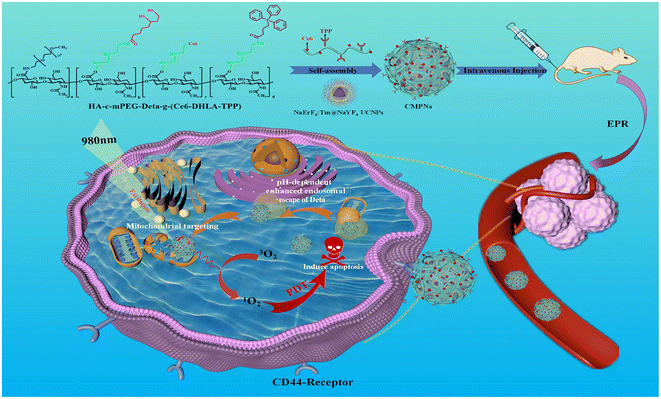
Multifunctional hyaluronic acid ligand-assisted construction of CD44- and mitochondria-targeted self-assembled upconversion nanoparticles for enhanced photodynamic therapy†
Ze Hao
Liu
a,
Xin Wang
Mo
a,
Wei
Jiang
a,
Changling
Liu
a,
Yue
Yin
*b,
Hong Yu
Yang
 *a and
Yan
Fu
*a
*a and
Yan
Fu
*a
aCollege of Materials Science and Engineering, Jilin Institute of Chemical Technology, Jilin City 132022, Jilin Province, PR China. E-mail: yanghy@jlict.edu.cn; fuyanqd@163.com
bSchool of Medical Technology, Beijing Institute of Technology, Beijing 100081, PR China. E-mail: yinyue@bit.edu.cn
First published on 27th September 2024
Abstract
Upconversion nanoparticles (UCNPs) have been used as a potential nanocarrier for photosensitizers (PSs), which have demonstrated a great deal of promise in achieving an effective photodynamic therapy (PDT) for deep-seated tumors. However, overcoming biological barriers to achieve mitochondria-targeted PDT remains a major challenge. Herein, CD44- and mitochondria-targeted photodynamic nanosystems were fabricated through the self-assembly of hyaluronic acid-conjugated-methoxy poly(ethylene glycol)-diethylenetriamine-grafted-(chlorin e6-dihydrolipoic acid-(3-carboxypropyl)triphenylphosphine bromide) polymeric ligands (HA-c-mPEG-Deta-g-(Ce6-DHLA-TPP)) and NaErF4:Tm@NaYF4 core–shell UCNPs (termed CMPNs). The CMPNs presented ideal physiological stability, a good drug loading capacity and an improved capacity for the generation of singlet oxygen (1O2) based on the FRET mechanism. Significantly, confocal images revealed that CMPNs not only facilitated cellular uptake through CD44-receptor-targeted endocytosis, subsequently enabling rapid evasion from endo-lysosomal sequestration, but also specifically targeted mitochondria, ultimately inducing a profound disruption of mitochondrial membrane potential, which triggered apoptosis upon laser irradiation, thereby significantly enhancing the therapeutic effect. Furthermore, in vitro antitumor experiments further confirmed the substantial enhancement in cancer cell killing efficiency achieved by treating with CMPNs upon near-infrared (NIR) laser irradiation. This innovative approach holds promise for the development of NIR-laser-activated photodynamic nanoagents specifically designed for mitochondria-targeted PDT, thus addressing the limitations of the current PDT treatments.
1. Introduction
Photodynamic therapy (PDT), which harnesses the cooperative effect of photosensitizers (PSs), molecular oxygen (O2) and targeted light wavelengths, has become a popular non-invasive cancer treatment option.1–3 This is mainly attributed to its numerous advantages, including cost-effectiveness, high spatiotemporal precision, minimal side effects in contrast to traditional chemotherapy and radiation therapy, and minimal damage to surrounding healthy tissues.4,5 Despite its promising potential, the widespread clinical adoption of PDT in the treatment of deep-seated tumors faces formidable challenges, primarily stemming from the inherent limitations of visible and ultraviolet (UV) light wavelengths in penetrating deeply into tissues, which significantly impairs the effectiveness of PDT in treating deep-seated tumors as these wavelengths are crucial for activating PSs.6–8 Currently, nanoparticles (NPs) employed in PDT have garnered significant attention due to their effective resolution of numerous crucial challenges associated with PDT efficiency.9,10 Recently, lanthanide-ion-doped UCNPs endowed with remarkable biocompatibility, a long luminescence lifetime and a low autofluorescence background have sparked considerable interest in deep-tissue PDT because they possess the capability to harness NIR light and subsequently convert it into visible light for activating PSs.11–13 Notably, the red emission band elicited by lanthanide-ion-doped UCNPs seamlessly aligns with the efficient absorption spectra of commonly utilized and readily accessible PSs. Consequently, the coupling of these PSs with UCNPs facilitates a more efficient fluorescence resonance energy transfer (FRET) process, which proficiently activates the PSs and subsequently enhances the generation of singlet oxygen.14,15 Given the aforementioned advantages, the advancement of a NIR-light-activated UCNP nanocarrier for the delivery of PSs holds significant promise in enhancing the efficiency of PDT.Although lanthanide-doped UCNPs have shown some advantages in biological applications,16,17 the construction of high-efficiency UCNPs and the development of an effective co-doping approach utilizing a sensitizer–activator pair of low concentration, including Yb3+–Er3+, Yb3+–Tm3+ or Yb3+–Ho3+, are typically deemed essential.18,19 Notably, the incorporation of sensitizers (exceeding 20 mol%) or activators (such as Er3+ exceeding 2%) of high concentrations is recognized to trigger intense luminescence quenching, stemming from cross-relaxation processes or the processes via which energy migrates to surface imperfections.20,21 Consequently, this inherent constraint necessitates the general employment of relatively low doping levels of lanthanide activators. To address the above shortcomings, we synthesized red-emitting NaErF4:Tm@NaYF4 UCNPs with a core/shell structure by enclosing a NaYF4 shell around the Er3+ and Tm3+ ion-doped NaErF4:Tm core, which could effectively alleviate luminescence concentration quenching within the nanoparticles by impeding energy migration towards surface defects.
Furthermore, the effectiveness of PDT is significantly constrained by the inherent limitations of reactive oxygen species (ROS). Specifically, the short half-life of ROS, often less than 40 nanoseconds, significantly impedes their lifespan and ability to reach distant targets.22,23 To address these limitations, enhancing the targeting ability of PDT nanoplatforms towards organelles is highly promising. Mitochondria, as pivotal organelles, not only function as the primary energy source within cells, but also significantly contribute to preserving cellular stability and balance.24,25 Increasing evidence indicates that mitochondria-targeting UCNP-based nanosystems can manipulate programmed cell deaths.26,27 Notably, excessive ROS accumulation within mitochondria can trigger mitochondrial depolarization, leading to disrupted homeostasis and ultimately inducing cell apoptosis.28 Therefore, the development of mitochondria-targeted PDT nanoplatforms holds immense potential for enhancing the therapeutic efficacy of PDT.
Additionally, the UCNP-based PDT nanosystems must effectively traverse the biological barriers before reaching the mitochondria of tumor cells,29 which requires not only an effective aggregation of NPs in tumor sites but also enhanced endocytosis and endo-lysosomal escape capabilities to ensure a successful delivery.30–33 However, when fabricating personalized nanosystems based on UCNPs, it is crucial to engineer their surface with precision-tailored capping ligands that endow them with an exceptional tumor-specific targeting ability, mitochondria-directed delivery functionality and endo-lysosomal evasion capability.
Using a PDT nanoplatform, we synthesized a photosensitizer-modified multifunctional hyaluronic acid polymeric ligand named HA-c-mPEG-Deta-g-(Ce6-DHLA-TPP) (Scheme 1). The hyaluronic acid (HA) scaffold serves as a potent tumor-specific ligand, demonstrating a robust binding capability towards cancer cells that exhibit an overexpression of CD44 receptors.34,35 Within its side-chain constituents, the mPEG moiety enhances the stability of nanosystems by forming a hydrophilic shell. The conjugated Deta components facilitate escape from endo-lysosomal confinement via a pH-responsive protonation mechanism through disrupting the endo/lysosomal membrane. The engineered TPP moiety serves as a mitochondria-targeted compound to migrate freely across the mitochondrial membrane, independent of specific uptake mechanisms,36,37 while the pendent LA groups function as anchors that drive the self-assembly of NaErF4:Tm@NaYF4 UCNPs into CD44- and mitochondria-targeted photodynamic nanosystems (CMPNs) (Scheme 1).
CMPNs exhibit excellent stability and ensure FRET from UCNPs to the modified Ce6, producing ROS upon 980 nm laser irradiation. Furthermore, CMPNs exhibit a remarkable capacity to significantly facilitate the uptake of CD44-overexpressing cancer cells, which substantially enhances their escape from the endo/lysosomal compartments (Scheme 1). Notably, confocal imaging reveals the colocalization of CMPNs with mitochondria, a pivotal subcellular organelle targeted in PDT. This colocalization is corroborated by the subsequent confirmation of mitochondrial damage caused by their interactions. This study introduced a comprehensively optimized UCNP-based PDT nanoplatform, which not only maintained exceptional biocompatibility but also demonstrated higher photocytotoxicity compared to non-mitochondria-targeted NPs under 980 nm laser irradiation. This advancement significantly elevates the biomedical significance of UCNPs as nanocarriers for mitochondria-targeted PDT nanoplatforms.
2. Materials and methods
2.1. Materials
Hyaluronic acid with a low molecular weight of 9100 g mol−1 (HA, repeating unit ∼23) was purchased from Freda Biopharm Co., Ltd (Shandong, China). N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC·HCl), N,N′-dicyclohexylcarbodiimide (DCC), methoxy-poly(ethylene glycol)-amine (mPEG–NH2, Mn = 5000), N-hydroxysuccinimide (NHS), diethylenetriamine (Deta), 1,3-diphenylisobenzofuran (DPBF), deuterium oxide (D2O), anhydrous dimethyl sulfoxide (DMSO), sodium tetrahydroborate (NaBH4), lipoic acid (LA), (3-carboxypropyl)triphenylphosphine (TPP), carbonyl diimidazole (CDI), deuterium oxide (D2O), phosphate buffered saline (PBS), 1-octadecene, oleic acid, sodium hydroxide (NaOH), ammonium fluoride (NH4F), anhydrous CH3OH, anhydrous ethanol, anhydrous chloroform, and 4% paraformaldehyde (PFA) were acquired from Energy Chemical Co., Ltd (Shanghai, China). ErCl3·6H2O, YCl3·6H2O and TmCl3·6H2O were purchased from Sigma-Aldrich Co., Ltd. MitoTracker Green, Hank's buffered salt solution, 2′,7′-dichlorofluorescin diacetate (DCFH-DA), chlorin e6 (Ce6), 10% fetal bovine serum (FBS), Hoechst 33342, RPMI-1640 medium, MTT reagent, LysoTracker Green and JC-1 (JC-1, mitochondrial membrane potential dye) were purchased from Beyotime Biotechnology Co., Ltd (Shanghai, China).2.2. Synthesis of NaErF4:Tm upconversion nanocrystal cores (UCNCs)
The synthesis of NaErF4:Tm UCNCs was adjusted based on previous studies conducted by other researchers.38 Tm3+ was doped into NaErF4 core upconversion nanocrystals as an energy trap center to effectively boost upconversion luminescence. Briefly, 0.995 mmol of ErCl3·6H2O and 0.005 mmol of TmCl3·6H2O were combined with 6 mL of OA and 15 mL of ODE at room temperature in a 50 mL 3-neck flask. Subsequently, the reaction was heated to 130 °C and held for 40 min to form an OA precursor solution. After allowing the mixture to cool to 50 °C and combining it with 10 mL of methanol solution (4 mmol of NH4F, 2.5 mmol of NaOH), it was agitated for 30 min and heated to 100 °C to eliminate methanol. Following this, the reaction mixture underwent degassing under an argon atmosphere for 30 min to purge residual methanol and oxygen. Subsequently, the nanocrystals were precipitated via centrifugation with ethanol at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. After 3 times washes with ethanol, the nanocrystals were dispersed in 10 mL of chloroform and made ready for further use.
000 rpm for 10 min. After 3 times washes with ethanol, the nanocrystals were dispersed in 10 mL of chloroform and made ready for further use.
2.3. The synthesis of NaErF4:Tm@NaYF4 core–shell UCNPs
The synthesis process involved a crucial step, wherein NaYF4 shells epitaxially grew around NaErF4:Tm cores, ultimately leading to the formation of core–shell-structured upconversion nanoparticles (UCNPs). Briefly, a mixture of 1 mmol of YCl3·6H2O, 6 mL of OA and 15 mL of ODE was added to a 50 mL 3-neck flask, which was then heated to 130 °C while constantly stirring for 40 min to produce an OA precursor solution. After cooling to 50 °C, the reaction mixture was supplemented with previously synthesized NaErF4:Tm core nanocrystals. After thoroughly heating the reaction mixture to 80 °C and maintaining this temperature for a duration of 30 min to ensure efficient elimination of cyclohexane, the resulting solution was subsequently combined with 10 mL of methanol solution containing 4 mmol of NH4F and 2.5 mmol of NaOH, preheated to 50 °C. The above-mentioned mixture was subjected to continuous agitation for 30 min, ensuring uniform dispersion and interaction of components. Following this, the reaction mixture was further heated to 100 °C and maintained at this elevated temperature for an additional period of 20 min. After a 10 minute degassing process, the solution was heated to 275 °C for 1 h while being shielded with argon gas. Subsequently, the temperature was once again elevated to 275 °C using argon gas for another hour. Notably, the centrifugation steps for NaErF4:Tm@NaYF4 core–shell nanocrystals aligned with the procedure employed for NaErF4 core nanocrystals. The final products were redispersed in anhydrous chloroform and kept for later use at 4 °C in a refrigerator.2.4. The fabrication of CD44- and mitochondria-targeted photodynamic nanosystems (CMPNs)
To fabricate CMPNs, the dual-solvent exchange strategy was employed. Briefly, 10 mL of DMSO was used to dissolve 100 mg of HA-c-mPEG-Deta-g-(Ce6-DHLA-TPP) polymer ligands containing the NaBH4 reductant (50 mM). Subsequently, this solution was mixed with a 5 mL chloroform solution containing UCNPs, adjusting the molar ratio of Ce6 to UCNPs, ranging from 5 to 1. Following the initial mixing, the mixture was subjected to sonication (1000 Hz, alternating 5 s on and 2 s off for a total of 20 min) to facilitate the self-assembly of polymer ligands and UCNPs. Subsequently, the mixture underwent rotary evaporation to effectively remove chloroform, while DMSO was incrementally substituted by deionized (DI) water. In parallel, to eliminate excess ligands, dialysis was employed using a membrane with a molecular weight cut-off of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da. Following dialysis, the mixture was subjected to centrifugation at 10
000 Da. Following dialysis, the mixture was subjected to centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min to facilitate the separation of solid components, which was followed by 3 rounds of rigorous washing with DI water to ensure purity. Finally, the product was lyophilized to yield the targeted CMPNs. Additionally, in a comparable preparation procedure, a non-mitochondria-targeting polymer ligand (HA-c-mPEG-Deta-g-(Ce6-LA)) was employed to substitute the mitochondria-targeting polymer ligand, resulting in the creation of CD44- and non-mitochondria-targeted photodynamic nanosystems (CNMPNs), designated as a control group for comparison purposes.
000 rpm for 15 min to facilitate the separation of solid components, which was followed by 3 rounds of rigorous washing with DI water to ensure purity. Finally, the product was lyophilized to yield the targeted CMPNs. Additionally, in a comparable preparation procedure, a non-mitochondria-targeting polymer ligand (HA-c-mPEG-Deta-g-(Ce6-LA)) was employed to substitute the mitochondria-targeting polymer ligand, resulting in the creation of CD44- and non-mitochondria-targeted photodynamic nanosystems (CNMPNs), designated as a control group for comparison purposes.
2.5. In vitro singlet oxygen (1O2) detection from CMPNs
To evaluate the efficiency of 1O2 generation using CMPNs, DPBF was utilized as a ROS indicator. This evaluation was conducted by closely monitoring the UV-vis absorption changes of DPBF at 420 nm. For the experimental setup, CMPNs (100 μg mL−1) were mixed with DPBF (50 μM) in a PBS buffer solution (10 mM) at a pH of 7.4. Subsequently, the solution was illuminated with a laser at either 660 nm or 980 nm, with a power intensity of 50 mW cm−2, for a predetermined duration of 0–240 seconds. The evaluation of singlet oxygen generation (SOG) was conducted by monitoring the terminal maximum absorption peak of DPBF at 420 nm. To establish a baseline, untreated PBS buffer solutions were utilized as the control group in this experimental setup.2.6. Cell culture
To evaluate CD44-mediated cell internalization, endo-lysosomal escape, in vitro ROS generation, mitochondria-targeting function, mitochondrial membrane potential, and in vitro cancer cell killing ability, the human lung cancer cell line A549 was employed. For comparison, the human hepatoblastoma cell line HepG2 was cultured as CD44-negative cells. The A549 cells and HepG2 cells were sourced from the Beijing Institute of Technology Cell Bank located in Beijing, China. Specifically, A549 cells or HepG2 cells were seeded onto 96-well plates at a density of 1 × 104 cells per well and incubated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. This incubation process was carried out for 24 hours at 37 °C under a humidified atmosphere containing 5% CO2 and 95% air.2.7. Analysis of cellular uptake and endo-lysosomal escape
The cellular uptake capacity and endosomal escape characteristics of CMPNs were investigated using CD44-positive A549 cells based on the high-resolution capability of an LSM700 confocal laser scanning microscope (CLSM, Carl Zeiss, magnification ×400) integrated with advanced 2.5D imaging technology. Initially, A549 cells were incubated with CMPNs at a concentration of 50 μg mL−1 for 4 h, following which the cells were washed 3 times with PBS to remove unbound nanoparticles. Subsequently, the nuclei were stained with Hoechst 33342 for 5 min to enable their visualization. For comparative purposes, HepG2 cells, serving as a CD44-negative cellular model, underwent an identical treatment protocol. To specifically visualize the endo/lysosomal compartment, the cells were labelled with a LysoTracker Green probe for 30 min, ensuring accurate targeting of these organelles. Subsequently, the cells were rinsed 3 times with PBS to eliminate any unbound probe. To capture the endo/lysosomal escape behaviour of CMPNs, a CLSM was employed by adjusting excitation wavelengths. Specifically, Hoechst 33342 was excited at 360 nm to visualize the nuclei, LysoTracker Green at 500 nm for endo/lysosomal compartments and Ce6 at 650 nm during image acquisition.2.8. In vitro 1O2 detection
The intracellular generation of 1O2 in A549 cells upon exposure to CMPNs was monitored using DCFH-DA as a ROS-sensitive probe. Initially, the A549 cells were seeded in a complete medium under standard incubation conditions for 24 h. Following this, the cells were exposed to a controlled concentration of 50 μg of CMPNs for 4 h. After this treatment period, the cells were gently rinsed with PBS to eliminate excess CMPNs. Subsequently, the cells were incubated with 10 μM DCFH-DA solution for 30 min to allow for the intracellular conversion of DCFH-DA into the fluorescent product DCF in the presence of ROS. After incubation, the cells were once again washed with PBS to remove any residual DCFH-DA. To trigger the generation of 1O2, the cells were irradiated with either a 660 nm laser or a 980 nm laser at an intensity of 50 mW cm−2 for 150 seconds. Immediately following irradiation, CLSM and 2.5D imaging were performed, with the excitation wavelength fixed at 488 nm, so as to visualize the intracellular fluorescence generated by the oxidation of DCF. Additionally, control experiments were conducted using non-irradiated A549 cells and cells treated with free Ce6 to assess the specific role of CMPNs in the generation of 1O2. For a flow cytometry analysis, the cells were collected, centrifuged for 5 min at 1000 rpm to pellet them, and resuspended in PBS for measuring the fluorescence intensity of individual cells.2.9. Analysis of mitochondria-targeting ability
For the purpose of assessing the mitochondrial-targeting potential of A549 cells, they were initially seeded onto confocal dishes and maintained in complete DMEM for 24 h. Subsequently, CMPNs were introduced to the cells and further incubated for an additional 4 h. To facilitate visualization, mitochondria and cell nuclei were stained with MitoTracker Green and Hoechst, respectively. Finally, fluorescence images were captured using CLSM and subsequently analyzed to evaluate the mitochondrial targeting efficiency of CMPNs in A549 cells.2.10. Analysis of the mitochondrial membrane potential
A549 cells were incubated in confocal dishes containing compete DMEM for a duration of 24 h. Subsequently, CMPNs and CNMNPs were introduced into these dishes and further incubated for 4 h. To assess the impact of laser irradiation, sets of cells were separated, both with and without exposure to laser radiation at an intensity of 1.0 W cm−2 for 10 min, after which they were maintained for an additional 12 h. Following this period, the cells were stained with JC-1 dye at a concentration of 10 μg mL−1 for 30 min to visualize the potential changes of mitochondrial membranes. Ultimately, fluorescence images were acquired using a CLSM.2.11. In vitro PDT efficacy
The in vitro PDT efficiency of CMPNs against A549 cells was comprehensively assessed both with and without laser irradiation using an MTT assay. Initially, A549 cells were cultured in a standardized growth medium within a Lab-Tek chamber slide for 24 h. Subsequently, under carefully controlled conditions, the cells were exposed to CMPNs or CNMNPs at varying concentrations ranging from 12.5 to 200 μg mL−1 for a duration of 4 h. This treatment was followed by irradiation with a 980 nm laser at an intensity of 1.0 W cm−2 for 10 min, aiming to investigate the cellular response to nanoparticles in the presence of laser exposure. Following the laser treatment, the cells were further incubated for an additional 24 h. To assess cell viability, the culture medium was then supplemented with 10 μL of MTT solution, and the cells were incubated at 37 °C for 4 h, enabling the formation of formazan crystals within viable cells. Ultimately, the absorbance of the formazan product at 540 nm was measured to quantitatively assess the relative cell viability. This metric served as a precise indicator of the cytotoxic effects imparted by CMPNs, both under laser-irradiated and non-irradiated conditions.3. Results and discussion
3.1. The synthesis of NaErF4:Tm@NaYF4 core–shell UCNPs
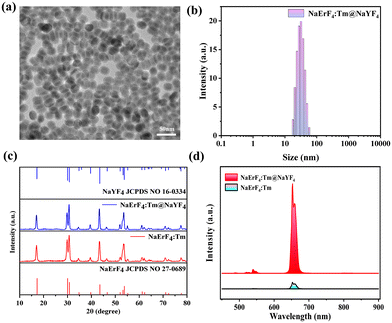
The synthesis procedure for NaErF4:Tm@NaYF4 core–shell-structured UCNPs is schematically depicted in Scheme S1.† Initially, NaErF4:Tm nanocrystals were synthesized, which were designed to act as core components within the UCNP architecture. TEM images showed that NaErF4:Tm nanocrystal cores exhibited uniform monodispersity and small particle size (Fig. S1†). Subsequently, a NaYF4 inert shell was meticulously deposited onto the surface of these cores, ultimately leading to the formation of the targeted NaErF:Tm@NaYF4 UCNPs with a well-defined core–shell structure. Fig. 1a shows a TEM analysis of NaErF4:Tm@NaYF4 UCNPs, revealing a remarkable uniformity and spherical morphology, with particles approximately 25 nm in size and a shell thickness of 4.5 nm, confirming the DLS analysis shown in Fig. 1b. As depicted in Fig. 1c, the X-ray diffraction (XRD) patterns of the synthesized NaErF4:Tm core nanocrystals and NaErF4:Tm@NaYF4 core–shell UCNPs aligned precisely with the standard diffraction peaks for NaErF4 (JCPDS no. 27-06**) and NaYF4 (JCPDS no. 27-06**) in the hexagonal phase, indicating the standard hexagonal phase structure and high crystallinity of the prepared nanocrystals. Furthermore, this consistency further verified that the NaErF4:Tm cores primarily comprised NaErF4, with a successful coating of the NaYF4 inert-shell on the surface of core nanocrystals. Moreover, NaErF4:Tm@NaYF4 UCNPs exhibited a substantial enhancement in upconversion luminescence compared to NaErF4:Tm core nanocrystals (Fig. 1d). This significant improvement can be attributed to Er3+-heavily-doped nanoparticles, which achieve energy condensation via the synergistic effects of Tm3+-driven fleeting energy sequestration and the shielding of an inert shell coating. Given the aforementioned exceptional properties of NaErF4:Tm@NaYF4 UCNPs, these UCNPs effectively serve as nanocarriers and donors, offering remarkable potential for FRET-enhanced PDT applications.3.2. Design and synthesis of the HA-c-mPEG-Deta-g-(Ce6-LA-TPP) polymer ligand
First of all, an mPEGylated HA-c-mPEG-Deta intermediate was synthesized via the sequential attachment of mPEG and Deta to a hydrophilic HA chain through an EDC/NHS-catalyzed amidation process. To validate the grafting degree of mPEG and Deta onto HA, the integrated peak intensities of methyl protons in acetyl groups (3H, ∼1.8 ppm) for HA, as well as the protons of mPEG (4H, ∼3.65 ppm) and Deta (8H, ∼3.1 to 3.5 ppm), were analyzed using 1H NMR spectroscopy (Fig. S2 and Table S1†). Subsequently, Ce6 (serving as a photosensitizer), LA (acting as an anchor) and TPP (serving as a mitochondria-targeting ligand) were individually conjugated to a HA-c-mPEG-Deta polymer (as depicted in Scheme S2†). The CDI-mediated coupled responses between the remaining amine groups of HA-c-mPEG-Deta and the carboxylic acid groups of Ce6, LA and TPP were utilized to synthesize the HA-c-mPEG-Deta-g-(Ce6-LA-TPP) precursor polymer ligand. The incorporation of LA and TPP into HA-c-mPEG-Deta was further authenticated via1H-NMR analysis (Fig. S3†). Moreover, the estimation of their grafting degrees was achieved by evaluating the integration ratio among the peaks of the TPP phenyl group (δ, ∼7.7–7.8 ppm), LA (δ, 1.5–2.5 ppm) and the methyl protons of the acetyl groups of HA (δ, ∼1.8 ppm) (depicted in Fig. S3 and summarized in Table S1†). The successful attachment of Ce6 to HA-c-mPEG-Deta was validated through a detailed comparison of the UV-vis absorption spectra of independent Ce6 and HA-c-mPEG-Deta-g-(Ce6-LA-TPP) (Fig. S4†). This comparative assessment not only confirmed the successful conjugation but also enabled an accurate determination of the number of Ce6 molecules conjugated to HA-c-mPEG-Deta-g-(Ce6-LA-TPP), as outlined in Table S1.† The HA-c-mPEG-Deta-g-(Ce6-DHLA-TPP) polymer ligand was ultimately obtained by reducing the dithiolane groups using NaBH4 as a reductant based on our previous investigation (Scheme S2†).393.3. Fabrication and characterization of CMPNs
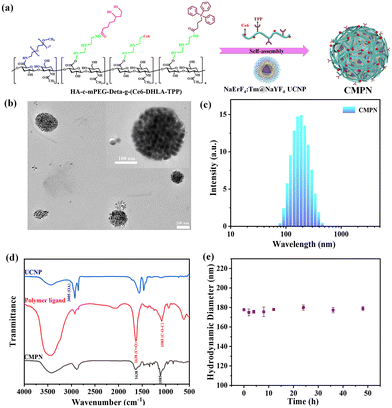
As schematically illustrated in Fig. 2a, hydrophilic and compact CMPNs were successfully constructed via the self-assembly of UCNPs with the HA-c-mPEG-Deta-g-(Ce6-DHLA-TPP) polymer ligand. The DHLA anchor within the polymer ligands could densely wrap around the periphery of small UCNPs, facilitating their assembly into nanoclusters with a distinct core–shell architecture. In these structures, the UCNPs constitute the core, while the PEG moiety within the polymer ligand functions as a hydrophilic exterior layer. Fig. 2b shows a HR-TEM image that clearly depicts the morphology of the CMPNs with a particle size of approximately 180 nm, which aligns well with the DLS measurements presented in Fig. 2c. The successful preparation of CMPNs was further substantiated through an observation of the overlapping UV-vis absorbance spectra exhibited by the polymer ligand and UCNPs within CMPNs (Fig. S4†). As depicted in Fig. 2d, the stark contrast between naked UCNPs and CMPNs lies in the disappearance of the peak of the latter at 3005 cm−1. This absence is attributed to the initial OA ligand of UCNPs being removed through the ligand exchange strategy. Consequently, new characteristic peaks at 1638 cm−1 and 1085 cm−1 emerge in CMPNs, aligning perfectly with the characteristic peaks of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O–C asymmetric stretching vibrations of the polymer ligand. This finding reinforces the notion that the self-assembly of UCNPs into CMPNs is indeed facilitated and driven by the ligand exchange strategy.
O and C–O–C asymmetric stretching vibrations of the polymer ligand. This finding reinforces the notion that the self-assembly of UCNPs into CMPNs is indeed facilitated and driven by the ligand exchange strategy.
Furthermore, the colloidal stability of CMPNs was investigated in a physiological environment, which was essential in determining their eligibility for delivering nanoagents in vivo. As illustrated in Fig. 2e, the CMPNs exhibited remarkable stability, maintaining their hydrodynamic diameter unchanged after 48 h of incubation in a simulated physiological setting (pH 7.4). The hydrophilic shell of PEG, which can effectively prevent aggregation and stabilize the nanoparticles by reducing their adsorption with other biological species, is most likely responsible for their stability.40,41
3.4. In vitro 1O2 generation
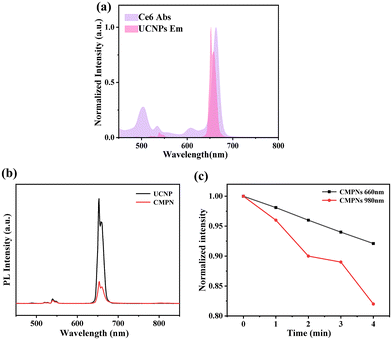
The fluorescence characteristics of UCNPs and CMPNs were further assessed to investigate 1O2 generation from CMPNs. Fig. 3a shows a significant spectral overlap between the prominent emission peak of UCNPs and the distinctive absorption peak of Ce6 at 660 nm, which is a critical factor for the FRET between UCNPs and Ce6-integrated CMPNs to produce 1O2. Furthermore, the emission intensity of CMPNs underwent a remarkable reduction compared to that of UCNPs, clearly indicating the effective FRET occurring from UCNPs to Ce6 (Fig. 3b and Fig. S5†). Moreover, we further investigated the 1O2 generating ability of CMPNs upon either 660 nm or 980 nm laser irradiation. DPBF, serving as a 1O2 detector, has the capability to specifically react with 1O2, causing its absorption peak at 420 nm to decline.42 Upon irradiation with a 980 nm laser, as depicted in Fig. 3c, when CMPNs are combined with DPBF in a solution, the decline rate of the DPBF absorption peak at 420 nm was significantly higher compared to that when a 660 nm laser was used to irradiate CMPNs with an extended irradiation time. These findings further indicated that the generation of 1O2 using CMPNs could be augmented via the FRET mechanism.3.5. CD44-mediated cellular uptake and pH-dependent endo-lysosomal escape of CMPNs
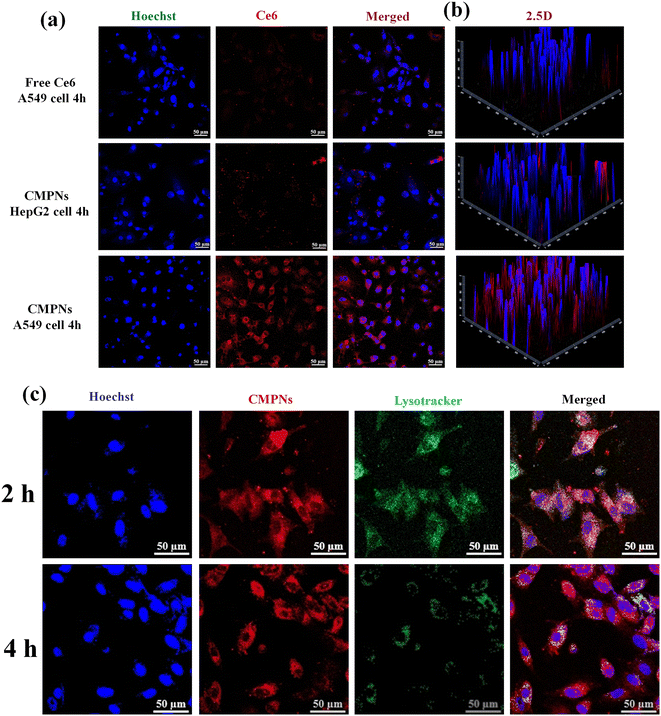
The endocytosis mediated by CD44 and the rapid escape behavior of CMPNs from endo-lysosomes were examined using CLSM in A549 cells expressing CD44, as opposed to CD44-deficient HepG2 cells.43 As illustrated in Fig. 4a, A549 cells treated with CMPNs displayed a notably intensified red signal in comparison with HepG2 cells exposed to an equivalent dosage of CMPNs. Remarkably, when A549 cells were administered with identical standalone Ce6, a negligible fluorescence signal was detected, which aligned with our previous results regarding the limited cellular uptake capacity of Ce6 alone.44 Furthermore, the reconstructed 2.5-dimensional (2.5D) fluorescence images provided further validation of these results without additional quantitative analyses (Fig. 4b). These findings not only highlighted the crucial role of CD44-mediated endocytosis in boosting cellular uptake but also indicated the potential of UCNPs as nanocarriers for the CD44-targeted delivery of Ce6.Exploring the evasion potential of CMPNs within the endo-lysosomal system has become a crucial factor in the realm of intracellular drug delivery. As depicted in Fig. 4c, it is evident that CMPNs were primarily confined within the endo/lysosomal compartments after 2 h treatments, as evidenced by the appearance of yellow regions stemming from the overlap of red-labeled NPs and green areas, representing colocalization. However, the majority of CMPNs were able to successfully evade from the endo/lysosomal compartment and migrate into the cytoplasm of A549 cells after 4 h of incubation. The accelerative endo/lysosomal escape capacity of CMPNs could be attributed to the pH-regulated protonation of the polymer-ligand-bearing Deta components within CMPNs, leading to destabilization of the endo/lysosomal membrane.45 Therefore, these findings suggested that internalization of CMPNs into A549 cells occurred predominantly through the CD44 endocytosis pathway and responded to the endosome environment to facilitate the escape of NPs.
3.6. Intracellular 1O2 generation
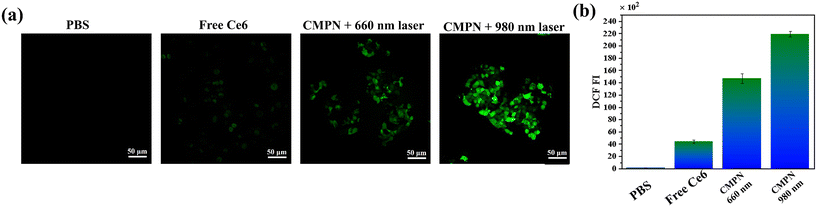
Considering the exceptional stability, FRET-mediated enhancement in the generation of 1O2, and the enhanced internalization and endo/lysosomal escape of CMPNs, the intracellular ROS generation capacity of CMPNs was further assessed through the incubation of A549 cells, employing DCF-DA as an indicator for 1O2 because it could undergo a rapid oxidation reaction, resulting in the production of green fluorescent dichlorofluorescein (DCF).46 As depicted in Fig. 5a and b, the A549 cells incubated with free Ce6 exhibit minimal green fluorescence signals of DCF, marginally exceeding those in the control group due to their limited internalization capability. However, upon exposure to 980 nm laser irradiation, the A549 cells treated with CMPNs displayed a significantly brighter green signal compared to cells incubated with the same NPs under 660 nm laser irradiation. These findings are consistent with the outcomes.3.7. Mitochondrial targeting and in vitro PDT
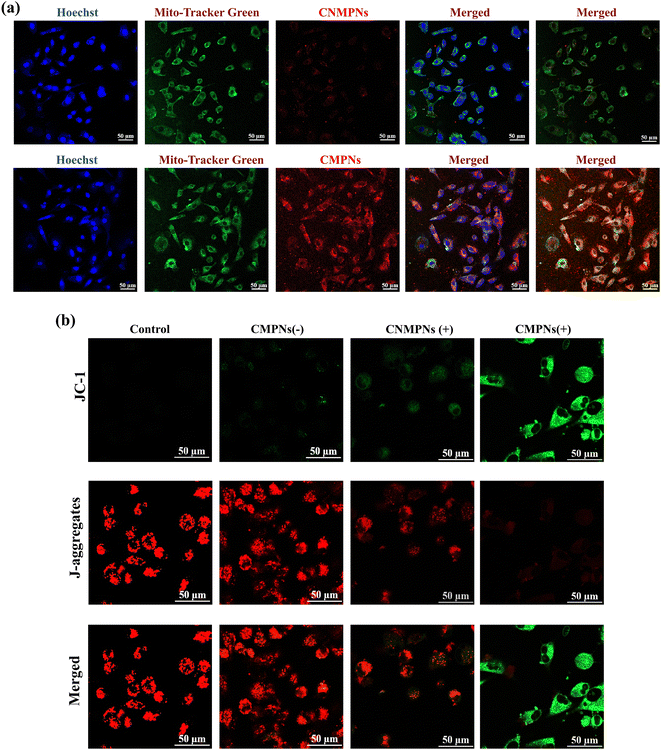
The capacity of CMPNs to specifically target the mitochondria of A549 cells was assessed using a CLSM. Both cell nuclei and mitochondria were differentially labelled with Hoechst and MitoTracker Green, respectively. A549 cells were assessed using a CLSM. Both cell nuclei and mitochondria were differentially labeled with Hoechst and MitoTracker Green, respectively. As depicted in Fig. 6a, the red fluorescence originating from the Ce6 component of CMPNs exhibits a substantial overlap with the green fluorescence of MitoTracker, indicating successful colocalization.In contrast, no significant yellow overlap was observed between the fluorescence signals of CNMNPs and MitoTracker, owing to the absence of mitochondria-targeting TPP ligands. These findings suggested that CMPNs possessed a high affinity for mitochondria, allowing them to effectively accumulate within A549 cells.
The diminution of mitochondrial membrane potentials (MMPs) can trigger mitochondrial depolarization, ultimately inducing cell apoptosis.47 To evaluate whether CMPNs induce alterations of MMPs upon laser irradiation, we employed CLSM to analyze the fluorescence signal variations of A549 cells labeled with JC-1 dye. JC-1 preferentially accumulates in healthy mitochondria, emitting a red fluorescent signal. Conversely, the prevalence of JC-1 monomers results in a green fluorescence signal, serving as a telltale sign of mitochondrial depolarization. As shown in Fig. 6b, in both the control group and the CMPN group without laser irradiation, conspicuous red signals accompanied by the minimal green fluorescence were detected, implying no alteration in MMPs when CMPNs were not exposed to laser irradiation. However, upon laser irradiation in the CNMNP group, a faint green fluorescence signal was discernible, pointing to a slight decrease in MMPs caused by the 1O2-producing capability of CNMNPs activated by laser light. In stark contrast, a prominent green signal was observed in the CMPN group following laser irradiation, which could be attributed to its targeted impact on mitochondria. These findings affirmed the exceptional mitochondrial targeting capability of CMPNs, resulting in the profound depolarization of mitochondria and subsequently causing their damage.
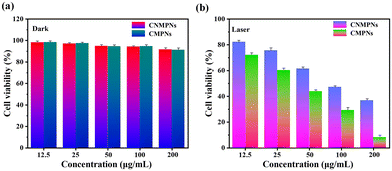
Given the crucial importance of nanoparticle biocompatibility in biomedicine, the biosafety of CMPNs and CNMNPs in darkness was assessed using an MTT assay. Notably, both CMPNs and CNMNPs, with a concentration extending up to 200 μg mL−1, exhibited substantial cell viability in A549 cells (Fig. 7a), indicating their favorable biocompatibility. However, upon exposure to 980 nm laser irradiation, the cytotoxicity observed in A549 cells treated with CMPNs was slightly elevated compared to that of those treated with CNMNPs (Fig. 7b). Such enhanced cytotoxicity of CMPNs can be attributed to the FRET-mediated augmentation of 1O2 generation and their targeted affinity towards mitochondria.
4. Conclusions
In summary, CD44- and mitochondria-targeted photodynamic nanoplatforms were fabricated through the self-assembly of HA-c-mPEG-Deta-g-(Ce6-DHLA-TPP) and UCNPs for FRET-mediated enhanced PDT. The incorporation of polymer ligands onto the surface of UCNPs imparted remarkable physiological stability, enhanced 1O2 production, and improved cellular internalization via CD44-mediated endocytosis, as well as an endo-lysosomal evasion mechanism. Furthermore, upon 980 nm laser irradiation, mitochondria-targeting CMPNs were able to efficiently induce mitochondrial depolarization, thereby significantly enhancing the efficacy of PDT. In vitro experiments validated the enhanced PDT effectiveness resulting from FRET mediation and mitochondria targeting, with no discernible cytotoxicity observed from the CMPN-treated A549 cells. The above combination of CD44 targeting with endo-lysosomal escape, mitochondrial targeting and FRET-mediated PDT in CMPNs offers a potent strategy for enhanced cancer therapies.Author contributions
Ze Hao Liu: Writing – review & editing, writing – original draft, methodology, and investigation. Xin Wang Mo: Methodology and investigation. Wei Jiang: Methodology and investigation. Changling Liu: Writing – original draft, methodology, and investigation. Yue Yin: Writing – original draft, methodology, and investigation. Hong Yu Yang: Writing – review & editing, supervision, project administration, funding acquisition, and conceptualization. Yan Fu: Writing – review & editing, supervision, project administration, funding acquisition, and conceptualization.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Science and Technology Development Plan Project of Jilin Province, China (No: YDZJ202201ZYTS269 and YDZJ202201ZYTS644) and by the Science and Technology Innovation Development Program of Jilin City (20240103009). This work was also supported by the National Natural Science Foundation of China (NSFC) (No. 21805108). NMR and TEM data were obtained using equipment maintained by the Jilin Institute of Chemical Technology Center of Characterization and Analysis.References
- S. S. Lucky, K. C. Soo and Y. Zhang, Chem. Rev., 2015, 115, 1990–2042 CrossRef CAS PubMed.
- H. Y. Yang, M.-S. Jang, Y. Li, Y. Fu, J. H. Lee and D. S. Lee, J. Controlled Release, 2019, 301, 157–165 CrossRef CAS PubMed.
- B. Zhang, L. Lin, J. Mao, W. Mo, Z. Li, S. Wang, Y. Tang, C. Cui, Y. Wu and Z. Yu, Chin. Chem. Lett., 2023, 34, 108518 CrossRef CAS.
- M. Lan, S. Zhao, W. Liu, C. S. Lee, W. Zhang and P. Wang, Adv. Healthcare Mater., 2019, 8, 1900132 CrossRef PubMed.
- C. Zhang, X. Hu, L. Jin, L. Lin, H. Lin, Z. Yang and W. Huang, Adv. Healthcare Mater., 2023, 12, 2300530 CrossRef CAS PubMed.
- Y. Tang, J. Feng, S. Li, G. Yang, Z. Tao, T. Xiao, F. Lu, B. Xie, Q. Fan and Q. Wang, J. Mater. Sci. Technol., 2025, 213, 196–212 CrossRef.
- S. Li, F. Yang, Y. Wang, T. Du and X. Hou, Chem. Eng. J., 2023, 451, 138621 CrossRef CAS.
- Y. Zhang, X. Gao, Y. Wu, J. Gui, S. Guo, H. Zheng and Z. L. Wang, Exploration, 2021, 1, 90–114 CrossRef PubMed.
- H. T. Yan, M.-S. Jang, C. Liu, Q. Fu, B. Wang, Y. Fu, J. H. Lee and H. Y. Yang, J. Colloid Interface Sci., 2024, 665, 188–203 CrossRef PubMed.
- Y. Shen, A. J. Shuhendler, D. Ye, J.-J. Xu and H.-Y. Chen, Chem. Soc. Rev., 2016, 45, 6725–6741 RSC.
- D. Kang, E. Jeon, S. Kim and J. Lee, BioChip J., 2020, 14, 124–135 CrossRef CAS.
- C. Yan, H. Zhao, D. F. Perepichka and F. Rosei, Small, 2016, 12, 3888–3907 CrossRef CAS PubMed.
- Y. I. Park, H. M. Kim, J. H. Kim, K. C. Moon, B. Yoo, K. T. Lee, N. Lee, Y. Choi, W. Park and D. Ling, Adv. Mater., 2012, 24, 5755–5761 CrossRef CAS PubMed.
- J. Li, Q. Zhao, F. Shi, C. Liu and Y. Tang, Adv. Healthcare Mater., 2016, 5, 2967–2971 CrossRef CAS PubMed.
- T.-H. Ho, S.-Y. Hong, C.-H. Yang, Y.-F. Chen, H.-Y. Lin and T.-L. Wang, J. Alloys Compd., 2022, 893, 162323 CrossRef CAS.
- C. Chen, C. Li and Z. Shi, Adv. Sci., 2016, 3, 1600029 CrossRef PubMed.
- A. Bagheri, H. Arandiyan, C. Boyer and M. Lim, Adv. Sci., 2016, 3, 1500437 CrossRef PubMed.
- D. Przybylska, A. Ekner-Grzyb, B. F. Grześkowiak and T. Grzyb, Sci. Rep., 2019, 9, 8669 CrossRef PubMed.
- R. Li, S. Gai, L. Wang, J. Wang and P. Yang, J. Colloid Interface Sci., 2012, 368, 165–171 CrossRef CAS PubMed.
- P. Thokwane and P. Mbule, Bull. Mater. Sci., 2023, 46, 140 CrossRef CAS.
- Q. Chen, X. Xie, B. Huang, L. Liang, S. Han, Z. Yi, Y. Wang, Y. Li, D. Fan and L. Huang, Angew. Chem., 2017, 129, 7713–7717 CrossRef.
- T. C. Pham, V.-N. Nguyen, Y. Choi, S. Lee and J. Yoon, Chem. Rev., 2021, 121, 13454–13619 CrossRef CAS PubMed.
- M. Sadraeian, L. Zhang, F. Aavani, E. Biazar and D. Jin, Mater. Today Phys., 2022, 28, 100882 CrossRef CAS.
- D. C. Wallace, Nat. Rev. Cancer, 2012, 12, 685–698 CrossRef CAS PubMed.
- J. Chen, Y. Zou, C. Deng, F. Meng, J. Zhang and Z. Zhong, Chem. Mater., 2016, 28, 8792–8799 CrossRef CAS.
- F. Yu, Y. Shao, X. Chai, Y. Zhao and L. Li, Angew. Chem., 2022, 134, e202203238 CrossRef.
- Y. Guan, H. Lu, W. Li, Y. Zheng, Z. Jiang, J. Zou and H. Gao, ACS Appl. Mater. Interfaces, 2017, 9, 26731–26739 CrossRef CAS PubMed.
- Y. Chen, Y. Yang, S. Du, J. Ren, H. Jiang, L. Zhang and J. Zhu, ACS Appl. Mater. Interfaces, 2023, 15, 35884–35894 CrossRef CAS PubMed.
- G. Yang, J. Ding and X. Chen, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2024, 16, e1985 CrossRef CAS PubMed.
- H. Y. Yang, M.-S. Jang, Y. Li, J. M. Du, C. Liu, J. H. Lee and Y. Fu, Colloids Surf., B, 2022, 217, 112638 CrossRef CAS PubMed.
- E. H. Seo, C. S. Lee and K. Na, Adv. Healthcare Mater., 2015, 4, 2822–2830 CrossRef CAS PubMed.
- X. Xue, H. Qu and Y. Li, Exploration, 2022, 2, 20210134 CrossRef PubMed.
- J. Sun, X. Huang, R. Shi, T. Ji, J. Ding and X. Chen, Nano Today, 2024, 56, 102222 CrossRef CAS.
- F. Dosio, S. Arpicco, B. Stella and E. Fattal, Adv. Drug Delivery Rev., 2016, 97, 204–236 CrossRef CAS PubMed.
- H. Hui, M.-S. Jang, C. Liu, Q. Fu, Y. Fu, J. H. Lee and H. Y. Yang, Eur. Polym. J., 2024, 210, 112955 CrossRef CAS.
- S. S. Liew, X. Qin, J. Zhou, L. Li, W. Huang and S. Q. Yao, Angew. Chem., Int. Ed., 2021, 60, 2232–2256 CrossRef CAS PubMed.
- S. Batheja, S. Gupta, K. K. Tejavath and U. Gupta, Drug Discovery Today, 2024, 29, 103983 CrossRef CAS PubMed.
- S. Han, A. Samanta, X. Xie, L. Huang, J. Peng, S. J. Park, D. B. L. Teh, Y. Choi, Y. T. Chang and A. H. All, Adv. Mater., 2017, 29, 1700244 CrossRef PubMed.
- G. Yang, X. Sun, J. Liu, L. Feng and Z. Liu, Adv. Funct. Mater., 2016, 26, 4722–4732 CrossRef CAS.
- H. Otsuka, Y. Nagasaki and K. Kataoka, Adv. Drug Delivery Rev., 2003, 55, 403–419 CrossRef CAS PubMed.
- Y. Liu, M. Li, J. Ding and X. Chen, Chin. Chem. Lett., 2024, 110146 CrossRef.
- Y. Fu, M.-S. Jang, C. Liu, Y. Li, J. H. Lee and H. Y. Yang, ACS Appl. Mater. Interfaces, 2023, 15, 36013–36024 CrossRef CAS PubMed.
- H. Y. Yang, M.-S. Jang, X. S. Sun, C. L. Liu, J. H. Lee, Y. Li and Y. Fu, Colloids Surf., B, 2023, 228, 113395 CrossRef CAS PubMed.
- Y. Fu, M.-S. Jang, N. Wang, Y. Li, J. H. Lee, D. S. Lee and H. Y. Yang, J. Controlled Release, 2020, 327, 129–139 CrossRef CAS PubMed.
- H. Y. Yang, J. Meng Du, M.-S. Jang, X. W. Mo, X. S. Sun, D. S. Lee, J. H. Lee and Y. Fu, Biomacromolecules, 2021, 22, 3590–3600 CrossRef CAS PubMed.
- M. Roesslein, C. Hirsch, J.-P. Kaiser, H. F. Krug and P. Wick, Int. J. Mol. Sci., 2013, 14, 24320–24337 CrossRef PubMed.
- X. Li, Y. Zhao, J. Yin and W. Lin, Coord. Chem. Rev., 2020, 420, 213419 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dt02399d |
| This journal is © The Royal Society of Chemistry 2024 |