Lignin as a MALDI matrix for small molecules: a proof of concept†
Xiaoyong
Zhao‡
a,
Huiwen
Wang‡
b,
Yilong
Liu
a,
Ruohan
Ou
a,
Yaqin
Liu
b,
Xian
Li
 *a and
Yuanjiang
Pan
*a and
Yuanjiang
Pan
 *b
*b
aZhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Zhejiang University, Hangzhou 310058, China. E-mail: xianli@zju.edu.cn; Fax: +86571-8898-2224; Tel: +86-571-8898-1263
bDepartment of Chemistry, Zhejiang University, Hangzhou 310027, China. E-mail: panyuanjiang@zju.edu.cn; Fax: +86571 87951629; Tel: +86 571 87951629
First published on 27th October 2021
Abstract
Driven by the interest in metabolomic studies and the progress of imaging techniques, small molecule analysis is booming, while it remains challenging to be realized by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). Herein, lignin, the second most abundant biomass in nature, was applied as a dual-ion-mode MALDI matrix for the first time to analyze small molecules. The low ionization efficiency and strong optical absorption properties make lignin a potential MALDI matrix in small molecule analysis. A total of 30 different small molecules were identified qualitatively and six kinds of representative molecules were detected quantitatively with a good linear response (R2 > 0.995). To verify the accuracy of our quantitative method in MALDI, myricitrin, a major bioactive component in Chinese bayberry, was analyzed in different cultivars and tissues. The myricitrin content in real samples detected by MALDI was highly consistent (R2 > 0.999) with that detected by high-performance liquid chromatography, thus indicating the applicability of the lignin matrix. Further characterization by ultraviolet and nuclear magnetic resonance spectroscopy was carried out to explain the possible mechanism of lignin as a matrix and provide more theories for a rational matrix design.
1. Introduction
Since its development in the 1980s, matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) has become an integral tool for analyzing high molecular weight biomolecules, owing to its soft ionization property, high sensitivity, high salt tolerance and high throughput.1,2 Currently, small molecule analysis is hyper emerging driven by the increasing interest in metabolomic studies, since the analysis of metabolites is crucial to the research of disease formation and progression, drug development, and the detection of environmental pollutants, and so forth,3 and MALDI-MS has been regarded as a powerful tool to achieve a rapid and convenient analysis with low sample consumption.4,5 Besides, the overwhelming growth of the mass spectrometry imaging (MSI) technique, which can achieve visualized analysis of molecules in tissue samples,6,7 also promotes the field of small molecule analysis. During MALDI-MS analysis, the appropriate choice of a matrix directly determines the detection results. The commonly used organic matrices, such as α-cyano-4-hydroxy-cinnamic acid (CHCA)8 and 2,5-dihydroxybenzoic acid (DHB),9 suffer from background interference due to the matrix-related fragments in the low-mass region.10 Therefore, exploring suitable MALDI matrices for small molecule analysis is necessary.To overcome the limits of detecting small molecules, several alternative matrices like inorganic materials11,12 polymers,13,14 reactive matrices,15,16etc. were employed constantly. Owing to the efficient desorption/ionization ability, universal optical absorption property, and selective enrichment capacity, inorganic materials such as carbon-based materials,17 porous silicon,18 and metal or metal oxide nanoparticles19,20 have been regarded as powerful matrices for the MALDI-MS analysis of small molecules. Meanwhile, the limitations of inorganic matrices include low solubility and increased analyte fragmentation caused by required high laser energy.4 Although several approaches like hybridization with organic matrices and hydrophilic modification were adopted to eliminate these limitations, the additional processing was time-consuming and tedious.21,22 Reactive matrices can work simultaneously as MALDI matrices and derivatization agents to increase the final mass to charge (m/z) value of small molecules with improved ionization efficiency,23 while the detection lacks universality because of the matrix reactivity only towards specific analytes with a corresponding functional group. Thus, a novel MALDI matrix material remains to be developed for better profiles of small molecules.
Lignin, the second most abundant plant biomass in nature after cellulose, is an amorphous, three-dimensional cross-linked heteropolymer consisting of phenylpropanoid units.24,25 Although knowledge of the macromolecular structure of lignin is still limited, it is composed of three main monomeric precursors: p-hydroxyphenyl (H), guaiacyl (G) and syringyl (S) monomers.26 As the only aromatic native biopolymer on the Earth, lignin has attracted much interest, which can be utilized as an energy source in the paper industry and a potential feedstock for valuable phenolic chemicals.27 In fact, the final application of lignin is significantly varied with the different extraction and isolation processes, including mechanical extraction, enzymolysis, and strong base depolymerization.28 Based on the structural feature and functional composition of lignin macromolecules, along with the sufficiently high hydrophobicity of the polymer, a low ionization efficiency is caused. As a result, lignin is quite difficult to produce signals under MALDI, especially within the low mass range, which exactly solves the problem of matrix interference during analyzing small molecules. Moreover, lignin inherently contains aromatic structures for effective laser energy absorption and transfer, and a favorable optical property is a crucial criterion for an ideal MALDI matrix. In addition, compared with traditional organic matrices, lignin with high molecular weight has lower saturated vapor pressure contributing to increased stability under high vacuum MALDI. Theoretically, these advantages and ‘disadvantages’ make lignin potentially act as a MALDI matrix for small molecules.
Myricitrin (Myr) is a naturally occurring flavonoid glycoside belonging to the flavonol subgroup, which is widely distributed in the fruits, roots, stems, bark, and leaves of various plants, such as Myrica esculenta, Ampelopsis grossedentata, and Chrysobalanus icaco.29,40 It has been shown that Myr possesses considerable bioactivities, including anti-oxidative, anti-inflammatory, anti-cancer, hypolipidemic, and hepatoprotective activities, thus used in clinical therapy as an important supplement in medicines for burns, skin diseases, and diabetes.30–32 The detection of Myr in plant organs is mainly based on high-performance liquid chromatography (HPLC) techniques such as HPLC-DAD33 and HPLC-UV,34 while these chromatographic methods were of low-resolution and time-consuming. Besides, Myr is widely distributed in nature, but not abundant in content, requiring an analytical tool with a higher sensitivity. As a result, based on the biological significance of Myr and the previous study in our group,35,36 establishing a fast and efficient method to directly quantify Myr from complex biological samples is essential to its further exploitation.
Herein, lignin was presented as a dual-ion mode MALDI matrix to analyze small molecules for the first time. At the same time, a total of 30 molecules were successfully detected and real samples were occupied for quantification verification, providing valuable information for effective application to natural small molecules further. Moreover, the exploitation of lignin as a MALDI matrix also provided more foundations for rational design of the MALDI matrix.
2. Materials and methods
2.1. Reagents and materials
Dealkaline lignin (DAL), stearic acid (SA), riboflavin (VB2), and vitamin-E (VE) were purchased from TCI Chemical Industrial Development Co., Ltd (Shanghai, China). Alkali lignin (AL), DHB, and CHCA were purchased from Sigma-Aldrich (St Louis, USA). Myr, quercitrin (Que), L-arabinose (Ara), D-glucose (G1), citric acid (CA), and diethyl malonate (DEM) were purchased from Aladdin (Shanghai, China). Maltose (G2), maltotriose (G3), maltohexose (G6), thymidine (T), 2′-deoxyadenosine (DOA), and 2′-deoxyguanosine (DOG) were purchased from Yuanye Biotechnology Co., Ltd (Shanghai, China). Palmitic acid (PA), linoleic acid (LA), dioctyl phosphonate (DOP), triacetin (TAN), tributyrin (TBN), and glyceryl monostearate (GMS) were purchased from Macklin (Shanghai, China). Acetylsalicylic acid (ASA), D-phenyllactic acid (PLA), uridine (U), 2′-deoxycytidine (DOC), and cholesterol (Chol) were purchased from Energy Chemical Co., Ltd (Shanghai, China). L-Valine (Val) was purchased from the Institute of Biochemistry, Chinese Academy of Sciences (Shanghai, China). L-Arginine (Arg) was purchased from Kangda Amino Acid Factory (Shanghai, China). L-Methionine (Met) and L-alanine (Ala) were purchased from Zhengxiang Medical Science and Technology Industrial Service Department, Second Military Medical University of China (Shanghai, China). L-Lysine (Lys), DL-malic acid (MA), NaCl, HPLC-grade methanol (MeOH), HPLC-grade ethanol (EtOH), and HPLC-grade dimethylsulfoxide (DMSO) were purchased from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). Deionized water (ddH2O) was purified by a Milli-Q system.Bayberry leaf and stem of three cultivars, i.e., ‘Wu Mei’ (WM), ‘Te Zao Mei’ (TZM) and ‘Ruan Si An Hai Bian’ (RSAHB), were collected from orchards of Fujian Academy of Agricultural Sciences in March 2019. All samples were ground into fine powder and stored at −80 °C until extraction and analysis.
2.2. Matrix preparation and MALDI-MS analysis
Matrix suspensions with different concentrations of DAL and AL were prepared in ddH2O (or MeOH), and matrix suspensions were sonicated for 15 min for further use. Furthermore, 15.4 mg mL−1 DHB and 18.9 mg mL−1 CHCA solutions were prepared in MeOH respectively as the control samples. The structures of DAL and AL were confirmed by 1H NMR and HSQC NMR on an Advance DMX 400 MHz spectrometer (Bruker, Germany), and the UV absorption spectra were recorded on an Ultraspect 3000 UV–vis spectrophotometer (Pharmacia Biotech, USA) equipped with a 1 cm quartz cell.The two-layer method (TL) was used to prepare the samples for MALDI-MS analysis. Briefly, 1 μL of the DAL matrix suspension was deposited on the sample plate first, and then it was dried on the sample plate followed by addition of 1 μL of the analyte solution. When the solution was air dried, the sample plate was sent to a Bruker UltrafleXtreme mass spectrometer (Bremen, Germany) equipped with a modified Nd:YAG laser (355 nm, 2000 Hz) in positive or negative ion reflector mode. In particular, the dried-droplet method (DD) was used for comparison during the matrix selection part. Briefly, 1 μL of sample was mixed with 1 μL of matrix on the polished target and dried at room temperature before detection.
2.3. Sample preparation for MALDI analysis
For qualitative analysis, in ddH2O, Ara, G1, Val, Met, Lys, Arg, MA, CA, PLA, ASA, U, DOA, DOG, and DOC were respectively dissolved to 1 mg mL−1. G2 (1.4 mg), G3 (2.1 mg), and G6 (3.9 mg) were respectively dissolved in 1 mL of ddH2O to 200 pmol μL−1. NaCl solution was used to dissolve VB2 to 1 mg mL−1. PA, SA, LA, DEM, VE, TAN, DOP, TBN, GMS, and Chol were dissolved in EtOH to 1 mg mL−1. All the solutions were stored at 4 °C for further use. Each kind of sample solution was detected three times randomly to exclude errors.For quantitative analysis, to investigate the limit of detections (LODs), all standard molecules were dissolved and diluted with serial (10-fold) concentrations in the range of 1 mg mL−1 to 1 ng mL−1. To obtain the standard quantitative curves, the standard small molecules were diluted to a series of concentrations with a coefficient of five, and the concentrations next mentioned were the final concentrations detected. A series of concentrations of standard substances G1 (from 0.0016 mg mL−1 to 1 mg mL−1), Val (from 0.008 mg mL−1 to 5 mg mL−1), and CA (from 0.008 mg mL−1 to 5 mg mL−1) were prepared in ddH2O and spiked with Ara (0.005 mg mL−1), Ala (0.1 mg mL−1), and MA (0.05 mg mL−1) as internal standards (ISs), respectively. A series of concentrations of standard substances SA (from 0.0016 mg mL−1 to 1 mg mL−1) and TBN (from 0.0016 mg mL−1 to 1 mg mL−1) were prepared in EtOH and spiked with PA (0.05 mg mL−1) and TAN (0.01 mg mL−1) as ISs, respectively. Using Que (0.06 mg mL−1) as an IS, a series of concentrations of standard substance Myr (from 0.025 mg mL−1 to 0.8 mg mL−1) were prepared in MeOH with two as the coefficient. All the solutions were stored at 4 °C for further use. Each sample with different concentrations was detected six times randomly for average.
The powder (0.15 g each) of leaf and stem of each bayberry cultivar was extracted with 1 mL of methanol by sonication in an ultrasonic cleaner for 30 min. The mixture was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and the samples were extracted another two times. All extracts were combined and filtered through a 0.22 μm membrane before injection. Extractions from stems and leaves of WM, TZM, and RSAHB were spiked with Que with a final concentration of 0.06 mg mL−1 and each kind of sample solution has quadruplicates and each sample was detected six times randomly for average. These real sample solutions should be used intraday.
000 rpm for 10 min and the samples were extracted another two times. All extracts were combined and filtered through a 0.22 μm membrane before injection. Extractions from stems and leaves of WM, TZM, and RSAHB were spiked with Que with a final concentration of 0.06 mg mL−1 and each kind of sample solution has quadruplicates and each sample was detected six times randomly for average. These real sample solutions should be used intraday.
2.4. HPLC analysis of real samples
Myr was analyzed according to our previous method.36 The HPLC system (Waters e2695, 2998 photodiode array detector, Waters, Milford, PA, USA) equipped with a Sunfire C18 analytical column (4.6 mm × 250 mm, 5 μm) was used with an Empower chromatography workstation. The mobile phase solutions were water with 0.1% formic acid (eluent A) and acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1% formic acid (1
0.1% formic acid (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) (eluent B) and were implemented in the following gradient: 0–40 min, 10–38% B; 40–60 min, 38–48% B; 60–70 min, 48–100% B; 70–75 min, 100–10% B; and 75–80 min, 10% B. The injection volume was 10 μL and the flow rate was 1 mL min−1. Myricitrin was detected under 350 nm. A standard solution at series concentrations from 1.95 to 1000 μg mL−1 was prepared to quantify the content of Myr in samples.
1, v/v) (eluent B) and were implemented in the following gradient: 0–40 min, 10–38% B; 40–60 min, 38–48% B; 60–70 min, 48–100% B; 70–75 min, 100–10% B; and 75–80 min, 10% B. The injection volume was 10 μL and the flow rate was 1 mL min−1. Myricitrin was detected under 350 nm. A standard solution at series concentrations from 1.95 to 1000 μg mL−1 was prepared to quantify the content of Myr in samples.
3. Results and discussion
3.1. Lignin DAL as a MALDI matrix for small molecules
Typically, an excellent MALDI matrix for small molecule analysis is evaluated by two criteria: fewer matrix background ions in the low m/z range and higher desorption/ionization efficiency toward analytes. Herein, the application of lignin is significantly varied with the extraction process, and thus, AL and DAL either pretreated by strong base or not were investigated to select a superior small molecule matrix and the traditional DHB and CHCA matrices were used as controls. After detecting blank matrices by MALDI (Fig. 1A), traditional matrices produced complex background interferences as previously mentioned, and the related ions produced by the AL matrix also was non-ignorable despite improvement. In contrast, the DAL matrix produced a clearer background with some easily attributed peaks, indicating its potential and feasibility in analyzing small molecules.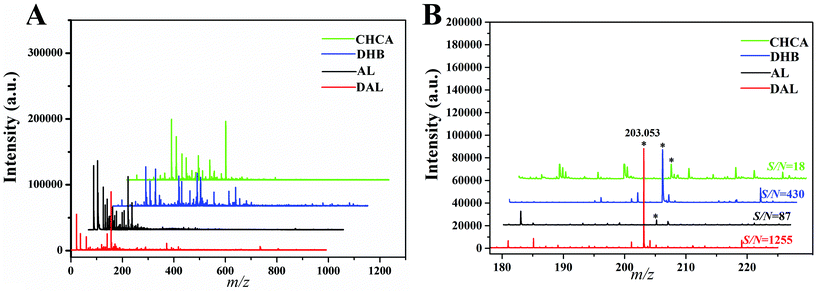 | ||
| Fig. 1 MALDI-TOF spectrum for different blank matrices (A) and G1 (B) detected by matrices (asterisks denote the alkali–metal ion adducts of G1). | ||
To compare the efficiency of matrices in assisting ionization towards analytes, glucose (G1) was used as a model chemical, and the signal-to-noise ratio (S/N) of its predominant alkali metal ion adducts [M + Na]+ at m/z 203.035 was used for evaluation. When this model standard substance was detected by DAL (Fig. 1B), an ideal result was obtained with the strongest signal intensity (S/N = 1255), compared with S/N = 430 and 18 detected by DHB and CHCA respectively. On the other hand, the S/N detected by AL was over ten times lower than that by DAL, indicating the necessity of selection between kinds of lignins. Overall, owing to the spectra and high sensitivity, it is obvious that DAL has superiority as a small molecule matrix during MALDI analysis.
3.2. Selection of the matrix preparation methods
During MALDI analysis progress, the analyte ions were produced by the energy and proton transfer between matrices and analytes, so the sample preparation method which influences the matrix–analyte cocrystals is important. Based on the physicochemical property of the adopted matrix, a specific preparation method should be optimized to ensure optimal detection results. In this study, MeOH and ddH2O were selected as solvents to disperse the DAL matrix particles, followed by different sample application methods, including TL and DD methods. After shooting 6 times on the same sample point for the relative standard deviation (RSD), the spectral results with error bars are shown in Fig. 2. Apparently, the two matrix suspensions prepared in ddH2O made G1 ion adducts have higher S/N compared with those in MeOH (Fig. 2A). Of these two, when using the TL method for the sample application, the S/N was nearly twice higher than the DD method. As a result, the following experiments adopted the TL method to prepare the matrix aqueous suspension.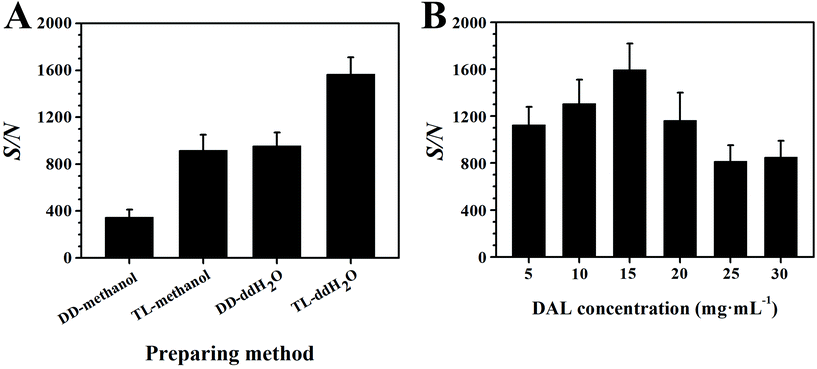 | ||
| Fig. 2 Detection effects of the preparation methods (A) and lignin concentrations (B) on the S/N of G1. | ||
A MALDI matrix takes a key role in protecting analytes in case of fragmentation under the laser, so it should be overwhelmingly excessive. But in fact, overly residual matrices tend to cause ionization suppression towards analytes resulting in low detection sensitivity. As a result, the concentration of the DAL matrix was also optimized in this study. G1 was the model chemical and the volume of matrix suspension pipetted on the target plate was kept constant at 1 μL level. After detection of G1 using the DAL matrix with different concentrations (Fig. 2B), the S/N increased first and then decreased with the increasing concentration of DAL, peaking at 15 mg mL−1. In general, the optimized sample preparation method was the TL method using ddH2O as a solvent to suspend the DAL matrix to a concentration of about 15 mg mL−1.
3.3. Qualitative analysis of small molecules
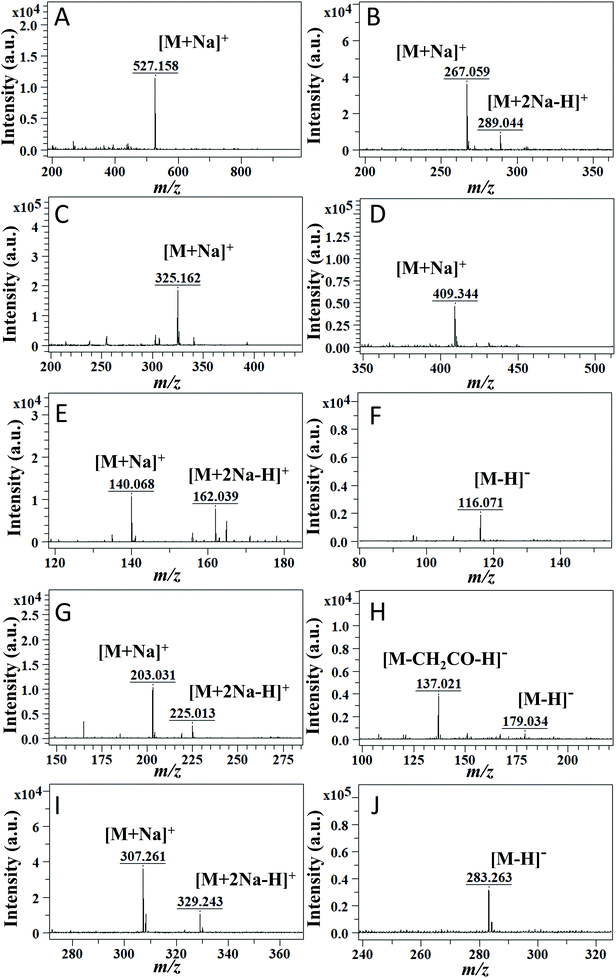
To investigate the applicability of the DAL matrix, a total of 29 common small molecules categorized into seven types were employed as target analytes for qualitative analysis, including oligosaccharides, (Ara, G1, G2, G3, and G6), glycosides (DOC, DOG, DOA, T, and U), esters (DEM, DOP, GMS, TBN, and TAN), vitamins (VE, Chol, and VB2), amino acids (Val, Met, Lys, and Arg), hydroxy acids (CA, MA, PLA, and ASA), and fatty acids (SA, PA, and LA), all of which actively participate in the metabolic process and are commonly used biomarkers during clinical diagnosis and therapy. For example, amino acid analysis by mass spectrometry for newborns was a powerful tool to screen and diagnose metabolic disorders.37 Detailed information about chemical structures and molecular weights (MW) can be seen in Fig. S1,† and their specific peak assignments are listed in Table S1.†As for the first four kinds of neutral small molecules, positive ion mode detection was carried out (Fig. 3A–D). The characteristic peaks of these compounds were mainly attributed to sodium ion adducts [M + Na]+. Taking G3 (MW = 504.438) as an example, the molecular ion peaks in the spectrum (Fig. 3A) could be assigned to alkali metal ion adducts [M + Na]+ at m/z 527.158. Occasionally, there were a few other types of ion adducts like [M + 2Na − H]+ along with the main ion adducts [M + Na]+. For instance, in Fig. 3B, U (MW = 244.203) produced abundant [M + Na]+ at m/z 267.059 and slight [M + 2Na − H]+ at m/z 289.044. In particular, DOG (MW = 267.245) and DOC (MW = 227.220) tended to form products from dehydration between monosaccharide and their nucleobases, such as [guanine + Na]+ at m/z 174.039 and [cytosine + Na]+ at m/z 134.046 as shown in Fig. S2B.† With the increase of laser energy, completely dehydrated products were observed.
As for the last three kinds of small molecules, considering the existence of easily lost protons, such as the hydrogen in carboxyl, a dual-ion-mode detection was carried out. In positive mode, differently from the results before, the ion adducts [M + 2Na − H]+ accounted for a large proportion (Fig. 3E, G and I), like ion adducts [M + 2Na − H]+ at m/z 162.039 of Val (MW = 117.148), and these multiple ions lead to a complex spectrum. However, when detecting these compounds in negative ion mode (Fig. 3F, H and J), considerable clean spectra were obtained, since they only exhibited peaks corresponding to deprotonated ions [M − H]−. An apparent comparison could be seen between Fig. 3I and J, and fatty acids such as SA (MW = 284.484) only produced [M − H]− at m/z 283.263. Furthermore, as the spectral result of ASA (MW = 180.159) (Fig. 3H), an unpredictable peak at m/z 137.021 was predominant, supposed to be due to a rearrangement of this molecule with a concomitant loss of CH2CO.
Overall, these accurate detections and rational assignments of signals among 29 different small molecules illustrated the universality and practicability of the DAL matrix; at the same time, the efficient ionization of analytes in both positive and negative modes further substantiates the potential of DAL as an excellent dual-ion-mode matrix.
3.4. Quantitative analysis of small molecules
Small molecule analysis is commonly applied to medical and health areas, especially used for biomarkers based on the content difference among cells and tissues. For example, the levels of G1 in blood and urine were common criteria in the diagnosis of hyperglycemia and diabetes, so quantification is vital for the efficient and reasonable application.38 In this work, five typical small molecules were quantified, including G1, Val, CA, SA, and TBN. The LODs of these representative standards are shown in Fig. S3,† which were estimated by S/N of predominant ion adducts. Apparently, a downward trend of the S/N ratio was observed with the decreasing concentrations of analytes, and the value of LOD was defined as the concentration with S/N equal to 3. To obtain the standard quantitative curves, the selection of internal standard (IS) plays a key role and the general selection principles are as follows.39 First, the IS should have similar chemical structures to the analyte resulting in similar ionization. Second, their mass spectra peaks should be neighboring but without interferences with each other. Third, the concentration of the IS should be the median of the analyte concentration range. Eventually, Ara, Ala, MA, PA, and TAN were selected as ISs for G1, Val, CA, SA, and TBN, respectively. As shown in Fig. S4A–S4E†, these selected ISs, which could meet all principles, were suitable for quantifying corresponding analytes, and their specific ionization results were clear to be calculated. The corresponding near dynamic curves are shown in Fig. 4A–E. Herein, the signal intensity of an analyte relative to that of IS was used as the response to the dynamic concentration of the analyte.40 The signal intensity for a specific analyte was acquired by calculation of the sum of the peak intensities of all types of ion adducts in the positive ion mode.41 For example, the intensity sum of Val was equal to the intensity of Val [M + Na]+ at m/z 140.068 plus that of Val [M + 2Na − H]+ at m/z 162.037 (Fig. S4B†). The regression curves are shown in the figures, equipped with correlation coefficients (R2) greater than 0.995, indicating the qualitative accuracy and availability for evaluating the level of target compounds.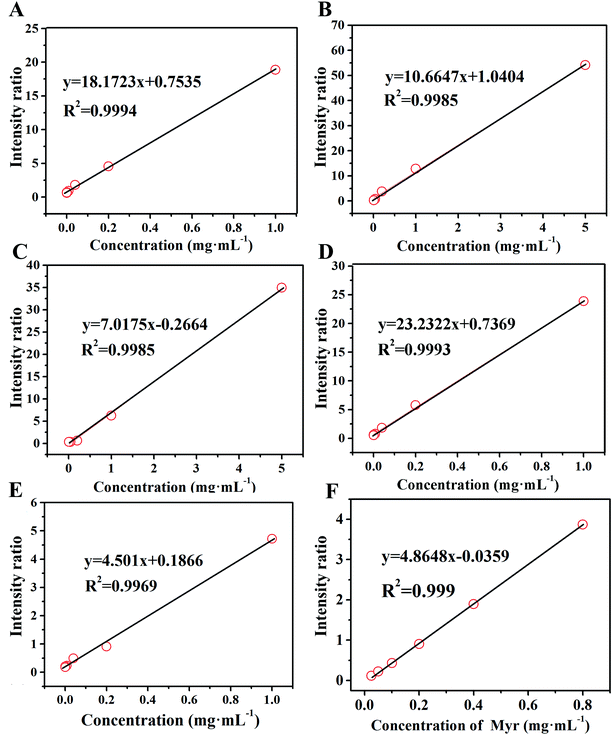 | ||
| Fig. 4 Linear dynamic ranges of G1 (A), Val (B), CA (C), SA (D), TBN (E) and Myr (F) using DAL as the MALDI matrix. | ||
3.5. Application in complex biological samples
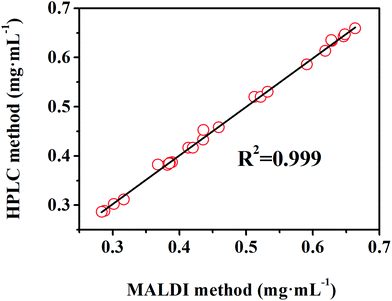
In this section, the feasibility of the DAL matrix in qualitative and quantitative analysis based on complex biological samples was verified. Myr, a widely employed supplement medicine extracted from plants, is not evenly distributed among plant organs, so rapid and high-throughput quality control is important for rational exploitation. Before application in real samples, the specific IS should be selected to ensure a regression equation with high linearity, and Que was finally screened out. As the chemical structures shown in Fig. S2F,† Myr and Que both are rhamnosides, and the rhamnose residues were easily lost during the in-source decay of hemiacetal O–C bonds in MALDI negative mode.42 After adjusting the laser energy, there were only corresponding aglycone anions produced, at m/z 317.230 and m/z 301.299 respectively. As a result, the Myr standard was quantified in negative mode, and the signal intensity was aimed at the intensity of the deprotonation peak of aglycone. Through the detection of Myr with different concentrations ranging from 0.025 mg mL−1 to 0.8 mg mL−1 with 0.06 mg mL−1 Que as the IS, the quantitation regression equation (R2 = 0.999) was obtained (Fig. 4F), feasibly used for the determination of its content in real samples.Next, extractions from leaves and stems of WM, TZ, and RSAHB were analyzed using the DAL matrix, followed by spiking into the Que standard as the IS. The contents of Myr in real samples were calculated by the obtained regression equation and compared with those obtained by the HPLC method, and the detailed results are listed in Table S2.† After detection in negative ion mode, the calculated concentrations of Myr in samples were varied from 0.2 mg mL−1 to 0.7 mg mL−1, within the effective linear range established, further proving the feasibility of calculation. Moreover, the excellent linear correlation (R2 = 0.999) in Fig. 5 evidenced the accuracy and applicability of the DAL matrix even used for quantitatively analyzing complex biological samples.
3.6. Possible mechanism of DAL as the MALDI matrix
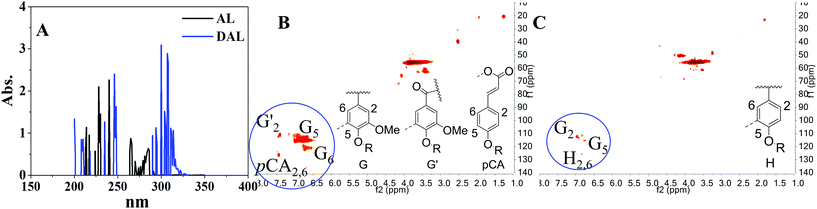
The excellent performance of DAL in assisting small molecule analysis by MALDI intrigued us to find out the possible mechanism underlying the performance, looking forward to providing more references for further matrix design and exploration. Since the ionization of analytes and matrix is initiated by the MALDI laser shot on the target plate, the optical property of the MALDI matrix is a valuable evaluation criterion.43 Currently, different UV lasers are equipped as a laser source for desorption and ionization on MALDI mass spectrometers, including a nitrogen laser (337 nm), a frequency-tripled Nd:YLF laser (349 nm), and frequency-tripled or quadrupled Nd:YAG lasers (355 or 266 nm, respectively).46 As expected, it was obviously shown in Fig. 6A, compared with AL, that DAL has a higher and broader UV absorbance near 355 nm, the MALDI laser wavelength used in the present study. As a result, the great performance of DAL in UV absorption efficiency corresponds to its superiority in using as the MALDI matrix. Moreover, the clean background of the DAL matrix (Fig. 1A) proved again that the strong UV-absorptivity of DAL did not exacerbate the formation of complex metastable matrix cluster ions during the laser ionization process, corresponding to the characteristics of small molecule analysis. To further understand the differences of DAL and AL in structural and optical properties, HSQC NMR was carried out for chemical structure identification. The HSQC NMR results of different lignins are shown in Fig. 6B and C. Compared with that in AL, there was much more and stronger signal response in the aromatic region in DAL. After comparing with the possible structural units of lignins reported,44 these HSQC signals were assigned in detail, proving that DAL was born with more conjugated structures than AL. So, the DAL matrix possibly has a higher performance in energy transfer during assisting ionization of analytes.45 Overall, the excellent performance of DAL is due to the superior capability in UV absorption and intrinsically equipping with a more conjugated structure, broadening more references for further matrix development.4. Conclusion
To break the limits of analyzing small molecules by MALDI, this study first develops lignin as a kind of universal small molecule matrix. Through selection of lignin varieties and sample preparation methods, we have established a dual-mode method based on the DAL matrix to achieve identification and quantitation of seven representative kinds of small molecules, including oligosaccharides, glycosides, esters, vitamins, amino acids, hydroxy acids, and fatty acids, and this method could achieve a simple, rapid, and accurate analysis even in the complex biological samples. Furthermore, the mechanism of DAL acting as a matrix was investigated and the performance was due to its superior optical property and abundant conjugated structures. In a word, DAL can act as a promising small molecule matrix for qualitative and quantitative high-throughput analysis and may have potential applications in medical research and development.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the National Basic Research Program of China, Grant No. 2016YFF0200503, the National Natural Science Foundation of China (21927810) and the Fundamental Research Funds for the Central Universities (2021QNA6016).References
- K. Tanaka, H. Waki, Y. Ido, S. Akita, Y. Yoshida, T. Yoshida and T. Matsuo, Protein and polymer analyses up to m/z 100000 by laser ionization time-of-flight mass spectrometry, Rapid Commun. Mass Spectrom., 1988, 2, 151–153, DOI:10.1002/rcm.1290020802.
- L. R. Ruhaak, G. G. Xu, Q. Y. Li, E. Goonatilleke and C. B. Lebrilla, Mass spectrometry approaches to glycomic and glycoproteomic analyses, Chem. Rev., 2018, 118, 7886–7930, DOI:10.1021/acs.chemrev.7b00732.
- G. J. Patti, O. Yanes and G. Siuzdak, Innovation: Metabolomics: the apogee of the omics trilogy, Nat. Rev. Mol. Cell Biol., 2012, 13, 263–269, DOI:10.1038/nrm3314.
- L. H. Cohen and A. I. Gusev, Small molecule analysis by MALDI mass spectrometry, Anal. Bioanal. Chem., 2002, 373, 571–586, DOI:10.1007/s00216-002-1321-z.
- C. D. Calvano, A. Monopoli, T. R. I. Cataldi and F. Palmisano, MALDI matrices for low molecular weight compounds: an endless story?, Anal. Bioanal. Chem., 2018, 410, 4015–4038, DOI:10.1007/s00216-018-1014-x.
- S. Schulz, M. Becker, M. R. Groseclose and S. Schadt, Advanced MALDI mass spectrometry imaging in pharmaceutical research and drug development, Curr. Opin. Biotechnol., 2019, 55, 51–59, DOI:10.1016/j.copbio.2018.08.003.
- I. S. Gilmore, S. Heiles and C. L. Pieterse, Metabolic imaging at the single-cell scale: recent advances in mass spectrometry imaging, Annu. Rev. Anal. Chem., 2019, 12, 201–224, DOI:10.1146/annurev-anchem-061318-115516.
- S. Madla, D. Miura and H. Wariishi, Potential applicability of MALDI-MS for low-molecular-weight pesticide determination, Anal. Sci., 2012, 28, 301–303, DOI:10.2116/analsci.28.301.
- B. Stahl, M. Steup, M. Karas and F. Hillenkamp, Analysis of neutral oligosaccharides by matrix-assisted laser desorption ionization mass spectrometry, Anal. Chem., 1991, 63, 1463–1466, DOI:10.1021/ac00014a022.
- J. J. A. Van Kampen, P. C. Burgers, R. De Groot, R. A. Gruters and T. M. Luider, Biomedical application of MALDI mass spectrometry for small-molecule analysis, Mass Spectrom. Rev., 2011, 30, 101–120, DOI:10.1002/mas.20268.
- M. Ronci, D. Rudd, T. Guinan, K. Benkendorff and N. H. Voelcker, Mass spectrometry imaging on porous silicon: investigating the distribution of bioactives in marine mollusc tissues, Anal. Chem., 2012, 84, 8996–9001, DOI:10.1021/ac3027433.
- Q. Min, X. Zhang, X. Chen, S. Li and J. J. Zhu, N-doped graphene: an alternative carbon-based matrix for highly efficient detection of small molecules by negative ion MALDI-TOF MS, Anal. Chem., 2014, 86, 9122–9130, DOI:10.1021/ac501943n.
- K. Horatz, M. Giampà, Y. Karpov, K. Sahre, H. Bednarz, A. Kiriy, B. Voit, K. Niehaus, N. Hadjichristidis, D. L. Michels and F. Lissel, Conjugated polymers as a new class of dual-mode matrices for MALDI mass spectrometry and imaging, J. Am. Chem. Soc., 2018, 140, 11416–11423, DOI:10.1021/jacs.8b06637.
- Q. Zhao, J. Xu, J. Yin and Y. Q. Feng, Humic acids as both matrix for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and adsorbent for magnetic solid phase extraction, Anal. Chim. Acta, 2015, 889, 138–146, DOI:10.1016/j.aca.2015.07.040.
- B. Flinders, J. Morrell, P. S. Marshall, L. E. Ranshaw and M. R. Clench, The use of hydrazine-based derivatization reagents for improved sensitivity and detection of carbonyl containing compounds using MALDI-MSI, Anal. Bioanal. Chem., 2015, 407, 2085–2094, DOI:10.1007/s00216-014-8223-8.
- M. Shariatgorji, A. Nilsson, E. Fridjonsdottir, T. Vallianatou, P. Kallback, L. Katan, J. Savmarker, I. Mantas, X. Q. Zhang, E. Bezard, P. Svenningsson, L. R. Odell and P. E. Andren, Comprehensive mapping of neurotransmitter networks by MALDI-MS imaging, Nat. Methods, 2019, 16, 1021–1028, DOI:10.1038/s41592-019-0551-3.
- S. Xu, Y. Li, H. Zou, J. Qiu, Z. Guo and B. Guo, Carbon nanotubes as assisted matrix for laser desorption/ionization time-of-flight mass spectrometry, Anal. Chem., 2003, 75, 6191–6195, DOI:10.1021/ac0345695.
- J. Wei, J. M. Buriak and G. Siuzdak, Desorption-ionization mass spectrometry on porous silicon, Nature, 1999, 399, 243–246, DOI:10.1038/20400.
- C.-T. Chen and Y.-C. Chen, Fe3O4/TiO2 core/shell nanoparticles as affinity probes for the analysis of phosphopeptides using TiO2 surface-assisted laser desorption/ionization mass spectrometry, Anal. Chem., 2005, 77, 5912–5919, DOI:10.1021/ac050831t.
- H. Wang, X. Zhao, Y. Huang, J. Liao, Y. Liu and Y. Pan, Rapid quality control of medicine and food dual purpose plant polysaccharides by matrix assisted laser desorption/ionization mass spectrometry, Analyst, 2020, 145, 2168–2175, 10.1039/C9AN02440A.
- P. C. Lin, M. C. Tseng, A. K. Su, Y. J. Chen and C. C. Lin, Functionalized magnetic nanoparticles for small-molecule isolation, identification, and quantification, Anal. Chem., 2007, 79, 3401–3408, DOI:10.1021/ac070195u.
- M. C. Tseng, R. Obena, Y. W. Lu, P. C. Lin, P. Y. Lin, Y. S. Yen, J. T. Lin, L. D. Huang, K. L. Lu, L. L. Lai, C. C. Lin and Y. J. Chen, Dihydrobenzoic acid modified nanoparticle as a MALDI-TOF MS matrix for soft ionization and structure determination of small molecules with diverse sctructures, J. Am. Soc. Mass Spectrom., 2010, 21, 1930–1939, DOI:10.1016/j.jasms.2010.08.001.
- X. Zhao, C. Guo, Y. Huang, L. Huang, G. Ma, Y. Liu, Q. He, H. Wang, K. Chen and Y. Pan, Combination strategy of reactive and catalytic matrices for qualitative and quantitative profiling of N-Glycans in MALDI-MS, Anal. Chem., 2019, 91, 9251–9258, DOI:10.1021/acs.analchem.9b02144.
- S. E. Bailey, T. J. Olin, R. M. Bricka and D. D. Adrian, A review of potentially low-cost sorbents for heavy metals, Water Res., 1999, 33, 2469–2479, DOI:10.1016/S0043-1354(98)00475-8.
- J. Zakzeski, P. C. A. Bruijnincx, A. L. Jongerius and B. M. Weckhuysen, The catalytic valorization of lignin for the production of renewable chemicals, Chem. Rev., 2010, 110, 3552–3599, DOI:10.1021/cr900354u.
- N. Balasundram, K. Sundram and S. Samman, Phenolic compounds in plants and agri-industrial by-products: antioxidant activity, occurrence, and potential uses, Food Chem., 2006, 99, 191–203, DOI:10.1016/j.foodchem.2005.07.042.
- A. J. Ragauskas, C. K. Williams, B. H. Davison, G. Britovsek, J. Cairney, C. A. Eckert, J. R. Frederick, W. J. Hallett, J. P. Leak, D. J. Liotta, C. L. Mielenz, J. R. Murphy, R. Templer and R. T. Tschaplinski, The path forward for biofuels and biomaterials, Science, 2006, 311, 484–489, DOI:10.1126/science.1114736.
- F. Lu and J. Ralph, Derivatization followed by reductive cleavage (DFRC Method), a new method for lignin analysis: protocol for analysis of DFRC monomers, J. Agric. Food Chem., 1997, 45, 2590–2592, DOI:10.1021/jf970258h.
- R. Domitrović, K. Rashed, O. Cvijanović, S. Vladimir-Knežević, M. Škoda and A. Višnić, Myricitrin exhibits antioxidant, anti-inflammatory and antifibrotic activity in carbon tetrachloride-intoxicated mice, Chem.-Biol. Interact., 2015, 230, 21–29, DOI:10.1016/j.cbi.2015.01.030.
- J. Kamoun, R. Rahier, M. Sellami, I. Koubaa, P. Mansuelle, R. Lebrun, A. Berlioz-Barbier, M. Fiore, K. Alvarez, A. Abousalham, F. Carriere and A. Aloulou, Identification of a new natural gastric lipase inhibitor from star anise, Food Funct., 2019, 10, 469–478, 10.1039/C8FO02009D.
- D. K. Emwal, R. B. Semwal, S. Combrinck and A. Viljoen, Myricetin: a dietary molecule with diverse biological activities, Nutrients, 2016, 8, 90, DOI:10.3390/nu8020090.
- H. Liu, X. Qi, S. Cao and P. Li, Protective effect of flavonoid extract from Chinese bayberry (Myrica rubra Sieb. et Zucc.) fruit on alcoholic liver oxidative injury in mice, J. Nat. Med., 2014, 68, 521–529, DOI:10.1007/s11418-014-0829-9.
- Q. Gao, R. Ma, L. Chen, S. Shi, P. Cai, S. Zhang and H. Xiang, Antioxidant profiling of vine tea (Ampelopsis grossedentata): Off-line coupling heart-cutting HSCCC with HPLC–DAD–QTOF-MS/MS, Food Chem., 2017, 225, 55–61, DOI:10.1016/j.foodchem.2016.11.122.
- S. Y. Mok and S. Lee, Identification of flavonoids and flavonoid rhamnosides from Rhododendron mucronulatum for. albiflorum and their inhibitory activities against aldose reductase, Food Chem., 2013, 136, 969–974, DOI:10.1016/j.foodchem.2012.08.091.
- Y. L. Liu, X. N. Zhang, L. H. Zhan, C. Xu, L. X. Sun, H. M. Jiang, C. D. Sun and X. Li, LC-Q-TOF-MS, characterization of polyphenols from white bayberry fruit and its antidiabetic effect in KK-Ay mice, ACS Omega, 2020, 5, 17839–17849, DOI:10.1021/acsomega.0c02759.
- Q. Lyu, X. Wen, Y. Liu, C. Sun, K. Chen, C. C. Hsu and X. Li, Comprehensive profiling of phenolic compounds in white and red Chinese bayberries (Morella rubra Sieb. et Zucc.) and their developmental variations using tandem Mass spectral molecular networking, J. Agric. Food Chem., 2021, 69, 741–749, DOI:10.1021/acs.jafc.0c04117.
- D. H. Chace, T. A. Kalas and E. W. Naylor, Use of tandem mass spectrometry for multianalyte screening of dried blood specimens from newborns, Clin. Chem., 2003, 11, 1797–1817, DOI:10.1373/clinchem.2003.022178.
- A. J. Atkinson Jr., W. A. Colburn, V. G. DeGruttola, D. L. DeMets, G. J. Downing, D. F. Hoth, J. A. Oates, C. C. Peck, R. T. Schooley, B. A. Spilker, J. Woodcock and S. L. Zeger, Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework, Clin. Pharmacol. Ther., 2001, 3, 89–95, DOI:10.1067/mcp.2001.113989.
- L. Sleno and D. A. Volmer, Assessing the properties of internal standards for quantitative matrix-assisted laser desorption/ionization mass spectrometry of small molecules, Rapid Commun. Mass Spectrom., 2006, 20, 1517–1524, DOI:10.1002/rcm.2498.
- P. Wang and R. W. Giese, Recommendations for quantitative analysis of small molecules by matrix-assisted laser desorption ionization mass spectrometry, J. Chromatogr. A, 2017, 1468, 35–41, DOI:10.1016/j.chroma.2017.01.040.
- J. Wang and P. Sporns, MALDI-TOF MS quantification of coccidiostats in poultry feeds, J. Agric. Food Chem., 2000, 48, 2807–2811, DOI:10.1021/jf000193+.
- V. Vukics and A. Guttman, Structural characterization of flavonoid glycosides by multi-stage mass spectrometry, Mass Spectrom. Rev., 2010, 29, 1–16, DOI:10.1002/mas.20212.
- X. Wang, J. Han, A. Chou, J. Yang, J. Pan and C. H. Borchers, Hydroxyflavones as a new family of matrices for MALDI tissue imaging, Anal. Chem., 2013, 85, 7566–7573, DOI:10.1021/ac401595a.
- X. J. Shen, T. Y. Chen, H. M. Wang, Q. Q. Mei, F. X. Yue, S. N. Sun, J. L. Wen, T. Q. Yuan and R. C. Sun, Structural and morphological transformations of lignin macromolecules during bio-based deep eutectic solvent (DES) pretreatment, ACS Sustainable Chem. Eng., 2020, 8, 2130–2137, DOI:10.1021/acssuschemeng.9b05106.
- L. Ling, C. Xiao, Y. Ma, L. Jiang, S. Wang, L. Guo, S. Jiang and X. Guo, 2-Phenyl-3-(p-aminophenyl) acrylonitrile: a reactive matrix for sensitive and selective analysis of glycans by MALDI-MS, Anal. Chem., 2019, 91, 8801–8807, DOI:10.1021/acs.analchem.9b01434.
- X. Wang, J. Han, A. Chou, J. Yang, J. Pan and C. H. Borchers, Hydroxyflavones as a New Family of Matrices for MALDI Tissue Imaging, Anal. Chem., 2013, 85(15), 7566–7573 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The abbreviations of all analytes. See DOI: 10.1039/d1an01632f |
| ‡ Equal contribution to this work. |
| This journal is © The Royal Society of Chemistry 2021 |