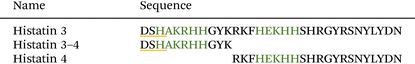Zn(II) coordination influences the secondary structure, but not antimicrobial activity of the N-terminal histatin 3 hydrolysis product†
Emilia
Dzień
 a,
Joanna
Wątły
a,
Joanna
Wątły
 *a,
Aleksandra
Hecel
a,
Aleksandra
Mikołajczyk
b,
Agnieszka
Matera-Witkiewicz
b,
Miquel
Adrover
cde,
Miquel
Barceló-Oliver
*a,
Aleksandra
Hecel
a,
Aleksandra
Mikołajczyk
b,
Agnieszka
Matera-Witkiewicz
b,
Miquel
Adrover
cde,
Miquel
Barceló-Oliver
 c,
Alicia
Domínguez-Martín
c,
Alicia
Domínguez-Martín
 *f and
Magdalena
Rowińska-Żyrek
*f and
Magdalena
Rowińska-Żyrek
 *a
*a
aFaculty of Chemistry, University of Wrocław, F. Joliot-Curie 14, 50-383 Wrocław, Poland. E-mail: magdalena.rowinska-zyrek@uwr.edu.pl
bScreening of Biological Activity Assays and Collection of Biological Material Laboratory, Wroclaw Medical Univeristy Biobank, Faculty of Pharmacy, Wroclaw Medical University, Borowska 211A, 50-556 Wrocław, Poland
cDepartment of Chemistry, University of Balearic Islands, Cra. de Valldemossa, km 7.7, 07122 Palma de Mallorca, Spain
dInstitut Universitari d'Investigació en Ciències de la Salut (IUNICS), Spain
eInstitut de Recerca en Ciències de la Salut (IdISBa), Spain
fDepartment of Inorganic Chemistry, Faculty of Pharmacy, University of Granada, E-18071 Granada, Spain
First published on 30th October 2024
Abstract
The relationship between the coordination chemistry and antimicrobial activity of Zn(II) and Cu(II)-bound histatins, salivary antimicrobial peptides, remains enigmatic. We focus on metal complexes of histatin 3 and its two products of hydrolysis: histatin 4 and its N-terminal fragment (histatin 3–4). The thermodynamic stability of these complexes is quite expected – the binding of Cu(II) via the ATCUN motif results in the formation of very stable complexes. In histatin-Zn(II) complexes, the {2Nim} type of coordination dominates, with polymorphic binding sites observed for histatin 3–4 and 5–8, resulting in their low thermodynamic stability compared to the complexes of histatin 3, 4, 5 and 8 with Zn(II), in which we observe a {2Nim, O−} type of coordination. Histatin 3, 3–4 and 4 have greater activity against Gram-positive bacteria than against Gram-negative ones, and Cu(II) or Zn(II) binding can, in some cases, moderately increase the antimicrobial activity of the native histatin 3 and 4, but not the remaining 3–4 fragment. The most probable reason for the metal-enhanced antimicrobial activity is, in this case, a local change of charge, while the chemically fascinating metal binding induced structural changes do not result in a change of biological activity. Neither histatin 3–4, the N-terminal fragment of histatin 3, which remains in solution after cleavage, nor its metal complexes have any antimicrobial activity, but histatin 3–4 presents intriguing Zn(II)-induced structural behavior, changing its secondary structure, with a tendency to form an α-helix.
Introduction
Antimicrobial peptides (AMPs) are a huge group of compounds that play an important role in the reactions of the innate immune system of all living organisms, including animals, plants, bacteria or fungi.1 Due to the large number of AMPs, a clear classification is not possible, however, peptides belonging to this group were classified taking into account several features: (i) net charge, their amphipathicity or anionic/cationic peptides, which constitute nearly 90% of all AMPs;2 (ii) three-dimensional structure, which includes α-helical and β-sheet peptides and peptides with a disordered structure;2 or (iv) the target of their antimicrobial activity, mainly antibacterial, antifungal, hemolytic and antiviral.3,4 This broad spectrum of activity is due to the fact that microorganisms have not yet developed an effective mechanism of resistance to these compounds.5,6The group of cationic AMPs, consisting of 24 peptides identified so far, are salivary histatins, produced in the parotid and submandibular glands of humans and some higher primates.7,8 A common feature of histatins is a large number of His residues in the amino acid sequence. As pointed out by Skog et al., the greater the number of His in the amino acid sequence, the deeper the peptide penetrates in the lipid bilayer, thus causing permanent damage to the cell membrane, and its further accumulation in the mitochondrium.9 This process, most likely, is the basis of the histatins’ mode of action.10,11 Interestingly, this mechanism has been proven for pathogens of fungal origin, including Candida albicans.12 However, for bacterial pathogens, the mechanism remains unclear. Another common feature of histatins is their ability to bind metal ions, including Cu(II), Ni(II), Fe(III) and Zn(II), which, in turn, may affect their antimicrobial activity by changing the secondary structure or net charge of the peptides.13,14 The ability to coordinate metal ions also causes histatins to participate in the so-called nutritional immunity, i.e. the process in which the host, in response to an infection, limits access to metal ions, necessary for the survival and virulence of the pathogen.15 Metal binding is possible due to the presence of specific metal ion-binding motifs in the peptide: (i) HEXXH16,17 and HXXXHH18 motifs, typical for Zn(II) binding, and (ii) the NH2-XXH motif, i.e. the amino terminal Cu(II)- and Ni(II) (ATCUN) motif, which binds Cu(II) and Ni(II) ions.19,20
The antimicrobial activity of histatins is widely described in the literature, with the best-studied histatin being histatin 5, which also has the greatest antimicrobial activity. It is the first line of defense against a pathogen causing, among others, oral candidiasis – the fungus C. albicans.10 On the other hand, histatin 3, which, next to histatin 1, is one of the two genetically encoded histatins (by HTN1 and HTN3 genes, respectively),21 also shows high activity against C. albicans.22,23 Tsai et al. indicated that the antifungal effect of histatin 3 is associated with effective inhibition of the germination of C. albicans blastoconidia.24 Reduced levels of histatin 3 are observed in patients infected with HIV (human immunodeficiency virus). This may lead to candidiasis, an infection caused by fungi of the Candida spp.25 In turn, increased levels of histatin 3 were observed in patients who experienced a short-term stressful episode. The study by Marvin et al. indicates the possible use of histatin 3 as a stress biomarker.26 Equally interesting is the fact that histatin 3, unlike histatin 5, has wound closing properties in vitro.27 This is most likely related to the differences in their amino acid sequences. Since histatin 5 is a proteolytic cleavage product of histatin 3, they differ in the 8-amino acid residues located at the C-terminus, and it is this specific fragment that most likely determines the wound-closing effect of histatin 3. Histatin 1 has a similar effect.28
Intrigued by our previous results, in which we described the coordination of Cu(II) and Zn(II) to: (i) histatin 5; (ii) to the N-terminal fragment of histatin 5 (called histatin 5–8); and (iii) to histatin 8, as well as the antimicrobial activity of both native peptides and their complexes,29 we decided to focus on the relationship between the metal binding ability, the structure, and microbiological activity of: (i) histatin 3; (ii) the N-terminal fragment of histatin 3 (histatin 3–4); and (iii) histatin 4 (Table 1). In addition, compelled by our recent findings proving the Zn(II)-induced structural rearrangment of DSHAKRHHGYKR (i.e. the N-terminal fragment of histatin 5, named ‘histatin 5–8’),29 we also used SAXS spectroscopy to study whether Zn(II) was also able to modify the conformational ensemble defining the structure of histatin 3–4.
Experimental
Materials
All peptides, histatin 3 (DSHAKRHHGYKRKFHEKHHSHRGYRSNYLYDN), histatin 3–4 (DSHAKRHHGYK), histatin 4 (RKFHEKHHSHRGYRSNYLYDN) and histatin 5–8 (DSHAKRHHGYKR) were purchased from KareBay Biochem (USA) (certified purity of 98%) and were used as received. The Cu(ClO4)2·6H2O and Zn(ClO4)2·6H2O were an extra pure product (Sigma-Aldrich), the concentrations of their stock solutions were determined by ICP-OES. The carbonate-free stock solution of 0.1 M NaOH was purchased from Sigma-Aldrich and then potentiometrically standardized with potassium hydrogen phthalate.Mass spectrometry
High-resolution mass spectra were obtained on an LCMS-9030 qTOF Shimadzu (Shimadzu, Kyoto, Japan) device, equipped with a standard ESI source and the Nexera X2 system. The mass spectrometer was used in the positive and negative ion modes. The instrumental parameters were as follows: scan range m/z 100–3000, nebulizing gas nitrogen, nebulizing gas flow 3.0 L min−1, drying gas flow 10 L min−1, heating gas flow 10 L min−1, interface temperature 300 °C, desolvation line temperature 400 °C, detector voltage 2.02 kV, interface voltage 4.0 kV, argon was used as collision gas, and mobile phase: (i) H2O + 0.1% HCOOH, and (ii) MeCN + 0.1% HCOOH at a flow of 0.3 mL min−1. The injection volume was optimized depending on the intensity of the signals observed on the mass spectrum within the range of 0.1 to 3 μL. The samples were prepared in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol–water mixture at a pH value of 7.4. The sample concentration was [ligand]tot = 10−4 M, and the M
1 methanol–water mixture at a pH value of 7.4. The sample concentration was [ligand]tot = 10−4 M, and the M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L molar ratio was 1
L molar ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The obtained data were analyzed by ACD/Spectrus Processor 220.2.0 (ACD/Labs, Ontario, Canada).
1. The obtained data were analyzed by ACD/Spectrus Processor 220.2.0 (ACD/Labs, Ontario, Canada).
Potentiometry
The stability constants for proton Cu(II) and Zn(II) complexes were calculated from titration curves carried out over the pH range 2–11 at 298 K under an argon atmosphere and ionic strength of 0.1 M (NaClO4) using a total volume of 2.8 mL. The potentiometric titrations were performed using a Metrohm Titrando 905 titrator and a Mettler Toledo InLab Micro combined pH electrode. The thermostabilized glass-cell was equipped with a magnetic stirring system and a microburet delivery tube. The solutions were titrated with 0.1 M carbonate-free NaOH. The electrodes were daily calibrated for hydrogen ion concentration by titrating HClO4 with NaOH under the same experimental conditions as mentioned above. The purities and the exact concentrations of the ligand solutions were determined using the Gran method.30 The ligand concentration was 0.5 mM, and the Cu(II)/Zn(II) to the ligand ratio was 0.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
The standard potential and the slope of the electrode couple were computed by means of the GLEE program.31 HYPERQUAD 2006 was used for the stability constant calculations.32 The standard deviations were computed by HYPERQUAD 2006 and refer to random errors only. The constants for the hydrolytic Zn(II) species were used in these calculations.33 The speciation and competition diagrams were computed with the HYSS program.34
Ultraviolet-visible (UV-Vis) and circular dichroism (CD) spectroscopy
The absorption UV-Vis spectra were recorded at 25 °C on a Varian Cary300 Bio spectrophotometer in the range of 200–800 nm, using a quartz cuvette with an optical path of 1 cm. Circular dichroism (CD) spectra were recorded on a Jasco J-1500 spectropolarimeter in the 200–800 nm range, using a quartz cuvette with an optical path of 1 cm in the visible and near-UV range and 0.01 cm in the spectral range of 180–250 nm to determine the secondary structure of ligands and their complexes. The concentrations of the solutions used for spectroscopic studies were similar to those employed in the potentiometric experiments: the metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand ratio was 0.8
ligand ratio was 0.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The UV-Vis and CD spectroscopy parameters were calculated from the spectra obtained at the pH values corresponding to the maximum concentration of each particular species, on the basis of potentiometric studies. OriginPro 8 was used to process and visualize the obtained spectra.
1. The UV-Vis and CD spectroscopy parameters were calculated from the spectra obtained at the pH values corresponding to the maximum concentration of each particular species, on the basis of potentiometric studies. OriginPro 8 was used to process and visualize the obtained spectra.
Nuclear magnetic resonance (NMR) spectroscopy
NMR spectra were recorded at 14.1 T on a Bruker Avance III 600 MHz spectrometer equipped with a Silicon Graphics workstation. The temperatures were controlled with an accuracy of ±0.1 K. Suppression of the residual water signal was achieved by excitation sculpting, using a selective square pulse on water 2 ms long. The samples were prepared in a 90% H2O and 10% D2O mixture. The proton resonance assignment was accomplished by 2D 1H–1H total correlation spectroscopy (TOCSY) and nuclear Overhauser effect spectroscopy (NOESY) experiments, carried out with standard pulse sequences. Samples of the complexes were prepared by adding Cu(II) (at the final M(II)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L molar ratio of 0.3
L molar ratio of 0.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to an acidic solution of a 1 mM ligand (pH = 3) and later on, the pH was increased to 7.4. The spectra processing and analysis were performed using Brucker TOPSPIN 2.1 and Sparky programs, respectively.35
1) to an acidic solution of a 1 mM ligand (pH = 3) and later on, the pH was increased to 7.4. The spectra processing and analysis were performed using Brucker TOPSPIN 2.1 and Sparky programs, respectively.35
The addition of paramagnetic Cu(II) to the solution leads to severe line broadening of NMR signals of those nuclei close to the paramagnetic ion because of the nuclear–electron dipolar couplings.36,37 To decipher the residues involved in the Cu(II) coordination, we monitored the binding of Cu(II) to histatin 4 by NMR. We performed Cu(II) measurements through the stepwise addition of a Cu(ClO4)2 stock solution to the solution of peptide, at Cu(II)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) histatin 4 molar ratios of 0.1
histatin 4 molar ratios of 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 0.2
1, 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 0.3
1 and 0.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. For each Cu(II)
1. For each Cu(II)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) histatin 4 ratio, we checked the pH and adjusted it to 7.4 (when necessary) with the addition of a 0.1 M NaOH stock solution. For each titration point, we recorded the same set of one-dimensional and 2D NMR spectra that were recorded for the free peptide.
histatin 4 ratio, we checked the pH and adjusted it to 7.4 (when necessary) with the addition of a 0.1 M NaOH stock solution. For each titration point, we recorded the same set of one-dimensional and 2D NMR spectra that were recorded for the free peptide.
SAXS spectroscopy
Solutions containing either histatin 3–4 or histatin 5–8 (10 mM) were prepared in 10 mM sodium cacodylate buffer at pH 7.6. In addition, these solutions were also prepared in the presence of 10 mM and 15 mM Zn(II) and afterwards, they were used for SAXS analysis. The experiments were carried out on a Xeuss 2.0 instrument (Xenocs, France) equipped with a microfocus Cu Kα source (λ 1.54 Å) and a Pilatus 300 k detector (Dectris, Switzerland). The distance between the detector and the sample was calibrated using silver behenate. Measurements were taken at two different sample-detector distances (i.e. 630 mm and 1100 mm). The measurements were carried out for 1 h under vacuum at 25 °C using a BioCUBE device (Xenocs, France), which consist into a temperature-controlled flow through cell. In addition, we also collected the scattering curve for the cacodylate buffer, which was used for buffer subtraction purposes. The averaged frames obtained for each peptide were buffer subtracted using RAW.38 In addition, RAW was used to perform the data processing and analysis. The data obtained when using a sample-detector distance of 1100 mm was used to obtain the radius of gyration (RG) of the peptides, which was derived by the Guinier approximation I(q) = I(0)exp(−q2RG2/3) for qRG < 1.3 using RAW. GNOM was used to compute de pair-distance distribution functions (Pr).39 Given that histatin 3–4 and histatin 5–8 peptides are supposed to be disordered, we used the scattering data acquired when using a sample-detector distance of 630 mm, together with EOM,40 to perform the ensemble optimization analysis. EOM generates a large ensemble of randomly generated conformers (represented as beads models) and using a genetic algorithm, subsets of this ensemble are selected that collectively reproduce the experimental solution data and represent the different conformations adopted by the peptide in solution. In our study, we generated a pool of over 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 random bead conformers for each peptide, and their representative theoretical scatter were calculated. Afterwards, the genetic algorithm identified the ensembles of conformers from this pool that best agree with the data. Distributions of RG and Dmax were calculated by the software. Low-resolution shapes were determined from the solution scattering data using the program DAMMIF41 which uses an algorithm to generate shape envelopes of the scattering molecule. Pymol was used to represent the bead models generated by EOM and the bead envelops generated by DAMMIF were represented using Chimera.
000 random bead conformers for each peptide, and their representative theoretical scatter were calculated. Afterwards, the genetic algorithm identified the ensembles of conformers from this pool that best agree with the data. Distributions of RG and Dmax were calculated by the software. Low-resolution shapes were determined from the solution scattering data using the program DAMMIF41 which uses an algorithm to generate shape envelopes of the scattering molecule. Pymol was used to represent the bead models generated by EOM and the bead envelops generated by DAMMIF were represented using Chimera.
Antimicrobial activity assay
Seven reference strains from ATCC (Enterococcus faecalis ATCC 29212, MRSA Staphylococcus aureus ATCC 43300, Klebsiella pneumoniae ATCC 700603, Acinetobacter baumannii ATCC 19606, Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922 and Candida albicans ATCC 10231) were used for antimicrobial activity assays. The minimum inhibitory concentration (MIC) was determined by a microdilution method with spectrophotometric measurements according to ISO 20776-1:201942 and ISO 16256:2012,43 whereas a modified Richard's method using the redox indicator (2,3,5-triphenyltetrazolium chloride, TTC) was used to determine the minimal bactericidal/fungicidal concentration (MBC/MFC).44–46Stock peptide solutions were prepared in 0.9% TSB four times concentrated. Serial dilutions of the peptides in Tryptic Soy Broth (TSB), with or without an equimolar concentration of Cu(II) or Zn(II), were made on 96-well microplates in the range between 0.5 and 256 μg mL−1. Tryptone Soya Agar (TSA) plates were inoculated with microbial strains from performed stocks. After 24 h per 37 °C incubation (for bacteria) or 24 h per 25 °C (for fungus), a proper density of bacterial and fungal suspension was prepared using a densitometer (final inoculum 5 × 105 CFU mL−1) was prepared in TSB. A positive (TSB + strain) and negative control (only TSB) were also included in the test. Spectrophotometric solubility control of each peptide and peptide-metal ion system was performed. To avoid using strains that had developed additional resistance, the following antibacterial/antifungal agents were used as a positive control, according to breakpoint values established by the EUCAST:47,48E. faecalis: 4 μg mL−1 levofloxacin, S. aureus: 1 μg mL−1 levofloxacin, K. pneumoniae: 4 μg mL−1 gentamicin, A. baumannii: 0.5 μg mL−1 levofloxacin, P. aeruginosa: 1 μg mL−1 levofloxacin, E. coli: 4 μg mL−1 gentamicin and C. albicans: 1 μg mL−1 amphotericin B. MIC was determined from a series of at least three experiments as the lowest concentration of an antimicrobial agent that decreased the measured microbial growth to 50% as referred to positive control.
Microplates were incubated at 37 ± 1 °C or 25 ± 1 °C for 24 h on the shaker (500 rpm). After this, the spectrophotometric measurement was performed at 580 nm and then 50 μL aliquots of 1% (m/v) 2,3,5-triphenyltetrazolium chloride (TTC) solution were added into each well. TTC is a commonly used indicator of cellular respiration. In oxidized form, TTC is colorless, while it turns pink after reduction due to reactions in the respiratory chain. MBC/MFC (Minimum Bactericidal/Fungicidal Concentration) can be observed as the lowest concentration required to kill a particular microbial strain, determined by visual analysis after 24 h incubation with TTC. The pink color indicates the presence of living microorganisms, while the lack of color indicates that the colonies do not survive. Thanks to both methods, MIC and MBC or MFC can be determined.
Results and discussion
Mass spectrometry proves the formation of the complexes of Cu(II) and Zn(II) with histatin 3, histatin 3–4 and histatin 4
Initially we used mass spectrometry to study whether histatin 3, 3–4 and 4 were able to form metal complexes with Cu(II) and Zn(II) cations. The mass spectrum obtained for Cu(II)–histatin 3 mixture (Fig. S1A†) revealed the appearance of signals corresponding to: the free ligand (m/z = 1016.50; z = 4+), its sodium (m/z = 1022.0; z = 4+) and potassium adducts (m/z = 1027.74; z = 4+), and the equimolar Cu(II) complex (m/z = 1031.48; z = 4+) and its sodium adducts containing one (m/z = 1037.47; z = 4+) and two sodium atoms (m/z = 1042.96; z = 4+). In the Zn(II)–histatin 3 mass spectra (Fig. S1B†) we detected signals from: the free ligand (m/z = 1016.50; z = 4+), its sodium (m/z = 1022.00; z = 4+) and potassium adducts (m/z = 1027.74; z = 4+), the histatin 3–Zn(II) complex (m/z = 1031.74; z = 4+), and its adducts with one sodium (m/z = 1038.22; z = 4+), one potassium atom (m/z = 1041.22; z = 4+) and two sodium atoms (m/z = 1042.97; z = 4+).In the mass spectrum obtained from the Cu(II)–histatin 3–4 mixture (Fig. S1C†), we could observe signals coming from the free ligand (m/z = 668.34; z = 2+), its sodium adduct (m/z = 679.33; z = 2+), the Cu(II)–histatin 3–4 complex (m/z = 698.79; z = 2+) and its sodium (m/z = 709.79; z = 2+) and potassium adducts (m/z = 717.77; z = 2+). In the spectra of the same ligand with Zn(II) (Fig. S1D†) we observed signals that can be assigned to the free ligand (m/z = 445.90; z = 3+), its complex with Zn(II) (m/z = 466.53; z = 3+) and the potassium adduct of its Zn(II) complex (m/z = 479.21; z = 3+).
The spectrum of Cu(II)–histatin 4 complex (Fig. S1E†) displayed the signal typical of the free ligand (m/z = 915.78; z = 3+), of its sodium (m/z = 923.12; z = 3+) and potassium adducts (m/z = 930.43; z = 3+), and that of the Cu(II)–histatin 4 complex (m/z = 936.42; z = 3+) and of its potassium adduct (m/z = 949.43; z = 3+). In the mass spectrum of Zn(II)–histatin 4 complex (Fig. S1F†), we detected signals coming from: the free ligand (m/z = 915.78; z = 3+), its sodium adduct (m/z = 923.11; z = 3+), the Zn(II)–histatin 4 complex (m/z = 936.75; z = 3+) and its sodium (m/z = 944.08; z = 3+) and dipotassium adducts (m/z = 964.42; z = 3+).
All simulated mass spectra for the Cu(II) and Zn(II) complexes are in perfect agreement with the experimental ones. Consequently, the obtained data proves that histatin 3, 3–4 and 4 form metal complexes with Zn(II) and Cu(II) cations.
Determination of the ionization constants of histatin 3, histatin 3–4 and histatin 4
Potentiometric measurements detected fifteen ionization constants for histatin 3 (DSHAKRHHGYKRKFHEKHHSHRGYRSNYLYDN). The most acidic constants with a pKa value of 2.59 and 3.32 come from the Asp and Glu side chain deprotonation. The next seven pKa values (4.08, 5.33, 5.73, 6.12, 6.42, 6.81 and 7.38) arise from the deprotonation of the imidazole groups of seven His. The next constant (pKa = 8.96) corresponds to N-terminal amino group. The following three pKa values (9.40, 9.82 and 10.02) are related to the deprotonation of the three Tyr, and the last two (10.30 and 10.53) come from the deprotonation of two Lys residues.Histatin 3–4 (DSHAKRHHGYK) behaves as an H8L acid with the deprotonating groups corresponding at first to the Asp side chain (pKa value 3.12). The next three acidic constants (pKa = 5.67, 6.31 and 7.08) are related to the deprotonation of the three imidazole groups of His residues. The following two pKa (9.19 and 10.01) come from the deprotonation of the N-terminal amino group and Tyr side chain. The last and the highest pKa constants (10.49 and 11.08) correspond to two Lys residues.
In the case of histatin 4 (RKFHEKHHSHRGYRSNYLYDN), eleven constants were detected. The first two acidic ones (pKa of 3.33 and 3.95) correspond to Asp and Glu side chains. The next four constants, with pKa values of 5.38, 6.01, 6.48 and 7.15, are related to the His imidazole groups, and the following one (pKa = 9.10) corresponds to the N-terminal amino group. The three higher constants (9.54, 10.12 and 10.17) come from the deprotonation of three Tyr residues. All the calculated constants are summarized in Table 2.
| Histatin 3 (DSHAKRHHGYKRKFHEKHHSHRGYRSNYLYDN) | Histatin 3–4 (DSHAKRHHGYK) | Histatin 4 (RKFHEKHHSHRGYRSNYLYDN) | ||||
|---|---|---|---|---|---|---|
| Species | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
pK![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ab ab |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
pK![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ab ab |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
pKab |
a
β(HjLk) = [HjLk]/([H]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) j[L]k), in which [L] is the concentration of the fully deprotonated peptide.
b logβ(HjLk) −logβ(Hj−1Lk) = pKa. j[L]k), in which [L] is the concentration of the fully deprotonated peptide.
b logβ(HjLk) −logβ(Hj−1Lk) = pKa.
|
||||||
| H17L | 128.87(3) | 2.59 (D) | ||||
| H16L | 126.28(4) | 3.32 (E) | ||||
| H15L | 122.96(4) | 4.08 (H) | ||||
| H14L | 118.88(3) | 5.33 (H) | ||||
| H13L | 113.55(4) | 5.73 (H) | ||||
| H12L | 107.82(5) | 6.12 (H) | 93.35(3) | 3.33 (D) | ||
| H11L | 101.70(6) | 6.42 (H) | 90.02(3) | 3.95 (E) | ||
| H10L | 95.28(5) | 6.81 (H) | 86.07(3) | 5.38 (H) | ||
| H9L | 88.47(5) | 7.38 (H) | 80.69(3) | 6.01 (H) | ||
| H8L | 81.09(4) | 8.96 (N-t) | 62.91(2) | 3.11 (D) | 74.68(3) | 6.48 (H) |
| H7L | 72.13(3) | 9.40 (Y) | 59.80(2) | 5.67 (H) | 68.20(3) | 7.15 (H) |
| H6L | 62.73(7) | 9.82 (Y) | 54.13(2) | 6.31 (H) | 61.05(3) | 9.10 (N-t) |
| H5L | 52.91(9) | 10.01 (Y) | 47.82(2) | 7.08 (H) | 51.95(2) | 9.54 (Y) |
| H4L | 42.90(9) | 10.30 (K) | 40.74(1) | 9.19 (N-t) | 42.41(3) | 10.12 (Y) |
| H3L | 32.60(7) | 10.53 (K) | 31.55(1) | 10.01 (Y) | 32.29(2) | 10.17 (Y) |
| H2L | 22.07(5) | — | 21.54(1) | 10.49 (K) | 22.12(3) | — |
| HL | — | — | 11.05(2) | 11.05 (K) | — | — |
| Cu(II) complexes | ||||||
| CuH12L | 114.03(1) | — | ||||
| CuH11L | 109.48(1) | 4.55 | — | — | — | — |
| CuH10L | 104.14(2) | 5.34 | — | — | — | — |
| CuH9L | 98.41(2) | 5.72 | — | — | — | — |
| CuH8L | 92.14(2) | 6.27 | — | — | 80.77(1) | — |
| CuH7L | 85.74(1) | 6.40 | — | — | 75.58(1) | 5.19 |
| CuH6L | 78.52(1) | 7.23 | — | — | 69.70(1) | 5.89 |
| CuH5L | 69.24(3) | 9.27 | — | — | 62.53(1) | 7.17 |
| CuH4L | 59.91(2) | 9.34 | 50.49(1) | — | 54.03(2) | 8.50 |
| CuH3L | 49.89(4) | 10.01 | 45.41(1) | 5.08 | 44.92(3) | 9.11 |
| CuH2L | 39.68(4) | 10.21 | 39.00(1) | 6.41 | 35.20(6) | 9.71 |
| CuHL | 29.28(3) | 10.40 | 29.44(1) | 9.56 | 25.44(5) | 9.77 |
| CuL | — | — | 19.27(1) | 10.16 | 15.12(5) | 10.32 |
| CuH−1L | — | — | 7.97(2) | 11.30 | 4.34(7) | 10.78 |
| CuH−2L | −3.90(6) | — | −3.41(3) | 11.38 | — | — |
| CuH−3L | — | — | — | — | −17.62(7) | — |
| Zn(II) complexes | ||||||
| ZnH11L | 106.08(1) | |||||
| ZnH10L | 100.23(1) | 5.85 | — | — | — | — |
| ZnH9L | 94.06(1) | 6.17 | — | — | — | — |
| ZnH8L | 87.10(1) | 6.96 | — | — | — | — |
| ZnH7L | 78.95(1) | 8.15 | — | — | 72.87(1) | — |
| ZnH6L | 69.94(2) | 9.01 | — | — | 66.78(1) | 6.09 |
| ZnH5L | 60.70(2) | 9.24 | — | — | 58.81(2) | 7.97 |
| ZnH4L | 50.96(2) | 9.74 | 44.74(1) | — | 49.49(1) | 9.32 |
| ZnH3L | 40.96(2) | 10.00 | 37.54(1) | 7.20 | 39.93(2) | 9.56 |
| ZnH2L | 29.45(1) | 8.09 | ||||
| ZnHL | 19.82(2) | 9.63 | ||||
| ZnL | 9.72(1) | 10.10 | ||||
The effect of pH on the formation of histatin 3, histatin 3–4 and histatin 4 complexes with Cu(II)
Cu(II) coordination to histatin 3 starts above pH 3.0 (Table 2, and Fig. S2A†). The first detected complex is CuH12L, with a maximum above pH 4.4. The stoichiometry of this species indicates that five acid–base sites are already deprotonated: it is reasonable to assume that the C-terminal, two Asp, one Glu and a His residue have lost a proton. The next deprotonation leads to the formation of the CuH11L complex with pKa value = 4.55 and a maximum at pH 4.9. UV-Vis (Fig. S3A†) band at 530 nm suggests that four nitrogen atoms are involved in Cu(II) binding.49 In the CD (Fig. S4A†) spectra, charge transfer bands indicate the involvement of the following nitrogen atoms in the Cu(II) binding: (i) at 308.6 nm the amide nitrogen (N−); (ii) at 270.0 nm the N-terminal amine (−NH2); and (iii) at 252.6 nm the imidazole nitrogen (Nim).50 Also, intensive CD bands for d–d transitions (with a positive band at 485.2 nm and a negative band at 569.0 nm appearing from Cotton effects), suggest the coordination of amide nitrogens and a square planar geometry, resulting in the typical “albumin-like coordination mode” {Nim, NH2, 2N−} (Fig. 2A). UV-Vis (Fig. S3A) and CD (Fig. S4A†) spectra do not change significantly within the pH range 5.5–11.0 (CuH10L, CuH9L, CuH8L, CuH7L, CuH6L, CuH5L, CuH4L, CuH3L, CuH2L, CuHL and CuH–2L), which means that the Cu(II) coordination mode is the same for all calculated species.The complex formed at lower pH (above pH 3.0) between Cu(II) and histatin 3–4 is CuH4L, which maximum occurs at pH 4.6 (Table 2, Fig. S2B†). As in histatin 3, an ATCUN motif is present, resulting in a {Nim, NH2, 2N−} binding mode at this pH (Fig. 2B). This is confirmed by its CD (Fig. S4B†) spectrum through the bands appearing at 310.0 nm (amide nitrogen), 270.4 nm (N-terminal amine) and 247.8 nm (imidazole nitrogen).50 In addition, the UV–Vis (Fig. S3B†) band observed at 520 nm, proves the presence of four nitrogen atoms in the coordination sphere.49 The presence of further forms (CuH3L, CuH2L, CuHL, CuL, CuH−1L and CuH−2L) in the 5.5–11 pH range, is related to the deprotonation of non-binding amino acid residues and does not affect the binding mode.
Cu(II) starts to interact with histatin 4, the only studied ligand without the typical ATCUN motif, above pH 3.0. The first observed complex is CuH8L, with a maximum population at pH 4.5 (Table 2, and Fig. S2A†). At this pH, most likely the C-terminal carboxylic group, the Asp, Glu and His residues are already deprotonated. A signal appearing at λmax = 630 nm in the corresponding UV-Vis spectrum (Fig. S3C†), suggests the participation of two nitrogens in Cu(II) coordination,49 most likely one from the His imidazole and the other from the amide group. The next two deprotonations lead to the formation of CuH7L and CuH6L complexes, with pKa values of 5.19 and 5.89, respectively, and are related to the deprotonation of the non-binding His residues and/or amide groups. The observed shift and increase in intensity in the UV-Vis spectra (from 630 nm to 590 nm, Fig. S3C†) indicate the participation of one more amide nitrogen atom in the metal coordination. Also a charge-transfer band near 296 nm and a d–d transition band at 551 nm in the CD spectra (Fig. S4C†) confirm the involvement of an imidazole ring and amide nitrogens in the coordination.50 Loss of one proton leads to the formation of CuH5L complex with pKa value of 7.17, which can be ascribed to the deprotonation of a non-binding His residue. Above pH 8.5, CuH4L (pKa = 8.50) becomes the most populated species, with the third amide group engaging in coordination. This binding mode is confirmed by a significant shift in the UV-Vis spectra (from 590 to 543 nm and then to 512 nm, Fig. S3C†) and the shift in the CD d–d transition band from 551 nm to 503 nm (Fig. S4C†). Also, the appearance of the band at 620 nm in the CD spectra (Fig. S4C†) indicates the formation of a square-planar complex above pH 8.5 with a {Nim, 3N−} binding mode (Fig. 2B). The remaining forms (CuH3L, CuH2L, CuHL, CuL, CuH−1L and CuH−3L) arise from the deprotonation of the N-terminal amine group, three Tyr and two Lys side chains, respectively. These residues have no further impact on the Cu(II) coordination mode.
NMR spectroscopy was employed to further investigate the Cu(II) binding mode, specifically to determine which of the four His in the sequence of histatin 4 is involved in the Cu(II) coordination. The NMR spectrum did not allow to univocally assign a typical sequential walk due to severe signal overlap; the identification of all four His of histatin 4 was neither possible. However, based on the broadening and shifts of the signals of those residues adjacent to His, it can be assessed what are the most probable histidyl residue involved in binding. Signals assigned to the Hα–Hβ correlations of Glu5 became broader and shift (Fig. 1A) and this, most probably, indicates the involvement of its neighboring His4 in Cu(II) coordination. In turn, the lack of shifts/broadening of those signals corresponding to residues near to His8 (e.g. Ser9; Fig. 1B) indicates the exclusion of the neighboring His8 from the Cu(II) coordination sphere.
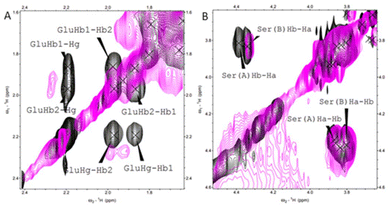 | ||
| Fig. 1 1H–1H TOCSY NMR spectra obtained for histatin 4 (black) and for the Cu(II)–histatin 4 complex (pink). Conditions: [histatin 4] = 1 mM, [Cu(II)] = 0.3 mM, pH = 7.4, T = 298 K (full spectra available in ESI Fig. S5†). | ||
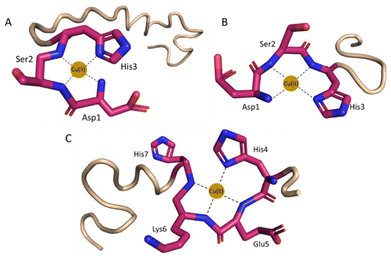 | ||
| Fig. 2 Proposed coordination mode of (A) Cu(II)–histatin 3 and (B) Cu(II)–histatin 3–4 complexes (Cu(II) ions are coordinated by the N-terminal amine, His3 imidazole and two amide nitrogen atoms) at pH 7.4. (C) Cu(II)–histatin 4 complex (Cu(II) ions are coordinated by one imidazole and three amide nitrogens) at pH 8.5. The structures are based on a Phyre251 simulation and the figure was generated using PyMOL.52 | ||
All these data point out His4 as the most probably His involved in coordination sphere of Cu(II).
Formation of histatin 3, histatin 3–4 and histatin 4 complexes with Zn(II)
The complex formed between Zn(II) and histatin 3 in acidic conditions is ZnH11L (Table 2, and Fig. S6A†), where most probably the carboxylate side chain of Glu and two His imidazole side chains coordinate Zn(II) ions. Due to the presence of the 15HEXXH19 motif, His15 and His19 must be involved in binding. The following three complexes: ZnH10L, ZnH9L and ZnH8L, with pKa values of 5.85, 6.17 and 6.96, respectively, come from the deprotonation of the non-coordinating His residues. Loss of the next proton leads to the formation of ZnH7L species (pKa = 8.15), which is related to the deprotonation of a water molecule bound to the central Zn(II) ion (Fig. 3A). The remaining complexes, ZnH6L, ZnH5L, ZnH4L and ZnH3L with a pKa values of 9.01, 9.24, 9.74 and 10.02 can be ascribed to the deprotonation of the N-terminal amine group and three Tyr residues, which non-binding character is confirmed by the lack of change in the binding constants (pKa = 8.96, 9.40, 9.82 and 10.01, respectively, in free ligand, Table 2).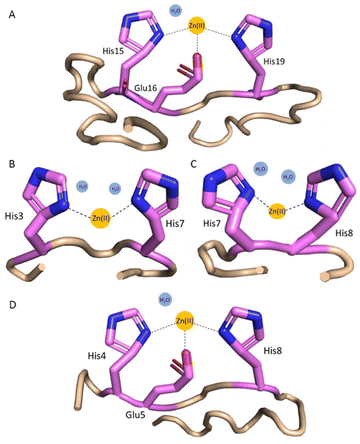 | ||
| Fig. 3 Proposed coordination modes at pH 7.4 for (A) Zn(II)–histatin 3 complex (Zn(II) is coordinated by His15 and His19 imidazoles and Glu16 carboxylate side chain); (B) Zn(II)–histatin 3–4 complex (Zn(II) is coordinated by His3 and His7 or His8 imidazoles); (C) Zn(II)–histatin 3–4 complex (Zn(II) ions are coordinated by His7 and His8 imidazoles); and (D) Zn(II)–histatin 4 complex (Zn(II) ions are coordinated by His4 and His8 imidazoles and Glu5 carboxylate side chain). The structures are based on a Phyre251 simulations and the figure was generated by PyMOL.52 | ||
The first observed complex formed between Zn(II) and histatin 3–4 is ZnH4L (Fig. S6B†), where most likely Asp side chain is already deprotonated and all the imidazoles (His at position 3, 7 and 8) are involved in coordination. Two histidyl residues are most likely involved in the coordination of Zn(II), but we are unable to clearly identify them. This indicates the presence of polymorphic binding sites, e.g. the state in which two different sets of two imidazole residues bind to the metal where one of the His residues is the same in both cases (two complexes in equilibrium are present). The loss of two protons leads to the formation of ZnH3L and ZnH2L complex forms with pKa values of 7.20 and 8.09, which can be related to the deprotonation of two water molecules bound to the central Zn(II) ion resulting in {2Nim} binding mode (Fig. 3B and C). The remaining forms (ZnHL and ZnL) come from the deprotonation of non-bonding N-terminal amine group and Tyr residue.
The first Zn(II)–histatin 4 complex (ZnH7L) appears around pH 4.50 and reaches its maximum population at pH 5.90 (Fig. S6C†). At this point, most probably, carboxylate side chain of Glu and two His imidazoles are involved in Zn(II) coordination. The following complex, ZnH6L, with pKa value of 6.09, comes from the deprotonation of a non-binding His imidazole. The ZnH5L form must arise from the deprotonation of a water molecule present in the vacant Zn(II) binding site. Loss of two additional protons results in the ZnH4L and ZnH3L form, with a pKa of 9.32 and 9.56 (the corresponding pKa in free ligand are 9.10 and 9.54, respectively; Table 2), which must correspond to the deprotonation of the N-terminal amine group and Tyr residue, without any further impact on the coordination mode. Due to the presence of a characteristic binding motif in the histatin 4 sequence (i.e.4HEXXH8), and taking into account the obtained data, it can be concluded that the Zn(II)–histatin 4 complex results in a {2Nim, O−} binding mode (Fig. 3D).
A comparison of metal binding abilities
To compare the affinity of the different histatins towards Cu(II) and Zn(II), we plot competition diagrams made based on the binding constants (Table 2).In the case of complexes where Cu(II) is coordinated by His3, the N-terminal amino group, and two amide nitrogen atoms, we observed a comparable stability (i.e. between histatin 3 and its N-terminal fragment). In the complex of Cu(II) with histatin 4, in which the metal ion is bound via an imidazole ring and three amide nitrogens, we observed the lowest stability in the entire studied pH range (Fig. 4A). This was quite expected, since histatin 4 does not contain the typical ATCUN motif, which binds Cu(II) in a very stable albumin-like binding mode (as in the case of histatin 3 and histatin 3–4).
 | ||
| Fig. 4 Competition plots between histatin 3, histatin 3–4, histatin 4 to bind (A) Cu(II) or (B) Zn(II). The plots describe the complex formation at different pH values in a hypothetical situation, in which equimolar amounts of reagents are mixed. Calculations are based on binding constants from Table 2. Conditions: T = 298 K, [Cu(II)]/[Zn(II)] = [histatin 3] = [histatin 3–4] = [histatin 4] = 0.001 M. | ||
Histatin 3 and histatin 4 coordinate Zn(II) via carboxylate side chain of Glu and two His imidazoles. These complexes show similar thermodynamic stability. In the case of the Zn(II)–histatin 3–4 complex, in which Zn(II) is bound by two different sets of two imidazole residues (polymorphic binding sites), we observe the lowest stability among all tested complexes (Fig. 4B). The lower stability of this complex is undoubtedly influenced by the lack of the typical Zn(II) binding – HEXXH-motif in the amino acid sequence of histatin 3–4.
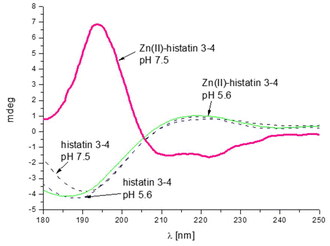
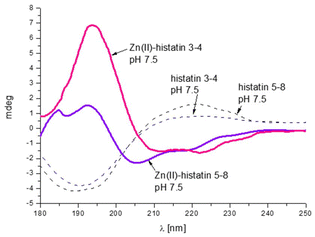
The presence of polymorphic binding sites also has consequences on the secondary structure. Similarly to the previous group of histatins, we observe a Zn(II)-induced structural change only for histatin 3–4 (as previously observed for histatin 5–8).29 Upon Zn(II) coordination, histatin 3–4 has a tendency to adopt an α-helical structure (Fig. 5), even greater than that of histatin 5–8 (Fig. 6).
The change in the secondary structure is most likely related to the movement of metal ions along the polypeptide chain, anchored to His, which leads to the formation of specific hydrogen bond networks that favor the formation of α-helical structures.53 The remaining peptides tested in this work do not change theirs secondary structures after binding metal ion (Fig. S7A–E†).
The effect of Zn(II) on the overall conformations of histatin 3–4 and histatin 5–8
We also used SAXS spectroscopy to better understand the Zn(II)-induced structural change on histatin 3–4 and histatin 5–8 (sequences: DSHAKRHHGYK and DSHAKRHHGYKR, respectively). We studied the holo structural ensembles of the peptides using two different Zn(II)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) peptide: ratios (i.e. 1
peptide: ratios (i.e. 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1.5
1 and 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to better assess the possible effect of Zn on the peptide conformation.
1) to better assess the possible effect of Zn on the peptide conformation.
The scattering curves obtained for histatin 3–4 already revealed subtle structural differences that occurred after having bound Zn(II) (Fig. S8A†) and therefore, they were used to determine the radius of gyration (Rg) from the Guinier plots of the low q region (Fig. S9A†) and in the real space (using GNOM) (Table 3). The obtained results clearly show that the presence of Zn(II) diminished the averaged size of the conformational ensemble of histatin 3–4 by 0.5 ± 0.1 Å and 0.9 ± 0.3 Å at Zn(II)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) histatin 3–4 ratios of 1
histatin 3–4 ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1.5
1 and 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. These results prove that Zn(II) induces a reduction of the overall size of the conformational ensemble of histatin 3–4.
1, respectively. These results prove that Zn(II) induces a reduction of the overall size of the conformational ensemble of histatin 3–4.
| Peptide | Rg (Guinier)a (A) | qR g | R g (Pr)b (A) | R g (EOM)c (A) | D max (EOM)c (A) |
|---|---|---|---|---|---|
| a Radius of gyration determined by the Guinier region in the reciprocal space. b Radius of gyration determined in the real space. c Average radius of gyration of the conformational ensemble calculated from the scattering data using EOM. | |||||
| Histatin 3–4 (Apo) | 8.38 ± 0.14 | 1.279 | 8.85 ± 0.09 | 9.00 | 22.05 |
Histatin 3–4 + Zn(II) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
7.99 ± 0.11 | 1.298 | 8.26 ± 0.08 | 8.80 | 21.13 |
Histatin 3–4 + Zn(II) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5) 1.5) |
7.79 ± 0.09 | 1.286 | 7.75 ± 0.17 | 8.76 | 20.01 |
| Histatin 5–8 (Apo) | 8.61 ± 0.05 | 1.287 | 8.95 ± 0.04 | 8.78 | 20.10 |
Histatin 5–8 + Zn(II) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
8.44 ± 0.04 | 1.291 | 8.81 ± 0.03 | 8.75 | 20.25 |
Histatin 5–8 + Zn(II) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5) 1.5) |
8.33 ± 0.05 | 1.284 | 8.67 ± 0.04 | 8.71 | 20.36 |
In addition, we studied the effect of Zn(II) on the conformational ensemble of histatin 5–8. The scattering curve obtained for the apo histatin 5–8 was slightly different to those obtained when Zn(II) was added (Fig. S8B†). These scattering curves were then also used to determine the radius of gyration (Rg) from the Guinier plots (Fig. S9B†) and in the real space (Table 3). In comparison with what it was observed for histatin 3–4, the addition of Zn(II) scarcely diminished the averaged size of the conformational ensemble of histatin 5–8. In fact, its Rg only decreased in 0.16 ± 0.2 Å and in 0.28 ± 0.1 Å when 1 and 1.5 equivalents of Zn(II) were added, respectively. Consequently, our data prove that the presence of Zn(II) induces a tiny (but noticeable) reduction or the size of the conformational ensemble of histatin 5–8, but this effect is much lower than that induced on the conformational ensemble of histatin 3–4.
SAXS data was also used to calculate the pair distribution functions (Pr) of holo and apo histatin 3–4 and histatin 5–8 (Fig. S10†). The apo histatin 3–4 displays a broad range of populated conformations whose maximum dimension (Dmax) was of ∼30 Å, and the most populated conformation was that having a Rg of ∼9 Å. Nevertheless, the maximum of the distribution function shifted towards lower Rg values at increasing Zn(II) concentrations, thus indicating a Zn-induced compaction of the conformational ensemble of histatin 3–4 (Fig. S10A†). In the case of histatin 5–8, both, apo and holo forms, display a broad range of populated conformations whose maximum dimensions (Dmax) were of ∼28 Å. While the most populated conformations of apo histatin 5–8 had a Rg of ∼8.5 Å, the distribution functions of holo histatin 5–8 slightly shifted towards lower Rg values. In fact, the distribution functions obtained for the peptide at Zn(II):histatin 5–8 ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1.5
1 to 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were almost identical, and their most populated conformations had a Rg of ∼7.0 Å (Fig. S10B†). This confirms that the presence of Zn induced a subtle compaction of the conformational ensemble of histatin 5–8 but again, this effect was lesser than that observed for histatin 3–4.
1 were almost identical, and their most populated conformations had a Rg of ∼7.0 Å (Fig. S10B†). This confirms that the presence of Zn induced a subtle compaction of the conformational ensemble of histatin 5–8 but again, this effect was lesser than that observed for histatin 3–4.
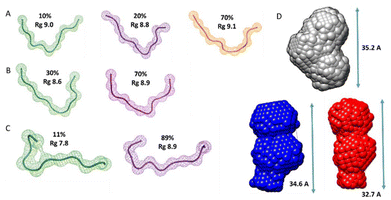
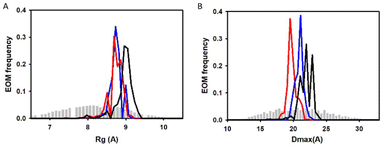
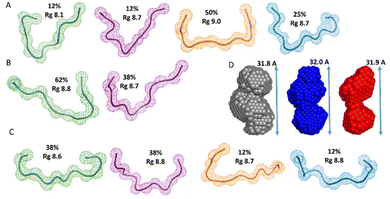
Additionally, we also used SAXS to carry out quantitative flexibility analysis using EOM approach. The conformational ensemble of apo histatin 3–4 was represented by three different bended conformers (Fig. 7A), which adopted Rg values between 8.8–9.1 Å. The addition of Zn(II) clearly shifted the pool of conformers towards lower Rg values (Fig. 8A). The pool of the holo histatin 3–4 was represented by two different conformers whose structure, was slightly more compact than that of the apo structure (Fig. 7B and C). This Zn(II)-induced compaction became evident when comparing the Dmax values (the maximum length that can the peptide can adopt), which were notably reduced in the presence of Zn(II) (Fig. 8B).
The conformational ensemble of the apo histatin 5–8 was represented by four bended conformers, which adopted Rg values between 8.1 and 9.0 Å. Nevertheless, the 50% of the overall conformational ensemble was represented by an extended conformer with a Rg of ∼9 Å (Fig. 9A). The addition of 1 equiv. of Zn(II) did not induce any change on the pool of conformers of histatin 5–8 (Fig. S11A†), and its overall conformational ensemble was represented by two fairly similar conformers, whose Rg values were similar to those obtained for the apo histatin 5–8 (Table 3, and Fig. 9B). The addition of 0.5 additional equiv. of Zn(II) slightly increased the heterogeneity of the pool of the conformers (Fig. S11A†) as a result of the appearance of four different conformers (two of them with a weight of 38% and two of them with a weight of 12%) representing the overall conformation of holo histatin 5–8 at 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Zn(II)
1 Zn(II)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) peptide ratio (Fig. 9C). In any case, all their Rg values were similar to those displayed by the conformers representing apo histatin 5–8. This indicates that the addition of Zn(II) to histatin 5–8 does not induce a remarkable compaction of its conformational ensemble, which also becomes evident when comparing the Dmax values (Fig. S11B†).
peptide ratio (Fig. 9C). In any case, all their Rg values were similar to those displayed by the conformers representing apo histatin 5–8. This indicates that the addition of Zn(II) to histatin 5–8 does not induce a remarkable compaction of its conformational ensemble, which also becomes evident when comparing the Dmax values (Fig. S11B†).
Finally, the scattering curves were also use to generate the DAMMIF ab initio models for the apo and holo histatin 3–4 and histatin 5–8. The model obtained for apo histatin 3–4 was slightly bended, and its length was larger than that of the ab initio models obtained for holo histatin 3–4 (Fig. 7D).
The obtained averaged models for apo and holo histatin 5–8 were all really similar and they all display a somehow lineal-like shape (Fig. 9D).
Antimicrobial activity
In vitro antimicrobial activities of all studied histatins and their Cu(II) and Zn(II) complexes were tested against an expanded panel of microorganisms: two Gram-positive (Enterococcus faecalis ATCC 29212 and MRSA Staphylococcus aureus ATCC 43300), four Gram-negative (Klebsiella pneumoniae ATCC 700603, Acinetobacter baumannii ATCC 19606, Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922) and one fungal strain (Candida albicans ATCC 10231).None of the tested histatins, nor their Cu(II) and Zn(II) complexes, show microbiological activity against the Gram-negative bacteria used in the tests: K. pneumoniae (ATCC 700603), A. baumannii (ATCC 19606), P. aeruginosa (ATCC 27853) and E. coli (ATCC 25922). The binding of Cu(II) to histatin 3 and histatin 4 triggers antibacterial activity against the Gram-positive bacterium S. aureus (ATCC 43300) (MIC = 64 μg mL−1 and 256 μg mL−1, respectively). A similar effect is observed in the case of the second Gram-positive bacterium E. faecalis (ATCC 29212) – the MIC value for the Cu(II)–histatin 4 complex is 128 μg mL−1. In the case of this strain, the binding of Zn(II) ions by histatin 4 also induces antimicrobial activity (MIC = 64 μg mL−1). Histatin 3 and its complexes with Cu(II) and Zn(II) showed activity against the fungus C. albicans (ATCC 10231), but its binding to these metal ions does not enhance its antifungal effect (MIC = 128 μg mL−1 for the native peptide, MIC = 256 μg mL−1 for the Cu(II)–histatin 3 complex and MIC = 128 μg mL−1 for the Zn(II)–histatin 3 complex; Table 4).
| Strain | E. faecalis (+) ATCC 29212 | S. aureus (+) ATCC 43300 | C. albicans ATCC 10231 |
|---|---|---|---|
| MIC (μg mL−1) | MIC (μg mL−1) | MIC (μg mL−1) | |
| histatin 3 | n/d | n/d | 128 |
| Cu(II)–histatin 3 | n/d | 64 | 256 |
| Zn(II)–histatin 3 | n/d | 128 | 128 |
| histatin 3–4 | n/d | n/d | n/d |
| Cu(II)–histatin 3–4 | n/d | n/d | n/d |
| Zn(II)–histatin 3–4 | n/d | n/d | n/d |
| histatin 4 | n/d | n/d | n/d |
| Cu(II)–histatin 4 | 128 | 256 | n/d |
| Zn(II)–histatin 4 | 64 | n/d | n/d |
Conclusions
Although nearly 40 years have passed since the discovery of histatins (the first histatin was isolated from human parotid secretion in 198854), the relationship between their coordination chemistry and their antimicrobial activity, as well as their particular mechanism of action, still remains unknown.The comparison of the six histatins that we have examined so far, clearly shows that the binding of Cu(II) ions via the ATCUN motif significantly enhances the thermodynamic stability of the metal complexes at pH between 3 and 9. Histatin 4, the only studied histatin that does not have the ATCUN motif, is able to coordinate Cu(II) through a histidyl imidazole ring and two amide nitrogens at physiological pH. Above pH 8.5, the coordination sphere is completed by a third amide nitrogen. This coordination mode is less stable than the so-called albumin-like one.
In histatin–Zn(II) complexes, the {2Nim} type of coordination dominates with polymorphic binding sites which we observed in the case of histatin 3–4 and 5–8. This results in a lower thermodynamic stability when compared to the complexes of histatin 3, 4, 5 and 8 with Zn(II), in which we observed a {2Nim, O−} type of coordination.
Our recently published work29 on Zn(II) and Cu(II) complexes with histatin 5, 5–8 and 8 shows that: (i) the antimicrobial activity of histatins and their complexes depends on the pH; (ii) histatins are more effective against Gram-positive bacteria compared to Gram-negative bacteria; and (iii) that the binding of Zn(II) in most cases increases the antimicrobial activity of histatins. Now, in this work, we also show that histatins 3, 3–4 and 4 have greater activity against Gram-positive bacteria, while the binding of Cu(II) or Zn(II) can, in some cases, moderately increase the antimicrobial activity of histatin 3 and 4, but not that of histatin 3–4. Interestingly, only one antimicrobial mode of action was observed in the absence of metal ions – histatin 3 is active against C. albicans.
The explanation of the metal-enhanced antimicrobial activity is most likely as trivial as a ‘local change of charge’. While the chemically fascinating metal binding induced structural changes, they do not result in a change of biological activity. Neither histatin 3–4, which remains in solution after cleavage, nor its metal complexes, had any antimicrobial activity. However, histatin 3–4 displayed intriguing Zn(II)-induced structural behavior – even more pronounced than that observed in the case of Zn(II)-bound histatin 5–8.
Both histatin 3–4 and histatin 5–8 displayed a higher propensity to adopt an α-helical-like conformation upon binding to Zn(II) and while increasing the compactness of their conformational ensemble. This effect, was much more evident in the case of the Zn(II)–histatin 3–4 complex, compared to the Zn(II)–histatin 5–8 one. However, in both cases, the Zn(II)-triggered structural change did not increase their antimicrobial activity.
The experimental results regarding thermodynamics, coordination mode, antimicrobial activity, as well as the influence of metal ions, especially Zn(II), on the secondary structure of subsequent peptides from the histatin family are another important step towards understanding the exact mechanism of action of histatins.
Author contributions
E.D., A.H., J.W., A.M., A.M-W., M.A., M.B-O., A.D-M and M.R-Ż. conceived and planned the experiments. E.D., A.H., A.M., M.A. and M.B-O. conducted the experiments. E.D., A.H., J.W., A.M., A.M-W., M.A., M.B-O., A.D-M and M.R-Ż. analyzed the results. All authors reviewed the manuscript.Data availability
The data supporting the article “Zn(II) coordination influences the secondary structure, but not antimicrobial activity of the N-terminal histatin 3 hydrolysis product” have been included as part of the ESI.† If any additional questions regarding experimental details may arise, the corresponding authors remain at the Readers’ disposal.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Support of the National Science Centre (UMO-2017/26/E/ST5/00364) is gratefully acknowledged. J. W. is supported by the National Science Centre (UMO-2021/41/B/ST4/02654). Funding which allowed staff exchange COST Action CA18202, NECTAR—Network for Equilibria and Chemical Thermodynamics Advanced Research, supported by COST (European Cooperation in Science and Technology) is gratefully acknowledged. The support of Spanish Ministerio de Ciencia e Innovación (MCIN) for funding the METALBIO Research Network (reference: RED 2022-134091-T) is also acknowledged.References
- J. L. Alexander, Z. Thompson and J. A. Cowan, ACS Chem. Biol., 2018, 13, 844–853 CrossRef CAS PubMed.
- G. Wang, X. Li and Z. Wang, Nucleic Acids Res., 2016, 44, D1087–D1093 CrossRef CAS PubMed.
- J. D. Steckbeck, B. Deslouches and R. C. Montelaro, Expert Opin. Biol. Ther., 2013, 14, 11–14 CrossRef PubMed.
- P. Dutta and S. Das, Curr. Top. Med. Chem., 2016, 16, 99–129 CrossRef CAS PubMed.
- M. R. Yeaman and N. Y. Yount, Pharmacol. Rev., 2003, 55(1), 27–55 CrossRef CAS PubMed.
- A. Moretta, C. Scieuzo, A. M. Petrone, R. Salvia, M. D. Manniello, A. Franco, D. Lucchetti, A. Vassallo, H. Vogel, A. Sgambato and P. Falabella, Front. Cell. Infect. Microbiol., 2021, 11, 668632 CrossRef CAS PubMed.
- M. Castagnola, R. Inzitari, D. V. Rossetti, C. Olmi, T. Cabras, V. Piras, P. Nicolussi, M. T. Sanna, M. Pellegrini, B. Giardina and B. I. Messana, J. Biol. Chem., 2004, 279, 41436–41443 CrossRef CAS PubMed.
- L. Padovan, L. Segat, A. Pontillo, N. Antcheva, A. Tossi and S. Crovella, Protein Pept. Lett., 2010, 17(7), 909–918 CrossRef CAS PubMed.
- A. E. Skog, G. Corucci, M. D. Tully, G. Fragneto, Y. Gerelli and M. Skepo, Langmuir, 2023, 39(22), 7694–7706 CrossRef CAS PubMed.
- S. Puri and M. Edgerton, Eukaryotic Cell, 2014, 13(8), 958–964 CrossRef PubMed.
- J. X. Campbell, S. Gao, K. S. Anand and K. J. Franz, ACS Infect. Dis., 2022, 8(9), 1920–1934 CrossRef CAS PubMed.
- E. J. Helmerhorst, P. Breeuwer, W. van't Hof, E. Walgreen-Weterings, L. C. Oomen, E. C. Veerman, A. V. Amerongen and T. Abee, J. Biol. Chem., 1999, 274, 7286–7291 CrossRef CAS PubMed.
- E. Kurowska, A. Bonna, G. Goch and W. Bal, J. Inorg. Biochem., 2011, 105, 1220–1225 CrossRef CAS PubMed.
- S. E. Conklin, E. C. Bridgman, Q. Su, P. Riggs-Gelasco, K. L. Haas and K. J. Franz, Biochemistry, 2017, 56(32), 4244–4255 CrossRef CAS PubMed.
- J. Wątły, S. Potocki and M. Rowińska-Żyrek, Chem. – Eur. J., 2016, 22, 15992–16010 CrossRef PubMed.
- J. Grogan, C. McKnight, R. F. Troxler and F. G. Oppenheim, FEBS Lett., 2001, 491, 76–80 CrossRef CAS PubMed.
- K. M. Fukasawa, T. Hata, Y. Ono and J. Hirose, J. Amino Acids, 2011, 2011, 574816 Search PubMed.
- C. Cragnell, L. Staby, S. Lenton, B. B. Kragelund and M. Skepo, Biomolecules, 2019, 9, 168 CrossRef CAS PubMed.
- R. Sankararamakrishnan, S. Verma and S. Kumar, Proteins, 2005, 58(1), 211–221 CrossRef CAS PubMed.
- P. Gonzalez, K. Bossak, E. Stefaniak, C. Hureau, L. Raibaut, W. Bal and P. Feller, Chem. – Eur. J., 2018, 24, 8029–8041 CrossRef CAS PubMed.
- L. Sabatini and E. Azen, Biochem. Biophys. Res. Commun., 1989, 160, 495–502 CrossRef CAS PubMed.
- H. Yamagishi, D. H. Fitzgerald, T. Sein, T. J. Walsh and B. C. O'Connell, FEMS Microbiol. Lett., 2005, 244(1), 207–212 CrossRef CAS PubMed.
- Y. Xu, I. Ambudkar, H. Yamagishi, W. Swaim, T. J. Walsh and B. C. O'Connell, Antimicrob. Agents Chemother., 1999, 43(9), 2256–2262 CrossRef CAS PubMed.
- H. Tsai and L. A. Bocek, Antimicrob. Agents Chemother., 1997, 41, 2224–2228 CrossRef CAS PubMed.
- K. Lal, J. J. Pollock, R. P. Santarpia, 3rd, H. M. Heller, H. W. Kaufman, J. Fuhrer and R. T. Steigbigel, J. Acquired Immune Defic. Syndr., 1992, 5, 904–914 CAS.
- R. K. Marvin, M. B. Saepoo, S. Ye, D. B. White, R. Liu, K. Hensley, P. Rega, V. Kazan, D. R. Giovannucci and D. Isailovic, Biomarkers, 2017, 22, 372–382 Search PubMed.
- M. J. Oudhoff, J. G. Bolscher, K. Nazmi, H. Kalay H, W. van ‘t Hof, A. V. Amerongen and E. C. Veerman, FASEB J., 2008, 22, 3805–3812 CrossRef CAS PubMed.
- X. Lei, L. Cheng, H. Lin, M. Pang, Z. Yao, C. Chen, T. Forouzanfar, F. J. Bikker, G. Wu and B. Cheng, Front. Bioeng. Biotechnol., 2020, 8, 999 CrossRef PubMed.
- E. Dzień, J. Wątły, A. Kola, A. Mikołajczyk, A. Miller, A. Matera-Witkiewicz, D. Valensin and M. Rowińska-Żyrek, Dalton Trans., 2024, 53, 7561–7570 RSC.
- G. Gran, Acta Chem. Scand., 1950, 4, 559–577 CrossRef CAS.
- P. Gans and B. O'Sullivan, Talanta, 2000, 51, 33–37 CrossRef CAS PubMed.
- P. Gans, A. Sabatini and A. Vacca, Talanta, 1996, 43, 1739–1753 CrossRef CAS PubMed.
- L. Petitt, IUPAC Stability Constants Database, Vol. 23, Chemistry International – Newsmagazine for IUPAC, 2001 Search PubMed.
- L. Alderighi, P. Gans, A. Ienco, D. Peters, A. Sabatini and A. Vacca, Coord. Chem. Rev., 1999, 184, 311–318 CrossRef CAS.
- W. Lee, M. Tonelli and J. L. Markley, Bioinformatics, 2015, 31(8), 1325–1327 CrossRef PubMed.
- E. Gaggelli, F. Bernardi, E. Molteni, R. Pogni, D. Valensin, G. Valensin, M. Remelli, M. Luczkowski and H. Kozlowski, J. Am. Chem. Soc., 2005, 127(3), 996–1006 CrossRef CAS PubMed.
- C. Migliorini, E. Porciatti, M. Luczkowski and D. Valensin, Coord. Chem. Rev., 2012, 256(1–2), 352–368 CrossRef CAS.
- J. B. Hopkins, J. Appl. Crystallogr., 2024, 57, 194–208 CrossRef CAS PubMed.
- D. I. Svergun, J. Appl. Crystallogr., 1992, 25, 495–503 CrossRef CAS.
- A. Sagar, C. M. Jeffries, M. V. Petoukhov, D. I. Svergun and P. Bernado, J. Chem. Theory Comput., 2021, 17(4), 2014–2021 CrossRef CAS PubMed.
- D. Franke and D. I. Svergun, J. Appl. Crystallogr., 2009, 42, 342–346 CrossRef CAS PubMed.
- International Organization for Standardization , Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices–Part 1: Broth micro-dilution reference method for testing the in vitro activity of antimicrobial agents against rapidly growing aerobic bacteria involved in infectious diseases (2019).
- International Organization for Standardization , Clinical laboratory testing and in vitro diagnostic test systems–Reference method for testing the in vitro activity of antimicrobial agents against yeast fungi involved in infectious diseases (2012).
- J. Gabrielson, M. Hart, A. Jarelov, I. Kuhn, D. McKenzie and R. Molby, J. Microbiol. Methods, 2002, 50(1), 63–73 CrossRef CAS PubMed.
- F. L. Francisco, A. M. Saviano, T. de J. Pinto and F. R. Lourenco, J. Microbiol. Methods, 2014, 103, 104–111 CrossRef CAS PubMed.
- P. Sabaeifard, A. Abdi-Ali, M. R. Soudi and R. Dinarvand, J. Microbiol. Methods, 2014, 105, 134–140 CrossRef CAS PubMed.
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 14.0, 2024. https://www.eucast.org.
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs for antifungal agents, version 10.0, 2020. https://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals/.
- L. Pettit, J. E. Gregor and H. Kozłowski, Complex formation between metal ions and peptides, in Perspectives on Bioinorganic Chemistry, ed. R. W. Hay, J. R. Dilworth and K. B. Nolan, JAI Press, London, 1991, Vol. 1, pp. 1–41 Search PubMed.
- T. Kowalik-Jankowska, M. Ruta-Dolejsz, K. Wiśniewska and L. Lankiewicz, J. Inorg. Biochem., 2002, 92, 1–10 CrossRef CAS PubMed.
- A. L. Kelley, S. Mezulis, C. M. Yates, M. N. Sternberg and M. J. E. Sternberg, Nat. Protoc., 2015, 10(6), 845–858 CrossRef PubMed.
- The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC Search PubMed.
- J. Watly, E. Simonovsky, N. Barbosa, M. Spodzieja, R. Wieczorek, S. Rodziewicz-Motowidlo, Y. Miller and H. Kozlowski, Inorg. Chem., 2015, 54, 7692–7702 CrossRef CAS PubMed.
- F. G. Oppenheim, T. Xu, F. M. McMillian, S. M. Levitz, R. D. Diamond, G. D. Offner and R. F. Troxler, J. Biol. Chem., 1988, 263, 7472–7477 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dt02274b |
| This journal is © The Royal Society of Chemistry 2024 |