Mass spectrometry-based techniques for single-cell analysis
Xiangyi
Xu†
,
Xuanxi
Jiang†
,
Meiyun
Shi
* and
Lei
Yin
 *
*
School of Life and Pharmaceutical Sciences, Dalian University of Technology, Panjin, 124221, PR China. E-mail: leiyin@dlut.edu.cn; shimy@dlut.edu.cn
First published on 29th June 2023
Abstract
The cell is the most basic structural unit and plays a vital role in the function of an organism. Studying the heterogeneity of cells, especially the qualitative and quantitative analyses of proteins and lipids at the cellular level and even at the subcellular level, is of great significance for the study of some important pathological or physiological processes. Due to the small size of a single cell, low content of analytes and large interference from the biological matrix within the single cell, analytical methods at the single cell level must be highly sensitive and selective. Mass spectrometry is a powerful technology for single-cell analysis, because it has high sensitivity, high selectivity and the ability to monitor multiple chemicals at the same time. In this review, four mass spectrometry-based methods applied to single-cell analysis are introduced and discussed in detail; these are electrospray ionization mass spectrometry (ESI-MS), laser desorption ionization mass spectrometry (LDI-MS), secondary ion mass spectrometry (SIMS) and inductively coupled plasma mass spectrometry (ICP-MS). The recent advances in single-cell analysis with these mass spectrometry-based techniques are summarized. We believe that this review can provide some help and reference for single-cell analysis by mass spectrometry.
1. Introduction
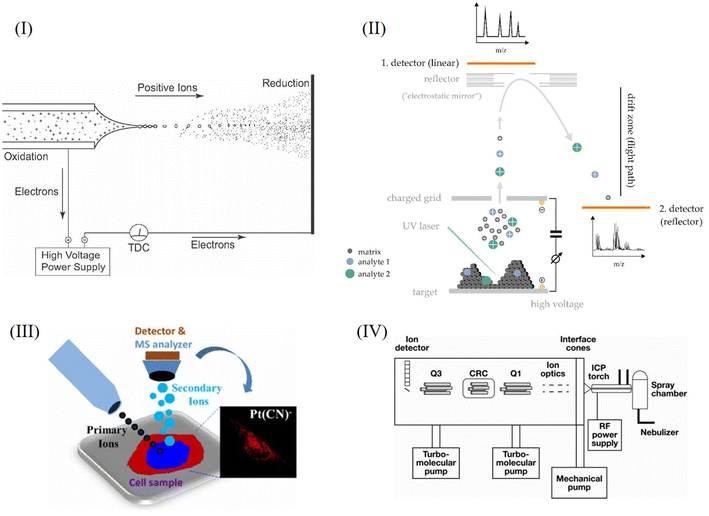
The cell is the most basic structural and functional unit of an organism. Cells have significant heterogeneity, which is of great significance. Specific cells of interest should be analyzed individually, such as rare cells, thus helping us to understand pathological processes such as the formation of tumors or important physiological processes.1–3 When studying the average level of cell populations, the analysis of small cell populations and rare cells may be neglected, and when there are subgroups with different dominant phenotypes, the average level of the population cannot reflect the characteristics of most cells.4–6 Therefore, research at the single-cell level is not only important but also necessary in some cases. However, single cell analysis faces many challenges. First of all, the volume of a single cell is small, which makes it difficult to separate or enrich the sample. Secondly, the substance content in a single cell is extremely low, so a high sensitivity level of the analytical method is essential. Thirdly, there are many kinds of metabolites in single cells, which change rapidly in cells, and different substances may interfere with each other during analysis, so high selectivity of the analytical methods is essential.7–10In recent years, many analytical techniques have been applied to single cell analysis, such as fluorescence imaging assays, electrochemical methods and mass spectrometry assays. During fluorescence assays, fluorescent markers are specifically combined with substances to be detected, so that the distribution and content of target compounds can be detected by detecting fluorescence.2 Fluorescence imaging usually requires the fluorescent labeling of target molecules and may change the activity of the molecules to be detected.11 Electrochemical methods have high sensitivity and can be used for the targeted analysis of electroactive substances, which can be used to monitor the occurrence of physiological processes, such as the release of neurotransmitters.3,12 During analysis, electrodes can be inserted into cells. However, electrochemical methods can only be used for electroactive substances, and the substances analyzed are limited. In addition, similar to fluorescence imaging assays, electrochemical methods are not suitable for the simultaneous detection of multiple chemicals.3,10 Mass spectrometry is a powerful technique for single cell analysis without labeling. It has extremely high sensitivity, high selectivity, the ability to detect multiple chemicals at the same time, and leads to the ability to identify the structures of molecules of interest.3,8,10,13 At present, many mass spectrometry analytical assays based on different ionization technologies have been developed for single cell analysis, including electrospray ionization mass spectrometry (ESI-MS), laser desorption ionization mass spectrometry (LDI-MS), secondary ion mass spectrometry (SIMS) and inductively coupled mass spectrometry (ICP-MS). These ionization methods can ionize different kinds of substances, such as proteins, peptides, esters, small molecule metabolites and elements. These methods have their own characteristics and can complement each other. Challenges of single cell analysis by mass spectrometry are summarized and shown in Fig. 1. Schematic diagrams of the above analysis methods are shown in Fig. 2. In the following sections, the characteristics of the methods and their applications are introduced in detail.
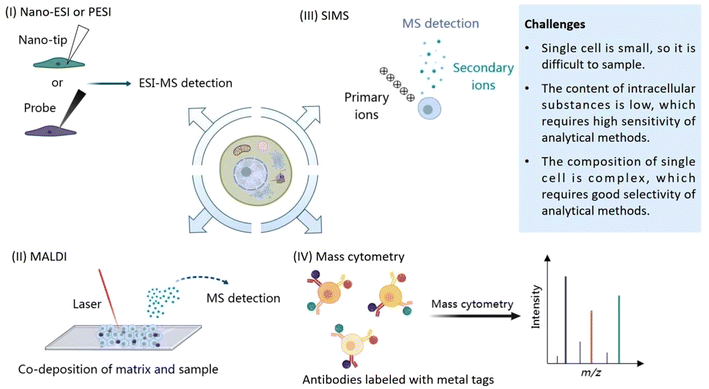 | ||
| Fig. 1 Challenges of single cell analysis by mass spectrometry: (I) Nano-ESI or PESI, (II) SIMS, (III) MALDI, (IV) Mass cytometry. | ||
2. ESI-MS
ESI is technology based on a “soft” ion source with high ionization efficiency, which is widely used in mass spectrometry. ESI-MS refers to electrospray ionization mass spectrometry. Taking the mode of positive ions as an example, under the action of an electric field, the sample to be tested first forms small droplets with a large number of positive charges. Due to the action of Coulomb forces, the droplets undergo Coulomb explosion and form gaseous ions. In other words, the generation of an electrospray is mainly divided into three processes: the formation of charged droplets, the disintegration of charged droplets, and the generation of gas-phase ions.14 The process is shown in Fig. 2I and it can be performed under atmospheric pressure. After the formation of gas-phase ions, they enter the vacuum region, and are purified by heating the capillary and applying a voltage. For protein analysis, ammonium acetate is often used as an additive to prevent Na+ from combining with the protein to form sodium adducts.The ionization efficiency of the electrospray changes due to the presence of substances coeluting with the substance to be tested, a phenomenon known as the matrix effect. The matrix effect may be related to competition between the substance being tested and the highly polar “matrix” in the gas phase. The matrix effect may affect the sensitivity and accuracy of ESI analysis. Eliminating the matrix effect in ESI is the key to improving the accuracy and reproducibility of the method.15 In addition, different from MALDI and SIMS, ESI is more suitable for the in situ analysis of living single cells.9 Five different kinds of ESI techniques, namely, Nano-ESI, probe-ESI (PESI-MS), desorption ESI (DESI), laser ablation ESI (LAESI) and capillary electrophoresis ESI (CE-ESI), are discussed in the following sections.
2.1. Nano-ESI
Nano-ESI is a widely used soft ion source for the detection of small molecules, proteins and lipids in single cells. Nano-ESI means that the experimenter takes samples directly from living cells using a Nano-tip under a microscope, and then applies a high voltage of 1–2 kV to the sample to form an electrospray.10 Compared with MALDI and SIMS methods, Nano-ESI has the characteristics that the ionization pressure is ambient pressure, the operation can be visualized, and the sampling of single cells can be completed directly. Direct sampling can minimize metabolite loss and metabolome disturbance.11This profile visualized during sampling is called live single-cell video mass spectrometry. Hajime Mizuno et al.16 performed single-cell video mass spectrometry under real-time microscopic observation, using a Nano-tip to suck cytoplasm and organelles from RBL-2H3 cells. By performing t-tests and principal component analysis (PCA) on peaks in the spectrograms, they selected meaningful peaks for MS/MS analysis to identify substances such as histamine, leukotriene, and serotonin. With the help of principal component analysis, the team completed the classification of seven cell types. The point of this technique is that accurate sampling enables the experimenter to accurately analyze the metabolic changes in different cells and thus classify cells by studying physiological processes at the molecular level, such as the gamma-glutamyl cycle, in this experiment. Hajime Mizuno et al.17 also used live single-cell video mass spectrometry to study the metabolic changes to histidine and tryptophan in RBL-2H3 cells. They also used a Nano-tip to absorb the cytoplasm or granules of cells when sampling, and through the t-test, the position distribution of histidine and tryptophan metabolite peaks could be measured. Naohiro Tsuyama et al.18 used nano-ESI to study single mouse-embryonic fibroblast cells, Swiss 3T3, and obtained hundreds of small molecular peaks, and selected cell-specific peaks by the t test. They chose acetonitrile with 0.5% formic acid as an ionization liquid. Dopamine and angiotensin I were used for calibration. They found that the proline content of Swiss 3T3 cells increased during mitosis and speculated that this might be related to the proline utilization gene. Mónica Lorenzo Tejedor et al.19 used nano-ESI to quantitatively analyze various organic compounds such as geranic acid and methyl citronellate in single cells from the flowers, stems and leaves of Pelargonium zonale. They used a Nano-tip to suck the cell's contents directly from an individual cell and then detected these chemicals by using a high-resolution Orbitrap mass spectrometer. They also used the t-test and PCA to compare and analyze the results of different detected compounds, the structures of which were further confirmed by MS/MS spectra. The nano-ESI-MS/MS method used in this study can be adopted to analyze the metabolites of single plant cells in different tissues, which is of great significance for plant disease detection, quality control and food analysis. Tumor recurrence and metastasis may be related to the heterogeneity of cancer cells. Yasmine Abouleila et al.20 combined live single-cell mass spectrometry and a microfluidics-based CTC enrichment technique to detect the metabolites of circulating tumor cells (CTCs) and analyzed them by the PCA-DA method. PCA simplifies data and makes it easy to visualize, while discriminant analysis (DA) mainly emphasizes the metabolic differences of different CTCs from different cancer phenotypes. They believe that the metabolic spectrum of CTCs is valuable for the diagnosis and prognosis of colorectal cancer (CRC) and gastric cancer (GC). Acyl carnitine metabolites, sterol lipids and eicosanoids are the possible characteristic metabolites of CRC-CTCs, while glycerophosphate is the possible characteristic metabolite of GC-CTCs. Xiaochao Zhang et al.21 combined droplet extraction and sampling technology with the pulsed direct current electrospray ionization mass spectrometry method (Pico-ESI-MS) to analyze the metabolites of single human glioblastoma cells (A172) and normal human astrocyte cells (HA) by tandem mass spectrometry (MS2). Droplet extraction can improve the sensitivity of the method, but the electrospray time is short, so it is difficult to obtain the MS2 spectrum. Pico-ESI-MS can prolong the time of the electrospray, but cannot guarantee the sensitivity and stability of the analysis of small volume samples. Therefore, the combination of the two provides complementary advantages. They found that the ratio of unsaturated phosphatidylcholines (PCs) to saturated PCs was larger in tumor cells. They used borosilicate glass capillary tubes to make Pico-tips with a diameter of ∼3 μm. Unlike other Nano-ESI techniques, their Pico-ESI analysis utilizes a homemade DC (direct current) power supply for ionization.22 Hongying Zhu et al.23 combined the patch clamp technique with ESI-MS to develop a method for the detection of urocanic acid (UCA) in a single hippocampal neuron. Through the combination of electrophysiology and electrospray ionization mass spectrometry, the group studied and monitored the changes to UCA, and discussed the effect of ultraviolet irradiation on the glutamate metabolism pathway in the brain. It is found that glutamate synthesis may be related to the His–UCA–Glu (histidine–UCA–glutamate) metabolic pathway in the nervous system. This study also proves that single-cell mass spectrometry is a powerful tool to study brain metabolic processes at the single-cell level. Later, Hongying Zhu et al.24 continued to combine the patch clamp technology with nano-ESI and established a single-lysosome mass spectrometry (SLMS) platform. According to different metabolic characteristics, lysosomes are divided into five subgroups.
Therefore, researchers often use a Nano-tip to suck cytoplasm or other contents from cells for single-cell level analysis. The spectra obtained are often processed by the t-test and PCA. PCA can also be used to distinguish cells. MS/MS is often used to further identify substances. Nano-ESI can also be combined with the patch clamp technique to study the metabolic physiology of the nervous system.23
2.2. PESI
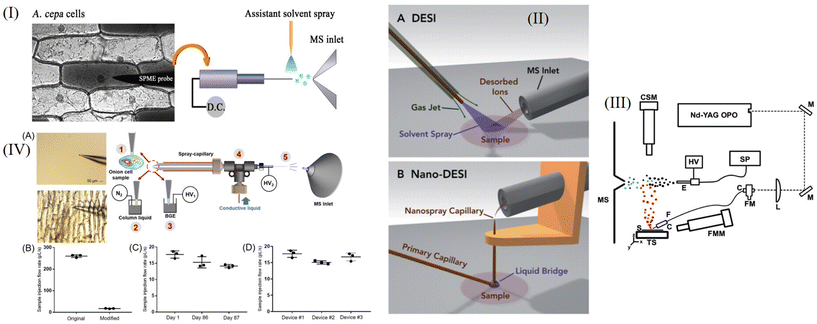
Probe-ESI is an ESI method developed by Kenzo Hiraoka et al.25 PESI is made by touching the sample with a solid needle probe and applying high pressure (about 3 kV) at the tip of the needle to form an electrospray and ionize the sample. PESI uses a probe instead of a capillary for the electrospray so that the sample does not clog the outlet during electrospraying. PESI is advantageous for the determination of peptides or proteins with molecular weights of several thousands, but the ion abundance decreases as the molecular weight increases. The research group demonstrated that the ion abundance could be increased dramatically by adding an acid or a salt to the aqueous solution of the substance to be tested.Xiaoyun Gong et al.26 proposed the use of PESI to detect metabolites such as fructans, lipids, and flavone derivatives in single Allium cepa cells. PESI uses a tungsten probe (≤1 μm in diameter) to collect viable intracellular material, which is then ionized at the tungsten probe tip (voltage ∼2500 V). They used PESI to test for angiotensin II in aqueous solution and found that PESI had good enrichment characteristics. Through the detection of egg yolk by PESI, it was found that PESI was very tolerant of high viscosity substances, while Nano-ESI did not respond to the substances in egg yolk. The team observed significant differences in metabolite types between inner and outer epidermis cells. They also performed subcellular analyses, taking samples from the nucleus and cytoplasm separately, and demonstrated that the material inside the cells was not evenly distributed. A schematic diagram of this process is shown in Fig. 3I. Fengming Chen et al.27 developed a single-cell lipid analysis method using PESI and drop-on-demand inkjet cell printing. The diameter of the tungsten probe is 2 μm. Using inkjet printing technology, single cells can be printed. They adjusted the position of the outlet of the inkjet printer and the tip of the needle so that the droplets from the inkjet could be ionized at the tip of the needle. After determination, PCA was used for cell classification. Eight cell types (U-87, U251, HepG2, McF-7, 293, Caco-2, HUVEC, and 3T3 cells) were analyzed and differentiated. At the same time, the device can also be used to detect cellular markers. PESI could detect single MCF-7 cells labeled with rhodamine. This experiment established a single-cell lipid analysis platform, which is of great significance for the study of diabetes and hypertension.
Overall, PESI has great advantages in the field of single-cell analysis. At present, PESI is used for single-cell analysis of lipids, metabolites, etc. PESI has good enrichment characteristics and high sensitivity, and subcellular detection can be performed by reducing the probe diameter.26 When PESI is combined with the inkjet printing of cells, labeled cells can be analyzed.27
2.3. DESI-MS
Desorption electrospray ionization (DESI) is an ionization method developed by Cooks et al.28 in 2004, which can be used for quantitative analysis and imaging analysis. DESI is essentially a process in which a charged particle flow and solvent collide with the surface of the sample plate to make the sample desorb and produce gaseous ions. The gaseous ions generated are transferred to the mass spectrometer by the atmospheric pressure ion transfer line for detection. The advantage of the DESI method is that the sample preparation process is simple, no substrate is required, and the sample can be directly sampled under ambient conditions, without vacuum conditions. This simple sampling method enables in situ imaging. This group also demonstrated that DESI could be used to sample tissue from a living organism and to detect loratadine from human fingers after the administration of 10 mg of the antihistamine loratadine.Nano-DESI is a modification of DESI that uses two capillaries to form a solvent bridge and is a liquid extraction–ionization technique.29 Ionization and desorption processes are separated, and splash loss of the sample is greatly reduced. Fig. 3II shows a comparison of Nano-DESI and DESI.
Nano-DESI can be used for high-throughput single-cell analysis. Hilde-Marléne Bergman et al.30 explored the detection of single-cell amino acids and phospholipids, and used Nano-DESI to perform quantitative analysis of phosphatidylcholine (PC) in single human cheek cells, the content of which was measured to be 1.2 pmol. By comparing data with the human metabolome database, peaks with signal to noise ratios (SNRs) greater than 2 were preliminarily determined. The metabolites and lipids were analyzed by tandem mass spectrometry, and the analysis was coordinated by two-dimensional non-metric multidimensional scaling (NMDS). Obtaining higher throughput single-cell analysis is the key problem of single-cell mass spectrometry. Two modes of low-throughput static detection and automatic high-throughput detection can be realized by Nano-DESI. The method with automatic surface scanning has many advantages such as high sensitivity and fast detection speed.
2.4. LAESI-MS
Laser ablation electrospray ionization mass spectrometry (LAESI-MS) means that the position of the optical fiber is adjusted and the tissue to be measured is desorbed through laser emission pulses; the product is ionized into ions after meeting the electrospray plume.31,32 Generally, a mid-infrared laser with a wavelength of 2.94 μm is used in LAESI. Considering the limitation of optical diffraction, the scale of infrared ablation performed by LAESI for single-cell analysis is generally between 10 and 100 μm.32 LAESI is suitable for the analysis of plant cells. However, LAESI has a high requirement for tissue water content, and samples with a low water content are not suitable for detection by this method.33Sylwia A. Stopka et al.31 used LAESI to analyze the primary metabolites and secondary metabolites of E. densa epidermal cells and infected G. max root nodule cells. They found that it was necessary to pre-target rare specialized excretory idioblasts in E. densa and compared other detected single cells with this cell to detect significant differences and explore the heterogeneity of cells. At the same time, they also analyzed the metabolites of single cells of E. densa and E. canadensis to distinguish between different plant subgroups. In the experiment, they used GeO2 fiber to directly and accurately act on a single cell and applied a 20 Hz laser to form a plume, which entered the mass spectrometer. This application of LAESI to single cells is helpful for future research on cell heterogeneity. However, it is worth noting that this technology can realize the analysis of single plant cells with a volume of 75–250 pL at present, but it is still challenging for the analysis of animal cells with a relatively small volume (low picoliters). Bindesh Shrestha et al.32 used the LAESI method to analyze single cells in Allium cepa, Narcissus pseudonarcissus and Lytechinus pictus in situ. Using the GeO2 fiber, the cells were cauterized by a laser. The positive ions produced by LAESI were analyzed by Q-TOF with a mass resolution of 8000 (fwhm). By analyzing the spectrogram, they may get information about anthocyanidins, oligosaccharides and other metabolites. A schematic diagram of the device is shown in Fig. 3III.
The above two examples are LAESI analysis of single plant cells, and GeO2 is used as an optical fiber. Sylwia A. Stopka et al.31 used LAESI single-cell analysis to study cell heterogeneity. Sometimes, the average level of cell population does not represent the level of all cells, which often hides the differences between different cells. However, the research group proposed that it was necessary to target a special cell first, and take this cell as the object of comparison before analyzing the heterogeneity of other cells.
2.5. CE-ESI-MS
CE-ESI-MS is an ESI method that combines capillary electrophoresis (CE) with ESI technology. The advantages of CE-ESI-MS are that it can elute and separate substances with high efficiency, be used to analyze complex contents and it has high sensitivity.3,13 Several single-cell analysis methods of CE-ESI-MS are introduced below.CE-ESI is also used in the study of cell heterogeneity. Rosemary M. Onjiko et al.34 used the CE-ESI method of a grounded sheath current, ionized in cation mode, to analyze ∼70 molecular characteristics to explore the cellular heterogeneity of the left ventral and left dorsal cells of the Xenopus laevis 8 cell embryo. The CE-MS device can undergo many changes, such as being combined with other technologies. Jordan T. Aerts et al.35 combined the patch-clamp technique of electrophysiology with CE-MS to detect γ-aminobutyric acid (GABA) in rat thalamic cytoplasm. In this technique, ∼3 pL of cytoplasm was removed using the patch-clamp technique and placed into PCR tubes for CE-ESI-MS analysis. This technique can be applied not only to neurons but also to other cells, such as astrocytes. J. Scott Mellors et al.36 designed an integrated microfluidic device, which separated individual red blood cells by capillary electrophoresis and lysed them one by one. Data on the heme group (m/z 616) and the α and β subunits of hemoglobin were obtained. The detection speed is about 12 cells per min, which means that the single-cell analysis by this technique is high-throughput. Theodore Lapainis et al.37 used capillary electrophoresis combined with time-of-flight electrospray mass spectrometry to develop a single-cell analysis method with high sensitivity and stability. They used the sheath flow mode, using a capillary with an inner diameter of 40 μm, and applied a voltage of 20 kV to the capillary to reduce foaming and ensure a more stable electrospray. They used CE-ESI to obtain the signal of ach and amino acids except cysteine in single R2 neurons, thus providing a useful tool for studying neurophysiology. Recently, Lushuang Huang et al.38 improved the spray capillary of traditional CE-ESI, as shown in Fig. 3IV. The wall thickness of the MS end of the modified capillary was 5–10 μm, and 0.1% FA was selected as the column liquid in the capillary. They used modified CE-ESI to analyze single cells of onion. They inserted capillaries into individual onion cells and performed repeated online analyses of their metabolites. This method is highly reproducible and the results of the metabolite contents in different single onion cells are similar. However, because the capillary wall is negatively charged, some positively charged substances may be lost.
2.6. Summary of ESI analysis methods
The advantages, disadvantages or future development directions of ESI are summarized and shown in Table 1.| ESI | Advantages | Disadvantages or future development directions | Ref. |
|---|---|---|---|
| Nano-ESI | The ionization pressure is ambient pressure, the operation is visual, and single-cell injection can be directly completed. Direct sampling can minimize metabolite loss. | The high-throughput capability of this technology is limited because its experimental operations require individual sampling. | 10 and 11 |
| PESI | It will not be blocked during the process of electrospray, and peptides with molecular weights of several thousand can be determined. It has good enrichment characteristics and high sensitivity, and subcellular detection can be carried out by reducing the probe diameter. | The future development direction is to further miniaturize the probe or modify the probe with specific materials. | 25 and 26 |
| DESI | The sample preparation process is simple, without interference from the matrix effect, and the sample can be directly sampled under environmental conditions without vacuum conditions. | The sampling area is not clear. | 28 and 33 |
| LAESI | It is suitable for the analysis of complex biological liquids and samples with high water content. | There are high requirements for tissue water content, and samples with low water content are relatively unsuitable for detection by this method. | 33 |
| CE-ESI | It can efficiently elute and separate substances, analyze complex contents and has high sensitivity. | It takes a long time to analyze, and the pretreatment of samples makes it unsuitable for in situ analysis. | 3 and 13 |
3. LDI-MS
Laser desorption ionization mass spectrometry (LDI-MS) can be divided into matrix-assisted laser desorption ionization (MALDI) and matrix-free LDI. In MALDI, it is necessary to use the matrix to co-crystallize with the sample. The matrix absorbs the energy of the laser and assists the phase transition and ionization of the sample.39 In single-cell MALDI analysis, the target cells are first deposited on a slide such as ITO and then co-crystallized with the substrate. CHCA and DHB are commonly used substrates in positive ion mode. Subsequently, under vacuum conditions, the crystals are treated with a laser to transform and ionize the sample.40–42 The basic principle of matrix-free LDI is similar to MALDI, but matrix-free LDI does not require the matrix to co-crystallize after mixing with the sample. Instead, it requires a surface that can transmit laser energy to the sample.39 A schematic diagram of LDI is shown in Fig. 2II.Compared with ESI introduced earlier, the advantage of LDI is that there is no need for manual sampling, and LDI has the function of imaging analysis. Unlike SIMS, MALDI is a soft ion source. The TOF analyzer has the characteristics of good sensitivity, wide detection quality range and high ion flux. MALDI-TOF-MS/MS is a powerful tool for single cell analysis.43 After the sample is prepared, it is often necessary to ionize it under vacuum, which is the limiting condition of MALDI.11 Compared with matrix-free LDI, MALDI, which needs matrix assistance, has some disadvantages. For example, the matrix itself may interfere with the detection of samples. Because of the use of a low molecular weight matrix, MALDI is rarely used for the analysis of low molecular weight substances. In contrast, matrix-free LDI is mainly suitable for low molecular weight substances.39 It is worth noting that MALDI-TOF is also of great significance in clinical research,44–46 for example, it can detect pathogenic bacteria in blood.47
Both of these LDI methods have their own advantages, disadvantages and application scope, and this information is summarized in the following sections.
3.1. Matrix-free laser desorption/ionization
MALDI needs to use a matrix for analysis, which leads to many problems for analysis. However, matrix-free LDI analysis does not need to use a matrix;39 therefore, matrix-free LDI is more suitable for analyzing small molecular substances than MALDI. In terms of imaging, compared with MALDI, matrix-free LDI can prevent interference from the matrix when processing samples, so it has a higher spatial imaging rate.48 It is of great significance to study matrix-free LDI.Matrix-free LDI analysis is usually divided into desorption ionization on silicon (DIOS), sol–gel, carbon-containing structures, nano-structures and so on.39 In single-cell analysis, matrix-free LDI sometimes takes advantage of the characteristics of cells or metabolites, does not need a matrix, and ionizes itself after laser irradiation.
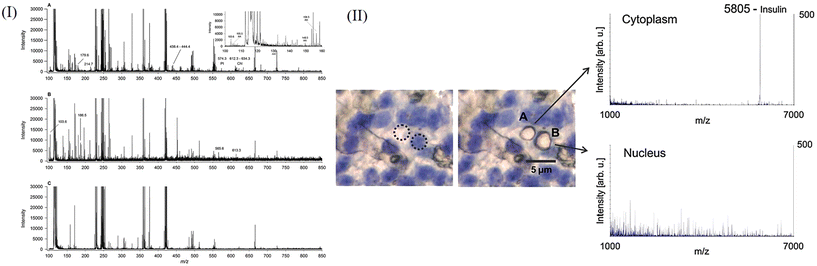
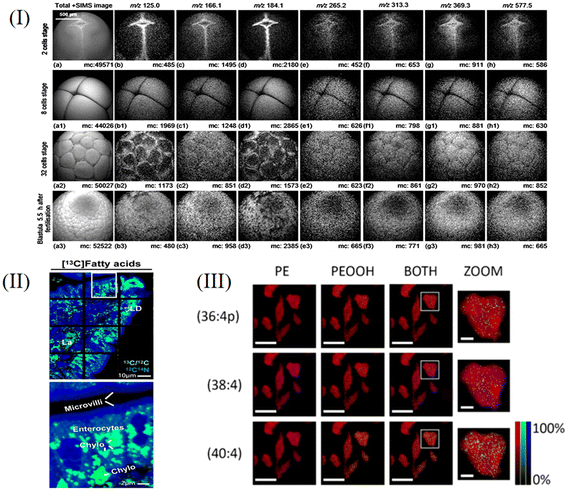
Tino Jaschinski et al.48 used matrix-free LDI to perform single-cell analysis on the diatoms Coscinodsicus granii and Thalassiosira pseudonana with special cell wall structures and detected chlorophyll, amino acids, phospholipids, etc. These algae have biomineralized nanostructured cell walls, the main component of which is SiO2. This structure facilitates ionization, allowing spatial resolution to depend only on the laser and its sensitivity. Another advantage is that relatively small molecules can be analyzed without interference from the matrix. They placed the diatoms in culture medium, incubated them at 14 °C, and sampled them when the diatoms were growing steadily. They used a 337 nm laser in positive ion mode. They achieved a spatial imaging rate of 20 μm. In the MSI experiment on Coscinodsicus granii, they were able to image using only one signal (m/z 177), whereas for more accurate characterization, other characteristic signals (m/z 214.7, 215.8, 265.6, 438.9, 443.3, 628.8) can also be combined for imaging. At present, the single-cell study of microalgae is still insufficient, so this work provides a new research method. Their spectra are shown in Fig. 4I. Dirk Hölscher et al.49 used matrix-free LDI to study UV-absorbing secondary metabolites in plant cells at the single-cell level. They selected Hypericum plants as the research object, and the spatial resolution of imaging could reach 10 μm, which was much higher than the usual resolution of MALDI. They combine laser microdissection (LMD) and LDI-TOF-MS techniques, in which LMD is useful for single-cell sampling. Usually, LDI analysis of substances needs matrix assistance, but the detected hypericin strongly absorbs ultraviolet rays, so it is suitable for direct matrix-free analysis. This research has made a great contribution to the single-cell imaging of plant secondary metabolites. In the future, this technology can expand the single-cell imaging analysis of compounds with strong ultraviolet absorption, such as flavonoids with condensed aromatic rings, anthocyanins, etc. Therefore, this technology has good application prospects.
The above two examples,48,49 respectively, use the special cell wall structure and metabolites that can absorb ultraviolet rays, that is, special structures or analytical substances, so that the sample can be ionized by itself without a matrix. Especially the work of Hölscher49 has opened up a new way of thinking, looking for metabolites with strong ultraviolet absorption, and using matrix-free LDI for single-cell level research. In addition, it is worth noting that matrix-free LDI also has advantages in imaging.
3.2. MALDI
Matrix-assisted laser desorption ionization (MALDI) is suitable for the analysis of biological macromolecules, such as proteins and peptides.43 For protein analysis, MALDI is more suitable for the analysis of high-abundance and high-mass proteins.50Many early single-cell MALDI analysis targets were peptides in neurons.43 Van Veelen et al.51 used MALDI to analyze the peptides of single Lymnaea stagnalis neurons. Stanislav S. Rubakhin et al.52,53 also used MALDI to analyze neuropeptides of single Aplysia californica neurons. Yousef Gholipour et al.54 improved the sampling process, so that the MALDI method could be used to analyze the plant cytoplasmic extract in subpicoliter volumes. They first extracted the cytoplasm (1–10 pL) of a single cell from the cells of a tulip leaf and bulb with a pressure probe, and then analyzed them with MALDI and compared the analysis signals of carbohydrates when three substrates (DHB, THAP, CNTs) were used to deposit samples. When using DHB, carbohydrates were often sodium addition ions; when using THAP and CNTs, carbohydrates were often potassium addition ions. MALDI can also be combined with other analysis techniques. Jason S. Page et al.55 used MALDI combined with CE to analyze the hormones and neuropeptides of single Aplysia californica neuronal cells and also performed 35S-MET radionuclide detection. For the first time, they developed a method to analyze hormones in individual cells by combining CE/MALDI with radionuclide detection. Yuchen Dai et al.56 used plasmonic colloidosomes and MALDI-TOF to detect the resistance of individual Escherichia coli cells to the antibiotic AMP. The traditional organic acid matrix may have the problem of uneven crystallization. D. C. Perdian et al.57 used colloidal silver as the matrix. They used LDI-MS combined with colloidal silver to analyze cholesterol levels in individual astrocytes. Colloidal particles provide a homogeneous matrix of nanometer size and do not crystallize, and at the same time, colloidal silver can form silver ion adducts with cholesterol. They sprayed only 12.5 μL of colloidal silver from an atomizer onto the sample eight times to evenly coat the sample with matrix. They also compared LDI-MS using the colloidal silver method with the traditional fluorescence detection method, and the two measurements were in good agreement. They performed an imaging analysis with a spatial resolution of 50 μm, which was larger than that of individual cells and therefore did not permit fine subcellular imaging. Akihito Korenaga et al.58 used a single-cell inkjet combined with MALDI to achieve phosphatidylcholine analysis of single HUVEC cells. The team also combined single-cell inkjet technology with PESI.27 The inkjet method provides a method to introduce a higher throughput of one or a few cells. In this study, they pretreated ITO glass slides and cleaned them with ultrasonic waves. They mixed 200 mM 2,5-dihydroxybenzoic acid, 50% acetonitrile, 10 mM NaI, and 0.1% trifluoroacetic acid as the matrix. For inkjet printing, a 30 V driving voltage was used to print the mixture of matrix and cells separately or all at once onto ITO glass slides for MALDI detection. They found that the PBS buffer interfered with ionization, so the cells were modified with glutaraldehyde and then ionized in pure water. Peter V. Shanta et al.59 used plasma gold film to replace the stainless steel plate, improving the MALDI detection device, and carried out a single-cell lipidomics study on algal cells from C. reinhardtii. Compared with the traditional method, the plasma gold film has the characteristic of enhancing fluorescence, and the advantage of using it is that metal-enhanced fluorescence (MEF) can be used to locate the cells, and the ionization is enhanced. They used electron beam deposition and photolithography to produce plasma gold films on BK-7 slides. A nanodelivery system was used to load cells onto gold films, and super-DHB was selected as the matrix for MALDI with a laser flux of 4500 au, and 200 shots were fired on the surface to obtain the spectra. They detected 54 lipids in C. reinhardtii and found that MGDG and DGDG were abundant in the algal plastidic membrane. This method provides the single-cell level detection of low abundance lipids and can be used for toxicological studies of herbicides in aquatic environments. M. Manikandan et al.60 used microwave digestion to analyze breast cancer cells at the single-cell level by MALDI. This method can be extended to clinical application. They developed a kind of FBS (80–120 nm) modified gold nanoparticle (10–17 nm) to modify the detected cancer cells. The existence of the gold particles can generate microwaves, crack the cell membrane, and enhance the MALDI signal of the detected substance by 60%, which is equivalent to a sensor. In the synthesis of AuNPs, they used HAuCl4 and mixed the prepared NPs with FBS. After 6 hours of AuNP simulation treatment, the cell structures of breast cancer cells, cancer stem cells and normal cells did not deform, which proved that the method had good biocompatibility. They measured the vitality of breast cancer cells, breast cancer stem cells and normal cells by MTT, and analyzed them by MALDI in positive ion mode. To sum up, when using MALDI single-cell analysis, researchers make improvements from different angles and aspects. In terms of the matrix, D. C. Perdian et al.57 used colloidal silver, which could provide a uniform matrix, instead of a traditional organic acid matrix. From the aspect of cell introduction, Akihito Korenaga et al.58 provided a high-throughput method of introducing single cells or several cells by the single-cell inkjet method. To improve the device, Peter V. Shanta et al.59 used a plasma gold film instead of a stainless steel plate to localize cells and enhance ionization. From the aspect of sample processing, M. Manikandan et al.60 used microwave digestion, and they designed gold nanoparticles, AuNPs, which acted as a “sensor” and enhanced the signal of the substance to be measured by 60%.
MALDI can also be used to perform mass spectrometry imaging (MSI). To image with MALDI, the mass spectrum array is collected by the pulsed laser, so as to image according to the signal of ions with a specific m/z at different spatial features.11 MALDI imaging can be used in the clinical diagnosis and treatment of cancer,61,62 such as detecting the distribution of irinotecan in rectal tumors.63
The main reasons that limit the imaging resolution are analyte displacement during substrate deposition, limited wavelength range of the laser, and the use of a substrate during crystallization.11,48 The lateral resolution of commercial instruments can reach about 10 μm.64 Using an ultraviolet laser to ionize the sample can improve the imaging resolution to the sub-micron level.11 In order to improve the resolution of imaging, a variety of substrate deposition methods have been developed to deposit the substrate uniformly.64
MALDI imaging analysis needs to select the appropriate stain and staining method. Commonly used stains are tissue annatto and eosin (H&E), but H&E is not suitable for the imaging analysis of MALDI, while toluenesulfonyl violet and methylene blue are suitable for MALDI staining. In addition, the sensitivity of the instrument is one of the limiting factors of imaging resolution for single-cell MALDI. Therefore, developing a mass spectrometer with higher resolution is one of the future development directions.64
High quality MSI images with micron-level lateral resolution can be obtained. Mario Kompauer et al.65 combined an Orbitrap mass spectrometer with their developed AP-MALDI MSI source. MALDI mass spectrometry imaging, which can be analyzed under atmospheric pressure, has been developed to monitor the distribution of phospholipids, peptides, and metabolites at the subcellular level with a lateral spatial resolution of 1.4 μm. They developed the MSI source based on the AP-MALDI source, using a new laser focusing objective with a numerical aperture (NA) of 0.9 and a free working distance of 18 mm at 337 nm, which reduced the ablation area to 1/13 of that of commercial instruments. Maria Emilia Dueñas et al.66 used MALDI with a spatial resolution of 5 μm to image 3D phospholipids (including PC, PE, and PI) of single zebrafish embryonic cells. This is the first application of MALDI 3D imaging to single cells. During imaging, 2D imaging was performed on 62 consecutive sections, and data were integrated and constructed to form 3D images. Bo Yang et al.67 also used imaging mass spectrometry to analyze lipids and metabolites in single cells. Tyler A. Zimmerman et al.68 used MALDI to perform imaging analysis on neuron cultures at the cellular level. They cultured Aplysia californica neurons on glass beads in the stretchable Parafilm M layer. By the stretch imaging method, different bead islands were formed in the tissue, with one or several cells in each bead island, and then MSI was performed on neuropeptides of neurons. The tensile imaging method does not consume samples significantly and is convenient for repeated detection. It is mainly used to detect small, spatially isolated samples. The analytes were identified by MS/MS in their study. Laura Capolupo et al.69 used MALDI to image the lipids of low-passage primary dermal human fibroblasts (dHFs) with an imaging resolution of 25 to 50 μm2 pixel size, and the characteristics of the lipids were verified by ESI-LC/MS, so as to study the lipidomes of single-cell heterogeneity. At the same time, they also sequenced single-cell mRNA and conducted transcriptome research. They found that sphingolipids were markers to distinguish the lipid state of cells and could be used as a regulator of fibroblast growth factor 2 (FGF2), among which globo-series sphingolipids and ganglio-series glycosphingolipids had positive and negative regulatory effects, respectively. The FGF2 signal also regulates the level of sphingolipids in turn. They believe that the lipid level may affect the transcription state, thus affecting the fate of cells. Gangqi Wang et al.70 used 13C isotope tracing and MALDI imaging to study the dynamic metabolic process in renal tissue slices with subcellular resolution (5 × 5 μm2 pixel size). When analyzing kidney tissue, they first used MALDI-MSI to obtain the characteristics of metabolites and lipids, stained the tissue, and then used multiplexed immunofluorescence (IF) imaging to distinguish cell types. They used the 13C labeling technique to study glycolysis and the TCA process in kidney cells. Through this method, the metabolism of the renal tissue microenvironment can be monitored at the single-cell level. This method can also be extended to other types of cells to explore the specific metabolic changes to cells in inflammation and cancer. Daniel C. Castro71 used single-cell MALDI imaging to characterize single organelles at the subcellular level, and analyzed the contents of lipids and peptides in 0.5 to 2 μm-diameter dense-core vesicles (DCVs) and electron lucent vesicles (LVs) of the exocrine atrial gland (AG). They used the sequence of image algorithms to identify the location of a single DCV without marking. They used LC-MS/MS to determine the sequence of the peptide. They found that there was a significant difference in lipids between the two vesicles, so they used a machine learning algorithm to distinguish DCVs from LVs according to lipid content. Andre Zavalin et al.72 proposed a new method to improve spatial resolution by optimizing the instrumental method of MALDI imaging. They developed a transmission geometric vacuum ion source that minimized the laser spot. They modified the light source of the laser imaging instrument, installed a window at the back of the chamber, cut the XY base, and mounted the support with the objective lens onto the sample holder. The objective lens can make the laser converge with a diameter less than 1 μm. They replaced the original optical encoder with a linear encoder with a resolution of 0.1 μm. For imaging, they selected RKO and HEK-293 cells as subjects to perform the subcellular imaging of their lipid profiles. By comparing data of single cells with the extract, it was verified that the detected spectra were from a single cell. Fig. 4II shows the spectrum obtained by MALDI. M. Niehaus et al.73 developed an optimized MALDI-MSI method in transmission-mode geometry (t-MALDI-MSI). The instrument adopts laser-induced post-ionization (MALDI-2), a t-MALDI-2 ion source and an Orbitrap mass analyzer to analyze, and the spatial resolution reaches 600 nm. Laser-induced ionization significantly enhanced the sensitivity of imaging. Vero B4 cells were imaged based on the m/z information of lipids. At the same time, the device can also contain a built-in optical microscope, which can simultaneously obtain optical images and ion images, providing a new idea for targeted imaging. Tanja Bien et al.74 combined an optical microscope with MALDI-2-MS imaging, and performed single-cell lipid analysis by overlapping and correlating data obtained from the two methods, and the imaging pixel was ≤2 μm. They shortened the sample preparation time by optimizing matrix preparation and using chamber slides. Single cell MS data are processed by machine learning, so that cell subtypes can be distinguished. They took Vero-B4 and Caki-2 as examples to explore the heterogeneity between cells, and THP-1 cells as examples to explore the heterogeneity changes in the process of cell differentiation.
In a word, a spatial imaging rate of 600 nm has been achieved by MALDI mass spectrometry, and subcellular imaging has been completed.72,73 Dueñas et al.66 also completed single-cell 3D lipid imaging. It is worth noting that Zimmerman et al.68 developed a unique sample preparation method, namely, the tensile imaging method, by forming pearl islands, allowing cells to separate. Machine learning algorithms are often used to study heterogeneity and distinguish cell types and organelle types.71,74 These studies provide valuable experience for the future field of MALDI single-cell imaging.
3.3. Summary of LDI analysis methods
The advantages and disadvantages of LDI are summarized and shown in Table 2.| LDI | Advantages | Disadvantages | Ref. |
|---|---|---|---|
| Matrix-free LDI | Mass spectrometry imaging can be performed. It can prevent interference from the matrix, so it has a higher spatial imaging rate than MALDI and is more suitable for analyzing small molecular substances. | The substances that can be analyzed have some limitations. It is relatively unsuitable for analyzing high molecular weight substances. | 39 and 48 |
| MALDI | Mass spectrometry imaging can be performed. It is suitable for analyzing biological macromolecules, such as protein and peptides. | The imaging resolution is relatively low. After sample preparation, it is often necessary to ionize it under vacuum. There is a matrix effect in the analysis, which is relatively unsuitable for the analysis of low molecular weight substances. | 11, 39 and 43 |
4. SIMS
Secondary ion mass spectrometry (SIMS) is a commonly used mass spectrometry method that bombards the surface of a sample with a specific ion beam under vacuum conditions to sputter partially charged secondary ions, which can be detected and analyzed by mass spectrometry.75,76In general, the surface of the sample is bombarded with a primary ion beam with high energy such as Ga, Ar, etc. Through elastic or inelastic collisions, the energy of these ions is transferred to atoms in the lattice, and after a series of cascade collisions, some atoms or atomic clusters on the surface of the sample will absorb energy and sputter, and ionization will occur during the process of leaving the surface. The low-energy secondary ion sputtering point is far away from the bombardment point of the primary ion (up to 10 nm). The final collision results in low-energy sputtering (∼20 eV) of secondary ions. More than 95% of the secondary ions originate from the top two layers of the solid, so SIMS is a highly sensitive mass spectrometry technique for surface analysis.77 In addition, SIMS can be used to analyze the molecular weight and molecular structure information of compounds in the sample. It has high sensitivity for the detection of many components and is the most sensitive technique for surface analysis.78 However, SIMS uses a relatively “hard” ion source. Using a high-energy primary ion beam to bombard the surface of the sample will generate many fragment ions of target molecules, which makes the analysis of biological macromolecules difficult. Therefore, SIMS is currently mainly used for lipids, small molecule metabolites, molecular fragments and elemental analysis. The process is shown in Fig. 2III. In this section, we mainly discuss the mass spectrometry analysis of single cells by time-of-flight secondary ion mass spectrometry (TOF-SIMS), Nano-SIMS, and gas cluster ion beam secondary ion mass spectrometry (GCIB-SIMS).
4.1. TOF-SIMS
TOF-SIMS uses a pulsed primary ion beam to bombard the surface of a sample and accelerate the sputtered secondary ions to a certain energy. The heavier ions reach the detector slowly and the lighter ions reach there quickly. According to the time of flight, the mass to charge ratio of secondary ions can be determined, and then the composition information of the solid surface can be obtained.77 The detection limit of TOF-SIMS can reach the order of ppb to ppm; it can isolate an individual cell layer and clearly display structural and chemical information from individual cells.79–82 TOF-SIMS can be used to perform 2D and 3D imaging to characterize the distribution of cellular components and some representative studies are described in subsequent paragraphs.T. L. Colliver et al.83 developed a complete cold chain freeze-fracture methodology to test the availability of using TOF-SIMS imaging for the molecular analysis of frozen hydrated biological samples. But water on the surface of the sample may be the main source of interference owing to the fact that the technique just samples the first few monolayers of a sample. They placed a cold trap (−196 °C for the fracture knife and housing) near the fractured sample at a higher temperature (−97 to −113 °C) to minimize this problem. By utilizing the above processing techniques, it was possible to demonstrate for the first time that TOF-SIMS imaging techniques could be used obtain images of the types of molecules on the cell surface using a submicron ion probe beam. T. P. Roddy et al.84 used a combination of bright field, scanning ion, and fluorescence microscopy to complement TOF-SIMS imaging of natural biomolecules. Bright-field microscopy provided a fuzzy visualization of cells in freeze–hydration samples, while scanning ion imaging provided morphological views of freeze-fractured cells after TOF-SIMS analysis was completed. By using selective fluorescent labels, fluorescence microscopy can locate individual mammalian cells in a complex ice matrix of freeze-fractured samples. They used this comprehensive approach to map lipids in a single cell membrane, helping to better understand the functional role of specific lipids in cell membrane function. Recent advances in SIMS technology have focused on generating new ion sources that can eject more complete molecular and biometric species from the sample in turn. The main disadvantage of conventional TOF-SIMS analysers is the requirement for short (ns) primary ion beam pulses. The ion beam is swept through a small aperture to generate the desired pulse. The very short pulses needed to produce high mass resolution are incompatible with obtaining the high spatial resolution of which the ion beam is capable. The very low duty cycle of the pulsed ion beam also means that imaging and depth profiling experiments will take a very long time. This may be exacerbated by the decrease in ion beam current after opening and focusing the beam to a fine point size. To overcome many shortcomings of current TOF-SIMS spectrometers, John S. Fletcher et al.85 developed an instrument that did not require a pulsed primary ion beam. The main aim of this development was to enable the use of a continuous primary ion beam while maintaining the advantages offered by time-of-flight analysers, and thus, generate continuous secondary ions from the sample using a 40 keV C60 primary ion beam in dc mode. The instrument provides higher throughput and greatly reduces the time of analysis, thus allowing 3D images to be generated on real timescales, which provides a further tool for species identification. Lu Huang et al.86 used TOF-SIMS to study the phenotypic changes to cisplatin-induced HeLa cells at the early stage of apoptosis, and analyzed the main molecular sources of variation through principal component analysis (PCA). They randomly inoculated HeLa cells onto ITO glass and cultured them for 24 hours prior to cisplatin induction. For high-throughput single-cell analysis, cell arrays were produced using ScP. They used a 25 kV Bi3+ beam for analysis and a 10 kV C60+ beam for etching, both of which bombarded the target at an incidence angle of 45°. TOF-SIMS was used to detect the cell populations treated with cisplatin over different time periods. In positive ion mode, both PC1 (66%) and PC2 (27%) captured a significant proportion of the population variance. Cells not induced by cisplatin are clearly separated from cells administered with cisplatin in PC1. The groups induced with cisplatin for 2 and 4 h overlapped with partial overlap with approximate PC2 values. They found significant reductions in ion species in cholesterol and fatty acids after cisplatin treatment, the marker of apoptosis, and consistent with results from other techniques. Moreover, the cells after 10 h of cisplatin induction showed significant differences from the other two groups in PC2. The results above suggest that TOF-SIMS can successfully detect cisplatin-induced time-dependent phenotypic changes in the early stages of apoptosis. A unique advantage of SIMS imaging is the ability to capture three-dimensional molecular distributions with very high spatial resolution, enabling subcellular level studies. Hua Tian et al.87 analyzed the spatial and temporal distribution of lipid debris on the surface of Xenopus embryos during embryo cleavage by TOF-SIMS 3D imaging, as shown in Fig. 5I. The results showed that TOF-SIMS was able to detect multiple components (such as glycerophosphocholine, cholesterol, vitamin E, diacylglycerol and triacylglycerol, etc.) in a single laevis embryo. The simple sample preparation for this research method does not require lipid extraction and preserves the original chemistry on the cell membrane, opening up the possibility to understand the function of biomolecules and advance biology using high-resolution imaging mass spectrometry. Evenbratt et al.88 used TOF-SIMS to detect biomolecule monolayers and multiple binding proteins on glass surfaces with high precision. The resulting TOF-SIMS imaging and line scan data provide detailed information about the distribution of adsorbed proteins, allowing microscopic research results to be used to open up the possibility of precisely controlling cellular responses.
4.2. Nano-SIMS
Nano-SIMS is a type of dynamic SIMS, which bombards the solid surface with a primary ion beam (such as Cs+ or O−), and then introduces the secondary ions sputtered from the surface into the magnetic field of the mass analyzer. Different secondary ions are separated in the electrostatic field area, detected, recorded and imaged by the mass spectrometer detector, so as to obtain the composition of the elements and compounds on the surface of the analyzed sample. It is capable of elemental and isotopic imaging at a lateral spatial resolution of about 50 nm.89 The ion species is mainly determined according to the different deflection radii of the secondary ions in the magnetic field; its spatial resolution can reach sub-micron or nanoscale levels, and the micron-level element distribution and composition information about the surface of the sample can be obtained. Nano-SIMS can be applied to track a variety of organic and inorganic compounds within cells.90 In order to improve the secondary ion yield, the development of new ion sources has become a research hotspot, and applications in biological science research are also maturing.91 Some representative studies are described in subsequent paragraphs.Vera I. Slaveykova et al.92 used the Cameca NanoSIMS50 ion microprobe to detect and image the distribution of copper ions in microalgal cells exposed to nanomolar and micromolar copper concentrations. The correlation of secondary ion maps of 63Cu− with those of 12C14N− and 31P− suggested that Cu may be related to cell components rich in protein and phosphorus. These results highlighted the potential of NanoSIMS for the intracellular tracking of essential trace elements, such as Cu, in Chlorella crusteri single cells. Mengyuan Su et al.93 combined Nano-SIMS with transmission electron microscopy (TEM) to study the morphology and monophyllium of specific microorganisms under pure bacterial culture conditions, and efficiently degraded polycyclic aromatic hydrocarbons with Image J software. The results of qualitative characterization of the elemental distribution and changes to 13C, 14N, 16O, and 12C in the BDG-3 monobacterium show the efficient degradation of polycyclic aromatic hydrocarbons by BDG-3, which can absorb and metabolize 13C-glucose, providing it with a carbon source of energy. This study provides a new analytical method for further exploring the degradation and metabolism of more complex environmental pollutants in microorganisms. Kai Chen et al.94 studied the take up of stable isotope-labeled lipids by intestinal enterocytes with correlative backscattered electron (BSE) and nanoscale secondary ion mass spectrometry (Nano-SIMS) imaging. Nano-SIMS images of the same section show the elemental and isotopic contents of the sections (e.g., 1H−, 2H−, 12C−, 13C−, 12C14N−, 32S−) at ∼50 nm resolution, making it possible to define both enterocyte morphology and the location of stable isotope-labeled lipids within enterocytes (Fig. 5II). Their correlative BSE/Nano-SIMS approach will be very useful for investigating the impact of genetic interventions and drug therapy on lipid absorption in the intestine.
4.3. GCIB-SIMS
GCIB is a high spatial resolution imaging mass spectrometry technique, the material processing of which is based on the use of gaseous materials using charged cluster ions composed of hundreds or thousands of atoms or molecules. Individual gas atoms are first condensed into neutral clusters, which are then ionized and accelerated.95–97 When the high-energy cluster ion hits the target surface, it interacts with many target atoms almost simultaneously and delivers high energy to the very small target. Simultaneous energetic interactions between the many atoms that make up the cluster and the target atoms in turn lead to highly nonlinear sputtering and implantation effects. There are other distinct advantages of using GCIB that are related to the low mass to charge ratios of cluster ions, which consist of up to thousands of atoms typically carrying only one charge or even less charge, so that cluster ion beams can transmit more atoms than monomer ion beams at the same current density. GCIB-SIMS has the ability to map intact lipids in biological systems with a lateral resolution of 10 μm and has corresponding applications in metabolomics.98 Some representative studies are described in subsequent paragraphs.Peroxidized phosphatidylethanolamine (PEox) species serve as predictive biomarkers of ferroptosis, a new procedure that regulates cell death; these have not been directly detected by imaging protocols in specific cell types and tissues. By applying GCIB-SIMS imaging, Sparvero et al.99 mapped phosphatidyl peroxide at the single-cell/subcellular level with a spatial resolution of 1.2 μm in ferrous H9c2 cardiomyocytes and cortical/hippocampal neurons after traumatic brain injury (Fig. 5III). This study was able to observe physiological levels of very low abundance lipid peroxides in subcellular compartments and their accumulation process under disease conditions. Pareek et al.100 used GCIB-SIMS to study the cytoplasmic distribution of intact molecular ions of purine biosynthesis pathway intermediates and terminal nucleotides in frozen hydration monolayer HeLa cells (Fig. 5IV). They observed the de novo biosynthesis of purines in multi-enzyme complex purine and found that multi-enzyme complex purine contained 9 enzymes with synergistic effects, which regulated the synthesis ratio of adenylate and guanylate while ensuring the high efficiency of the synthesis chain. This study highlights the application of high-resolution GCIB-SIMS for multiplexed biomolecular analysis at the level of single cells and these findings have important implications for the course of cancer treatment.
4.4. Summary of SIMS analysis methods
The advantages and disadvantages of SIMS are summarized and shown in Table 3.| SIMS | Advantages | Disadvantages | Ref. |
|---|---|---|---|
| TOF-SIMS | The limit of detection can reach ppb–ppm. The distribution of cell components can be characterized by 2D and 3D imaging. | Sputtering will cause minor damage to the sample. It is difficult to guarantee both high mass resolution and high spatial resolution. | 79–88 |
| Nano-SIMS | It has high sensitivity, high mass resolution and high spatial resolution. It is often used to track trace elements in a single cell. | Generally used in conjunction with isotope labeling techniques, the process of preparing samples for analysis may have an impact on the isotopic composition of cells, or may affect the correct analysis of single cells if the selection of the region reflecting isotopic enrichment is inappropriate. | 89 and 91–94 |
| GCIB-SIMS | The ability to map complete lipids with a lateral resolution of 10 μm. It is often used in metabolomics studies. | For the practical analysis of trace components in complex biomaterials, the sensitivity of this technique is still insufficient. | 97–100 |
5. ICP-MS
Inductively coupled plasma mass spectrometry (ICP-MS) uses the high temperature of the plasma torch to gasify and ionize metal elements under normal pressure conditions, and convert them into positively charged ions. Finally, the separation was carried out according to different mass to charge ratios by using a mass spectrometer.101,102 ICP-MS has the advantages of high precision, sensitivity and accuracy, a wide linear range, multi-element measurement capability, and isotopic analysis capability.103 It is also used for single-cell analysis and the identification of ppb-scale nanoparticle concentrations, particle size, and particle size distribution.104,105 The process is shown in Fig. 2IV.5.1. Mass cytometry
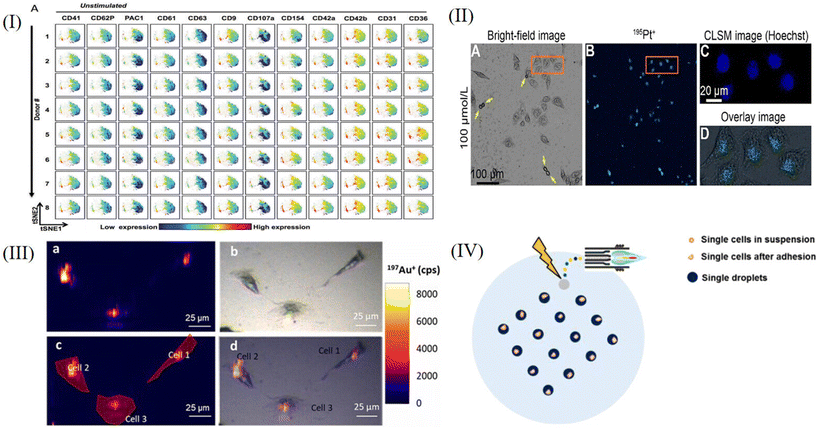
Mass cytometry is a flow technology that uses mass spectrometry to perform multi-parameter detection of single cells. It is a recently developed method that merges time-of-flight ICP-MS with flow cytometry. It can simultaneously measure more than 50 compounds at the single cell level, significantly enhancing the ability to evaluate cellular metabolic processes and complex cellular systems.106–109 Some representative studies are described in subsequent paragraphs.O. Ornatsky et al.110 developed a novel application of ICP-MS-linked metal-tagged immunophenotyping with great potential for highly multiproteomic analysis. The expression of BCR/Abl, myeloid surface antigen CD33, human stem cell factor receptor c-Kit and integrin receptor VLA-4 were studied in human leukemia model cell lines. They combined four commercially available labels (Au, Sm, Eu, and Tb) with the secondary antibody to perform a 4-plex analysis on the condition that the primary antibodies were not cross-reactive. The results suggested that as an analytical detector, ICP-MS could improve the immunoassay performance, and while research and feasibility studies for multiplex using metal-coupled reagents were at a very early stage, it was clear that more than four antigens could be accurately detected simultaneously using the ICP-MS instrument. D. Majonis et al.111 described the synthesis of metal-chelated polymers (MCPs) with an orthogonal end-group and four different pendant polyaminocarboxylate ligands (EDTA, DTPA, TTHA, DOTA), a fluorescein molecule or a bismaleimide ligand for antibody attachment. The polymer samples were loaded with three different types of lanthanide ions as well as palladium and platinum ions. The number of metal atoms in each chain was determined by a combination of UV/visible and conventional ICP-MS measurements. When they performed large-scale cell bioassays using P(EDTA)-maleimide loaded with palladium or platinum, they obtained strange results. The secondary antibody–polymer conjugate lost its affinity for the target primary antibody and selectively bound to dead cells. They held the view that it was because the soft metal atoms were not sufficiently isolated by their ligands, so these palladium or platinum atoms formed new bonds with the soft ligands found inside and outside the dead cells. This was an unexpected result, but it could effectively identify dead cells and provide a new, reliable assay for distinguishing between living and dead cells. Studies by G. K. Behbehani et al.112 described methods for delineating cell cycle phases, using 5-iodo-2-deoxyuridine (IdU) to label S-phase cells, while using antibodies against cyclin B1, cyclin A, and phosphorylated histone H3 (S28) to characterize other cell cycle phases. They developed an antibody against phosphorylated retinoblastoma protein (Rb) serines 807 and 811 to isolate cells in the G0 and G1 phases of the cell cycle. The cell cycle distribution of normal and cancer cell populations produced by this large-scale cytology method was comparable to that obtained by conventional fluorescent cytology techniques. They applied it to map the phases of the cell cycle across the hematopoietic level in healthy human bone marrow as a prelude to subsequent research into cancer and other diseases in this lineage. Blair et al.113 described a group of 14 platelet-specific metal-conjugated antibodies (targeting clusters of differentiation CD9, CD29, CD31, CD36, CD41, CD42a, CD42b, CD61, CD62P, CD63, CD107a, CD154, glycoprotein [GP]VI and activated integrin αIIbβ3), using large-scale cytometry staining and analysis of platelets. The levels of all 14 compounds detected in each cell by cytometry could be used to identify the consistency of platelet immunophenotypes (Fig. 6I). The analysis showed that the surface expression levels of activated GPIIb-IIIa and P-selectin were significantly increased due to the presence of the platelet subset, suggesting that this subset might play a greater role in thrombosis than other platelet subsets. A. L. S. Oliver et al.114 intended to establish a new MC-based quantification procedure to receive absolute numbers of NPs per single cell by using a calibration that considered the specific transmission efficiency (TE) of suspended NPs. In their study, they provided an MC-specific calibration procedure to precisely calculate absolute numbers of NPs per single cell. Combined with its unique feature of multiplexing up to 50 parameters, MC provides much information at the single cell level. It provides a new option for nanotoxicological analysis of NP–cell interactions in cell lines and complex biological samples such as blood or other body fluids.
5.2. LA-ICP-MS
Laser ablation (LA) can be combined with inductively coupled plasma mass spectrometry (ICP-MS). In biological applications, LA-ICP-MS can be applied to create images of metal distributions in specimens. The whole cell sample is progressively laser ablated before being introduced into the mass spectrometer, and this special sampling mode provides high spatial resolution and flexibility for the sample, greatly improving the detection sensitivity.115–119 Some representative studies are described in subsequent paragraphs.Y. F. Meng et al.120 incubated HeLa cells with different concentrations of carboplatin on quartz wafers and used the nanoscale LA-ICP-MSI technique to help visualize the drug target in a cell. They observed cell contraction and apoptosis (shown by yellow arrows) in bright-field microscopic images of cells cultured with the drug for 12 hours (Fig. 6IIA) and the MS image of 195Pt+ in the same area (Fig. 6IIB). By comparing the nuclear dye Hoechst 33342 (Fig. 6IIC) with the 195Pt+ image (Fig. 6IID) for the regions outlined, carboplatin is shown to be primarily located in the nucleus. This results verifies the mechanism of carboplatin binding with DNA inducing apoptosis of cancer cells, causing cross-linking, and thus, destroying the function of DNA and inhibiting cell mitosis. Quantitative analysis of elements and biomolecules in biological samples by LA-ICP-MS is still hindered by the lack of sufficient matrix-matched reference materials. To overcome the current limitations regarding LA-ICP-MS quantification in cultured cells, A. Lores-Padin et al.121 proposed a new matrix-matched calibration strategy, which fully mimicked the matrix of cultured cells. Quantitative imaging of two cytoplasmic proteins (MT2A and APOE) in single HRPEsv cells was performed by LA-ICP-MS, both in cytokine interleukin-1α (IL-1α) inflammatory response cells and in control cells (CT). The selected proteins in single cells were analyzed by LA-ICP-MS using Au nanoclusters (AuNCs) as specific antibody labels (Fig. 6III). The average results of MT2A and APOE quantities obtained by the proposed methodology were consistent with quantitative results obtained by using commercial ELISA kits, indicating that APOE levels decreased after proinflammatory therapy, while MT2A showed protein overexpression after treatment. It should be emphasized that using the proposed methodology for LA-ICP-MS, it is possible to determine not only the total mass of proteins in each cell, but also the quantitative distribution of target proteins. In order to overcome the two main obstacles of LA-ICP-MS of low analytical throughput and lack of commercial reference materials, Ling-Na Zheng et al.122 demonstrated the principles of a new approach termed “single-cell isotope dilution analysis” (SCIDA) to remove these two obstacles. They studied the take up of silver NPs (AgNPs) at the single-cell level using the LA-ICP-MS method and chose macrophages as models. In their study, the accurate quantification of AgNPs in single cells was achieved by using isotope dilution LA-ICP-MS (Fig. 6IV). The average Ag mass of 1100 single cells, 396 ± 219 fg Ag per cell, was in good accord with the average of the population of cells determined by solution ICP-MS analysis. The detection limit was 0.2 fg Ag per cell. The SCIDA approach is expected to be widely applied for the study of cell–NP interactions and the biological effects of NPs at the single-cell level.
5.3. Summary of ICP-MS analysis methods
The advantages and disadvantages of ICP-MS are summarized and shown in Table 4.| ICP-MS | Advantages | Disadvantages | Ref. |
|---|---|---|---|
| Mass cytometry | The presence of the metal element as a label in the cell is extremely low, eliminating inherent background problems, and more than 50 markers can be measured simultaneously at the single-cell level. | Each probe is strictly required to have a separate metal isotope. Small molecule metabolites cannot be detected and continuous dynamic monitoring of a cell is not possible. | 106–114 |
| LA-ICP-MS | Low sample consumption; preparation is simple. In situ, real-time, fast analysis. | Complex isotope fractionation behavior, significant matrix effects and mass spectrum interference limit the accuracy and precision of analysis. | 115–119, 121 and 122 |
6. Conclusions and future perspectives
With the continuous development of cell biology, the requirement for single-cell analysis technology is also increasing. Higher spatial resolution and detection sensitivity have become the development trends for single-cell mass spectrometry. It mainly involves investigating subcellular compartments and precisely analyzing metabolites corresponding to specific organelles and pushing the limit of detection sensitivity to broaden the detection spectrum of cellular metabolites, especially for those with low abundance or low ionization efficiency.8 Existing mass spectrometry techniques based on ESI-MS, LDI-MS, SIMS, and ICP-MS are also being continuously improved. We introduce the principles of these analytical methods and summarize the latest advances in single-cell analysis and imaging studies of these different mass spectrometry techniques. ESI-based single-cell mass spectrometry methods can be studied at the subcellular level and can be observed under microscopy to precisely control the sampling process, but this also limits the throughput of such research methods to some extent. Although LDI-based single-cell mass spectrometry methods have high throughput and can detect more kinds of metabolites, such methods are limited by the spatial resolution of laser light sources and it is difficult to perform imaging analysis at the subcellular level. SIMS-based single-cell mass spectrometry analysis methods have high spatial resolution. SIMS is a mass spectrometry imaging method that can achieve three-dimensional imaging, but it uses a relatively “hard” ion source, and a primary ion beam with high energy will produce fragmented ions of many target molecules after bombardment, so it is difficult to achieve the imaging of macromolecules. ICP-based single-cell mass spectrometry methods have high sensitivity, but it is difficult to ensure the stability of the morphology of each element during sample preparation.123Now single-cell-based mass spectrometry is developing towards methodological diversification, because any method has certain flaws. It should also be emphasized that different methods can complement each other and jointly provide a more comprehensive analysis environment for single-cell mass spectrometry, thereby obtaining more comprehensive qualitative and quantitative data for single-cell multi-omics. Single-cell mass spectrometry technology is continuously being improved and perfected for biological research; this can further promote the development of biological research and ultimately provide technical support for drug development, medical research, ecological protection and other important scientific fields.
Author contributions
Lei Yin and Meiyun Shi conceived the idea for the article. Xiangyi Xu wrote the abstract, introduction, ESI-MS and LDI-MS sections. Xuanxi Jiang wrote the SIMS, ICP-MS and conclusions and future perspectives sections. Lei Yin and Meiyun Shi revised the article. All the authors agreed on the final version of the manuscript.Conflicts of interest
All the authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant Nos. 81603182 and 81703607), the Fundamental Research Funds for the Central Universities DUT21RC (3)057, and the Science and Technology Innovation Fund of Dalian (2022JJ13SN075).References
- L. F. Vistain and S. Tay, Trends Biochem. Sci., 2021, 46, 661–672 CrossRef CAS PubMed.
- X. Shao, L. X. Weng, M. X. Gao and X. M. Zhang, TrAC, Trends Anal. Chem., 2019, 120, 115666 CrossRef CAS.
- A. Amantonico, P. L. Urban and R. Zenobi, Anal. Bioanal. Chem., 2010, 398, 2493–2504 CrossRef CAS PubMed.
- L. P. de Souza, M. Borghi and A. Fernie, Int. J. Mol. Sci., 2020, 21, 8987 CrossRef CAS PubMed.
- S. J. Altschuler and L. F. Wu, Cell, 2010, 141, 559–563 CrossRef CAS PubMed.
- J. E. Ferrell and E. M. Machleder, Science, 1998, 280, 895–898 CrossRef CAS PubMed.
- L. W. Zhang and A. Vertes, Angew. Chem., Int. Ed., 2018, 57, 4466–4477 CrossRef CAS PubMed.
- R. Liu and Z. Yang, Anal. Chim. Acta, 2021, 1143, 124–134 CrossRef CAS PubMed.
- G. Z. Zhu, Y. L. Shao, Y. X. Liu, T. Pei, L. J. Li, D. T. Zhang, G. S. Guo and X. Y. Wang, TrAC, Trends Anal. Chem., 2021, 143, 116351 CrossRef CAS.
- J. D. Wang, J. F. Song, C. Li, X. H. Dai, X. Fang, X. Y. Gong and Z. H. Ye, Chin. J. Anal. Chem., 2020, 48, 969–980 CAS.
- R. Hu, Y. Li, Y. H. Yang and M. L. Liu, Mass Spectrom. Rev., 2023, 42, 67–94 CrossRef CAS PubMed.
- Y. L. Wang, R. Jin, N. Sojic, D. C. Jiang and H. Y. Chen, Angew. Chem., Int. Ed., 2020, 59, 10416–10420 CrossRef CAS PubMed.
- S. T. Xu, C. Yang, X. P. Yan and H. W. Liu, Anal. Bioanal. Chem., 2022, 414, 219–233 CrossRef CAS PubMed.
- P. Kebarle and U. H. Verkerk, Mass Spectrom. Rev., 2009, 28, 898–917 CrossRef CAS.
- P. J. Taylor, Clin. Biochem., 2005, 38, 328–334 CrossRef CAS PubMed.
- H. Mizuno, N. Tsuyama, T. Harada and T. Masujima, J. Mass Spectrom., 2008, 43, 1692–1700 CrossRef CAS PubMed.
- H. Mizuno, N. Tsuyama, S. Date, T. Harada and T. Masujima, Anal. Sci., 2008, 24, 1525–1527 CrossRef CAS PubMed.
- N. Tsuyama, H. Mizuno, E. Tokunaga and T. Masujima, Anal. Sci., 2008, 24, 559–561 CrossRef CAS PubMed.
- M. L. Tejedor, H. Mizuno, N. Tsuyama, T. Harada and T. Masujima, Anal. Chem., 2012, 84, 5221–5228 CrossRef PubMed.
- Y. Abouleila, K. Onidani, A. Ali, H. Shoji, T. Kawai, C. T. Lim, V. Kumar, S. Okaya, K. Kato, E. Hiyama, T. Yanagida, T. Masujima, Y. Shimizu and K. Honda, Cancer Sci., 2019, 110, 697–706 CrossRef CAS PubMed.
- X. C. Zhang, Q. C. Zang, H. S. Zhao, X. X. Ma, X. Y. Pan, J. X. Feng, S. C. Zhang, R. P. Zhang, Z. Abliz and X. R. Zhang, Anal. Chem., 2018, 90, 9897–9903 CrossRef CAS PubMed.
- Z. Wei, X. Xiong, C. Guo, X. Si, Y. Zhao, M. He, C. Yang, W. Xu, F. Tang, X. Fang, S. Zhang and X. Zhang, Anal. Chem., 2015, 87, 11242–11248 CrossRef CAS PubMed.
- H. Zhu, N. Wang, L. Yao, Q. Chen, R. Zhang, J. Qian, Y. Hou, W. Guo, S. Fan, S. Liu, Q. Zhao, F. Du, X. Zuo, Y. Guo, Y. Xu, J. Li, T. Xue, K. Zhong, X. Song, G. Huang and W. Xiong, Cell, 2018, 173, 1716–1727.e17 CrossRef CAS PubMed.
- H. Y. Zhu, Q. Q. Li, T. P. Liao, X. Yin, Q. Chen, Z. Y. Wang, M. F. Dai, L. Yi, S. Y. Ge, C. J. Miao, W. P. Zeng, L. L. Qu, Z. Y. Ju, G. M. Huang, C. L. Cang and W. Xiong, Nat. Methods, 2021, 18, 788–798 CrossRef CAS PubMed.
- K. Hiraoka, K. Nishidate, K. Mori, D. Asakawa and S. Suzuki, Rapid Commun. Mass Spectrom., 2007, 21, 3139–3144 CrossRef CAS.
- X. Y. Gong, Y. Y. Zhao, S. Q. Cai, S. J. Fu, C. D. Yang, S. C. Zhang and X. R. Zhang, Anal. Chem., 2014, 86, 3809–3816 CrossRef CAS PubMed.
- F. Chen, L. Lin, J. Zhang, Z. He, K. Uchiyama and J. M. Lin, Anal. Chem., 2016, 88, 4354–4360 CrossRef CAS PubMed.
- Z. Takats, J. M. Wiseman, B. Gologan and R. G. Cooks, Science, 2004, 306, 471–473 CrossRef CAS PubMed.
- P. J. Roach, J. Laskin and A. Laskin, Analyst, 2010, 135, 2233–2236 RSC.
- H. M. Bergman and I. Lanekoff, Analyst, 2017, 142, 3639–3647 RSC.
- S. A. Stopka, R. Khattar, B. J. Agtuca, C. R. Anderton, L. Pasa-Tolic, G. Stacey and A. Vertes, Front. Plant Sci., 2018, 9, 13 CrossRef PubMed.
- B. Shrestha and A. Vertes, Anal. Chem., 2009, 81, 8265–8271 CrossRef CAS PubMed.
- P. Nemes and A. Vertes, Anal. Chem., 2007, 79, 8098–8106 CrossRef CAS PubMed.
- R. M. Onjiko, D. O. Plotnick, S. A. Moody and P. Nemes, Anal. Methods, 2017, 9, 4964–4970 RSC.
- J. T. Aerts, K. R. Louis, S. R. Crandall, G. Govindaiah, C. L. Cox and J. V. Sweedler, Anal. Chem., 2014, 86, 3203–3208 CrossRef CAS PubMed.
- J. S. Mellors, K. Jorabchi, L. M. Smith and J. M. Ramsey, Anal. Chem., 2010, 82, 967–973 CrossRef CAS PubMed.
- T. Lapainis, S. S. Rubakhin and J. V. Sweedler, Anal. Chem., 2009, 81, 5858–5864 CrossRef CAS PubMed.
- L. S. Huang, M. L. Fang, K. A. Cupp-Sutton, Z. Wang, K. Smith and S. Wu, Anal. Chem., 2021, 93, 4479–4487 CrossRef CAS PubMed.
- D. S. Peterson, Mass Spectrom. Rev., 2007, 26, 19–34 CrossRef CAS PubMed.
- Y. Luo, C. Liu, Y. Qu and N. Fang, Bioanalysis, 2012, 4, 453–463 CrossRef CAS.
- M. Feucherolles and G. Frache, Cells, 2022, 11, 3900 CrossRef CAS PubMed.
- J. Leopold, Y. Popkova, K. M. Engel and J. Schiller, Biomolecules, 2018, 8, 173 CrossRef PubMed.
- L. J. Li, R. W. Garden and J. V. Sweedler, Trends Biotechnol., 2000, 18, 151–160 CrossRef CAS PubMed.
- A. van Belkum, S. Chatellier, V. Girard, D. Pincus, P. Deol and W. M. Dunne Jr., Expert Rev. Proteomics, 2015, 12, 595–605 CrossRef CAS PubMed.
- M. van Nuenen, D. Kirk, B. Gu and A. van Belkum, Chin. J. Lab. Med., 2015, 38, 64–66 CAS.
- M. Z. Israr, D. Bernieh, A. Salzano, S. Cassambai, Y. Yazaki and T. Suzuki, Clin. Chem. Lab. Med., 2020, 58, 883–896 CrossRef CAS PubMed.
- S. Tsuchida, H. Umemura and T. Nakayama, Molecules, 2020, 25, 4775 CrossRef CAS PubMed.
- T. Jaschinski, E. J. N. Helfrich, C. Bock, S. Wolfram, A. Svatos, C. Hertweck and G. Pohnert, J. Mass Spectrom., 2014, 49, 136–144 CrossRef CAS PubMed.
- D. Hölscher, R. Shroff, K. Knop, M. Gottschaldt, A. Crecelius, B. Schneider, D. G. Heckel, U. S. Schubert and A. Svatos, Plant J., 2009, 60, 907–918 CrossRef PubMed.
- J. Albrethsen, Clin. Chem., 2007, 53, 852–858 CrossRef CAS.
- P. A. Van Veelen, C. R. Jimenez, K. W. Li, W. C. Wildering, W. P. M. Geraerts, U. R. Tjaden and J. Vandergreef, Org. Mass Spectrom., 1993, 28, 1542–1546 CrossRef CAS.
- S. S. Rubakhin, W. T. Greenough and J. V. Sweedler, Anal. Chem., 2003, 75, 5374–5380 CrossRef CAS PubMed.
- S. S. Rubakhin and J. V. Sweedler, Anal. Chem., 2008, 80, 7128–7136 CrossRef CAS PubMed.
- Y. Gholipour, H. Nonami and R. Erra-Balsells, J. Am. Soc. Mass Spectrom., 2008, 19, 1841–1848 CrossRef CAS PubMed.
- J. S. Page, S. S. Rubakhin and J. V. Sweedler, Anal. Chem., 2002, 74, 497–503 CrossRef CAS PubMed.
- Y. C. Dai, C. Y. Li, J. Yi, Q. Qin, B. H. Liu and L. Qiao, Anal. Chem., 2020, 92, 8051–8057 CrossRef CAS PubMed.
- D. C. Perdian, S. Cha, J. Oh, D. S. Sakaguchi, E. S. Yeung and Y. J. Lee, Rapid Commun. Mass Spectrom., 2010, 24, 1147–1154 CrossRef CAS PubMed.
- A. Korenaga, F. Chen, H. Li, K. Uchiyama and J.-M. Lin, Talanta, 2017, 162, 474–478 CrossRef CAS PubMed.
- P. V. Shanta, B. Li, D. D. Stuart and Q. Cheng, Anal. Chem., 2020, 92, 6213–6217 CrossRef CAS PubMed.
- M. Manikandan and H.-F. Wu, Sens. Actuators, B, 2016, 231, 154–165 CrossRef CAS.
- J. Kriegsmann, M. Kriegsmann and R. Casadonte, Int. J. Oncol., 2015, 46, 893–906 CrossRef CAS PubMed.
- A. Nikitina, D. N. Huang, L. Li, N. Peterman, S. E. Cleavenger, F. M. Fernandez and M. L. Kemp, J. Am. Soc. Mass Spectrom., 2020, 31, 986–989 CrossRef CAS PubMed.
- X. Liu, C. Flinders, S. M. Mumenthaler and A. B. Hummon, J. Am. Soc. Mass Spectrom., 2018, 29, 516–526 CrossRef CAS PubMed.
- K. J. Boggio, E. Obasuyi, K. Sugino, S. B. Nelson, N. Y. R. Agar and J. N. Agar, Expert Rev. Proteomics, 2011, 8, 591–604 CrossRef CAS PubMed.
- M. Kompauer, S. Heiles and B. Spengler, Nat. Methods, 2017, 14, 90–96 CrossRef CAS PubMed.
- M. E. Dueñas, J. J. Essner and Y. J. Lee, Sci. Rep., 2017, 7, 14946 CrossRef PubMed.
- B. Yang, T. Tsui, R. M. Caprioli and J. L. Norris, Methods Mol. Biol., 2020, 2064, 125–134 CrossRef CAS PubMed.
- T. A. Zimmerman, S. S. Rubakhin and J. V. Sweedler, J. Am. Soc. Mass Spectrom., 2011, 22, 828–836 CrossRef CAS PubMed.
- L. Capolupo, I. Khven, A. R. Lederer, L. Mazzeo, G. Glousker, S. Ho, F. Russo, J. P. Montoya, D. R. Bhandari, A. P. Bowman, S. R. Ellis, R. Guiet, O. Burri, J. Detzner, J. Muthing, K. Homicsko, F. Kuonen, M. Gilliet, B. Spengler, R. M. A. Heeren, G. P. Dotto, G. La Manno and G. D'Angelo, Science, 2022, 376, eabh1623 CrossRef CAS PubMed.
- G. Q. Wang, B. Heijs, S. Kostidis, A. Mahfouz, R. G. J. Rietjens, R. Bijkerk, A. Koudijs, L. A. K. Van der Pluijm, C. W. Van den Berg, S. J. Dumas, P. Carmeliet, M. Giera, B. M. Van den Berg and T. J. Rabelink, Nat. Metab., 2022, 4, 1109–1118 CrossRef CAS PubMed.
- D. C. Castro, Y. R. Xie, S. S. Rubakhin, E. V. Romanova and J. V. Sweedler, Nat. Methods, 2021, 18, 1233–1238 CrossRef CAS PubMed.
- A. Zavalin, E. M. Todd, P. D. Rawhouser, J. H. Yang, J. L. Norris and R. M. Caprioli, J. Mass Spectrom., 2012, 47, 1473–1481 CrossRef PubMed.
- M. Niehaus, J. Soltwisch, M. E. Belov and K. Dreisewerd, Nat. Methods, 2019, 16, 925–931 CrossRef CAS PubMed.
- T. Bien, K. Koerfer, J. Schwenzfeier, K. Dreisewerd and J. Soltwisch, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2114365119 CrossRef CAS PubMed.
- H. Zhu, L. Yi, Z. Wang and W. Xiong, J. Univ. Sci. Technol. China, 2018, 48, 842–852 CAS.
- C.-F. Shao, Y. Zhao, K. Wu, F.-F. Jia, Q. Luo, Z. Liu and F.-Y. Wang, Chin. J. Anal. Chem., 2018, 46, 1005–1016 CAS.
- Z. Li, Bull. Mineral., Petrol. Geochem., 2020, 39, 1173–1190 Search PubMed.
- A. Benninghoven, Surf. Sci., 1994, 299, 246–260 CrossRef.
- D. Graham and L. Gamble, Microsc. Microanal., 2020, 26, 78–78 CrossRef.
- M. L. Kraft and H. A. Klitzing, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2014, 1841, 1108–1119 CrossRef CAS PubMed.
- L. Sun, J. Chin. Mass Spectrom. Soc., 2014, 35, 385–396 CAS.
- C. Peihong, L. I. Tongsheng, L. I. U. Xujun and M. Shigeyuki, Tribology, 2007, 27, 592–599 Search PubMed.
- T. L. Colliver, C. L. Brummel, M. L. Pacholski, F. D. Swanek, A. G. Ewing and N. Winograd, Anal. Chem., 1997, 69, 2225–2231 CrossRef CAS PubMed.
- T. P. Roddy, D. M. Cannon, C. A. Meserole, N. Winograd and A. G. Ewing, Anal. Chem., 2002, 74, 4011–4019 CrossRef CAS PubMed.
- J. S. Fletcher, S. Rabbani, A. Henderson, P. Blenkinsopp, S. P. Thompson, N. P. Lockyer and J. C. Vickerman, Anal. Chem., 2008, 80, 9058–9064 CrossRef CAS PubMed.
- L. Huang, Y. Chen, L. T. Weng, M. Leung, X. Xing, Z. Fan and H. Wu, Anal. Chem., 2016, 88, 12196–12203 CrossRef CAS PubMed.
- H. Tian, J. S. Fletcher, R. Thuret, A. Henderson, N. Papalopulu, J. C. Vickerman and N. P. Lockyer, J. Lipid Res., 2014, 55, 1970–1980 CrossRef CAS PubMed.
- H. Evenbratt, M. Munem and P. Malmberg, Biointerphases, 2020, 15, 061014 CrossRef CAS PubMed.
- Z. Gajdosechova and Z. Mester, Anal. Bioanal. Chem., 2019, 411, 4277–4292 CrossRef CAS PubMed.
- P. L. Clode, R. A. Stern and A. T. Marshall, Microsc. Res. Tech., 2007, 70, 220–229 CrossRef CAS PubMed.
- H. Hu, L. Zhang and J. He, Acta Ecol. Sin., 2013, 33, 348–357 CrossRef CAS.
- V. I. Slaveykova, C. Guignard, T. Eybe, H.-N. Migeon and L. Hoffmann, Anal. Bioanal. Chem., 2009, 393, 583–589 CrossRef CAS PubMed.
- M. Su, Y. Li, H. Wang, S. Diao and M. Wang, J. Beijing Norm. Univ., Nat. Sci., 2016, 52, 223–227 Search PubMed.
- K. Chen, W. Song, R. Russell, A. Ferrari, T. Darwish, P. Tontonoz, S. G. Young and H. Jiang, J. Lipid Res., 2022, 63, 100290 CrossRef CAS PubMed.
- T. B. Angerer, P. Blenkinsopp and J. S. Fletcher, Int. J. Mass Spectrom., 2015, 377, 591–598 CrossRef CAS.
- N. Winograd, in Annual Review of Analytical Chemistry, ed. P. W. Bohn and J. E. Pemberton, 2018, vol. 11, pp. 29–48 Search PubMed.
- M. Fujii, S. Nakagawa, K. Matsuda, N. Man, T. Seki, T. Aoki and J. Matsuo, Rapid Commun. Mass Spectrom., 2014, 28, 917–920 CrossRef CAS PubMed.
- T. Hua, Microsc. Microanal., 2020, 26, 810–810 CrossRef.
- L. J. Sparvero, H. Tian, A. A. Amoscato, W.-Y. Sun, T. S. Anthonymuthu, Y. Y. Tyurina, O. Kapralov, S. Javadov, R.-R. He, S. C. Watkins, N. Winograd, V. E. Kagan and H. Bayir, Angew. Chem., Int. Ed., 2021, 60, 11784–11788 CrossRef CAS PubMed.
- V. Pareek, H. Tian, N. Winograd and S. J. Benkovic, Science, 2020, 368, 283–290 CrossRef CAS PubMed.
- Y. Z. Hu, Anal. Sci., 2021, 37, 1653–1654 CrossRef PubMed.
- T. Pluhacek and V. Maier, Chem. Listy, 2020, 114, 239–243 Search PubMed.
- H. S. Yang, D. R. LaFrance and Y. Hao, Am. J. Clin. Pathol., 2021, 156, 167–175 CrossRef CAS PubMed.
- O. Chahrour, D. Cobice and J. Malone, J. Pharm. Biomed. Anal., 2015, 113, 2–20 CrossRef CAS PubMed.
- G. K. Zoorob, J. W. McKiernan and J. A. Caruso, Mikrochim. Acta, 1998, 128, 145–168 CrossRef CAS.
- R. D. Devine and G. K. Behbehani, Clin. Lab. Med., 2021, 41, 297–308 CrossRef PubMed.
- M. H. Spitzer and G. P. Nolan, Cell, 2016, 165, 780–791 CrossRef CAS PubMed.
- Y. Yang, C. Sun, Z. Gong, R. Yang and Y. Wang, Chin. J. Cell Biol., 2020, 42, 358–368 Search PubMed.
- L. R. Olsen, M. D. Leipold, C. B. Pedersen and H. T. Maecker, Cytometry, Part A, 2019, 95A, 156–172 CrossRef PubMed.
- O. Ornatsky, V. Baranov, D. R. Bandura, S. D. Tanner and J. Dick, J. Immunol. Methods, 2006, 308, 68–76 CrossRef CAS PubMed.
- D. Majonis, O. Ornatsky, R. Kinach and M. A. Winnik, Biomacromolecules, 2011, 12, 3997–4010 CrossRef CAS PubMed.
- G. K. Behbehani, S. C. Bendall, M. R. Clutter, W. J. Fantl and G. P. Nolan, Cytometry, Part A, 2012, 81A, 552–566 CrossRef PubMed.
- T. A. Blair and A. L. Frelinger, Platelets, 2020, 31, 633–640 CrossRef CAS PubMed.
- A. L. S. Oliver, A. Haase, A. Peddinghaus, D. Wittke, N. Jakubowski, A. Luch, A. Grutzkau and S. Baumgart, Anal. Chem., 2019, 91, 11514–11519 CrossRef PubMed.
- M. Ohata, H. Yasuda, Y. Namai and N. Furuta, Anal. Sci., 2002, 18, 1105–1110 CrossRef CAS PubMed.
- W. Guo, X. Lin and S. Hu, Earth Sci., 2020, 45, 1362–1374 Search PubMed.
- J. S. Becker, A. Matusch and B. Wu, Anal. Chim. Acta, 2014, 835, 1–18 CrossRef CAS PubMed.
- D. Pozebon, G. L. Scheffler and V. L. Dressler, J. Anal. At. Spectrom., 2017, 32, 890–919 RSC.
- J. H. Chen, R. X. Wang, M. H. Ma, L. R. Gao, B. Zhao and M. Xu, Anal. Bioanal. Chem., 2022, 414, 7023–7033 CrossRef CAS PubMed.
- Y. F. Meng, C. H. Gao, Q. Lu, S. Y. Ma and W. Hang, ACS Nano, 2021, 15, 13220–13229 CrossRef CAS PubMed.
- A. Lores-Padin, B. Fernandez, M. Garcia, H. Gonzalez-Iglesias and R. Pereiro, Anal. Chim. Acta, 2022, 1221, 340128 CrossRef CAS PubMed.
- L. N. Zheng, L. X. Feng, J. W. Shi, H. Q. Chen, B. Wang, M. Wang, H. F. Wang and W. Y. Feng, Anal. Chem., 2020, 92, 14339–14345 CrossRef CAS PubMed.
- X. Gong, X. Xiong, S. Zhang, X. Fang and X. Zhang, Sci. Sin.: Chim., 2016, 46, 133–152 CAS.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |