Metabolomics-based mass spectrometry methods to analyze the chemical content of 3D organoid models
Shannon E.
Murphy
 and
Jonathan V.
Sweedler
and
Jonathan V.
Sweedler
 *
*
Department of Chemistry, University of Illinois at Urbana-Champaign, 600 South Mathews Avenue, Urbana, Illinois 61801, USA. E-mail: jsweedle@illinois.edu
First published on 30th May 2022
Abstract
Metabolomics, the study of metabolites present in biological samples, can provide a global view of sample state as well as insights into biological changes caused by disease or environmental interactions. Mass spectrometry (MS) is commonly used for metabolomics analysis given its high-throughput capabilities, high sensitivity, and capacity to identify multiple compounds in complex samples simultaneously. MS can be coupled to separation methods that can handle small volumes, making it well suited for analyzing the metabolome of organoids, miniaturized three-dimensional aggregates of stem cells that model in vivo organs. Organoids are being used in research efforts to study human disease and development, and in the design of personalized drug treatments. For organoid models to be useful, they need to recapitulate morphological and chemical aspects, such as the metabolome, of the parent tissue. This review highlights the separation- and imaging-based MS-based metabolomics methods that have been used to analyze the chemical contents of organoids. Future perspectives on how MS techniques can be optimized to determine the accuracy of organoid models and expand the field of organoid research are also discussed.
1. Introduction
Organoids are three-dimensional (3D) aggregates of stem cells that differentiate to form organ-like structures that model some of the characteristics of in vivo organs, such as the gut, liver, brain, retina, and kidney, among others.1 While many organoids are created using cells from animal models, they can also can be grown using induced pluripotent stem (iPSCs) cells derived from patients, providing great potential for disease modeling, drug discovery, development of personalized therapeutics, and drug toxicity testing.2 In addition, organoids can model biological processes related to brain development and diseases, such as microcephaly, that are specific to the human body and difficult to accurately represent using rodent models.3,4 The 3D structure of organoids enables them to better recapitulate the developmental biology, human physiology, and complexity of an in vivo organ compared to using two-dimensional cell culture models.3,5One commonly used culturing technique to obtain 3D organoid structures is the extracellular matrix (ECM) scaffolding method (Fig. 1a). In this method, cells are plated on an ECM (Matrigel is commonly used) and maintained under culture conditions.5 The PSC are then expanded and differentiated through multistep protocols via the addition of growth factors into the cell culture media.6,7 These growth factors resemble developmental cues needed to differentiate the PSCs into the organoid model of interest.7 Other methods for culturing organoids include the spinning bioreactor method, hanging drop method, low-adherent cell culture plate method, and magnetic levitation method (Fig. 1). One drawback of the ECM scaffolding method is that the use of Matrigel introduces variability as not every batch is the same.5 Research has shown that organoids demonstrate batch-to-batch variability,8,9 and this variability could be related to the variance of the Matrigel composition.10 Additionally, Matrigel has several components that can interfere with mass spectrometry (MS) analysis.11,12 Sample preparation methods to reduce the interference from Matrigel components are discussed in Sections 2.1 and 2.3.
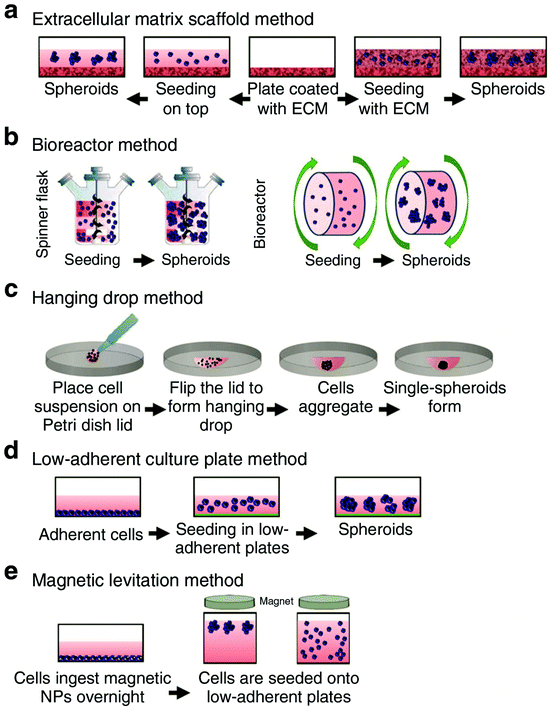 | ||
| Fig. 1 Schematic illustrating several of the methods used for culturing organoids. These methods include (a) extracellular matrix scaffold, (b) bioreactor, (c) hanging drop, (d) low-adherent culture plate, and (e) magnetic levitation method. Adapted from ref. 5 with permission under Creative Commons Attribution (CC-BY 4.0) license (https://creativecommons.org/licenses/by/4.0/). | ||
Organoids refer to aggregates of organ-specific cells, whereas similar 3D cell culture models known as spheroids refer to clusters of free-floating cells that typically mimic tumor organization and are often established from immortalized cell lines. Even though spheroids are generally lower in complexity than organoid models, the terms organoids, spheroids, and 3D cell cultures are often used interchangeably in the literature.10 We focus our discussion on studies that used the term organoids to describe their 3D cell culture systems. Previous research has shown that organoids can accurately model parent tissue in terms of anatomical structure and aspects of human development.4,13–15 However, examining the morphology of organoids does not provide insight into their chemical composition and the chemical interplay within these tissue-like structures. Can metabolomics techniques be used to characterize the complex intercellular chemistry of organoids?
Metabolomics investigations analyze the flux and chemical movement of metabolites in a biological sample, producing a comprehensive representation of its composition.16,17 Global metabolomics analysis can provide insights into biological changes caused by disease or environmental interactions and reveal information about the potential mechanisms involved, which can help to identify possible biomarkers that may improve diagnosis and treatment of diseases.18 MS is a commonly used technique for metabolomics analysis due to its sensitivity, high throughput, and ability to detect a large number of molecules in complex biological samples.19 MS-based metabolomics analysis has been applied to biomarker discovery,20 drug development,21 embryonic development,22 and toxicology.23
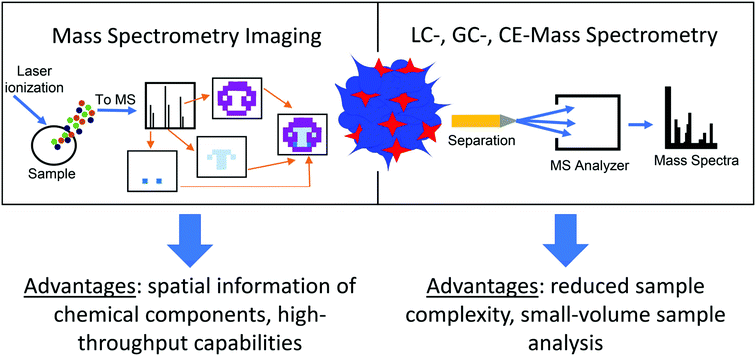
While organoids include the various cell types found in the organ being replicated, they are rarely at the same scale and are usually much smaller, typically less than 1 millimeter in diameter.5,24 Their small size poses a difficult analytical challenge when measuring their chemical contents, as discussed in a recent review.24 MS is an advantageous approach for organoid analysis because it can be coupled to separation techniques, such as nano-liquid chromatography (nano-LC) or capillary electrophoresis (CE), which can handle small-volume samples.24,25 Additional benefits include the ability to measure many compounds simultaneously without analyte preselection while providing low limits of detection and high sensitivity, making MS particularly well suited to detection of low amounts of analytes in organoid samples.26 Additionally, MS imaging (MSI) techniques, such as matrix-assisted laser desorption/ionization (MALDI), can provide spatial details about the chemical content of organoids, offering another layer of information in measuring the metabolome of organoid models (Fig. 2).27 While several reviews covering the use of MS for organoid analysis have been published,12,28–30 this work focuses on metabolomics measurements in organoids using separation- and imaging-based MS methods. We describe applications beyond cancer research and drug discovery, and present some of the latest research in this rapidly evolving field. We conclude our discussion with future perspectives on how MS-based technologies can be used to further explore the chemical content of organoids.
2. Metabolomics methods for the analysis of organoids
2.1. Mass spectrometry coupled with separation techniques
Separation methods, such as gas chromatography (GC) or liquid chromatography (LC), are used to fractionate the chemical components of a sample mixture and are commonly coupled to information-rich detection method such as MS for analyte identification and quantification. Sample components are separated from each other reducing the chemical complexity in a sample and isolating molecular interferants from signals of interest. Coupling GC or LC to MS leverages the advantages of both techniques. Performing the separation before MS detection makes it easier to identify the components of interest in a complex sample, and improves the analytical figures of merit for both qualitative and quantitative experiments.31,32 Perhaps one of the first examples of combining a separation step with MS for analyzing the chemical content of 3D cell culture models was performed in 2008 by van Vliet et al.33 The authors used LC-MS to measure metabolic alterations in an in vitro 3D cell culture model of the developing brain in response to methyl mercury chloride and caffeine treatment. By performing metabolomics measurements to detect neurotoxicity and discover new biomarkers, this innovative approach paved the way for future metabolomics studies using in vitro 3D cell culture models. In this section we examine some recent works that have applied LC-MS and GC-MS metabolomics methods to analyze organoids used to investigate disease development, drug exposure, variability based on the source of the cells, and the impacts of environmental exposure to healthy development. We focus our discussion on new insights learned from these measurements; for technical details about the measurements themselves, we refer the reader to the cited literature.An intriguing application of LC- and GC-MS metabolomics-based methods is to determine how the chemistry of organoids tracks the development of disease, and how this differs from healthy development. Gomez-Giro et al.34 used GC-MS to study the metabolic differences between juvenile neuronal ceroid lipofuscinosis (JNCL, also referred to as CLN3 disease) and control cerebral organoids to understand how fundamental neurodevelopmental mechanisms are altered in the CLN3 mutant organoids. Thirty-one metabolites were found to be significantly different between the control and the CLN3 mutant organoids, with most of them being significantly decreased in the CLN3 mutant organoid (Fig. 3a). Neurotransmitter production in the CLN3 mutant organoid was also found to be downregulated compared to control organoids (Fig. 3b). These results demonstrate how the global measurement of metabolites in organoids advances our understanding of the development of a rare genetic disorder like CLN3 when patient tissue is hard to obtain. In a similar study,35 GC-MS was used to examine metabolomic differences between early (within six months) and late (later than six months) recurrent pancreatic ductal adenocarcinoma (PDAC) organoid models. Using patient-derived organoids (PDOs), partial least squares discriminant analysis showed significant global alterations between early- and late-recurrent PDOs; nine metabolites were increased in the early-recurrent compared to late-recurrent PDOs. Understanding metabolomic differences between early- and late-recurrent PDAC provides insights into creating therapeutic interventions to delay PDAC recurrence and prolong patient survival. Additionally, these results help guide the development of biomarkers that could predict disease recurrence in patients, further illustrating how global metabolomics measurements can aid in understanding disease progression using organoid models.
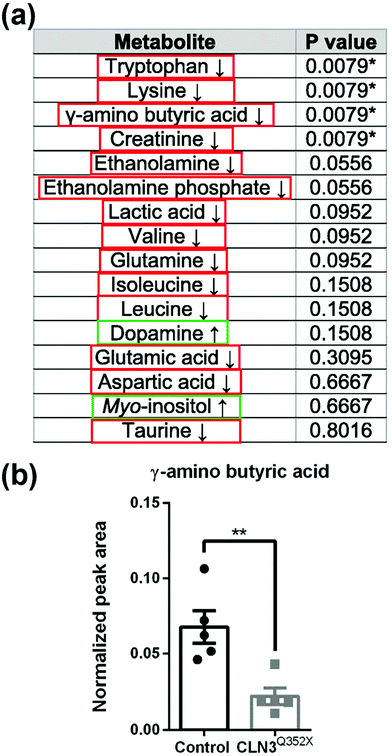 | ||
| Fig. 3 Detection of metabolites in organoids using GC-MS. (a) Metabolites detected in cerebral organoids with arrows indicating relative increase or decrease in CLN3 mutant organoids compared to the controls. Red = relative decrease compared to control. Green = relative increase compared to control. Asterisks indicate significantly deregulated metabolites. (b) The neurotransmitter GABA (γ-amino butyric acid) is significantly downregulated in CLN3 mutant organoids compared to control organoids. Adapted from ref. 34 with permission under Creative Commons Attribution (CC-BY 4.0) license (https://creativecommons.org/licenses/by/4.0/). | ||
LC-MS methods have also been used to analyze the metabolic response of organoids to drug exposure. Duong et al.36 performed LC-MS metabolomics measurements on control and drug-resistant colorectal cancer organoids to understand the signaling pathways that produce mutations in colorectal cancer. Severe metabolic reprogramming was observed in the drug-resistant compared to control organoids. Drug resistance was found to be influenced by the proteins CEMIP (cell migration-inducing and hyaluronan-binging protein) and Myc (transcription factors which play an important role in regulating cell proliferation and apoptosis). This understanding that CEMIP can regulate the levels of specific metabolites through the Myc-associated signaling pathway and cause drug resistance has great therapeutic potential for treating drug-resistant colorectal cancers. Neef and colleagues37 also performed metabolic profiling of colorectal cancer organoids in response to 5-fluorouracil treatment using LC-MS. Dose-dependent changes in the metabolomic profiles of treated organoids were mapped and the upregulated metabolites found in response to drug treatment were mainly involved in purine and pyrimidine metabolism, aligning with the mechanism of action of 5-fluorouracil. In addition, the elevated levels of 2′-deoxyuridine and depleted levels of 2′-deoxyadenosine found in treated organoids conformed with the cellular mechanism of 5-fluorouracil. The authors also developed a novel protocol for metabolomic profiling of organoids that helped overcome the challenge of dealing with small sample amounts (<500 cells per injection) in the presence of an ECM (Matrigel). Sample preparation consisted of rapid on-plate washing followed by cold methanol extraction without the removal of the ECM, which allowed for rapid quenching of metabolic reactions and extraction of metabolites with minimal cell manipulation. They also implemented a data filtering procedure based on fold change and p-value cut offs of ECM peaks in biological vs. ECM blank samples to eliminate ECM-derived background signals that interfere with metabolite analysis. This filtering method has the advantage of removing features with high variability, improving the repeatability of organoid sample analysis. This newly developed protocol reduces sample preparation time and can be applied to high-throughput drug screening, biomarker discovery, and personalized therapies.
The potential of using liver organoids with LC-MS measurements to perform drug metabolism studies has been shown by Skottvoll et al.38 To study the phase I metabolism of heroin to 6-monoacetylmorphine (6-MAM) and morphine, liver organoids were exposed to the drug for 1, 3, 6, and 24 hours, with the concentrations of heroin and 6-MAM tracked during those timepoints. The authors developed an electromembrane extraction approach to separate drug metabolites from components of cell culturing media, allowing for simple and repeatable measurements. Parallel electromembrane extractions were performed in a 96-well plate, illustrating the high-throughput nature of this extraction method. Collectively, these studies demonstrate how MS-based metabolomics can be used to understand metabolic pathways in cancer in response to drug treatments, develop therapeutic approaches to treat these cancers, and optimize sample preparation methods for high-throughput analysis of organoid samples.
MS-based methods have also been used to provide insight into the chemical variability of organoid models created from different cell sources. Organoid cultures often contain multiple cell types, so it is important to understand their variability as organoids are being used to model disease, drug screening, and drug testing. Human gut adult stem cell (ASC) organoids were created from six different donors to better understand donor-to-donor variability in human organoids, which has not been well studied.39 In this work, 108 central carbon metabolites were profiled at multiple developmental timepoints in the ASC organoids. Differences in metabolism between the timepoints were found because donors from the same timepoint tended to cluster together, but there was minimal donor-to-donor variability. The authors also found that metabolic profiles in the ASC organoids were consistent with what is known about their physiological function, indicating that the organoids were recapitulating key features of the originating tissue. The simultaneous measurement of hundreds of compounds in these ASC organoids demonstrates the advantage of using MS for analyzing biological variability in a model system to answer fundamental biological questions and advance therapeutic discovery.
Insight into how environmental and hormonal factors can influence organoid development can also be attained using MS technologies. One example is the work by Notaras et al.,40 where the authors created human-derived forebrain organoids and used LC-MS metabolomics methods to understand the prenatal effects of drugs and neuropsychiatric risk factors on early brain development. Using organoid models for this type of research overcomes the ethical and technical challenges of obtaining human prenatal brain tissue. Forebrain organoids were exposed over a seven-day period to chemically defined enviromimetic agents to mimic exposure to opiates (μ-opioid agonist endomorphin), cannabinoids (WIN 55212-2), alcohol (ethanol), smoking (nicotine), chronic stress (human cortisol), and maternal immune activation (human IL 17a). The metabolome of the exposed organoids was found to be subtly altered compared to vehicle-treated controls where most enviromimetic treatment groups exhibited at least one unique metabolome alteration. Common metabolome alterations in L-phenylalanine and guanosine triphosphate were detected in all but one of the treatment groups, supporting the idea that prenatal exposure to a variety of factors can alter the metabolome and other cellular processes that are critical for normal cortical development. Employing an integrative, multi-omics approach, the authors also performed proteomics measurements on exposed organoids to gain a more complete understanding of how environmental factors affect brain development. Multi-omics analyses are discussed in depth in Section 2.2.
To better understand how estrogens influence metabolism in the mammary gland and impact organoid development, mammary epithelial cell organoids were exposed to the hormone estradiol (E2).41 The E2 treatment was found to induce significant alterations in metabolite levels from several metabolic pathways, including the tricarboxylic acid cycle, urea cycle, and amino acid metabolism. These results provide insight into the relationship between hormones and metabolism in the mammary gland, with applications to studying how hormones reprogram metabolism during lactation and breast cancer. The ability to identify significantly altered metabolites of interest without analyte preselection demonstrates the advantage of using MS-based methods to analyze metabolic changes in organoids when exposed to chemical compounds and their impact on biological development.
Together these studies demonstrate the use of GC- and LC-MS methods for understanding the biology of organoids and how disease and exposure to drugs, hormones, or environmental factors alters metabolic pathways in these model systems. This information has applications for developing therapeutic approaches, personalized medicine, and studying normal and disease development in organs where patient tissue is difficult to access. Well-established LC-MS sampling preparation and techniques needed to be adapted to handle the small-volume requirements of organoid analysis. This was achieved through the novel sample preparation method developed by Neef et al.37 and the parallel electromembrane extraction approach developed by Skottvoll et al.38 Their work demonstrated how new high-throughput sample preparation methods allowed for the analysis of hundreds of organoids in a single experiment, enabling a more complete understanding of the metabolites present in organoid models.
2.2 Multi-omics analysis
Multi-omics integrates different-omics characterization methods (e.g., metabolomics and transcriptomics) to gain enhanced information on a sample. Each-omics technique provides distinct details and key insights into biological pathways that would not be discovered with single-omics methods.42,43 Though the application of multi-omics approaches to organoid research are not common, we describe two studies that suggest their potential. In the first, Lindeboom et al.44 performed multi-omics analysis—genomics, epigenomics, transcriptomics, proteomics, and metabolomics—on stem cell-enriched and stem cell-depleted mouse intestinal organoids to obtain a holistic understanding of the mechanisms that influence differential gene expression during intestinal stem cell differentiation (Fig. 4a). Integrating data from MS-based metabolomics and lipidomics analysis with gene expression dynamics revealed a correlation between the abundance of enzymatic activities and their involved metabolites. Results showed that a strong upregulation of glutathione in the stem cell-depleted organoids correlated with an upregulation of the enzymes related to glutathione metabolism (Fig. 4b). In the second, Shechter and colleagues45 combined transcriptomics, MS-based lipidomics and metabolomics, and DNA methylomics measurements to better understand the mechanisms involved with tumor relapse from treatment-resistant cells (minimal residual disease, or MRD). By combining transcriptomic and metabolic data and flux modeling, not only were metabolic alterations in the MRD organoids revealed, these MRD organoids held a metabolic memory of the prior tumor state. This multi-omics approach allowed for metabolic distinctions between normal, tumor, and MRD organoids to be made in a holistic fashion. Multi-omics analysis provides a strong framework for obtaining system-wide overview of molecular mechanisms in cultured organoids. However, there are some drawbacks, such as needing large quantities of cellular material for high-resolution data generation and requiring multiple areas of expertise to integrate, visualize, and interpret the collected data. Nonetheless, these studies illustrate the utility of MS-based metabolomics in a multi-omics context to deepen the understanding of mechanisms involved in healthy cell development and the development of disease using organoid models.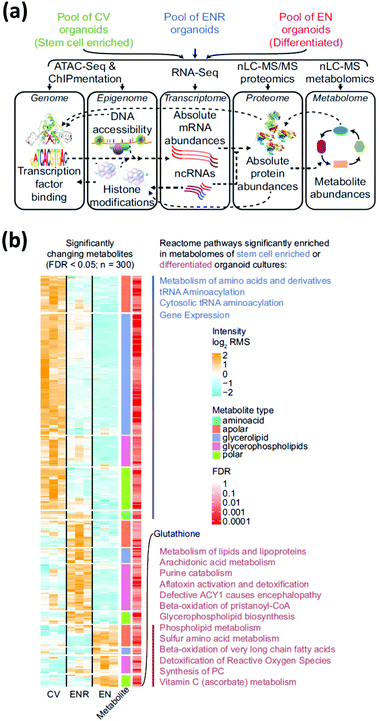 | ||
| Fig. 4 Multi-omic analysis of intestinal organoids provides a complete profile of cell-type enriched and depleted organoid cultures. (a) Multi-omic approaches require the combination of several assays. (b) Relative abundance of significantly changing metabolites in stem-cell enriched vs. stem-cell depleted intestinal organoids measured using MS metabolomics and lipidomics measurements. Significantly enriched pathways are indicated next to the corresponding cluster of metabolites. CV = stem-cell enriched organoids; ENR = organoids grown in regular culture media; EN = stem-cell depleted organoids. Adapted from ref. 44 with permission under Creative Commons Attribution (CC-BY 4.0) license (https://creativecommons.org/licenses/by/4.0/). | ||
2.3 Mass spectrometry imaging
MSI is an ionization approach in which a probe is rastered across a sample surface to allow untargeted examination of the spatial distribution of chemical species in the sample. Thousands of molecules can be imaged in a single, label-free experiment in a spatially resolved manner, providing added dimension to the MS data that is especially valuable when studying heterogenous samples. Several recent reviews highlight the details of the measurement process and information that can be obtained from MSI analysis.46–49 Although the spatial information can be useful, the depth of molecular coverage is less than that of separations-based MS approaches, which is why both approaches are often used on the same sample.50–52Researchers have applied MSI in areas ranging from pharmaceutical drug discovery to basic biomedical research.53,54 In one of the first examples to use MSI to analyze the chemical composition of a 3D cell culture system, MALDI-MSI was used in combination with nano-LC-tandem MS to map and identify the protein distribution in colon carcinoma spheroid models.27 The MALDI-MSI approach was developed to overcome limitations of other visualization methods, such as immunohistochemistry, where prior knowledge of the analytes of interest is needed. Because MALDI is the most common ionization approach used with MSI to analyze organoids,55 this section focuses on MALDI-MSI approaches.
MALDI-MS has been used to track drug distribution in organoids, allowing for the potential to track patient-specific responses to drug treatments. The Hummon group56 used MALDI-MSI to study the behavior of colorectal tumor organoids (CTOs) created from two patients to analyze how the CTOs responded to the anti-cancer drug irinotecan and its metabolites. Building on prior work57 using MALDI-MSI to examine the spatial distribution of irinotecan and its metabolites in colon carcinoma multicellular spheroid models, they used organoids instead of spheroids because spheroids demonstrate a limited morphological, phenotypic, and genetic heterogeneity compared to in vivo tumors. Spheroids tend to be less complex than organoids and are typically spherical in shape, perhaps limiting their ability to fully capture the complex morphological structures found in patient tumors.10,56 By studying the distribution of irinotecan and its metabolites in CTOs, Hummon and colleagues mapped drug uptake over time and in a concentration-dependent manner in the organoid models (Fig. 5). The high-throughput nature of MALDI-MS allowed them to analyze thousands of CTOs in a single experiment, illustrating how MALDI-MS methods are helpful for collecting and evaluating multiple aspects of therapeutic drug responses by mapping the distribution in both treatment-time and concentration-dependent manners.
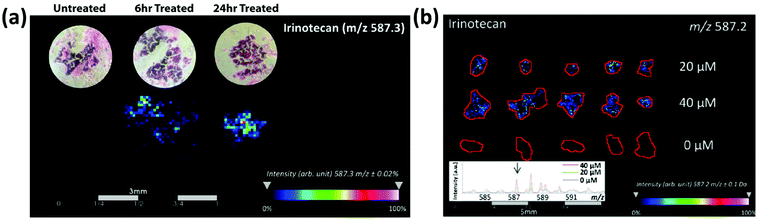 | ||
| Fig. 5 MALDI-MSI of colorectal tumor organoids reveals the time and concentration distribution dynamics of irinotecan. (a) MALDI-MSI intensity maps showing the time-dependent distribution of irinotecan in colorectal tumor organoid samples. (b) MALDI-MSI ion density maps showing the concentration-dependent distribution of 0, 20, and 40 μM irinotecan-treated colorectal tumor organoid samples. Adapted from ref. 56 with permission from the American Chemical Society, copyright 2018. | ||
In another interesting application, the technique has been used to track drug transport in organoids that do not model tumors, such as blood–brain barrier (BBB) organoids. Bergmann et al.58 illustrated the use of MALDI-MS for high-throughput analysis of the permeability of the drugs BKM-120 and dabrafenib across BBB organoids. High intensities of BKM-120 were detected in BBB organoids whereas no levels of dabrafenib were found. This correlates with the known permeability of these compounds; BKM-120 is known to cross the BBB but dabrafenib has limited BBB penetration. These results demonstrate the ability of MALDI-MS analyses to track drugs of interest in organoid models and contribute to the development of therapeutics for disease treatments. Interestingly, though this work is based on previously published work by Cho et al.59 who used the term spheroid to describe the 3D BBB model, Bergmann and colleagues used the term organoid to describe the same 3D model. This incongruity in terminology highlights how researchers use the terms spheroid and organoid interchangeably.
MALDI-MS can also be used to study the effects of drug treatment on organoid models. Using primary tumor organoids, Johnson et al.60 demonstrated the use of MALDI-MSI for studying drug response. Tumor organoids were treated with the drugs 5-fluorouracil (5-FU) and gemcitabine (GEM), a common combination therapy given to pancreatic cancer patients. While the parent drugs 5-FU and GEM and their metabolites were not detected in the organoids, the authors did identify metabolites that were significantly different between the control and treated organoids. These identified metabolites were correlated with the effectiveness of the drug treatment and drug excretion products surrounding the organoids. The authors also developed new sample preparation methods for organoids that improved MALDI-MSI analysis, such as the use of a gelatin microarray technology (trade name Matrigel), which enabled tens to hundreds of organoids to be aligned for high-throughput imaging. Metabolite interference peaks seen in the MALDI-MSI resulting from the Matrigel that the organoids were cultured in were removed by centrifuging the organoids out of the gel. These methods help reduce sample preparation time and improve throughput, demonstrating how MALDI-MSI can be applied for drug development research with organoid models.
Another sample preparation methods using MALDI-MSI to analyze the chemical content of organoids have been presented is the protocol by Bakker et al.61 that allows for accurate molecular identification based on tandem MS in organoids <600 μm in diameter while also preserving the 3D structure and the spatiotemporal distribution of molecules in the organoids. In this sample preparation method, organoids are centrifuged into a gelatin microarray that contains holes of either 200 or 800 μm, aligning the organoids into a pattern amenable to MSI. While this protocol was designed to detect lipids in organoid samples, it can be adjusted to detect other types of small molecules by modifying the MALDI matrix. The improved sample preparation methods presented by Johnson et al. and Bakker et al. will facilitate the use of MSI methods for organoid analysis beyond the research discussed here, and could easily be adapted to perform other types of MS analysis with organoid samples; e.g., LC-, CE-, or GC-MS.
Overall, the studies discussed here demonstrate the potential of using MALDI-MSI to evaluate the therapeutic response of drug treatments in organoid models. MALDI-MSI can measure drug response in single organoids as well as track the location of the drug and its metabolites over time. Additionally, the high-throughput capabilities of MALDI-MS allow for the analysis of hundreds to thousands of organoids in a single experiment,56,60 with implications for pharmaceutical and basic research applications. While the focus of these papers has been on the use of MALDI-MSI for analyzing drug responses in organoids, these methods are adaptable to other studies using organoid models. One such example is to measure the distribution of the chemical contents in organoids to determine how similar their chemical composition is to in vivo tissue and therefore, how accurately they model normal function and health. Using techniques such as MALDI-MS to analyze the chemical contents of organoids will help guide researchers in choosing the most applicable model system to answer their research questions related to disease and development, as detailed below.
3. Future perspectives
Organoid characterization using MS-based technologies has advanced rapidly over the past several years and this trend is expected to accelerate. Recent metabolomics studies demonstrate the advantages of using organoid models to increase our understanding of the molecular mechanisms involved in human development and disease, including rare diseases where patient tissue is difficult to obtain, including applications to personalized medicine. New methods have been developed to overcome the challenges of analyzing small organoid samples grown in an extracellular matrix, which can introduce a chemical background and to enable analysis of hundreds of organoids in a single experiment in a high-throughput manner. The impact of recent studies is expected to grow as the reported metabolomic information is used for follow-up efforts.What future developments are expected? In addition to MALDI, other MSI ionization approaches, such as secondary ionization mass spectrometry (SIMS) and desorption electrospray ionization (DESI), can be adapted for organoid analysis. SIMS, which offers high spatial resolutions of less than 500 nm, but typically provides less information on chemical content, is advantageous when finer spatial information is needed.62,63 Unlike SIMS and MALDI, DESI does not need to be performed under high vacuum and therefore allows for the analysis of releasates from hydrate samples under atmospheric conditions. Additionally, sample preparation for DESI is simpler because no matrix application is needed, and the sample does not need to be dried before analysis.64 Thus far, DESI methods have been used to analyze 3D cell culture systems and not organoids, but should be adaptable for this application.65
In the area of separations-based MS chemical characterization, it appears that no studies have yet applied CE-MS to the metabolomics analysis of organoids. Nonetheless, the low-volume capabilities of CE-MS make it well suited for analyzing the chemical contents of small organoid samples and even single organoids. With the current methods used to create organoids, there is a high amount of heterogeneity between organoids grown in the same batch, leading to high inter-experimental variation.66 CE-MS and MALDI-MS techniques can be used to analyze individual organoids to further understand these variations.
There has been a large effort to adapt both CE-MS and MALDI-MS for single cell tissue studies,67–72 and these methods can also be adapted for organoid studies; for example, to determine which specific types of cells are present in organoids. To ensure accuracy when using organoids, it is important to fully understand how well they model the parent tissue. Detailed MSI and single cell studies have the potential to provide this information.
Even if organoids have the appropriate morphology and other physical characteristics, more research is needed to determine if organoids contain the correct molecular content and chemical dynamics to accurately emulate the parent tissue. One potential option is to assess this in a functional context, for example, by performing stimulation experiments where both organoids and the parent tissue are stimulated and their chemical releasates compared. Though researchers have analyzed the neurotransmitter release of organoids,73,74 no studies to date have analyzed organoid release after stimulation in order to compare their function to parent tissue. If a discrepancy is found between the chemical content of organoids and the parent tissue, this information would help bioengineers and neuroscientists develop new protocols for generating improved organoid models.
Overall, the perspectives presented in this review offer insight on how MS-based methods can play an important role in the optimization of organoid model systems for the study of human development, disease, and drug response.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors acknowledge support by Award Number P30 DA018310 from the National Institute on Drug Abuse (J. V. S.), and the National Science Foundation under grant 1735252 (S. E. M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.References
- M. A. Lancaster and J. A. Knoblich, Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies, Science, 2014, 345, 1247125, DOI:10.1126/science.1247125.
- S. Bartfeld and H. Clevers, Stem Cell-Derived Organoids and Their Application for Medical Research and Patient Treatment, J. Mol. Med., 2017, 95, 729–738, DOI:10.1007/s00109-017-1531-7.
- J. Kim, B. K. Koo and J. A. Knoblich, Human Organoids: Model Systems for Human Biology and Medicine, Nat. Rev. Mol. Cell Biol., 2020, 21, 571–584, DOI:10.1038/s41580-020-0259-3.
- M. A. Lancaster, M. Renner, C. A. Martin, D. Wenzel, L. S. Bicknell, M. E. Hurles, T. Homfray, J. M. Penninger, A. P. Jackson and J. A. Knoblich, Cerebral organoids model human brain development and microcephaly, Nature, 2013, 501, 7467, DOI:10.1038/nature12517.
- V. Velasco, S. A. Shariati and R. Esfandyarpour, Microtechnology-Based Methods for Organoid Models, Microsyst. Nanoeng., 2020, 6, 76, DOI:10.1038/s41378-020-00185-3.
- F. Schutgens and H. Clevers, Human Organoids: Tools for Understanding Biology and Treating Diseases, Anu. Rev. Pathol. Mech. Dis., 2020, 15, 211–243, DOI:10.1146/annurev-pathmechdis.
- C. Corrò, L. Novellasdemunt and V. W. Li, A Brief History of Organoids, Am. J. Physiol. Cell Physiol., 2020, 319, 151–165, DOI:10.1152/ajpcell.00120.2020.
- G. Quadrato, T. Nguyen, E. Z. Macosko, J. L. Sherwood, S. M. Yang, D. R. Berger, N. Maria, J. Scholvin, M. Goldman, J. P. Kinney, E. S. Boyden, J. W. Lichtman, Z. M. Williams, S. A. McCarroll and P. Arlotta, Cell Diversity and Network Dynamics in Photosensitive Human Brain Organoids, Nature, 2017, 545(7652), 48–53, DOI:10.1038/nature22047.
- B. Phipson, P. X. Er, A. N. Combes, T. A. Forbes, S. E. Howden, L. Zappia, H. J. Yen, K. T. Lawlor, L. J. Hale, J. Sun, E. Wolvetang, M. Takasato, A. Oshlack and M. H. Little, Evaluation of Variability in Human Kidney Organoids, Nat. Methods, 2019, 16(1), 79–87, DOI:10.1038/s41592-018-0253-2.
- S. Gunti, A. T. K. Hoke, K. P. Vu and N. R. London, Organoid and Spheroid Tumor Models: Techniques and Applications, Cancers, 2021, 13(4), 874, DOI:10.3390/cancers13040874.
- M. Wang, H. Yu, T. Zhang, L. Cao, Y. Du, Y. Xie, J. Ji and J. Wu, In-Depth Comparison of Matrigel Dissolving Methods on Proteomic Profiling of Organoids, Mol. Cell. Proteomics, 2022, 21(1), 100181, DOI:10.1016/j.mcpro.2021.100181.
- Y. Wang and A. B. Hummon, MS Imaging of Multicellular Tumor Spheroids and Organoids as an Emerging Tool for Personalized Medicine and Drug Discovery, J. Biol. Chem., 2021, 297(4), 101139, DOI:10.1016/j.jbc.2021.101139.
- C. A. Trujillo, R. Gao, P. D. Negraes, J. Gu, J. Buchanan, S. Preissl, A. Wang, W. Wu, G. G. Haddad, I. A. Chaim, A. Domissy, M. Vandenberghe, A. Devor, G. W. Yeo, B. Voytek and A. R. Muotri, Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development, Cell Stem Cell, 2019, 25(4), 558–569, DOI:10.1016/j.stem.2019.08.002 .e7.
- J. M. Nascimento, V. M. Saia-Cereda, R. C. Sartore, R. M. da Costa, C. S. Schitine, H. R. Freitas, M. Murgu, R. A. de Melo Reis, S. K. Rehen and D. Martins-de-Souza, Human Cerebral Organoids and Fetal Brain Tissue Share Proteomic Similarities, Front. Cell Dev. Biol., 2019, 7, 303, DOI:10.3389/fcell.2019.00303.
- M. Takasato, P. X. Er, H. S. Chiu, B. Maier, G. J. Baillie, C. Ferguson, R. G. Parton, E. J. Wolvetang, M. S. Roost, S. M. C. de Sousa Lopes and M. H. Little, Kidney Organoids from Human IPS Cells Contain Multiple Lineages and Model Human Nephrogenesis, Nature, 2015, 526(7574), 564–568, DOI:10.1038/nature15695.
- E. Riekeberg and R. Powers, New Frontiers in Metabolomics: From Measurement to Insight, F1000Research, 2017, 6, 1148, DOI:10.12688/f1000research.11495.1.
- C. B. Clish, Metabolomics: An Emerging but Powerful Tool for Precision Medicine, Mol. Case Stud., 2015, 1(1), a000588, DOI:10.1101/mcs.a000588.
- C. H. Johnson and F. J. Gonzalez, Challenges and Opportunities of Metabolomics, J. Cell Physiol., 2012, 227(8), 2975–2981, DOI:10.1002/jcp.24002.
- X. W. Zhang, Q. H. Li, Z. D. Xu and J. J. Dou, Mass Spectrometry-Based Metabolomics in Health and Medical Science: A Systematic Review, RSC Adv., 2020, 10, 3092–3104, 10.1039/C9RA08985C.
- Y. Fan, Y. Li, Y. Chen, Y. J. Zhao, L. W. Liu, J. Li, S. L. Wang, R. N. Alolga, Y. Yin, X. M. Wang, D. S. Zhao, J. H. Shen, F. Q. Meng, X. Zhou, H. Xu, G. P. He, M.-D. Lai, P. Li, W. Zhu and L. W. Qi, Comprehensive Metabolomic Characterization of Coronary Artery Diseases, J. Am. Coll. Cardiol., 2016, 68(12), 1281–1293, DOI:10.1016/j.jacc.2016.06.044.
- D. S. Wishart, Emerging Applications of Metabolomics in Drug Discovery and Precision Medicine, Nat. Rev. Drug Discovery, 2016, 15, 473–484, DOI:10.1038/nrd.2016.32.
- R. M. Onjiko, E. P. Portero, S. A. Moody and P. Nemes, In Situ Microprobe Single-Cell Capillary Electrophoresis Mass Spectrometry: Metabolic Reorganization in Single Differentiating Cells in the Live Vertebrate (Xenopus Laevis) Embryo, Anal. Chem., 2017, 89(13), 7069–7076, DOI:10.1021/acs.analchem.7b00880.
- B. G. J. Dekkers, M. Bakker, K. C. M. van der Elst, M. G. G. Sturkenboom, A. Veringa, L. F. R. Span and J. W. C. Alffenaar, Therapeutic Drug Monitoring of Posaconazole: An Update, Curr. Fungal Infect. Rep., 2016, 10, 51–61, DOI:10.1007/s12281-016-0255-4.
- A. Lin, F. Sved Skottvoll, S. Rayner, S. Pedersen-Bjergaard, G. Sullivan, S. Krauss, S. Ray Wilson and S. Harrison, 3D Cell Culture Models and Organ-on-a-Chip: Meet Separation Science and Mass Spectrometry, Electrophoresis, 2020, 41, 56–64, DOI:10.1002/elps.201900170.
- J. L. Ren, A. H. Zhang, L. Kong and X. J. Wang, Advances in Mass Spectrometry-Based Metabolomics for Investigation of Metabolites, RSC Adv., 2018, 8(40), 22335–22350, 10.1039/c8ra01574k.
- P. Nemes, A. M. Knolhoff, S. S. Rubakhin and J. V. Sweedler, Single-Cell Metabolomics: Changes in the Metabolome of Freshly Isolated and Cultured Neurons, ACS Chem. Neurosci., 2012, 3(10), 782–792, DOI:10.1021/cn300100u.
- H. Li and A. B. Hummon, Imaging Mass Spectrometry of Three-Dimensional Cell Culture Systems, Anal. Chem., 2011, 83(22), 8794–8801, DOI:10.1021/ac202356g.
- P. V. Migisha Ntwali, C. E. Heo, J. Y. Han, S. Y. Chae, M. Kim, H. M. Vu, M. S. Kim and H. I. Kim, Mass Spectrometry-Based Proteomics of Single Cells and Organoids: The New Generation of Cancer Research, TrAC, 2020, 130, 116005, DOI:10.1016/j.trac.2020.116005.
- P. H. Lindhorst and A. B. Hummon, Proteomics of Colorectal Cancer: Tumors, Organoids, and Cell Cultures—A Minireview, Front. Mol. Biosci., 2020, 7, 604492, DOI:10.3389/fmolb.2020.604492.
- A. Gonneaud, C. Asselin, F. Boudreau and F. M. Boisvert, Phenotypic Analysis of Organoids by Proteomics, Proteomics, 2017, 17, 1700023, DOI:10.1002/pmic.201700023.
- P. Nemes, A. M. Knolhoff, S. S. Rubakhin and J. V. Sweedler, Metabolic Differentiation of Neuronal Phenotypes by Single-Cell Capillary Electrophoresis-Electrospray Ionization-Mass Spectrometry, Anal. Chem., 2011, 83(17), 6810–6817, DOI:10.1021/ac2015855.
- K. Patel, J. Patel, M. Patel, G. Rajput and H. Patel, Introduction to Hyphenated Techniques and Their Applications in Pharmacy, Pharm. Methods, 2010, 1(1), 2, DOI:10.4103/2229-4708.72222.
- E. van Vliet, S. Morath, C. Eskes, J. Linge, J. Rappsilber, P. Honegger, T. Hartung and S. Coecke, A Novel in Vitro Metabolomics Approach for Neurotoxicity Testing, Proof of Principle for Methyl Mercury Chloride and Caffeine, NeuroToxicology, 2008, 29(1), 1–12, DOI:10.1016/j.neuro.2007.09.007.
- G. Gomez-Giro, J. Arias-Fuenzalida, J. Jarazo, D. Zeuschner, M. Ali, N. Possemis, S. Bolognin, R. Halder, C. Jäger, W. F. E. Kuper, P. M. van Hasselt, H. Zaehres, A. del Sol, H. van der Putten, H. R. Schöler and J. C. Schwamborn, Synapse Alterations Precede Neuronal Damage and Storage Pathology in a Human Cerebral Organoid Model of CLN3-Juvenile Neuronal Ceroid Lipofuscinosis, Acta Neuropathol. Commun., 2019, 7(1), 222, DOI:10.1186/s40478-019-0871-7.
- L. M. Braun, S. Lagies, R. F. U. Klar, S. Hussung, R. M. Fritsch, B. Kammerer and U. A. Wittel, Metabolic Profiling of Early and Late Recurrent Pancreatic Ductal Adenocarcinoma Using Patient-Derived Organoid Cultures, Cancers, 2020, 12(6), 1440, DOI:10.3390/cancers12061440.
- H. Q. Duong, I. Nemazanyy, F. Rambow, S. C. Tang, S. Delaunay, L. Tharun, A. Florin, R. Buttner, D. Vandaele, P. Close, J. C. Marine, K. Shostak and A. Chariot, The Endosomal Protein CEMIP Links WNT Signaling to MEK1−ERK1/2 Activation in Selumetinib-Resistant Intestinal Organoids, Cancer Res., 2018, 78(16), 4533–4548, DOI:10.1158/0008-5472.CAN-17-3149.
- S. K. Neef, N. Janssen, S. Winter, S. K. Wallisch, U. Hofmann, M. H. Dahlke, M. Schwab, T. E. Mürdter and M. Haag, Metabolic Drug Response Phenotyping in Colorectal Cancer Organoids by LC-QTOF-Ms, Metabolites, 2020, 10(12), 1–17, DOI:10.3390/metabo10120494.
- F. S. Skottvoll, F. A. Hansen, S. Harrison, I. S. Boger, A. Mrsa, M. S. Restan, M. Stein, E. Lundanes, S. Pedersen-Bjergaard, A. Aizenshtadt, S. Krauss, G. Sullivan, I. L. Bogen and S. R. Wilson, Electromembrane Extraction and Mass Spectrometry for Liver Organoid Drug Metabolism Studies, Anal. Chem., 2021, 93(7), 3576–3585, DOI:10.1021/acs.analchem.0c05082.
- S. Mohammadi, C. Morell-Perez, C. W. Wright, T. P. Wyche, C. H. White, T. R. Sana and L. A. Lieberman, Assessing donor-to-donor variability in human intestinal organoid cultures, Stem Cell Rep., 2021, 16, 2364–2378, DOI:10.1016/j.stemcr.2021.07.016.
- M. Notaras, A. Lodhi, E. Barrio-Alonso, C. Foord, T. Rodrick, D. Jones, H. Fang, D. Greening and D. Colak, Neurodevelopmental Signatures of Narcotic and Neuropsychiatric Risk Factors in 3D Human-Derived Forebrain Organoids, Mol. Psychiatry, 2021, 26, 7760–7783, DOI:10.1038/s41380-021-01189-9.
- A. Lacouture, C. Jobin, C. Weidmann, L. Berthiaume, D. Bastien, I. Laverdière, M. Pelletier and É. Audet-Walsh, A FACS-Free Purification Method to Study Estrogen Signaling, Organoid Formation, and Metabolic Reprogramming in Mammary Epithelial Cells, Front. Endocrinol., 2021, 12, 672466, DOI:10.3389/fendo.2021.672466.
- E. J. Koh and S. Y. Hwang, Multi-Omics Approaches for Understanding Environmental Exposure and Human Health, Mol. Cell. Toxicol., 2019, 15, 1–7, DOI:10.1007/s13273-019-0001-4.
- Y. Hasin, M. Seldin and A. Lusis, Multi-Omics Approaches to Disease, Genome Biol., 2017, 18, 83, DOI:10.1186/s13059-017-1215-1.
- R. G. Lindeboom, L. Voorthuijsen, K. C. Oost, M. J. Rodríguez-Colman, M. Luna-Velez, C. V. Furlan, F. Baraille, P. W. Jansen, A. Ribeiro, B. M. Burgering, H. J. Snippert and M. Vermeulen, Integrative Multi-omics Analysis of Intestinal Organoid Differentiation, Mol. Syst. Biol., 2018, 14(6), e8227, DOI:10.15252/msb.20188227.
- K. R. Shechter, E. Kafkia, K. Zirngibl, S. Gawrzak, A. Alladin, D. Machado, C. Lüchtenborg, D. C. Sévin, B. Brügger, K. R. Patil and M. Jechlinger, Metabolic Memory Underlying Minimal Residual Disease in Breast Cancer, Mol. Syst. Biol., 2021, 17((10)), 310141, DOI:10.15252/msb.202010141.
- E. K. Neumann, K. V. Djambazova, R. M. Caprioli and J. M. Spraggins, Multimodal Imaging Mass Spectrometry: Next Generation Molecular Mapping in Biology and Medicine, J. Am. Soc. Mass Spectrom., 2020, 31, 2401–2415, DOI:10.1021/jasms.0c00232.
- E. Davoli, M. Zucchetti, C. Matteo, P. Ubezio, M. D'Incalci and L. Morosi, The Space Dimension at the Micro Level: Mass Spectrometry Imaging of Drugs in Tissues, Mass Spectrom. Rev., 2021, 40, 201–214, DOI:10.1002/mas.21633.
- A. Ajith, Y. Sthanikam and S. Banerjee, Chemical Analysis of the Human Brain by Imaging Mass Spectrometry, Analyst, 2021, 146, 5451–5473, 10.1039/d1an01109j.
- D. Unsihuay, D. Mesa Sanchez and J. Laskin, Quantitative Mass Spectrometry Imaging of Biological Systems, Annu. Rev. Phys. Chem., 2020, 72, 307–329, DOI:10.1146/annurev-physchem-061020-053416.
- T. J. Comi, M. A. Makurath, M. C. Philip, S. S. Rubakhin and J. V. Sweedler, MALDI MS Guided Liquid Microjunction Extraction for Capillary Electrophoresis-Electrospray Ionization MS Analysis of Single Pancreatic Islet Cells, Anal. Chem., 2017, 89(14), 7765–7772, DOI:10.1021/acs.analchem.7b01782.
- W. M. Bodnar, R. K. Blackburn, J. M. Krise and M. A. Moseley, Exploiting the Complementary Nature of LC/MALDI/MS/MS and LC/ESI/MS/MS for Increased Proteome Coverage, J. Am. Soc. Mass Spectrom., 2003, 14(9), 971–979, DOI:10.1016/S1044-0305(03)00209-5.
- M. Poetzsch, A. E. Steuer, A. T. Roemmelt, M. R. Baumgartner and T. Kraemer, Single Hair Analysis of Small Molecules Using MALDI-Triple Quadrupole MS Imaging and LC-MS/MS: Investigations on Opportunities and Pitfalls, Anal. Chem., 2014, 86(23), 11758–11765, DOI:10.1021/ac503193w.
- A. R. Buchberger, K. DeLaney, J. Johnson and L. Li, Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights, Anal. Chem., 2018, 90, 240–265, DOI:10.1021/acs.analchem.7b04733.
- T. P. Siegel, G. Hamm, J. Bunch, J. Cappell, J. S. Fletcher and K. Schwamborn, Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues, Mol. Imaging Biol., 2018, 20, 888–901, DOI:10.1007/s11307-018-1267-y.
- C. E. Spencer, L. E. Flint, C. J. Duckett, L. M. Cole, N. Cross, D. P. Smith and M. R. Clench, Role of MALDI-MSI in Combination with 3D Tissue Models for Early Stage Efficacy and Safety Testing of Drugs and Toxicants, Expert Rev. Proteom., 2020, 11–12, 827–841, DOI:10.1080/14789450.2021.1876568.
- X. Liu, C. Flinders, S. M. Mumenthaler and A. B. Hummon, MALDI Mass Spectrometry Imaging for Evaluation of Therapeutics in Colorectal Tumor Organoids, J. Am. Soc. Mass Spectrom., 2018, 29(3), 516–526, DOI:10.1007/s13361-017-1851-4.
- X. Liu, E. M. Weaver and A. B. Hummon, Evaluation of Therapeutics in Three-Dimensional Cell Culture Systems by MALDI Imaging Mass Spectrometry, Anal. Chem., 2013, 85(13), 6295–6302, DOI:10.1021/ac400519c.
- S. Bergmann, S. E. Lawler, Y. Qu, C. M. Fadzen, J. M. Wolfe, M. S. Regan, B. L. Pentelute, N. Y. R. Agar and C. F. Cho, Blood–Brain-Barrier Organoids for Investigating the Permeability of CNS Therapeutics, Nat. Protoc., 2018, 13(12), 2827–2843, DOI:10.1038/s41596-018-0066-x.
- C. F. Cho, J. M. Wolfe, C. M. Fadzen, D. Calligaris, K. Hornburg, E. A. Chiocca, N. Y. R. Agar, B. L. Pentelute and S. E. Lawler, Blood-Brain-Barrier Spheroids as an in Vitro Screening Platform for Brain-Penetrating Agents, Nat. Commun., 2017, 8, 15623, DOI:10.1038/ncomms15623.
- J. Johnson, J. T. Sharick, M. C. Skala and L. Li, Sample Preparation Strategies for High-Throughput Mass Spectrometry Imaging of Primary Tumor Organoids, J. Mass Spectrom., 2020, 55(4), e4452, DOI:10.1002/jms.4452.
- B. Bakker, R. D. W. Vaes, M. R. Aberle, T. Welbers, T. Hankemeier, S. S. Rensen, S. W. M. Olde Damink and R. M. A. Heeren, Preparing Ductal Epithelial Organoids for High-Spatial-Resolution Molecular Profiling Using Mass Spectrometry Imaging, Nat. Protoc., 2022, 17, 962–979, DOI:10.1038/s41596-021-00661-8.
- J. Hanrieder, N. T. N. Phan, M. E. Kurczy and A. G. Ewing, Imaging Mass Spectrometry in Neuroscience, ACS Chem. Neurosci., 2013, 4, 666–679, DOI:10.1021/cn400053c.
- D. Touboul and A. Brunelle, What More Can TOF-SIMS Bring than Other MS Imaging Methods?, Bioanalysis, 2016, 8(5), 367–369, DOI:10.4155/bio.16.11.
- D. Parrot, S. Papazian, D. Foil and D. Tasdemir, Imaging the Unimaginable: Desorption Electrospray Ionization – Imaging Mass Spectrometry (DESI-IMS) in Natural Product Research, Planta Med., 2018, 84, 584–593, DOI:10.1055/s-0044-100188.
- L. E. Flint, G. Hamm, J. D. Ready, S. Ling, C. J. Duckett, N. A. Cross, L. M. Cole, D. P. Smith, R. J. A. Goodwin and M. R. Clench, Characterization of an Aggregated Three-Dimensional Cell Culture Model by Multimodal Mass Spectrometry Imaging, Anal. Chem., 2020, 92(18), 12538–12547, DOI:10.1021/acs.analchem.0c02389.
- E. Garreta, R. D. Kamm, S. M. Chuva de Sousa Lopes, M. A. Lancaster, R. Weiss, X. Trepat, I. Hyun and N. Montserrat, Rethinking Organoid Technology through Bioengineering, Nat. Mater., 2021, 20, 145–155, DOI:10.1038/s41563-020-00804-4.
- K. DeLaney, C. S. Sauer, N. Q. Vu and L. Li, Recent Advances and New Perspectives in Capillary Electrophoresis-Mass Spectrometry for Single Cell “Omics”, Molecules, 2019, 24, 42, DOI:10.3390/molecules24010042.
- T. Kawai, N. Ota, K. Okada, A. Imasato, Y. Owa, M. Morita, M. Tada and Y. Tanaka, Ultrasensitive Single Cell Metabolomics by Capillary Electrophoresis-Mass Spectrometry with a Thin-Walled Tapered Emitter and Large-Volume Dual Sample Preconcentration, Anal. Chem., 2019, 91(16), 10564–10572, DOI:10.1021/acs.analchem.9b01578.
- S. Wu, L. Huang, M. Fang, K. A. Cupp-Sutton, Z. Wang and K. Smith, Spray-Capillary-Based Capillary Electrophoresis Mass Spectrometry for Metabolite Analysis in Single Cells, Anal. Chem., 2021, 93(10), 4479–4487, DOI:10.1021/acs.analchem.0c04624.
- E. K. Neumann, T. J. Comi, S. S. Rubakhin and J. V. Sweedler, Lipid Heterogeneity between Astrocytes and Neurons Revealed by Single-Cell MALDI-MS Combined with Immunocytochemical Classification, Angew. Chem., Int. Ed., 2019, 58(18), 5910–5914, DOI:10.1002/anie.201812892.
- T. Yang, D. Gao, F. Jin, Y. Jiang and H. Liu, Surface-Printed Microdot Array Chips Coupled with Matrixassisted Laser Desorption/Ionization Mass Spectrometry for Highthroughput Single-Cell Patterning and Phospholipid Analysis, Rapid Commun. Mass Spectrom., 2016, 30, 73–79, DOI:10.1002/rcm.7628.
- J. Krismer, J. Sobek, R. F. Steinhoff, R. Brönnimann, M. Pabst and R. Zenobi, A MALDI-MS Methodology for Studying Metabolic Heterogeneity of Single Cells in a Population, in Single Cell Metabolism Methods and Protocols Methods in Molecular Biology, Humana, New York, NY, 2020, ch. 9, vol. 2064, pp. 113–124 Search PubMed.
- A. D. Mandić, A. Woting, T. Jaenicke, A. Sander, W. Sabrowski, U. Rolle-Kampcyk, M. von Bergen and M. Blaut, Clostridium Ramosum Regulates Enterochromaffin Cell Development and Serotonin Release, Sci. Rep., 2019, 9(1), 1177, DOI:10.1038/s41598-018-38018-z.
- L. M. Smits, L. Reinhardt, P. Reinhardt, M. Glatza, A. S. Monzel, N. Stanslowsky, M. D. Rosato-Siri, A. Zanon, P. M. Antony, J. Bellmann, S. M. Nicklas, K. Hemmer, X. Qing, E. Berger, N. Kalmbach, M. Ehrlich, S. Bolognin, A. A. Hicks, F. Wegner, J. L. Sterneckert and J. C. Schwamborn, Modeling Parkinson's Disease in Midbrain-like Organoids, NPJ Parkinsons Dis., 2019, 5(1), 5, DOI:10.1038/s41531-019-0078-4.
| This journal is © The Royal Society of Chemistry 2022 |