Nondestructive molecular imaging by Raman spectroscopy vs. marker detection by MALDI IMS for an early diagnosis of HCC†
Tatiana
Kirchberger-Tolstik‡
ab,
Oleg
Ryabchykov‡
bc,
Thomas
Bocklitz
 *bc,
Olaf
Dirsch
d,
Utz
Settmacher
a,
Juergen
Popp§
*bc,
Olaf
Dirsch
d,
Utz
Settmacher
a,
Juergen
Popp§
 bc and
Andreas
Stallmach§
a
bc and
Andreas
Stallmach§
a
aJena University Hospital, Department of Internal Medicine IV, Gastroenterology, Hepatology, Infectious Disease, Am Klinikum, 1, 07747 Jena, Germany
bLeibniz Institute of Photonic Technology, Albert-Einstein-Straße 9, 07745 Jena, Germany. E-mail: thomas.bocklitz@uni-jena.de
cFriedrich Schiller University of Jena, Institute of Physical Chemistry, Helmholtzweg 4, 07743 Jena, Germany
dKlinikum Chemnitz gGmbH, Institute of Pathology, Chemnitz, Flemmingstraße 2, 09116 Chemnitz, Germany
First published on 27th November 2020
Abstract
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related deaths worldwide with a steadily increasing mortality rate. Fast diagnosis at early stages of HCC is of key importance for the improvement of patient survival rates. In this regard, we combined two imaging techniques with high potential for HCC diagnosis in order to improve the prediction of liver cancer. In detail, Raman spectroscopic imaging and matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI IMS) were applied for the diagnosis of 36 HCC tissue samples. The data were analyzed using multivariate methods, and the results revealed that Raman spectroscopy alone showed a good capability for HCC tumor identification (sensitivity of 88% and specificity of 80%), which could not be improved by combining the Raman data with MALDI IMS. In addition, it could be shown that the two methods in combination can differentiate between well-, moderately- and poorly-differentiated HCC using a linear classification model. MALDI IMS not only classified the HCC grades with a sensitivity of 100% and a specificity of 80%, but also showed significant differences in the expression of glycerophospholipids and fatty acyls during HCC differentiation. Furthermore, important differences in the protein, lipid and collagen compositions of differentiated HCC were detected using the model coefficients of a Raman based classification model. Both Raman and MALDI IMS, as well as their combination showed high potential for resolving concrete questions in liver cancer diagnosis.
Introduction
Hepatocellular carcinoma (HCC), the most common type of primary liver cancer, is the second cause of cancer-related deaths in the world and is generally diagnosed at an advanced stage with poor prognosis.1 Conventionally, the diagnosis of HCC includes blood and imaging tests and, in some cases, a biopsy is performed to define the type and grade of liver cancer and identify the tumor stage. During the last few years, histopathological diagnosis of liver cancer has been improved by techniques such as immunohistochemistry and confocal and electron microscopy in addition to classical H&E staining.2 Nevertheless, the stratification of different tumor types and grades is not possible based on histology alone.3 Therefore progress is needed in the development of new molecular methods and automation of cancer diagnosis that will supplement histological techniques in order to save time, solve unclear cases and reduce human errors.4 Particularly important is the molecular analysis based on both qualitative and quantitative characteristics of tissues and the identification of tumor grades, which continuously change during carcinogenesis and treatment.In the past few years, Raman spectroscopy and matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI IMS) have been proved to be highly promising techniques for the diagnosis of HCC.5–9 The identification, classification and prediction of liver cancer cell lines (an accuracy of 93%), their organelles (an accuracy of 90.5% for nucleus, 86.5% for cytoplasm and 96.5% for lipid droplets) and cell proliferation states (accuracy of 99%) were successfully performed using Raman spectroscopy by our group.5 Moreover, the investigation of the liver cancer tissue sections from patients with HCC was performed by Raman spectroscopy, which allowed cancer identification with an accuracy of 86%.6 At the same time, MALDI IMS showed its capability as a method for liver cancer diagnosis in various studies, which were mostly based on the investigation of proteins rather than lipids.7–9 MALDI allowed the identification of thirteen m/z values of protein markers, which were differentially expressed in HCC and cirrhosis by Le Faouder et al.7 Later, additional identification of these markers was performed and it was concluded that HCC is associated with early ubiquitin post-translational modifications.8 In parallel, significantly higher expression of four more proteins was found in HCC tissues by MALDI IMS and these signals allowed the classification of liver cancer with a 90% accuracy.9
In other studies the aberrant liver metabolism has been investigated, and it was found that it causes fibrosis and tissue inflammation leading to cancer and complete liver dysfunction.1,10–13 Recently, it has been published that the progression of HCC leads to lipid accumulation and high expression of fatty acids in the liver.1,10 Based on MALDI IMS it was shown that the lipidomic fingerprinting of plasma and serum can be successfully applied for the diagnosis of hepatitis B and C related HCC.12,13 In our previous studies we were able to detect significant variations in the lipid composition of HCC cells and tissues using Raman spectroscopy.5,6 Using a combination of these two complementary techniques, we aimed to investigate the improvement in liver cancer diagnosis. Furthermore, we focused on the investigation of molecular markers of HCC by Raman imaging and MALDI IMS based lipidomics.
The carcinogenesis of HCC begins with the formation of a regenerative nodule followed by differentiation into a low-grade dysplastic nodule, a high-grade dysplastic nodule, small HCC (<2 cm) and large HCC (≥2 cm). The last two steps in HCC carcinogenesis are the progression from well- (HCC at the early stage of development) to moderately- and poorly-differentiated HCC (HCC in progression). The histopathological parameters for HCC diagnosis are well established, but the detection of small and early HCC is usually a complicated task compared to progressed HCC, which can be easily identified by H&E based histopathological diagnosis.14 Therefore, we were interested in the prediction of the degree of cell differentiation in HCC and the identification of molecular markers and pattern variations in the liver tissue during cancer development.
Material and methods
Sample collection and selection
The ethics committee of the Jena University Hospital (Jena, Germany) approved the study (number 2158-11/07) and informed consent was obtained for any experimentation with human subjects and patient samples. The tissue collection from patients diagnosed with HCC was carried out at the Department of General, Visceral and Vascular Surgery (Jena University Hospital, Germany). Directly after liver resection, transplantation or hemihepatectomy tissue specimens of the tumor margins were shock frozen in liquid nitrogen and stored at −80 °C. The histological examination and diagnosis of the tissue sections were performed at the Institute of Pathology (Jena University Hospital, Germany). The tissue collection included 34 males and 14 females with a median age of 76 years. All tissue samples obtained during liver resection were cut, stained with hematoxylin and eosin (H&E) and evaluated in a blind manner (with unknown patient information) by an experienced pathologist (OD). In our collection of tissue samples we found 36 HCC and 12 samples with a combination of other liver cancer types or liver cirrhosis. In order to obtain a large dataset, all 36 HCC samples were chosen for investigation by Raman and MALDI IMS. Various tissue and disease types such as HCC, fibrosis, hepatic nodules, adenocarcinoma, neoplasia, necrosis and healthy liver tissue as well as lymphocytic infiltration, cholestasis, cirrhosis and metastases in the liver were found and annotated in each tissue sample. Patient information together with the pathological characteristics of HCC tissue samples are summarized in Table 1, including the annotations made by the pathologist on the cryo-sections involved in this study.| No | Age | Gender | Tumor type | Multifocal | Grade of differentiation | pT | pN | pM | L | V | Size of the tumor (cm) | Operation | Hepatopathological annotations of the tissue sections |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| The abbreviations are summarized as follows: Gender: M – Male, F – Female. HCC – hepatocellular carcinoma, CCC – cholangiocellular carcinoma. TNM classification for hepatocellular carcinoma: primary tumor (pT): pTX – primary tumor cannot be assessed, pT0 – no evidence of primary tumor, pT1 – solitary tumor without vascular invasion, pT2 – solitary tumor with vascular invasion or multiple tumors, none >5 cm, pT3 – multiple tumors >5 cm and single tumor or multiple tumors of any size involving a major branch of the portal or hepatic vein, pT4-tumor(s) with direct invasion of adjacent organs other than gallbladder or with visceral peritoneum. Regional lymph nodes (pN): pNX – regional lymph nodes cannot be assessed, pN0 – no regional lymph node metastasis, pN1 – regional lymph node metastasis. Distant metastasis (pM): pM0 – no distant metastasis, pM1 – distant metastasis, X – unknown. L – invasion into lymphatic vessels: X – unknown, 0 – no, 1 – yes. V – presence of microvascular invasion: X – unknown, 0 – no, 1 – yes. LTP – liver transplantation, HH – hemihepatectomy, LR – liver resection, Reg Nod – regenerative nodules, Necr – necrosis, Neopl – neoplasia, and Norm – normal (healthy) liver tissue. | |||||||||||||
| 1 | 85 | M | HCC | Unifocal | Well to moderate | 2 | 0 | 0 | 0 | X | 11 cm | LR | HCC, Fibrosis |
| 2 | 86 | M | HCC | Unifocal | Well | 1 | X | X | X | 0 | 13 × 11 | Right HH | HCC, Reg Nod |
| 3 | 69 | F | HCC | Unifocal | Unclear | X | X | X | X | X | 5 × 4 × 4 | LR | HCC, Norm |
| 4 | 76 | M | HCC | Unifocal | Well | 3 | X | 0 | X | X | 10 × 6 | LR | HCC, Fibrosis, Reg Nod |
| 5 | 88 | M | HCC | Unifocal | Well to moderate | 3 | X | 0 | 0 | 0 | 10 × 8 × 67 | HH | HCC, Norm, Neopl |
| 6 | 79 | M | HCC | Unifocal | Well | 2 | 0 | X | 0 | 1 | 3 × 2 × 2 | LTR | Norm |
| 7 | 82 | M | HCC | Unifocal | Moderate | 3 | 0 | X | 0 | 1 | 16 × 15 × 9 | Right HH | HCC, Fibrosis |
| 8 | 76 | M | HCC | Multifocal | Well | 2 | 0 | 0 | 0 | 0 | 5 × 4 × 4 | LTR | Fibrosis, Reg Nod, Necr |
| 9 | 69 | M | HCC | Multifocal | Poor | 4 | 0 | 0 | X | 1 | X | LTR | HCC, Fibrosis |
| 10 | 52 | M | HCC | Unifocal | Unclear | 1 | 0 | X | 0 | 0 | X | LTR | Norm |
| 11 | 76 | M | HCC | Multifocal | Well to moderate | 2 | 0 | 0 | X | 1 | X | LTR | Fibr, Reg Nod, |
| 12 | 71 | M | HCC | Multifocal | Unclear | 2 | X | X | X | 1 | 4 × 4 | LR | HCC, Fibrosis |
| 13 | 70 | M | HCC | Unifocal | Moderate | 2 | 0 | X | 0 | 1 | 3 × 3 × 2 | LTR | HCC, Fibrosis, Reg Nod |
| 14 | 77 | M | HCC | Multifocal | Moderate | 3 | 0 | X | X | 1 | X | LTR | HCC, Fibrosis |
| 15 | 92 | M | HCC | Unifocal | Moderate | 1 | 0 | 0 | 0 | 0 | 8 × 7 × 6 | HH | HCC, Fibrosis, Norm |
| 16 | 62 | M | HCC | Multifocal | Unclear | 2 | 0 | X | 0 | 0 | 6 × 5 × 4 | LTR | HCC, Fibrosis, Reg Nod |
| 17 | 81 | F | HCC | Multifocal | Unclear | X | 0 | X | X | X | 9 × 8 × 6 | LTR | Fibrosis, Necr |
| 18 | 69 | M | HCC | Unifocal | Moderate | 2 | 0 | X | 0 | 1 | 8 × 8 × 7 | LTR | HCC, Fibrosis |
| 19 | 83 | M | HCC | Unifocal | Moderate | 3 | 0 | X | 0 | 1 | 9 × 6 × 4 | LTR | HCC, Fibrosis, Reg Nod |
| 20 | 73 | M | HCC & CCC | Unifocal | Moderate | 3 | 0 | 1 | X | 1 | 6 × 5 × 3 | LTR | HCC, Fibrosis, Reg Nod |
| 21 | 87 | F | HCC | Unifocal | Poor | 3 | 0 | X | 0 | 1 | 14 × 14 × 9 | HH | HCC, Fibrosis |
| 22 | 54 | F | HCC | Unifocal | Moderate | 3 | 0 | X | 0 | 0 | 14 × 13 × 8 | Right HH | HCC, Fibrosis |
| 23 | 68 | M | HCC | Multifocal | Unclear | X | 0 | X | X | X | 5 × 5 × 5 | LTR | HCC, Fibrosis, Reg Nod |
| 24 | 88 | M | HCC | Unifocal | Poor | 2 | 0 | X | 0 | 1 | 15 × 10 × 10 | HH | HCC |
| 25 | 57 | F | HCC | Unifocal | Moderate | 3 | 0 | X | 0 | 0 | 15 × 14 × 11 | HH | HCC, Fibrosis |
| 26 | 29 | M | HCC | Unifocal | Moderate | 3 | 1 | X | 1 | 0 | 18 × 13 × 8 | Left HH | HCC, Fibrosis, Reg Nod |
| 27 | 88 | F | HCC | Multifocal | Unclear | 1 | X | X | 0 | 0 | 3 × 2 × 2 | LR | HCC, Fibrosis |
| 28 | 80 | M | HCC | Unifocal | Moderate | 2 | 0 | X | 0 | 0 | 6 × 6 × 6 | LTR | HCC |
| 29 | 95 | M | HCC | Unifocal | Moderate to poor | 3 | 0 | X | 0 | 1 | 9 × 8 × 7 | Right HH | HCC, Fibrosis, Norm |
| 30 | 80 | M | HCC | Unifocal | Moderate | 1 | 0 | 0 | 0 | 0 | X | Left HH | HCC, Fibrosis, Norm |
| 31 | 76 | M | HCC | Unifocal | Moderate | 2 | X | 0 | 0 | 1 | 3 × 2 × 2 | Left HH | HCC, Fibrosis |
| 32 | 82 | F | HCC | Unifocal | Moderate | 1 | 0 | 0 | 0 | 0 | 4 × 2 × 2 | Right HH | HCC, Fibrosis, Norm |
| 33 | 72 | M | HCC | Unifocal | Unclear | 3 | X | X | X | 1 | 14 × 13 × 14 | HH | HCC, Fibrosis |
| 34 | 69 | M | HCC | Multifocal | Moderate | 2 | 0 | X | 0 | 0 | 4 × 3 × 3 | LTR | HCC, Fibrosis |
| 35 | 49 | M | HCC | Unifocal | Poor | 2 | 0 | X | 0 | 1 | 15 × 11 × 7 | Right HH | HCC, Necr |
| 36 | 56 | M | HCC | Unifocal | Moderate | 1 | X | X | 0 | 0 | X | Right HH | HCC, Norm |
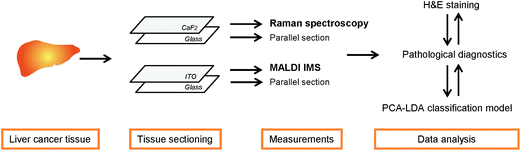
Tissue sample preparations
The frozen tissue samples of HCC were cut into two parallel tissue sections (10 and 5 μm thickness) and they were prepared two times (Cryostat Leica 3050 S, Leica Biosystems, Germany). One cryo-section (10 μm thick) was mounted onto a calcium fluoride slide (CaF2; Vacuum-UV quality, Crystal GmbH, Berlin) for the Raman measurements. The other 10 μm tissue section was deposited onto an electrically conductive indium-tin oxide (ITO)-coated slide for MALDI imaging. The two 5 μm thick cryo-sections were placed on a microscope glass slide and stained with standard H&E in order to identify the areas of interest for both measurements. According to the earlier defined regions of interest or the complete area, Raman and MALDI images of the corresponding regions were obtained using a light microscope. After the measurements, the tissue sections were still intact and stained with H&E to further obtain the correlation with the obtained Raman maps, MALDI images and pathological features.Further sample preparation was performed for the MALDI IMS measurements. In order to co-register mass spectrometric and microscopic images, reference points with a water-based correction fluid were made around the tissue section. Afterwards, the ITO slide with the tissue sections was placed on the ImagePrep device (ImagePrep station, Bruker Daltonik GmbH, Bremen, Germany) where the matrix application was performed. For the investigation of lipids in the liver tissue sections the 7 g l−1 α-cyano-4-hydroxycinnamic acid (HCCA) (C8982, Sigma-Aldrich, Germany) matrix was dissolved in a mixture of acetonitrile (50%), trifluoroacetic acid (0.2%) and water, which was sprayed on the tissue sections. The matrix type for the investigation of lipids by using an UltrafleXtreme MALDI-TOF/TOF mass spectrometer was chosen based on the requirements of Bruker Daltonics (Workshop April 2013 in Bremen, Germany).
After the measurements were performed, the slides with tissue sections were stained with H&E and the stained sections were assigned by an experienced pathologist to different tissue type regions. The complete workflow of the experiments, e.g. the liver section measurements by Raman spectroscopic imaging and MALDI imaging mass spectrometry, is presented in Fig. 1.
Raman imaging experiments
The Raman maps were obtained using a confocal Raman microscope (WITec, Ulm, Germany, Model CRM 2000). A 785 nm diode laser and grating of 300 lines per mm were selected for these experiments. The laser was connected to the microscope by using a single mode optical fiber. Measurements were performed with a 50× NA 0.95 objective (EC Epiplan-Apochromat, Zeiss, Germany). A back-illuminated deep-depletion CCD camera operating at −65 °C was used to detect the Raman scattered photons. Before each experiment the Raman system was calibrated first to the 520.7 cm−1 spectral line of silicon. Afterwards the spectrum of 4-acetamidophenol (paracetamol) (A3035, Sigma-Aldrich, Germany) was used as the reference spectrum for further calibration of the Raman spectra acquired later.The Raman spectral maps (n = 47) of the selected tissue regions were acquired. The measurement area of the tumor center, fibrotic nodules and regenerative nodules, the most common tissue types, was 75 × 75 μm2. The measurement spectral region of 3200–200 cm−1 was chosen in the mapping mode of WITec Control. A lateral step size of 1 μm was utilized in order to obtain precise information about the molecular composition of the tissue areas. In order to suppress the auto-fluorescence from the liver tissue 2 s pre-bleaching followed by an integration time of 5 s per spectra was chosen. As specific CaF2 slides were used for mounting the tissue sections the substrate background was reduced compared with the glass slides. To remove the remaining fluorescence background (Fig. S1†), a baseline correction was applied during data preprocessing (see the Data analysis subsection).
MALDI IMS experiments
All MALDI IMS measurements were performed using an UltrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany) after the tissue sections were placed on the ITO slides and a matrix was applied. For lipid detection the MALDI measurements were performed with the HCCA matrix in the 200–1500 Da mass range. The mass-to-charge ratios (m/z) of the molecules were detected using a time-of-flight analyzer (TOF) in the reflector positive ion mode. A sampling rate of 1.25 GS/sample was used. 500 shots per spectra were acquired with a 50 μm step size for each measured point within the tissue sample. Depending on the size of the ROI, different number of spectra were acquired, ranging from 23 to over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 spectra. The median number of spectra per ROI was 558, which corresponds to about 1.4 mm2.
000 spectra. The median number of spectra per ROI was 558, which corresponds to about 1.4 mm2.
For the calibration of MALDI IMS, the Peptide Calibration Standard II (222570, Bruker, Germany) was prepared according to the manufacturer's instructions. This calibration standard is recommended for peptide investigation by MALDI IMS, but it covers only the m/z range of 750–3200 m/z. Therefore, some variations in the low m/z values may be introduced due to this standard choice. A mixture of the calibration standard and matrix was applied on the ITO slide next to the tissue section and measured for the calibration of the m/z axis.
Data analysis
The Raman images of the liver tissue sections were acquired with the WiTec control software (WITec, Ulm, Germany). The acquired maps were correlated with the H&E stained tissue sections to assign every region to a tissue or disease type (HCC, regeneration nodules or fibrosis based). Finally, the spectral data were exported and further analyzed with statistical programming language R.15Similarly, each of the acquired datasets from the MALDI measurements was first imported into the FlexImaging software (Bruker Daltonik GmbH, Bremen, Germany) and co-registered with an image of the H&E stained tissue section. Based on the histological annotation made by a pathologist, regions showing specific tissue or disease types such as HCC, regenerative nodules, normal tissue, necrosis, neoplasia and fibrosis were marked and exported for further analysis with statistical programming language R.
For the interpretation of the identified Raman bands and MALDI IMS peaks different literature sources were referred and they are presented in the ESI.† Nevertheless, here we need to admit that in MALDI IMS, depending on the presence of e.g. salts in the HCCA matrix, adduct formation can occur during the ionization of analytes. Therefore, a shift like mass + 1 Da = H + adduct, mass + 23 Da = Na + adduct or mass + 39 Da = K + adduct can be expected and need to be considered.16
All data were preprocessed and analyzed using the statistical programming language R. The preprocessing of the spectral data was performed in order to avoid variations based on the measurement artifacts. Prior to the preprocessing of the MALDI data, every ROI was divided into 15 subareas, from which the mean spectrum was obtained for each subarea. This allowed for a decrease in noise and equalization of the number of observations obtained from every ROI. The baseline correction of the averaged MALDI spectra was performed using the sensitive nonlinear iterative peak (SNIP) clipping algorithm.17 After the baseline correction, spectral warping, peak picking, merging of peaks, and total ion count (TIC) normalization procedures were applied. The peak binning tolerance value at the peak picking step was set to 0.002. Within the analyzed range, this tolerance value corresponds to the binning window between 0.2 and 1 m/z.
The baseline correction of the Raman spectra was also performed using the SNIP background correction algorithm. After the baseline correction, each Raman scan was divided into 15 subareas and the mean spectra were calculated to obtain the same number of Raman and MALDI spectra per ROI (see Fig. S2†). In the last step of the preprocessing vector normalization of the mean Raman spectra was performed.
To avoid overfitting of the classification models, a dimension reduction by principal component analysis (PCA) was performed for both Raman spectroscopic data and MALDI spectrometric data. Subsequently, linear discriminant analysis (LDA) models were built on the PCA scores for the classification between fibrosis and HCC regions. These models were validated by applying the two-layer leave-one batch-out cross-validation (LBOCV) procedure with a selection of number of PCs within the internal loop of LBOCV.18 As a batch, the spectra of the samples obtained from the same patient were considered. For performing a 2-level cross-validation, one ‘test’ batch is excluded from the data set and the LBOCV procedure is applied on the remaining data for parameter optimization, then the ‘test’ batch is predicted based on the model trained with the chosen parameter. This process is repeated until all batches are predicted once. This type of validation makes it possible to estimate the model performance in a way similar to the testing of an independent data set.
For a combined data analysis, PCA scores of the two data types were concatenated based on the patient IDs and sample labels. Then the combined data set was used to construct a combined LDA model. Unfortunately, the combined data set had a lot of missing data, due to the fact that high quality spectral data of some tissue types were acquired only by one of the measurement techniques. To overcome this problem, we imputed missing values using regularized iterative expectation maximization PCA.19 To simplify the processing workflow, the combined analysis was performed by applying the one-layer LBOCV procedure, using a number of principal components (PCs) which were found to be optimal for the separate analysis of the Raman data and MALDI data.
Besides LDA classification models, a linear regression was built for discrimination between the grades of HCC differentiation for both the Raman data and the MALDI data. In order to ensure the correctness of the assigned grades, only the HCC areas of the samples with clearly identified grades (poor, moderate, or well) were used. The unclear samples (e.g. identified as ‘poor to moderate’) were skipped. As the number of samples that were labeled accordingly was smaller than the data for the classification task, it was decided to use the one-layer LBOCV with a fixed number of principal components, so that a more stable model could be obtained. Five PCs were preselected for the regression analysis of the Raman spectral data and MALDI spectral data.
Furthermore, a model interpretation was carried out. To do so, for every model the scaling vector or the model coefficients were calculated according to the published literature.5,6,12,20,21
At this point it is important to admit that the number of patient samples is always an issue due to every patient's personal decision, the availability of the medical staff and correct sample collection. Moreover, different tissue types can only be identified after the pathological evaluation of H&E stained cryo-sections, which is performed after Raman or MALDI measurements. All of these facts limit the final number of samples, which can be used for our investigations. In order to obtain statistically relevant data, we chose the patient samples with tissue types found in at least 10 samples. Therefore, from our HCC tissue collection of 36 samples only 21 were included in the data analysis based on the presence of at least two tissue types in one section (HCC and fibrosis), tissue conditions (mostly depend on the tissue collection procedure) and spectral quality (depends on the auto-fluorescence of the sample and tissue architecture). In this analysis, we investigated HCC and fibrosis as two distinct groups without considering the differentiation grades of HCC or locations of fibrosis within the sample. Due to the low number of patient samples with the tissue types regenerative nodules (n = 8), normal (n = 5), necrosis (n = 3) and neoplasia (n = 1), these samples were not utilized in the analysis. Nevertheless, we want to note that the Raman spectra of the regions of regenerative nodules had high spectral similarities to the HCC spectra in both MALDI and Raman datasets (seen in Fig. S3 in the ESI†). The analysis of other regions was not reasonable from the statistical point of view due to their small sample size.
Results
A combination of Raman imaging and MALDI IMS for HCC diagnosis
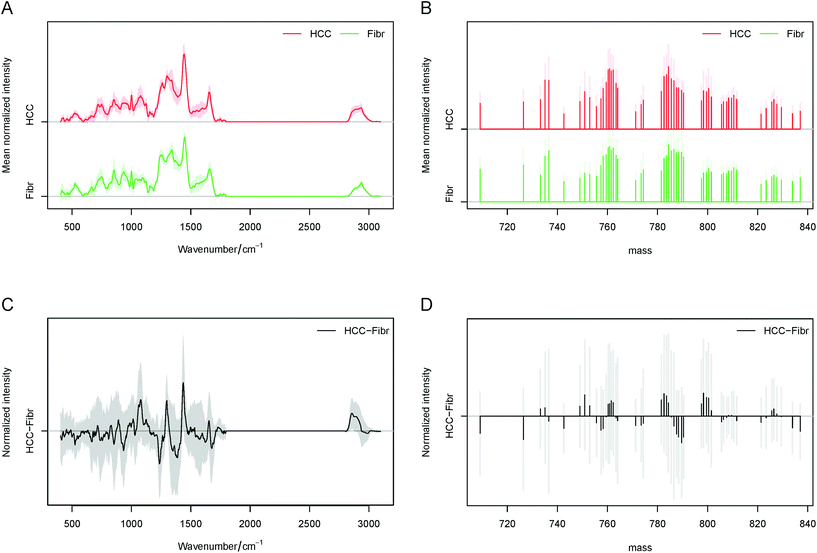
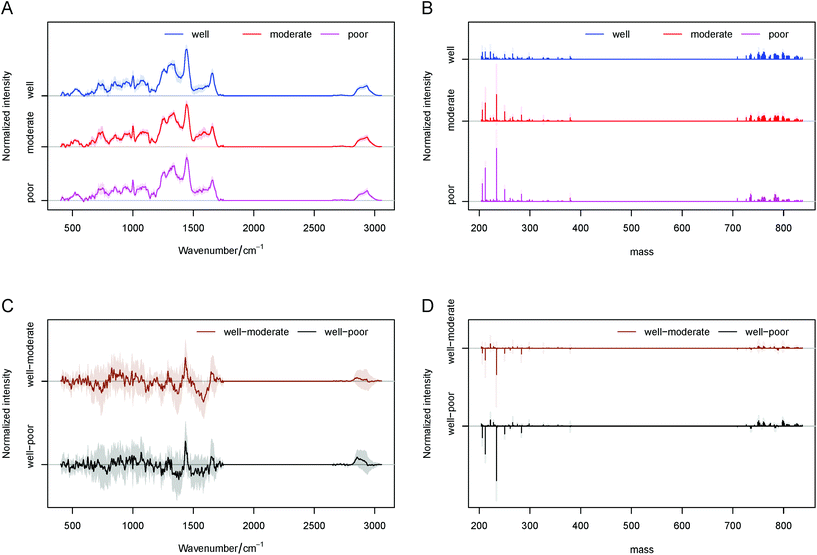
Consequently, the average spectra obtained by Raman and MALDI imaging for each liver tissue type are presented in Fig. 2A and B. By subtraction of the fibrosis (fibrotic tissues surrounding liver cancer) the average spectra of HCC (liver cancer tissues), and the difference spectra for Raman and MALDI datasets can be calculated (Fig. 2C and D). In these plots, higher intensities of lipids in the Raman spectra of HCC were identified at 717, 1059, 1077, 1299, 1440, 1653, 1743, 2856, 2880, 2889 and 2896 cm−1. Furthermore, the spectral bands at 999, 1569, 1587, 2880, 2889 and 2896 cm−1 that represent the molecular vibration of proteins were seen in the cancer tissue. In the surrounding tissues various bands of collagen at 810, 852, 918, 933, 1032, 1239, 1275, 1341, 1401 and 1677 cm−1 were found, and they can be correlated with ongoing liver fibrosis. A more detailed identification of the Raman bands based on several references can be found in Table S1 in the ESI.† Essentially the main variations seen in HCC correspond to lipids and proteins, while collagen, glycogen, DNA as well as proteins and lipids were visible in the surrounding fibrotic tissues. The MALDI data showed significant differences in the lipid types between HCC and fibrotic tissues. Within the measured ranges of 200–400, 700–900 and up to 1000 m/z, the identification of fatty acyls (e.g. subclass of fatty acids), phospholipids and complex lipids such as sphingolipids can be achieved, respectively.
In this study, no significant differences and high variations were observed in the range of 200–400 m/z whereas significant differences could be detected in the range of 700–1000 m/z. Therefore, the 700–850 m/z region was selected for the development of a LDA classification model. We used the LIPID MAPS® Lipidomics Gateway database for the identification of lipids based on their m/z ratios, as it was published in other studies.12,13,22 The found lipid classes and subclasses are provided in Table S2 in the ESI.† Roughly, main differences could be identified in Glycerophosphocholines (GP01) at 783.56 and 789.61 m/z and in Glycerophosphoinositols (GP06) at 799.56, 790.38 and 800.38 m/z for both HCC and fibrosis. In addition, the following changes could be found: in Glycerophosphoserines (GP03) at 799.56 m/z, in Glycerophosphates (GP10) at 782.59 m/z and mostly in Glycerophosphoglycerols (GP04) at 750.94, 784.35 and 798.36 m/z for only HCC and in Glycerophosphoethanolamines (GP02) at 709.12 m/z for only fibrosis. The role of glycerophospholipids in the metastasis development of HCC and as a diagnostic marker was already published and will be described later in the Discussion section.11,23
| A | Raman data LBOCV | Prediction | Acc | Sens | Spec | ||
|---|---|---|---|---|---|---|---|
| Fibrosis | HCC | ||||||
| True | Fibrosis | 15 | 2 | 84.4% | 88.2% | 80.0% | |
| HCC | 3 | 12 | 80.0% | 88.2% | |||
| B | MALDI data LBOCV | Prediction | Acc | Sens | Spec | ||
|---|---|---|---|---|---|---|---|
| Fibrosis | HCC | ||||||
| True | Fibrosis | 15 | 6 | 68.2% | 71.4% | 65.2% | |
| HCC | 8 | 15 | 65.2% | 71.4% | |||
| C | Combined data LBOCV | Prediction | Acc | Sens | Spec | ||
|---|---|---|---|---|---|---|---|
| Fibrosis | HCC | ||||||
| True | Fibrosis | 17 | 8 | 66.7% | 68.0% | 65.5% | |
| HCC | 10 | 19 | 65.5% | 68.0% | |||
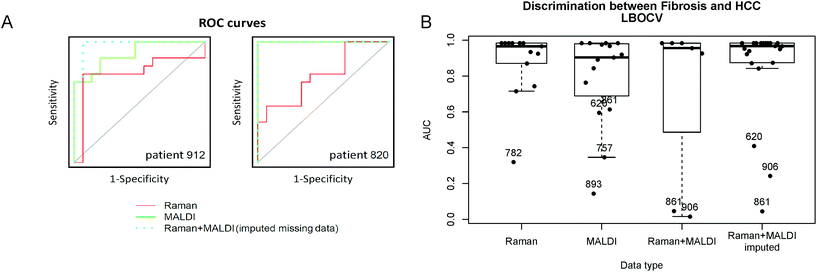
Besides comparing the two techniques separately we also performed a combined analysis. Although multiple samples were measured using Raman spectroscopy and MALDI imaging, some patients had data only for one technique due to the limitations described above. Therefore, a direct combination of the data resulted in a number of observations with missing data that cannot be used in the analysis. The fact of missing data leads to a dramatic decrease of the training data, e.g. patients, thus lowering the analysis efficiency. However, when the missing data are imputed artificially based on the available data as described in the Data analysis subsection, the resulting amount of training data increases and this can increase the performance of the model using the combined data. The calculated ROC curves for 2 patients can be seen in Fig. 3A as examples. In these cases the combination of the Raman and MALDI IMS data provided the best prediction result. The area under the ROC curve (AUC) for all data of the combined model is comparable to the AUC for the Raman spectroscopic data (Fig. 3B) even for the samples that were not measured with Raman imaging.
Although the AUC values above 0.5 in Fig. 3B prove that the MALDI imaging data, Raman spectroscopic data, and the combined data reflect differences between the fibrosis and HCC regions, the patient-to-patient variations complicate the task of predicting the tissue regions in the test data. Thus, Table 2 clearly shows that the prediction efficiency for the MALDI data, as well as for the combined analysis remained low with an accuracy of 68.2% and 66.7%, respectively. This can be related to the large patient-to-patient variations, which are more pronounced in the MALDI imaging data, than in the Raman spectra. The variations may be more prominent in MALDI IMS due to the technical difficulties in the slide-to-slide reproducibility of the technique and sample preparation variations. Nevertheless, the prediction of the tissue type by using the LDA model for the Raman data showed promising results with an accuracy of 84.4% with only 2–3 misclassified patients (Table 2A).
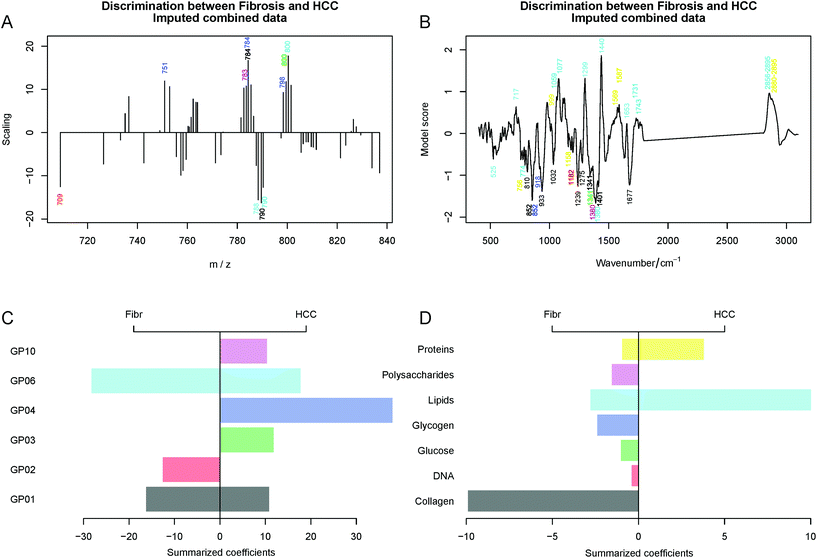
The LDA model loadings (Fig. 4A and B) for both MALDI imaging data and Raman spectroscopic data reveal significant differences between the fibrosis and HCC regions. Therefore, the peaks highlighted by the LDA scaling vectors for the MALDI data were assigned and grouped into classes of GP and the peaks of the LDA scaling vectors for the Raman data were grouped according to the compounds that can be found in the tissue. This was performed based on ref. 5, 6, 12 and 20–22.
The positive and negative peaks of the LDA scaling vectors are related to the HCC and fibrosis regions, respectively. Therefore, we summarized the positive and negative intensities separately within each group. The results of this condensing reflect the contribution of different types of substances and classes of lipids and this is depicted in Fig. 4C and D. Moreover, the exact band identification of the Raman spectra was based on four references and the band assignment can be seen in Table S1.† In addition, the identification of lipid types based on the MALDI m/z values is presented in Table S2 in the ESI.† In summary, the MALDI data allowed us to detect high expression profiles of GP01, GP03, GP06, GP10 and especially GP04 in HCC and high expression profiles of GP01, GP02 and especially GP06 in the fibrotic tissue. Lastly, GP03, GP04 and GP10 were specifically expressed only in HCC and GP02 was expressed in fibrosis. The loadings for the LDA model for the combined imputed Raman and MALDI data can be found in Fig. 4.
In conclusion, Raman spectroscopy alone and in combination with MALDI has high potential for the prediction of HCC within fibrotic liver tissues. Here, Raman imaging can predict HCC with an accuracy of 84% and MALDI IMS with an accuracy of 68%. The combination of both unfortunately did not improve the prediction accuracy which was 67%, a really low value. Moreover, Raman imaging detected significant variations in the expression of lipids, proteins and collagen. In addition, MALDI IMS could highlight variations in the expression of GP which will be discussed later.
A combination of Raman imaging and MALDI IMS for the classification of the HCC differentiation grades
For all experiments, the annotations for each patient sample were generated by an experienced pathologist and are provided in Table 1. First, the spectral differences for each of the three grades were analyzed. Fig. 5 shows the average spectra of well-, moderately- and poorly differentiated HCC and their standard deviations for Raman and MALDI imaging. In the same figure the difference spectra of the three grades are shown. The largest variations in the Raman spectra were identified between well- and moderately-differentiated HCC as they can be seen in Fig. 5A and C. The main differences in the moderate grades were found in the lipid associated peaks at 776, 1304, 1436, 1446, 1716, 2817, and 2850 cm−1 and the protein based bands at 753, 850, 873, 1002, 1008, 1104, 1155, 1209, 1230, 1552, 1586, 1676 and 2928 cm−1. Interestingly collagen was seen only in the Raman spectra of the moderate differentiation grade at 859, 1035, 1246, 1280, 1335, and 1635 cm−1. In the MALDI data significant variations in the lipid expression were found in two classes: phospholipids, especially glycerophospholipids (GP) at 200–400 m/z and fatty acyls (FA) (e.g. subclass of fatty acids) at 700–850 m/z. These m/z regions were further analyzed and a visualization of the MALDI spectra can be seen in Fig. 5B and D. The band annotations for the Raman and MALDI spectra can be found in Tables S3 and S4 in the ESI,† respectively.As there were low number of poor differentiation grade patient samples, we focused on whether we could predict well- and moderately-differentiated HCC by both measurement techniques. This is in-line with the major interest in the histopathology of HCC. For a statistically significant comparison of Raman spectroscopy and MALDI spectrometry for HCC differentiation, we used a linear regression model. PCA was used for data dimensionality reduction prior to regression. The response values for the moderately-differentiated HCC samples were set negative whereas the values for the well-differentiated samples were set positive. Thus, to maintain the unit difference, the values of −1/2 and +1/2 were set, respectively. Moreover, the sign of the predicted value was used to predict if the tissue region belongs to the moderately- or well-differentiated HCC region. Table 3 shows the LBOCV prediction of HCC differentiation based on the MALDI data. We found a high sensitivity and specificity resulting in an accuracy of 90%. This is especially astonishing, because the number of patients in each group was quite low. This result supports our concept of the importance of the lipid types for the development of HCC in the human liver.
| MALDI data LBOCV | Prediction | Acc | Sens | Spec | ||
|---|---|---|---|---|---|---|
| Moderate | Well | |||||
| True | Moderate | 4 | 0 | 89.9% | 100% | 80.0% |
| Well | 1 | 4 | 80.0% | 100% | ||
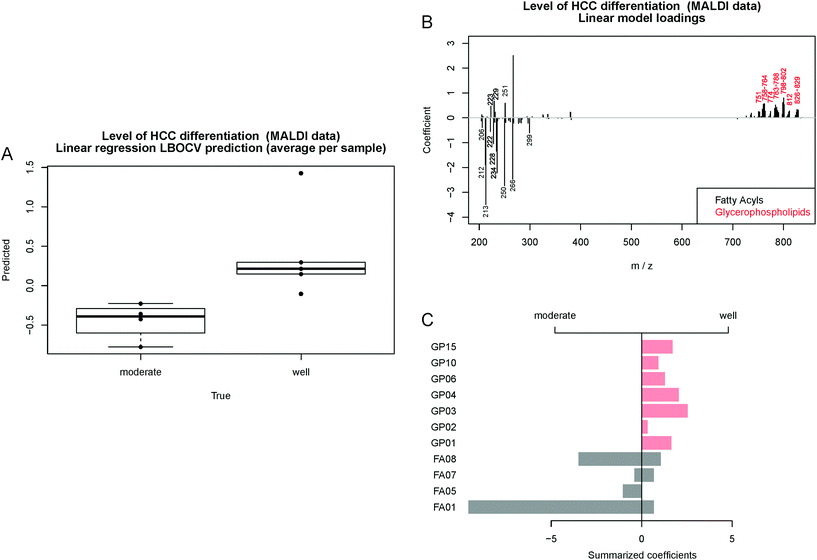
Another representation of the regression results is shown in the form of a boxplot showing the predicted values obtained by LBOCV (Fig. 6A). In contrast to Table 3, the figure shows that even though one of the well-differentiated HCC samples is predicted to be moderate, its prediction is located near the threshold and it is clearly separated from the moderately-differentiated samples. To analyze the model we assigned the MALDI peaks related to the model coefficients and grouped them to the classes FA and GP based on the reference database LIPID MAPS22 (Fig. 6B). Then, the positive and negative coefficients were summarized within each class and visualized in a barplot (Fig. 6C). One of the most interesting findings was that significant differences in the lipid expression in moderately- and well-differentiated HCC were found by MALDI IMS. Moderately differentiated HCC showed only the expression of FA, especially FA01 and FA08. In well-differentiated HCC lower amounts of FA and mostly GP01, GP03, GP04 and GP15 were found to be significant for the classification model. The identified lipid classes are listed in Table S3 in the ESI.† These results show the high potential of MALDI IMS for lipid marker identification for early (well-differentiated) HCC diagnosis. The roles of GP and FA in HCC have already been investigated by other groups and will be described later in the Discussion section.12,13,24–29
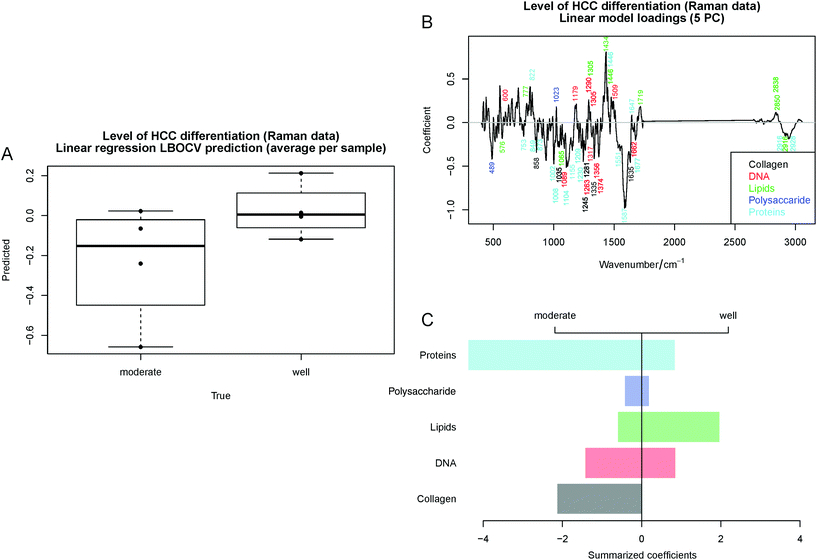
Although the prediction of HCC differentiation using a linear regression model based on the Raman spectroscopic data (Table 4) is less promising than that based on the MALDI data, the model shows a trend for HCC differentiation (Fig. 7A). Therefore, the investigation of the model coefficients was performed similar to those shown in Fig. 4 and 6. The labeled coefficients and the summarized results are depicted in Fig. 7B and C. The following components were detected in HCC: proteins, lipids, collagen, DNA and polysaccharides. Moderately-differentiated HCC had higher protein content than well-differentiated liver cancer and collagen was seen only in this tumor grade. In contrast, lipids were highly expressed in well-differentiated HCC, but lower amounts of polysaccharides and DNA could be seen in this tissue. The annotation of the Raman bands can be found in Table S4 in the ESI.† Based on the second study, MALDI IMS showed a higher prediction potential (accuracy of 90%) for the detection of HCC differentiation at early stages based on the GP and FA lipid profiles. Moreover, significant changes in the molecular composition of proteins, lipids and collagen in HCC during carcinogenesis can be detected by Raman spectroscopy with a prediction accuracy of 63%. Unfortunately, for this task the combination of the two methods was not possible from the analytical point of view due to the low number of samples per group.
| Raman data LBOCV | Prediction | Acc | Sens | Spec | ||
|---|---|---|---|---|---|---|
| Moderate | Well | |||||
| True | Moderate | 3 | 1 | 62.5% | 75.0% | 50.0% |
| Well | 2 | 2 | 50.0% | 75.0% | ||
Discussion and conclusion
The purpose of this study was to compare two molecular imaging methods (Raman imaging and MALDI IMS) with high potential for the diagnosis of HCC in order to improve the diagnosis of liver cancer. As we can see in Tables 2 and 3, the combination of these two techniques was not the best to complete this task. However, we found optimal application scenarios for each technique. Raman spectroscopy can predict liver cancer against non-cancerous tissue with an accuracy of 84% (a sensitivity of 88% and a specificity of 80%) and MALDI IMS is good at predicting moderately- and well-differentiated HCC with an accuracy of 90% (a sensitivity of 100% and a specificity of 80%).Moreover a precise analysis of lipidomic changes taking place during cancer development in the human liver was performed based on the Raman and MALDI imaging data. In the first part of our study, we were able to show that Raman spectroscopy provides complex molecular information about lipids, proteins, polysaccharides, glycogen, glucose, DNA, collagen and others. This information could be used to investigate the difference between the tumor and the surrounding tissue. In addition, the investigation of the differentiation grades of HCC by Raman imaging provided important information about the molecular composition of the samples, but the Raman based classifier showed a low performance. Furthermore, lipids and proteins were detected in the HCC tissue and collagen, glycogen, proteins and lipids were detected in the fibrotic tissue. Lipid markers such as GP03, GP10 and especially GP04 were found in HCC, GP02 in fibrosis and GP01 and GP06 in both tissue regions by MALDI IMS. In the second part of our study, Raman imaging allowed the detection of a decrease in protein and collagen and in parallel an increase in lipids when HCC evolved from the well- to moderate-differentiation grade. At the same time MALDI IMS showed the same behavior as well-differentiated HCC had high expression of lipids in comparison with that of moderately-differentiated HCC. The most interesting results in the application of MALDI IMS were found for lipidomics of the HCC differentiation grades. Here, particular GPs (GP01, GP02, GP03, GP04, GP06, GP10 and GP15) were highly significant for the classification model of well-differentiated HCC and not for the moderate grade. In addition, significantly higher expressions of FA01 and FA08 were identified in moderately differentiated HCC.
The results are in-line with the current state of research on fatty acids (subclass of class FA) and GP expression during HCC development in the human liver.1,11,23,30 The role of fatty acids in HCC as important molecules for providing energy and metabolites for other anabolic pathways in hepatocytes has been under investigation for a long time.1 Alterations in the fatty acid translocase protein CD36 expression were also linked to the increased uptake of fatty acids by obese patients with HCC.30 Moreover, Nath et al. demonstrated that the increased fatty acids are crucial to the activation of the pro-inflammatory pathways or lipotoxicity in HCC.1 Besides this, fatty acid elongation in the livers of patients with non-alcoholic steatohepatitis and HCC was found to occur. The role of glycerophospholipid as a structural component in biological membranes and its effects on signaling and transport molecules were studied intensively as well. Lin et al. identified a correlation between the decreased palmitic acyl (C16:0)-containing glycerophospholipids and HCC metastasis. Furthermore, several other lipid types were studied in the liver cancer tissue as markers of HCC. For example, Patterson et al. showed that the levels of glycodeoxycholate, deoxycholate 3-sulfate, biliverdin and fetal bile acids increased and lysophosphocholines, lignoceric acid and nervonic acid decreased in the plasma of HCC patients.
One major challenge in our study is the precise identification of lipids found by MALDI IMS and Raman spectroscopic imaging in HCC tissues. We were able to assign each band that played a significant role in the classification of HCC and fibrosis as well as moderately- and well-differentiated HCC.5,6,12,20–22 Subsequent literature research allowed us to confirm our findings based on the MALDI IMS bands from already published articles about the main classes of lipids specifically expressed in HCC.12,13 Passos-Castilho et al. showed the role of sterols (ST01), Glycerophosphocholines (GP01), Glycerophosphates (GP10), fatty acids and conjugates (FA01), Glycerophosphoserines (GP03) and Glycerophosphoinositols (GP06) in hepatitis C infected HCC. These data are in accordance with our results, as the following lipid classes were found by MALDI IMS: GP01 at 784 m/z found in the HCC tissue and 764, 784, 788, 812 and 828 m/z found in well-differentiated HCC; GP10 at 783 m/z in HCC and 761 and 783 m/z in well-differentiated HCC; FA01 at 212, 222, 234, 250, 266, and 299 m/z in moderately-differentiated HCC and 229 m/z in well-differentiated HCC; GP03 at 800 m/z in HCC and 760, 762, 800, 802, and 830 m/z in well-differentiated HCC; and GP06 at 789, 791, and 801 m/z in HCC and 789, 801, and 827 m/z in well-differentiated HCC. Furthermore, Thomas et al. found the band at 790 m/z in the normal liver tissue and in our study it was found in the fibrotic tissue surrounding HCC.27 The abnormal distribution of phospholipids such as sphingomyelin (16:0) was previously reported, but was not seen in our data because this complex lipid belongs to the class phosphosphingolipids (SP03) and it can be seen in the range from 1000 to 2000 m/z.26 Data on the investigation of HCC by Raman imaging are more limited, but our data showed the expected correlation with the composition of HCC tissue and adjacent tissue (fibrosis). In the cancer regions we expected proteins and lipids to be the main liver cell components. At the same time the fibrotic regions mostly consist of collagens and proteins (glycoproteins and proteoglycans).31 Furthermore, lipids in hepatic stellate cells in the fibrotic tissue, as well as glucose (glycogen) and polysaccharides as storage components can be seen in the liver. In our previous article we were able to detect higher expression of unsaturated fatty acids in the HCC cells by Raman imaging5 and we were able to find 4 main chemical components by using the N-FINDR algorithm: protein, collagen, triglycerides and cholesterol ester. Moreover, we were able to show the selective expression of triglycerides, as a major form of storage and transport of fatty acids in 5 of 23 patients’ HCC samples.6,32 In the second part of our study, during which the HCC grades were investigated, we were able to correlate the findings of high expression of collagen in moderately-differentiated HCC in comparison with that in well-differentiated HCC (Fig. 7C) with the results of the second harmonic generation analysis of the HCC grades published by Lin et al. in 2018.33
In conclusion, our study has demonstrated the potential of Raman spectroscopy and MALDI IMS for liver cancer classification in specific applications. Raman spectroscopy is a fast, label-free and non-destructive technique that can differentiate cancer from non-cancerous tissues. Although this is a trivial task for in vitro investigations in clinical routine, Raman spectroscopy can potentially be implemented in vivo34 to define the regions of interest for biopsy.
In addition, MALDI IMS can predict the differentiation grade of HCC based on the lipid expression with high accuracy. Moreover, significant lipid changes, especially in GP and FA (class of subclass fatty acids), were found in the liver carcinogenesis and in the differentiation of HCC from early well-differentiated HCC to moderately-differentiated HCC. The knowledge of these processes during HCC development is limited, and therefore further investigation into this research area is of high importance for diagnostics and therapy. Here, we were able to highlight the potential of Raman spectroscopic imaging not only for the classification and prediction of HCC tumor (a sensitivity of 88% and a specificity of 80) but also as a novel technique for studying the molecular composition of HCC and HCC differentiation grades. Furthermore, we showed that MALDI IMS exhibits high predictive performance for the classification of early and progressed HCC with a sensitivity of 100% and a specificity of 80%. The two investigated techniques demonstrated their potential for application in two different levels of tumor diagnosis: HCC detection and cancer grade identification. It could be possibly applied as a 2-step diagnostic process based on Raman and subsequent MALDI IMS investigation in order to obtain precise results.
Moreover, new lipid markers: GP03, GP10 and especially GP04 for HCC; GP02 for fibrosis; GP01, GP02, GP03, GP04, GP06, GP10 and GP15 for well-differentiated HCC and FA01 and FA08 for moderately-differentiated HCC were identified by MALDI IMS. Both techniques have potential for cancer prediction and lipidomics studies of HCC. Particularly, the in vivo application of Raman spectroscopy and the direct identification of biomarkers by MALDI IMS are optimal application scenarios. Nevertheless, their advantages and disadvantages in clinical diagnostics need to be recognized and are summarized in Table 5.
| Raman spectroscopy | MALDI IMS | ||
|---|---|---|---|
| Advantages | Disadvantages | Advantages | Disadvantages |
| + Non-destructive | − Weak Raman signal | + Fast | − Sample preparation is complex and expensive |
| + Label-free | − Auto-fluorescence | + Label-free | − Limited reproducibility |
| + Complex “fingerprint” molecular information | − Lack of sensitivity for low concentrations | + Selective analysis of lipids, proteins or nanoparticles | − Limited spatial resolution |
| + Easy sample preparation and small sample size | − Need for chemometric data analysis | + Analysis of samples with large size | − Limited detectable mass range |
| + Suitable for fiber optic probes and in vivo application | − Long measurement time | + Suitable for large number of samples | − Not suitable for in vivo application |
| − Direct identification of individual substances is not possible | + Direct identification of the analyte is possible | ||
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to the co-workers from the gastroenterology laboratory for support, Dr med. Claudio Marquardt for MALDI IMS expertise and Univ. Prof. Dr med. Tony Bruns for scientific discussions. Financial support from the German Research Foundation (DFG) providing research funding to TB (BR 4182/3-1), to TWB (BO 4700/1-1), to JP (PO 563/30-1) and to STA (295/11-1) is gratefully acknowledged. The project on which these results are based was supported by the Free State of Thuringia under the number 2019 FGR 0083 and co-financed by European Union funds within the framework of the European Social Fund (ESF).References
- A. Nath, I. Li, L. R. Roberts and C. Chan, Sci. Rep., 2015, 5, 14752 CrossRef CAS.
- C. I. Theodore and M. Irena, Liver Biopsy. InTech, 2011, 109–136 Search PubMed.
- E. S. Chan and M. M. Yeh, Clin. Liver Dis., 2010, 14, 687–703 CrossRef.
- M. Mireskandari and I. Petersen, in Clinical Pathology, ed. J. Popp, Wiley-VCH Verlag GmbH & Co. KGaA, 2014, ch. 1, pp. 215–248 Search PubMed.
- T. Tolstik, C. Marquardt, C. Matthäus, N. Bergner, C. Bielecki, C. Krafft, A. Stallmach and J. Popp, Analyst, 2014, 139, 6036–6043 RSC.
- T. Tolstik, C. Marquardt, C. Beleites, C. Matthäus, C. Bielecki, M. Bürger, C. Krafft, O. Dirsch, U. Settmacher, J. Popp and A. Stallmach, J. Cancer Res. Clin. Oncol., 2015, 141, 407–418 CrossRef CAS.
- J. Le Faouder, S. Laouirem, M. Chapelle, M. Albuquerque, J. Belghiti, F. Degos, V. Paradis, J.-M. Camadro and P. Bedossa, J. Proteome Res., 2011, 10, 3755–3765 CrossRef CAS.
- S. Laouirem, J. Le Faouder, T. Alexandrov, D. Mestivier, T. Léger, X. Baudin, M. Mebarki, V. Paradis, J.-M. Camadro and P. Bedossa, J. Pathol., 2014, 234, 452–463 CrossRef CAS.
- C. Marquardt, T. Tolstik, C. Bielecki, R. Kaufmann, A. Crecelius, U. Schubert, U. Settmacher, A. Stallmach and O. Dirsch, Z. Gastroenterol., 2015, 53, 33–39 CrossRef CAS.
- J. Liu, P. Yang, G. Zuo, S. He, W. Tan, X. Zhang, C. Su, L. Zhao, L. Wei, Y. Chen, X. Ruan and Y. Chen, Lipids Health Dis., 2018, 17, 153 CrossRef.
- A. D. Patterson, O. Maurhofer, D. Beyoğlu, C. Lanz, K. W. Krausz, T. Pabst, F. J. Gonzalez, J.-F. Dufour and J. R. Idle, Cancer Res., 2011, 71, 6590–6600 CrossRef CAS.
- A. M. Passos-Castilho, E. Lo Turco, M. L. Ferraz, C. Matos, I. Silva, E. Parise, E. Pilau, F. Gozzo and C. Granato, J. Gastrointestin. Liver Dis., 2015, 24, 43–49 CrossRef.
- A. M. Passos-Castilho, V. M. Carvalho, K. H. M. Cardozo, L. Kikuchi, A. L. Chagas, M. S. Gomes-Gouvêa, F. Malta, A. C. S. S. Nastri, J. R. R. Pinho, F. J. Carrilho and C. F. H. Granato, BMC Cancer, 2015, 15, 985 CrossRef.
- M. Schlageter, L. M. Terracciano, S. D'Angelo and P. Sorrentino, World J. Gastroenterol., 2014, 20, 15955 CrossRef.
- R Core Team, R Foundation for Statistical Computing, Vienna, Austria, 2018 Search PubMed.
- B. O. Keller and L. Li, J. Am. Soc. Mass Spectrom., 2000, 11, 88–93 CrossRef CAS.
- C. G. Ryan, E. Clayton, W. L. Griffin, S. H. Sie and D. R. Cousens, Nucl. Instrum. Methods Phys. Res., Sect. B, 1988, 34, 396–402 CrossRef.
- S. X. Guo, T. Bocklitz, U. Neugebauer and J. Popp, Anal. Methods, 2017, 9, 4410–4417 RSC.
- J. Josse and F. Husson, J. Soc. Fr. Statistique, 2012, 153, 79–99 Search PubMed.
- Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2007, 42, 493–541 CrossRef CAS.
- A. Bonifacio, C. Beleites, F. Vittur, E. Marsich, S. Semeraro, S. Paoletti and V. Sergo, Analyst, 2010, 135, 3193–3204 RSC.
- LIPID MAPS Lipidomics Gateway, https://www.lipidmaps.org/, (accessed October 11, 2019).
- L. Lin, Y. Ding, Y. Wang, Z. Wang, X. Yin, G. Yan, L. Zhang, P. Yang and H. Shen, Hepatology, 2017, 66, 432–448 CrossRef CAS.
- S. Jiang, Y. Li, S. Lin, H. Yang, X.-Y. Guan, H. Zhou, T. Luan and Z. Cai, Anal. Methods, 2015, 7, 8466–8471 RSC.
- X. Chen, J. Gao, T. Wang, X. Jiang, J. Chen, X. Liang and J. Wu, Anal. Chem., 2019, 91, 10376–10380 CrossRef CAS.
- S. Shimma, Y. Sugiura, T. Hayasaka, Y. Hoshikawa, T. Noda and M. Setou, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 855, 98–103 CrossRef CAS.
- A. Thomas, N. H. Patterson, M. M. Marcinkiewicz, A. Lazaris, P. Metrakos and P. Chaurand, Anal. Chem., 2013, 85, 2860–2866 CrossRef CAS.
- J. Wattacheril, E. H. Seeley, P. Angel, H. Chen, B. P. Bowen, C. Lanciault, R. M. Caprioli, N. Abumrad and C. R. Flynn, PLoS One, 2013, 8, e57165 CrossRef CAS.
- Y. Morita, T. Sakaguchi, K. Ikegami, N. Goto-Inoue, T. Hayasaka, V. T. Hang, H. Tanaka, T. Harada, Y. Shibasaki, A. Suzuki, K. Fukumoto, K. Inaba, M. Murakami, M. Setou and H. Konno, J. Hepatol., 2013, 59, 292–299 CrossRef CAS.
- D. P. Y. Koonen, R. L. Jacobs, M. Febbraio, M. E. Young, C.-L. M. Soltys, H. Ong, D. E. Vance and J. R. B. Dyck, Diabetes, 2007, 56, 2863–2871 CrossRef CAS.
- M. F. Sorrell, E. R. Schiff and W. C. Maddrey, Schiff's Diseases of the Liver, Lippincott-Raven Publishers, 1999 Search PubMed.
- M. Alves-Bezerra and D. E. Cohen, Comprehensive Physiology 2018, pp. 1–22 Search PubMed.
- H. Lin, L. Lin, G. Wang, N. Zuo, Z. Zhan, S. Xie, G. Chen, J. Chen and S. Zhuo, Biomed. Opt. Express, 2018, 9, 3783–3793 CrossRef CAS.
- E. Cordero, I. Latka, C. Matthäus, I. Schie and J. Popp, J. Biomed. Opt., 2018, 23, 071210 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0an01555e |
| ‡ Equal first-author contribution. |
| § Equal senior-author contribution. |
| This journal is © The Royal Society of Chemistry 2021 |