explored to develop selective polymers for cancer therapy that could provide increased cellular internalisation of the drugs and decreased toxicity to healthy cells. Encouraging results have been obtained using folic acid,46,55 arginine–glycine–aspartic acid,56 galactose,55 hyaluronidase57 and arginine–glycine–aspartic acid-D-phenylalanine-lysine54 to target different cancer cell lines. The decoration of PDPA-based nanoparticles with glycopolymers (synthetic polymers containing sugar molecules) has also been reported for the preparation of probes to detect and treat cancer.58–61 Targeting moieties in the nanocarrier's structure can also be effective if the target is inside the cell. For example, micelles composed of PEG-b-PDPA block copolymers and D-α-tocopheryl polyethylene glycol succinate (vitamin E derivative) proved to target mitochondrial organelles.62 This strategy effectively decreased DOX resistance in breast cancer by increasing the drug cellular uptake and reducing the mitochondrial transmembrane potential. The prepared nanocarrier with targeting ability exhibited an interesting 23-fold or 4.4-fold reduction in the IC50 of the drug for DOX-resistant MCF-7/ADR cells when compared to DOX-free or DOX-loaded PEG-b-PDPA control micelles, respectively. The functionalisation of a PDPA-based nanocarrier with a fluorescent molecule for effective encapsulation of DOX for cancer diagnosis and treatment has also been reported,63 showing higher drug loading, sharper pH response to endosomes and greater fluorescence performance than analogous systems based on other pH-responsive segments.64
Generally, the literature indicates that the use of multiple molecules (e.g., target ligands and sensitive molecules) is a powerful strategy to improve the selectivity and efficiency of therapeutic nanosystems for cancer treatment by synergistic effects. In this context, a very elegant multifunctional PDPA-based nanosystem that combined folic acid and galactose active targets with dual-responsive behaviour (pH/redox), to potentiate the delivery of DOX into hepatocarcinoma cells has been reported (Fig. 3).55 PDPA was employed as a pH-responsive block, while poly(pyridyl disulphide methylacrylate) (PDEMA) was used as the redox-sensitive block. The PDEMA segment also allowed the preparation of core-crosslinked nanocarriers, which could potentially provide higher drug stability during blood circulation than the non-crosslinked analogues. In vitro DOX release studies showed that the combination of mild acidic and reductive conditions, mimicking the microenvironment of cancer cells, potentiated drug release from PDPA-based nanoparticles (77.6% in 20 h at pH 5.0 vs. 95.5% in 11 h at pH 5.0 and 10 mM DL-dithiothreitol). In addition, the presence of folic acid improved the selectivity of the nanocarriers, as demonstrated by a 3.54-fold increase in the cellular uptake efficiency by HepG2 cells when both active targets were used instead of galactose alone.
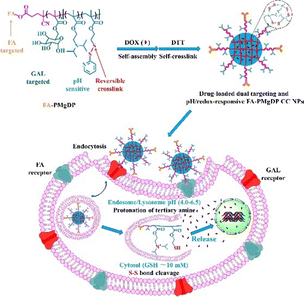 | ||
| Fig. 3 PDPA-based nanosystem with pH/redox responsive behaviour and folic acid and galactose dual-targeting properties for enhanced intracellular delivery of DOX into hepatoma cells. Reproduced from,55 Copyright (2025), with permission from Elsevier. | ||
In another contribution, the efficacy of β-lapachone in combination with PEG-b-PDPA micelles loaded with superparamagnetic iron oxide nanoparticles (SPIONs) has been demonstrated.65 β-Lapachone can generate a high concentration of superoxide and hydrogen peroxide inside cancer cells that overexpress NAD(P)H quinone oxidoreductase 1, such as breast, prostate, lung and pancreatic cancer cells, ultimately causing cell death.66,67 The authors demonstrated that the presence of PDPA in the nanosystem led to a pH-triggered release of iron cations inside cancer cells, from SPION-loaded PEG-b-PDPA micelles, potentiating the activity of β-lapachone by increasing reactive oxygen species (ROS) generation, due to the Fenton reaction between iron cations and the hydrogen peroxide produced by the anti-cancer drug. Interestingly, these nanoparticles could also potentially be used in theragnostics due to the presence of SPIONs, which can be used as imaging agents.68 In another contribution, pH-sensitive PEG-b-PDPA-based assemblies were stabilised in the form of microgels by a bioreducible crosslinker, providing dual pH/redox-responsive DOX nanocarriers that showed superior in vivo tumour growth inhibition using a subcutaneous 4T1 tumour-bearing Balb/c mice model, compared to control groups (DOX-free, phosphate-buffered saline and DOX-loaded non-crosslinked PEG-b-PDPA-based assemblies).69 Specifically, 14 days after drug injection, the tumour volume treated with the dual-responsive systems was about 500 mm3, whereas the one treated with the pH-responsive system was about 700 mm3. The developed nanosystem was found to be safe, as the mice maintained their body weight during treatment.
Temperature70 and light71 are two stimuli that can be easily and precisely manipulated outside the body, offering great spatiotemporal control over drug delivery by hyperthermia and light irradiation, respectively. Temperature-sensitive poly(N-isopropylacrylamide-co-dimethylacrylamide)72 and light-responsive azobenzene73 or photoacid generators74 have been combined with pH-sensitive PDPA-based carriers to enhance the therapeutic efficacy of DOX in HeLa cells. Very recently, a more complex multi-responsive PDPA-containing nanosystem was reported for precision therapy against glioblastoma.75 The concept (Fig. 4) involved the preparation of gold nanoparticles that were further functionalised with a well-defined PEG-b-PDPA-b-PS (PS, polystyrene) triblock copolymer and finally assembled and loaded with both DOX and ammonium bicarbonate to form vesicles (GVND). After entering the cancer cells, GVND swelled because of the protonation of the PDPA segment under acidic conditions. In the second stage, a mild hyperthermia treatment (laser irradiation) was applied, which induced the partial disassembly of GVND and in situ thermal decomposition of ammonium bicarbonate (NH4HCO3) to form CO2 bubbles. The CO2-responsive behaviour of GVND was confirmed by the 4-fold higher DOX release into tumour tissue compared to that observed with control vesicles (GV-DOX) without loaded NH4HCO3. Notably, the U87MG tumour-bearing mice treated with this PDPA-containing nanosystem showed no tumour growth or recurrence.
 | ||
| Fig. 4 Multi-responsive (pH, light and CO2) PDPA-containing GVND for programmed release of DOX against glioblastoma. Reproduced from,75 Copyright (2025), with permission from Elsevier. | ||
When designing polymeric drug carriers, the use of synergistic interactions between drugs and polymers, such as hydrophobic and electrostatic interactions, could be an interesting strategy to increase drug loading and circulation time of the resulting nanocarriers.76 An alternative method relies on the design of prodrugs, for example polymer–drug conjugates. Unlike common nanosystems that are loaded with anticancer drugs by coprecipitation during the self-assembly of amphiphilic block copolymers, prodrugs are expected to exhibit higher drug loading and lower drug leakage during blood circulation, as the therapeutic molecules are covalently linked to the polymer.77 The use of chemical linkages that can be easily cleaved in an intracellular environment, such as acid–labile, bioreducible, ROS-sensitive or enzyme-sensitive, is preferred to afford rapid drug release inside tumour cells. For example, an amphiphilic PDPA-b-poly-(4-formylphenyl methacrylate-co-polyethylene glycol monomethyl ether methacrylate) (PDPA-b-P(FPMA-co-OEOMA)) copolymer was prepared by 2-step RAFT polymerisation, followed by functionalisation with DOX (10% molar drug content) to produce a prodrug. This was further assembled into micelles, through thin-film hydration in phosphate buffer saline (pH 7.4), in the presence and in the absence of nifuroxazide (anti-metastasis agent). The pH-sensitive imine linkage formed between DOX and PFPMA allowed the preservation of the drug under physiological conditions (pH 7.4, 20% drug release after 30 h), while 80% of DOX was released at pH 5.5 after 4 h.78 More recently, the importance of having PDPA as a pH-sensitive segment in amphiphilic block copolymer prodrugs to allow the formation of PEG-b-PDPA-b-hydrophobic block-DOX core–shell–corona structures has been demonstrated.79,80 By having the PDPA segment between the core and the shell of the micelles, lower drug leakage at physiological pH (5% vs. 10%) and higher in vitro drug release (80% vs. 50%) in the tumour environment were achieved compared to those observed for the core–shell PEG-b-hydrophobic block-DOX analogues. In these studies, DOX was introduced in the polymeric structure either by post-polymerisation functionalisation of the hydrophobic block (PFPMA) via acid–labile imine bond80 or by the direct polymerisation of a DOX-containing vinyl monomer,79 obtained by the reaction between thiolated DOX and pyridyldisulphide ethyl methacrylate. Under similar acidic conditions, the cumulative drug release was higher for the nanosystem functionalised with DOX (80%) compared to the one derived from the DOX-based vinyl monomer (50%), which was attributed to the low solubility of the drug released from the latter nanosystem. In fact, by adding 0.1% of a surfactant to the releasing medium, the authors were able to improve drug release, reaching 85%, which is comparable to the one observed for the DOX-functionalised micelles. However, it took longer for this nanosystem to achieve the release plateau (20 h compared to 48 h).
The development of PDPA-containing prodrugs for combinational chemotherapy with DOX and camptothecin (CPT) aiming to achieve superior performance has also been reported.81 In this study, the research team took advantage of the multifunctional and biocompatible β-cyclodextrin (β-CD) molecule to prepare amphiphilic star-shaped β-CD-P(DPA-co-OEOMA-co-CPT) copolymers by ATRP. While CPT was introduced into the copolymer through the polymerisation of a CPT-based vinyl monomer containing a bioreducible disulphide linkage, DOX was encapsulated in the hydrophobic β-CD cavity during the self-assembly of the copolymers in water. This elegant design allowed the efficient release of both drugs under tumour environment-mimicking conditions, owing to the dual-responsive nature (bioreduction and pH) of the nanocarrier. Cell viability assays also showed that encapsulation of DOX by the β-CD-P(DPA-co-OEOMA-co-CPT) copolymer potentiated the therapeutic effect of this nanosystem against HeLa and MCF-7 cancer cells. In fact, combinational chemotherapy has been recognised as a useful strategy to achieve synergistic effectiveness of drugs and reduce side effects.82 In support of this, studies have shown that PDPA-containing vesicles could be used for the simultaneous encapsulation and delivery of hydrophobic and hydrophilic drugs for the treatment of oral head and neck squamous cell carcinoma83 and endometrial carcinoma,84 showing superior performance than the single-drug loaded vesicles. For example, cell survival of an in vitro 3D tumour model of FaDu squamous cell carcinoma was about 50% when treated with PDPA-based vesicles loaded with DOX and PTX, whereas about 70% or 60% of cell survival were verified using monotherapy of PTX or DOX, respectively.83 Regarding the endometrial carcinoma, in vivo studies with mice revealed impressive results when using combined therapy of navitoclax and DOX delivered by PEG-b-PDPA block copolymers. Tumour volume increased about 50% 35 days after treatment with combinational therapy, whereas the same increased about 650% and 1000% using navitoclax or DOX monotherapy, respectively.84
This section shows that DOX has been usually selected as an anticancer model drug, while PEG-b-PDPA copolymers and their variations have been the most investigated polymers for drug delivery to prolong the circulation life of the resulting carriers. There are a variety of polymeric structures that can be efficiently used to produce PDPA-based carriers, such as microgels, vesicles, micelles and crosslinked micelles. Fortunately, many studies have demonstrated the applicability of PDPA-based copolymers in different types of specific cancers, such as breast cancer, lung cancer and hepatocellular carcinoma (Table 1), with few of them also presenting valuable in vivo investigation. This shows the increasing interest in PDPA as pH-sensitive segment for cancer therapy.
| Polymer | Drug | Type of carrier | Cancer type (cells) | Ref. |
|---|---|---|---|---|
| a Prodrug. b No drug was used. The authors studied the internalisation and biodistribution of the polymersomes in cancer cells. | ||||
| PEG-b-PDPA | DOX | Micelle | Breast cancer (MCF-7/ADR cells and 4T1 cells) | 62 |
| Dextran-g-P(DPA-co-TIBMA) | DOX | Micelle | 43 | |
| PDPA-DSDMA-PEG | DOX | Microgel | 69 | |
| β-CD-P(DPA-co-OEOMA-co-CPT)a | CPT + DOX | Unimolecular micelle | 81 | |
| PEG-b-PDPA | DOX + Navitoclax | Vesicle | Endometrial carcinoma (Ishikawa cell line) | 84 |
| PEG-b-PDPA | TMZ | Micelle | Glioma (C6 cells and U87MG cells) | 56 |
| PEG-b-PDPA-b-PS | DOX | Vesicle | 75 | |
| PEG-b-PDPA-b-PFPMA-DOXa | DOX | Core–shell–corona NP | Hepatocellular carcinoma (HepG 2 cells) | 80 |
| PEG-b-PDPA-b-MALDOXa | DOX | Core–shell–corona NP | 79 | |
| PEG-b-PDPA-b-PG | EPI | Micelle | 57 | |
| PEG-b-P(DPA-co-APMA)-DOXa | DOX | Micelle | 76 | |
| FA-PMAgGP-b-P(DPA-co-PDEMA) | DOX | Core-crosslinked NP | 55 | |
| FA-PMPC-b-PDPA | TAM + PTX | Micelle | Leukaemia (K-562 cells) colon carcinoma (Caco-2 cells) | 46 |
| PEG-b-PDPA | β-lap | SPION-micelle | Lung cancer (A549 cells) | 65 |
| PEG-P(DPA-co-GlyMA) | DOX | Crosslinked vesicle | 52 | |
| PEG-b-P(DPA-co-DTM) | DOX | Micelle | 63 | |
| PHPMA-b-PDPA | DOX | Polymersome | Lymphoma (EL4 cells) | 85 and 86 |
| PMPC-b-PDPA | PTX + DOX | Polymersome | Oral head and neck squamous cell carcinoma (SCC4 and HNSCC cells) | 83 |
| PEG-b-PDPA and PMPC-b-PDPA | Polymersome | 53 | ||
2.2. PDPA-based gene carriers for cancer treatment
Because of the ability of cationic PDPA to bind anionic nucleic acids via electrostatic interactions to form polyplexes (Fig. 1), several authors have reported the synthesis and characterisation of different PDPA-based copolymers, such as folic acid-PMPC-b-PDPA,27 PDPA-co-PDEAEMA (PDEAEMA, poly(2-diethylamino ethyl methacrylate)),87 PMPC-b-PDPA,28 PHPMA-b-PDPA,88 and an alkyne-functionalised PDPA,89 as potential carriers for gene therapy.Recently, Grimme and co-authors demonstrated the utility of PDPA to achieve gene silencing via transfection of antisense oligonucleotides, by preparing and evaluating a series of self-assembled amphiphilic PDMAEMA-b-poly((hydrophobic monomer)-co-DPA) block copolymers, using copolymers without the PDPA segment as control samples.90In vitro tests with a human embryonic kidney cell line that expresses a destabilised green fluorescent protein (deGFP-HEK) revealed that the presence of PDPA in the resulting nanocarriers increased gene silencing up to six times (silencing in PDPA-free micelles ranged from 15% to 68%, while in PDPA-containing micelles it ranged from 64% to 90%). However, this performance was dependent on the copolymer composition, with PDMAEMA-co-PDPA-co-poly(lauryl methacrylate) and PDMAEMA-co-PDPA-co-poly(stearyl methacrylate) performing the best. The cellular viability observed with the PDPA-containing micelles was lower (<40%) than that of the PDPA-free micelles (up to 65%), suggesting that this pH-responsive polymer could contribute to some degree of cytotoxicity. Nevertheless, the former exhibited lower cytotoxicity than Lipofectamine 2000 (94% silencing, 2% viability), which is a commercial transfection reagent for siRNA and pDNA, considered an industry-standard in the work reported.
PDPA-based polymers have also shown the ability to transfect different types of DNA. For example, plasmid DNA (pDNA) was encapsulated by polymersomes composed of POEOMA-b-PDPA block copolymers (up to 60% encapsulation) and then assembled into multilayered capsules. The deprotonation of the PDPA segment due to the increase in the surrounding medium's pH (in vitro) allowed the delivery of the nucleic acid, although complete release was not observed, which was attributed to the inability of pDNA to penetrate the layers forming the shell of the capsules.91 Another work shows that in human fibroblasts and HeLa cancer cells, no cytotoxicity was observed for polyplexes with this copolymer, even at N/P ratios as high as 50 (N/P, molar ratio between the nitrogen groups from the polymer and the phosphate groups from the nucleic acid).29 Higher encapsulation efficiencies were also achieved (55% to 95%) in this study and live cell imaging revealed that, in in vitro tests with HeLa cells, the genetic material was able to reach the cell's nucleus.29 Studies have also demonstrated that polymersomes composed of PMPC-b-PDPA copolymers can also encapsulate pDNA with acceptable efficiency (50–64%),92 and deliver it into HEK293T cells.93 The encapsulation of salmon DNA was also reported using PEG-b-PDMAEMA-b-PDPA94 and hyperbranched POEOMA-co-PDPA49 copolymer-based carriers. Polyplexes from the latter copolymer revealed to be sufficiently non-toxic towards the HEK293 healthy cell line, with cell viability ≈100% for polymer concentrations up to 100 μg mL−1 and PDPA content in the copolymers up to 29 wt%. Higher content of PDPA in the copolymers (54 wt%) led to decreased cell viability, even for the lowest concentration investigated (25 μg mL−1, cell viability around 85%). A POEOMA-b-P(OEOMA-co-DEA-co-DPA) (DEA, 2-(diethyl amino)ethyl methacrylate) copolymer was investigated as a carrier for calf thymus DNA95,96 delivery in the Telo-RF cell line. The resulting polyplexes presented high cytocompatibility (cell viability ≈100%) but the DNA delivery was very low (up to 35% cell uptake and 4% gene expression). Adding the gene delivery “gold standard” PEI to these polyplexes improved cellular uptake and gene expression (by up to 40% and 10%, respectively) at the expense of a slight increase in cytotoxicity (cell viability ≈80%).
The possibility of using PDPA copolymers for RNA-based therapies has also been reported with great success using micro RNAs24 and small interfering RNAs,97,98 showing the versatility of this pH-responsive polymer. For example, POEOMA-b-PDPA block copolymers have been prepared for the delivery of pre-miRNA-29b and the results showed high RNA encapsulation (≈90%) and polyplexes with high cytocompatibility (cell viability ≈100%) towards human skin fibroblasts and A549 cell lines.24 In addition, the gene transfection was highly efficient (97%) and a significant reduction in gene expression (51%) was observed in A549 cancer cells. Likewise, the delivery of siRNA-FITC by PEO-b-PDPA-b-PAA (PAA, poly(acrylic acid)) block copolymers inhibited the proliferation of cancer stem cells very efficiently (close to 100%).98 In this work, the PAA segment was used to link streptavidin, allowing to produce EpCAM-Ab-labelled vesicles, to target the delivery of RNA.
Besides studies dealing with encapsulation and delivery of different types of nucleic acids using PDPA-based copolymers, there are also a few works investigating the utility of this pH-responsive polymer for the treatment of specific types of cancer, namely breast cancer and prostate cancer. For example, the downregulation of ATP6, a protein coded in mitochondrial DNA, in MDA-MB-231 breast cancer cells has been studied.97 To achieve so, the authors prepared nanoparticles (NPs) of PEG-b-PDPA loaded with MT-NPs(siATP6) (Fig. 5). The efficacy of the proposed therapy was investigated in vitro (down-regulation of 80%) and results also showed that the blood circulation of siRNA in vivo was prolonged (3 times) in healthy BALB/c female mice. The anticancer effect in mice with 4T1 orthotopic tumours was verified by an impressive 5-fold decrease in tumour size. The simultaneous delivery of siRNA and drugs, such as DOX,99,100 amphotericin B101 and cisplatin produg102 have also been explored with PDPA-based copolymers to treat breast cancer with promising results. The rationale behind this co-delivery strategy is that these molecules can promote RNA endosomal escape synergistically, thus enhancing the therapeutic effect of the nanosystems.99,101 The successful co-delivery of Bcl-2-siRNA and DOX into MDA-MB-231 cells for the treatment of triple-negative breast cancer using a PEG-b-PDMAEMA-b-PDPA block copolymer has been reported.100 The developed polymer exhibited suitable cytocompatibility (cell viability >80%) and the delivery of siRNA by the carriers efficiently suppressed antiapoptotic mechanisms by downregulating Bcl-2 mRNA expression by 50% with a 100 nM concentration of siRNA. The therapy based on Bcl-2-siRNA and DOX decreased the viability of the treated cancer cells significantly, to as low as 40%. For the treatment of metastatic breast cancer, siRNA-p65 and alkylated cisplatin prodrug were co-delivered by PEG-b-PEDAGA-b-PDPA-based polyplexes (PEDAGA, poly(glycidyl methacrylate) modified with ethylene diamine).102 The in vitro transfection of siRNA promoted silencing up to 80% of the expression of green fluorescent protein in A549-GFP cells, while cell viability remained above 90%. When transfecting siRNA-p65 in vitro into 4T1 cancer cells, silencing of NF-κB subunit p65 reached 50% and expression of matrix metalloproteinase-9 was inhibited by 40%. The co-delivery of genetic material and drug via polyplexes induced 95% cellular apoptosis in vitro, with great reduction of tumour growth (more than 4 times less tumour volume) and metastasis in vivo with 4T1 tumour bearing nude mice (Fig. 6). The authors also verified a satisfactory biosafety of the developed nanosystem.
 | ||
| Fig. 5 Schematic representation of (A) preparation and (B) transfection mechanism of polyplexes of PEG-b-PDPA loaded with MT-NPs(siATP6). (a) polyplex uptake by the cell, (b) RNA endosomal escape, (c) transfection into the mitochondria, (d) gene transduction, (e) mitochondrial damage due to gene expression, (f) leakage of mitochondrial DNA and (g), (h) tumour growth suppression. Copyright (2025) Wiley. Used with permission from.97 | ||
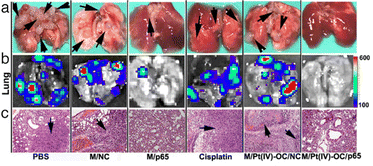 | ||
| Fig. 6 The effect of polyplexes co-delivering siRNA-p65 and Pt(IV)-OC (hydrophobic cisplatin prodrug) in suppressing lung tumour growth metastasis. In this Fig., M denotes the PDPA copolymer polyplex and different acronyms are used to describe what is being loaded into the polyplex: NC denotes siRNA-NC; p65 denotes siRNA-965; Pt(IV)-OC denotes the hydrophobic cisplatin prodrug. Lung metastasis of 4T1 breast tumour recorded after 24 days via photo (a), BLI imaging (b) and H&E staining (c). (The black arrows indicate the location of metastasis nodules in the lung) (200× for H&E and tunnel staining images). The black arrows in a and c indicate the presence of metastasis nodules in the lungs. Adapted from102 with permission from Ivyspring. | ||
For the treatment of prostate cancer, only two different PDPA-based carriers for gene delivery have been reported. In one study, a POEOMA-co-PDPA copolymer was investigated as a carrier for the pro-apoptotic BikDDA gene delivery and no cytotoxicity was observed in both healthy PNT1A and LNCaP prostate cancer cell lines.103 The prepared polyplexes were functionalised with a targeting moiety for LNCaP cells, which enhanced cellular uptake, doubling Bik gene expression and decreasing cancer cell viability by 50%. With the goal of downregulating Arf6 protein expression in PC-3 human prostate cancer cells, mPEG-b-poly(L-lysine)-b-PDPA-based polyplexes were used to co-deliver DOX with either siRNA against Arf6 or scrambled siRNA.99 The delivery of scrambled siRNA did not influence the downregulation of the desired protein, which was successfully achieved with siRNA against Arf6. The presence of the latter in the polyplex enhanced DOX cytotoxicity in cancer cells, generating apoptosis rates of 61%.
The different studies reviewed in this section and summarised in Table 2 demonstrate that PDPA-based copolymers have been successfully employed as vehicles for the transfection of genetic material into human cells. In particular, the copolymers are amphiphilic and the PDPA segment is responsible for the interaction with nucleic acids, forming the core of the polyplexes. As reported, PDPA-based copolymers can exhibit good encapsulation and release capacities (reported values can reach 90% RNA encapsulation) and the transfection of nucleic acids into tumour cells showed promising results for the treatment of cancer. Not only did the viability of tumour cells decrease, but in vivo tests also proved that tumour growth was slower than that of control samples. Although the studies reveal that multiple PDPA-based copolymers are adequate to deliver both RNA and DNA molecules, opening the possibility to treat different types of cancers, most of the proposed polymers are still very far from being used in human trials. Nevertheless, research on PDPA-based gene carriers seems to be progressing favourably, given that some studies have already reported in vivo results, albeit in their early stages of development. In summary, although there are only a handful of studies dealing with this topic, it can be stated that this approach to treat cancer seems extremely promising and will certainly see a surge in interest in the short-term.
| Polymer | Genetic material | Cells transfected (cancer type) | Ref. |
|---|---|---|---|
| PDMAEMA-co-PDPA-co-poly(lauryl methacrylate) | deGFP knockdown ASOs | deGFP-HEK | 90 |
| PDMAEMA-co-PDPA-co-poly(stearyl methacrylate) | |||
| POEOMA-b-PDPA | Plasmid DNA | HeLa (cervical cancer) | 29 |
| PMPC-b-PDPA | Plasmid DNA | HEK293T | 93 |
| POEOMA-co-PDPA | Salmon DNA | HEK293 | 49 |
| POEOMA-b-P(OEOMA-co-DEA-co-DPA) | Calf thymus DNA | Telomerase immortalised rhesus fibroblasts – Telo-RF | 95 and 96 |
| POEOMA-b-PDPA | Pre-miRNA-29b | Human skin fibroblasts A549 (lung cancer) | 24 |
| PEO-b-PDPA-b-PAA | siRNA-FITC | EpCAM + cancer stem cells (liver cancer) | 98 |
| PEG-b-PDPA | siATP6 | MDA-MB-231 (breast cancer) | 97 |
| PEG-b-PDMAEMA-b-PDPA | Bcl-2-siRNA | MDA-MB-231 (breast cancer) | 100 |
| PEG-b-PEDAGA-b-PDPA | siRNA-p65 | A549-GFP; 4T1 (breast cancer) | 102 |
| POEOMA-co-PDPA | BikDDA | PNT1A LNCaP (prostate cancer) | 103 |
| mPEG-b-poly(L-lysine)-b-PDPA | siRNA against Arf6 | PC-3 (prostate cancer) | 99 |
2.3. PDPA-based carriers for photodynamic and photothermal therapies for cancer treatment
Therapies based on the use of light in combination with nanoparticles loaded with photosensitive agents can be employed for cancer diagnostics and/or treatment. Irradiated photosensitive molecules emit fluorescence, which can be used for imaging to detect tumours. On the therapeutic side, photoactive agents can act in two different ways: (i) generation of radicals that induce oxidative stress and destroy cancer cells (photodynamic therapy, PDT)74,104 or (ii) increase of tumour temperature to kill cancer cells via hyperthermia (photothermal therapy, PTT).37,105 Photo-based cancer therapies offer the advantage of being more localised in the irradiation site, causing minimal damage to surrounding healthy tissues (Fig. 7).106 Most of the photosensitive agents are water-insoluble and unspecific.31 Therefore, one of the strategies used to improve the efficacy of these treatments is the encapsulation/incorporation of the therapeutic agents into nanoparticles that can respond to physiologic stimuli (e.g., pH), such as PDPA, allowing the fine-tuning of their release.31,37,105,107–110 | ||
| Fig. 7 Schematic illustration of cell damage by PTT and PDT after polymeric nanocarrier endosomal escape and photosensitiser release. | ||
Amphiphilic PEG-b-PLA and PEG-b-PDPA block copolymers were synthesised and self-assembled into micelles, to better understand the effects of micelle core environment in the generation of either singlet oxygen (1O2) or radical and radical anion species (HO˙ and O2˙−) by 5,10,15,20-tetrakis(meso-hydroxyphenyl) porphyrin (mTHPP) under light irradiation (532 nm laser).109 The authors concluded that in an electron-rich environment (due to the presence of the PDPA segment in the nanocarrier) there was an increase in O2˙− generation and a decrease of 1O2 production, being this generation further increased under hypoxic conditions. It was also shown that the PDPA-based carrier was significantly more phototoxic to cancer cells than the PEG-b-poly(D,L-lactide)-based analogue in the absence of air. This work demonstrated the importance of the PDPA-segment in the enhancement of PDT effects under hypoxic conditions, which are present in the inner regions of many cancers.109 POEOMA-co-PDPA nanoparticles were developed to deliver meta tetra(hydroxyphenyl)chlorin (mTHPC) to HT-29 cancer cells.31In vitro studies revealed that 58% of mTHPC was released after 48 hours at pH 5.0, while only 10% was released at pH 7.0, as a result of the pH-sensitive nature of PDPA. NIH/3T3 normal cells viability remained unaffected when treated with either unloaded nanocarriers or photosensitiser-loaded nanoparticles without light irradiation. However, under 435 nm light irradiation, the loaded nanoparticles induced cytotoxicity in HT-29 colorectal adenocarcinoma cells, achieving 50% mortality at mTHPC concentrations of 0.22 and 0.08 μg mL−1 with respective light doses of 0.36 and 1.43 J cm−2.
The use of monotherapy has its downfalls. Lately, most studies have investigated the use of combined therapies (such as chemotherapy and photothermal/photodynamic therapies) in hopes of enhancing therapeutic efficacy and minimising limitations by synergistic effects.104 For example, poly(aspartic acid-butanediamine) (PAsp(DAB))-b-PDPA-based micelles coated with a discontinuous gold nano shell (termed GNS@PDPM) were prepared for the delivery of DOX to C6 glioma cells.107 Importantly, the authors demonstrated that at neutral pH (7.4) less than 10% of DOX was released in 24 h in the presence or absence of near-infrared (NIR) laser irradiation, confirming the high stability of the nanosystem. In contrast, under acidic conditions mimicking the tumour environment (pH = 5.0), 65.9% of the drug was released in 24 h when no laser was used. This value further increased to 71% with the use of NIR laser irradiation for 5 min within the same time frame, possibly due to laser-induced deformation of the nano shell. Similarly, another research team synthesised unimolecular micelles composed of β-CD-P(DPA-co-OEOMA) random copolymers obtained by ATRP for the delivery of DOX and BBT-2FT to HeLa cells.37 This carrier was designed to avoid premature release of both chemotherapeutic drug and BBT-2FT photothermal agent. However, this system showed inferior performance compared to the previous example,107 as approximately 20% of DOX was released at physiological pH with and without NIR laser irradiation, whereas at acidic pH with laser treatment, this percentage increased to 52%. These two examples of combinational therapy also showed the capability of the nanocarriers to release DOX intracellularly, as after just 6 h107 and 2 h37 this drug was detected by fluorescence inside the cell nuclei, a site of action of this drug. Both reports demonstrated that the combination of chemo-photothermal therapy with a PDPA-based nanocarrier allowed a controlled and targeted release of the drug in tumour-like conditions, which could lower the risk of side effects.
Recently, a library of very complex nanosystems for the delivery of small PROTACs (proteolysis targeting chimeras) for cancer therapy has been reported (Fig. 8).110 The purpose of these carriers was to degrade protein BRD4 in a more efficient and tumour-specific way. Generically, the poly-PROTAC NPs were composed of self-assembled block copolymers in the form of micelles, with an extracellular matrix metalloproteinase-2 (MMP-2)-labile PEG corona and a PDPA-based core bearing redox-labile PROTAC molecules (via disulphide bond) (Fig. 8(a)). The presence of the cleavable MMP-2 spacer allowed better cellular uptake and improved deep tumour penetration (approximately 2.8- and 3.0-fold higher fluorescence, respectively, compared to control NPs (without the MMP-2 labile segment) in MDA-MB-231 breast cancer cells). Owing to the pH-sensitivity of PDPA moieties and the presence of disulphide bonds in the nanocarrier, therapeutic agents (PROTACs) were released inside the acidic and reductive environment of the tumour. The developed nanoplatform was further functionalised with an azide group to promote an in situ biorthogonal click reaction with dibenzocyclooctyne-loaded pretargeted nanoparticles, which are composed of a PEG corona and a poly(2-(ethyl(propyl)amino)ethyl methacrylate)-based core. This strategy resulted in a 3.9-fold increase in nanoplatform accumulation inside the tumour (Fig. 8(b)). Additionally, the synergistic effect of this “click” strategy with PDT (671 nm laser) afforded a more efficient BRD4 degradation, caspase-3 activation and subsequent apoptosis induction (Fig. 8(c)), leading to a 40% increase in mouse survival while ensuring good biosafety.
 | ||
| Fig. 8 (a) Chemical structure of POLY-PROTACs present in the produced NPs, (b) the involvement of these carriers in a biorthogonal “click” reaction and their proposed disassembly process. (c) Schematic illustration of POLY-PROTAC NPs mechanism of action after the injection of a first carrier, followed by irradiation and the injection of a second carrier (distinct from the first one). Reproduced from,110 with permission from Springer Nature. | ||
The synthesis of pH and redox-sensitive PCL-SS-b-P(DPA-co-GMA-co-MPC) block copolymers has been reported.108 These polymers self-assembled into micelles and were crosslinked with amino-PEG-decorated copper sulphide nanoparticles (termed DGM-CuS crosslinked NPs) to deliver DOX.98 Non-crosslinked micelles released about 50% of the drug in 24 h under physiological conditions, while crosslinked nanocarriers released approximately 30%, which reveals an inferior performance compared to aforementioned strategies. This suggests that these nanocarriers may eventually have an increased risk of causing side effects, due to early drug release. Additionally, DGM-CuS crosslinked NPs DOX release was evaluated in acidic environment, in reductive environment or under irradiation. In acidic or reductive environments, DOX release after 6 h was nearly 100%, while laser irradiation at physiological pH increased drug release of the crosslinked carrier to nearly 40%. These data demonstrate the carrier's multi-responsive nature, which allows a precise drug release to the tumour site. This study also validated the nanocarrier's safety and efficacy in vivo. Although tumour targeting properties were unsatisfactory, their antitumour effect upon irradiation was significant. Specifically, tumours treated with DGM-CuS loaded crosslinked NPs (DGM-CuS@DOX) and DGM-CuS@DOX-P micelles (carriers modified by peptides to enhance targeting) and irradiated with a NIR laser showed minimal growth or regression by the end of treatment. Ex vivo analysis revealed almost no tumour tissue at inoculation sites and the mice maintained stable weight, indicating the carriers’ safety and efficacy.
Identical to the aforementioned study, a nanocarrier based on dual temperature- and pH-responsive poly(N,N-diethyl acrylamide)-b-PDPA block copolymers and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-PEG for the combined transport of a chemotherapeutic agent (CLT: celastrol) and indocyanine green (a photosensitiser that generates heat when stimulated by NIR irradiation) has been developed.105 This nanoplatform, termed CIPP, was designed to respond to changes in pH and temperature by varying its size, aiming for better penetration into deep tumour tissue and prolonged accumulation in the tumour to improve therapeutic efficiency. It was demonstrated that at pH 7.4 after 48 h, CLT release reached about 55% while at acidic pH (5.5) the drug release increased to 93% without irradiation and 97% with light treatment, suggesting that pH could be a more appropriate stimulus than light in this case. The authors observed that this nanocarrier improved the internalisation of a fluorescent probe (coumarin) in B16F10 melanoma cells when compared to the free probe. The results also confirmed that the nanoplatform accumulated in the lysosomes after 1 h of incubation and that successful lysosomal escape occurred after 5 h. In vivo and ex vivo studies showed that tumour accumulation (3.3-fold higher fluorescence) and penetration were superior in the laser-treated CIPP group than in the control without radiation. Overall, the designed nanosystem was able to eradicate tumours within 12 days in a safe manner, as the mice maintained their body weight and did not present lesions in the main organs. Supramolecular micelles (SPNs) composed of 6-arm star-shaped P(DAPA-co-DPA-co-OEOMA)6 (DAPA: diaminopyridine acrylamide) copolymers capable of carrying both 1-hexylcarbamoyl-5-fluorouracil (HCFU) as a chemotherapeutic agent and thymine-functionalised tetraphenylporphyrin (TTPP) as a photosensitiser have also been explored for combination phototherapy.111 This study showed that after 96 h at physiological pH, the platform released approximately 10% of each drug, while at lysosomal pH (5.0), it surpassed 80%. Moreover, the loaded carrier was able to kill about 90% of the MCF-7 breast cancer cells upon irradiation (655 nm laser), contrary to what was observed without laser treatment (near 40%). In vivo studies revealed that 24 h after intravenous injection of HCFU(72%)-TTPP(28%)-SPNs, there was an accumulation of the nanoplatform in the tumour site, preferentially and in the liver. Upon treatment with three injections of this nanosystem followed by 655 nm laser irradiation, the tumour inhibition growth rate was almost 98%, while the mice did not present significant weight changes or abnormalities in the main organs (e.g., heart and liver).
The different studies on PDT and PTT using PDPA as a segment of the nanocarriers demonstrate the potential of this polymer in multiple cancer treatment applications. Specifically, the pH-sensitivity of PDPA can extend the circulation time of drug-loaded carriers, allowing better tumour accumulation and penetration while decreasing the probability of side effects. Nevertheless, some of the platforms tested still lack tumour specificity, accumulating in the liver. The use of PDPA-based platforms in combinational therapies using light seems to be in an exploratory phase, judging by the lack of focus of the few published studies on a specific cell line and drug (Table 3).
| Polymer | Drug | Photosensitiser/photothermal agent | Cancer type (cells) | Ref. |
|---|---|---|---|---|
| PCL-SS-P(DPA/GMA/MPC) | DOX | Copper sulphide NPs | Breast cancer (4T1 cells) | 108 |
| 6 arm-P(DAPA-co-DPA-co-OEGMA) | HCFU | TTPP | Breast cancer (MCF-7 cells) | 104 |
| PEG-GALGLPG-b-P(DPA-co-PROTAC) | PROTACs | Pheophorbide A | Breast cancer (MDA-MB-231 cells) | 110 |
| β-CD-P(OEGMA-DPA) | DOX | BBT-2FT | Cervical cancer (HeLa cells) | 37 |
| PAsp(DAB)-b-PDPA | DOX | Gold nano shell | Glioma (C6 cells) | 107 |
| PEG-co-PDPA | — | mTHPC | Colorectal adenocarcinoma (HT-29 cells) | 31 |
| PEG-b-PDPA | — | mTHPP | Lung cancer (A549 and H2009 cells) | 109 |
| Prostate cancer (PC-3 cells) | ||||
| PDEAA-b-PDPA | CLT | ICG | Melanoma (B16F10 cells) | 105 |
3. Conclusions
Oncological diseases affect millions of people and constitute one of the primary leading causes of mortality worldwide. Extensive research on this topic has directed the attention of the scientific community toward the use of PNPs as promising vehicles for therapeutic molecules to treat cancer, with the objective of increasing therapeutic efficacy and reducing adverse side effects when compared to free drug administration. Indeed, polymeric nanocarriers are very versatile, easy to prepare and can efficiently encapsulate and deliver various types of therapeutic molecules, such as small drugs, genetic material and photosensitisers, to target cancer cells, providing a wide range of available treatments. One of the most extensively investigated strategies is the use of pH-responsive polymers, such as PDPA, that abruptly alter their solubility upon minor changes in the pH of the surrounding aqueous environment. Compared with normal cells, the acidic pH gradient observed in cancer cells has been employed to trigger the release of the carried therapeutic molecules at the tumour site. Various PDPA-based copolymers have been engineered and have also demonstrated promising in vitro and in vivo results in the treatment of several types of cancer through drug delivery, gene therapy or photodynamic therapies. The versatility of this polymer is noteworthy and numerous potential nanoparticle delivery systems can be developed. The studies indicate that the combination of PDPA with other functionalities, such as targeting molecules or redox-sensitive segments, in the same copolymer can enhance the resulting nanocarriers’ therapeutic efficacy. Furthermore, the use of combinational therapy (e.g., simultaneous drug delivery and photodynamic therapy) with PDPA-based multivalent systems has resulted in higher cellular uptake and improved tumour penetration of the nanocarriers. Unfortunately, most of the studies reported have been conducted with model cancer cells, such as HeLa cells, which may not translate the performance of the nanocarriers in real application. Therefore and due to the scarce promising in vivo antitumour effects reported, we consider that future directions should be focused on studies involving in vivo models. Moreover, there is a need to evaluate the biocompatibility of the PDPA homopolymer to better assist the development of new drug/gene carriers. The investigation of the yet unexplored temperature-responsive character of PDPA could also open new perspectives on the application of this polymer in cancer therapy. Undoubtedly, macromolecular engineering is a powerful tool that will continue to be employed for the design of more potent and personalised treatments based on polymeric drug nanocarriers for the treatment of challenging diseases. From a materials science perspective, it is anticipated that the interest in PDPA-based copolymers for medical applications will increase and significant attention will be given to the development of biocompatible and multifunctional polymers.Abbreviations
| APMA | Aminopropyl methacrylamide |
| ATRP | Atom transfer radical polymerisation |
| BBT-2FT | Benzo[1,2-c;4,5-c′]bis[1,2,5]thiadiazole-4,7-bis(9,9-dioctyl-9H-fluoren-2-yl)thiophene |
| β-CD | β-Cyclodextrin |
| CLT | Celastrol |
| CPT | Camptothecin |
| DAPA | Diaminopyridine acrylamide |
| DEA | 2-((Diethyl amino)ethyl methacrylate) |
| DOX | Doxorubicin |
| DSDMA | Bis(2-methacryloyloxyethyl)disulphide |
| DTM | Dithiomaleimide |
| EPI | Epirubicin |
| FA | Folic acid |
| GlyMA | Glycidyl methacrylate |
| HCFU | 1-Hexylcarbamoyl-5-fluorouracil |
| ICG | Indocyanine green |
| MALDOX | DOX-based vinyl monomer |
| MMP-2 | Metalloproteinase-2 |
| mTHPC | Meta tetra(hydroxyphenyl)chlorin |
| mTHPP | 5,10,15,20-Tetrakis(meso-hydroxyphenyl)porphyrin |
| NPs | Nanoparticles |
| PAA | Poly(acrylic acid) |
| PAE | Poly(γ-L-glutamic acid) |
| PAsp(DAB) | Poly(aspartic acid-butanediamine) |
| PBAEs | Poly(β-amino ester)s |
| PCBMA | Poly(carboxybetaine methacrylate) |
| PCL | Poly(ε-caprolactone) |
| PDEAEMA | Poly(2-diethylamino ethyl methacrylate) |
| PDEMA | Poly(pyridyl disulphide methylacrylate) |
| PDMAEMA | Poly(2-(dimethylamino)ethyl methacrylate) |
| pDNA | plasmid DNA |
| PDPA | Poly(2-(diisopropylamino)ethyl methacrylate) |
| PDT | Photodynamic therapy |
| PEDAGA | Poly(glycidyl methacrylate) modified with ethylene diamine |
| PEI | Poly(ethyleneimine) |
| PFPMA | Poly(4-formylphenyl methacrylate) |
| PHPMA | Poly(N-(2-hydroxypropyl) methacrylamide) |
| PG | Poly(2-guanidinoethylmethacrylate) |
| PGA | Poly(γ-L-glutamic acid) |
| PMAgGP | Poly(6-O-methacryloyl-D-galactopyranose) |
| PMPC | Poly(2-(methacryloyloxy)ethyl phosphorylcholine) |
| PNPs | Polymeric nanoparticles |
| POEOMA | Poly(oligo(ethylene oxide) methyl ether methacrylate) |
| PROTACs | Proteolysis targeting chimeras |
| PS | Polystyrene |
| PTT | Photothermal therapy |
| PTX | Paclitaxel |
| RAFT | Reversible addition–fragmentation chain transfer polymerisation |
| RDRP | Reversible deactivation radical polymerisation |
| SPION | Superparamagnetic iron oxide nanoparticles |
| SPNs | Supramolecular micelles |
| TAM | Tamoxifen |
| TIBMA | 2-(2′,3′,5′-Triiodobenzoyl)ethyl methacrylate |
| TMZ | Temozolomide |
| TTPP | Thymine-functionalised tetraphenylporphyrin |
Author contributions
Data collection: Patrícia V. Mendonça; funding acquisition: A. C. Fonseca; project administration: Patrícia V. Mendonça and A. C. Fonseca; writing – original draft: Joana S. Ferreira, João. P. Vareda, A. S. Oliveira, Jéssica S. Barbosa and Patrícia V. Mendonça; writing – review & editing: Francisca Bastos, Patrícia V. Mendonça and A. C. Fonseca. All authors have read and approved the final manuscript.Data availability
No new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has received funding under the Portuguese Resilience and Recovery Plan, through the NextGenerationEU Fund (ref. no. C644865576-00000005).Notes and references
- S. Chakraborty and T. Rahman, Ecancermedicalscience, 2012, 6, ed16 Search PubMed.
- S. Zhu, T. Zhang, L. Zheng, H. Liu, W. Song, D. Liu, Z. Li and C. Pan, J. Hematol. Oncol., 2021, 14, 156 CrossRef PubMed.
- M. Arruebo, N. Vilaboa, B. Sáez-Gutierrez, J. Lambea, A. Tres, M. Valladares and Á. González-Fernández, Cancers, 2011, 3, 3279–3330 CrossRef CAS PubMed.
- D. Debela, S. Muzazu, K. Digamo, M. Ndalama, B. Mesele, D. Haile, S. Kitui and T. Manyazewal, SAGE Open Med, 2021, 9, 20503121211034370 CrossRef PubMed.
- L. Ding, P. Agrawal, S. K. Singh, Y. S. Chhonker, J. Sun and D. J. Murry, Polymers, 2024, 16, 843 CrossRef CAS PubMed.
- C. Wang, C. Pan, H. Yong, F. Wang, T. Bo, Y. Zhao, B. Ma, W. He and M. Li, J. Nanobiotechnol., 2023, 21, 272 CrossRef PubMed.
- R. S. Riley, C. H. June, R. Langer and M. J. Mitchell, Nat. Rev. Drug Discovery, 2019, 18, 175–196 CrossRef CAS PubMed.
- J. D. Torres-Vanegas, J. C. Cruz and L. H. Reyes, Pharmaceutics, 2021, 13, 428 CrossRef CAS PubMed.
- X. Han, M. J. Mitchell and G. Nie, Matter, 2020, 3, 1948–1975 CrossRef.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed.
- S. Sur, A. Rathore, V. Dave, K. R. Reddy, R. S. Chouhan and V. Sadhu, Nano-Struct. Nano-Objects, 2019, 20, 100397 CrossRef CAS.
- A. M. Bodratti and P. Alexandridis, Expert Opin. Drug Delivery, 2018, 15, 1085–1104 CrossRef CAS PubMed.
- F. Perin, A. Motta and D. Maniglio, Mater. Sci. Eng., C, 2021, 123, 111952 CrossRef CAS PubMed.
- K. Kuperkar, D. Patel, L. I. Atanase and P. Bahadur, Polymers, 2022, 14, 4702 CrossRef CAS PubMed.
- M. Fonseca, I. Jarak, F. Victor, C. Domingues, F. Veiga and A. Figueiras, Materials, 2024, 17, 319 CrossRef CAS PubMed.
- L. Osorno, A. Brandley, D. Maldonado, A. Yiantsos, R. Mosley and M. Byrne, Nanomaterials, 2021, 11, 278 CrossRef CAS PubMed.
- W. Yang, L. Mixich, E. Boonstra and H. Cabral, Adv. Healthcare Mater., 2023, 12, 2202688 CrossRef CAS PubMed.
- R. Rai, S. Alwani and I. Badea, Polymers, 2019, 11, 745 CrossRef CAS PubMed.
- M. F. Attia, N. Anton, J. Wallyn, Z. Omran and T. F. Vandamme, J. Pharm. Pharmacol., 2019, 71, 1185–1198 CrossRef CAS PubMed.
- S. Chu, X. Shi, Y. Tian and F. Gao, Front. Oncol., 2022, 12, 855019 CrossRef CAS PubMed.
- M. Alsehli, Saudi Pharm. J., 2020, 28, 255–265 CrossRef CAS PubMed.
- N. Deirram, C. Zhang, S. S. Kermaniyan, A. P. R. Johnston and G. K. Such, Macromol. Rapid Commun., 2019, 40, 1800917 CrossRef PubMed.
- S. Agarwal, Y. Zhang, S. Maji and A. Greiner, Mate. Today, 2012, 15, 388–393 CrossRef CAS.
- B. Baptista, A. S. R. Oliveira, P. Mendonça, A. C. Serra, J. F. J. Coelho and F. Sousa, Biomater. Adv., 2023, 145, 213267 CrossRef CAS PubMed.
- L. Salminen, E. Karjalainen, V. Aseyev and H. Tenhu, Langmuir, 2022, 38, 5135–5148 CrossRef CAS PubMed.
- J. S. Correia, S. Mirón-Barroso, C. Hutchings, S. Ottaviani, B. Somuncuoğlu, L. Castellano, A. E. Porter, J. Krell and T. K. Georgiou, Polym. Chem., 2023, 14, 303–317 RSC.
- M. Licciardi, Y. Tang, N. C. Billingham, S. P. Armes and A. L. Lewis, Biomacromolecules, 2005, 6, 1085–1096 CrossRef CAS PubMed.
- Y. Ma, Y. Tang, N. C. Billingham, S. P. Armes, A. L. Lewis, A. W. Lloyd and J. P. Salvage, Macromolecules, 2003, 36, 3475–3484 CrossRef CAS.
- J. R. Góis, F. Reis, A. M. Almeida, P. Pereira, F. Sousa, A. C. Serra and J. F. J. Coelho, Colloids Surf., B, 2018, 169, 107–117 CrossRef PubMed.
- F. C. Giacomelli, P. Stepánek, C. Giacomelli, V. Schmidt, E. Jäger, A. Jäger and K. Ulbrich, Soft Matter, 2011, 7, 9316–9325 RSC.
- C.-L. Peng, L.-Y. Yang, T.-Y. Luo, P.-S. Lai, S.-J. Yang, W.-J. Lin and M.-J. Shieh, Nanotechnology, 2010, 21, 155103 CrossRef PubMed.
- J. Hu, G. Zhang, Z. Ge and S. Liu, Prog. Polym. Sci., 2014, 39, 1096–1143 CrossRef CAS.
- M. Ouchi and M. Sawamoto, Polym. J., 2018, 50, 83–94 CrossRef CAS.
- J. R. Góis, N. Rocha, A. V. Popov, T. Guliashvili, K. Matyjaszewski, A. C. Serra and J. F. J. Coelho, Polym. Chem., 2014, 5, 3919–3928 RSC.
- J. R. Góis, D. Konkolewic, A. V. Popov, T. Guliashvili, K. Matyjaszewski, A. C. Serra and J. F. J. Coelho, Polym. Chem., 2014, 5, 4617–4626 RSC.
- P. Maximiano, P. V. Mendonça, J. R. C. Costa, N. L. Haworth, A. C. Serra, T. Guliashvili, M. L. Coote and J. F. J. Coelho, Macromolecules, 2016, 49, 1597–1604 CrossRef CAS.
- T. Jia, S. Huang, C. Yang and M. Wang, J. Mater. Chem. B, 2017, 5, 8514–8524 RSC.
- X. Wang, Y. Liu and L. Yan, ChemistrySelect, 2021, 6, 6260–6267 CrossRef CAS.
- Y. Kotsuchibashi, Y. Wang, Y.-J. Kim, M. Ebara, T. Aoyagi and R. Narain, ACS Appl. Mater. Interfaces, 2013, 5, 10004–10010 CrossRef CAS PubMed.
- L. A. Fielding, S. Edmondson and S. P. Armes, J. Mater. Chem., 2011, 21, 11773–11780 RSC.
- D. Selianitis and S. Pispas, Polym. Chem., 2023, 14, 587–599 RSC.
- D. Selianitis and S. Pispas, Polym. Chem., 2021, 12, 6582–6593 RSC.
- P. Liu, P. Huang and E.-T. Kang, Langmuir, 2021, 37, 12990–12999 CrossRef CAS PubMed.
- J. R. Góis, D. Konkolewic, A. V. Popov, T. Guliashvili, K. Matyjaszewski, A. C. Serra and J. F. J. Coelho, Polym. Chem., 2014, 5, 4617–4626 RSC.
- J. R. Góis, N. Rocha, A. V. Popov, T. Guliashvili, K. Matyjaszewski, A. C. Serra and J. F. J. Coelho, Polym. Chem., 2014, 5, 3919–3928 RSC.
- M. Licciardi, E. F. Craparo, G. Giammona, S. P. Armes, Y. Tang and A. L. Lewis, Macromol. Biosci., 2008, 8, 615–626 CrossRef CAS PubMed.
- Z. Xu, P. Xue, Y. E. Gao, S. Liu, X. Shi, M. Hou and Y. Kang, J. Colloid Interface Sci., 2017, 490, 511–519 CrossRef CAS PubMed.
- A. Jäger, E. Jäger, F. Surman, A. Höcherl, B. Angelov, K. Ulbrich, M. Drechsler, V. M. Garamus, C. Rodriguez-Emmenegger, F. Nallet and P. Štěpánek, Polym. Chem., 2015, 6, 4946–4954 RSC.
- D. Selianitis, H. Katifelis, M. Gazouli and S. Pispas, Pharmaceutics, 2023, 15, 1627 CrossRef CAS PubMed.
- S. H. Min, S. K. Kwak and B.-S. Kim, Polymer, 2017, 124, 219–225 CrossRef CAS.
- T. Thavanesan, C. Herbert and F. A. Plamper, Langmuir, 2014, 30, 5609–5619 CrossRef CAS PubMed.
- F. Zhang, Q. Yao, X. Chen, H. Zhou, M. Zhou, Y. Li and H. Cheng, Drug Delivery, 2023, 30, 2162626 CrossRef PubMed.
- C. Murdoch, K. J. Reeves, V. Hearnden, H. Colley, M. Massignani, I. Canton, J. Madsen, A. Blanazs, S. P. Armes, A. L. Lewis, S. MacNeil, N. J. Brown, M. H. Thornhill and G. Battaglia, Nanomedicine, 2010, 5, 1025–1036 CrossRef CAS PubMed.
- F. Ding, S. Yang, G. Zhiliang, J. Guo, P. Zhang, X. Qiu, Q. Li, M. Dong, J. Hao, Q. Yu and J. Cui, Front. Chem., 2019, 7, 770 CrossRef CAS PubMed.
- J. Zhao, C. Yan, Z. Chen, J. Liu, H. Song, W. Wang, J. Liu, N. Yang, Y. Zhao and L. Chen, J. Colloid Interface Sci., 2019, 540, 66–77 CrossRef CAS PubMed.
- J. Tan, X. Duan, F. Zhang, X. Ban, J. Mao, M. Cao, S. Han, X. Shuai and J. Shen, Adv. Sci., 2020, 7, 2003036 CrossRef CAS PubMed.
- E. Chen, S. Han, B. Song, L. Xu, H. Yuan, M. Liang and Y. Sun, Int. J. Nanomed., 2020, 15, 6311–6324 CrossRef CAS PubMed.
- K. Bhattacharya, M. Kundu, S. Das, S. Samanta, S. S. Roy, M. Mandal and N. K. Singha, Macromol. Rapid Commun., 2023, 44, 2200594 CrossRef CAS PubMed.
- J. De Mel, M. Hossain, O. Shofolawe-Bakare, S. A. Mohammad, E. Rasmussen, K. Milloy, M. Shields, E. W. Roth, K. Arora, R. Cueto, S.-C. Tang, J. T. Wilson, A. E. Smith and T. A. Werfel, Mol. Pharmaceutics, 2022, 19, 4705–4716 CrossRef CAS PubMed.
- K. Hulugalla, O. Shofolawe-Bakare, V. B. Toragall, S. A. Mohammad, R. Mayatt, K. Hand, J. Anderson, C. Chism, S. K. Misra, T. Shaikh, E. E. L. Tanner, A. E. Smith, J. S. Sharp, N. C. Fitzkee and T. Werfel, ACS Nano, 2024, 18, 30540–30560 CrossRef CAS PubMed.
- O. Shofolawe-Bakare, V. B. Toragall, K. Hulugalla, R. Mayatt, P. Iammarino, J. P. Bentley, A. E. Smith and T. Werfel, ACS Appl. Nano Mater., 2024, 7, 28851–28863 CrossRef CAS.
- P. Yu, H. Yu, C. Guo, Z. Cui, X. Chen, Q. Yin, P. Zhang, X. Yang, H. Cui and Y. Li, Acta Biomater., 2015, 14, 115–124 CrossRef CAS PubMed.
- T. Bai, D. Shao, J. Chen, Y. Li, B. Bin Xu and J. Kong, J. Colloid Interface Sci., 2019, 552, 439–447 CrossRef CAS PubMed.
- K. Zhou, Y. Wang, X. Huang, K. Luby-Phelps, B. D. Sumer and J. Gao, Angew. Chem., Int. Ed., 2011, 50, 6109–6114 CrossRef CAS PubMed.
- G. Huang, H. Chen, Y. Dong, X. Luo, H. Yu, Z. Moore, E. A. Bey, D. A. Boothman and J. Gao, Theranostics, 2013, 3, 116–126 CrossRef CAS PubMed.
- E. Blanco, E. A. Bey, C. Khemtong, S.-G. Yang, J. Setti-Guthi, H. Chen, C. W. Kessinger, K. A. Carnevale, W. G. Bornmann, D. A. Boothman and J. Gao, Cancer Res., 2010, 70, 3896–3904 CrossRef CAS PubMed.
- E. Bey, M. Srougi, K. Reinicke, Y. Dong, C.-R. Yang, L. Girard, J. Minna, W. Bornmann, J. Gao and D. Boothman, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11832–11837 CrossRef CAS PubMed.
- J. Sikorski, M. Matczuk, M. Stępień, K. Ogórek, L. Ruzik and M. Jarosz, Nanotechnology, 2024, 35, 212001 CrossRef CAS PubMed.
- D. Huang, J. Zhu, M. F. Xu, J. Chen, X. Gao, L. Zhao, F. Ding and C. Z. Wu, Colloids Surf., A, 2024, 685, 133320 CrossRef CAS.
- M. Jaymand, J. Drug Delivery Sci. Technol., 2024, 95, 105581 CrossRef CAS.
- A. Raza, U. Hayat, T. Rasheed, M. Bilal and H. M. N. Iqbal, J. Mater. Res. Technol., 2019, 8, 1497–1509 CrossRef CAS.
- Y. Hiruta, K. Yuki, N. Katsuyama and H. Kanazawa, RSC Adv., 2017, 7, 29540–29549 RSC.
- W. Hao, D. Liu, Y. Wang, X. Han, S. Xu and H. Liu, Colloids Surf., A, 2018, 537, 446–451 CrossRef CAS.
- T.-Y. Wang and C.-Y. Chen, ACS Appl. Bio. Mater., 2019, 2, 3659–3667 CrossRef CAS PubMed.
- M. R. Younis, Y. He, X. Yao, G. He, H. Liu, P. Huang and J. Lin, Acta Biomater., 2023, 157, 442–450 CrossRef CAS PubMed.
- P. Huang, W. Wang, J. Zhou, F. Zhao, Y. Zhang, J. Liu, J. Liu, A. Dong, D. Kong and J. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 6340–6350 CrossRef CAS PubMed.
- A. Xie, S. Hanif, J. Ouyang, Z. Tang, N. Kong, N. Y. Kim, B. Qi, D. Patel, B. Shi and W. Tao, EBioMedicine, 2020, 56, 102821 CrossRef PubMed.
- J. Mao, Y. Li, T. Wu, C. Yuan, B. Zeng, Y. Xu and L. Dai, ACS Appl. Mater. Interfaces, 2016, 8, 17109–17117 CrossRef CAS PubMed.
- Y. Dong and P. Liu, Langmuir, 2021, 37, 7356–7363 CrossRef CAS PubMed.
- Y. Dong and P. Liu, Int. J. Pharm., 2020, 589, 119796 CrossRef CAS PubMed.
- Y. E. Gao, S. Bai, X. Ma, X. Zhang, M. Hou, X. Shi, X. Huang, J. Chen, F. Wen, P. Xue, Y. Kang and Z. Xu, Colloids Surf., B, 2019, 183, 110428 CrossRef CAS PubMed.
- B. Shrestha, L. Wang, E. M. Brey, G. R. Uribe and L. Tang, Pharmaceutics, 2021, 13, 853 CrossRef CAS PubMed.
- H. E. Colley, V. Hearnden, M. Avila-Olias, D. Cecchin, I. Canton, J. Madsen, S. MacNeil, N. Warren, K. Hu, J. A. McKeating, S. P. Armes, C. Murdoch, M. H. Thornhill and G. Battaglia, Mol. Pharmaceutics, 2014, 11, 1176–1188 CrossRef CAS PubMed.
- J. Ding, X. Zhang, C. Chen, Y. Huang, X. Yu and X. Li, Biomater. Sci., 2020, 8, 2264–2273 RSC.
- L. J. C. Albuquerque, V. Sincari, A. Jäger, J. Kucka, J. Humajova, J. Pankrac, P. Paral, T. Heizer, O. Janouškova, I. Davidovich, Y. Talmon, P. Pouckova, P. Štěpánek, L. Sefc, M. Hruby, F. C. Giacomelli and E. Jäger, J. Controlled Release, 2021, 332, 529–538 CrossRef CAS PubMed.
- E. Jäger, P. Černoch, M. Vragovic, L. J. Calumby Albuquerque, V. Sincari, T. Heizer, A. Jäger, J. Kučka, O. Š. Janoušková, E. Pavlova, L. Šefc and F. C. Giacomelli, Biomacromolecules, 2024, 25, 4192–4202 CrossRef PubMed.
- M. A. Beach, S. L. Y. Teo, M. Z. Chen, S. A. Smith, C. W. Pouton, A. P. R. Johnston and G. K. Such, ACS Appl. Mater. Interfaces, 2022, 14, 3653–3661 CrossRef CAS PubMed.
- F. A. de Oliveira, C. C. da, S. Batista, P. Černoch, V. Sincari, A. Jäger, E. Jäger and F. C. Giacomelli, Biomacromolecules, 2023, 24, 2291–2300 CrossRef CAS PubMed.
- S. L. Ng, J. P. Best, K. Kempe, K. Liang, A. P. R. Johnston, G. K. Such and F. Caruso, Biomacromolecules, 2014, 15, 2784–2792 CrossRef CAS PubMed.
- C. J. Grimme, M. G. Hanson and T. M. Reineke, Bioconjugate Chem., 2023, 34, 1244–1257 CrossRef CAS PubMed.
- H. Lomas, A. P. R. Johnston, G. K. Such, Z. Zhu, K. Liang, M. P. Van Koeverden, S. Alongkornchotikul and F. Caruso, Small, 2011, 7, 2109–2119 CrossRef CAS PubMed.
- H. Lomas, J. Du, I. Canton, J. Madsen, N. Warren, S. P. Armes, A. L. Lewis and G. Battaglia, Macromol. Biosci., 2010, 10, 513–530 CrossRef CAS PubMed.
- L. Wang, L. Chierico, D. Little, N. Patikarnmonthon, Z. Yang, M. Azzouz, J. Madsen, S. P. Armes and G. Battaglia, Angew. Chem., Int. Ed., 2012, 51, 11122–11125 CrossRef CAS PubMed.
- Y. Zhang, X. Li, W. Wei and X. Liu, Colloid Interface Sci. Commun., 2021, 41, 100366 CrossRef CAS.
- L. J. C. Albuquerque, C. E. de Castro, K. A. Riske, M. C. C. da Silva, P. I. R. Muraro, V. Schmidt, C. Giacomelli and F. C. Giacomelli, Biomacromolecules, 2017, 18, 1918–1927 CrossRef CAS PubMed.
- L. E. Prevette, M. L. Lynch, K. Kizjakina and T. M. Reineke, Langmuir, 2008, 24, 8090–8101 CrossRef CAS PubMed.
- R. Xu, L. Huang, J. Liu, Y. Zhang, Y. Xu, R. Li, S. Su and X. Xu, Small, 2024, 20, 2305923 CrossRef CAS PubMed.
- J. Chen, Q. Liu, J. Xiao and J. Du, Biomacromolecules, 2015, 16, 1695–1705 CrossRef CAS PubMed.
- Y. Yuan, Y. Wang, H. Huang, S. Tao and J. Huang, Macromol. Biosci., 2023, 23, 2200529 CrossRef CAS PubMed.
- H.-H. Lu, H. W. Liu, T. K. Dinh, C.-H. Huang, H.-C. Huang, Y.-C. Tseng, M.-H. Ku, F.-S. Wang, Y. Chen and C.-H. Peng, Polym. Chem., 2022, 13, 5568–5578 RSC.
- H. Yu, Y. Zou, Y. Wang, X. Huang, G. Huang, B. D. Sumer, D. A. Boothman and J. Gao, ACS Nano, 2011, 5, 9246–9255 CrossRef CAS PubMed.
- H. Yu, C. Guo, B. Feng, J. Liu, X. Chen, D. Wang, L. Teng, Y. Li, Q. Yin, Z. Zhang and Y. Li, Theranostics, 2016, 6, 14–27 CrossRef CAS PubMed.
- U. C. Oz, Z. B. Bolat, A. Poma, L. Guan, D. Telci, F. Sahin, G. Battaglia and A. Bozkır, Appl. Nanosci., 2020, 10, 3389–3401 CrossRef CAS.
- Y. Wu, S. Chen and J. Zhu, ACS Nano, 2024, 18, 4104–4117 CrossRef CAS PubMed.
- Y. Li, L. Yang, X. Xu, M. Li, Y. Zhang, Q. Lin, T. Gong, X. Sun, Z. Zhang and L. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 46361–46374 CrossRef CAS PubMed.
- P. Husni, Y. Shin, J. C. Kim, K. Kang, E. S. Lee, Y. S. Youn, T. Rusdiana and K. T. Oh, Biomedicines, 2020, 8, 618 CrossRef CAS PubMed.
- Y. Huang, X.-X. Li, L. Zhang, X.-Y. Chen, C.-B. Liu, J.-Q. Chen, Y. Wang and X.-T. Shuai, Chin. J. Polym. Sci., 2018, 36, 1139–1149 CrossRef CAS.
- Z. Wu, P. Zhang, P. Wang, Z. Wang and X. Luo, Nanoscale, 2021, 13, 3723–3736 RSC.
- H. Ding, H. Yu, Y. Dong, R. Tian, G. Huang, D. A. Boothman, B. D. Sumer and J. Gao, J. Controlled Release, 2011, 156, 276–280 CrossRef CAS PubMed.
- J. Gao, B. Hou, Q. Zhu, L. Yang, X. Jiang, Z. Zou, X. Li, T. Xu, M. Zheng, Y.-H. Chen, Z. Xu, H. Xu and H. Yu, Nat. Commun., 2022, 13, 4318 CrossRef CAS PubMed.
- Y. Wu, S. Chen and J. Zhu, ACS Nano, 2024, 18, 4104–4117 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally as first authors. |
| This journal is © The Royal Society of Chemistry 2025 |