Metabolism in action: stable isotope probing using vibrational spectroscopy and SIMS reveals kinetic and metabolic flux of key substrates†
Malama
Chisanga
ab,
Howbeer
Muhamadali
c,
Danielle
McDougall
d,
Yun
Xu
 c,
Nicholas
Lockyer
c,
Nicholas
Lockyer
 d and
Royston
Goodacre
d and
Royston
Goodacre
 *c
*c
aDepartment of Chemistry, Manchester Institute of Biotechnology, University of Manchester, Manchester, M1 7DN, UK
bSchool of Mathematics and Natural Sciences, Department of Chemistry, Copperbelt University, Kitwe, Zambia
cDepartment of Biochemistry and Systems Biology, Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Liverpool, L69 7ZB, UK. E-mail: roy.goodacre@liverpool.ac.uk; Tel: +44 (0) 151 795 7689
dDepartment of Chemistry, Photon Science Institute, University of Manchester, Manchester, M13 9PL, UK
First published on 11th January 2021
Abstract
Microbial communities play essential functions which drive various ecosystems supporting animal and aquatic life. However, linking bacteria with specific metabolic functions is difficult, since microbial communities consist of numerous and phylogenetically diverse microbes. Stable isotope probing (SIP) combined with single-cell tools has emerged as a novel culture-independent strategy for unravelling microbial metabolic roles and intertwined interactions in complex communities. In this study, we applied Raman and Fourier-transform infrared (FT-IR) spectroscopies, secondary ion mass spectrometry (SIMS) with SIP to probe the rate of 13C incorporation in Escherichia coli at 37 and 25 °C. Our results indicate quantitative enrichment and flow of 13C into E. coli at various time points. Multivariate and univariate analyses of Raman and FT-IR data demonstrated distinctive 13C concentration-dependent trends that were due to vibrational bands shifting to lower frequencies and these shifts were a result of incubation time and metabolic rate. SIMS results were in complete agreement with the spectroscopy findings, and confirmed the detected levels of 13C incorporation into microbial biomass at the investigated conditions. Having established that FT-IR and Raman spectroscopy with SIP can measure metabolism kinetics in this simple system, we have applied the kinetics concept to study the metabolism of phenol by Pseudomonas putida and metabolic interactions within a two-species consortia with E. coli that could not degrade phenol. Raman spectroscopy combined with SIP identified quantitative shifts in P. putida due to temporal assimilation of phenol. Although E. coli was unable to grow on phenol, in co-culture with P. putida, general metabolic probing using deuterated water for SIP revealed that E. coli displayed increasing metabolic activity, presumably due to cross feeding from metabolites generated by P. putida. This study clearly demonstrates that Raman and FT-IR combined with SIP provide rapid and sensitive detection of carbon incorporation rates and microbial interactions. These novel findings may guide the identification of primary substrate consumers in complex microbial communities in situ, which is a key step towards the characterisation of novel genes, enzymes and metabolic flux analysis in microbial consortia.
Introduction
Microbial communities play vital ecological functions which support essential environmental and mammalian biological systems. The fundamental theme of contemporary microbial ecology and functional genomics is to obtain detailed understanding of functions for individual members of a multi-species consortium and link them to phylogeny;1 with a view to being able to modulate these functions for industrial application. Stable isotopic probing (SIP) has become a powerful tool to achieve this goal, since it discerns functions of metabolically active microbes directly within complex environments. In the past decade, SIP has been extensively applied in parallel with single-cell tools to detect microbes linked with the metabolism of a particular substrate (usually labelled with 13C or 15N isotopes).2 High-throughput molecular techniques combined with density-gradient centrifugation are perhaps the most commonly applied approaches for identification of DNA/RNA biomarkers of labelled and unlabelled cells.3,4 Whilst delivering a wealth of valuable information about phylogeny and metabolic genes, DNA and RNA SIP tools exhibit high detection limit for cellular isotopic content (>50 and >15 atom%, respectively).3,5 Thus, long incubation times are needed for cell replication and optimal isotopic enrichment of DNA.3 By contrast, Raman spectroscopy provides rapid and sensitive measurements with detection limit of ∼10 atom%,6 and physiological heterogeneity of intact bacterial cells in situ.Raman spectroscopy is a high-throughput technique where Stokes scattered light is measured to detect biomolecular dynamics7 and isotopic content8 of polarisable molecular bonds. The application of FT-IR as a complementary tool to identify environmental microbes has also been proved useful.9,10 FT-IR generates spectral information from chemical bonds whose permanent dipole moments change upon absorption of infrared light. Combining Raman and FT-IR with SIP offers comprehensive whole-organism biochemical fingerprints which are used to unravel intertwined metabolic networks and functions of microbes in complex consortia, since Raman and FT-IR bands shift to lower wavenumber (redshift) upon assimilation of heavy atoms. When secondary ion mass spectrometry (SIMS) (detection limit of 0.1 atom%)11 is used in parallel, heavy atoms assimilated by bacteria can be probed to supplement Raman and FT-IR data.12,13
To date however, FT-IR and Raman-based SIP studies have mainly focused on functional characterisation of fully or quantitatively labelled microbes;14,15 the latter based on growing microbes on predefined mixtures of natural and label substrates.16 Although this discerns niche roles such as nutrient fixation,17,18 and degradation of pollutants,19,20 it does not provide clear insights into rates of substrate turnover and metabolic activity with respect to time. Moreover, lengthy incubation time required to ensure full isotopic labelling complicates phylogenetic resolution of key substrate degraders, due to excessive cross-feeding of labelled metabolites from primary consumers to the rest of the habitat.8 Thus, neighboring microbes assimilating labelled metabolic products may also display redshifted bands. To overcome this challenge, there is a need to perform a systematic, quick and robust time-course SIP study to allow for sampling of bacteria over time and to determine kinetics of isotopic labelling.8,14,21
In this study, we first investigate the kinetics of 13C isotope incorporation by E. coli cultivated in minimal medium containing 13C-glucose as a sole carbon source under aerobic conditions. For the first time we demonstrate novel utility of Raman, FT-IR and SIMS combined with SIP to assess and quantify the incorporation rate of 13C by E. coli, and to monitor the flow of 13C into biomolecules at various time points. The Raman and FT-IR bands related to carbon bond vibrations are observed to be redshifted progressively due to cumulative assimilation of 13C between 0 and 4 h at 37 and 25 °C. SIMS data also displayed concentration-dependent trends along a time gradient that were wholly consistent with quantitative 13C level detected by Raman and FT-IR. This initial study was designed to establish that the quantitative kinetic uptake of 13C substrates was possible using this simple model system, before extending this novel kinetic approach to investigating cross-feeding, via reverse SIP (rSIP) and deuterated water (D2O) labelling,8 in a two-component community where P. putida was able to catabolise phenol whilst E. coli did not grow when phenol was the sole carbon source. Reverse labelling was achieved by exposing 13C-labelled cells to minimal medium containing 12C-phenol (MMP) as the only carbon source, with or without 30% D2O. Co-labelling with D2O was applied to probe the general metabolic activity22 and interaction between P. putida and E. coli in phenolic media.
Experimental methods
Growth conditions
Unless otherwise stated, all chemicals were purchased from Fisher (Fisher Scientific, UK).Kinetic measurements of 13C incorporation in E. coli
Wild type E. coli K-12 MG1655 was grown in minimal medium (MM) supplemented with 5 g L−1 of 99 atom% uniformly labelled 13C-glucose (Sigma Aldrich, UK), as a sole carbon source. Bacterial growth were performed in replicates (n = 4) in 5 mL volumes in sterile Falcon tubes (Greiner Bio-One, Germany). Prior to incubation, 200 μL of cell samples for each condition were transferred to a clean and sterile 96-well plate to monitor the growth profiles via optical density at 600 nm (OD600). All cells were incubated at 37 and 25 °C in a Multitron shaker incubator (INFORS-HT Bottmingen, Switzerland) shaking at 170 rpm. Cells were collected at 20 min intervals, between 0 and 4 h at 37 and 25 °C to probe 13C assimilation at various time points. In quantitative SIP study, E. coli was grown in MM containing varying ratios of 12C/13C-glucose and harvested in the early stationary growth phase after 16 h as reported previously.23Study of phenol metabolism by bacteria
E. coli K-12 MG1655 and P. putida ATCC 700369 were grown axenically in minimal medium (MM) containing 5 g L−1 of 99 atom% uniformly labelled 13C-glucose (Sigma Aldrich, UK) (MMG). Cells were harvested after 16 h of incubation to ensure complete incorporation of 13C-glucose and utilised as inoculant for subsequent experiments. After normalising OD600, cells were inoculated as 5 mL aliquots (n = 4) in MM supplemented with industrially relevant concentration1 (0.2 g L−1) of 12C-phenol (Sigma Aldrich, UK) (MMP), as the only carbon source at pH of 7, as mono and co-cultures with or without 30% D2O. Here, 30% D2O was used since this optimal D2O level does not significantly affect metabolism.22 For co-culture analysis, the inoculum was prepared in sterile deep 6-well plates in which E. coli and P. putida shared the same MMP, although the cells were physically separated by means of sterile culture inserts having 0.4 μm pore size (BD Biosciences, USA). E. coli and P. putida were incubated at 37 and 30 °C, respectively, and sampled at 0, 1, 2, 4, 8 and 18 h time intervals.Sample preparation, spectral measurements and data processing for Raman and FT-IR
After incubation, all sample aliquots were collected by centrifugation for 5 min at 4000g and 4 °C using a bench top Eppendorf Centrifuge 5810 R (Hamburg, Germany). After discarding the supernatant, the collected biomass was washed twice using 5 mL of physiological saline (0.9% NaCl) to remove any remaining media components following the above centrifugation parameters. The cell pellets were collected and stored at −80 °C prior to further investigation. Detailed instrumental, sample measurement parameters and data processing steps for Raman and FT-IR are similar to our previously published protocols,16 and are clearly presented in the (ESI†) section.SIMS analysis for kinetics measurements of 13C incorporation in E. coli
For SIMS analysis, 1 × 1 cm Si wafers (Agar Scientific, UK) were prepared by sonication in hexane followed by methanol and stored in glass vial containing 80% ethanol. Each pelleted sample of E. coli collected at 0, 1, 2, 3, 4 and 16 h time points at 25 and 37 °C was resuspended in ammonium formate solution (150 mM, 50 μL) and the resulting solution was pipetted onto clean Si wafers. Prior to SIMS analysis, Si wafers containing cellular biomaterial were placed in a desiccator under vacuum to remove any water and traces of ammonium formate.Quantitative ToF-SIMS spectra of cells were acquired using a BioToF-SIMS instrument (Kore Technology Ltd, Ely, UK). The primary ion beam of choice was a Wein filtered Au3+ liquid metal ion gun (LMIG) operated at 20 keV with a continuous beam flux of 0.6 nA and a pulse time of 100 ns. Data were acquired in delayed-extraction mode, providing mass resolving power of ∼1000. The mass range within 0–1000 Da consisting of metabolites of interest was selected. In order to acquire representative information from the whole sample on each Si wafer, SIMS spectra were measured from three different regions of each sample. This allowed for detailed information through the layers of biomass. All SIMS spectra were collected in positive polarity mode using a post-acceleration voltage of 7 kV.
Quantitative analysis of 13C incorporation by E. coli via PLSR for kinetic measurements
To effect quantitative analysis, univariate and partial least-squares regression (PLSR)24 were utilised. PLSR model was trained with Raman and FT-IR data measured from E. coli grown for 16 h in varying 12C/13C-glucose ratios, and subsequently employed to predict the amount of 13C in E. coli cultivated at different sampling time points at 37 and 25 °C temperatures.Results and discussion
Analysis of kinetic measurements of 13C incorporation in E. coli
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch in amide I (ref. 26), shifted to 1620 cm−1 on complete 13C substitution by cells. It is worth mentioning that the Raman biomarker band for phenylalanine and amide I at 1002 and 1654 cm−1 displayed quantitative redshifts due to gradual 13C substitution over time (Fig. S1a and b†). This observation was supported by PC1 loadings plots (Fig. S2c and d†) that revealed these bands (phenylalanine and amide I) to be the major input wavenumber shifts affected by 13C labelling, and hence influenced the clustering patterns observed on PCA scores plots in Fig. 2a – where the trends in these ordination plots were entirely related to incubation time. Several other important diagnostic vibrations of lower intensities at 1573, 1338, 1250, 781, 828 and 642 cm−1, ascribed to ring breathing in guanine, adenine, amide III, cytosine, uracil and tyrosine, respectively, were also affected by isotope labelling of E. coli cells as detailed in Table S1† (ref. 27 and 28). Though dominated by carbon vibrations (Fig. 1a) in biomolecules, some bands such as: 622 cm−1 (N–H stretch) and 1100 cm−1 (>PO2− symmetric stretch)29,30 did not display detectable redshifts irrespective of the level of 13C uptake since they are not linked to carbon atom(s).
O stretch in amide I (ref. 26), shifted to 1620 cm−1 on complete 13C substitution by cells. It is worth mentioning that the Raman biomarker band for phenylalanine and amide I at 1002 and 1654 cm−1 displayed quantitative redshifts due to gradual 13C substitution over time (Fig. S1a and b†). This observation was supported by PC1 loadings plots (Fig. S2c and d†) that revealed these bands (phenylalanine and amide I) to be the major input wavenumber shifts affected by 13C labelling, and hence influenced the clustering patterns observed on PCA scores plots in Fig. 2a – where the trends in these ordination plots were entirely related to incubation time. Several other important diagnostic vibrations of lower intensities at 1573, 1338, 1250, 781, 828 and 642 cm−1, ascribed to ring breathing in guanine, adenine, amide III, cytosine, uracil and tyrosine, respectively, were also affected by isotope labelling of E. coli cells as detailed in Table S1† (ref. 27 and 28). Though dominated by carbon vibrations (Fig. 1a) in biomolecules, some bands such as: 622 cm−1 (N–H stretch) and 1100 cm−1 (>PO2− symmetric stretch)29,30 did not display detectable redshifts irrespective of the level of 13C uptake since they are not linked to carbon atom(s).
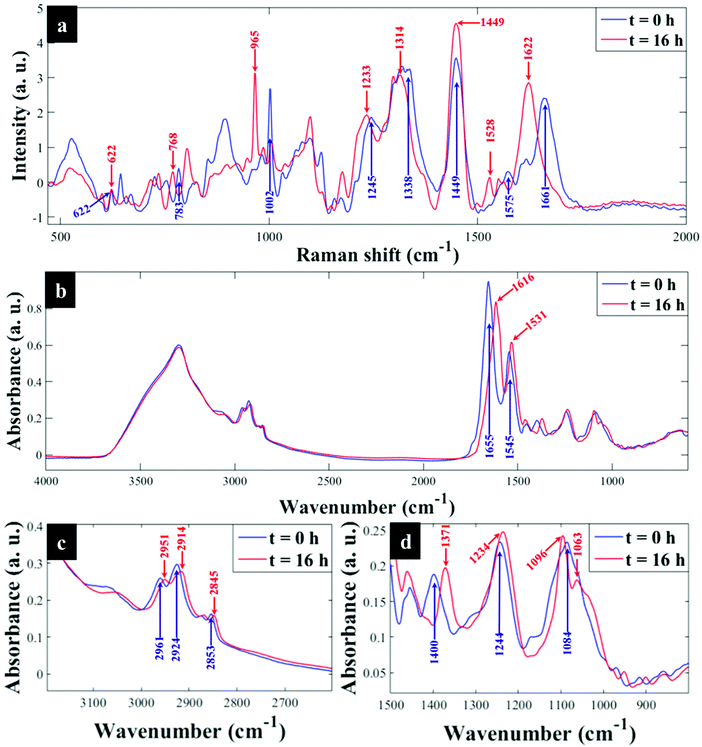 | ||
| Fig. 1 Typical (a) Raman and (b) FT-IR spectra of E. coli grown on 13C-glucose as the only source of carbon and sampled at t = 0 h (blue trace) and t = 16 h (red trace), (c and d) illustrate magnified spectral regions for FT-IR bands. After 16 h of incubation full incorporation of 13C by cells has resulted in characteristic carbon-related vibrations being redshifted. In this figure the major 12C to 13C shifted band positions are indicated by blue (t = 0 h or 0% 13C) and red (t = 16 h or 100% 13C) arrows, respectively. All major Raman and FT-IR bands affected by 13C incorporation and their corresponding molecular assignments are summarised in Tables S1 and S2.† | ||
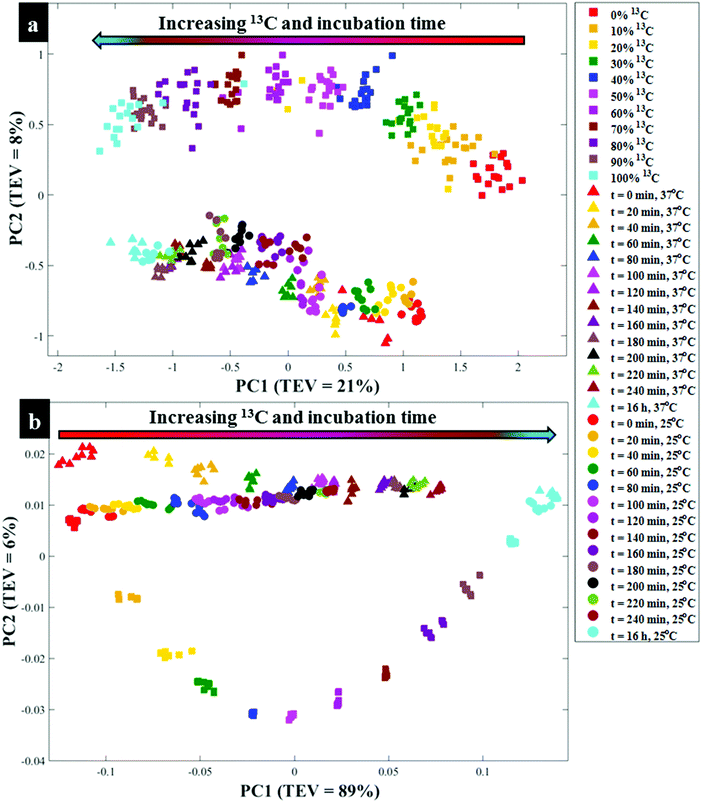
Next all Raman data were subjected to PCA to visualise any cellular 13C isotope-driven patterns over time and quantitative 13C assimilation as shown in Fig. 2a. The PCA scores illustrated in Fig. 2a are plotted from the Raman data measured from quantitatively enriched E. coli that were harvested in the early stationary growth phase (t = 16 h) to achieve complete 13C enrichment, and E. coli samples for kinetics experiments that were mainly collected in the lag and early log phases. As can be seen in Fig. 2a and Fig. S2(a, b),† PCA scores plots show time and concentration-dependent trends for Raman spectra of cells sampled at various time points and those cultivated on increasing 12C/13C ratios. Clustering trends on PCA scores indicate that 13C incorporation was quantitative and so should be predictable. The first principal component (PC1) revealed a linear increase in 13C incorporation from right (t = 0 h and 0% 13C) to left (t = 16 h and 100% 13C) as the most dominant effect. PC1 is extracted to contain the most natural variance in the data, and this observation is not surprising as carbon is considered the most abundant element in cells, and thus the majority of the detected band shifts are carbon-related modes. As can be seen in Fig. 2a, E. coli harvested at t = 0 h and those cells incubated on 0% 13C, similar to cells collected at t = 16 h and 100% 13C-glucose assimilation, clustered in the same space, whilst cells labelled with intermediate 13C levels also clustered accordingly. This suggests that cells harvested at t = 0 h were mainly composed of ∼100% natural 12C-molecules, and the level of 13C in cells increased linearly with sampling time at 20 min intervals, achieving maximum (100%) 13C incorporation by t = 16 h (Fig. S1†). The second PC (PC2) on Fig. S2(c and d)† explained a minimal effect compared to PC1 which only separated the samples based on metabolic states, and thus the PC2 loadings are not discussed in detail.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch), amide II (C–N stretch and N–H bending) and lipids (COO− stretch) vibrations,33,34 showed clear peak shifts from 1654, 1544 and 1400 cm−1 to 1616, 1531 and 1373 cm−1, respectively, after assimilating 13C atom into biomolecules over time (Fig. S1c and d†). It is also important to note that the amide I and lipid bands indicated complete redshifts within 20 min of cell growth, suggesting quick uptake of 13C, which supports the Raman finding. For clearer visualisation of redshifts on FT-IR spectra for E. coli sampled at 0 and 20 min, separate PCA plots were constructed (Fig. S3†). Several carbon-related vibrations of lower intensity including aromatic C
O stretch), amide II (C–N stretch and N–H bending) and lipids (COO− stretch) vibrations,33,34 showed clear peak shifts from 1654, 1544 and 1400 cm−1 to 1616, 1531 and 1373 cm−1, respectively, after assimilating 13C atom into biomolecules over time (Fig. S1c and d†). It is also important to note that the amide I and lipid bands indicated complete redshifts within 20 min of cell growth, suggesting quick uptake of 13C, which supports the Raman finding. For clearer visualisation of redshifts on FT-IR spectra for E. coli sampled at 0 and 20 min, separate PCA plots were constructed (Fig. S3†). Several carbon-related vibrations of lower intensity including aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H (lipids) detected around 3100 cm−1, C–O and N–H bending coupled with C–N stretch (amide III)30,34 (Fig. 1c and d), were also affected by 13C enrichment as shown in Table S2.† Some FT-IR bands were unaffected by 13C uptake, confirming that such modes are not related to carbon vibrations as explained above. When investigated by PCA scores (Fig. 2b and S4a, b†) and loadings, FT-IR data exhibited similar 13C isotope-induced clustering trends to the Raman data. PC1 showed specific time and concentration gradients from left to right for cells collected at various time points and those incubated for 16 h on varying amounts of 12C/13C-glucose. As one may have predicted, PC1 loadings (Fig. S4c and d†) for FT-IR data confirmed the amide I and amide II to be the main inputs linked with PCA (Fig. 2b), whereas PC2 only distinguished between different E. coli growth phases at 37 and 25 °C as already explained on the Raman data above.
C–H (lipids) detected around 3100 cm−1, C–O and N–H bending coupled with C–N stretch (amide III)30,34 (Fig. 1c and d), were also affected by 13C enrichment as shown in Table S2.† Some FT-IR bands were unaffected by 13C uptake, confirming that such modes are not related to carbon vibrations as explained above. When investigated by PCA scores (Fig. 2b and S4a, b†) and loadings, FT-IR data exhibited similar 13C isotope-induced clustering trends to the Raman data. PC1 showed specific time and concentration gradients from left to right for cells collected at various time points and those incubated for 16 h on varying amounts of 12C/13C-glucose. As one may have predicted, PC1 loadings (Fig. S4c and d†) for FT-IR data confirmed the amide I and amide II to be the main inputs linked with PCA (Fig. 2b), whereas PC2 only distinguished between different E. coli growth phases at 37 and 25 °C as already explained on the Raman data above.
Kinetics of 13C isotope incorporation by E. coli
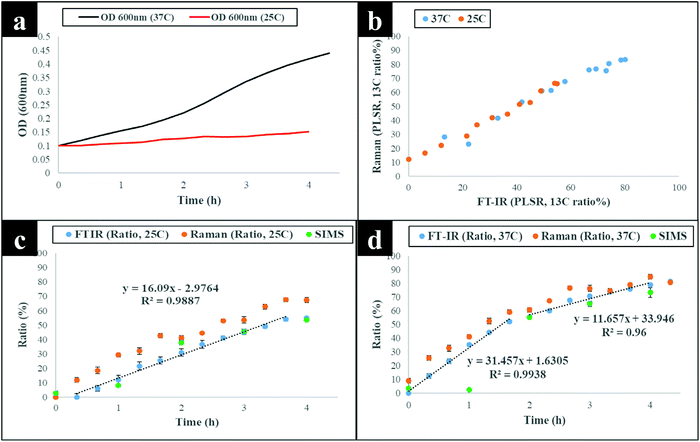
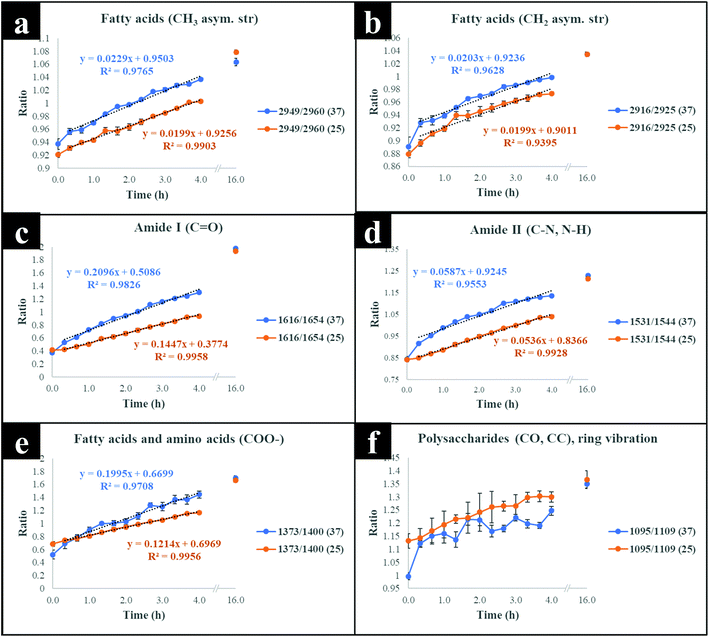
The next stage of the analysis was to investigate the kinetics of isotope assimilation which was readily monitored by band redshifts. In order to achieve this, PLSR multivariate models were generated, where the input data consisted of Raman and FT-IR data from E. coli which were cultured for 16 h on varying ratios of 12C/13C as the training set, with the 12C/13C ratio as the target output. These models were then applied to predict the levels of 13C enrichment in cells harvested over time (the independent hold out dataset). Univariate analysis based on the ratio of 12C/13C was also calculated via regression of peak intensities of the amide I, fatty and amino acids, and polysaccharides for Raman and FT-IR data. These bands were employed to generate separate standard curves to determine the level of 13C incorporation into various biomolecules as a function of time (13C incorporation rate) and metabolic activity (37 vs. 25 °C). The outputs from PLSR and univariate standard curves were compared with growth rates and OD600 of cells harvested at different time points at 37 and 25 °C (Fig. 3). Fig. 3(a and b) demonstrates 13C incorporation trends for cells grown at 37 °C that closely mimicked growth profiles; although the OD600 of cells is not changing significantly and the cells are still in the lag phase at 25 °C, the incorporation of 13C occurred at a high rate and was not limited by cell replication.It is clear on Fig. 3(b–d) that the 13C content of cells increased linearly with incubation time (correlation coefficient (R2) close to 1 on Fig. 3c and d), and that 13C levels in E. coli grown in varying ratios of 13C-glucose precisely coincided with predicted 13C amounts in E. coli sampled at various time points. The concentration of incorporated 13C predicted by PLSR (Fig. 3b) were in complete agreement with univariate analysis based on amide I for FT-IR and Raman, and SIMS intensity ratios (Fig. 3c and d). The trend lines (dotted black lines) fitted on the amide I and SIMS intensity data indicate continuous linear increase in 13C levels in the cells grown at 25 °C over time (Fig. 3c). By comparison, these ratios display decreasing gradients and 13C enrichment when cells entered the log phase at 37 °C (Fig. 3d) which entirely agrees with the observed 13C clustering trends on PCA scores from 2 h onwards (Fig. 2). The line gradients were also employed to monitor the rate of 13C incorporation in cellular fatty acids, proteins and polysaccharides for FT-IR data (Fig. 4), and in proteins and nucleic acids for Raman data (Fig. S5†). The results clearly demonstrate that whilst 13C was assimilated in biomolecules at increasing rates, the amide I (FT-IR data Fig. 4) were labelled much more rapidly compared to amide II and fatty acids as indicated by larger slope. The Raman data also revealed similar trends where the amide I and phenylalanine were labelled faster (Fig. S5†). The Raman data were collected in a limited spectral range (unlike FT-IR data) which did not cover lipid bands and so 13C incorporation rate in fatty acids/lipids is not included on Fig. S5.† The quicker rate of 13C incorporation in proteins is not surprising since proteins are synthesised faster in the lag phase of bacterial growth to facilitate cell growth and division.35 Importantly, there is a good match between the rate and order of 13C flow into amides and amino acids and Raman and FT-IR spectral redshifts detected in 20 min of cell incubation at 37 °C in this study. These results agree with recent SIP studies which have demonstrated redshifts for amides and phenylalanine are observable for E. coli grown in as low as 10% (v/v) of 13C-glucose.14,16
Although cells cultivated at different time intervals at 37 °C exhibited higher 13C incorporation rates at all other time points, they appeared to have assimilated same levels of 13C atom as cells grown at 25 °C on the last time point (16 h), as indicated by intersected curves on Fig. 4. In addition, E. coli exhibited the highest metabolic rate and 13C uptake in the lag phase between 0 and 20 min at 37 °C, which is entirely consistent with observed redshifts for phenylalanine (in Raman; Fig. S1a†) and amide I (in FT-IR; Fig. S1c and S3†) bands. It is postulated that at this adaptation stage, cells produce enzymes that facilitate rapid uptake and utilisation of available energy sources to replenish nutrients and prepare for long term survival.35 Therefore, bacteria generally accumulate sufficient cellular biomass to support cell division.36 However, cells exhibited lower rate of 13C incorporation between 2 and 4 h at 37 and 25 °C. The slower rate of 13C incorporation is also reflected by similarity in quantified levels of 13C-glucose by HPLC (Fig. S6†), and slower growth rates between 2 and 4 h.
SIMS spectral analysis
To confirm and supplement the Raman and FT-IR spectral findings, SIMS was applied to probe cellular 13C composition and time-dependent isotope uptake by E. coli. Quantitative levels of 13C in cells were calculated by generating [13CH3/(12CH3 + 13CH3)] intensity ratios for clearly resolved biomolecular ion peaks at mass-to-charge ratio (m/z) of 16 (13CH3) and 15 (12CH3) in SIMS spectra from the same bacterial samples analysed above (Fig. 3c and d). At t = 0 h, the 13CH3 signal was below the limit of detection (LOD) at both 37 and 25 °C. Whilst at t = 1 h, the 13CH3 signal was above the LOD for both temperatures but below the limit of quantification for cells grown at 37 °C. After 1 h, quantifiable SIMS data were generated which showed an increase of the [13CH3/(12CH3 + 13CH3)] relative ratio up to 16 h of growth at both 37 and 25 °C (Fig. S7†) due to gradual accumulation of 13C by E. coli. Again 13C enrichment at various time points (t > 2 h) were consistently higher at 37 °C compared to cells incubated at 25 °C. The calculated median percentages of 13C for SIMS data precisely matched cellular 13C quantities predicted by PLSR and univariate tools for Raman and FT-IR data (Fig. 3 and 4), indicating high sensitivity and robustness of all metabolic profiling tools applied in this study.Linking bacterial identity to phenol metabolism via reverse 12C labelling in co-cultures
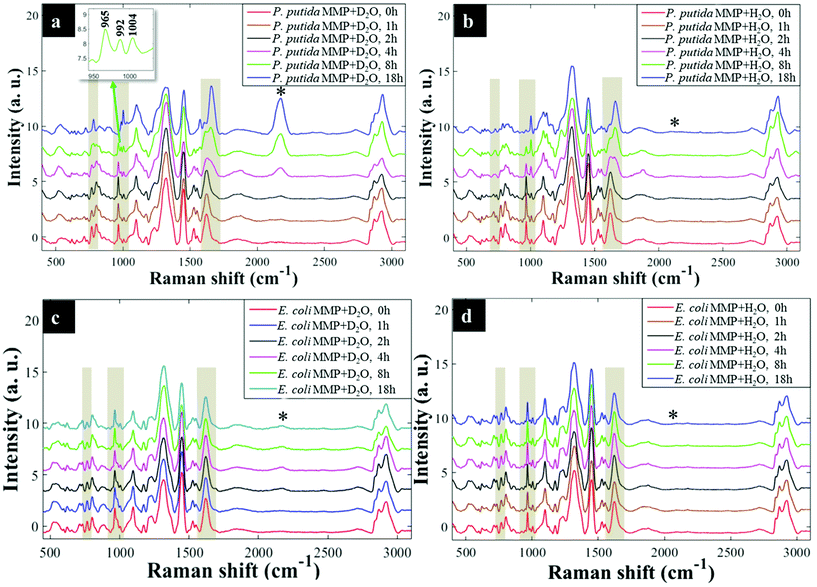
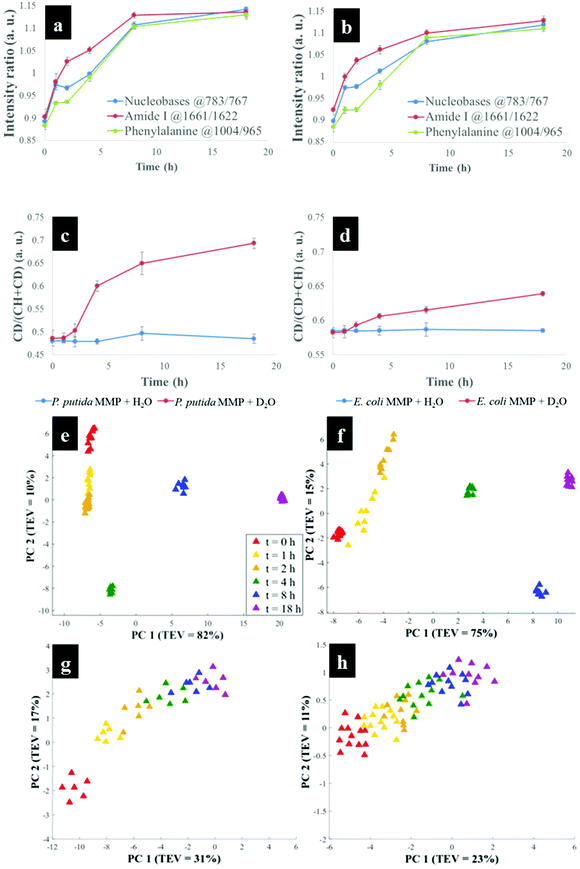
Having established that Raman and FT-IR can both provide data on the differential kinetic incorporation of 13C into differing cellular biomass, a more complex two-component bacterial system was investigated that included co-culture of P. putida, which could catabolise phenol, with E. coli, which was unable to grow when phenol was the sole carbon source. In addition, this experiment used D2O as an isotope labelling strategy to provide evidence of cross feeding from one bacteria to another as reported recently.22,37 D2O labelling highlights the general metabolism of bacteria and cross-feeding in microbial consortia regardless of whether bacteria are able or unable to breakdown substrates they are grown in. Though P. putida and E. coli rarely co-exist, they are usually found in the same environmental niches such as the soil and water.38 Previous research has shown that P. putida and E. coli species interact with each other in co-cultures and such interaction results in transcriptional dynamics associated with central carbon substrate metabolism and activation of efflux pumps.39 There is also evidence that TOL catabolic plasmid encoding pollutant-degrading enzymes can be transferred from P. putida to E. coli in co-cultures.40 These reported microbial interactions motivated this study aimed at probing phenotypes of P. putida and E. coli, and how these bacteria influence each other's metabolic capability in co-cultures containing phenol as a single carbon source.Initially P. putida and E. coli were grown on 13C-glucose as the sole carbon supply in order isotopically to label both bacteria with 13C isotope. Fig. 5a shows carbon-based Raman bands for P. putida that shifted from lower (13C) to higher (12C) wavenumber (so-called reverse stable isotope probing (rSIP)8 after 18 h of cell growth on 12C-phenol. Since phenol is present as the sole carbon source in co-cultures, the observed blueshifts were driven by catabolism of phenol and subsequent uptake of 12C isotope, which was the basis of identifying the phenol degrading P. putida. Examination of the Raman spectra in Fig. 5(a and b) shows that the band ascribed to phenylalanine modes, which was initially at 965 cm−1 (due to all 6 carbons in the benzene ring being 13C), underwent a large (Δ + 37 cm−1) shift back to 1002 cm−1 on complete incorporation with phenol-derived 12C. In addition to displaying time-related blueshifts, phenylalanine band (965 → 1002 cm−1) displayed splitting patterns for co-cultures of P. putida harvested after 8 h in MMP with 30% D2O due to the general metabolic activity of cells (Fig. 5a). The three peaks detected at 965, 992 and 1002 cm−1 highlight the formation of isotopomers of phenylalanine, consisting of varying levels of 13C and/or 1H substitution by 12C and/or D isotopes in benzene during anabolic bioprocesses,14,41 similar to the Raman findings discussed already. Band shift associated with D assimilation in lipids8 was also detected at 1104 cm−1 from 1124 cm−1. The reverse isotopic effect was also observed in C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch in amide I (1622 → 1662 cm−1) and ring modes in nucleobases (766 → 783 cm−1).29 According to multivariate analysis of Raman spectra for P. putida from co-culture (Fig. S8†), bands for amide I and nucleobases reverted to 12C positions within 4 h, whilst complete blueshift for phenylalanine was not apparent until 8 h. This signifies faster incorporation of 12C into amides and nucleobases than phenylalanine, that was confirmed by plotting peak intensity ratios with incubation time (Fig. 6a and b). Previous research has demonstrated the applicability of RNA-SIP for the detection of phenol-degrading P. putida under similar conditions to those applied in the present study.1 Given that 13C-RNA fractions from P. putida were identified after 24 h of exposure to 13C-phenol, compared to 4 h at which this organism was detected via rSIP, our results provide complementary or alternative protocol for phenol metabolism studies. Low intensity bands in the fingerprint region exhibited reverse shifts to 12C positions (Fig. 5a and b). All of these carbon-mediated Raman modes moved progressively back to standard wavenumber positions due to cumulative assimilation of 12C, further confirming step-wise turnover of phenol by P. putida.
O stretch in amide I (1622 → 1662 cm−1) and ring modes in nucleobases (766 → 783 cm−1).29 According to multivariate analysis of Raman spectra for P. putida from co-culture (Fig. S8†), bands for amide I and nucleobases reverted to 12C positions within 4 h, whilst complete blueshift for phenylalanine was not apparent until 8 h. This signifies faster incorporation of 12C into amides and nucleobases than phenylalanine, that was confirmed by plotting peak intensity ratios with incubation time (Fig. 6a and b). Previous research has demonstrated the applicability of RNA-SIP for the detection of phenol-degrading P. putida under similar conditions to those applied in the present study.1 Given that 13C-RNA fractions from P. putida were identified after 24 h of exposure to 13C-phenol, compared to 4 h at which this organism was detected via rSIP, our results provide complementary or alternative protocol for phenol metabolism studies. Low intensity bands in the fingerprint region exhibited reverse shifts to 12C positions (Fig. 5a and b). All of these carbon-mediated Raman modes moved progressively back to standard wavenumber positions due to cumulative assimilation of 12C, further confirming step-wise turnover of phenol by P. putida.
By contrast, Raman bands for E. coli from MMP axenic and co-cultures remained at 13C points (Fig. 5c and d), and their corresponding intensity ratios did not change over time. This agrees with OD600 readings that remained unchanged throughout the incubation time (Fig. S9†), reflecting the inability of E. coli to mineralise phenol, which was the only carbon source.
D2O labelling unravels metabolic activity of P. putida and E. coli in co-cultures
D2O labelling is considered a reliable means to probe the general metabolic activity of bacteria, and this is proved to be valuable for probing response of microbes to contaminants.8,42 Here, we use this strategy to assess metabolic behaviour of P. putida and E. coli in MMP co-cultures. Although peak blueshifts due to 13C → 12C rSIP incorporation for metabolically active P. putida from co-culture were only distinguishable after 4 h of exposure to phenol, metabolic activity due to the incorporation of D2O could be detected after 2 h (Fig. 5a). This metabolic activity was revealed by the appearance of a broad band in the “silent region” of a Raman spectrum at 2172 cm−1 for P. putida from MMP with 30% D2O (Fig. 5a). This peak is assigned to C–D stretch resulting from intracellular H → D reactions on aliphatic backbone chains of molecules produced in lipid synthetic pathways.22,43 Thus, Raman spectroscopy sensitively elucidated the metabolic action of P. putida before reverse carbon-based spectral blueshifts were detectable.On the other hand, E. coli was quiescent in axenic mono-cultures, as there was no reverse labelling with natural carbon isotopes, but became metabolically active after 2 h when co-cultivated with P. putida in MMP with 30% D2O, as evidenced by the C–D band at 2172 cm−1 that can be seen in Fig. 5c. Additional Raman peaks derived from C–D stretch modes22 were detected at 992 and 1101 cm−1. The C–D bands were absent from the Raman spectra of E. coli harvested from MMP with H2O only co-cultures (Fig. 5d). The resuscitation of metabolic activity in E. coli could be plausibly explained by the cross-feeding of metabolic degradation products from P. putida as illustrated previously using different bacteria.8 Nevertheless, one would expect distinctive carbon-based band blueshifts as a consequence of cross-feeding, but regardless of incubation time no cell growth (Fig. S9a and b†) or blueshifts (Fig. 5c and d) were detected for E. coli despite being metabolically activity in co-culture. That is to say, there is no evidence of 12C assimilation; however, the incorporation of D atom suggests metabolic activity in E. coli. This was perhaps due to the assimilated 12C being below the detection limit of Raman spectroscopy.11 Likewise, OD600 remained constant at all time points (Fig. S9a and b†), depicting independence of metabolic behaviour from cell replication.22 Therefore, probing bacterial cell metabolism with D2O labelling overcame the shortfall in Raman sensitivity at our investigated conditions, and revealed metabolic behaviour of bacteria when the level of incorporated 12C was perhaps too weak and thus below detection limits.
The incorporation rate and level of D2O in P. putida and E. coli were quantified based on the intensity ratio plot [C–D/(C–H + C–D)] for Raman bands detected at 2172 cm−1 (C–D) and 2930 cm−1 (C–H) in MMP co-cultures at different time points. It is clear that the increase in intensity ratios for C–D and C–H spectral bands over time observed on Fig. 6(c and d) indicates increasing incorporation rate of D and metabolic activity. This observation proposed increased cellular activity in co-cultures of P. putida and E. coli, since water is involved in nearly all metabolic processes. As a negative control, the C–D bands were not observed in cells grown in MMP with H2O co-cultures (Fig. 5a and d).
To explore the reverse 12C and D effects and induced functional phenotypes for P. putida in co-cultures further, PCA was used. Fig. 6(e and f) shows PCA scores plots, which differentiate P. putida based on the quantity of 12C, D and sampling time. As expected at t = 0 h, P. putida biomass was dominated by ∼100% 13C from growth on 13C-glucose, whilst the cells sampled between 1 and 8 h absorbed increasing levels of 12C from phenol until complete assimilation (∼100% 12C) was detected after 18 h. According to the PC1, P. putida sampled between 0 and 4 h were discriminated from those collected at 8 and 18 h based on cumulative uptake of 12C and D (Fig. 6e). The PC1 loadings (Fig. S10†) identified nucleobases (762 → 783 cm−1), phenylalanine (965 → 1004 cm−1), amide I (1620 → 1661 cm−1) and C–D (2172 cm−1) peaks, as major features linked with clustering patterns on Fig. 6e. PC2 showed similar distribution of data for P. putida assimilating 12C and D, and sampling time, increasing from top (t = 0 h) to bottom (t = 4 h), which was clearly confirmed by PC2 loadings (Fig. S10b†). The PCA scores (Fig. 6f) and loadings (Fig. S11†) plots for Raman data of P. putida from MMP with H2O co-cultures exhibited identical clustering trends to those grown in MMP with 30% D2O co-cultures (Fig. 6e and S10†), except those patterns caused by incorporation of D isotope.
The PCA scores for mono cultures of E. coli (Fig. S12†) did not display any recognisable trends pertaining to rSIP and D2O labelling. By comparison, E. coli from MMP with 30% D2O co-culture (Fig. 6g) revealed a clustering pattern dominated by C–D vibrations in lipids,22 consistent with the spectral finding in Fig. 5c. The PC1 loadings in Fig. S13a† demonstrates this to be the case by revealing a unique band at 992 cm−1 and a clear redshift from 1124 cm−1 to 1101 cm−1, and band intensity ratio dynamics, though no spectral blueshifts were detected. However, PC2 loadings plot identified C–D peaks as the main variables differentiating E. coli at t = 0 h from E. coli cells harvested at successive time points. This observation is perhaps not surprising, considering that E. coli became active but did not appear to assimilate adequate levels of the 12C available in MMP to initiate detectable blueshifts. Similarly, PCA scores for E. coli from MMP with H2O co-cultures (Fig. 6h) indicated revived metabolic activity, however this finding was more distinct with D2O labelling as can be seen on Fig. 6g. The clustering profiles along PC1 (Fig. 6g and h) were mainly induced by peak intensity dynamics rather than by 12C isotope incorporation, and this reasoning was clearly supported by the PC loadings plots on Fig. S13 and S14.† While Raman, FT-IR and SIMS phenotypic fingerprints have demonstrated the capability to track 13C-glucose intake in E. coli and phenol metabolism overtime in axenic and co-cultures of P. putida and E. coli, this has been performed on cell colonies, and hence this needs to be translated to single cell metabolism analysis. Raman spectroscopy and nano-SIMS can probe bacterial metabolism at the single cell level, however, FT-IR can only analyse cell colonies due to poor spatial resolution though optical photothermal infrared44 and atomic force microscopy-infrared45 are allowing single cell phenotypic studies.
Conclusions
We have demonstrated the application of highly sensitive and reproducible Raman, FT-IR and SIMS-SIP – augmented with chemometrics – for rapid detection of 13C incorporation of E. coli along a time gradient; which, to the best of our knowledge, has never been reported before. High-throughput Raman, FT-IR and SIMS spectral profiles provided distinct bacterial identity, functional activity and 13C incorporation rate in E. coli at different time points. Kinetic analysis of isotope incorporation elucidated cross-feeding process and allowed the detection of primary consumers of phenol in a two-species community of P. putida and E. coli. Importantly, isotopic labelling has revealed quicker incorporation of 13C from glucose in amide I, phenylalanine and fatty acids whilst phenol-derived 12C were assimilated quickly into amides and nucleobases. Reverse and D2O SIP of community bacteria highlights the utility of low-cost tools for studying the metabolism of noxious compounds whose labelled analogues are costly or synthetically unamenable. Combining time course SIP and metagenomics may open up new avenues for probing the flow of essential nutrients in individual cells and within multi-species consortia.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
MC thanks the Commonwealth Scholarship Commission, UK for PhD funding (Grant No. ZMCA-2016-152). HM and RG thank the European Commission's Seventh Framework Program for funding (STREPSYNTH; Project No. 613877). HM also thanks the University of Liverpool for funding and support. RG is indebted to UK BBSRC (BB/L014823/1) for funding for Raman spectroscopy. We thank Rehana Sung for assistance with HPLC instrument. DM gratefully acknowledges studentship funding from EPSRC and Waters Corp.References
- M. Manefield, A. S. Whiteley, R. I. Griffiths and M. J. Bailey, Appl. Environ. Microbiol., 2002, 68, 5367–5373 CrossRef CAS.
- W. E. Huang, M. Q. Li, R. M. Jarvis, R. Goodacre and S. A. Banwart, in Advances in Applied Microbiology, ed. A. I. Laskin, S. Sariaslani and G. M. Gadd, Elsevier Academic Press Inc, San Diego, 2010, vol. 70, pp. 153–186 Search PubMed.
- S. Radajewski, I. R. McDonald and J. C. Murrell, Curr. Opin. Biotechnol., 2003, 14, 296–302 CrossRef CAS.
- J. D. Neufeld, J. Vohra, M. G. Dumont, T. Lueders, M. Manefield, M. W. Friedrich and J. C. Murrell, Nat. Protoc., 2007, 2, 860–866 CrossRef CAS.
- M. Manefield, A. S. Whiteley, N. Ostle, P. Ineson and M. J. Bailey, Rapid Commun. Mass Spectrom., 2002, 16, 2179–2183 CrossRef CAS.
- W. E. Huang, K. Stoecker, R. Griffiths, L. Newbold, H. Daims, A. S. Whiteley and M. Wagner, Environ. Microbiol., 2007, 9, 1878–1889 CrossRef CAS.
- P. Heraud, J. Beardall, D. McNaughton and B. R. Wood, FEMS Microbiol. Lett., 2007, 275, 24–30 CrossRef CAS.
- Y. Wang, Y. Z. Song, Y. F. Tao, H. Muhamadali, R. Goodacre, N. Y. Zhou, G. M. Preston, J. Xu and W. E. Huang, Anal. Chem., 2016, 88, 9443–9450 CrossRef CAS.
- E. S. Wharfe, R. M. Jarvis, C. L. Winder, A. S. Whiteley and R. Goodacre, Environ. Microbiol., 2010, 12, 3253–3263 CrossRef CAS.
- L. Corte, R. di Cagno, M. Groenewald, L. Roscini, C. Colabella, M. Gobbetti and G. Cardinali, Food Microbiol., 2015, 48, 206–215 CrossRef CAS.
- W. R. Abraham, Appl. Microbiol. Biotechnol., 2014, 98, 4817–4828 CrossRef CAS.
- S. A. Eichorst, F. Strasser, T. Woyke, A. Schintlmeister, M. Wagner and D. Woebken, FEMS Microbiol. Ecol., 2015, 91, 106–119 CrossRef.
- S. E. McGlynn, G. L. Chadwick, C. P. Kempes and V. J. Orphan, Nature, 2015, 526, 531–535 CrossRef CAS.
- M. Q. Li, W. E. Huang, C. M. Gibson, P. W. Fowler and A. Jousset, Anal. Chem., 2013, 85, 1642–1649 CrossRef CAS.
- L. Cui, K. Yang, G. W. Zhou, W. E. Huang and Y. G. Zhu, Anal. Chem., 2017, 89, 5794–5801 Search PubMed.
- H. Muhamadali, M. Chisanga, A. Subaihi and R. Goodacre, Anal. Chem., 2015, 87, 4578–4586 CrossRef CAS.
- L. Cui, K. Yang, H. Z. Li, H. Zhang, J. Q. Su, M. Paraskevaidi, F. L. Martin, B. Ren and Y. G. Zhu, Anal. Chem., 2018, 90, 5082–5089 CrossRef CAS.
- T. Jochum, A. Fastnacht, S. E. Trumbore, J. Popp and T. Frosch, Anal. Chem., 2017, 89, 1117–1122 CrossRef CAS.
- B. N. V. Kumar, S. Guo, T. Booklitz, P. Rosch and J. Popp, Anal. Chem., 2016, 88, 7574–7582 CrossRef.
- J. B. Xu, D. Zhu, A. D. Ibrahim, C. C. R. Allen, C. M. Gibson, P. W. Fowler, Y. Z. Song and W. E. Huang, Anal. Chem., 2017, 89, 13305–13312 CrossRef CAS.
- S. Pawelczyk, D. Bumann and W. R. Abraham, J. Appl. Microbiol., 2011, 110, 1065–1073 CrossRef CAS.
- D. Berry, E. Mader, T. K. Lee, D. Woebken, Y. Wang, D. Zhu, M. Palatinszky, A. Schintimeister, M. C. Schmid, B. T. Hanson, N. Shterzer, I. Mizrahi, I. Rauch, T. Decker, T. Bocklitz, J. Popp, C. M. Gibson, P. W. Fowler, W. E. Huang and M. Wagner, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E194–E203 CrossRef CAS.
- M. Chisanga, H. Muhamadali, R. Kimber and R. Goodacre, Faraday Discuss., 2017, 205, 331–343 RSC.
- S. Wold, M. Sjostrom and L. Eriksson, Chemom. Intell. Lab. Syst., 2001, 58, 109–130 CrossRef CAS.
- R. Goodacre, D. Graham and K. Faulds, TrAC, Trends Anal. Chem., 2018, 102, 359–368 CrossRef CAS.
- M. Chisanga, H. Muhamadali, D. I. Ellis and R. Goodacre, Appl. Spectrosc., 2018, 72, 987–1000 CrossRef CAS.
- Y. Wang, Y. T. Ji, E. S. Wharfe, R. S. Meadows, P. March, R. Goodacre, J. Xu and W. E. Huang, Anal. Chem., 2013, 85, 10697–10701 CrossRef CAS.
- J. De Gelder, K. De Gussem, P. Vandenabeele and L. Moens, J. Raman Spectrosc., 2007, 38, 1133–1147 CrossRef CAS.
- C. Xie, J. Mace, M. A. Dinno, Y. Q. Li, W. Tang, R. J. Newton and P. J. Gemperline, Anal. Chem., 2005, 77, 4390–4397 CrossRef CAS.
- K. Maquelin, C. Kirschner, L. P. Choo-Smith, N. van den Braak, H. P. Endtz, D. Naumann and G. J. Puppels, J. Microbiol. Methods, 2002, 51, 255–271 CrossRef CAS.
- M. J. Baker, J. Trevisan, P. Bassan, R. Bhargava, H. J. Butler, K. M. Dorling, P. R. Fielden, S. W. Fogarty, N. J. Fullwood, K. A. Heys, C. Hughes, P. Lasch, P. L. Martin-Hirsch, B. Obinaju, G. D. Sockalingum, J. Sule-Suso, R. J. Strong, M. J. Walsh, B. R. Wood, P. Gardner and F. L. Martin, Nat. Protoc., 2014, 9, 1771–1791 CrossRef CAS.
- M. Giordano, M. Kansiz, P. Heraud, J. Beardall, B. Wood and D. McNaughton, J. Phycol., 2001, 37, 271–279 CrossRef CAS.
- A. Barth, Biochim. Biophys. Acta, Bioenerg., 2007, 1767, 1073–1101 CrossRef CAS.
- D. Naumann, D. Helm and H. Labischinski, Nature, 1991, 351, 81–82 CrossRef CAS.
- J. Zhu and C. B. Thompson, Nat. Rev. Mol. Cell Biol., 2019, 20, 436–450 CrossRef CAS.
- D. Schultz and R. Kishony, BMC Biol., 2013, 11, 120–122 CrossRef.
- Y. Wang, J. B. Xu, L. C. Kong, T. Liu, L. B. Yi, H. J. Wang, W. E. Huang and C. M. Zheng, Microb. Biotechnol., 2020, 13, 572–583 CrossRef CAS.
- L. Gonzales-Siles and A. Sjoling, Environ. Microbiol., 2016, 18, 741–751 CrossRef CAS.
- C. Molina-Santiago, Z. Udaondo, B. F. Cordero and J. L. Ramos, Environ. Microbiol. Rep., 2017, 9, 441–448 CrossRef CAS.
- M. I. Ramosgonzalez, M. A. Ramosdiaz and J. L. Ramos, J. Bacteriol., 1994, 176, 4635–4641 CrossRef CAS.
- P. Kubryk, J. S. Kolschbach, S. Marozava, T. Lueders, R. U. Meckenstock, R. Niessner and N. P. Ivleva, Anal. Chem., 2015, 87, 6622–6630 CrossRef CAS.
- O. O. Olaniyi, K. Yang, Y. G. Zhu and L. Cui, Appl. Microbiol. Biotechnol., 2019, 103, 1455–1464 CrossRef CAS.
- S. H. Kopf, A. L. Sessions, E. S. Cowley, C. Reyes, L. Van Sambeek, Y. Hu, V. J. Orphan, R. Kato and D. K. Newman, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E110–E116 CrossRef CAS.
- D. L. Zhang, C. Li, C. Zhang, M. N. Slipchenko, G. Eakins and J. X. Cheng, Sci. Adv., 2016, 2, 7 Search PubMed.
- A. Dazzi, C. B. Prater, Q. C. Hu, D. B. Chase, J. F. Rabolt and C. Marcott, Appl. Spectrosc., 2012, 66, 1365–1384 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0an02319a |
| This journal is © The Royal Society of Chemistry 2021 |