Paper spray mass spectrometry utilizing Teslin® substrate for rapid detection of lipid metabolite changes during COVID-19 infection†
Imesha W.
De Silva
 a,
Subhayu
Nayek
a,
Subhayu
Nayek
 b,
Vijay
Singh
c,
Jay
Reddy
c,
John K.
Granger
c and
Guido F.
Verbeck
b,
Vijay
Singh
c,
Jay
Reddy
c,
John K.
Granger
c and
Guido F.
Verbeck
 *a
*a
aUniversity of North Texas, Department of Chemistry, 1155 Union Circle, #305070, Denton, TX 76203, USA. E-mail: Guido.Verbeck@unt.edu
bUniversity of North Texas, Department of Biology, 1155 Union Circle, #305220, Denton, TX 76203, USA
cHealth TrackRx™, 1500 Interstate 35W, Denton, TX 7620, USA
First published on 16th July 2020
Abstract
The SARS-CoV-2 virus is known as the causal agent for the current COVID-19 global pandemic. The majority of COVID-19 patients develop acute respiratory distress syndrome (ARDS), while some experience a cytokine storm effect, which is considered as one of the leading causes of patient mortality. Lipids are known to be involved in the various stages of the lifecycle of a virus functioning as receptors or co-receptors that controls viral propagation inside the host cell. Therefore, lipid-related metabolomics aims to provide insight into the immune response of the novel coronavirus. Our study has focused on determination of the potential metabolomic biomarkers utilizing a Teslin® Substrate in paper spray mass spectrometry (PS-MS) for the development of a rapid detection test within 60 seconds of analysis time. In this study, results were correlated with PCR tests to reflect that the systemic responses of the cells were affected by the COVID-19 virus.
Multiple patients were diagnosed with a case of unexplained pneumonia in December 2019 in Wuhan, China.1 After scientific research and analysis of the disease, it was concluded to be caused by a virus that was identified as a new strain of beta coronavirus and officially named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).2 One of the hallmark characteristics of this disease is pulmonary inflammation.3 Another feature of this disease is hyper inflammation, which is primarily caused by a mechanism called “cytokine storm”. Cytokine storm is a pathological phenomenon which results in the release of a plethora of pro-inflammatory cytokines (e.g., tumor necrosis factor-α, interferon-γ, interleukins like IL-1β, IL-2, IL-6, IL-7, IL-8, granulocyte colony-stimulating factor, monocyte chemoattractant protein-1 and macrophage inflammatory protein 1-α).2 A large majority of COVID-19 patients develop a condition called acute respiratory distress syndrome (ARDS) because of the cytokine storm and is considered one of the main causes of patient mortality.4 Another under-recognized symptom of the disease is haemophagocytic lymphohistiocytosis (sHLH), which is characterized by a severe and lethal hypercytokinaemia that eventually leads to multi-organ failure. In adults, most viral infections activate the sHLH response by the immune system.5 One of the significant factors in the activation of the cytokine storm is increased activation of IL-6, which can result in vascular leakage, activation of the complement system, and induction of a coagulation cascade pathway, which can cause symptoms such as diffuse intravascular coagulation (DIC).6,7 IL-6 is also considered to be the causative agent of the cardiac ailments in COVID-19 patients by inducing myocardial dysfunction, a significant outcome of the cytokine storm.8 Almost all primary immune system cells can produce IL-6, like B lymphocytes, T lymphocytes, macrophages, monocytes, dendritic cells, mast cells. These components can also be released by non-lymphocytic cells such as fibroblasts, endothelial cells, and glomerular Mesangial cells.9 Moreover, IL-6 production is primarily induced by expressed activators like IL-1β and TNF-α.9 Some other pathways that promote the production of IL-6 involve Toll-like receptors, prostaglandins, and adipokines.7
One of the commonalities in the mechanism of infection in both the betacoronavirus SARS-CoV and SARS-CoV-2 is the utilization of an angiotensin-converting enzyme related carboxypeptidase (ACE2) receptor to gain entry into the cells.10 Cardiopulmonary and hematopoietic cells like macrophages and monocytes have this receptor expressed in a large quantity, which explains why these viruses have deleterious effects in the lungs and other associated organs. Another essential feature of this COVID-19 viral infection is a decreasing blood lymphocyte count (lymphopenia), which has a linear correlation with the severity of the disease.10 Apart from cytokines, a large number of chemokines are also activated in SARS-CoV infections such as CCL2, CCL-3, CCL-5, CCL-9, CCL-10, and IP-10. These chemokines in conjugation with reactive oxygen species (ROS) lead to acute respiratory distress syndrome ARDS and pulmonary fibrosis, ultimately leading to death.11 In a recent study, biopsy examination and histological analysis of tissue samples procured from a SARS-CoV-2 infected individual who died from the disease revealed an increased concentration of pro-inflammatory molecules such as CCR4+, CCR6+, Th17+, CD4 T-cells, indicating that hyperactivation of T-cells is the probable cause of fatality.12 It has also been shown that patients with viral infections who exhibit symptoms of hyper-inflammation have elevated levels of C-Reactive Protein (CRP) and hyperferritinaemia,13,14 which explains similar observations in COVID-19 patients.
In addition to the enzymes and proteins previously discussed, one of the major classes of biomolecules considered to be involved in the various stages of the lifecycle of a virus is the lipids. Lipids are known to function as receptors or co-receptors that control the surface attachment and entry of the virus inside the host cell or the endosomes.15 It was proposed that heterogeneous classes of lipids such as cholesterol and sphingolipids form discrete lipid domains known as microdomains. These domains referred to lipid rafts located on the host plasma membrane, which can be utilized by the virus for signal transduction, entry into the host by endocytosis, and virion assembly and budding.16,17 The formation and function of the viral replication complex involves various categories of lipids, and lipid synthesis processes.18
Furthermore, viral replication requires a tremendous amount of ATP expenditure, which is aided by lipid metabolism.19 Lastly, lipids can control the proper cellular distribution of viral proteins, as well as the trafficking, assembly, and release of virion particles, as shown in the case of the Hepatitis C virus (HCV).20,21 Hence, it's evident that host lipid biogenesis pathways play crucial roles in guiding the viral propagation. Like in the case of other viruses, lipids, and small metabolites play a major role in the life cycle of enveloped, (+) sense, ssRNA viruses like SARS-CoV-2, the causal agent for the COVID-19 global pandemic. Viruses produce double-membrane vesicles (DMVs), by hijacking the intracellular membranes of the host cells. These vesicles not only contain viral proteins but also include a plethora of confiscated host lipids and proteins which aid in viral replication.22,23 In the case of most viral infections, modulations in blood plasma metabolites and lipid levels are frequently observed, which can manifest themselves in the form of disease symptoms.
However, the higher emerging transmission rates and the mortality rate of the COVID-19 has become a severe public health concern worldwide. Due to the fact of the long incubation period of the virus, prior to visualizing symptoms, has made the diagnosis process more complicated. According to the Centers for Disease Control and Prevention (CDC), two major tests are considered for the identification of COVID-19. One is the Polymerase Chain Reaction (PCR) test used for the identification of current viral infected individuals and for the detection of antibodies from the previous infection. However, it is known that depending on the timing of the test from the point infection, and the antibody test is not the best indicator of a current viral infection since it would take up to 1 to 3 weeks for the body to produce the antibodies. Remarkably, the negative identification for the antibody test primarily does not provide a definite indication of the current infection and requires additional follow-up testing with PCR analysis for the confirmation of viral infection. Even though the PCR tests do indicate the status of the currently infected individuals, complexity, time, backlog of testing kits, and the cost of testing has become more challenging in the diagnosis process.24,25 Apart from the challenges mentioned above, some researchers have pointed out an essential risk with considerable uncertainty with PCR analysis by eliciting false-negative and false-positive results.24–27 The main causatives for these risks may occur due to the cross-contamination, carryover contamination, mutation of the primer, and improper sampling procedures and handling.24,25 Therefore, neither the PCR nor antibody tests are considered to be perfect but reliable enough for the current diagnosis process.
Recent studies have suggested that membrane cholesterol controls the entering of pathogenic viruses into the host cell by maintaining signaling, transporting, and adhesion by inducing oligomerization of the fusion peptide of a virus. The infection of cells leads to elevated amounts of cytokines in the blood serum, which causes the systemic inflammation.28 It has been found that systemic inflammation causes lipid oxidation and becomes cellular dysfunctional on lipid profile including phospholipids, free cholesterol, cholesteryl ester, and triglycerides.28 Therefore, total cholesterol, including high-density lipoproteins (HDL) and low-density lipoproteins (LDL), was considered to be more responsive to the infection as they are involved in the immunomodulation.28 Meanwhile, if an individual is infected with this virus, the metabolomic profile would indicate a marked difference in the lipid profile of this person versus that of someone who is healthy, warranting further investigation. Therefore, it is permitted that a real-time, rapid, sensitive, and specific method of analysis is required in the diagnostic prosses during the recent SARS-CoV-2 outbreak for faster identification of virally infected individuals.
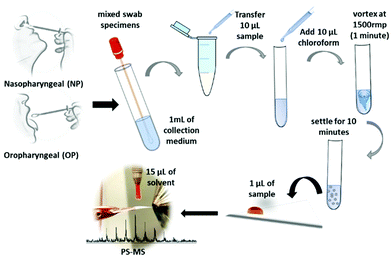
Paper spray mass spectrometry (PS-MS) is known as a quick and convenient method that can be applied for detecting various forms of compounds such as hormones, lipids,29 and drugs,30,31 and many other small molecules. PS-MS is not only applicable to the biofluids such as urine, serum, and entire blood30–32 but also with the whole tissue biopsy for clinical diagnosis within a short period of time.29 Furthermore, high-quality analytical data have shown that the PS-MS can be a feasible alternative for the monitoring of complex mixtures33 as a rapid and point-of care34 monitoring in many applications. Therefore, our method for using paper spray mass spectrometry (PS-MS) with a Teslin® substrate,35 which requires little or no sample preparation, is used as a novel approach in this study. Teslin® is a single-layered, microporous polyolefin-silica matrix that is a durable, waterproof, breathable, and lightweight, synthetic paper, which can be used for direct, sensitive, selective, and fast analysis of the sample using PS-MS.35 It has been proven previously that the Teslin® substrate allowed the samples to have a longer activation time and, therefore, active signal, resulting in a higher amount of ion formation, and with less interference of molecules from the substrate in comparison to the cellulose paper.35 For this analysis, COVID-19 positive and negative samples were obtained from the Health TrackRx™ company (Denton, TX) as a correlation to the PCR test results. Ten individual samples for both COVID-19 positive and negative lysed cell samples, previously identified from PCR tests, were obtained. Subsequently, ten samples of blind results were obtained and analyzed to validate the analytical method of PS-MS. All the lysed samples, including COVID-positive, negative and blind test samples, were recieved from COVID-19 symptomatic individuals consisting of fever, cough, and breathing difficulty. More representative samples for both upper and lower respiratory tract infections were obtained by mixing nasopharyngeal (NP), oropharyngeal (OP), and sputum specimen swabs (Fig. 1). This research was approved by the University of North Texas Institutional Biosafety Committee (IBC) under IBC# 05-2020. Therefore, the safety protocols were carried out in accordance with IBC regulations. Specifically, preparation and handling of specimens were performed in a biosafety level 2 (BSL-2), and appropriate personal protective equipment (PPE) was worn by the personal during all tasks to be performed from sample handling to the PS-MS testing. The mixed swab specimens were suspended in 1mL of proprietary collection medium created by the Health TrackRx™ company. After that, 10 μL of the above samples were mixed into 10 μL of optimum chloroform solution, and the cellular fluid was extracted from the chloroform layer by vortexing at the speed of 1500 rpm for 1 minute. Then the solution was left to settle at 25 °C for 10 minutes, and 1 μL of the extracted chloroform layer was placed on to the paper triangle for PS-MS analysis (Fig. 1). Most importantly, all the 30 samples, including COVID-19 positive, negative, and blind tests, were collected, treated, and analyzed with the exact similar conditions to avoid occurrences of any uncertain matrix effects in the final test results. However, apart from the above method, no additional time-related chemical treatment was done for the sample preparation during this study. Here, the paper spray is performed by mounting a sharp cut triangle (10 mm base, 20 mm height) on an applied high voltage (3.5 kV) to a copper alligator clip, which is mounted on to an XYZ adjustable stage in front of the heated mass spectrometer inlet with a temperature set point of 180 °C degrees. Fifteen microliters (15 μL) of optima methanol to water 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 v/v ratios with 0.1% acetic acid was used as the spray solvent for the analysis. Each sample was analyzed for 60 seconds over an m/z 90–1000 mass range in positive ionization mode on a Thermo LTQ XL Linear Ion Trap Mass Spectrometer (Thermo Scientific™, Carlsbad, CA). Resultant mass spectral data are shown in Fig. 2. Peak values are determined based on the LIPID MAPS online search tools for lipid research (http://www.lipidmaps.org/).
10 v/v ratios with 0.1% acetic acid was used as the spray solvent for the analysis. Each sample was analyzed for 60 seconds over an m/z 90–1000 mass range in positive ionization mode on a Thermo LTQ XL Linear Ion Trap Mass Spectrometer (Thermo Scientific™, Carlsbad, CA). Resultant mass spectral data are shown in Fig. 2. Peak values are determined based on the LIPID MAPS online search tools for lipid research (http://www.lipidmaps.org/).
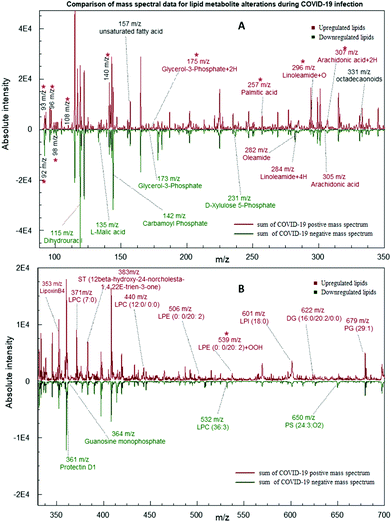
According to our analysis, it is proven that there were significant differences between the positive and negative test samples. As shown in Fig. 2, a comparison of summed mass spectral data for ten test samples of each category depicts nine downregulated metabolites and twenty-two upregulated metabolites in COVID-19 infected samples. As shown in Table 1, the expressions of each type of metabolites were determined by comparing the normalized averaged peak intensities of the COVID-19 positives and the negative samples. The normalized peak intensities of positives higher than the average value for both positives and the negatives are considered as an upregulation expression, while lower than the average is considered as a downregulation (Calculated log values are indicated in the ESI 1; S1-Fig. 1†). The five metabolites L-Malic acid (m/z 135), Carbamoyl phosphate (m/z 142), D-Xylulose 5-phosphate (m/z 173), Guanosine monophosphate (m/z 364), and Dihydrouracil (m/z 115) were matched with the results of a recent publication of downregulation in COVID-19 patient's blood plasma levels compared to healthy individual samples.36 Among the predominant changes in the swab samples, seventeen lipids were significantly elevated in the positive group. These lipid biomarkers belonged primarily to Phosphatidyl Choline (PC), Phosphatidyl Serine (PS), Phosphatidyl Ethanolamine (PE), Sterol Lipids (ST) and Diglycerides (DG) as shown in Table 1 (Identification of each metabolite with ID numbers and MS/MS mass spectral data are shown in the ESI: S1 and S2†).
| Metabolite | m/z [M + H]+ | Expression during COVID-19 infection |
|---|---|---|
| a Metabolites that expressed with oxidative degradation indicating 2, 4, 16, or 32 m/z mass shifts and used as the variables in the statistical analysis. LPC – Lysophosphatidylcholines/lysoPC; ST – Sterol Lipids; LPE – Lysophosphatidylethanolamine; LPI – Lysophosphatidylinositol; PS – Phosphatidylserine; PG – Phosphatidylglycerol. | ||
| Aminooxyacetic acida | 92 | Downregulated |
| Hydroxyethyl methyl sulfidea | 93 | Upregulated |
| 3-Hydroxypyridinea | 96 | Upregulated |
| N-Pyrrylcarbinola | 98 | Upregulated |
| Benzylaminea | 108 | Upregulated |
| Dihydrouracil | 115 | Downregulated |
| L-Malic acid | 135 | Downregulated |
| 3-Hydroxypicolinic acida | 140 | Upregulated |
| Carbamoyl phosphate | 142 | Downregulated |
| Glycerol-3-phosphate | 173 | Downregulated |
| Glycerol-3-phosphate + 2Ha | 175 | Upregulated |
| D-Xylulose 5-phosphate | 231 | Downregulated |
| Palmitic acid | 257 | Upregulated |
| Oleamide | 282 | Upregulated |
| Linoleamide + 4Ha | 284 | Upregulated |
| Linoleamide + Oa | 296 | Upregulated |
| Arachidonic acid | 305 | Upregulated |
| Arachidonic acid + 2Ha | 307 | Upregulated |
| LipoxinB4 | 353 | Upregulated |
| Protectin D1 | 361 | Downregulated |
| Guanosine monophosphate | 364 | Downregulated |
| LPC (7:0) | 371 | Upregulated |
| ST (12β-hydroxy-24-norcholesta-1,4,22E-trien-3-one) | 383 | Upregulated |
| LPC (12:0/0:0) | 440 | Upregulated |
| LPE (0:0/20:2) | 506 | Upregulated |
| LPC (36:3) | 532 | Upregulated |
| LPE (0:0/20:2) + OOHa | 539 | Upregulated |
| LPI (18:0) | 601 | Upregulated |
| DG (16:0/20:2/0:0) | 622 | Upregulated |
| PS (24:3; O2) | 650 | Downregulated |
| PG (29:1) | 679 | Upregulated |
It's fascinating to note that most of the metabolites mentioned above are involved in the major biochemical processes of an organism like the Krebs cycle, Pentose Phosphate Pathway (PPP), and synthesis of Purines and Pyrimidines. This is a clear indication of the fact that the virus effectively manipulates the host to produce more of these molecules to aid in its DNA replication and metabolism.36
All these findings indicate the crucial role played by lipids and small metabolites in the proliferation and propagation of these viruses inside the host cell. Furthermore, upregulation of Glycerol-3-Phosphate + 2H (m/z 175), Linoleamide + 4H (m/z 284), Linoleamide + O (m/z 296), Arachidonic acid + 2H (m/z 307), LPE (0:0/20:2) + OOH (m/z 539) peak values demonstrate that the COVID-19 infection causes an increase in lipid peroxidation which are shown in Fig. 2 and Table 1 with 2, 4, 16, or 32 m/z mass shifts with respect to substitution of double bonds with hydrogen and oxygen molecules. This mechanism can cause, in turn, impaired assembly and secretion of very-low-density lipoproteins(VLD), decreased export of cholesterol, and increased intracellular accumulation of lipid droplets (Steatosis).37,38 It has been shown that HCV infection has also been linked to the activation of an innate pathway involving IkB kinase-α (IKK-α), which regulates lipogenesis and viral assembly. This intrinsic pathway helps to activate lipogenic genes and promotes lipid droplet formation, which aids in virion particle assembly.39 The key to the successful infection of the majority of ssRNA viruses is linked to modulation of lipid components in the various organelle membranes as well as in the replication complex function.40
In addition to long-chain lipid molecules, free fatty acids (FFA) are also shown to be altered by virus infection. Upregulation in levels of oleamide (m/z 282), arachidonic acid (AA) (m/z 305), and Palmitic acid (PA) (m/z 257) have also agreed with a recent publication demonstration, which indicated the Middle East Respiratory Syndrome Coronavirus (MERS-CoV)-Infected and Human Coronavirus (HCoV-229E)-Infected Huh-7 cells. It was also noted that coronavirus-infected cells externally supplemented with additional LA and AA inhibited viral proliferation of both HCoV-229E and MERS-CoV.41 It has also been proposed that there is a crosstalk between Type I interferon response and lipid homeostasis during infection Flavivirus infection, which leads to the downregulation of intracellular production of fatty acids and cholesterol.42 omega-3 polyunsaturated fatty acid (PUFA)-derived lipid mediator protectin D1 (PD1) (m/z 361) is also shown to be a potent inhibitor of viral replication in the case of Influenza A infection. A decrease in PD1 is noticed in the case of strong pathogenicity.43
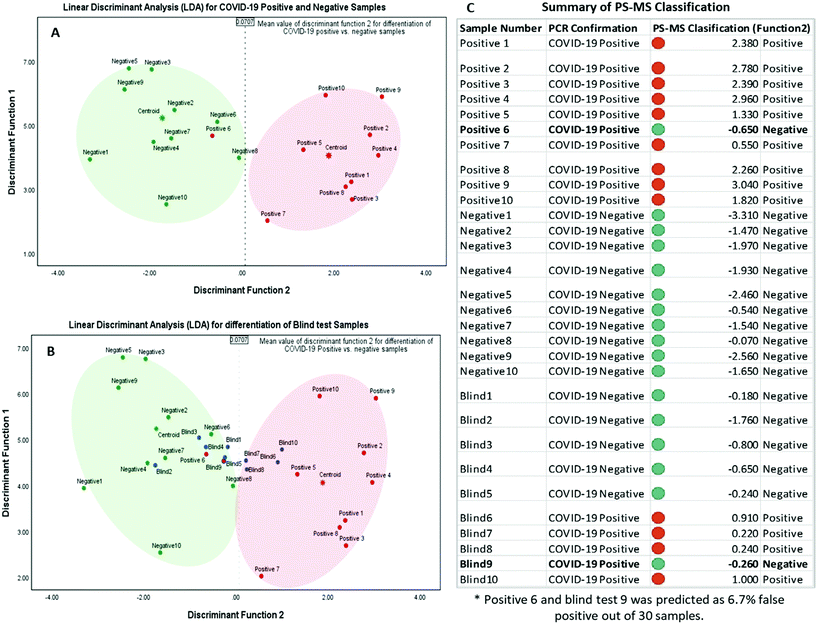
Each of the resultant mass spectral data for individual samples was extracted and compiled with nominal m/z values for a 1amu difference for statistical analysis. The absolute intensity counts were normalized individually with respect to the two peaks m/z 157 and m/z 330 that are nearly equal distribution for all the 20 test samples, and 10 blind samples, m/z 157 peaks for protonated unsaturated fatty acid and m/z 331 peak for protonated octadecanoid subclasses are used as endogenous biomarkers for each sample. Normalized mass spectral data were analyzed using statistical software, IBM SPSS Statistics for Windows, Version 25.0 (Armonk, NY: IBM Corp. Released 2017). There were 910 m/z values used as the total number of variables in this analysis. Initially, a p-value associated statistic (p < 0.05) test was performed in the evaluation of the correlation of 910 variables. The output showed 11 m/z variable values that are significantly different among COVID-19 positive and negative samples. The resulted peak values of m/z 92, 93, 96, 98, 108, 140, 175, 257, 296, 307, and 539 were used as predictor variables in grouping each category of samples (MS/MS identification for each peak values are shown in the ESI: S2 and S3†). In here, Linear Discriminant Function Analysis (LDA) was performed as a multivariate test for the determination of categorical variables (different m/z values) to separate COVID-19 positive and negative samples. This discriminant analysis was carried out with two functions or dimensions for three group discriminant analysis for positive, negative, and ungrouped samples. According to the Standardized canonical discriminant function coefficients, the function scores can be calculated using the following equations for each case:
After obtaining each score for all the 30 samples, results were plotted on a graph of individuals on the discriminant function 1 vs. function 2, as shown in Fig. 3. This distribution indicated one false-negative classification of a positive sample in PS-MS analysis versus the results of the PCR observations. Furthermore, it is determined that this model was able to predict higher scores above the mean value of 0.0707 in function 2 to be considered as COVID-19 positive samples. Overall, one positive sample and one positive blind sample was classified as a negative sample. 28 samples out of 30 samples were correctly classified with respect to the PCR analysis. Therefore, based on 10 blind tests, it is possible to determine that the PS-MS technique was able to correctly classify 93.3% of the original identification of COVID-19 Positive from negative samples. Even though our tests indicate only one false negative sample based on 10 blind tests, a higher population of clinical samples has to be recruited in order for further validation of this method compared to the PCR identification.
Conclusions
The main purpose of this study was to develop an identification methodology that could greatly aid in detecting viral infection. It has been made clear that this method can be utilized as a sensitive and rapid technique to obtain vastly different lipid information for the chemical changes in lipid metabolites and small molecules present in the human body fluids with a minimum effort in sample preparation. Although thirty-one metabolites were (nine downregulated, twenty-two upregulated) detected with the PS-MS, only eleven of the metabolites were eligible (m/z 92, 93, 96, 98, 108, 140, 175, 257, 296, 307, and 539) in implementing significant statistical dataset with the integration of symptomatic PCR analysis. It is because individuals may express a high degree of heterogeneity in metabolomic responses under different symptomatic conditions. Overall, based on the statistical analysis PS-MS has proved a 93.3% correlation to the PCR classification. Nonetheless, consideration needs to be given to the potential uncertainty with respect to the PCR study. Because the accuracy of the predictive PCR value depends on the patient's period of exposure and on the onset of symptoms, it has been shown that 39% of false-negatives26 and 16.7% of false-positives25 can be reported with PCR analysis without any other clinical support. Despite the current uncertainties, we had to correlate our dataset with the PCR because it is the viral identification “gold standard” currently in operation.24 That being said, considering follow up clinical and epidemiological character assays as standard confirmatory tests may minimize the impact of false identity. Importantly, a combination of PCR and external clinical features will make it easier to mitigate the risk and the false impression of diagnosis. However, PS-MS is not the ultimate method for the detection of COVID-19, but we believe the demonstrative results urge new methodologies to be implemented along with machine learning. This experiment can be further beneficial for expanding and developing for better and fast real-time monitoring of the diseased stage using biofluid samples over time-consuming extensive laboratory testing methods.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work presented here was supported by the Group of PPG Industries (Pittsburgh, Pennsylvania) and Health TrackRX™ (Denton, Texas) by providing valuable samples for the analysis. Specifically, authors would like to acknowledge Dr Jae Schwartz at the ThermoFisher Scientific for all the mass Spectrometry support and expertise.References
- J. She, J. Jiang, L. Ye, L. Hu, C. Bai and Y. Song, 2019 novel coronavirus of pneumonia in Wuhan, China: emerging attack and management strategies, Clin. Transl. Med., 2020, 9(1), 19–19 CrossRef PubMed.
- C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang and B. Cao, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet, 2020, 395(10223), 497–506 CrossRef CAS.
- P. Mehta, D. F. McAuley, M. Brown, E. Sanchez, R. S. Tattersall and J. J. Manson, COVID-19: consider cytokine storm syndromes and immunosuppression, Lancet, 2020, 395(10229), 1033–1034 CrossRef CAS.
- Q. Ruan, K. Yang, W. Wang, L. Jiang and J. Song, Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China, Intensive Care Med., 2020, 46(5), 846–848 CrossRef CAS.
- M. Ramos-Casals, P. Brito-Zerón, A. López-Guillermo, M. A. Khamashta and X. Bosch, Adult haemophagocytic syndrome, Lancet, 2014, 383(9927), 1503–1516 CrossRef.
- T. Tanaka, M. Narazaki and T. Kishimoto, Immunotherapeutic implications of IL-6 blockade for cytokine storm, Immunotherapy, 2016, 8(8), 959–970 CrossRef CAS.
- C. A. Hunter and S. A. Jones, IL-6 as a keystone cytokine in health and disease, Nat. Immunol., 2015, 16(5), 448–457 CrossRef.
- N. Pathan, C. A. Hemingway, A. A. Alizadeh, A. C. Stephens, J. C. Boldrick, E. E. Oragui, C. McCabe, S. B. Welch, A. Whitney, P. O'Gara, S. Nadel, D. A. Relman, S. E. Harding and M. Levin, Role of interleukin 6 in myocardial dysfunction of meningococcal septic shock, Lancet, 2004, 363(9404), 203–209 CrossRef.
- S. A. Jones and B. J. Jenkins, Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer, Nat. Rev. Immunol., 2018, 18(12), 773–789 CrossRef CAS.
- X. Yang, Y. Yu, J. Xu, H. Shu, J. A. Xia, H. Liu, Y. Wu, L. Zhang, Z. Yu, M. Fang, T. Yu, Y. Wang, S. Pan, X. Zou, S. Yuan and Y. Shang, Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study, Lancet Respir. Med., 2020, 8(5), 475–481 CrossRef CAS PubMed.
- R. Reghunathan, M. Jayapal, L. Y. Hsu, H. H. Chng, D. Tai, B. P. Leung and A. J. Melendez, Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome, BMC Immunol., 2005, 6, 2 CrossRef.
- Z. Xu, L. Shi, Y. Wang, J. Zhang, L. Huang, C. Zhang, S. Liu, P. Zhao, H. Liu, L. Zhu, Y. Tai, C. Bai, T. Gao, J. Song, P. Xia, J. Dong, J. Zhao and F.-S. Wang, Pathological findings of COVID-19 associated with acute respiratory distress syndrome, Lancet Respir. Med., 2020, 8(4), 420–422 CrossRef CAS.
- K. Sharif, V. Vieira Borba, G. Zandman-Goddard and Y. Shoenfeld, Eppur Si Muove: ferritin is essential in modulating inflammation, Clin. Exp. Immunol., 2018, 191(2), 149–150 CrossRef CAS PubMed.
- S. Colafrancesco, R. Priori, C. Alessandri, E. Astorri, C. Perricone, M. Blank, N. Agmon-Levin, Y. Shoenfeld and G. Valesini, sCD163 in AOSD: a biomarker for macrophage activation related to hyperferritinemia, Immunol. Res., 2014, 60(2), 177–183 CrossRef CAS PubMed.
- S. Taube, M. Jiang and C. E. Wobus, Glycosphingolipids as receptors for non-enveloped viruses, Viruses, 2010, 2(4), 1011–1049 CrossRef CAS PubMed.
- Y. Lu, D. X. Liu and J. P. Tam, Lipid rafts are involved in SARS-CoV entry into Vero E6 cells, Biochem. Biophys. Res. Commun., 2008, 369(2), 344–349 CrossRef CAS.
- G. M. Li, Y. G. Li, M. Yamate, S. M. Li and K. Ikuta, Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle, Microbes Infect., 2007, 9(1), 96–102 CrossRef CAS PubMed.
- A. M, A. Vazquez-Calvo, F. Caridi, J.-C. Saiz and F. Sobrino, Lipid Involvement in Viral Infections: Present and Future Perspectives for the Design of Antiviral Strategies, in Lipid Metabolism, 2013 Search PubMed.
- E. Ketter and G. Randall, Virus Impact on Lipids and Membranes, Annu. Rev. Virol., 2019, 6(1), 319–340 CrossRef CAS.
- J. A. Del Campo and M. Romero-Gómez, Modulation of host lipid metabolism by hepatitis C virus: Role of new therapies, World J. Gastroenterol., 2015, 21(38), 10776–10782 CrossRef CAS.
- C. I. Popescu and J. Dubuisson, Role of lipid metabolism in hepatitis C virus assembly and entry, Biol. Cell, 2009, 102(1), 63–74 CrossRef PubMed.
- M. Lorizate and H. G. Krausslich, Role of lipids in virus replication, Cold Spring Harbor Perspect. Biol., 2011, 3(10), a004820 Search PubMed.
- C. Yin, Genotyping coronavirus SARS-CoV-2: methods and implications, Genomics, 2020, 112(5), 3588–3596 CrossRef CAS PubMed.
- A. Tahamtan and A. Ardebili, Real-time RT-PCR in COVID-19 detection: issues affecting the results, Expert Rev. Mol. Diagn., 2020, 20(5), 453–454 CrossRef CAS PubMed.
- A. N. Cohen and B. Kessel, False positives in reverse transcription PCR testing for SARS-CoV-2, medRxiv, 2020, 2020.04.26.20080911.
- L. Kucirka, S. Lauer, O. Laeyendecker, D. Boon and J. Lessler, Variation in False Negative Rate of RT-PCR Based SARS-CoV-2 Tests by Time Since Exposure, medRxiv, 2020, 2020.04.07.20051474.
- C. P. West, V. M. Montori and P. Sampathkumar, COVID-19 Testing: The Threat of False-Negative Results, Mayo Clin. Proc., 2020, 95(6), 1127–1129 CrossRef CAS PubMed.
- X. Hu, D. Chen, L. Wu, G. He and W. Ye, Low Serum Cholesterol Level Among Patients with COVID-19 Infection in Wenzhou, China, 2020, p. 21 Search PubMed.
- H. Wang, N. E. Manicke, Q. Yang, L. Zheng, R. Shi, R. G. Cooks and Z. Ouyang, Direct Analysis of Biological Tissue by Paper Spray Mass Spectrometry, Anal. Chem., 2011, 83(4), 1197–1201 CrossRef CAS.
- H. Wang, Y. Ren, M. N. McLuckey, N. E. Manicke, J. Park, L. Zheng, R. Shi, R. G. Cooks and Z. Ouyang, Direct Quantitative Analysis of Nicotine Alkaloids from Biofluid Samples using Paper Spray Mass Spectrometry, Anal. Chem., 2013, 85(23), 11540–11544 CrossRef CAS PubMed.
- N. E. Manicke, P. Abu-Rabie, N. Spooner, Z. Ouyang and R. G. Cooks, Quantitative Analysis of Therapeutic Drugs in Dried Blood Spot Samples by Paper Spray Mass Spectrometry: An Avenue to Therapeutic Drug Monitoring, J. Am. Soc. Mass Spectrom., 2011, 22(9), 1501–1507 CrossRef CAS.
- C. Vega, C. Spence, C. Zhang, B. J. Bills and N. E. Manicke, Ionization Suppression and Recovery in Direct Biofluid Analysis Using Paper Spray Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2016, 27(4), 726–734 CrossRef CAS PubMed.
- H. Wang, J. Liu, R. G. Cooks and Z. Ouyang, Paper Spray for Direct Analysis of Complex Mixtures Using Mass Spectrometry, Angew. Chem., Int. Ed., 2010, 49(5), 877–880 CrossRef CAS PubMed.
- R. D. Espy, N. E. Manicke, Z. Ouyang and R. G. Cooks, Rapid analysis of whole blood by paper spray mass spectrometry for point-of-care therapeutic drug monitoring, Analyst, 2012, 137(10), 2344–2349 RSC.
- D. Silva, W. Imesha, D. T. Converse, L. A. Juel and G. F. Verbeck, A comparative study of microporous polyolefin silica-based paper and cellulose paper substrates utilizing paper spray-mass spectrometry in drug analysis, Anal. Methods, 2019, 11(24), 3066–3072 RSC.
- D. Wu, T. Shu, X. Yang, J.-X. Song, M. Zhang, C. Yao, L. Wen, M. Huang, Y. Yu, Q. Yang, T. Zhu, J. Xu, J. Mu, Y. Wang, H. Wang, T. Tang, Y. Ren, Y. Wu, S.-H. Lin, Y. Qiu, D.-Y. Zhang, Y. Shang and X. Zhou, Plasma Metabolomic and Lipidomic Alterations Associated with COVID-19, 2020 Search PubMed.
- G. H. Syed, Y. Amako and A. Siddiqui, Hepatitis C virus hijacks host lipid metabolism, Trends Endocrinol. Metab., 2010, 21(1), 33–40 CrossRef CAS.
- J. Ye, Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus, PLoS Pathog., 2007, 3(8), e108 CrossRef.
- Q. Li, V. Pene, S. Krishnamurthy, H. Cha and T. J. Liang, Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly, Nat. Med., 2013, 19(6), 722–729 CrossRef CAS.
- K. A. Stapleford and D. J. Miller, Role of cellular lipids in positive-sense RNA virus replication complex assembly and function, Viruses, 2010, 2(5), 1055–1068 CrossRef CAS PubMed.
- B. Yan, H. Chu, D. Yang, K.-H. Sze, P.-M. Lai, S. Yuan, H. Shuai, Y. Wang, R. Y.-T. Kao, J. F.-W. Chan and K.-Y. Yuen, Characterization of the Lipidomic Profile of Human Coronavirus-Infected Cells: Implications for Lipid Metabolism Remodeling upon Coronavirus Replication, Viruses, 2019, 11(1), 73 CrossRef CAS.
- J. P. Pombo and S. Sanyal, Perturbation of Intracellular Cholesterol and Fatty Acid Homeostasis During Flavivirus Infections, Front. Immunol., 2018, 9, 1276 CrossRef PubMed.
- M. Morita, K. Kuba, A. Ichikawa, M. Nakayama, J. Katahira, R. Iwamoto, T. Watanebe, S. Sakabe, T. Daidoji, S. Nakamura, A. Kadowaki, T. Ohto, H. Nakanishi, R. Taguchi, T. Nakaya, M. Murakami, Y. Yoneda, H. Arai, Y. Kawaoka, J. M. Penninger, M. Arita and Y. Imai, The lipid mediator protectin D1 inhibits influenza virus replication and improves severe Influenza, Cell, 2013, 153(1), 112–125 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/D0AN01074J |
| This journal is © The Royal Society of Chemistry 2020 |