Multiple ion isolation and accumulation events for selective chemical noise reduction and dynamic range enhancement in MALDI imaging mass spectrometry†
Troy R.
Scoggins
IV
 ,
Jonathan T.
Specker
and
Boone M.
Prentice
,
Jonathan T.
Specker
and
Boone M.
Prentice
 *
*
Department of Chemistry, University of Florida, Gainesville, FL, USA. E-mail: booneprentice@chem.ufl.edu
First published on 20th March 2024
Abstract
Abundant chemical noise in MALDI imaging mass spectrometry experiments can impede the detection of less abundant compounds of interest. This chemical noise commonly originates from the MALDI matrix as well as other endogenous compounds present in high concentrations and/or with high ionization efficiencies. MALDI imaging mass spectrometry of biological tissues measures numerous biomolecular compounds that exist in a wide range of concentrations in vivo. When ion trapping instruments are used, highly abundant ions can dominate the charge capacity and lead to space charge effects that hinder the dynamic range and detection of lowly abundant compounds of interest. Gas-phase fractionation has been previously utilized in mass spectrometry to isolate and enrich target analytes. Herein, we have characterized the use of multiple continuous accumulations of selected ions (Multi CASI) to reduce the abundance of chemical noise and diminish the effects of space charge in MALDI imaging mass spectrometry experiments. Multi CASI utilizes the mass-resolving capability of a quadrupole mass filter to perform multiple sequential ion isolation events prior to a single mass analysis of the combined ion population. Multi CASI was used to improve metabolite and lipid detection in the MALDI imaging mass spectrometry analysis of rat brain tissue.
Introduction
Matrix-assisted laser desorption/ionization (MALDI) imaging mass spectrometry is a valuable tool for biological tissue analysis, enabling the detection of a wide variety of metabolites, lipids, and proteins over a range of concentrations.1–5 Extending the limit of detection and dynamic range for the spatial mapping of lowly abundant compounds in tissue is a major analytical challenge in imaging mass spectrometry experiments.6–9 Oftentimes, MALDI analyses of biological tissues produce highly abundant chemical noise, which suppresses the detection of lowly abundant ions of interest. Chemical noise can originate from the MALDI matrix and/or from abundant endogenous compounds in the tissue environment.10–13 This issue is exacerbated in instrument configurations that utilize ion trapping devices that have finite charge capacities. The presence of highly abundant ions can introduce space charge effects that detrimentally affect the limits of detection and dynamic range for more lowly abundant compounds.14,15A variety of approaches have been developed to mitigate chemical noise encountered during biological tissue analysis. For example, orthogonal separation using liquid chromatography (LC) prior to mass spectrometry is routinely used to increase the peak capacity of analysis.16–23 However, LC separation technologies are largely not directly compatible with imaging mass spectrometry due the lengthy timescales needed for each chromatographic separation.24 Condensed-phase tissue washing and chemical derivatization approaches are performed prior to subjection to in vacuo conditions of the mass spectrometer and can be used to selectively remove undesired compounds that may suppress ion signals of interest and selectively improve the ionization efficiency of targeted analytes, respectively. For example, tissue washing using volatile buffers at different pH ranges has been used to enhance the detection of neutral lipids and proteins.25–27 However, harsh solvent conditions can remove tissue content and further distort the natural morphology of the tissue environment.27 Chemical derivatization with reagents, such as 1-(4-(aminomethyl)phenyl)pyridin-1-ium chloride (AMPP), can be performed on tissue, but can suffer from incomplete and side reactions that complicate the chemical milieu.28,29 These condensed-phase approaches also require additional time-consuming steps to sample preparation protocols.
Gas-phase separation techniques offer rapid and flexible means of fractionating ion populations after ionization, but prior to detection, without manipulating the original tissue sample. Ion mobility-mass spectrometry (IM-MS) is widely recognized for its ability to separate ions based on their collisional cross section (CCS) and is routinely applied to imaging mass spectrometry studies.30–33 This method of gas-phase separation greatly increases the peak capacity of the analysis, but requires the presence of an ion mobility device on the mass spectrometer. Other gas-phase fractionation strategies include the use of selective ion/ion reactions that allow separation of isobaric (i.e., same nominal m/z) compounds on the basis of gas-phase chemical reactivity.34,35 More broadly applicable gas-phase separation strategies involve the use of selective ion isolation using a mass resolving quadrupole on a hybrid mass spectrometer.36–39 The instrument used herein is a quadrupole-hexapole Fourier transform ion cyclotron resonance (QhFT-ICR) hybrid mass spectrometer, which accumulates ions in the hexapole ion trap prior to transfer to the ICR for mass analysis. RF/DC isolation can be performed as ions pass through the quadrupole mass filter (QMF) to provide for mass-selective ion accumulation in the hexapole ion trap.40,41 More recently, mass-selective ion accumulation has been termed continuous accumulation of selected ions (CASI) and used to improve limit of detection and dynamic range in imaging mass spectrometry experiments.36,42–44 Conversely, selected ion ejection (SIE) can be performed by applying a supplemental alternating current (AC) waveform to the QMF to eject a small m/z interval as the ions transmit through the device.45 SIE has been employed to selectively exclude highly abundant chemical noise and the subsequent broadband enrichment of ions of interest in imaging mass spectrometry experiments.36
Most applications of CASI in imaging mass spectrometry have used a singular mass isolation window for ion accumulation.36,43 However, multiple ion isolation windows can be used to selectively target multiple analytes simultaneously.46,47 For example, broadband dipolar AC isolation waveforms that contain multiple frequency notches matched with the secular frequencies of ions of interest can be applied to quadrupole ion traps and quadrupole mass filters to effect multiple mass isolation windows.48,49 While these approaches allow for the simultaneous isolation of multiple windows, a broadband AC waveform is required. Multiple sequential isolation windows can also be affected by altering the RF/DC settings of the QMF. The resulting ion populations can then be collected in a downstream ion trap and analyzed in a single mass analysis step; a process that has been termed Multi CASI. Multi CASI can be used to selectively exclude one or more interfering ions, while still allowing for broadband enrichment at m/z values below and above the excluded m/z window. This process has recently been used to quantify pharmaceuticals in MALDI analysis of biological samples,12 but has not seen broader adoption in imaging mass spectrometry and its analytical performance has not been rigorously characterized. Herein, we have used Multi CASI to exclude the signal from the MALDI matrix (i.e., between m/z 150 and 250), while still simultaneously capturing metabolite ion signals below m/z 150 as well as metabolite and lipid ion signals above m/z 250. This reduces chemical noise and increases the limits of detection and dynamic ranges of ions of interest. The figures of merit of full scan, single CASI, and Multi CASI workflows are compared here to assess their analytical utility in imaging mass spectrometry experiments.
Experimental
Materials
1,5-Diaminonapthalene (DAN), hematoxylin, eosin, ammonium acetate, and chloroform were purchased from Sigma-Aldrich Chemical Co. (St Louis, MO, USA). 2-Propanol and methanol were purchased from Fisher Scientific (Pittsburgh, PA, USA). Rat brain (Brown Norway) was purchased from BioIVT (Westbury, NY, USA) and stored at −80 °C until analysis.Tissue extraction
Microscope slides with uniform metabolite and fatty acid coatings were prepared using an automated robotic sprayer. Metabolites and fatty acids were extracted from rat brain tissue (∼300 mg) using the Folch method.50 Briefly, tissue was homogenized using a FastPrep-24 Classic (MP Biomedicals, LLC, Irvine, CA, USA) with 50 μL of 5 mM ammonium acetate per 10 mg of tissue. The homogenate was centrifuged and 1 mL of supernatant was collected. To the supernatant, 4 mL of methanol and 8 mL of chloroform were added. Aqueous and organic layers were separated and dried with nitrogen gas. The aqueous portion was reconstituted in 600 μL of methanol and the organic portion was reconstituted in 600 μL of 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 v/v methanol
20 v/v methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-propanol. These solutions were mixed and 1 mL of the mixture was applied to a glass slide using an M5 TM Sprayer (flow rate: 0.15 mL min−1, nitrogen flow: 10 psi, spray temperature: 40 °C, four passes with offsets and rotations, spray velocity: 1300 mm min−1, dry time: 2 seconds) (HTX Technologies, LLC, Chapel Hill, NC, USA). A DAN MALDI matrix layer was then applied using a custom-built sublimation apparatus (120 °C, 8 min, <70 mTorr).51,52 Approximately 2 mg of DAN was sublimated onto each microscope slide. DAN was used as the MALDI matrix for all experiments due to its broad compatibility for many analytes, including the low mass metabolites and fatty acids studied herein. The most abundant of chemical noise from this matrix is in the low m/z mass range, which typically complicates analysis of low mass metabolites and nicely demonstrates the capabilities of Multi CASI. However, we have also performed Multi CASI using other MALDI matrices (data not shown) and have observed similar improvements in performance to those described herein.
2-propanol. These solutions were mixed and 1 mL of the mixture was applied to a glass slide using an M5 TM Sprayer (flow rate: 0.15 mL min−1, nitrogen flow: 10 psi, spray temperature: 40 °C, four passes with offsets and rotations, spray velocity: 1300 mm min−1, dry time: 2 seconds) (HTX Technologies, LLC, Chapel Hill, NC, USA). A DAN MALDI matrix layer was then applied using a custom-built sublimation apparatus (120 °C, 8 min, <70 mTorr).51,52 Approximately 2 mg of DAN was sublimated onto each microscope slide. DAN was used as the MALDI matrix for all experiments due to its broad compatibility for many analytes, including the low mass metabolites and fatty acids studied herein. The most abundant of chemical noise from this matrix is in the low m/z mass range, which typically complicates analysis of low mass metabolites and nicely demonstrates the capabilities of Multi CASI. However, we have also performed Multi CASI using other MALDI matrices (data not shown) and have observed similar improvements in performance to those described herein.
Mass spectrometry
All mass spectrometry experiments were performed on a commercial hybrid 7T solariX XR FT-ICR mass spectrometer equipped with an Apollo II dual MALDI/ESI source along with a dynamically harmonized ParaCell (Bruker Daltonics, Billerica, MA).53 The MALDI ion source utilizes a Smartbeam II Nd:YAG laser system (2 kHz, 355 nm). Mass analysis was performed using a 0.4893-second time-domain transient length, resulting in a mass resolving power of ∼200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at m/z 130.069. Mass spectra were recorded from m/z 100 to 600 and were visualized in Compass DataAnalysis 5.0 (Bruker Daltonics, Billerica, MA).
000 at m/z 130.069. Mass spectra were recorded from m/z 100 to 600 and were visualized in Compass DataAnalysis 5.0 (Bruker Daltonics, Billerica, MA).
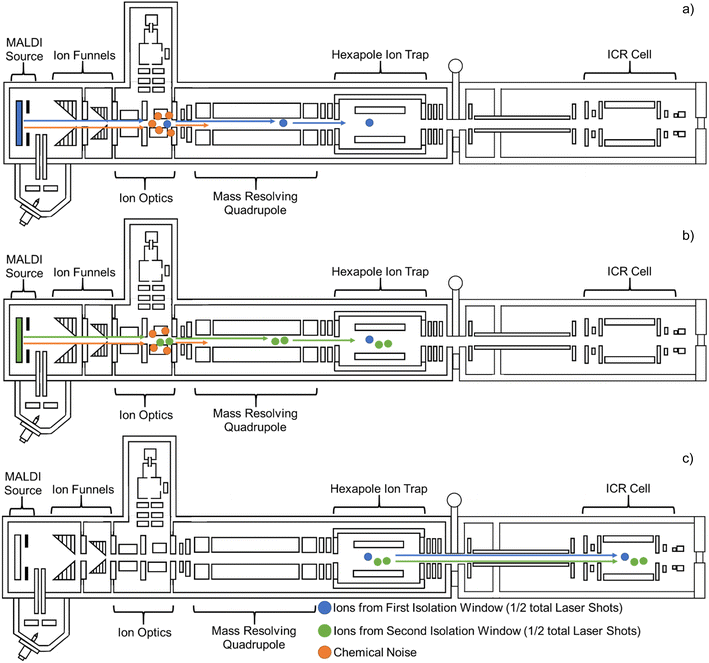
Full scan and Multi CASI analyses were compared using the same acquisition conditions, where the only difference between the two experiments was the addition of Multi CASI. During full-scan analysis, no mass isolation is performed. Multi CASI utilizes multiple sequential mass isolation windows to isolate ion populations of disparate m/z prior to collection of the ion populations in a downstream hexapole ion trap (Fig. 1). For MALDI analysis, the total number of laser shots defined for the acquisition is divided evenly between the sequential mass isolation windows. Following accumulation in the hexapole ion trap, the mass selected ions are jointly transferred to the ICR cell for mass analysis. Full scan and single CASI methods using a defined number of laser shots are compared with Multi CASI methods using double the number of laser shots. This ensures that each isolation window has the same number of laser shots as the full scan and single CASI methods.
Imaging mass spectrometry
Transverse sections of a control Brown Norway rat brain were collected at 10 μm thickness using a Leica CM 3050S Research Cryostat (Leica Biosystems, Buffalo Grove, IL, USA), thaw mounted onto indium tin oxide (ITO)-coated slides (Delta Technologies, Loveland, CO, USA), and warmed to room temperature in a desiccator for 30 minutes. Following desiccation, a 1,5-diaminonaphthalene (DAN) MALDI matrix layer was applied using a custom-built sublimation apparatus (120 °C, 8 min, <70 mTorr).51,52 Approximately 2 mg of DAN was sublimated onto each slide.Metabolite and free fatty acid imaging mass spectrometry of rat brain tissue was performed in negative ion mode. Full scan and Multi CASI experiments were performed to deplete chemical noise and to enrich selected analyte ions. Images were acquired using pixel spacings of 150 or 300 μm in both the x and y dimensions (1000 laser shots, 135 μm smart walk raster). Serial tissue sections were stained using hematoxylin and eosin (H&E) and scanned at 10× magnification using a Zeiss Axio Imager M2 Microscope (Carl Zeiss Microscopy LLC, White Plains, NY, USA). Data analysis of individual spectra was performed using DataAnalysis (Bruker Daltonics, Billerica, MA, USA). Analyte identifications were assigned using high resolution accurate mass measurements (HRAM) with a tolerance of less than 5 ppm error along and searched against The Human Metabolome Database (https://hmdb.ca/). Data analysis of imaging experiments was performed using flexImaging (Bruker Daltonics, Billerica, MA, USA).
Results
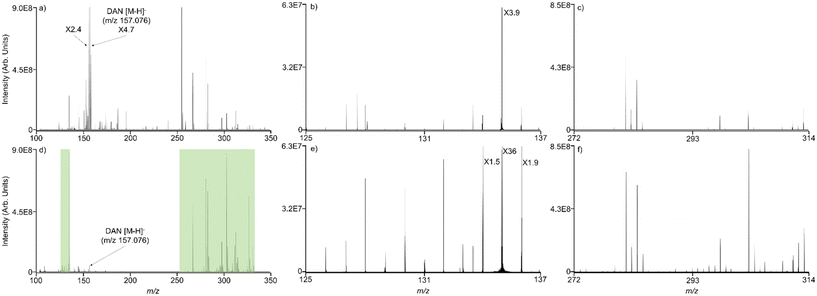
CASI using a single mass isolation window has previously been shown to improve the limit of detection in imaging mass spectrometry experiments.36 Multiple continuous accumulations of selected ions (Multi CASI) uses multiple sequential RF/DC isolation windows in a QMF to selectively accumulate disparate m/z ion populations in a downstream hexapole ion trap. The resulting ion population is then transferred to the ICR cell and mass analyzed in a single acquisition (Fig. 1). When using MALDI, the total number of laser shots for Multi CASI acquisition is divided evenly between the successive mass isolation windows. Multi CASI methods generally enrich populations of selected ions, while preventing the accumulation of ions outside of the selected m/z windows through quadrupole isolation events. This more efficiently fills the charge capacity of the ion trap with analytes of interest, improving the limit of detection and dynamic range. Both the hexapole and ICR cells are ion trap devices with finite charge capacities. When the space charge limit is reached, the confinement of ions can be diminished, and peak abundances may be distorted in the mass spectrum. Space charge artifacts in the ICR cell may manifest via peak coalescence and peak shifting.54–57 The multiple mass isolation windows employed during Multi CASI reduces the number of ions transmitted to the hexapole and ICR cell, which significantly reduces space charge effects and allows for greater accumulation of the ions within the selected m/z windows.In MALDI mass spectrometry, Multi CASI can be used to improve the limit of detection and dynamic range for lowly abundant compounds of interest. MALDI mass spectrometry analysis was performed on a complex lipid mixture extracted from rat brain tissue. Full scan and Multi CASI spectra were collected to demonstrate the depletion of chemical noise and enrichment of selected m/z regions (Fig. 2). The Multi CASI method excluded much of the chemical noise originating from the DAN matrix peak (m/z 157.007, 0.172 ppm) and related isotopic peaks. The depletion of this chemical noise led to enrichment of the ions in the m/z 131 ± 5 Da and 239 ± 40 Da mass regions selected by the Multi CASI windows. These mass windows were selected due to their proximity to the DAN [M − H]− peak (m/z 157.007). The proximity of these isolation windows to the primary noise peak allows for a rigorous evaluation of chemical noise exclusion. However, Multi CASI is also effective at other mass ranges (e.g., m/z 700–1000 to sample phospholipids, data not shown), depending on the chemical noise to be excluded.
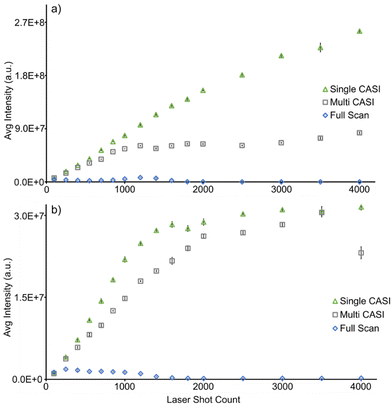
The extent of signal enrichment was monitored as a function of the number of MALDI laser shots accumulated in full scan and Multi CASI methods (Fig. 3). Ion abundance is expected to increase linearly with the number of accumulated laser shots, until space charge effects are encountered, which will result in a disruption of the linear trend. Space charge is apparent with as few as 250 laser shots in the full scan experiment, as detected by a decrease in signal intensity for aspartate (m/z 132.030, 0.041 ppm) and eicosadienoate (m/z 307.264, 0.586 ppm) ions of interest (Fig. 3). Conversely, Multi CASI allowed for improvement of over 500-fold in signal intensity using 4000 laser shots for aspartate (present in the first Multi CASI window, Fig. 3a) and over 180-fold improvement in signal intensity using 3500 laser shots for eicosadienoate (present in the second Multi CASI window, Fig. 3b) compared to the full scan measurements. This is due to the reduction in ion population through chemical noise exclusion. The ion population reduction then allows for the traps to be filled with larger amounts of the selected ions. Multi CASI is also able to reduce variance in the data. The average relative standard deviation was 6.35% for full scan, 1.85% for single CASI, and 1.64% for Multi CASI (Fig. 3). Space charge effects cause ions to behave unpredictably in ion traps at capacity, leading to detrimental effects such as reduced dynamic range, higher variability, and peak shifting.14,58,59 Multi CASI is able to exclude chemical noise and therefore reduce the ion population in the instrument. A lower ion population in the ion trap minimizes the repulsion effects of space charge, lowering the overall standard deviation of the data. A reduction in the accumulation of chemical noise also allows for the detection of additional analytes. Multi CASI allowed for the detection of nearly 5-fold more peaks than that in the full scan analysis. While space charge effects are reduced, they are not eliminated. This is evident at 1200 laser shots in the Multi CASI experiment, where space charge effects are observed (Fig. 3a). Space charge in the ICR cell commonly manifests as peak shifting and peak coalescence.60 As the laser shot count increases, the peaks for aspartate (m/z 132.030) and eicosadienoate (m/z 307.264) shift to higher m/z values (ESI Fig. 1†).60 Ion cyclotron frequency decreases with increasing ion population in the ICR cell, which results in a shift to higher m/z values.61,62 Space charge affects ions in the m/z 131 ± 5 Da window at a lower number of laser shots when compared to ions in the m/z 293 ± 40 Da window (i.e., 1200 versus 2000 laser shots in Fig. 3), and does so independent of the order of the two isolation windows (data not shown). This disparity may be due to a low-mass cutoff (LMCO) or pseudopotential trapping well depth effect in the hexapole ion trap.
Another method for limit of detection and dynamic range enhancement in MALDI mass spectrometry is single CASI.19 The signal enrichment capabilities of Single CASI and Multi CASI were compared to quantify the improvements in dynamic range (Fig. 3). The two methods demonstrate comparable improvements in signal enrichment until ICR the charge capacity limit is reached in the Multi CASI experiment around 2000 laser shots (ESI Fig. 2†). Space charge effects will generally appear with fewer numbers of laser shots in the Multi CASI methods, as multiple isolation windows are included in the analysis, though this will depend on the width of the mass windows as well as the number and abundances of peaks in those windows. Increasing the quantity and width of isolation windows allows for more ions to be added to the ion trap and therefore reach the charge capacity limit with a fewer number of laser shots (ESI Fig. 3†). Even with this downside, Multi CASI still is able to detect more analytes as it can cover multiple mass ranges. Multi CASI allowed for the detection of 1.7-fold more peaks than single CASI analysis. Subsequent Multi CASI experiments are all performed using 1000 laser shots per window to minimize the effects of space charge.
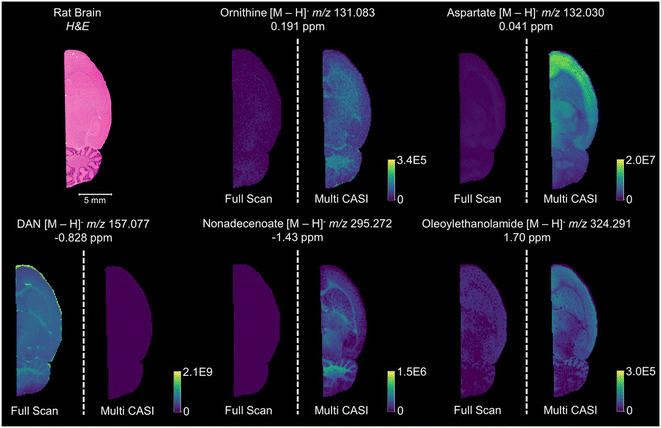
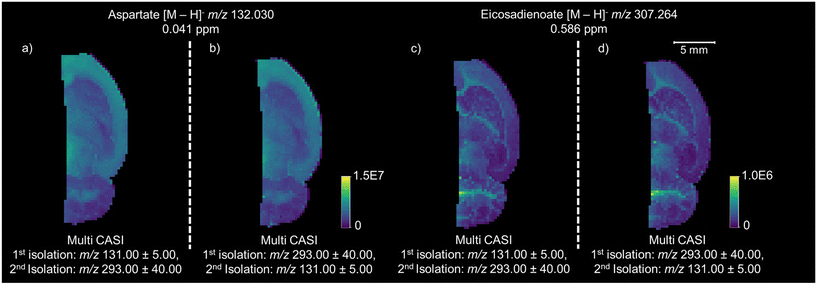
Multi CASI can be used to improve the limits of detection and dynamic ranges for lowly abundant compounds in imaging mass spectrometry experiments. Multi CASI windows centered at m/z 131 ± 5 Da and m/z 293 ± 40 Da are used to acquire an image of rat brain tissue at 150 μm spatial resolution (Fig. 4). Full scan and Multi CASI utilized 1000 laser shots per window for both methods for comparison. Many ions that were not detected or detected at low abundance in the full scan experiment are readily detected across the rat brain tissue in the Multi CASI experiment. An increase in LOD is observed as an increase in the brightness of the image and an increase in dynamic range is observed as an increase in the contrast of the image. For example, a 6.5-fold increase in aspartate ion intensity was observed and 4.5-fold more peaks were detected in Multi CASI analysis when compared to full scan analysis, and DAN matrix intensity decreased by over 50-fold in Multi CASI analysis. Multi CASI is performed by ablating the same pixel in multiple (i.e., two) discrete MALDI ablation steps. Each MALDI ablation step then uses separate quadrupole mass filter settings to accumulate ions from disparate mass isolation windows. As such, MALDI laser power and shot count need to be optimized to ensure sufficient material is sampled in each Multi CASI ablation event and that signal is not diminished in the subsequent isolation windows. Laser power was optimized for each experiment to yield maximum signal in the first ablation event, while not diminishing analyte signal in the second ablation event. Two Multi CASI images were acquired using different sequential orders of the two isolation windows to verify that the sampling and accumulation conditions used here had no effect on the imaging mass spectrometry results (Fig. 5). For example, the tissue pixel is ablated and this first packet of ions is isolated with a mass isolation window of m/z 131 ± 5 Da. The same pixel is then ablated a second time and this packet of ions is isolated with a mass isolation window of m/z 293 ± 40 Da. Both ion populations are accumulated in the hexapole ion trap and then mass analyzed together in the ICR cell (Fig. 5a and c). In a second image, the mass isolation windows are then switched so that the first packet of ions is isolated using a mass window of m/z 293 ± 40 Da, followed by a second mass window of m/z 131 ± 5 Da (Fig. 5b and d). The distributions and intensity for aspartate and eicosadienoate are consistent across the tissue when comparing the two methods, indicating that the order of the ablation events does not have an impact on the quality of the imaging data using the settings describe here.
Conclusions
Herein, we have described a method for gas-phase ion enrichment in imaging mass spectrometry experiments. Multiple continuous accumulations of selected ions (Multi CASI) utilizes multiple sequential RF/DC mass isolation windows in a quadrupole mass filter (QMF) to selectively accumulate ions in a downstream hexapole ion trap. In this study, Multi CASI can provide for nearly 500-fold enrichment in targeted ion signals, resulting in substantial improvements in the limits of detection and dynamic ranges of selected ions within the mass isolation windows. The ion enrichment provided by Multi CASI improves the MALDI analyses of biological tissue. Signal for lowly abundant compounds, such as the amino acids and lipids observed in our analyses, is greatly enriched. Multi CASI methods applied to several other workflows in our lab have also resulted in similar improvements in performance. Multi CASI improves upon typical CASI methods by allowing for the selection of multiple mass windows for analysis, enabling a broader view of compounds in a single image. This has a multiplexing effect compared to other CASI and selected ion monitoring (SIM) configurations. Multi CASI is also enabled without the addition of a supplemental waveform to the quadrupole, as has been reported in workflows that leverage multiple frequency notches. Accurate use of Multi CASI requires sufficient sample at each pixel for multiple laser ablation steps (i.e., for multiple sequential mass isolation windows) on the same spot. Space charge effects can persist at higher laser shot counts due to abundant endogenous molecules, though these effects are observable at far fewer number of laser shots in full scan experiments. Herein, Multi CASI allowed for the detection of 4.6-fold more peaks in imaging mass spectrometry analysis of rat brain when compared to full scan analysis, and 1.7-fold more peaks than single CASI analysis. This leads to an increase in throughput, reducing the number of targeted windows needed to enrich lowly abundant analytes in imaging mass spectrometry experiments.Abbreviations
| MALDI | Matrix-assisted laser desorption ionization |
| Multi CASI | Multiple continuous accumulations of selected ions |
| LC-MS | Liquid chromatography-mass spectrometry |
| DAN | 1,5-Diaminonaphthalene |
Author contributions
Troy R. Scoggins IV – data collection, data analysis, experimental design, and manuscript authorship. Jonathan T. Specker – training of primary author, experimental design, and manuscript review. Boone M. Prentice – data analysis, experimental design, manuscript review, and funding acquisition.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Institutes of Health (NIH) under award R01 GM138660 (National Institute of General Medical Sciences [NIGMS]) and by a contribution from Eli Lilly and Company.References
- M. Aichler and A. Walch, MALDI Imaging mass spectrometry: current frontiers and perspectives in pathology research and practice, Lab. Invest., 2015, 95(4), 422–431, DOI:10.1038/labinvest.2014.156.
- R. Casadonte and R. M. Caprioli, Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry, Nat. Protoc., 2011, 6(11), 1695–1709, DOI:10.1038/nprot.2011.388.
- D. S. Cornett, M. L. Reyzer, P. Chaurand and R. M. Caprioli, MALDI imaging mass spectrometry: molecular snapshots of biochemical systems, Nat. Methods, 2007, 4(10), 828–833, DOI:10.1038/nmeth1094.
- M. M. Gessel, J. L. Norris and R. M. Caprioli, MALDI imaging mass spectrometry: spatial molecular analysis to enable a new age of discovery, J. Proteomics, 2014, 107, 71–82, DOI:10.1016/j.jprot.2014.03.021.
- C. J. Good, E. K. Neumann, C. E. Butrico, J. E. Cassat, R. M. Caprioli and J. M. Spraggins, High Spatial Resolution MALDI Imaging Mass Spectrometry of Fresh-Frozen Bone, Anal. Chem., 2022, 94(7), 3165–3172, DOI:10.1021/acs.analchem.1c04604.
- R. D. Addie, B. Balluff, J. V. Bovée, H. Morreau and L. A. McDonnell, Current State and Future Challenges of Mass Spectrometry Imaging for Clinical Research, Anal. Chem., 2015, 87(13), 6426–6433, DOI:10.1021/acs.analchem.5b00416.
- L. Morosi, M. Zucchetti, M. D'Incalci and E. Davoli, Imaging mass spectrometry: challenges in visualization of drug distribution in solid tumors, Curr. Opin. Pharmacol., 2013, 13(5), 807–812, DOI:10.1016/j.coph.2013.06.003.
- B. Prideaux and M. Stoeckli, Mass spectrometry imaging for drug distribution studies, J. Proteomics, 2012, 75(16), 4999–5013, DOI:10.1016/j.jprot.2012.07.028.
- E. H. Seeley and R. M. Caprioli, MALDI imaging mass spectrometry of human tissue: method challenges and clinical perspectives, Trends Biotechnol., 2011, 29(3), 136–143, DOI:10.1016/j.tibtech.2010.12.002.
- C. D. Calvano, A. Monopoli, T. R. I. Cataldi and F. Palmisano, MALDI matrices for low molecular weight compounds: an endless story?, Anal. Bioanal. Chem., 2018, 410(17), 4015–4038, DOI:10.1007/s00216-018-1014-x.
- A. N. Krutchinsky and B. T. Chait, On the nature of the chemical noise in MALDI mass spectra, J. Am. Soc. Mass Spectrom., 2002, 13(2), 129–134, DOI:10.1016/s1044-0305(01)00336-1.
- Z. Liang and B. M. Prentice, Quantification of pharmaceutical compounds in tissue and plasma samples using selective ion accumulation with multiple mass isolation windows, J. Mass Spectrom., 2023, 58(7), e4958, DOI:10.1002/jms.4958.
- I. P. Smirnov, X. Zhu, T. Taylor, Y. Huang, P. Ross, I. A. Papayanopoulos, S. A. Martin and D. J. Pappin, Suppression of alpha-cyano-4-hydroxycinnamic acid matrix clusters and reduction of chemical noise in MALDI-TOF mass spectrometry, Anal. Chem., 2004, 76(10), 2958–2965, DOI:10.1021/ac035331j.
- K. Busch, Space Charge in Mass Spectrometry, 2004 Search PubMed.
- M. L. Easterling, T. H. Mize and I. J. Amster, Routine Part-per-Million Mass Accuracy for High- Mass Ions: Space-Charge Effects in MALDI FT-ICR, Anal. Chem., 1999, 71(3), 624–632, DOI:10.1021/ac980690d.
- M. A. Baldwin and F. W. McLafferty, Liquid chromatography–mass spectrometry interface–I: The direct introduction of liquid solutions into a chemical ionization mass spectrometer, Org. Mass Spectrom., 1973, 7(9), 1111–1112 CrossRef CAS.
- S. J. Blanksby and T. W. Mitchell, Advances in mass spectrometry for lipidomics, Annu. Rev. Anal. Chem., 2010, 3, 433–465, DOI:10.1146/annurev.anchem.111808.073705.
- K. Dettmer, P. A. Aronov and B. D. Hammock, Mass spectrometry-based metabolomics, Mass Spectrom. Rev., 2007, 26(1), 51–78, DOI:10.1002/mas.20108.
- R. S. Gohlke, Time-of-Flight Mass Spectrometry and Gas-Liquid Partition Chromatography, Anal. Chem., 1959, 31(4), 535–541 CrossRef CAS.
- J. C. Holmes and F. A. Morrell, Oscillographic Mass Spectrometric Monitoring of Gas Chromatography, Appl. Spectrosc., 1957, 11(2), 86–87 CrossRef CAS.
- J. A. Olivares, N. T. Nguyen, C. R. Yonker and R. D. Smith, On-Line Mass Spectrometric Detection for Capillary Zone Electrophoresis, Anal. Chem., 1987, 59(8), 1230–1232 CrossRef CAS.
- P. Schmitt-Kopplin and M. Frommberger, Capillary electrophoresis-mass spectrometry: 15 years of developments and applications, Electrophoresis, 2003, 24(22–23), 3837–3867, DOI:10.1002/elps.200305659.
- J. R. Yates, C. I. Ruse and A. Nakorchevsky, Proteomics by mass spectrometry: approaches, advances, and applications, Annu. Rev. Biomed. Eng., 2009, 11, 49–79, DOI:10.1146/annurev-bioeng-061008-124934.
- F. Pu, S. Chiang, W. Zhang and Z. Ouyang, Direct sampling mass spectrometry for clinical analysis, Analyst, 2019, 144(4), 1034–1051, 10.1039/c8an01722k.
- P. M. Angel, J. M. Spraggins, H. S. Baldwin and R. Caprioli, Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry, Anal. Chem., 2012, 84(3), 1557–1564, DOI:10.1021/ac202383m.
- J. Yang and R. M. Caprioli, Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution, Anal. Chem., 2011, 83(14), 5728–5734, DOI:10.1021/ac200998a.
- E. H. Seeley, S. R. Oppenheimer, D. Mi, P. Chaurand and R. M. Caprioli, Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections, J. Am. Soc. Mass Spectrom., 2008, 19(8), 1069–1077, DOI:10.1016/j.jasms.2008.03.016.
- C. Harkin, K. W. Smith, F. L. Cruickshank, C. Logan Mackay, B. Flinders, R. M. A. Heeren, T. Moore, S. Brockbank and D. F. Cobice, On-tissue chemical derivatization in mass spectrometry imaging, Mass Spectrom. Rev., 2022, 41(5), 662–694, DOI:10.1002/mas.21680.
- I. Kaya, L. S. Schembri, A. Nilsson, R. Shariatgorji, S. Baijnath, X. Zhang, E. Bezard, P. Svenningsson, L. R. Odell and P. E. Andrén, On-Tissue Chemical Derivatization for Comprehensive Mapping of Brain Carboxyl and Aldehyde Metabolites by MALDI-MS Imaging, J. Am. Soc. Mass Spectrom., 2023, 34(5), 836–846, DOI:10.1021/jasms.2c00336.
- M. Kliman, J. C. May and J. A. McLean, Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry, Biochim. Biophys. Acta, 2011, 1811(11), 935–945, DOI:10.1016/j.bbalip.2011.05.016.
- J. A. McLean, W. B. Ridenour and R. M. Caprioli, Profiling and imaging of tissues by imaging ion mobility-mass spectrometry, J. Mass Spectrom., 2007, 42(8), 1099–1105, DOI:10.1002/jms.1254.
- E. S. Rivera, K. V. Djambazova, E. K. Neumann, R. M. Caprioli and J. M. Spraggins, Integrating ion mobility and imaging mass spectrometry for comprehensive analysis of biological tissues: A brief review and perspective, J. Mass Spectrom., 2020, 55(12), e4614, DOI:10.1002/jms.4614.
- M. Sans, C. L. Feider and L. S. Eberlin, Advances in mass spectrometry imaging coupled to ion mobility spectrometry for enhanced imaging of biological tissues, Curr. Opin. Chem. Biol., 2018, 42, 138–146, DOI:10.1016/j.cbpa.2017.12.005.
- X. Diao, N. R. Ellin and B. M. Prentice, Selective Schiff base formation via gas-phase ion/ion reactions to enable differentiation of isobaric lipids in imaging mass spectrometry, Anal. Bioanal. Chem., 2023, 415(18), 4319–4331, DOI:10.1007/s00216-023-04523-y.
- J. T. Specker and B. M. Prentice, Separation of Isobaric Lipids in Imaging Mass Spectrometry Using Gas-Phase Charge Inversion Ion/Ion Reactions, J. Am. Soc. Mass Spectrom., 2023, 34(9), 1868–1878, DOI:10.1021/jasms.3c00081.
- B. M. Prentice, D. J. Ryan, K. J. Grove, D. S. Cornett, R. M. Caprioli and J. M. Spraggins, Dynamic Range Expansion by Gas-Phase Ion Fractionation and Enrichment for Imaging Mass Spectrometry, Anal. Chem., 2020, 92(19), 13092–13100, DOI:10.1021/acs.analchem.0c02121.
- T. L. Quenzer, M. R. Emmett, C. L. Hendrickson, P. H. Kelly and A. G. Marshall, High sensitivity Fourier transform ion cyclotron resonance mass spectrometry for biological analysis with nano-LC and microelectrospray ionization, Anal. Chem., 2001, 73(8), 1721–1725, DOI:10.1021/ac001095q.
- M. W. Senko, C. L. Hendrickson, M. R. Emmett, S. D. H. Shi and A. G. Marshall, External accumulation of ions for enhanced electrospray ionization fourier transform ion cyclotron resonance mass spectrometry, J. Am. Soc. Mass Spectrom., 1997, 8(9), 970–976 CrossRef CAS.
- Y. Wang, X. Zhang, Y. Zhai, Y. Jiang, X. Fang, M. Zhou, Y. Deng and W. Xu, Mass selective ion transfer and accumulation in ion trap arrays, Anal. Chem., 2014, 86(20), 10164–10170, DOI:10.1021/ac502583b.
- M. E. Belov, E. N. Nikolaev, G. A. Anderson, K. J. Auberry, R. Harkewicz and R. D. Smith, Electrospray ionization-Fourier transform ion cyclotron mass spectrometry using ion preselection and external accumulation for ultrahigh sensitivity, J. Am. Soc. Mass Spectrom., 2001, 12(1), 38–48, DOI:10.1016/S1044-0305(00)00198-7.
- Y. Wang, S. D. H. Shi, C. L. Hendrickson and A. G. Marshall, Mass-selective ion accumulation and fragmentation in a linear octopole ion trap external to a Fourier transform ion cyclotron resonance mass spectrometer, Int. J. Mass Spectrom., 2000, 198(1–2), 113–120 CrossRef CAS.
- C. J. Thompson, M. Witt, S. Forcisi, F. Moritz, N. Kessler, F. H. Laukien and P. Schmitt-Kopplin, An Enhanced Isotopic Fine Structure Method for Exact Mass Analysis in Discovery Metabolomics: FIA-CASI-FTMS, J. Am. Soc. Mass Spectrom., 2020, 31(10), 2025–2034, DOI:10.1021/jasms.0c00047.
- N. Sun and A. Walch, Qualitative and quantitative mass spectrometry imaging of drugs and metabolites in tissue at therapeutic levels, Histochem. Cell Biol., 2013, 140(2), 93–104, DOI:10.1007/s00418-013-1127-4.
- C. D. Huffstutler, D. M. Sanchez, M. R. Weigand, H. Hu, X. Li, A. J. Chegwidden, K. O. Nagornov, A. N. Kozhinov, Y. O. Tsybin and J. Laskin, Multiple selected ion monitoring mode for sensitive imaging of eicosanoids in tissues using nanospray desorption electrospray ionization (nano-DESI) mass spectrometry, Int. J. Mass Spectrom., 2023, 491, 117101 CrossRef CAS.
- M. E. Belov, G. A. Anderson, N. H. Angell, Y. Shen, N. Tolic, H. R. Udseth and R. D. Smith, Dynamic range expansion applied to mass spectrometry based on data-dependent selective ion ejection in capillary liquid chromatography fourier transform ion cyclotron resonance for enhanced proteome characterization, Anal. Chem., 2001, 73(21), 5052–5060, DOI:10.1021/ac010733h.
- B. M. Prentice, C. W. Chumbley and R. M. Caprioli, Absolute Quantification of Rifampicin by MALDI Imaging Mass Spectrometry Using Multiple TOF/TOF Events in a Single Laser Shot, J. Am. Soc. Mass Spectrom., 2017, 28(1), 136–144, DOI:10.1007/s13361-016-1501-2.
- B. M. Prentice, J. C. McMillen and R. M. Caprioli, Multiple TOF/TOF Events in a Single Laser Shot for Multiplexed Lipid Identifications in MALDI Imaging Mass Spectrometry, Int. J. Mass Spectrom., 2019, 437, 30–37, DOI:10.1016/j.ijms.2018.06.006.
- B. K. Erickson, M. P. Jedrychowski, G. C. McAlister, R. A. Everley, R. Kunz and S. P. Gygi, Evaluating multiplexed quantitative phosphopeptide analysis on a hybrid quadrupole mass filter/linear ion trap/orbitrap mass spectrometer, Anal. Chem., 2015, 87(2), 1241–1249, DOI:10.1021/ac503934f.
- G. C. McAlister, D. P. Nusinow, M. P. Jedrychowski, M. Wühr, E. L. Huttlin, B. K. Erickson, R. Rad, W. Haas and S. P. Gygi, MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes, Anal. Chem., 2014, 86(14), 7150–7158, DOI:10.1021/ac502040v.
- J. Folch, M. Lees and G. H. Sloane Stanley, A simple method for the isolation and purification of total lipides from animal tissues, J. Biol. Chem., 1957, 226(1), 497–509 CrossRef CAS PubMed.
- J. A. Hankin, R. M. Barkley and R. C. Murphy, Sublimation as a method of matrix application for mass spectrometric imaging, J. Am. Soc. Mass Spectrom., 2007, 18(9), 1646–1652, DOI:10.1016/j.jasms.2007.06.010.
- A. Thomas, J. L. Charbonneau, E. Fournaise and P. Chaurand, Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition, Anal. Chem., 2012, 84(4), 2048–2054, DOI:10.1021/ac2033547.
- Y. I. Kostyukevich, G. N. Vladimirov and E. N. Nikolaev, Dynamically harmonized FT-ICR cell with specially shaped electrodes for compensation of inhomogeneity of the magnetic field. Computer simulations of the electric field and ion motion dynamics, J. Am. Soc. Mass Spectrom., 2012, 23(12), 2198–2207, DOI:10.1007/s13361-012-0480-1.
- A. G. Marshall, C. L. Hendrickson and G. S. Jackson, Fourier transform ion cyclotron resonance mass spectrometry: a primer, Mass Spectrom. Rev., 1998, 17(1), 1–35, DOI:10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K.
- D. W. Mitchell and R. D. Smith, Prediction of a space charge induced upper molecular mass limit towards achieving unit mass resolution in Fourier transform ion cyclotron resonance mass spectrometry, J. Mass Spectrom., 1996, 31(7), 771–790 CrossRef CAS.
- E. N. Nikolaev, Y. I. Kostyukevich and G. N. Vladimirov, Fourier transform ion cyclotron resonance (FT ICR) mass spectrometry: Theory and simulations, Mass Spectrom. Rev., 2016, 35(2), 219–258, DOI:10.1002/mas.21422.
- L. Paša-Tolić, Y. Huang, S. Guan, H. S. Kim and A. G. Marshall, Ultrahigh–resolution matrix–assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectra of peptides, J. Mass Spectrom., 1995, 30(6), 825–833 CrossRef.
- K. Cox, C. Cleven and R. Cooks, Mass shifts and local space charge effects observed in the quadrupole ion trap at higher resolution, Int. J. Mass Spectrom. Ion Processes, 1995, 144(1–2), 47–65 CrossRef CAS.
- D. Guo, Y. Wang, X. Xiong, H. Zhang, X. Zhang, T. Yuan, X. Fang and W. Xu, Space charge induced nonlinear effects in quadrupole ion traps, J. Am. Soc. Mass Spectrom., 2014, 25(3), 498–508 CrossRef CAS PubMed.
- R. L. Wong and I. J. Amster, Experimental Evidence for Space-Charge Effects between Ions of the Same Mass-to-Charge in Fourier-Transform Ion Cyclotron Resonance Mass Spectrometry, Int. J. Mass Spectrom., 2007, 265(2–3), 99–105, DOI:10.1016/j.ijms.2007.01.014.
- T. J. Francl, M. G. Sherman, R. L. Hunter, J. M. Locke, W. D. Bowers and R. T. McIver, Experimental determination of the effects of space charge on ion cyclotron resonance frequencies, Int. J. Mass Spectrom. Ion Processes, 1983, 54(1–2), 189–199 CrossRef CAS.
- J. B. Jeffries, S. E. Barlow and G. H. Dunn, Theory of space-charge shift of ion cyclotron resonance frequencies, Int. J. Mass Spectrom. Ion Processes, 1983, 54(1–2), 169–187 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an00160e |
| This journal is © The Royal Society of Chemistry 2024 |