Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Management of phyto-parasitic nematodes using bacteria and fungi and their consortia as biocontrol agents
Shashi Prabha
Yadav
 a,
Chhavi
Sharma
a,
Chhavi
Sharma
 *bc,
Puneet
Pathak
*bc,
Puneet
Pathak
 *a,
Anil
Kanaujia
d,
Mohan Ji
Saxena
d and
Anup
Kalra
d
*a,
Anil
Kanaujia
d,
Mohan Ji
Saxena
d and
Anup
Kalra
d
aAgriliv Research Foundation, Chidana, Sonipat-131306, Haryana, India. E-mail: pathakpuneet26aug@gmail.com
bUniversity Institute of Biotechnology, Chandigarh University, Mohali-140413, Punjab, India. E-mail: chhavisharma19@gmail.com
cDepartment of Biotechnology, University Centre of Research and Development, Chandigarh University, Mohali-140413, Punjab, India
dAgriliv Research Foundation, New Delhi-110092, India
First published on 23rd December 2024
Abstract
Phyto-parasitic nematodes are the main risks to the agroecosystem that cause agricultural output to decline in a variety of crops around the world. An intriguing and promising substitute for the chemical practice of shielding plants against the growing hazards of these pathogens lies in biological plant protection. This approach focuses on using biological control agents (BCAs) using microbial-based biocontrol techniques to inhibit the growth of phytopathogens responsible for plant diseases. Microbial BCAs interact with pathogens or plant hosts to increase their resistance, which may be a useful way to control the development of agricultural diseases. However, in comparison to a single strain, microbial consortia with distinct modes of action might exhibit a multifunctional and more resilient effect as a biocontrol. The market is currently offering only a small number of microbial consortia-based biocontrol interventions as these products are still in their infancy of development and demand substantial research to avert phyto-parasitic nematodes. The employment of BCAs to combat phytopathogens will become an increasingly vital component of sustainable agriculture in the future. Thus, this article provides a thorough review of the current status of bacteria and fungi and their microbial consortia-based biocontrol for plant protection research through a biological manner considering upcoming and advanced technological developments. Commercialization of biocontrol products and associated challenges and ways to overcome these hurdles are also discussed as future perspectives. The present review also summarizes the latest research done (particularly the past five years' data) on the activity of BCAs bacteria, fungi and their consortium against various plant pathogens with their enormous benefits for upgrading plant growth and defense mechanisms. The present review efficiently contributes to sustainable development goal 2, which is concerned with food security and sustainable agriculture.
Environmental significanceThe major source of our food system is contributed by the plants and a substantial proportion of plant-based food losses occurs yearly due to agricultural pests including plant parasitic nematodes. The substantial and perpetuated overuse of agrochemicals to control these pests poses serious threats to the environment by reducing the fertility and quality of soil, contamination of water sources and affecting health. Therefore, investigating feasible and secure agrochemical substitutes for combating plant diseases is crucial to preserving soil and crop quality and attaining the sustainable development goal 2 (SDG 2) related to food security and sustainable agriculture and ‘One Health Program’. One such approach is the use of biocontrol agents (bacteria and fungi or their consortia) using different microorganisms to control nematodes. It is vital to comprehend the importance of potential microbes and their synergistic association for future commercialization as a potent biocontrol product to mitigate crop losses and boost agricultural productivity, thereby safeguarding the environment (soil, water) and improving health by avoiding the harmful impact of conventionally used agrochemicals. |
1. Introduction
Food quality and quantity are continuously being depleted during production, which is defined as food loss, negatively impacting food security and safety globally. According to the United Nations, the 8 billion world population in 2023 is predicted to increase up to 9.7 billion by 2050. The growing global population is expected to boost food demand as well.1 The decline of food production is one of the primary causes of famine and malnutrition.2 There is a growing need for food as the world's population rises, with plants making up the primary food source globally.Plants cover approximately 80% of the food we consume. However, agricultural pests, including plant parasitic nematodes (PPNs), cause around 40% of food crop losses yearly.3 Plant diseases, both endemic and newly surfacing, are spreading and getting worse because of several factors, including pathogen repercussions, the creation of novel pathogen lineages, transmission through international food trade networks, and climate change.4
Though agrochemicals have boosted agricultural output and quality to fulfill the world's expanding requirement for food security,5 the agricultural soil system has become contaminated because of the substantial and perpetuated use of agrochemicals.5 Artificial compounds (synthetic chemicals) can reduce the fertility and quality of soil, increase the risk of sickness from heavy metal intake and reduce the nutritional value of plants when used consistently or arbitrarily to encourage plant growth.6,7 Agrochemical contamination poses serious threats to the environment.8–10 To achieve Sustainable Development Goal 2 (SDG 2), which is concerned with food security, sustainable agriculture, and the “One Health Program,” it is imperative that viable and secure agrochemical alternatives be looked into for combatting plant diseases. This will preserve the quality of the soil and crops.11
PPNs, or roundworms, are large organisms belonging to the phylum Nematoda. Nematodes are present in both parasitic and free-living forms in plants and animals and are adapted to survive in different habitats. PPNs are a pathogenic group found in many plants and crops worldwide, decelerating agricultural yield by restricting plant growth and lowering crop productivity.12 In total, 4100 PPNs species were considered a severe constraint for global food security.13
Nematode assault in crops manifests as leaf chlorosis, stunted growth, sluggish growth rate, and falling of leaves. Nevertheless, these symptoms can vary according to the type of nematode attacking the plant. According to Rosmiza et al.14 there are four types of nematodes: (1) lesion nematode (Pratylenchus sp.), (2) leaf nematode (Aphelenchoides sp.), (3) root-knot nematodes (RKNs) (Meloidogyne sp.), and (4) rice root nematode (Hirschmanniella sp.).14
The European and Mediterranean Plant Protection Organization (EPPO) proposed member states of the EU regulate nematodes, which are Radopholus similis (Cobb, 1893, Thorne, 1949, Tylenchida: Pratylenchidae), Aphelenchoides besseyi (Christie, 1942, Tylenchida: Aphelenchoididae), Meloidogyne enterolobii (Yang & Eisenback, 1983, Tylenchida: Heteroderidae), Globodera pallida (Stone, 1973, Tylenchida: Heteroderidae), Heterodera glycines (Ichinohe, 1952, Tylenchida: Heteroderidae), Globodera rostochiensis (Wollenweber, 1923, Tylenchida: Heteroderidae), Meloidogyne fallax (Karssen, 1996, Tylenchida: Meloidogynidae), Meloidogyne mali (Ito, Oshima & Ichinohe, 1969, Tylenchida: Meloidogynidae), Xiphinema rivesi (Cobb, 1913, Dorylaimida: Longidoridae), Bursaphelenchus xylophilus (Steiner & Buhrer 1934, Nickle 1981, Parasitaphelenchidae: Bursaphelenchus), Meloidogyne chitwoodi (Golden, O'Bannon, Santo, & Finley, 1980, Tylenchida: Heteroderidae) and Ditylenchus dipsaci (Kuhn, 1857, Tylenchida: Anguinidae).15–17
The United States reports that the global economic loss in crop yield due to PPNs infection is estimated to be over USD 173 billion. According to the global index, PPNs account for 12.3% of losses annually in 40 important crops, and 14.6% in developed nations.18 Recent studies show the annual losses to crops considering all pests and diseases in India estimated at Rs. 500 billion, of which 20.4% of losses in crop production caused by nematodes reported by many Centres of All India Coordinated Research Project (AICRP) (Nematodes).19 According to reports, pine nematodes cause an average annual total economic loss of CNY 7.17 billion in China, out of which CNY 1.53 billion are direct economic losses and CNY 5.64 billion are indirect economic losses.20 A study shows pests and diseases are responsible for losses of 10–28% worldwide in soybeans, 25–41% in rice, 20–41% in maize, 8–21% in potatoes, and 10–28% in wheat.21 Crop losses each year due to the problem of rice pests, such as nematode assaults, have reached 25%.22 It attacks vegetable crops such as tomato, cucumber, lettuce, zucchini and watermelon causing yield loss of about 85%, 59%, 40%, 36%, and 29% respectively.23 More information about the percentage yield loss due to PPNs in different crops is covered in Table 1.
| S. no. | Crops | Nematodes species | Part affected | Yield loss (%) | References |
|---|---|---|---|---|---|
| Vegetables | |||||
| 1. | Tomato | Meloidogyne incognita | 40 | 27 | |
| 2. | Okra | M. incognita, Meloidogyne javanica and Meloidogyne arenaria | Root-knot | 38 | 28 |
| Cucumber | 31 | ||||
| Tomato | 17 | ||||
| Eggplant/beans | 8 | ||||
| 3. | Onion | Pratylenchus | Root-knot | 29.7 | 29 |
| Meloidogyne spp. | 95.7 | ||||
| Scutellonema | 51.5 | ||||
| Helicotylenchus | 81.6 | ||||
| 4. | Bitter gourd | M. incognita | Root-knot | 13.5 | 19 |
| Bottle gourd | 22 | ||||
| Brinjal | Meloidogyne spp. | 21 | |||
| Capsicum | 10 | ||||
| Carrot | 34 | ||||
| Chilli | 15 | ||||
| Okra | 19.5 | ||||
| Cucumber | 12 | ||||
| Tomato | 23 | ||||
| Potato | Globodera spp. | Cyst | 26 | ||
| 5. | Radish | Meloidogyne spp. | Different parts | 8–20 | 30 |
| 6. | Pumpkin | M. incognita | Different parts | 27 | 27 |
| Sponge gourd | M. incognita | 15 | |||
| Bottle gourd | M. incognita | 35 | |||
| Bean | M. incognita | 27 | |||
| Pea | M. incognita | 20 | |||
| Eggplant | Pratylenchus spp. and M. incognita | 43 | |||
| Carrot | M. incognita | 25 | |||
| 7. | Pointed gourd | M. incognita | Different parts | 20–50 | 31 |
| 8. | Ivy gourd | M. incognita | Different parts | 35.09 | 32 |
| Fruits | |||||
| 1. | Pomegranates | Helicotylenchus digitus and M. incognita | Root-knot | 40.2 | 33 |
| 2. | Pineapple | Rotylenchulus reniformis | Different parts | 86.4 | 34 |
| 3. | Watermelon | M. incognita & M. javanica | Different parts | 37 | 35 |
| Spices | |||||
| 1. | Black pepper | Meloidogyne spp. | Different parts | 52–65 | 35 |
| 2. | Turmeric | M. incognita | 33 | 19 | |
| 3. | Ginger | M. incognita | 29–33 | ||
| Pulses | |||||
| 1. | Chickpea | M. incognita | Different parts | 40–55 | 36 |
| 2. | Pigeon pea | M. javanica, M. incognita and M. javanica | Different parts | 14.2–29.4 and 27.5 | |
| 3. | Mung bean | M. incognita and R. reniformis | Different parts | 19–49 | |
| 4. | Urd bean | M. incognita, M. javanica and R. reniformis | Different parts | 29–49 | |
| 5. | Field bean | M. javanica | Different parts | 15.2 | |
| 6. | Lentil | M. javanica | Different parts | 15.4 | |
| Rice | Meloidogyne graminicola | Different parts | 42 | 37 | |
Such a huge agriculture productivity loss must promptly be mitigated through various environmentally friendly approaches. One such approach is the use of biocontrol agents (BCAs) having different microorganisms to mitigate the crop productivity losses due to nematodes. The BCAs, including bacteria and fungi or both, could be employed to minimize the agricultural losses caused by these nematodes. Moreover, single-strain inoculants could often is not a potent biocontrol due to low competitiveness against native microorganisms and changing environmental conditions.24 Integrating multiple strains that encompass an expanded spectrum of target organisms and environments has been suggested as a strategy for navigating these problems.25 However, there are not many effective examples of microbial consortia functioning effectively.
Nowadays, the study is much more focused on the consortia of different microorganisms that might safeguard to protect from pests in an eco-friendly way. Biological defense against plant disease is a feasible and intriguing approach to chemical approaches for protecting plants against the growing threat of plant diseases. BCAs are used in this method to lower the activity of important plant diseases. Microbial BCAs are an efficient way to control the spread of crop diseases.26 However, the existing efficacy of biological approaches is insufficient, opening up novel avenues towards environmentally friendly plant monitoring. There are currently very few readily available microbial consortia-based biological control formulations, despite scientific efforts to find or create microbial consortia that can help with crop protection. As a result, the focus of the present article is to provide an in-depth assessment of the present status of bacteria, fungi, and microbial consortium-based biocontrol and to explore potential future research avenues for biological protection of plant research with new technical breakthroughs. Thus, this review explains the role of BCAs against PPNs and provides a glimpse of the effective use of bacterial and fungal strains singly or in combination including emerging novel microbial consortia. It provides the knowledge of possible routes by which nematodes infect plants. It also elaborates on different biological control mechanisms of bacteria and fungi, alone and in consortia, with their significant roles, such as hyper-parasitism, lytic enzyme production, induced systematic resistance, antibiosis, siderophores, phytohormone, volatile compound, and nematicidal toxin synthesis, potassium, phosphate solubilization and nitrogen-fixing capabilities as BCAs to fix the nematodes effects on plant and improves its disease resistance. This review also provides information on the commercially available BCAs, challenges associated with their formulations and futuristic approaches to make BCAs a feasible sustainable option to mitigate phytoparasites. Thus, this review contributes efficiently towards sustainable development goal 2 which relates to food security and sustainable agriculture.
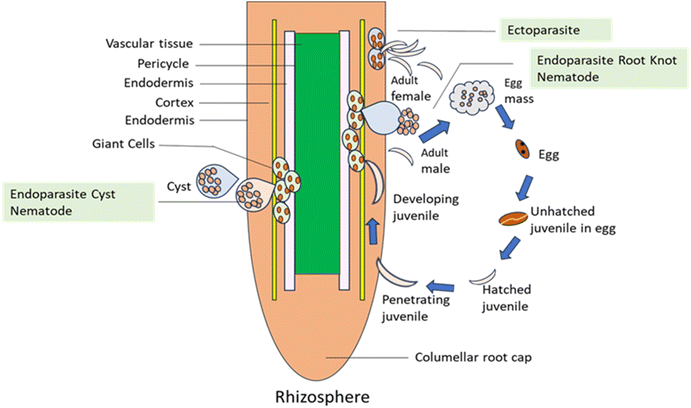
2. Life cycle and mechanism of infection of plant parasite nematodes
PPNs are nematodes that consume plant components and are commonly found in soil used for agriculture. There are three life phases for nematodes: eggs, juveniles, and adults. The environment, the toxicity of the nematode species, the plant's tolerance to nematode feeding, and the initial worm numbers upon planting all interact complexly to cause crop harm.PPNs fall into three primary categories: endoparasitic, semi-endoparasitic and ectoparasitic, based on their feeding habits. Ectoparasitic nematodes only come in physical touch with their hosts. They live their complete life cycle outside the host during the implantation of a protracted, strict feeding style. The posterior of semi-endoparasitic nematodes stays in the soil while they eat by penetrating roots. Endoparasitic nematodes consume interior tissues after fully penetrating the root. These feeding categories are further separated into sedentary and migratory lifestyles. As they move through root tissues to feed on plant cells, migratory endoparasites (such as the burrowing nematodes Radopholus spp. and the root-lesion nematodes Pratylenchus spp.) inflict tissue damage on their host cells. In opposition to this, sedentary endoparasites drive host cells to develop into multinucleate, hypertrophic feeding cells when they enter the vascular cylinder. The two main PPNs in the sedentary group are RKNs, Meloidogyne sp., and the cyst nematodes (CNs), which comprise the genera Heterodera sp., Globodera sp.38
The life cycle of RKNs can last 20–40 days at 27 °C; however, the duration of each life stage varies according to temperature, soil moisture content, and, to a lesser extent, host species. The female deposits about 500 eggs into a viscous matrix that is produced by her six anal glands. The 25–30 days process that separates the egg from the adult is divided into six stages: the egg stage, four juvenile or larval phases, and the adult stage.39
Initial phase juveniles (J1s) are created inside the egg during embryogenesis, and they molt to become juveniles in the second stage (J2s), which are contagious. J2 enters the plant using the root zone, moves through the cell elongation site, and begins feeding by injecting secretions from the esophagus into the root cells, forming multinucleate large cells (Fig. 1). RKNs and CNs both release virulence effectors with the help of stylet. PPNs release effectors such as proteins needed for developing feeding cells, enzymes that disrupt cellular barriers, and inhibitors of anti-nematicidal plant enzymes. J2 enters the plant through the root zone, moves through the cell elongation site, and begins feeding by infusing secretions from the esophageal gland into the root cells, forming ‘giant cells’.
The nematode takes on a sausage-like shape and becomes sedentary following the creation of large cells. The J2s molt into third-stage juveniles (J3s) under optimum conditions. Another molt produces fourth-stage juveniles (J4s), which include root galling. J4s also exhibit sexual dimorphism, while J3s lack a functioning stylet. The males are vermiform, emerge from the stem, and become free to live in the soil if they are found. The stationary adult female resumes eating and has a sausage or pear-like shape.
The adult female keeps getting bigger and laying eggs. The generation of eggs leads to a large population of nematodes, which restricts plant growth and development thereby leading to substantial economic loss. The majority of PPNs feed by using structures called stylets, which resemble needles, to pierce and kill root cells. This kind of feeding is used by lesion, lance, needle, sting, stunt, and sting nematodes, among other nematodes. Some nematodes, such as the RKNs and the soybean cyst nematode, enter roots and form persistent feeding sites where they complete their life cycles without damaging the surrounding cells. These nematodes can cause enormous economic loss. Nematode infection symptoms can mimic both biotic causes like stem and root rots and abiotic stressors like nutritional deficits and drought because they are comparable to symptoms of reduced root growth and function. Nematodes generally cause wilting, yellowing, and stunting along with a decrease in production. When it comes to SCN, visible to the unaided sight female bodies that range in color from white to pale yellow are indicators of infection.
3. Different aspects to reduce plant parasitic nematodes pathogenicity
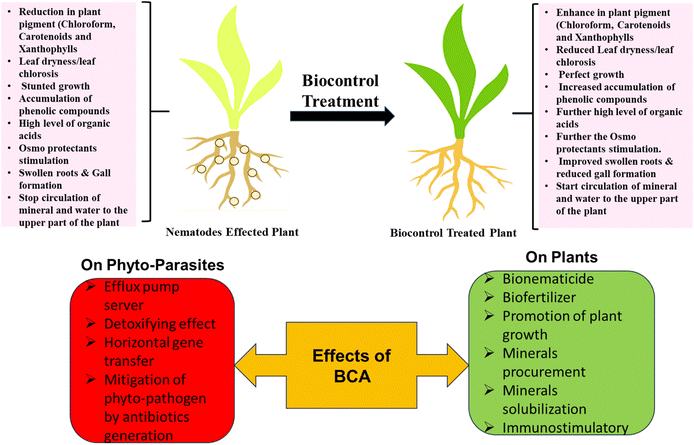
Sustainable and agrosystem-friendly action must be taken to reduce these economic losses caused by PPNs. Various well-verified methods are being used to control plant parasite nematodes, such as chemical, crop rotation, host plant resistance, Soil amendments, irrigation and soil solarization, but all these methods are more laborious, high in cost and without long-term solutions. Sometimes, chemical methods are very effective, but they are not available everywhere, and day by day, PPNs are becoming more resistant to synthetic chemical nematicides.41 It influences farmers to increase doses and quantities of it in their agricultural fields, negatively impacting the soil and its microflora in the long run, creating ecological disbalance and a toxic environment. Crop rotation, soil solarization amendments, and irrigation are also good alternatives to chemical methods, but these methods need adequate land and induce more labor costs.In the present scenario, BCAs play a magnificent role in suppressing the PPNs in crop ecosystems by their direct or indirect mechanisms Fig. 3. BCAs can combat their natural enemies by providing plant growth-promoting effects to enhance their defense mechanism to decrease formed diseases through nematodes.12 It also shows antagonistic effects, including parasitisation and the secretion of molecules or substances to inhibit nematodes.42 Several positive effects of using BCA to invade PPNs are shown in Fig. 2. It is one of the best pest management methods that may be opted against PPNs, which sounds environment-friendly and economically sustainable.
4. Biocontrol agents
Microorganisms are essential for controlling plant diseases as well as enhancing the health and profile of the soil, which promotes plant development and growth. By their inherent mode of action, microorganisms can be used as BCAs to treat the majority of plant diseases that are transmitted through the soil.43Bacillus thuringiensis was the first microbe noticed for biological plant protection, and it is used to fight insect pests like the silk moth (Bombyx mori Linnaeus, 1758, Lepidoptera: Bombycidae).44 There was 100% inhibition of J2 juvenile, reduction in egg masses, female population and decline of root nodule problem studied against M. incognita infected tomato plant under greenhouse when B. thuringiensis isolates were applied through their spore crystal production mechanism.45 Similarly, in Aphelenchoides besseyi infested rice, B. thuringiensis strain GBAC46 and NMTD81 were applied and showed 88.80% and 82.65% nematicidal activity by producing novel Cry 31A protein which acts as pore formation toxin and also activates systematic resistance in rice.46 According to recent studies, Dactylaria brochopaga and Drechslerella dactyloides caused a decrease in the infection rates of M. incognita in tomato roots-knots (67.2–70.4%), females (80.5–85.8%), eggs, and juveniles (84.9–88.8%). Their primary output was conidial traps that lacked a mycelial phase.47 The grape pathogenic nematode Xiphinema index is significantly reduced by nematode-trapping fungi (NTF) Arthrobotrys flagrans (Duddingtonia flagrans).48 In the infected tomato plant, Bacillus megaterium C3, B. safensis VW3, Lysinibacillus sp. VW6, L. fusiformis C1, Pseudomonas resinovorans VW4, and Sphingobacterium daejeonense LV1 demonstrated their potential to biocontrol the RKN M. javanica.49 The bacterium Pseudomonas simiae MBS751 showed strong nematicidal activity and performance against M. incognita by producing cyclic dipeptides that act as potential BCA.50 An endospore-forming bacteria Pasteuria penetrans parasitizes the J2s of RKNs. When J2s bear bacterial spores, they become immobile and are not able to pass through the roots. After adhering to the nematode's cuticle, bacterial spores emerge from the germination tube, develop into endospores within the body, and ultimately cause the nematode to die.51 Furthermore, it has been shown that 88–93% of M. incognita J2s die, while 88–83% of Serratia plymuthica and Pseudomonas protegens eggs hatch in greenhouse tomatoes. The metabolites of both bacteria reduced the number of egg masses and root galls, but the P. protegens enhanced plant development and seed germination.52 Thus, using advantageous microorganisms (BCAs) upon chemical pesticides would remove the negative consequences of pesticides and reduce the detrimental impact of chemicals on the environment. BCAs have several beneficial responses as they increase plant productivity qualitatively and quantitatively for a long duration without any environmental threat.534.1. Bacterial and fungal interaction (BFIs)
Bacteria and fungi frequently coexist in microhabitats where they come together to form vibrant, coevolving ecosystems. These bacterial–fungal communities comprise microorganisms from a broad range of fungal and bacterial groups and have been found to exist in almost all habitats. Numerous ecosystems depend on the interactions between fungi and bacteria to function properly. These microorganisms are essential to communities that drive biochemical cycles and are involved in plant and animal health sickness.54 In agricultural settings, bacterial fungal interactions (BFIs) are significant since they can determine the health of crops. Expanding our understanding of the mechanisms behind the interactions between fungi and bacteria would give agricultural biological control over pests and diseases a stronger foundation.55 Utilizing nematophagous fungi as endophytes, or having BCAs colonize the rhizosphere, is a viable approach for plant-parasitic nematode biocontrol. Endophytes could establish themselves in the root system before nematodes are drawn to roots since they are reasonably simple to introduce as inoculants to seeds or seedlings.56 Both internally and externally, bacteria and fungi can interact, and the nature of the relationship is influenced by substances released by the bacteria and fungal and bacterial surface molecules and the morphology of the fungal cells. Furthermore, BFIs are exhibiting facultative endofungal activity. Ascomycete and Basidiomycete fungi that interact with Alphaproteobacteria and Gammaproteobacteria are involved in the majority of these facultative partnerships. An endosymbiont of the fungus Piriformspora indica called Rhizobium radiobacter helps the fungus and a variety of plants build a beneficial symbiotic relationship.57 When subjected to nematode-derived cues, such as a conserved family of pheromones called the ascarosides, Arthrobotrys oligospora, and other nematode-trapping fungi develop specialized structures for parasite capture. Through conserved MAPK and cAMP-PKA pathways, A. oligospora detects ascarosides.58 A stable interaction between bacteria and fungi in soil may invade the growth of nematode. A tailor-made consortium of bacteria and fungi by checking their coordination and stability can evolve a better biocontrol agent. Understanding the signaling pattern of bacteria and fungi is an important factor in this regard. BCAs can control plant diseases in a variety of ways. An understanding of the mechanisms underlying the beneficial role of microorganisms will facilitate the optimization of biocontrol. In contrast to pests, this will encourage the development of BCAs in the rhizosphere, resulting in the formation of biofilms that stop pathogens from invading the roots, the release of vital micronutrients like siderophores, and an efficient system for absorbing micronutrients.59 BFIs are a crucial factor in consortium development which decides the faith of the plant growth and development. An analyzed well-developed bacterial and fungal consortium with a proper understanding of their work pattern may be much more effective than one species. As bacteria make food and fungi give shelter the mutualistic behaviour can change the soil quality and reduce the pathogen activities.4.2. Microbial consortia as potent BCA
Microbial associations can contain various microorganisms with varying environmental habits including host plant and soil type, preferred colonized areas, and action against different pathogen species.60 The spectrum of microbial strains' efficacy against a variety of plant diseases can be expanded by assembling many microorganisms into consortia, even if individual strains may have different modes of action.61 Additionally, the microorganisms in biocontrol consortia may enhance the growth of plants and other bacteria, boosting the potential of such products.62 Consortia construction requires an understanding of the roles played by different single strains and the potential interactions between different inoculants within a consortium. To develop microbial consortium there is a need to incorporate functional characteristics like redundancy, composition and variety with metabolic complementarity.11 The consortium's protective effect exceeds single-strain inoculations, despite a meta-analysis showing that consortium activities are higher in greenhouses than in outdoor settings.11 Regardless of the extensive possibilities of the microbial consortium, the market currently lacks biocontrol products in microbial-based consortia.63 The subsequent discovery of penicillin a few years later astonished the scientific community.64 As a result, it seemed sensible to examine possible microbes in agricultural practices, specifically to look for strains that produce antibiotics effective against essential plant infections.Indeed, this has led to the identification of numerous bacterial strains from the genera Bacillus and Pseudomonas, which are well-known for producing a wide range of antibiotics and antimicrobials.65 Consequently, the first biocontrol product based on living microorganisms (Galltrol) was enhanced with Agrobacterium radiobacter K84, the source of the antibiotic agrocin 84 against A. tumefaciens.66 In liquid, powder, and isotonic solution (0.01 M MgSO4) formulations containing three, four, or five bacteria in mixtures of the bacteria, Bacillus amyloliquefaciens FR203A, B. megaterium FB133M, B. thuringiensis FS213P, B. thuringiensis FB833T, Bacillus weihenstephanensis FB25M, Bacillus frigoritolerans FB37BR, and Bacillus fluorescens FP805PU were assessed in the concentrations of 106, 108, and 109 colony-forming units per milliliter. These three created consortia demonstrate a notable decline in the populations of Meloidogyne ethiopica and Xiphinema index vineyard nematode.67 Similarly, combining strains of Fusarium oxysporum using the parasite egg fungus Purpureocillium lilacinus decreased populations of Radopholus similis by 68.5%, whereas in greenhouse circumstances, combining Fusarium oxysporum with Bacillus firmus decreased PPNs populations by 86% in bananas.68 Consortium of Bacillus subtilis, Pseudomonas fluorescens, Beauveria bassiana, Metarhizium anisopliae, Trichoderma harzianum and P. lilacinum lowers M. incognita egg hatching up to 55.65% during 96 hours of exposure.69 However, the “magic bullet” approach of finding a single strain that could protect different crops from different diseases in different types of soil tends to fail.70 Three biocontrol bacterial strains Providenciavermicola AAU PR1 (NCBI ACCN: KJ161325), Pseudomonas putida AAU PR2 (NCBI ACCN: KJ161326), and P. fluorescens AAU PR3 (NCBI ACCN: KJ161327) were developed and found to be effective in controlling M. incognita in laboratory settings by the plant growth-promoting rhizobacterial (PGPR) consortium in a study.71 Following 24, 48, and 72 hours of exposure, the strains that had been fortified with botanical extracts—specifically, Azadirachta indica, Ipomoea carnea, and Brassica juncea (mustard cake)—showed the largest reduction in egg hatching.65
The activity of biocontrol strains was shown to be regulated by the setting in which they were deployed.66 As a result, microbial mixes with varied modes of action were offered as a strategy to get beyond the obstacles associated with substandard colonization circumstances and improve the durability of biocontrol products' shielding impact.62,72 Potential bacterial and fungal strains against specific PPNs are highlighted in Table 2.
| S. no. | Biological control agents | Mode of action (nematicidal compound) | Target PPNs | Crop | Reduction, % | References |
|---|---|---|---|---|---|---|
| Bacteria as BCAs | ||||||
| 1. | P. fluorescens (pf1) | Antibiotics, enzymes and toxins | M. incognita | Cowpea | 69.8 | 73 |
| B. subtilis (bs2) | 82 | |||||
| Bacillus pumilus (bp2) | 81.8 | |||||
| 2. | B. megaterium DS9 | Enzymatic activity (protease and chitinase) | Meloidogyne sp. | Black pepper | Soil area-81.86 | 74 |
| Root area- 73.11 | ||||||
| 3. | Streptomyces sp. | Parasitism | M. javanica | Banana | In vitro->50 | 75 |
| Sterile soil-70.7 | ||||||
| 4. | Streptomyces CBG9 | Parasitism | M. incognita | Coffee | 85.8 | 76 |
| 5. | Bacillus altitudinis (AMCC1040) | Colonization, parasitism | M. incognita | Ginger | Soil area-84.48 and root area-93.68 | 77 |
| 6. | Bacillus cereus S2 | Sphingosine (lipid acts as toxin) | Caenorhabditis elegans | Tomato | 77.89 | 78 |
| M. incognita | 99.96 | |||||
| 7. | Paenibacillus polymyxa | Extracellular enzymatic activity (protease, chitosanase and chitinase) | M. incognita | In vitro | 100 (juvenile mortality), 77 (eggs hatching) | 79 |
| B. subtilis | 97 (juvenile fatality), 78 (eggs hatching) | |||||
| 8. | Bacillus halotolerans, Bacillus kochii, Bacillus. Oceanisediminis, Bacillus pumilus, Bacillus toyonensis, B. cereus, Pseudomonas aeruginosa and Bacillus pseudomycoides | Growth-promoting activity & antimicrobial compound production | M. incognita | Tomato | Up to 69.96 | 80 |
| 9. | Pseudomonas rhodesiae GC-7 | Activities of the enzymes phenylalanine ammonia-lyase, polyphenol oxidase, and peroxidase | M. graminicola | Rice | 95.82 (juvenile mortality in 24 h) and 60.65 (eggs hatching in 96 h) | 81 |
| 10. | Bacillus aryabhattai Sneb517 | Parasitism (antagonistic activity) | H. glycines | Soybean | 70 (juvenile fatality) and 60 (cyst reduction in soil) | 82 |
| 11. | B. aryabhattai A08 | Induced systematic resistance | M. incognita | Tomato & carrot | 85.2 | 83 |
| Paenibacillus alvei T30 | 81.5 | |||||
| 12. | B. subtilis B10 | Secondary metabolites, hydrolytic enzymes and lipopeptide antibiotics | M. incognita | Tomato | 69.7 (in gall formation) and 71.2 (egg masses) | 84 |
| Fungi as BCAs | ||||||
| 1. | Fusarium moniliforme Fe14 | Parasitism | M. graminicola | Rice | 55 | 85 |
| 2. | Trichoderma asperellum T00 | Enzymatic action involving chitinase, peroxidase, and β-1,3-glucanase | Pratylenchus brachyurus | Soybean | 41 | 86 |
| T. harzianum ALL42 | 65 | |||||
| 3. | Aspergillus japonicus | 1,5-Dimethyl citrate hydrochloride ester compound | M. incognita | Tomato | 51.8 (eggs) and 47.3 (gall formation) | 87 |
| 4. | Penicillium chrysogenum | Induced systematic resistance | M. incognita | Okra | 97.67 | 88 |
| Trichoderma spp. | 95 | |||||
| 5. | Acremonium sclerotigenum | Specialized metabolites/molecular signaling pathway | M. incognita | Tomato | 95.5 (juvenile mortality) and up to 43 (eggs hatching) | 89 |
| 6. | Metarhizium anisopliae | Antagonistic activity, enzyme and toxin production | M. incognita | Cowpea | 83.6 | 90 |
| Purpureocillium platinum | 85.3 | |||||
| Beauveria bassiana | 77.1 | |||||
5. Mechanism of BCAs towards plant protection
Different mechanisms underlie the activity of BCAs, contributing to plant protection from nematodes as summarized in Fig. 3.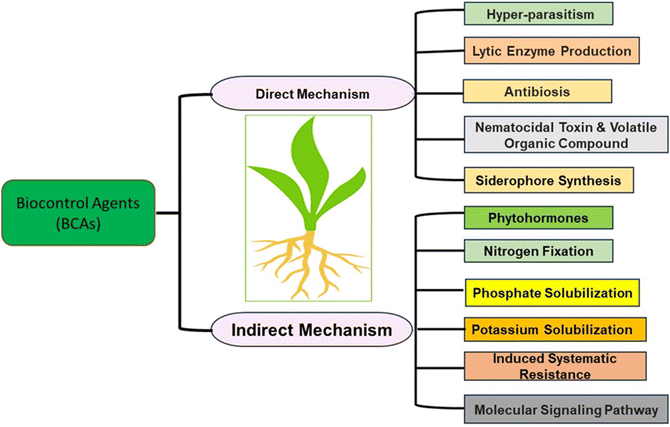 | ||
| Fig. 3 Direct and indirect methods of BCAs to prevent PPNs [modified from ref. 91]. | ||
5.1. Direct mechanisms
Different BCAs activities have come under direct mechanisms as follows:Low molecular weight substances with high vapor pressure found in plants and microbes are known as volatile organic compounds or VOCs. It is considered a direct method as it readily diffuses through air, water and soil. Chemically VOCs are alkenes, alkynes, alkyl compounds, ketones, esters, ethers, phenols, and aldehydes which help with nematicidal activity.106P. polymyxa KM2501-1 shows volatile compound characteristics, reducing M. incognita juveniles by 87.66% in vitro and 82.61% in tomatoes under greenhouse conditions.107
Iron is insoluble in soil; therefore, plants cannot absorb it directly. The insoluble form of iron can be bound by microorganisms through the production of low molecular weight compounds siderophore.109Streptomyces sp. (CMU-LPS003), Actinomycetes and Aspergillus sp. (CMU-LPS019) fungi produce siderophore and show nematicidal activity.110T. asperellum (FbMi6) is also reported as a siderophores producer and nematophagous fungus against M. incognita.111 Similarly, P. aeruginosa AC17 strain showed a nematicidal effect against M. hapla in strawberry plants.112
5.2. Indirect mechanism
Using damage-associated molecular patterns (DAMPs) and NAMPs, PPNs can activate several signaling pathways that facilitate advantageous interactions with plants.136 To overcome the plant defense system, they are released at the early stages of intracellular root invasion.135,136 PPNs' effector proteins enable them to trigger and regulate several signaling pathways in plants. These effectors can induce auxin and cytokinin signaling for the synthesis of NFSs, as well as block the pathways of salicylic acid (SA) and jasmonic acid (JA) signaling to elude host defense mechanisms.135
PPNs emit NAMPs that initiate signaling for plants to develop NAMPs-triggered immunity (NTI) based on camalexin. The BCAs, like GPCRs/GTPases (G-Protein-coupled receptors) and MAPK-cascade (Mitogen-Activated-Protein Kinases) trap nematodes via the signal transduction pathway. GPCRs/GTPases generate the protein to increase nematicidal activity, and MAPK retains the cell wall's integrity.137 A study found that the nematodes of plant pathogenic roots, M. hapla, and sugar beet cysts, Heterodera schachtii, are inhibited by the genetic expression of A. oligospora, Monacrosporium cionopagum, and Arthrobotrys dactyloides.138
Similarly, the secretions of nematodes can start several signaling cascades in plants, activating genes necessary for the creation of PPNs in those plants. In addition, the innate defense mechanism is triggered by the host plants' identification of these NAMPs as a defense mechanism to protect them from nematode invasion.139 Additionally, it has been suggested that nematodes could use their ability to create effector proteins to alter the cytoskeleton, small RNA synthesis, and cell cycle of plants to get beyond the host's basic defenses against parasitism.135,140
Moreover, PPNs can influence several signaling pathways in their hosts, including post-transcriptional alterations, gibberellin (GA), cytokinin, jasmonic acid–salicylic acid (JA–SA), and gene silencing pathways. Receptors for pattern recognition (PRRs) concentrated in membranes are found next to the cell wall. They are capable of identifying conserved pathogen/microbe-associated molecular patterns (PAMPs/MAMPs), which include cell wall derivatives, proteins, lipids, and carbohydrates.141 Following PAMP/MAMP recognition, PRRs start a conserved downstream cellular signaling cascade inside the host cell's cytoplasm known as PAMP-triggered immunity (PTI).141 Reactive oxygen species (ROS) are produced, mitogen-activated protein kinases (MAPKs) are activated, and signaling pathways are induced by jasmonic acid (JA) and salicylic acid (SA).135
In a different investigation, it was discovered that whilst a T-DNA-insertion mutant of PAD3 resulted in increased susceptibility, the overexpression of both WRKY33 and MKK4 boosted resistance against H. schachtii in Arabidopsis.142 Members of the CLAVATA/ESR (CLE) family of signaling peptides have also been found to be present in PPNs. It was found in a different study that CLE peptides have a dual role in plant meristematic tissues: they stimulate and inhibit cell development. CLE peptides are involved in CLE signaling, which promotes plant nematode parasitism, and they are essential for meristem development.
A different study claims that host plants use venom allergen-like proteins (Vap), which are essential PPN components, to detect nematode infections and mount a defense.143 To cause PCD and increase resistance to the infection by G. rostochiensis, there are strong connections between the G. rostochiensis Vap protein (Gr-Vap1) and the tomato cysteine protease, Rcr3pim, and immune receptor protein, Cf-2.135 These two genes seem to play a significant role in tomatoes' ability to perceive signals and how those signals combine with unknown R proteins to produce resistance responses. There have been suggestions that the nematode effector protein Mj-FAR-1 from M. javanica may be involved in inhibiting JA-mediated signaling and regulating lipid-based molecular signaling in plant–nematode interactions.
A small quantitative trait locus (QTLs) controlling nematode resistance was additionally documented, in addition to the well-characterized R genes. Among these, Rhg and Cre are well-known examples that give defense against soybean cyst nematodes (SCN) and cereal cyst nematodes (CCN) in soybean and wheat, respectively. Naturally occurring QTLs called Rhg1 and Rhg4 have been found in soybeans to regulate resistance reactions to SCN.144 Finding resistant QTLs may also be accomplished with the help of genome-wide association studies (GWAS). For example, 13 single nucleotide polymorphisms (SNPs) were shown to be linked to soybean cyst nematode resistance in soybeans, and 3 of these SNPs were connected to the Rhg1 and Rhg4 QTLs that have been described before.145
Studies reveal that nematodes alter RNA silencing pathways to facilitate the infection process: Plant responses to abiotic and biotic stressors often involve RNA silencing, a tactic in which microRNAs (miRNAs) play a key role as modulators. It was discovered that the Arabidopsis microRNA mi396 interacts with transcription factors like Growth-Regulating Factor, GRF1/GRF3 to control how the infection caused by H. schachtii reprogrammes root growth.138 Recognizing molecular signaling in interactions between plants and nematodes may be aided by in-plant RNA silencing of PPNs and host plant genes implicated in different signaling pathways.146 To identify genes that interact in different signaling pathways linked to plant defense systems, new transcriptomic technologies like high-throughput RNA-seq will be useful.146
Many differently regulated plant genes have been found in PPNs secretomes and diverse host plant transcriptomes146 and these genes may be useful in understanding how syncytia silencing is site-specific and enhances plant resistance to nematodes.144 New molecular methods, for high throughput, techniques like deep sequencing could be helpful in research to give a comprehensive picture of the signaling pathways that control plant–nematode interactions.143 Sequencing of RNA from gland cells may be used to comprehend how different effectors are connected to the signaling cascade function.147 Analogously, genome editing techniques such as CRISPR-Cas9 can be utilized to investigate the roles of different genes in plants and nematodes that are associated with both positive and negative interactions. The way of interaction between the nematode effector proteins and plant R proteins (GETs) may be more fully understood with the aid of these genome editing techniques.135
6. Market size and commercialization of bacterial and fungal BCAs
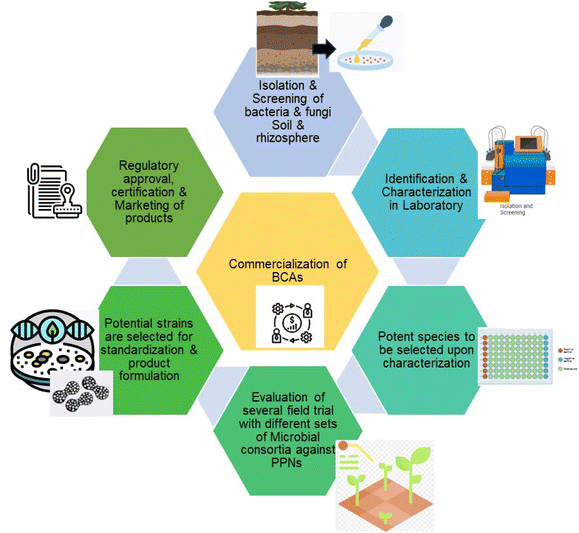
The market for biological control agents is presently estimated to be worth more than USD 5.23 billion, and by the end of 2032, it is expected to have grown to USD 20.4 billion, with a compound yearly growth rate (CAGR) of more than 16.4% from 2024 to 2032.148 The market is divided into categories based on crops: fruit and vegetables, grains and cereals, pulses, and others (such as mushrooms and oilseeds). In 2023, fruits and vegetables accounted for a significant 42% of the market, and through 2032, growth is predicted to be more profitable.148 In India, the market for BCAs is projected to be worth 1.95 billion USD in 2024 and 2.57 billion USD by 2029, expanding at a CAGR of 5.67% from 2024 to 2029.149 This data show the upcoming market opportunity and the demand for biological control in the future. Day by day public demand is increasing for organic farming and production for better health perspective. Thus, in the current situation, the addition of BCAs used in the agriculture field to chemicals is a safer approach towards the health of soil and plants which is directly and indirectly associated with all living aspects on earth.BCAs are suitable candidates that can be used as an alternative to chemical nematicides. It can fulfill all the needs of plants by enhancing plant growth and development, soil fertility also fighting against disease. To achieve eco-friendly sustainable agriculture, biocontrol may be formulated for commercialization. For the formulation process, it's essential to understand the microflora and their symbiotic association for consortium development. Proper isolation, screening, and several field trials can scale up the potential microbes from the lab to the agriculture field. Various effective BCAs are available commercially (Table 3).
| S. no. | Biocontrol agent strain | Mode of action | Commercial products | Country | Target PPN | References |
|---|---|---|---|---|---|---|
| 1. | Metarhizium anisopliae strain F52 | Penetration inside the cuticle or exoskeleton and begin to grow inside, causing the insect to die | MET52® Novozymes | The root weevils, (black vine weevils and strawberry root weevils) infesting ornamental plants grown in pots | 158 | |
| 2. | B. subtilis | Production of chitinolytic enzymes, reduction of the activity of egg hatching factors, alteration of root exudates and inhibition of nematode penetration into the roots as well as improving the plant growth | Stanes Sting (Gaara Company) | Egypt | M. arenaria | 159 |
| P. lilacinus | Parasitizing nematode eggs, juveniles and females | Stanes Bio-Nematon (Gaara Company) | Egypt | |||
| Bacillus circulans | Potassium solubilizing bacteria | Potassium (Agric. Res. Centre, Giza) | Egypt | |||
| Azospirillum sp. + Azotobacter sp. | N2-fixing bacteria | Nitrobein (Agric. Res. Centre, Giza) | Egypt | |||
| Glomus fasciculatum | Phosphate solubilizing fungus VAM | Stanes Symbion VAM plus (Gaara Company) | Egypt | |||
| 3. | T. harzianum IIHR-Th-2 | Antagonistic activity | Ecosom-TH Agri Life SOM Phytopharma Limited | India | M. incognita | 160 |
| 4. | Bacillus firmus I-1582 | Induced systematic resistance & colonization | Poncho Votivo Crop Science, Raleigh NC | U.S. | Meloidogyne spp. & H. glycines | 161 , 162 |
| 5. | B. megaterium | Plant promoting activity (phosphorus solubilizing bacterium) | Bio-Arc | London, England | Meloidogyne spp. | 163 |
| Induced systematic resistance | ||||||
| 6. | P. fluorescens | — | Sheathgua (Sudozone) | India | Meloidogyne spp. & Cyst nematodes | 164 |
| Agriland Biotech | ||||||
| Aspergillus niger | Kalisena, Cadila Pharmaceutical Limited | India | Nematodes | |||
| Burkholderia cepacia | Deny Blue Circle, Stine Microbial Products | USA | Nematodes | |||
| 7. | T. harzianum | Different nematicidal activity | Multiplex Safe Root, Bio Nematicide | India | Cyst and RKNs | 165 |
| 8. | Bacillus licheniformis, B. subtilis and P. lilacinum, B. amyloliquefaciens, Pseudomonas oryzihabitans | Different nematicidal activity | Profix, Agrivalle, Corteva, Lumialza, Indigo, Biotrinsic™ | Brazil | Meloidogyne spp. and Pratylenchus spp. | 166 |
| 9. | Bacillus spp. | Obligate parasitism, antibiosis associated with bioactive metabolites | Biotech Nemato-Cure | India | Cyst, root-knot, Meloidogyne spp., Heterodera spp., Helicotylenchus spp., Hoplolaimus spp. | 167 |
| 10. | Verticillium chlamydosporium | Covering the egg mass by the hyphal network, egg penetration, and production of chymoelastase-(protease enzyme) | Katyayani Nemotode Plus | India | Root-knot, reniform, and citrus nematodes | 168 |
| 11. | P. lilacinus | Parasitism & enzymatic activity | T Stanes Bio-Nematon Nematicide | India | Lesion nematodes, burrowing nematodes, cyst nematodes, and nematodes with knotted roots | 169 |
| Bionemat and Nemator | Knots formation in roots (Meloidogyne spp.) and cysts formation (Heterodera spp. and Globodera spp.) | 170 | ||||
| Produces antibiotics (toxin) beauvericin | Paecinemo, Vijay agro industries | Nematodes | 171 |
The delay in the registration process of products based on many strains of microorganisms can also be attributed to several issues. These include the difficulties in developing and sustaining products that contain live organisms, as well as the gradual adoption of technology and regulations intended for chemical products.150 Nonetheless, there is hope for improvement in this situation primarily because of scientific efforts to enhance the functionality of such drugs and identify the cause of unstable action.151,152
Moreover, the agricultural sector acknowledges biological control as a possible source of cutting-edge “pesticides” for use in organic (green) farming.153 The simplicity and speed with which microorganism-based agricultural commodities can now be registered is an observable trend; this opens the door for a variety of microbial consortia-based products to reach the market.154 The benefits of biocontrol consortia over single species are more efficient, robust, and modular methods. The capacity of microorganisms, particularly their mixes, to protect plants from illness and stimulate plant growth is a significant benefit of pathogen biological management.155 This makes it possible for a microbial consortium to be approved as a biofertilizer, which frequently happens through a less complicated product registration procedure.156
Farmers can be encouraged by biofertilizers that shield plants against illness, particularly those who are interested in conventional, sustainable, or organic farming, to adopt biological approaches.157 It is a continuous process in which new strains need to be identified to lower the resistant power of PPNs in laboratories, and the best candidate should be taken to the pilot level for commercialization Fig. 4.
7. Challenges and future prospectives
Utilizing bacteria, fungi, and their consortia as BCAs to manage phyto-parasitic nematodes is a viable strategy for reducing the need for chemical nematodes and mitigating the detrimental effects of nematodes on crop productivity. However, there are still several challenges that need to be addressed to fully utilize these BCAs effectively and sustainably. The first and foremost among these issues is that, akin to other pest management approaches, biological control offers numerous advantages; however, since it impacts the natural environment, we must preferably predict, any possible risks that could result from its implementation. Therefore, before using BCAs, a comprehensive analysis of the advantages and hazards should be conducted to give stakeholders the knowledge they need for effective, secure, and long-term pest control and production.64Another obstacle is that while many microbial strains and isolates show promise when evaluated in vitro for the biological control of PPNs, not all of them can be effectively transformed into bio-nematicides since not all of them consistently produce positive results when used in field settings. For instance, there aren't many commercially available plant growth-promoting rhizobacteria that serve as BCAs against PPNs. Similarly, not all BCAs, may be used as independent tools for PPN management.172 Conversely, microbial consortia are at the cutting edge of extensive study and have considerable potential for biocontrol; yet, barriers to BCA registration may deter the use of commercial plant protection solutions.
As BCAs are typically selective and only kill the intended organisms; their application is highly biodegradable and profitable to improve crops and crop quality, understanding the epidemiology, survival, and molecular mechanism of action of BCA is an essential part of delivering an effective nematicidal activity. Recently, attention has turned to the molecular underpinnings of the interactions between nematodes and microbes.173 Knowing the molecular mechanisms behind these interactions is a crucial part of understanding how the plant host reacts to this interaction as well as how nematodes react to BCAs and vice versa.172 Similarly, there is growing interest globally in the identification and application of metabolites from bacteria and fungi that have nematostatic and nematicidal properties.174 To properly understand the scope of these interactions, biological control, an ecologically grounded field, should thus constantly take an evolutionary stance, taking into account the innate diversity in behavior, phenotype, and genetics of BCAs and their targets.175 Naturally, new avenues for biological control are being opened up by the growing accessibility and affordability of DNA sequencing, as well as the capacity to modify an organism's genome and create new traits in it.176 Using multi-omics data, which is increasing in number, could be an intriguing route for such a study.177 By employing openly accessible genomes, transcriptomes, phenomes, metabolomes, and proteomes in the meta-analysis, we can identify broad trends in scientific experimentation.178
The process by which plants discern between good and harmful bacteria is still unknown. However, it has been proposed that plants can adjust their microbiome by releasing certain nutrients to increase competitiveness.179 Plant-beneficial microbes produce and withstand a variety of antimicrobials, giving them an edge in competitive environments.179 Furthermore, microbial strains can encourage the production of metabolites required for their protective function against plants and outcompete or inhibit them.180,181 Because of its intricacy, microbial consortia composition for biological plant protection is more challenging. Therefore, examining how extremely hostile strains interact in biocontrol is essential to guiding the generation of successful consortia. Climate and environmental factors can impact the effectiveness of biological control agents in the domain. Temperature, humidity, and the availability of suitable habitats are some of the characteristics that affect the adaptation and establishment of an imported species.182 Understanding suitable strains according to different zone climates is an important factor that can be acquired by analyzing the soil properties. More focused management approaches could be developed by looking into PPNs that have been neglected. Moreover, much recent research lacks realistic applicability in natural settings and is conducted in vitro using single inoculants or by-products (lysates and filtrates).183 Teaching farmers about the application of soil organic amendments is also necessary., which promotes the establishment of soil microbes capable of reducing PPN populations and strengthening plant and soil fertility development.184 Along with other control methods, this also lowers labor costs.185–187
Nevertheless, insufficient knowledge sharing between industry and academics may also be the reason for the comparatively low number of biocontrol formulations.188 Therefore, it is imperative that scientists, companies that make plant protection products, farmers, and regulatory bodies communicate better. When managing dangerous pests, in particular, common knowledge, experience, and resources can be used to successfully undertake a global biocontrol program. It is vital to comprehend the importance of potential microbes and their synergistic association for future commercialization as a potent biocontrol product to mitigate crop losses and boost agricultural productivity.
Although it takes much budget and planning to accumulate a biological control system, biological control can be costly initially but can end up being cost-effective in the long term because BCA is self-sustaining in the environment.189–191
However, no matter how promising, BCAs still have a lot of obstacles to overcome. The biology of the BCA might not be appropriate for the location in which it would be best used or compatible with the application method and formulation.15 However, the bulk manufacture of microbial agents, their storage and release techniques, their long-term effects, and their potential to harm non-target creatures continue to be some of the major barriers to the effective application of biocontrol measures.
Similarly, microbial BCAs can lose their effectiveness in unfavorable climatic settings, and although it is difficult to forecast, climate change may damage natural adversaries and impair their capacity to control pests in otherwise mild climate circumstances.15
8. Conclusions
Soil microorganisms have emerged as vital components of crop management approaches that attempt to contribute to agricultural sustainability. However, farmers tend to advocate the implementation of chemical-based fertilizers regardless of the vast quantity and diversity of microorganisms considered to be BCAs. Chemical preference is the reason for the limited efficacy of microbial-based treatments as well as the challenges posed by cropping system variety. The biological control of pests and plant diseases in agriculture by the use of helpful microorganisms is a feasible substitute for conventional pesticides. Single microorganism application was the traditional method of crop protection using microbe-based biocontrol techniques. Although the efficacy of microorganisms in managing pests and illnesses has been extensively studied, the significant obstacle to their broader implementation in agriculture is the unpredictability of outcomes frequently observed under field conditions. Their intricate relationships with the soil microbiota and the surrounding environment have a significant impact on the durability and functionality of the inoculant. Variable environmental circumstances and low competitiveness against native bacteria may contribute to the inconsistent or inefficient performance of single-strain inoculants. Combining diverse strains to cover a larger range of target organisms and environments is one of the best ways to address these problems. Therefore, a major biotechnology development that is being exploited for economic gain in the context of sustainable agriculture is the formation of microbial consortia to boost the dependability of already utilized biological control approaches. However, there are very few examples of microbial consortia performing better and they are often related to growth or yield promotion and generally involve Aureobasidium, Trichoderma, Bacillus and Pseudomonas. Combining beneficial microorganisms that are compatible and have complementary impacts on a variety of targets, direct and indirect ways of control, and effectiveness in a range of conditions would produce biocontrol products with greater versatility. For the products to be commercialized, they must first undergo testing and validation through repeated field trials in the intended geographic areas. This might be a crucial step in successfully using this environmentally-friendly technology in farming. This is because microbial-based plant protection solutions, particularly those containing numerous species, will likely remain challenging and need further research. By examining unexplored microbial diversity, future studies must concentrate on finding novel strains with strong biocontrol characteristics. Finding bacteria and fungi with broad-spectrum capability to invade various plant diseases may entail a thorough sample and screening of varied habitats. Researchers might benefit more from learning about the interactions between diseases and beneficial microorganisms by combining contemporary omics and metagenomic techniques. The transcriptome, proteome, and genome profiles of microbes and plants may provide important insights into the mechanisms and variables influencing disease suppression during biocontrol interactions.It is essential to evaluate the different roles of single bacterial and fungal strains for novel consortia development. Finally, for the adoption of biological control agents over chemical methods, there is an utmost requirement that industry, government and academia should come forward to dig out the various beneficial synergistic combinations of microorganisms that can surpass chemical pesticides for mitigating the plant pathogens thus providing plant protection in a greener way. Eventually, the productivity and sustainability of agriculture may be greatly impacted by the development of efficient microbial consortium strategies for mitigating phyto-nematode infestations.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
Conceptualization: S. P. Y., P. P., formal analysis: S. P. Y., C. S., P. P., investigation: S. P. Y., methodology: S. P. Y., P. P., C. C., supervision: P. P., A. K., M. J. S., A. K. validation; visualization: S. P. Y., P. P., roles/writing – original draft: S. P. Y., C. S., and writing – review & editing: C. S., P. P.Conflicts of interest
There is no conflict of interest to declare.References
- United Nation, World Population Day, 2023, https://www.un.org/en/observances/world-population-day, accessed 21 December 2023 Search PubMed.
- S. Vero, G. Garmendia, E. Allori, J. M. Sanz, M. Gonda, T. Alconada, I. Cavello, J. R. Dib, M. A. Diaz, C. Nally and R. S. Pimenta, Diversity, 2023, 15(3), 457, DOI:10.3390/d15030457.
- FAO, International Year of Plant Health, 2020: Communication Guide (fao.org), Rome, 2019, https://www.fao.org/publications/card/en/c/CA7186E, accessed 6 December 2023 Search PubMed.
- J. B. Ristaino, P. K. Anderson, D. P. Bebber, K. A. Brauman, N. J. Cunniffe, N. V. Fedoroff, C. Finegold, K. A. Garrett, C. A. Gilligan, C. M. Jones and M. D. Martin, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(23), e2022239118, DOI:10.1073/pnas.2022239118.
- P. D. Gnanaprakasam and A. J. Vanisree, Environ. Sci. Pollut. Res., 2022, 29, 75103–75112, DOI:10.1007/s11356-022-22750-1.
- A. Farnia and K. Hassanpour, Indian J. Nat. Sci., 2015, 5, 7792–7808 Search PubMed.
- S. Mariyam, S. K. Upadhyay, K. Chakraborty, K. K. Verma, J. S. Duhan, S. Muneer, M. Meena, R. K. Sharma, G. Ghodake and C. S. Seth, Sci. Total Environ., 2024, 912, 169097, DOI:10.1016/j.scitotenv.2023.169097.
- X. Z. Meng, A. K. Venkatesan, Y. L. Ni, J. C. Steele, L. L. Wu, A. Bignert and R. U. Halden, Environ. Sci. Technol., 2016, 50(11), 5454–5466 CrossRef PubMed.
- A. Sharma, V. Kumar, B. Shahzad, M. Tanveer, G. P. S. Sidhu, N. Handa, S. K. Kohli, P. Yadav, A. S. Bali, R. D. Parihar and O. I. Dar, SN Appl. Sci., 2019, 1, 1446, DOI:10.1007/s42452-019-1485-1.
- S. K. Upadhyay and S. A. Edrisi, Land Degrad. Dev., 2021, 32, 4817–4831, DOI:10.1002/ldr.4090.
- X. Liu, S. Mei and J. F. Salles, Appl. Soil Ecol., 2023, 190, 105011, DOI:10.1016/j.apsoil.2023.105011.
- C. M. Mesa-Valle, J. A. Garrido-Cardenas, J. Cebrian-Carmona, M. Talavera and F. Manzano-Agugliaro, Agronomy, 2020, 10(8), 1148, DOI:10.3390/agronomy10081148.
- A. Pulavarty, A. Egan, A. Karpinska, K. Horgan and T. Kakouli-Duarte, Plants, 2021, 10(11), 2352, DOI:10.3390/plants10112352.
- M. Z. Rosmiza, Z. S. Muhammad, Z. Milah and N. M. Mohd, Asian J. Agric. Rural Dev., 2021, 11(1), 105–112, DOI:10.18488/journal.ajard.2021.111.105.112.
- D. Pires, C. S. Vicente, E. Menéndez, J. M. Faria, L. Rusinque, M. J. Camacho and M. L. Inácio, Pathogens, 2022, 11(10), 1178, DOI:10.3390/pathogens11101178.
- G. M. Fusco, R. Nicastro, Y. Rouphael and P. Carillo, Foods, 2022, 11(17), 2656, DOI:10.3390/foods11172656.
- E. Muñoz-Carvajal, J. P. Araya-Angel, N. Garrido-Sáez, M. González and A. Stoll, Microorganisms, 2023, 11(4), 1061, DOI:10.3390/microorganisms11041061.
- A. A. Elling, Phytopathology, 2013, 103, 1092–1102, DOI:10.1094/phyto-01-13-0019-rvw.
- V. Kumar, M. R. Khan and R. K. Walia, Natl. Acad. Sci. Lett., 2020, 43(5), 409–412, DOI:10.1007/s40009-020-00895-2.
- J. Zhao, J. Huang, J. Yan and G. Fang, Forests, 2020, 11(10), 042, DOI:10.3390/f11101042.
- M. Gonzalez-Chang, S. Tiwari, S. Sharma and S. D. Wratten, Ann. Entomol. Soc. Am., 2019, 112(4), 302–317, DOI:10.1093/aesa/saz020.
- N. Van Nguyen and A. Ferrero, Paddy Water Environ., 2006, 4, 1–9, DOI:10.1007/s10333-005-0031-5.
- M. L. Gullino, R. Albajes and P. C. Nicot, Integrated Pest and Disease Management in Greenhouse Crops, Springer International Publishing, New York, NY, USA, 2020, vol. 9, DOI:10.1007/978-3-030-22304-5.
- P. Trivedi, J. E. Leach, S. G. Tringe, T. Sa and B. K. Singh, Nat. Rev. Microbiol., 2020, 18, 607–621, DOI:10.1038/s41579-020-0412-1.
- K. Faust, Trends Biotechnol., 2019, 37, 123–125, DOI:10.1016/j.tibtech.2018.11.004.
- T. Maciag, E. Kozieł, P. Rusin, K. Otulak-Kozieł, S. Jafra and R. Czajkowski, Int. J. Mol. Sci., 2023, 24(15), 12227, DOI:10.3390/ijms241512227.
- R. Singh and U. Kumar, Int. J. Sci. Res., 2015, 4(5), 2812–2816 Search PubMed.
- R. A. A. Khan, S. Najeeb, Z. Mao, J. Ling, Y. Yang, Y. Li and B. Xie, Microorganisms, 2020, 8(3), 401, DOI:10.3390/microorganisms8030401.
- Z. Tileubayeva, A. Avdeenko, S. Avdeenko, N. Stroiteleva and S. Kondrashev, Saudi. J. Biol. Sci., 2021, 28(9), 5428–5433, DOI:10.1016/j.sjbs.2021.05.075.
- T. M. Gowda, A. B. Rai and B. Singh, Technical Bulletin No. 76 IIVR, Varanasi, 2017, p. 32 Search PubMed.
- M. Sehgal, D. S. Srivastava, M. Malik and A. Singh, Int. J. Adv. Appl. Sci., 2021, 2(1), 123–125, DOI:10.52804/ijaas2021.2113.
- B. Basumatary, B. Mahanta, A. Borah and P. Dutta, Indian J. Nematol., 2018, 48(1), 119–121 Search PubMed.
- A. Singh, R. K. Kaul, P. S. Khapte, K. S. Jadon, Y. Rouphael, B. Basile and P. Kumar, Horticulturae, 2022, 8(2), 160, DOI:10.3390/horticulturae8020160.
- A. Soler, P. A. Marie-Alphonsine, P. Quénéhervé, Y. Prin, H. Sanguin, P. Tisseyre, R. Daumur, C. Dorey, E. Pochat, R. G. Rodriguez and N. Portal, Crop Prod., 2021, 141, 105446, DOI:10.1016/j.cropro.2020.105446.
- A. M. Fullana, A. Expósito, N. Escudero, M. Cunquero, P. Loza-Alvarez, A. Giné and F. J. Sorribas, Front. Plant Sci., 2023, 14, 1133095, DOI:10.3389/fpls.2023.1133095.
- B. Singh and M. Devindrappa, in Plant Pathogen, Pathogenesis and Management, ed. S. Nehra, 2021, pp. 305–315 Search PubMed.
- Z. Haque, M. R. Khan and F. Ahamad, Biol. Control., 2018, 126, 109–116, DOI:10.1016/j.biocontrol.2018.07.018.
- K. Sato, Y. Kadota and K. Shirasu, Front. Plant Sci., 2019, 1165, DOI:10.3389/fpls.2019.01165.
- A. Khan, M. Haris, T. Hussain, A. A. Khan, S. E. Laasli, R. Lahlali and F. Mokrini, Heliyon, 2023, 9(11), 1–17, DOI:10.1016/j.heliyon.2023.e21653.
- I. Tapia-Vázquez, A. C. Montoya-Martínez, S. De los Santos-Villalobos, M. J. Ek-Ramos, R. Montesinos-Matías and C. Martínez-Anaya, World J. Microbiol. Biotechnol., 2022, 38(2), 26, DOI:10.1007/s11274-021-03211-2.
- P. Sharma and R. Pandey, Afr. J. Agric. Res., 2009, 4, 564–567 Search PubMed.
- G. R. Stirling, Biological Control of Plant-Parasitic Nematodes: Building Coherence between Microbial Ecology and Molecular MechanismsSpringer, Berlin, Germany, 2011, pp. 1–38, DOI:10.1007/978-1-4020-9648-8_1.
- J. Zarb, R. Ghorbani, A. Koocheki and C. Leifert, Outlooks. Pest. Manag., 2005, 16, 52–55, DOI:10.1564/16apl02.
- S. Ishiwatari, Dainihon. Sanshi. Kaiho., 1901, 114, 1–5 Search PubMed.
- A. Ramalakshmi, R. Sharmila, M. Iniyakumar and V. Gomathi, Egypt. J. Biol. Pest. Control., 2020, 30, 90, DOI:10.1186/s41938-020-00293-2.
- Z. Liang, Q. Ali, Y. Wang, G. Mu, X. Kan, Y. Ren, H. Manghwar, Q. Gu, H. Wu and X. Gao, Int. J. Mol. Sci., 2022, 23(15), 8189, DOI:10.3390/ijms23158189.
- D. Kumar, Rhizosphere, 2024, 29, 100845, DOI:10.1016/j.rhisph.2023.100845.
- V. Wernet and R. Fischer, Environ. Microbiol., 2023, 25(2), 283–293, DOI:10.1111/1462-2920.16282.
- M. Rostami, A. Karegar and S. M. Taghavi, Egypt. J. Biol. Pest. Control., 2021, 31, 36, DOI:10.1186/s41938-021-00383-9.
- X. Sun, R. Zhang, M. Ding, Y. Liu and L. Li, Pest. Manag. Sci., 2021, 77(10), 4365–4374 CrossRef PubMed.
- A. Khan, S. Chen, S. Fatima, L. Ahamad and M. A. Siddiqui, Plants, 2023, 12(12), 2387, DOI:10.3390/plants12122387.
- J. Zhao, S. Wang, X. Zhu, Y. Wang, X. Liu, Y. Duan, H. Fan and L. Chen, Appl. Soil Ecol., 2021, 164, 103924, DOI:10.1016/j.apsoil.2021.103924.
- D. B. Collinge, D. F. Jensen, M. Rabiey, S. Sarrocco, M. W. Shaw and R. H. Shaw, Plant Pathol., 2022, 71(5), 1024–1047, DOI:10.1111/ppa.13555.
- A. Deveau, G. Bonito, J. Uehling, M. Paoletti, M. Becker, S. Bindschedler, S. Hacquard, V. Herve, J. Labbé, O. A. Lastovetsky and S. Mieszkin, FEMS Microbiol. Rev., 2018, 42(3), 335–352, DOI:10.1093/femsre/fuy008.
- Y. Zhou, H. Wang and S. Xu, Stress. Biology., 2022, 2, 22, DOI:10.1007/s44154-022-00046-1.
- Y. Zhang, S. Li, H. Li, R. Wang, K. Q. Zhang and J. Xu, J. Fungi., 2020, 6(4), 206, DOI:10.3390/jof6040206.
- B. N. Steffan, N. Venkatesh and N. P. Keller, J. Fungi., 2020, 6(4), 243, DOI:10.3390/jof6040243.
- C. Y. Kuo, R. J. Tay and H. C. Lin, Nat. Microbiol., 2024, 9, 1738–1751, DOI:10.1038/s41564-024-01679-w.
- M. Ayaz, C. H. Li, Q. Ali, W. Zhao, Y. K. Chi, M. Shafiq, F. Ali, X. Y. Yu, Q. Yu and J. T. Zhao, Molecules, 2023, 28(18), 6735, DOI:10.3390/molecules28186735.
- M. Dinis, J. R. Vicente, N. Cesar de Sa, F. A. López-Núñez, E. Marchante and H. Marchante, Front. Ecol. Evol., 2020, 8, 576667, DOI:10.3389/fevo.2020.576667.
- E. Kozieł, J. J. Bujarski and K. O. Kozieł, in Plant RNA Viruses Molecular Pathogenesis and Management, ed. R. K. Gaur, B. E. Patil and R. Selvarajan, Academic Press, London, UK, 1st edn, 2023, pp. 311–328, DOI:10.1016/b978-0-323-95339-9.00008-9.
- B. Niu, W. Wang, Z. Yuan, R. R. Sederoff, H. Sederoff, V. L. Chiang and R. Borriss, Front. Microbiol., 2020, 11, 585404, DOI:10.3389/fmicb.2020.585404.
- R. M. Ram, A. Debnath, S. Negi and H. B. Singh, Biopestic. Int., 2022, 2, 319–335, DOI:10.1016/b978-0-12-823355-9.00017-1.
- A. Fleming, Bull. World Health Organ., 1929, 79, 780–790 Search PubMed.
- I. Dimkić, T. Janakiev, M. Petrović, G. Degrassi and D. Fira, Physiol. Mol. Plant. Pathol., 2022, 117, 101754, DOI:10.1016/j.pmpp.2021.101754.
- D. R. Fravel, Annu. Rev. Phytopathol., 2005, 43, 337–359, DOI:10.1146/annurev.phyto.43.032904.092924.
- E. Aballay, S. Prodan, P. Correa and J. Allende, Crop. Prot., 2020, 131, 105103, DOI:10.1016/j.cropro.2020.105103.
- A. R. Mendoza and R. A. Sikora, BioControl, 2009, 54, 263–272, DOI:10.1007/s10526-008-9181-x.
- Nivedita, S. Koulagi, A. Prashantha, V. K. Rathod, Vijaymahantesh and H. Iranna, Biol. Forum., 2023, 15(12), 01–04 Search PubMed.
- M. Wisniewski, S. Droby, J. Norelli, J. Liu and L. Schena, Postharvest. Biol. Technol., 2016, 122, 3–10, DOI:10.1016/j.postharvbio.2016.05.012.
- D. G. Panpatte, H. N. Shelat, Y. K. Jhala and R. V. Vyas, Biol. Control., 2021, 155, 104528, DOI:10.1016/j.biocontrol.2020.104528.
- F. Mascher, C. Hase, M. L. Bouffaud, G. Défago and Y. Moënne-Loccoz, Microbiol. Ecol., 2014, 87(2), 441–450, DOI:10.1111/1574-6941.12234.
- H. Abd-El-Khair, W. El-Nagdi, M. Youssef, M. M. Abd-Elgawad and M. G. Dawood, Bull. Natl. Res. Cent., 2019, 43, 1–7, DOI:10.1186/s42269-019-0108-8.
- T. P. H. Tran, S. L. Wang, V. B. Nguyen, D. M. Tran, D. S. Nguyen and A. D. Nguyen, Agronomy, 2019, 9(11), 714, DOI:10.3390/agronomy9110714.
- L. Su, Z. Shen, Y. Ruan, C. Tao, Y. Chao, R. Li and Q. Shen, Front. Microbiol., 2017, 8, 20–70, DOI:10.3389/fmicb.2017.02070.
- H. Hoang, L. H. Tran, T. H. Nguyen, D. A. Nguyen, H. H. Nguyen, N. B. Pham, P. Q. Trinh, T. de Boer, A. Brouwer and H. H. Chu, Symbiosis, 2020, 80, 75–84, DOI:10.1007/s13199-019-00649-9.
- J. Y. Wang, C. Guo, P. Zhao, F. Y. Yu, Y. Su, J. P. Qu, J. L. Wang, R. S. Lin, B. Wang, Z. Gao and Z. Y. Yang, Biol. Control., 2021, 158, 104598, DOI:10.1016/j.biocontrol.2021.104598.
- H. Gao, G. Qi, R. Yin, H. Zhang, C. Li and X. Zhao, Sci. Rep., 2016, 6(1), 28756, DOI:10.1038/srep28756.
- G. M. Soliman, H. H. Ameen, S. M. Abdel-Aziz and G. M. El-Sayed, Bull. Natl. Res. Cent., 2019, 43, 1–7, DOI:10.1186/s42269-019-0200-0.
- G. Liu, X. Lin, S. Xu, G. Liu, F. Liu and W. Mu, Pest. Manag. Sci., 2020, 76(6), 2217–2224, DOI:10.1002/ps.5759.
- S. Ye, R. Yan, X. Li, Y. Lin, Z. Yang, Y. Ma and Z. Ding, Front. Microbiol., 2022, 13, 1025727, DOI:10.3389/fmicb.2022.1025727.
- J. Zhao, D. Liu, Y. Wang, X. Zhu, L. Chen and Y. Duan, Biol. Control., 2020, 142, 104147, DOI:10.1016/j.biocontrol.2019.104147.
- J. J. Viljoen, N. Labuschagne, H. Fourie and R. A. Sikora, Trop. Plant. Pathol., 2019, 44, 284–291, DOI:10.1007/s40858-019-00283-2.
- A. G. Basyony and G. A. Abo-Zaid, Egypt. J. Biol. Pest. Control., 2018, 28(1), 1–3, DOI:10.1186/s41938-018-0094-4.
- H. T. Le, J. L. Padgham, M. H. Hagemann, R. A. Sikora and A. Schouten, Ann. Appl. Biol., 2016, 169(1), 134–143, DOI:10.1111/aab.12287.
- C. M. de Oliveira, N. O. Almeida, M. V. Côrtes, M. L. Júnior, M. R. da Rocha and C. J. Ulhoa, Biol. Control., 2021, 152, 104425, DOI:10.1016/j.biocontrol.2020.104425.
- Q. He, D. Wang, B. Li, A. Maqsood and H. Wu, Agronomy, 2020, 10(9), 1222, DOI:10.3390/agronomy10091222.
- W. M. Ali, M. A. Abdel-Mageed, M. G. Hegazy, M. K. Abou-Shlell, S. M. Sultan, E. A. Salama and A. F. Yousef, Sci. Rep., 2023, 13(1), 11103, DOI:10.1038/s41598-023-37837-z.
- Y. Yao, J. Huo, H. Ben, W. Gao, Y. Hao, W. Wang and J. Xu, Biologia, 2023, 78(11), 3305–3313, DOI:10.1007/s11756-023-01505-4.
- M. M. Youssef, W. M. El-Nagdi and D. E. Lotfy, Bull. Natl. Res. Cent., 2020, 44, 112, DOI:10.1186/s42269-020-00367-z.
- S. Antil, R. Kumar, D. V. Pathak and A. Kumari, Biol. Control, 2023, 183, 105244, DOI:10.1016/j.biocontrol.2023.105244.
- K. G. Davies, Adv. Parasitol., 2009, 68, 211–245, DOI:10.1016/S0065-308X(08)00609-X.
- X. Yao, H. Guo, K. Zhang, M. Zhao, J. Ruan and J. Chen, Front. Microbiol., 2023, 14, 1160551, DOI:10.3389/fmicb.2023.1160551.
- P. H. Mhatre, C. Karthik, K. Kadirvelu, K. L. Divya, E. P. Venkatasalam, S. Srinivasan, G. Ramkumar, C. Saranya and R. Shanmuganathan, Biocatal. Agric. Biotechnol., 2019, 17, 119–128, DOI:10.1016/j.bcab.2018.11.009.
- S. J. Won, J. H. Moon, H. B. Ajuna, S. I. Choi, C. E. Maung, S. Lee and Y. S. Ahn, Int. J. Mol. Sci., 2021, 22(20), 11296, DOI:10.3390/ijms222011296.
- S. Sharma, S. Kumar, A. Khajura, P. Ohri, R. Kaur and R. Kaur, World J. Microbiol. Biotechnol., 2020, 36, 1–5, DOI:10.1007/s11274-020-02864-9.
- S. Martínez-Servat, L. Pinyol-Escala, O. Daura-Pich, M. Almazán, I. Hernández, B. López-García and C. Fernández, AIMS Microbiol., 2023, 9(1), 151–176, DOI:10.3934/microbiol.2023010.
- J. Poveda, P. Abril-Urias and C. Escobar, Front. Microbiol., 2020, 11, 992, DOI:10.3389/fmicb.2020.00992.
- T. Cao, Y. Fang, Y. Chen, X. Kong, J. Yang, H. Alharbi, Y. Kuzyakov and X. Tian, Geoderma, 2022, 410, 115662, DOI:10.1016/j.geoderma.2021.115662.
- C. Vos, S. Claerhout, R. Mkandawire, B. Panis, D. De Waele and A. Elsen, Plant Soil, 2012, 354, 335–345, DOI:10.1007/s11104-011-1070-x.
- J. M. Raaijmakers and M. Mazzola, Annu. Rev. Phytopathol., 2012, 50(1), 403–424, DOI:10.1146/annurev-phyto-081211-172908.
- P. Subedi, K. Gattoni, W. Liu, K. S. Lawrence and S. W. Park, Plants, 2020, 9(9), 1167, DOI:10.3390/plants9091167.
- J. Lee, S. Kim, H. Jung, B. K. Koo, J. A. Han and H. S. Lee, J. Plant Biol., 2023, 66(6), 485–498, DOI:10.1007/s12374-023-09404-6.
- G. S. Jouzani, E. Valijanian and R. Sharafi, Appl. Microbiol. Biotechnol., 2017, 101, 2691–2711, DOI:10.1007/s00253-017-8175-y.
- Z. Yu, J. Xiong, Q. Zhou, H. Luo, S. Hu, L. Xia, M. Sun, L. Li and Z. Yu, J. Invertebr. Pathol., 2015, 125, 73–80, DOI:10.1016/j.jip.2014.12.011.
- X. Deng, X. Wang and G. Li, Microorganisms, 2022, 10(6), 1201, DOI:10.3390/microorganisms10061201.
- W. Cheng, J. Yang, Q. Nie, D. Huang, C. Yu, L. Zheng, M. Cai, L. S. Thomashow, D. M. Weller, Z. Yu and J. Zhang, Sci. Rep., 2017, 7(1), 16213, DOI:10.1038/s41598-017-16631-8.
- B. Khasheii, P. Mahmoodi and A. Mohammadzadeh, Microbiol. Res., 2021, 250, 126790, DOI:10.1016/j.micres.2021.126790.
- R. Schwabe, C. H. Senges, J. E. Bandow, T. Heine, H. Lehmann, O. Wiche, M. Schlömann, G. Levicán and D. Tischler, Microbiol. Res., 2020, 238, 126481, DOI:10.1016/j.micres.2020.126481.
- P. Ruanpanun, N. Tangchitsomkid, K. D. Hyde and S. Lumyong, World J. Microbiol. Biotechnol., 2010, 26, 1569–1578, DOI:10.1007/s11274-010-0332-8.
- R. Saharan, J. A. Patil, S. Yadav, A. Kumar and V. Goyal, Sci. Rep., 2023, 13(1), 6603, DOI:10.1038/s41598-023-33669-z.
- M. Camacho, B. de los Santos, M. D. Vela and M. Talavera, Horticulturae, 2023, 9(3), 346, DOI:10.3390/horticulturae9030346.
- K. Tsukanova, J. Meyer and T. Bibikova, S. Afr. J. Bot., 2017, 113, 91–102, DOI:10.1016/j.sajb.2017.07.007.
- M. S. Khan, A. Zaidi, P. Wani, M. Ahemad and M. Oves, Microbial Strategies for Crop Improvement, Springer, New York, 2009, pp. 105–132, DOI:10.1007/978-3-642-01979-1_6.
- D. Egamberdieva, S. J. Wirth, A. A. Alqarawi, E. F. Abd Allah and A. Hashem, Front. Microbiol., 2017, 8, 2104, DOI:10.3389/fmicb.2017.02104.
- U. Chakraborty, B. Chakraborty and M. Basnet, J. Basic Microbiol., 2006, 46(3), 186–195, DOI:10.1002/jobm.200510050.
- M. A. Pandit, J. Kumar, S. Gulati, N. Bhandari, P. Mehta, R. Katyal, C. D. Rawat, V. Mishra and J. Kaur, Pathogens, 2022, 11(2), 273, DOI:10.3390/pathogens11020273.
- M. Vocciante, M. Grifoni, D. Fusini, G. Petruzzelli and E. Franchi, Appl. Sci., 2022, 12(3), 1231, DOI:10.3390/app12031231.
- M. Liu, J. Philp, Y. Wang, J. Hu, Y. Wei, J. Li, M. Ryder, R. Toh, Y. Zhou, M. D. Denton, Y. Wu and H. Yang, Sci. Rep., 2022, 12(1), 8381, DOI:10.1038/s41598-022-12472-2.
- M. El-Hadad, M. Mustafa, S. M. Selim, T. El-Tayeb, A. Mahgoob and N. H. A. Aziz, Braz. J. Microbiol., 2011, 42, 105–113, DOI:10.1590/s1517-83822011000100014.
- G. Kalayu, Int. J. Agron., 2019, 1–7, DOI:10.1155/2019/4917256.
- A. Suleimanova, D. Bulmakova, L. Sokolnikova, E. Egorova, D. Itkina, O. Kuzminova, A. Gizatullina and M. Sharipova, Microorganisms, 2023, 11(5), 1136, DOI:10.3390/microorganisms11051136.
- M. Satyaprakash, T. Nikitha, E. U. Reddi, B. Sadhana and S. S. Vani, Int. J. Curr. Microbiol. Appl. Sci., 2017, 6(4), 2133–2144, DOI:10.20546/ijcmas.2017.604.251.
- M. Vera-Morales, S. E. López Medina, J. Naranjo-Morán, A. Quevedo and M. F. Ratti, Microorganisms, 2023, 11(1), 137, DOI:10.3390/microorganisms11010137.
- S. K. Gupta, R. K. Singh, A. K. Patel and U. Banjare, Biosci. Biotechnol. Res. Commun., 2021, 14, 4, DOI:10.21786/bbrc/14.4.29.
- P. Parmar and S. S. Sindhu, J. Microbiol. Res., 2013,(1), 25–31, DOI:10.1080/01490451.2018.1514442.
- F. T. Olaniyan, E. T. Alori, A. O. Adekiya, B. B. Ayorinde, F. Y. Daramola, O. O. Osemwegie and O. O. Babalola, Ann. Microbiol., 2022, 72(1), 45, DOI:10.1186/s13213-022-01701-8.
- R. Devi, T. Kaur, D. Kour, A. Yadav, A. N. Yadav, A. Suman, A. S. Ahluwalia and A. K. Saxena, J. Appl. Microbiol., 2022, 133(3), 1245–1272, DOI:10.1111/jam.15627.
- M. Kurjogi, K. N. Basavesha and V. P. Savalgi, in Biocontrol Agents and Secondary Metabolites, Woodhead Publishing, 2021, pp. 23–39, DOI:10.1016/b978-0-12-822919-4.00002-8.
- K. Yu, C. M. J. Pieterse, P. A. Bakker and R. L. Berendsen, Plant Cell Environ., 2019, 42(10), 2860–2870, DOI:10.1111/pce.13632.
- M. L. Tonelli, M. S. Figueredo, J. Rodríguez, A. Fabra and F. Ibañez, Plant Soil, 2020, 448(1–2), 1–14, DOI:10.1007/s11104-020-04423-5.
- Y. Yu, Y. Gui, Z. Li, C. Jiang, J. Guo and D. Niu, Plants, 2022, 11(3), 386, DOI:10.3390/plants11030386.
- C. Jiang, Z. Fan, Z. Li, D. Niu, Y. Li, M. Zheng, Q. Wang, H. Jin and J. Guo, Mol. Plant Pathol., 2020, 21(6), 854–870, DOI:10.22271/09746315.2020.v16.i2.1351.
- C. M. Vos, A. N. Tesfahun, B. Panis, D. De Waele and A. Elsen, Appl. Soil Ecol., 2012, 61, 1–6, DOI:10.1016/j.apsoil.2012.04.007.
- M. A. Ali, M. S. Anjam, M. A. Nawaz, H. M. Lam and G. Chung, Int. J. Mol. Sci., 2018, 19(6), 1648, DOI:10.3390/ijms19061648.
- Q. Zheng, V. Putker and A. Goverse, Front. Plant Sci., 2021, 12, 641582, DOI:10.3389/fpls.2021.641582.
- K. Khanna, S. K. Kohli and P. O. R. Bhardwaj, Microbiol. Res., 2021, 248, 126755, DOI:10.1016/j.micres.2021.126755.
- K. M. Andersson, D. Kumar, J. Bentzer, E. Friman, D. Ahrén and A. Tunlid, BMC Genom., 2014, 15(1), 1–5, DOI:10.1186/1471-2164-15-968.
- H. W. Choi and D. F. Klessig, BMC Plant Biol., 2016, 16, 1–10, DOI:10.1186/s12870-016-0921-2.
- T. Hewezi, Plant Physiol., 2015, 169, 1018–1026, DOI:10.1104/pp.15.00923.
- S. Stael, P. Kmiecik, P. Willems, K. Van Der Kelen, N. S. Coll, M. Teige and F. Van Breusegem, Trends Plant Sci., 2015, 20(1), 3–11, DOI:10.1016/j.tplants.2014.10.002.
- J. Carpentier, E. Grenier, M. Esquibet, L. P. Hamel, P. Moffett, M. J. Manzanares-Dauleux and M. C. Kerlan, BMC Evol. Biol., 2013, 13, 1–14, DOI:10.1186/1471-2148-13-87.
- M. A. Ali, K. Wieczorek, D. P. Kreil and H. Bohlmann, PLoS One, 2014, 9(7), e102360, DOI:10.1371/journal.pone.0102360.
- J. Carpentier, E. Grenier, M. Esquibet, L. P. Hamel, P. Moffett, M. J. Manzanares-Dauleux and M. C. Kerlan, BMC Evol. Biol., 2013, 13, 1–14, DOI:10.1186/1471-2148-13-87.
- J. Zhang, Z. Wen, W. Li, Y. Zhang, L. Zhang, H. Dai, D. Wang and R. Xu, Mol. Breed., 2017, 37, 1–10, DOI:10.1007/s11032-017-0665-1.
- G. Gheysen and B. Vanholme, Trends Biotechnol., 2007, 25(3), 89–92, DOI:10.1016/j.tibtech.2007.01.007.
- M. A. Ali, F. Azeem, H. Li and H. Bohlmann, Front. Plant Sci., 2017, 8, 287402, DOI:10.3389/fpls.2017.01699.
- Global Market Insight, 2024, https://www.gminsights.com/industry-analysis/biocontrol-agents-market, accessed 21 March 2024 Search PubMed.
- Mordor Intelligence, India Biocontrol Agents Market Size & Share Analysis – Growth Trends & Forecast Up to 2029, https://www.mordorintelligence.com/industry-reports/india-biocontrol-agents-market, accessed 21 March 2024 Search PubMed.
- R. Lahlali, S. Ezrari, N. Radouane, J. Kenfaoui, Q. Esmaeel, H. El Hamss, Z. Belabess and E. A. Barka, Microorganisms, 2022, 10(3), 596, DOI:10.3390/microorganisms10030596.
- I. Arif, M. Batool and P. M. Schenk, Trends Biotechnol., 2020, 38, 1385–1396, DOI:10.1016/j.tibtech.2020.04.015.
- S. F. Ab Rahman, E. Singh, C. M. Pieterse and P. M. Schenk, Plant Sci., 2018, 267, 102–111, DOI:10.1016/j.plantsci.2017.11.012.
- P. G. Marrone, Pest Manag. Sci., 2019, 75(9), 2325–2340, DOI:10.1002/ps.5433.
- P. H. Togni, M. Venzon, A. C. Lagôa and E. R. Sujii, Neotrop. Entomol., 2019, 48(2), 175–185, DOI:10.1007/s13744-019-00675-8.
- R. Seenivasagan and O. O. Babalola, Biology, 2021, 10(11), 1111, DOI:10.3390/biology10111111.
- R. Czajkowski, T. Maciag, D. M. Krzyzanowska and S. Jafra, in How Research Can Stimulate the Development of Commercial Biological Control against Plant Diseases, Springer, Cham, Switzerland, 2020, pp. 183–202, DOI:10.1007/978-3-030-53238-3_12.
- A. S. Elnahal, M. T. El-Saadony, A. M. Saad, E. S. Desoky, A. M. El-Tahan, M. M. Rady, S. F. AbuQamar and K. A. El-Tarabily, Eur. J. Plant Pathol., 2022, 162(4), 759–792, DOI:10.1007/s10658-021-02393-7.
- M. S. Tranier, J. Pognant-Gros, R. D. Quiroz, C. N. González, T. Mateille and S. Roussos, Braz. Arch. Biol. Technol., 2014, 57, 831–841, DOI:10.1590/s1516-8913201402540.
- H. H. Ameen, H. A. Osman, A. M. S. Lashein, S. A. Hasabo and F. H. Koura, Middle East J. Agric. Res., 2015, 4, 37–41 Search PubMed.
- M. R. Moosavi and T. H. Askary, in Biocontrol Agents of Phytonematodes, Wallingford, UK, 2015, pp. 187–202, DOI:10.1079/9781780643755.0187.
- N. Xiang, K. S. Lawrence, J. W. Kloepper, P. A. Donald and J. A. McInroy, PLoS One, 2017, 12(7), e0181201, DOI:10.1371/journal.pone.0181201.
- N. Susič, U. Žibrat, L. Sinkovič, A. Vončina, J. Razinger, M. Knapič, A. Sedlar, S. Širca and B. Gerič Stare, Plants, 2020, 9(5), 592, DOI:10.3390/plants9050592.
- F. A. Mostafa, A. E. Khalil, A. H. Nour El-Deen and D. S. Ibrahim, Egypt J. Biol. Pest Control, 2018, 28, 1–6, DOI:10.1186/s41938-018-0068-6.
- M. M. M. Abd-Elgawad and T. H. Askary, Egypt J. Biol. Pest Control, 2018, 28(1), 1–24, DOI:10.1186/s41938-018-0080-x.
- Agriplex India, https://www.agriplexindia.com/products/multiplex-safe-root-bio-nematicide-1kg, accessed 21 December 2023.
- Agropages Brazil, Agnews, The Rise of Bionematicides: Tackling the Multi-Billion Dollar Threat of Plant-Parasitic Nematodes, 2023, https://www.news.agropages.com/News/NewsDetail---47987.htm, accessed 21 December 2023 Search PubMed.
- IndiaMart. Biotech., International Limited, Delhi, https://www.m.indiamart.com/proddetail/biotech-nemato-cure-500-ml-as-bionematicides-21364413673.html, accessed 21 December 2023.
- Krishi Seva, Kendra, India, https://www.krishisevakendra.in/products/katyayani-nemotode-plus?_pos=1%26_sid=e79a1f629%26_ss=r, accessed 21 December 2023.
- Agriplex, India, https://www.agriplexindia.com/products/t-stanes-bio-nematon?_pos=1%26_sid=ed14b04ce%26_ss=r, accessed 21 December 2023.
- Biotech., Bionematicide, International Ltd, https://www.biotech-int.com/BIONEMATNEMATORpopup.html, accessed 21 December 2023 Search PubMed.
- Vijaya, Agro. Industries, India, https://www.vijayaagro.com/bio-nematicides.html, accessed 21 December 2023.
- J. Köhl, R. Kolnaar and W. J. Ravensberg, Front. Plant Sci., 2019, 10, 845, DOI:10.3389/fpls.2019.00845.
- V. R. Messa, Rhizosphere, 2021, 17, 100323, DOI:10.1016/j.rhisph.2021.100323.
- R. Liu, Z. X. Bao, G. H. Li, C. Q. Li., S. L. Wang, X. R. Pan, K. Q. Zhang and P. J. Zhao, Microorganisms, 2022, 10(7), 1343, DOI:10.3390/microorganisms10071343.
- A. Sentis, J. L. Hemptinne, A. Magro and Y. Outreman, Evol. Appl., 2022, 15(10), 1537–1554, DOI:10.1111/eva.13457.
- I. P. Sarethy and A. Saharan, Indian Phytopathol., 2021, 74(1), 3–12, DOI:10.1007/s42360-020-00302-2.
- S. G. Crandall, K. M. Gold, M. D. M. Jiménez-Gasco, C. Camila Filgueiras and D. S. Willett, PLoS One, 2020, 15(9), e0237975, DOI:10.1371/journal.pone.0237975.
- J. Nelkner, G. Torres Tejerizo, J. Hassa, T. W. Lin, J. Witte, B. Verwaaijen, A. Winkler, B. Bunk, C. Spröer, J. Overmann and R. Grosch, Genes, 2019, 10(8), 601, DOI:10.3390/genes10080601.
- G. Santoyo, J. Adv. Res., 2022, 40, 45–58, DOI:10.1016/j.jare.2021.11.020.
- M. Kai and B. Piechulla, FEMS Microbiol. Lett., 2018, 365(22), fny253, DOI:10.1093/femsle/fny253.
- N. Manzar, A. S. Kashyap, R. S. Goutam, M. V. Rajawat, P. K. Sharma, S. K. Sharma and H. V. Singh, Sustainability, 2022, 14(19), 12786, DOI:10.3390/su141912786.
- F. V. Ferreira and M. A. Musumeci, World J. Microbiol. Biotechnol., 2021, 37(5), 90, DOI:10.1007/s11274-021-03058-7.
- M. Kang, E. Ko and T. B. Mersha, Briefings Bioinf., 2022, 23(1), bbab454, DOI:10.1093/bib/bbab454.
- N. Sasanelli, A. Konrat, V. Migunova, I. Toderas, E. Iurcu-Straistaru, S. Rusu, A. Bivol, C. Andoni and P. Veronico, Agriculture, 2021, 11(7), 602, DOI:10.3390/agriculture11070602.
- T. H. Askary, in Biocontrol Agents of Phytonematodes, CABI, Wallingford, UK, 2015, pp. 446–454, DOI:10.1079/9781780643755.0446.
- P. Sharma, Indian Phytopathol., 2023, 76(1), 47–59, DOI:10.1007/s42360-023-00601-4.
- L. G. Silva, R. C. Camargo, G. M. Mascarin, P. S. Nunes, C. Dunlap and W. Bettiol, Front. Plant Sci., 2022, 13, 983127, DOI:10.3389/fpls.2022.983127.
- M. O'Callaghan, R. A. Ballard and D. Wright, Soil Use Manag., 2022, 38(3), 1340–1369, DOI:10.1111/sum.12811.
- P. Guzmán-Guzmán, A. Kumar, S. de Los Santos-Villalobos, F. I. Parra-Cota, M. D. Orozco-Mosqueda, A. E. Fadiji, S. Hyder, O. O. Babalola and G. Santoyo, Plants, 2023, 12(3), 432, DOI:10.3390/plants12030432.
- R. Tyśkiewicz, A. Nowak, E. Ozimek and J. Jaroszuk-Ściseł, Int. J. Mol. Sci., 2022, 23(4), 2329, DOI:10.3390/ijms23042329.
- H. del Carmen, M. Rodríguez, H. C. Evans, L. M. de Abreu, D. M. de Macedo, M. K. Ndacnou, K. B. Bekele and R. W. Barreto, Sci. Rep., 2021, 11(1), 5671, DOI:10.1038/s41598-021-84111-1.
| This journal is © The Royal Society of Chemistry 2025 |