Microfluidic filter device coupled mass spectrometry for rapid bacterial antimicrobial resistance analysis†
Dongxue
Zhang
 a,
Yijie
Zhang
a,
Fan
Yin
a,
Yijie
Zhang
a,
Fan
Yin
 a,
Qin
Qin
b,
Hongyan
Bi
c,
Baohong
Liu
a,
Qin
Qin
b,
Hongyan
Bi
c,
Baohong
Liu
 a and
Liang
Qiao
a and
Liang
Qiao
 *a
*a
aDepartment of Chemistry, Shanghai Stomatological Hospital, Fudan University, Shanghai 200000, China. E-mail: liang_qiao@fudan.edu.cn
bChanghai Hospital, The Naval Military Medical University, Shanghai 200433, China
cCollege of Food Science and Engineering, Shanghai Ocean University, 999 Hucheng Ring Road, Pudong New District, Shanghai 201306, China
First published on 9th November 2020
Abstract
The problem of antimicrobial resistance (AMR) is becoming increasingly serious. Bacteria producing extended spectrum beta-lactamase (ESBL), which can hydrolyze beta-lactam antibiotics, are among the most important drug resistant bacteria. Rapid AMR analysis methods are essential for identifying antibiotic resistant bacteria, which is of significant positive value to the clinical therapy of infectious disease. We developed a platform which integrates a sandwich microfluidic filter device with electrospray ionization mass spectrometry (ESI-MS). Bacterial cells were loaded in the sandwich microfluidic chip and antibiotic drugs were injected to pass through the blocked bacterial cells. By online ESI-MS analysis of the antibiotic drugs and their hydrolysis products, the AMR of the bacteria can be assessed within 30 minutes. Four Escherichia coli strains, namely two ESBL-positive and two ESBL-negative, were successfully discriminated using ampicillin and the third generation cephalosporin ceftriaxone. Considering the simplicity and high efficiency of the assay, the microfluidic chip integrated online ESI-MS system is promising in the rapid clinical diagnosis of ESBL-producing bacteria.
Introduction
Antimicrobial resistance (AMR) is a major problem threatening public safety and inflicting finance damage. The abusive and excessive usage of antibiotics in medicine,1 agriculture2 and the food industry3 results in and exacerbates this serious situation. Bacteria resist antibiotics via mainly the following mechanisms: by synthesizing enzymes to inactivate antibiotics,4 by reducing the membrane permeability or using efflux pumps to prevent antibiotics from accessing the target sites,5 by modifying target sites,6 and by alternating metabolic pathways to bypass the action of antibiotics.7 Extended spectrum beta-lactamase (ESBL) producing bacteria are one important class of antibiotic resistant bacteria that synthesize enzymes to inactivate a number of beta-lactam antibiotics, including the third generation cephalosporins, such as ceftazidime, cefotaxime, ceftriaxone, etc., leading to serious AMR problems.8,9 These antibiotics kill bacteria by impeding the enzymes required for peptidoglycan formation, thereby inhibiting the bacterial cell wall synthesis, which is essential for maintaining cellular activity.8 However, ESBL can make the antibiotics out of work by hydrolyzing and degrading the drugs.10 The first ESBL-producing bacterium was found in Europe in 1983.11 In the 1980s and 1990s, ESBL-producing Klebsiella pneumoniae was the main culprit giving rise to nosocomial infection outbreaks.12 According to the 2019 antibiotic resistance (AR) threats report, 197![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 estimated cases and 9100 estimated deaths in the United States were triggered by ESBL-producing Enterobacteriaceae.13 Rapid AMR analysis methods are essential for identifying antibiotic resistant bacteria, and are of significant positive value to the clinical therapy of infectious disease.
400 estimated cases and 9100 estimated deaths in the United States were triggered by ESBL-producing Enterobacteriaceae.13 Rapid AMR analysis methods are essential for identifying antibiotic resistant bacteria, and are of significant positive value to the clinical therapy of infectious disease.
Traditional AMR detection methods include disk diffusion, broth dilution, and E-test. Disk diffusion, as the “gold standard”, is simple and cost efficient, however it is influenced by experimental factors, e.g. pH, temperature, solvent, etc.14 Broth dilution is laborious and consumes a great amount of reagents. As for the outcomes, the traditional AMR detection methods can report false positive results.15 Moreover, these methods consume 16 to 24 hours, limiting fast bacterial AMR analysis and hence delaying the clinical therapy. Molecular biology techniques, e.g. PCR and loop-mediated isothermal amplification (LAMP), are fast and sensitive in the detection of bacterial antimicrobial resistant genes. However, resistant genes don't necessarily express, and thus the correlation between the bacterial AMR phenotype and the existence of resistant genes is low.14 Matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been widely used for bacterial identification and is also applied to AMR detection. By comparing the bacterial mass fingerprints, susceptible and resistant bacterial strains can be discriminated.16 Proteins expressed by antimicrobial resistant genes can be detected by MALDI-TOF MS to identify bacterial AMR.17 TiO2-assited laser desorption/ionization MS has been used to detect bacterial intracellular metabolites to identify bacterial AMR.18 MALDI-TOF MS can also be used to analyze antibiotic drugs co-cultured with bacteria to assess the bacterial susceptibility. For example, by detecting the signal of meropenem, a carbapenem, carbapenemase-producing bacteria can be identified.19 Identification of ESBL-producing bacteria has been achieved in a similar way.20
For fast and high throughput screening of ESBL-producing bacteria, we combined microfluidic chips and mass spectrometry to analyze antibiotic drugs co-cultured with bacteria. Microfluidics has diverse advantages, e.g. low reagent consumption, high throughput, etc. Compared to traditional AMR detection methods, analysis on a microfluidic chip is more convenient, safer and faster. Using microfluidic chips, antimicrobial susceptibility testing (AST) was achieved within 150 min by detecting the sample capacitance to obtain the bacterial cell count in the presence of different concentrations of antibiotics.21 A droplet based microfluidic chip was used for rapid Escherichia coli AST by observing the bacterial morphology and cell number.22 Utilizing a 96 cell plate-like microfluidic chip, imaging-based single-cell morphological analysis (SCMA) was realized for fast AST that showed the same accuracy as traditional methods.23
In this work, we developed a sandwich microfluidic filter device coupled with electrospray ionization mass spectrometry (ESI-MS) for on-line rapid detection of ESBL-producing bacteria. Bacterial cells were loaded in the sandwich microfluidic chip and antibiotic drugs were injected to pass through the bacterial cells. By detecting the intensity of antibiotics and the corresponding hydrolysis peaks, bacterial AMR analysis could be accomplished within 30 min. Two strains of ESBL-positive E. coli and two strains of ESBL-negative E. coli were used as model samples to illustrate the performance of the method. Under the treatments of two beta-lactam antibiotics, i.e. ampicillin (AMP) and ceftriaxone (CEF), ESBL-positive E. coli strains were well distinguished from the ESBL-negative ones.
Results and discussion
A microfluidic filter device with three components, i.e. a holder, two PMMA slices and a piece of polycarbonate (PC) membrane, was modified from our previously reported microchips that were used for protein sample desalting for on-line ESI-MS analysis.24 As shown in Fig. 1(A), symmetrical serpentine channels were engraved on the two polymethyl methacrylate (PMMA) slides, which were fixed with the steel holder to clamp the PC membrane. The serpentine channels were located in an area of 12 mm × 12 mm on the PMMA slices. The width of the channels and the distance between two adjacent channels were both 500 μm (Fig. 1(B)). The depth of the channels was 80 μm. The PC membrane had uniform pores with a diameter of 220 nm and a density of 3 × 108 pores per cm2 (Fig. 1(C)).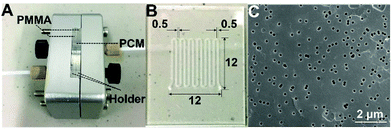 | ||
| Fig. 1 (A) The whole microfluidic filter device with a holder, two PMMA slices and a piece of PC membrane (PCM); (B) a PMMA slice with a serpentine channel; (C) a SEM image of the PC membrane. | ||
Bacteria were cultured overnight and washed with sterile water twice. The washed bacterial pellets were resuspended in sterile water (∼108 CFU mL−1, 30 μL) and loaded into the microfluidic filter device. During bacterial loading, inlets b and c were blocked, and the bacterial sample was injected via inlet a at a speed of 5 μL min−1. The bacterial cells were blocked by the PC membrane, and water was wasted from outlet d. Then, antibiotic solution was injected into the chip from inlet a continuously at 5 μL min−1. Outlet d and a syringe filled with ESI buffer (MeOH, 4% acetic acid) were both connected to a polyethylene terephthalate (PET) microfluidic chip for on-line ESI-MS analysis25 of the antibiotic drugs and their hydrolysis products (Scheme 1). The flow rate of the ESI buffer was equal to that of the antibiotic solution. The mass spectra of 5 μg mL−1 AMP aqueous solution mixed with the ESI buffer on or off the PET chip are shown in ESI Fig. S1.† Similar mass spectra were obtained, indicating that the sample was well mixed with the ESI buffer in the PET chip for ESI-MS. The whole bacterial AMR analysis took <30 min. With the microfluidic filter device, the bacterial cells cannot enter the PET microfluidic chip for ESI, thereby avoiding the influence from most bacterial components in the analysis of the hydrolysis of the antibiotic drugs.
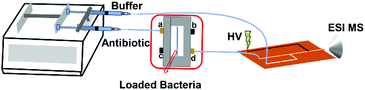 | ||
| Scheme 1 Schematic diagram of the microfluidic filter device coupled ESI-MS for on-line detection of bacterial AMR. HV: high voltage. | ||
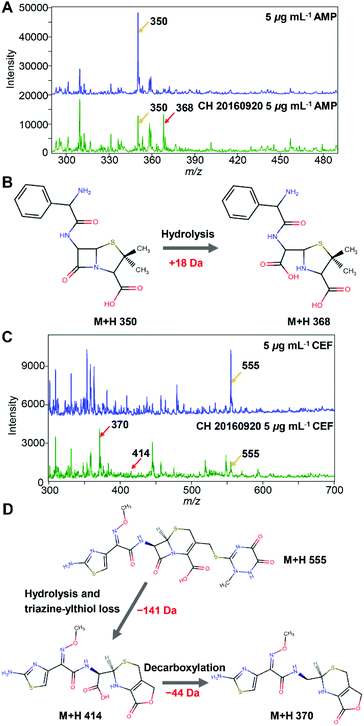
Two beta-lactam antibiotics, i.e. AMP and CEF, were utilized. The method was firstly demonstrated with an ESBL-positive E. coli strain (CH 20160920) clinically isolated by Changhai hospital. The identification and AST results of the isolate are shown in ESI Fig. S2, S3, and Table S1.† The mass spectra of 5 μg mL−1 AMP before and after being hydrolysed by the ESBL-positive E. coli strain are shown in Fig. 2A. The peak at m/z = 350 corresponds to protonated AMP ions [AMP + H]+. After passing through the ESBL-positive E. coli loaded microfluidic filter device, a hydrolysis product of AMP ([AMPhydro + H]+) was observed at m/z = 368, Fig. 2B, consistent with the previous report.26 The absolute intensity of the peak [AMP + H]+ of 5 μg mL−1 AMP was close to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. After being hydrolysed by ESBL, the intensity of [AMP + H]+ reduced to less than 15
000. After being hydrolysed by ESBL, the intensity of [AMP + H]+ reduced to less than 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. At the same time, the signal of [AMPhydro + H]+ increased tremendously and surpassed the signal of [AMP + H]+.
000. At the same time, the signal of [AMPhydro + H]+ increased tremendously and surpassed the signal of [AMP + H]+.
Similarly, before the hydrolysis, a strong peak at m/z = 555 was observed for protonated CEF ions [CEF + H]+ (Fig. 2C). After passing through the ESBL-positive E. coli loaded microfluidic filter device, two hydrolysis products of CEF were observed at m/z = 414 and m/z = 370, corresponding to the loss of a triazine-thiol group and a CO2, respectively, Fig. 2D.27–29 As shown in Fig. 2C, the intensity of [CEF + H]+ (m/z 555) decreased from ∼5000 to <2000 after the hydrolysis. In contrast, the intensity of the peak at m/z = 370 increased dramatically to >4000. From the results, E. coli (CH 20160920) can hydrolyse both AMP and third generation cephalosporins, clearly demonstrating that it is an ESBL-producing strain, consistent with the disk diffusion test (Fig. S3 and Table S1†).
To demonstrate that the method can be applied to different strains, four E. coli strains, namely two ESBL-positive (CICC 10663, CH 20160920) and two ESBL-negative (CICC 10661, ATCC 25922) were further tested. The identification and AST results of the isolates are shown in ESI Fig. S2, and S3, and Table S1.†Fig. 3A shows the hydrolysis results of 5 μg mL−1 AMP by the four different strains. Compared to the blank control wherein no bacteria were loaded into the microfluidic chip, the E. coli strain CICC 10661 and E. coli strain ATCC 25922 hardly hydrolysed any AMP. The weak signal of m/z = 368 for AMP hydrolysis products in the control group was from the baseline rather than a real MS peak. In contrast, the two ESBL-positive E. coli hydrolysed AMP obviously. The E. coli strain CH 20160920 showed the strongest hydrolysis effect against AMP, wherein the hydrolysis products gave an MS signal as strong as that of AMP itself.
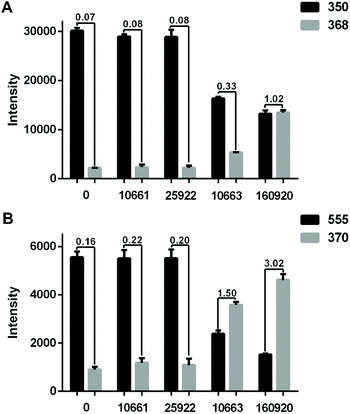 | ||
| Fig. 3 (A) Bar plots of [AMP + H]+ (m/z 350) and [AMPhydro + H]+ (m/z 368) after passing 5 μg mL−1 AMP through the microfluidic filter chip without any bacteria or loaded with the strain CICC 10661, ATCC 25922, CICC 10663 or CH 20160920 (3 × 106 bacterial cells each). (B) The results obtained with 5 μg mL−1 CEF. The intensity ratio of hydrolysates/substrates in different groups was labeled. The error bar shows the standard deviation from three replicates. The relative standard deviations (RSD) of the MS signal intensity from a single experiment during the period of MS signal recording and three replicates are shown in ESI Table S2.† | ||
In order to verify that beta-lactamase is an extended spectrum beta-lactamase, a third generation cephalosporin, CEF, was applied for AMR detection likewise. The result was similar to that of the analysis using AMP. As shown in Fig. 3B, compared to the blank control, the signals of CEF did not change significantly after passing through the microfluidic filter device loaded with E. coli strain CICC 10661 or E. coli strain ATCC 25922. However, significant hydrolysis of CEF was observed with E. coli strain CH 20160920 and E. coli strain CICC 10663. Combining the results in Fig. 3, it is clear that E. coli strain CH 20160920 and E. coli strain CICC 10663 produced ESBL, and E. coli strain CH 20160920 has a stronger AMR compared to the strain CICC 10663, consistent with the disk diffusion test (Fig. S3 and Table S1†).
According to the literature, at least 200 different types of ESBL enzymes have been identified, e.g. TEM-beta-lactamase, SHV-beta-lactamase, CTX-M beta-lactamase, etc.30 The production of ESBL is due to mutations in the beta-lactamase encoding genes, such as blaTEM, or gene mobilization from other bacteria producing ESBL in the environment.31 Jaen-Luchoro et al. characterized the ESBL-producing E. coli strain CCUG 73778 by genomics and proteomics.32 They identified 47 potential antibiotic resistance determinant genes localized on chromosomes, including 17 efflux pump or transport protein encoding genes and the beta-lactamase encoding gene ampC. Ma et al. studied the differences between 10 ESBL-producing and 10 susceptible E. coli strains and revealed potential mechanisms by combining proteomics and metabolomics.33 They found that the purine metabolism pathway showed a high enrichment in ESBL-E. coli and might participate in the antibiotic resistance. Consequently, the phenotype of antibiotic resistance by ESBL-producing strains can be from a combined effect of multiple genes. Herein, the strains CH 20160920 and CICC 10663 showed strong hydrolysis of AMP and CEF, indicating possibly that the two strains could express a high amount of ESBL with high activity.
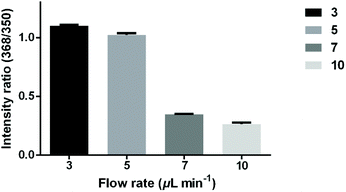
To study the relationship between the hydrolysis efficiency and reaction time, different flow rates of antibiotics were set to control the reaction time. The microfluidic filter chip was loaded with 3 × 106E. coli strain CH 20160920 bacterial cells. As shown in Fig. 4, as the flow rate increased from 3 μL min−1 to 10 μL min−1, the reaction time gradually decreased, leading to a lower hydrolysis efficiency. When the flow rate was 5 μL min−1, the intensity ratio of AMPhydro/AMP was 1.02 on average. As the flow rate was reduced to 3 μL min−1, the hydrolysis efficiency was not significantly improved. To balance the analysis time and analysis sensitivity, a flow rate of 5 μL min−1 was chosen for this study.
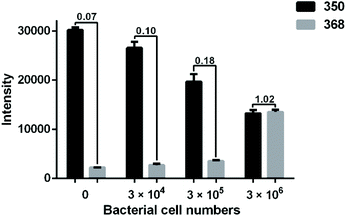
To assess the sensitivity of the method, different amounts of E. coli strain CH 20160920 (0, 3 × 104, 3 × 105 and 3 × 106 cells) were loaded into the microfluidic filter chip. As shown in Fig. 5, it is clear that the intensity of [AMP + H]+ gradually decreased as the bacterial count increased from 3 × 104 to 3 × 106 cells. Meanwhile, the intensity of [AMPhydro + H]+ was increasing. Compared to the blank control, there was a significant difference in the MS signals of both AMP and its hydrolysis products when the number of bacterial cells was lowered to 3 × 104 in the microfluidic filter chip. A lower bacterial count would lead to no significant signal change. It is possible to achieve a higher sensitivity in bacterial AMR detection using a stop-flow strategy, where antibiotic solution is filled in the microfluidic filter chip and maintained with the bacterial cells for a certain period of time.
Conclusions
We have developed microfluidic filter device coupled ESI-MS for rapid bacterial AMR analysis. On the basis of antibiotic hydrolysis, ESBL-positive E. coli and ESBL-negative E. coli were successfully discriminated by the platform. Compared with traditional methods, our assay is rapid and accurate. The whole volume, including the channels in the PMMA chip, connection microtubes between PMMA and PET chips, and channels in the PET chip, was about 50 μL. At the speed of 5 μL min−1, it took only ∼10 min from the antibiotic drug loading to MS signal generation for bacterial AMR detection. On adding the time of bacteria loading (6 min) and assembling the PMMA chip, the whole analysis time was <30 min. Such an assay can be run in parallel to achieve a higher throughput and can be applied to other classes of drug resistant bacteria, e.g. carbapenemase-producing bacteria. Consequently, it has the potential to be applied in rapid AMR analysis for clinical usage.Experimental
Chemicals and materials
A polycarbonate membrane (220 nm) was purchased from Wuwei Kejin Xinfa Technology Co., Ltd (Lanzhou, China). Ampicillin sodium (AMP) was from Sungon Biotech Co., Ltd (Shanghai, China). Ceftriaxone sodium (CEF) was from CNW, ANPEL Laboratory Technologies Inc. (Shanghai, China). Methanol (MeOH, HPLC grade) and acetonitrile (CH3CN, HPLC grade) were from Merck (Darmstadt, Germany). Trypticase soy broth (TSB) was from Beijing Land Bridge Technology CO., Ltd (Beijing, China). Trifluoroacetic acid (TFA) and α-cyano-4-hydroxycinnamic acid (HCCA) were purchased from Sigma-Aldrich (St Louis, MO, USA). Acetic acid (AR) was from Titan Scientific Co., Ltd (Shanghai, China). Test papers for the microbial susceptibility test were from Bio-Kont (Wenzhou, China). The E. coli strain CH 20160920 was from Changhai hospital in Shanghai, China. The E. coli strain CICC 10663 and strain CICC 10661 were from the China Center of Industrial Culture Collection (CICC, Beijing, China). The E. coli strain ATCC 25922 was from the American Type Culture Collection (ATCC, Manassas, VA, USA). The ultrapure water (18.2 MΩ cm) used in all experiments was obtained from a Super-Genie E 125 water system (RephiLe Bioscience, Shanghai, China). All chemicals were used as received without further purification.Microfluidic chip design and fabrication
The PMMA microchip and stainless-steel holder were fabricated by Wenhao Co., Ltd (Suzhou, China).24 The PET chip for ESI-MS was with three channels, 100 μm in width and 50 μm in depth, as shown in ESI Fig. S4.† The structure was designed by AutoCAD. To fabricate the chip, a piece of PET slice was engraved using a laser engraving machine (Han's Laser, Guangdong, China).34 After being engraved, one channel was filled with electric carbon paste to work as an electrode for ESI. Then, the PET slice was laminated with a laminating film. One of the other two channels in the PET chip was connected with the microfluidic filter device. The last channel was used to inject the ESI buffer (MeOH, 4% acetic acid) to assist in ESI.ESI-MS analysis
The antibiotic drugs flew through the PC membrane, passed the PET chip, and finally were ionized by ESI for MS analysis using a linear ion trap mass spectrometer (LTQ, Thermo Fisher Scientific, USA). The mass spectrometer was run in the positive mode with the instrumental parameters optimized as: 3.7 kV electrospray voltage, 275 °C capillary temperature, 19 V capillary voltage, 55 V tube lens voltage, 3 microscans and 100 ms max injection time. The scan range was from 50 to 2000 Da.MALDI-TOF MS identification of bacterial isolates
Four E. coli strains (CH 20160920, ATCC 25922, CICC 10661 and CICC 10663) were cultured overnight. For each bacterial sample, 1 μL of solutions containing ∼105 bacterial cells after being washed with sterile water was deposited on a well of a MALDI target plate. After solvent evaporation, 1 μL of 10 mg mL−1 HCCA matrix dissolved in 50% CH3CN, 47.5% H2O and 2.5% TFA was added to the same well. The dried samples were analyzed by MALDI-TOF MS (Clin-TOF-II, Bioyong Technology Co. Ltd, Beijing, China) in linear positive mode with the optimized instrumental parameters (laser type: free-hand; laser firing power: 95; profiles: 26; shots: 20; laser rep rate: 40 Hz; ion gate: 3000 Da; pulsed extraction optimized at: 6000 Da) and the m/z range of 3000–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. E. coli (8739) was used as the calibration sample. The identification of the isolates was realized by searching the obtained MALDI-TOF MS mass spectra against a built-in library using BioExplorer (v3.2, Bioyong Technology Co. Ltd, Beijing, China).
000. E. coli (8739) was used as the calibration sample. The identification of the isolates was realized by searching the obtained MALDI-TOF MS mass spectra against a built-in library using BioExplorer (v3.2, Bioyong Technology Co. Ltd, Beijing, China).
Antimicrobial susceptibility testing of bacterial isolates
Four E. coli strains (CH 20160920, ATCC 25922, CICC 10661 and CICC 10663) were cultured overnight. For each bacterial sample, a bacterial suspension (∼1.5 × 108 CFU mL−1) in water was dipped with sterile cotton swabs and spread on Mueller–Hinton (MH) agar plates uniformly. Two groups of tablets including cefotaxime (CTX) and cefotaxime/clavulanic acid (CTC), and ceftazidime (CAZ) and ceftazidime/clavulanic acid (CAC) were put on the MH plates and cultured under 37 °C for 18 to 24 h. By measuring and comparing the diameter of the bacterial inhibition ring of these two groups of tablets, ESBL producing strains could be determined.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22074022, 22022401), the Science and Technology Commission of Shanghai Municipality (18441901000, 18050502200), and the Ministry of Science and Technology (MOST) of the People's Republic of China (SQ2020YFF0426500). The authors acknowledge Bioyong Technology Co. Ltd for the help in the MALDI-TOF identification of bacterial isolates.References
- M. Ferri, E. Ranucci, P. Romagnoli and V. Giaccone, Crit. Rev. Food Sci., 2017, 57, 2857–2876 CrossRef CAS.
- P. Buchy, S. Ascioglu, Y. Buisson, S. Datta, M. Nissen, P. A. Tambyah and S. Vong, Int. J. Infect. Dis., 2020, 90, 188–196 CrossRef CAS.
- O. O. Adebowale, F. A. Adeyemo, N. Bankole, M. Olasoju, H. K. Adesokan, O. Fasanmi, O. Adeyemo, O. Awoyomi, O. Kehinde and F. O. Fasina, Int. J. Environ. Res. Public Health, 2020, 17, 3579 CrossRef CAS.
- G. D. Wright, Adv. Drug Delivery Rev., 2005, 57, 1451–1470 CrossRef CAS.
- S. Santajit and N. Indrawattana, BioMed Res. Int., 2016, 2016, 2475067 Search PubMed.
- P. A. Lambert, Adv. Drug Delivery Rev., 2005, 57, 1471–1485 CrossRef CAS.
- F. C. Tenover, Am. J. Med., 2006, 119, S3–S10 CrossRef CAS.
- S. Shaikh, J. Fatima, S. Shakil, S. M. D. Rizvi and M. A. Kamal, Saudi J. Biol. Sci., 2015, 22, 90–101 CrossRef CAS.
- A. Shrestha, A. M. Bajracharya, H. Subedi, R. S. Turha, S. Kafle, S. Sharma, S. Neupane and D. K. Chaudhary, BMC Res. Notes, 2017, 10, 574 CrossRef.
- L. A. Dever and T. S. Dermody, Arch. Intern. Med., 1991, 151, 886–895 CrossRef CAS.
- R. Canton, A. Novais, A. Valverde, E. Machado, L. Peixe, F. Baquero and T. M. Coque, Clin. Microbiol. Infect., 2008, 14, 144–153 CrossRef CAS.
- Y. Chong, Y. Ito and T. Kamimura, Infect., Genet. Evol., 2011, 11, 1499–1504 CrossRef CAS.
- CDC, Antibiotic Resistance Threats in the United States, 2019, U.S. Department of Health and Human Services, CDC, Atlanta, GA, 2019 Search PubMed.
- Z. A. Khan, M. F. Siddiqui and S. Park, Diagnostics, 2019, 9, 49 CrossRef CAS.
- H. Sammer-ul and X. L. Zhang, Curr. Anal. Chem., 2020, 16, 41–51 CrossRef.
- J. M. Kim, I. Kim, S. H. Chung, Y. Chung, M. Han and J. S. Kim, Pathogens, 2019, 8, 214 CrossRef CAS.
- Y. Zhu, N. Gasilova, M. Jovic, L. Qiao, B. Liu, L. T. Lovey, H. Pick and H. H. Girault, Chem. Sci., 2018, 9, 2212–2221 RSC.
- R. Zhang, Q. Qin, B. Liu and L. Qiao, Anal. Chem., 2018, 90, 3863–3870 CrossRef CAS.
- J. Hrabak, R. Walkova, V. Studentova, E. Chudackova and T. Bergerova, J. Clin. Microbiol., 2011, 49, 3222–3227 CrossRef CAS.
- M. Oviano, B. Fernandez, A. Fernandez, M. J. Barba, C. Mourino and G. Bou, Clin. Microbiol. Infect., 2014, 20, 1146–1157 CrossRef CAS.
- H. Jeon, Z. A. Khan, E. Barakat and S. Park, Antibiotics, 2020, 9, 348 CrossRef.
- P. Sabhachandani, S. Sarkar, P. C. Zucchi, B. A. Whitfield, J. E. Kirby, E. B. Hirsch and T. Konry, Microchim. Acta, 2017, 184, 4619–4628 CrossRef CAS.
- J. Choi, J. Yoo, M. Lee, E. G. Kim, J. S. Lee, S. Lee, S. Joo, S. H. Song, E. C. Kim, J. C. Lee, H. C. Kim, Y. G. Jung and S. Kwon, Sci. Transl. Med., 2014, 6, 267ra174 CrossRef.
- W. Wu, D. Zhang, K. Chen, P. Zhou, M. Zhao, L. Qiao and B. Su, Anal. Chem., 2018, 90, 14395–14401 CrossRef CAS.
- L. Qiao, Y. Lu, B. H. Liu and H. H. Girault, J. Am. Chem. Soc., 2011, 133, 19823–19831 CrossRef CAS.
- K. Sparbier, S. Schubert, U. Weller, C. Boogen and M. Kostrzewa, J. Clin. Microbiol., 2012, 50, 927–937 CrossRef CAS.
- C. Li, S. Ding, Y. Huang, Z. Wang, J. Shen, H. Ling and Y. Xu, J. Hosp. Infect., 2018, 99, 200–207 CrossRef CAS.
- M. Oviano, M. Gomara, M. J. Barba, M. J. Revillo, L. P. Barbeyto and G. Bou, J. Antimicrob. Chemother., 2017, 72, 2259–2262 CrossRef CAS.
- J. M. Indelicato, G. L. Engel and J. L. Occolowitz, J. Pharm. Sci., 1985, 74, 1162–1166 CrossRef CAS.
- S. Ghafourian, N. Sadeghifard, S. Soheili and Z. Sekawi, Curr. Issues Mol. Biol., 2015, 17, 11–21 Search PubMed.
- Overdevest and Ilse, Emerging Infect. Dis., 2011, 17, 1216–1222 CrossRef.
- D. Jaen-Luchoro, A. Busquets, R. Karlsson, F. Salva-Serra, C. Ahren, N. Karami and E. R. B. Moore, Microorganisms, 2020, 8, 893 CrossRef CAS.
- H. Ma, B. Lai, Y. Jin, C. Tian, J. Liu and K. Wang, RSC Adv., 2020, 10, 26862–26873 RSC.
- S. J. Liu, Z. W. Yu, L. Qiao and B. H. Liu, Sci. Rep., 2017, 7, 46669 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Identification and antimicrobial susceptibility testing results of the clinical isolates. See DOI: 10.1039/d0an01876g |
| This journal is © The Royal Society of Chemistry 2021 |